Abstract
Purpose
ORM1-like 3 (ORMDL3) belongs to a highly conserved protein family which is anchored as transmembrane protein in the endoplasmic reticulum. Gasdermin B (GSDMB) is adjacent to ORMDL3 on chromosome 17q21.2 and belongs to the gasdermin-domain containing the protein family (GSDM family). Recent reports suggest that GSDMB and ORMDL3 are associated with asthma in several populations. However, genetic association studies that examined the association of GSDMB and ORMDL3 gene variants with asthma showed conflicting results. To assess whether combined evidence shows the association between GSDMB/ORMDL3 polymorphism and asthma.
Methods
A bibliographic search from MEDLINE identified 13 original articles using the search keywords 'GSDMB', 'ORMDL3', and 'asthma'. An updated literature-based meta-analysis involving 6,691 subjects with asthma, 9,281 control individuals, and 1,360 families were conducted. Meta-odds ratios (ORs) and 95% confidence intervals (CIs) based on the fixed effects model or the random effects model depended on Cochran's Q-statistic and I2 values. Data from case-control and TDT studies were analyzed in an allelic model using the Catmap software.
Results
We selected and identified 3 SNPs of ORMDL3 associated with asthma (rs8076131: OR=1.10; 95% CI, 1.02-1.20; P=0.012. rs12603332: OR=1.15; 95% CI, 1.05-1.25; P=0.002. rs3744246: OR=1.10; 95% CI, 1.02-1.17; P=0.008) and 1 SNP of GSDMB associated with asthma (rs7216389: OR=1.37; 95% CI, 1.27-1.47; P<0.01). Publication bias was estimated using modified Egger's linear regression test proposed by Harbordetal and revealed no evidence of biases. Furthermore, cumulative meta-analysis in chronological order showed the inclination toward significant association for rs7216389 and rs12603332 with continually adding studies, and the inclination toward null-significant association for rs3744246 and rs8076131.
Conclusions
Moderate evidence exists for associations of the ORMDL3 rs8076131, rs12603332, and rs3744246 and GSDMB rs7216389 variants with asthma. Large sample size and representative population-based studies and TDT studies with homogeneous asthmatic patients and well-matched controls are warranted to confirm this finding.
Keywords: GSDMB, ORMDL3, polymorphism, asthma, meta-analysis
INTRODUCTION
Asthma is a chronic immunological disorder of the lung characterized by reversible airway obstruction, airway inflammation, and increased airway hyperresponsiveness in response to provocative challenge. It is a complex disease that involves the interplay between multiple physiological processes.1 Physiological changes of the disease include accumulation of inflammatory cells, especially eosinophils, and goblet cell metaplasia of lung epithelium with a mucus-secreting phenotype.2 To reduce the disease burden of asthma, it is critical to understand its genetic and environmental risk factors. It is well known that the genetic background for asthma susceptibility is strong, with estimates of heritability up to 75%.3 Genome-wide linkage and genome-wide association (GWA) studies have found that more than 100 genes are associated with the development of asthma and asthma-related phenotypes.4 Several studies showed that variation in 17q12-21 locus harboring ORM1-like 3 (ORMDL3) and gasdermin B (GSDMB) is associated with asthma. ORMDL3 is a gene encoding ORM1-like 3, a transmembrane protein of the endoplasmatic reticulum, and a member of the family of orosomucoid-like proteins, which is produced in a number of cells, including lymphocytes and liver cells. ORM proteins regulate sphingolipid production, and altered expression of ORM genes or mutations affecting their phosphorylation sites can result in deregulation of sphingolipid production.5 ORMDL3 is also involved in the development of the unfolded protein response, a process that can initiate inflammation, which may explain the reported association between ORMDL3 with asthma.6 Multiple SNPs within the ORMDL3 gene, including rs8076131 and rs12603332, have been reported to be associated with asthma in different populations.7,8,9,10 In a study by Galanter et al.8 African Americans and Mexicans showed a positive association between rs12603332 and asthma, and Puerto Ricans showed a negative association. Contradictory results had also been reported for rs3744246 polymorphism of ORMDL3.7,11 Moffatt et al.12 genotyped healthy children and patients with childhood-onset asthma from Germany and the UK. In their study, German subjects showed a positive association of rs3744246 and asthma while UK subjects showed a negative association.
GSDMB encodes gasdermin B, a member of the family of gasdermin domain containing proteins. These proteins participate in numerous cell processes which are associated with tumor growth and progression, such as cell differentiation, cell cycle control, and apoptosis.13 The role of gasdermin B in bronchial asthma (BA) is still not clear, but it is known that gasdermin B participates in terminal differentiation of epithelial cells.14 GSDMB may also have a role in stem cell proliferation.15 Studies showed both positive and negative associations between rs7216389 of GSDMB and asthma.7,12,16,17,18,19,20,21 In the study by Galanter et al.,8 Puerto Ricans showed a positive association of rs7216389 and asthma, while African Americans and Mexicans showed a negative association. The Costa Rican Hispanic subjects showed a positive association of rs7216389 and asthma and the Non-Hispanic White subjects from the Childhood Asthma Management Program (CAMP) showed a negative association in the study of Verlaan et al.22
These conflicting results about the association of GSDMB and ORMDL3 gene polymorphisms with asthma raised the question regarding the significance of these SNPs in asthma pathogenesis. Obviously, the statistical power of an individual study could be very limited for the efficient assessment of these variants. Integration of these datasets may provide improved statistical power to detect the significance. The transmission/disequilibrium test (TDT) based on family is particularly advantageous with less confounding caused by population admixture and has the same importance as case-control study in genetic association analysis.23 Therefore, we have electronically searched all genetic association studies published in the field of asthma and GSDMB or ORMDL3 and conducted a meta-analysis of published studies to integrate the results from both case-control and TDT studies to provide more precise evaluation for the association of rs8076131, rs12603332, and rs3744246 of ORMDL3 and rs7216389 of GSDMB with susceptibility of asthma.
MATERIALS AND METHODS
Study identification and data extraction
We searched the PubMed biomedical database (US National Library of Medicine, Bethesda, Maryland) for all English-language articles related to asthma and GSDMB and ORMDL3 genetic polymorphisms that had been published through January 24, 2014.
We used the following search criterion: 'GSDMB' or 'gasdermin B', 'ORMDL3' or 'ORM1-like 3' in combination with 'asthma'. Eligible studies fulfilled the following inclusion criteria: (1) either case-control or TDT study design; (2) data on either or both of GSDMB and ORMDL3 polymorphisms; (3) presentation of data necessary for calculating odds ratios (ORs); and (4) studies including either children or adult (>18-year-old) patients with clear definition of asthma. Case reports, editorials, nonepidemiologic studies (e.g. studies on animals or cell culture), treatment outcome studies and review articles were excluded. The full texts of the candidate articles were examined to determine whether they contained sufficient information on the GSDMB or ORMDL3 gene polymorphism and asthma. Furthermore, reference lists were also reviewed to trace further relevant studies. In total 17 studies (13 case-control studies and 4 TDT studies) with 6,691 asthma subjects and 9,281 healthy controls, and 1,360 nuclear families were included in this review.
The following data were extracted using a piloted data extraction form: name of the first author, year of publication, ethnicity, type of study design, and diagnostic criteria for asthma. Information about the counts of alleles in case and control groups in case-control studies and numbers of transmitted alleles from heterozygous parents to affected offspring in family-based studies was also extracted.
Statistical analysis
We estimated unadjusted odds ratios (OR) for published genotype frequencies. The associations were indicated as pooled odds ratios with corresponding 95% confidence intervals. For meta-analysis, the overall or pooled estimate of risk was obtained by using the Mantel-Haenszel method in the fixed effects model24 or by the DerSimonian and Laird method in the random effect model.25 In the absence of heterogeneity, the random-effects and fixed-effects models will provide similar results. We assessed the within- and between-study variation or heterogeneity by testing Cochran's Q-statistic.26 This heterogeneity test assessed the null hypothesis that all studies were evaluating the same effect. When a significant Q-statistic (P<0.10) indicated heterogeneity across studies, the random effect model was used for meta-analysis to take into account the possibility of heterogeneity between studies. Otherwise, the fixed effects model was used. Fixed effects model assumes all of the studies are estimating the same underlying effect and considers only within-study variation. As studies in this review were from different populations and heterogeneity was inherent, we chose to use random effect model first. However, in the meta-analyses of rs3744246 and rs8076131, the fixed effects model was used given the fact that tau2 was less than or equal to 0 and no random effect estimates could be calculated.
We used funnel plot and Egger's test to detect publication bias. A funnel plot is a useful graph designed to check the existence of publication bias in meta-analyses. Publication bias can be visualized with a funnel plot which is a scatter plot of sample size and effect size. We also evaluated publication bias using Egger's linear regression test,27 which measures funnel plot asymmetry on the natural logarithm scale of the odds ratio. The Chi-square goodness of fit test was used to test if observed frequencies of genotypes in controls conformed to Hardy-Weinberg (HWE) expectations.
For the synthesis of case-control and TDT studies, the method described by Kazeem et al.28 was used. After obtaining the estimate of logarithm of the OR and its associated SE in each case-control or TDT study, the estimate of combined OR and its associated SE can be calculated by a weighted analysis method. The Catmap software implemented this method for the fix-effects model and extended this method for the random-effects model of DerSimonian and Laird to conduct case-control and TDT meta-analysis, which could be downloaded from the comprehensive R network (http://www.r-project.org).29 Additionally, sensitivity analysis was performed to assess the influence of each study on the overall estimate. Cumulative meta-analysis was also conducted via the assortment of studies by publication time. All P values were 2-tailed with a significant level at 0.05. All statistical analyses were carried out in Catmap software V1.6.
RESULTS
Studies included in the meta-analysis
An initial screening of all abstracts produced 24 articles containing information on either GSDMB or ORMDL3 polymorphisms and asthma. Eligible studies were genotype-based case-control studies and TDT studies that reported associations of rs8076131, rs12603332, and rs3744246 of ORMDL3 and rs7216389 of GSDMB polymorphisms with asthma. Both hospital-based and population-based studies were included in the analysis. Eleven studies were excluded because they did not provide the data necessary for calculating odds ratios or they did not provide the data of the 4 SNPs of this review.30,31,32,33,34,35,36,37,38,39,40 According to the prespecified inclusion criteria, in total 13 reports7,8,9,10,11,12,16,17,18,19,20,21,22 were included in this review, including 17 studies (13 case-control studies and 4 TDT studies) with 6,691 subjects with asthma and 9,281 controls. Most of the studies were about childhood asthma (12 childhood asthma studies, 4 adult asthma studies, and 1 study with both childhood and adult asthma subjects).
These 17 studies had been carried out in various countries, including China, Scotland, Russia, Korea, the United Kingdom, Germany, Australia, Canada, Costa Rica, Japan, the USA, Portugal, Hungary, Greece, Slovakia, and Czech Republic. A description of these studies is given in Table 1. The selected papers assessed the association of GSDMB and ORMDL3 gene polymorphisms with asthma. Of these papers, 37,8,9 evaluated the association of the rs8076131 polymorphism with asthma risk; 47,8,9,10 the association of the rs12603332 polymorphism with asthma, 47,8,11,12 the association of the rs3744246 polymorphism with asthma, and 117,8,11,12,16,17,18,19,20,21,22 the association of the rs7216389 polymorphism with asthma.
Table 1. Characteristics of studies included in a meta-analysis of ORMDL3 and GSDMB polymorphisms, and asthma.
| First author, year (Reference No.) | Country (ethnicity) | Sample size (cases/controls or family) | Design type | Gene | Polymorphisms evaluated | Risk allele | OR (95% CI)/χ2 (P value) | Genotype and method | PHWE control |
|---|---|---|---|---|---|---|---|---|---|
| Tavendale R, 200816 | Scotland (E) | 1,279/1,541 | Case-control | GSDMB | rs7216389 | T | 1.46 (1.31-1.62) | TaqMan | >0.05 |
| Yang FF, 20127 | China (Asia) | 152/190 | Case-control | GSDMB | rs7216389 | T | 1.653 (1.170-2.333) | PCR | >0.001 |
| Yang FF, 20127 | China (Asia) | 152/190 | Case-control | ORMDL3 | rs8076131 | A | 1.2 (0.843-1.708) | PCR | >0.001 |
| Yang FF, 20127 | China (Asia) | 152/190 | Case-control | ORMDL3 | rs12603332 | C | 1.2 (0.843-1.708) | PCR | >0.001 |
| Yang FF, 20127 | China (Asia) | 152/190 | Case-control | ORMDL3 | rs3744246 | C | 1.243 (0.868-1.779) | PCR | >0.001 |
| Karunas AS, 201117 | Russia (E) | 358/369 | Case-control | GSDMB | rs7216389 | T | 1.8 (1.45-2.23) | Illumina | NS |
| Russians | |||||||||
| Tatars | |||||||||
| Bashkirs | |||||||||
| Hrdlickova B, 20119 | Czech Republic (E) | 337/331 | Case-control | ORMDL3 | rs12603332 | C | 1.118 (0.902-1.387) | TaqMan | >0.05 |
| Hrdlickova B, 20119 | Czech Republic (E) | 337/331 | Case-control | ORMDL3 | rs8076131 | A | 1.077 (0.866-1.338) | TaqMan | >0.05 |
| Yu J, 201118 | Korea (Asia) | 786/522 | Case-control | GSDMB | rs7216389 | T | 1.264 (1.055-1.514) | PCR | >0.05 |
| Galanter J (A), 20088 | America (AA) | 261/176 | Case-control | GSDMB | rs7216389 | T | 1.21 (0.79-1.93) | PCR | NS |
| ORMDL3 | rs8076131 | A | 1.20 (0.76-1.90) | PCR | NS | ||||
| ORMDL3 | rs12603332 | C | 1.74 (1.24-2.44) | PCR | NS | ||||
| ORMDL3 | rs3744246 | C | 1.40 (0.96-2.04) | PCR | NS | ||||
| Moffatt MF (G), 200712 | German (E) | 728/694 | Case-control | GSDMB | rs7216389 | T | 1.475 (1.272-1.710) | Illumina | NS |
| Moffatt MF (G), 200712 | German (E) | 728/694 | Case-control | ORMDL3 | rs3744246 | C | 1.222 (1.01-1.479) | Illumina | NS |
| Moffatt MF (B), 200712 | British (E) | 306/1,041 | Case-control | GSDMB | rs7216389 | T | 1.634 (1.359-1.965) | Illumina | NS |
| Moffatt MF (B), 200712 | British (E) | 306/1,041 | Case-control | ORMDL3 | rs3744246 | C | 1.246 (0.986-1.576) | Illumina | NS |
| Li FX, 201210 | China (Asia) | 241/212 | Case-control | ORMDL3 | rs12603332 | C | 1.217 (0.903-1.641) | Sequenom MassARRAY | >0.05 |
| iPLEX platform | |||||||||
| Binia AD, 201119 | UK (E) | 397/1,429 | Case-control | GSDMB | rs7216389 | T | 1.42 (1.21-1.67) | TaqMan | >0.05 |
| Leung TF, 200911 | China (Asia) | 315/192 | Case-control | GSDMB | rs7216389 | T | 1.231 (0.905-1.675) | PCR | >0.05 |
| Leung TF, 200911 | China (Asia) | 315/192 | Case-control | GSDMB | rs3744246 | C | 1.191 (0.874-1.622) | PCR | >0.05 |
| Ferreira MA, 201120 | Australia (E) | 986/1,846 | Case-control | GSDMB | rs7216389 | T | 1.25 (1.12-1.40) | Illumina | NS |
| Hirota T, 200821 | Japan (Asia) | 545/738 | Case-control | GSDMB | rs7216389 | T | 1.44 (1.2-1.73) | TaqMan | NS |
| Verlaan DJ (CR), 200922 | Costa Rica (LA) | 382 | Family | GSDMB | rs7216389 | T | 19.6 (9.32×310-6) | TaqMan | NS |
| Verlaan DJ (CA), 200922 | CAMP (NA) | 278 | Family | GSDMB | rs7216389 | T | 2.24 (0.130) | TaqMan | NS |
| Galanter J (P), 20088 | Puerto Ricans (LA) | 399 | Family | GSDMB | rs7216389 | T | 1.35 (1.07-1.70) | PCR | NS |
| Galanter J (P), 20088 | Puerto Ricans (LA) | 399 | Family | ORMDL3 | rs8076131 | A | 1.29 (1.03-1.62) | PCR | NS |
| Galanter J (P), 20088 | Puerto Ricans ( LA) | 399 | Family | ORMDL3 | rs12603332 | C | 1.22 (0.98-1.52) | PCR | NS |
| Galanter J (P), 20088 | Puerto Ricans ( LA) | 399 | Family | ORMDL3 | rs3744246 | C | 1.10 (0.86-1.41) | PCR | NS |
| Galanter J (M), 20088 | Mexican (NA) | 301 | Family | GSDMB | rs7216389 | T | 1.26 (0.95-1.65) | PCR | NS |
| Galanter J (M), 20088 | Mexican (NA) | 301 | Family | ORMDL3 | rs8076131 | A | 1.38 (1.05-1.81) | PCR | NS |
| Galanter J (M), 20088 | Mexican (NA) | 301 | Family | ORMDL3 | rs12603332 | C | 1.36 (1.04-1.78) | PCR | NS |
| Galanter J (M), 20088 | Mexican (NA) | 301 | Family | ORMDL3 | rs3744246 | C | 1.35 (1.01-1.81) | PCR | NS |
OR, Odds ratio; 95% CI, 95% confidence intervals; A, America; G, German; B, British; CR, Costa Rica; CA, CAMP; P, Puerto Ricans; M, Mexican; E, European; AA, African American; EA, European American; SA, South American; NA, North America; LA, Latin America; PHWE, P value for Hardy-Weinberg equilibrium test; NS, not stated.
Combining results of case-control and TDT studies
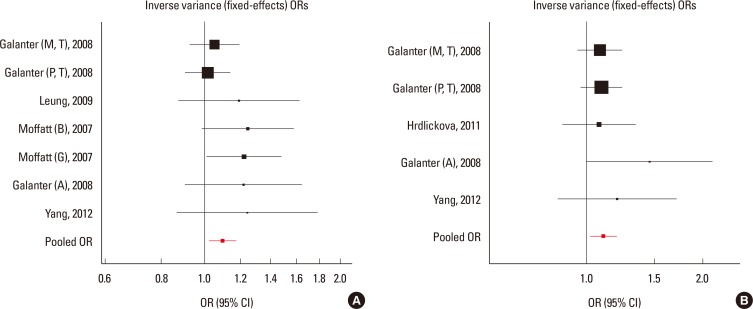
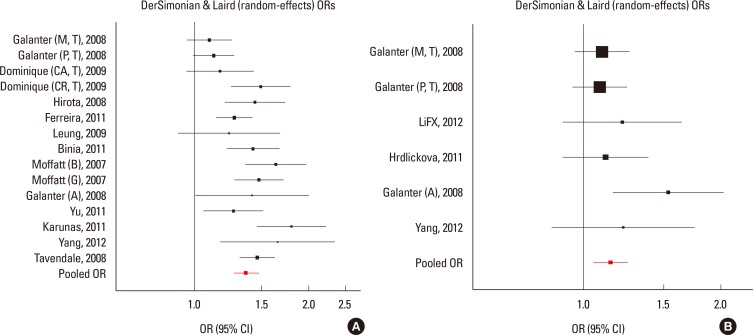
Meta-analyses for GSDMB and ORMDL3 were conducted using 4 SNPs in 20,052 participants. Three SNPs (rs8076131, rs12603332, and rs3744246) of ORMDL3 and 1 SNP (rs7216389) of GSDMB were all associated with asthma at the P<0.05 level (Figs. 1, 2, Tables 2 and 3). For the most significant marker, rs7216389 of GSDMB, the T allele was consistently less common in controls than in asthma cases. Random effects (DerSimonian-Laird) pooled odds ratio was 1.37 (95% confidence intervals (CIs), 1.27-1.47), with evidence of heterogeneity (χ2=40.04, P<0.01). We subsequently obtained results for associations of rs8076131 and rs3744246 of ORMDL3 with asthma. The A allele of rs8076131 was significantly higher in asthma cases compared to controls (OR=1.10; 95% CIs, 1.02-1.20) by the fixed effects model. The random-effects model (DerSimonian-Laird) showed apparent evidence of association between the C allele of rs12603332 and asthma (OR=1.15; 95% CIs, 1.05-1.25). The odds ratio of the C allele of rs3744246 for asthma was 1.10 (95% CIs, 1.02-1.17) by the fixed effects model.
Fig. 1. Results of meta-analysis of associations between ORMDL3 variants and asthma risk. (A) Forest plot for rs3744246 using the random-effects model. (B) Forest plot for rs8076131 using the fixed effects model.
Fig. 2. Results of meta-analysis of associations between GSDMB and ORMDL3 variants and asthma risk. (A) Forest plot for rs7216389 using the random-effects model. (B) Forest plot for rs12603332 using the fixed effects model.
Table 2. Random-effects odds ratios for asthma and heterogeneity test results for the risk allele of GSDMB (rs7216389) gene polymorphisms in relation to asthma.
| Study ID | Case-control | TDT | OR (95% CI) | |||
|---|---|---|---|---|---|---|
| Cases | Controls | Transmitted T | Untransmitted T | Case-control | TDT | |
| T/C | T/C | |||||
| Tavendale R, 2008 | 1,429/1,129 | 1,431/1,651 | 1.46 (1.31-1.62) | |||
| Yang FF, 2012 | 237/67 | 259/121 | 1.65 (1.16-2.34) | |||
| Karunas AS, 2011 | 426/290 | 332/406 | 1.80 (1.46-2.21) | |||
| Yu J, 2011 | 1,216/356 | 762/282 | 1.26 (1.06-1.51) | |||
| Galanter J (A), 2008 | 437/85 | 276/76 | 1.42 (1.00-2.00) | |||
| Moffatt MF (G), 2007 | 830/626 | 657/731 | 1.48 (1.27-1.71) | |||
| Moffatt MF (B), 2007 | 378/234 | 657/731 | 1.63 (1.36-1.96) | |||
| Binia AD, 2011 | 432/338 | 1,352/1,506 | 1.42 (1.21-1.67) | |||
| Leung TF, 2009 | 506/124 | 295/89 | 1.23 (0.90-1.67) | |||
| Ferreira MA, 2011 | 1,045/927 | 1,735/1,957 | 1.27 (1.14-1.42) | |||
| Hirota T, 2008 | 852/238 | 1,052/424 | 1.44 (1.20-1.73) | |||
| Verlaan DJ (CR), 2009 | 299 | 200 | 1.50 (1.25-1.79) | |||
| Verlaan DJ (CA), 2009 | 202 | 173 | 1.17 (0.95-1.43) | |||
| Galanter J (P), 2008 | 533 | 474 | 1.12 (0.99-1.27) | |||
| Galanter J (M), 2008 | 441 | 403 | 1.09 (0.96-1.25) | |||
| Total | 7,788/4,414 | 9,186/8,290 | 1,475 | 1,250 | 1.44 (1.35-1.53)* | 1.20 (1.05-1.37)† |
| Total | 1.37 (1.27-1.47)‡ | |||||
*Random-effects pooled OR, P<0.01; χ2=119.58, Pheterogeneity=0.142; †Random-effects pooled OR, P<0.01; χ2=7.38, Pheterogeneity=0.037; ‡Random-effects pooled OR of case-control and TDT studies, P<0.01; χ2=119.58, Pheterogeneity=0.142.
A, America; G, German; B, British; CR, Costa Rica; CA, CAMP; P, Puerto Ricans; M, Mexican.
Table 3. Odds ratios for asthma and heterogeneity test results for the risk allele of ORMDL3 (rs3744246, rs12603332, rs8076131) gene polymorphisms in relation to asthma.
| Study ID | Case-control | TDT | OR (95% CIs) | |||
|---|---|---|---|---|---|---|
| Cases | Controls | Transmitted C/A | Untransmitted C/A | Case-control | TDT | |
| C/T (A/G) | C/T (A/G) | |||||
| rs3744246 | ||||||
| Yang FF, 2012 | 239/65 | 284/96 | 1.24 (0.87-1.78) | |||
| Galanter J (A), 2008 | 383/139 | 244/108 | 1.22 (0.91-1.64) | |||
| Moffatt MF (G), 2007 | 1,213/243 | 1,115/273 | 1.22 (1.01-1.48) | |||
| Moffatt MF (B), 2007 | 506/106 | 1,651/431 | 1.25 (0.99-1.58) | |||
| Leung TF, 2009 | 502/126 | 291/87 | 1.19 (0.87-1.62) | |||
| Galanter J (P), 2008 | 592 | 583 | 1.02 (0.91-1.14) | |||
| Galanter J (M), 2008 | 494 | 470 | 1.05 (0.93-1.19) | |||
| Total | 2,843/679 | 3,585/995 | 1086 | 1053 | 1.10 (1.02-1.17)* | |
| rs12603332 | ||||||
| Yang FF, 2012 | 235/69 | 281/99 | 1.22 (0.85-1.75) | |||
| Galanter J (A), 2008 | 239/283 | 125/227 | 1.53 (1.16-2.03) | |||
| Hrdlickova B, 2011 | 372/302 | 347/315 | 1.12 (0.90-1.39) | |||
| Li FX, 2012 | 350/116 | 295/119 | 1.22 (0.90-1.64) | |||
| Galanter J (P), 2008 | 428 | 394 | 1.09 (0.95-1.25) | |||
| Galanter J (M), 2008 | 430 | 391 | 1.10 (0.96-1.26) | |||
| Total | 1.15 (1.05-1.25)† | |||||
| rs8076131 | ||||||
| Yang FF, 2012 | 235/69 | 281/99 | 1.20 (0.84-1.71) | |||
| Galanter J (A), 2008 | 176/454 | 68/289 | 1.46 (1.00-2.11) | |||
| Hrdlickova B, 2011 | 397/277 | 378/284 | 1.08 (0.87-1.34) | |||
| Galanter J (P), 2008 | 535 | 490 | 1.09 (0.97-1.23) | |||
| Galanter J (M), 2008 | 461 | 426 | 1.08 (0.95-1.23) | |||
| Total | 1,616/800 | 727/672 | 996 | 916 | 1.10 (1.02-1.20)‡ | |
*Fixed-effects pooled OR, P=0.008; χ2=6.85, Pheterogeneity=0.45; †Random-effects pooled OR, P=0.002;χ2=9.94, Pheterogeneity=0.36; ‡Fixed-effects pooled OR, P=0.012; χ2=6.22, Pheterogeneity=0.65.
A, America; G, German; B, British; P, Puerto Ricans; M, Mexican.
Stratified analysis
Stratified analysis was performed by the type of study design for rs7216389 of GSDMB. In case-control studies, evidence of between-study heterogeneity was not found for rs7216389 of GSDMB (χ2=14.74, Pheterogeneity=0.14). The pooled allelic OR in the random-effects model was 1.44 (95% CI, 1.35-1.53, P<0.01). In TDT studies, the heterogeneity test showed positive results (χ2=8.50, Pheterogeneity=0.04). In the random-effects model, this variant was associated with asthma risk (OR=1.20; 95% CIs, 1.05-1.37; P=0.007) (Table 2). In the stratified analysis by age at onset of the disease, 12 studies7,11,12,16,17,18,19,20,21 provided age at onset of the disease subgroup information, and significant associations between rs7216389 polymorphism and asthma risk were found for both the subgroup of adults (OR=1.50; 95% CIs, 1.38-1.63) and the subgroup of children (OR=1.38; 95% CIs, 1.02-1.87). Because there were less than 3 studies of the subgroup of TDT or adults for rs8076131, rs12603332, and rs3744246 of ORMDL3 in this review, we did not perform stratified analysis.
Sensitivity analyses for combined studies of GSDMB and ORMDL3 polymorphism
Given the significant between-study heterogeneity for GSDMB rs7216389 polymorphism, we conducted a sensitive meta-analysis to assess the effect of each individual study on the combined OR. A random-effects model was employed since heterogeneity was indicated. A series of combined OR with 95% CIs produced repeatedly after the removal of each individual study was greater than 1.0, suggesting the stability of the outcome that rs7216389 was associated with asthma risk (Table 4). Additionally, sensitivity analyses indicated that all studies contributed to the heterogeneity for rs7216389 equally. The P values for the Q test were all not less than 0.1 after deletion of each individual study, and the effect of rs7216389 was steadily significant.
Table 4. Sensitivity analysis of pooled OR combining family-based and case-control studies for GSDMB rs7216389 polymorphism.
| Study omitted | OR (95% CIs) | χ2 | P* | P†heterogeneity |
|---|---|---|---|---|
| Tavendale R, 2008 | 1.36 (1.25-1.47) | 54.72 | <0.01 | <0.01 |
| Yang FF, 2012 | 1.36 (1.26-1.47) | 61.83 | <0.01 | <0.01 |
| Karunas AS, 2011 | 1.34 (1.25-1.44) | 64.91 | <0.01 | <0.01 |
| Yu J, 2011 | 1.38 (1.27-1.49) | 61.36 | <0.01 | <0.01 |
| Galanter J (A), 2008 | 1.37 (1.26-1.48) | 62.04 | <0.01 | <0.01 |
| Moffatt MF (G), 2007 | 1.36 (1.25-1.47) | 56.69 | <0.01 | <0.01 |
| Moffatt MF (B), 2007 | 1.35 (1.25-1.45) | 60.29 | <0.01 | <0.01 |
| Binia AD, 2011 | 1.36 (1.26-1.48) | 57.07 | <0.01 | <0.01 |
| Leung TF, 2009 | 1.37 (1.27-1.48) | 64.06 | <0.01 | <0.01 |
| Ferreira MA, 2011 | 1.38 (1.27-1.50) | 57.84 | <0.01 | <0.01 |
| Hirota T, 2008 | 1.36 (1.26-1.47) | 57.97 | <0.01 | <0.01 |
| Verlaan DJ (CR), 2009 | 1.36 (1.26-1.47) | 57.92 | <0.01 | <0.01 |
| Verlaan DJ (CA), 2009 | 1.38 (1.28-1.49) | 66.35 | <0.01 | <0.01 |
| Galanter J (P), 2008 | 1.39 (1.29-1.50) | 78.57 | <0.01 | <0.01 |
| Galanter J (M), 2008 | 1.39 (1.30-1.50) | 82.63 | <0.01 | <0.01 |
*DerSimonian and Laird random-effects model used to determine the significance of the overall OR; †Cochran's χ2-based Q statistic test used to assess the heterogeneity.
TDT, transmission/disequilibrium test; A, America; G, German; B, British; CR, Costa Rica; CA, CAMP; P, Puerto Ricans; M, Mexican.
Cumulative meta-analysis
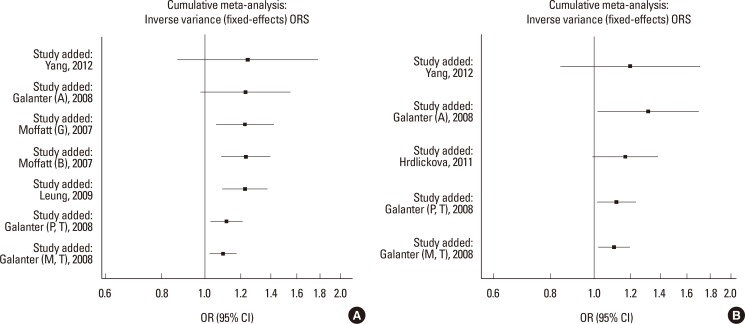
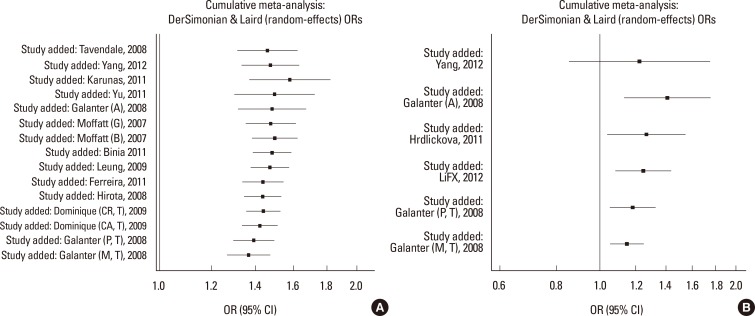
Cumulative meta-analyses of these 4 variants were also conducted via assortment of studies in chronological order. Figs. 3, 4 showed the results from the cumulative meta-analyses for rs7216389 and rs12603332 in the random-effects model and rs8076131 and rs3744246 in the fixed effects model. Rs3744246 and rs8076131 both tended to have no significant association over time, and rs7216389 and rs12603332 tended to have significant associations over time. Moreover, with increasing data the 95% CIs became increasingly narrower, suggesting that the precision of the estimates was progressively enhanced by continually adding more studies.
Fig. 3. Results of cumulative meta-analysis of associations between ORMDL3 variants and asthma risk. (A) Cumulative forest plot for rs3744246 using the random-effects model. (B) Cumulative forest plot for rs8076131 using the fixed effects model.
Fig. 4. Results of cumulative meta-analysis of associations between GSDMB and ORMDL3 variants and asthma risk. (A) Cumulative forest plot for rs7216389 using the random-effects model. (B) Cumulative forest plot for rs12603332 using the fixed effects model.
Publication bias
We assessed potential publication bias by examining funnel plots and using Egger's test. Egger's test detects whether the intercept deviates significantly from zero in a regression of the standardized effect estimates against their precision. The sensitivity of the regression method is generally low in meta-analysis based on less than 20 trials.41 There was no publication bias for rs8076131, rs7216389, and rs12603332 (P=0.106, 0.289, and 0.591, respectively). The result of Egger's test of rs3744246 was significant (P=0.011). We neglected publication bias because the small number of studies was included in this review.
DISCUSSION
This current meta-analysis, to the best of our knowledge, is the first study to integrate the case-control and TDT studies to reflect the precision effect of GSDMB and ORMDL3 variants in asthma risk. Our meta-analysis showed that moderate evidence existed for associations of the ORMDL3 rs8076131, rs12603332 and rs3744246 and GSDMB rs7216389 variants with asthma.
In the past few years, precise definition of asthma phenotypes has become increasingly important in the study of the genetic architecture and disease triggers of these phenotypes. Based on twin studies, estimates of the heritability of asthma ranged between 75%42 and 92%.43 Advances in technology have allowed genome-wide association studies to replace candidate gene studies and the susceptibility locus for asthma was identified12,22,44 as 17q12-21.1, which contained the GSDMB and ORMDL3 genes. 17q12-21 polymorphisms were found to be associated with the presence and severity of asthma across different populations. The gasdermin gene was originally identified as a candidate gene responsible for the phenotype of a mouse skin mutant, namely recombination-induced mutation 3 (Rim3).45 GSDMB is expressed as an oncogene product in stem cell-resident regions of normal epithelium.46 ORMDL3 is the third member of the gene family that encodes trans-membrane proteins that are anchored in the endoplasmic reticulum.47 ORMDL3 binds and inhibits the sarcoendoplasmic reticulum Ca2+ pump, resulting in a reduced ER Ca2+ concentration and an increased unfolded-protein response, which is relevant in chronic inflammatory diseases, such as asthma.6 Although the present knowledge of the ORMDL3 function is limited, a study in yeast has shown that the gene product may be involved in protein folding.6 Studies of ORMDL3 in Puerto Rican asthmatics have demonstrated a significant association between SNP rs12603332 and IgE levels,8 whereas subgroup analysis showed important associations between SNPs rs4378650 and rs12603332 in patients with allergic asthma (IgE >100 IU/mL). Associations between these SNPs and asthma became stronger in patients with IgE level >100 IU/mL.8 Although the function of GSDMB is not clear, the polymorphisms of GSDMB and ORMDL3 which are located in 17q gene were associated with early-onset asthma, smoking exposure,44 and several asthma-related phenotypes, including IgE11 and change in FEV1 in response to albuterol.8
Tavendale et al.16 confirmed that the rs7216389 C/T polymorphism at locus 17q21 controlling ORMDL3 gene expression was strongly associated with the occurrence of childhood asthma, which showed that the effect was dose-dependent with each C to T substitution at the SNP site, and the odds of the occurrence of asthma was increased by about 50%, with the homozygous T allele being associated with doubled odds of the homozygous C allele. In addition to its effects on asthma susceptibility, the study from Tavendale et al.16 also showed that the rs7216389 polymorphism was associated with the risk of asthma exacerbations.
The rs12603332 (C allele) and rs8076131 (A allele) of ORMDL3 are both in the intronic region. The rs12603332 SNP lies in a highly conserved element across species with a high correlation to the consensus target sequence for the transcription factor E47.48,49 E47, a member of the "E protein" family, a subset of helix-loop-helix proteins, has been linked to T- and B-cell development,50,51 and the C allele substitution in the position of rs12603332 reduces the transcription factor score of this region. Hrdlickova et al.9 reported that high-risk allele A of rs8076131 is significantly associated with hypersensitivity to pollen, and also reported a marginal significant relationship between polymorphism rs8076131 and hypersensitivity to birch antigen. Genetic variant rs3744246 of ORMDL3 was also significantly associated with allergic rhinitis in the Japanese population.52 The expression level of the ORMDL3 transcript was significantly correlated with the genotype of rs3744246, and ORMDL3 mRNA was highly expressed in nasal epithelium.
ORMDL3 is expressed in multiple cell types important to the pathogenesis of asthma (i.e., epithelial cells, macrophages, eosinophils, and T cells),53,54 and increased expression of ORMDL3 in multiple cell types contributes to the pathogenesis of asthma. Recent work has suggested that ORMDL3 may play a role in viral respiratory infections.21 A study by Miller et al.55 showed that ORMDL3 plays an important role in the activation of the ATF6 UPR pathway in vivo and that expression of ORMDL3 in vivo regulates airway remodeling (smooth muscle, fibrosis, and mucus). In addition, the ORM protein is believed to be critical mediators of sphingolipid homeostasis that may contribute to the development of childhood asthma.5 The human GSDM family consists of GSDMA, GSDMB, GSDMC, and GSDMD. The GSDMA and GSDMB genes are located at 17q21.2, and the GSDMC and GSDMD genes are located at 8q24.13 GSDMB was revealed to have several polymorphisms associated with asthma susceptibility and asthma related phenotypes.8,11,12,16,21,31,37,40,44 Among the genes, GSDMB and ORMDL3 have received the most attention, and their genotype-mediated expression is also affected by rhinovirus infection, one of the most common and powerful triggers for asthma exacerbations.56 Rs7216389 in GSDMB located in an intron was also strongly associated in cis with transcript levels of ORMDL3 in EBV-transformed lymphoblastoid cell lines from children with asthma.12 It has further been suggested that these 2 genes might be co-regulated, as their transcript levels seem connected.22 The study of Bouzigon et al.44 also showed an interaction between 17q21 variants and exposure to environmental tobacco smoke in early life. Among the offspring, an association between early-onset asthma and 11 SNPs (including rs7219923 of GSDML [or GSDMB] and rs8076131 of ORMDL3) was highly significant in the smoke-exposed families. Under the best-fitting recessive model, the overall risk of early-onset asthma when smoke exposure was not taken into account was increased for subjects who were homozygous for the risk alleles, as compared to those with other genotypes. However, exposure to environmental tobacco smoke in early life is associated with an even greater risk. Moreover, for SNPs showing significant heterogeneity, the SNP effect on early-onset asthma was significant in subjects with early exposure, as compared to those who did not have such exposure.
Our results confirm the role of the ORMDL3 and GSDMB genomic area as a locus conferring susceptibility to asthma. In combination with the recent published studies in ethnically diverse populations, it highlights the importance of ORMDL3 and GSDMB from the chromosome 17q21 region in the development of this complex disease. Despite the heterogeneity in biological function, further studies to examine the functions of GSDMB and ORMDL3 should focus on their role in asthma pathogenesis because of strong correlations observed between SNPs in these genes and asthma in many different ethnic groups.
Asthma is a complex condition with several contributing pathologic processes. Genetic variation in these processes and individual variation in response to pharmacologic therapies combine to form the range of phenotypes seen in the condition. It is likely that deep resequencing of portions of ORMDL3 and GSDMB will be necessary to uncover untypical or novel variants that contribute to associations between these genes and asthma. Asthma has very variable clinical subphenotypes, and genetic differences may explain the high heterogeneity of disease manifestations. Therefore, we encourage larger studies being conducted in this area. Larger, more comprehensive studies would make meaningful stratification possible, and would also permit evaluation of gene-gene and gene-environment interactions, and factors that are clearly important in complex diseases, such as asthma.
ACKNOWLEDGMENTS
We acknowledge financial support from the National Natural Science Foundation of China (NO. 81072385 and NO. 81402773).
Footnotes
There are no financial or other issues that might lead to conflict of interest.
References
- 1.Lemanske RF, Jr, Busse WW. Asthma: clinical expression and molecular mechanisms. J Allergy Clin Immunol. 2010;125:S95–S102. doi: 10.1016/j.jaci.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laitinen LA, Heino M, Laitinen A, Kava T, Haahtela T. Damage of the airway epithelium and bronchial reactivity in patients with asthma. Am Rev Respir Dis. 1985;131:599–606. doi: 10.1164/arrd.1985.131.4.599. [DOI] [PubMed] [Google Scholar]
- 3.Koppelman GH, Los H, Postma DS. Genetic and environment in asthma: the answer of twin studies. Eur Respir J. 1999;13:2–4. doi: 10.1183/09031936.99.13100299. [DOI] [PubMed] [Google Scholar]
- 4.Zhang G, Goldblatt J, LeSouëf P. The era of genome-wide association studies: opportunities and challenges for asthma genetics. J Hum Genet. 2009;54:624–628. doi: 10.1038/jhg.2009.97. [DOI] [PubMed] [Google Scholar]
- 5.Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–121. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 7.Yang FF, Huang Y, Li QB, Dai JH, Fu Z. Single nucleotide polymorphisms in the ORM1-like 3 gene associated with childhood asthma in a Chinese population. Genet Mol Res. 2012;11:4646–4653. doi: 10.4238/2012.October.19.1. [DOI] [PubMed] [Google Scholar]
- 8.Galanter J, Choudhry S, Eng C, Nazario S, Rodríguez-Santana JR, Casal J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–1200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrdlickova B, Holla LI. Relationship between the 17q21 locus and adult asthma in a Czech population. Hum Immunol. 2011;72:921–925. doi: 10.1016/j.humimm.2011.07.309. [DOI] [PubMed] [Google Scholar]
- 10.Li FX, Tan JY, Yang XX, Wu YS, Wu D, Li M. Genetic variants on 17q21 are associated with asthma in a Han Chinese population. Genet Mol Res. 2012;11:340–347. doi: 10.4238/2012.February.10.5. [DOI] [PubMed] [Google Scholar]
- 11.Leung TF, Sy HY, Ng MC, Chan IH, Wong GW, Tang NL, et al. Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy. 2009;64:621–628. doi: 10.1111/j.1398-9995.2008.01873.x. [DOI] [PubMed] [Google Scholar]
- 12.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 13.Tamura M, Tanaka S, Fujii T, Aoki A, Komiyama H, Ezawa K, et al. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics. 2007;89:618–629. doi: 10.1016/j.ygeno.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Carl-McGrath S, Schneider-Stock R, Ebert M, Röcken C. Differential expression and localisation of gasdermin-like (GSDML), a novel member of the cancer-associated GSDMDC protein family, in neoplastic and non-neoplastic gastric, hepatic, and colon tissues. Pathology. 2008;40:13–24. doi: 10.1080/00313020701716250. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Moffatt MF, Cookson WO. Genetic and genomic approaches to asthma: new insights for the origins. Curr Opin Pulm Med. 2012;18:6–13. doi: 10.1097/MCP.0b013e32834dc532. [DOI] [PubMed] [Google Scholar]
- 16.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CN. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–863. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Karunas AS, Iunusbaev BB, Fedorova I, Gimalova GF, Ramazanova NN, Gur'eva LL, et al. Genome-wide association study of bronchial asthma in the Volga-Ural region of Russia. Mol Biol (Mosk) 2011;45:992–1003. [PubMed] [Google Scholar]
- 18.Yu J, Kang MJ, Kim BJ, Kwon JW, Song YH, Choi WA, et al. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR. Pediatr Pulmonol. 2011;46:701–708. doi: 10.1002/ppul.21424. [DOI] [PubMed] [Google Scholar]
- 19.Binia A, Khorasani N, Bhavsar PK, Adcock I, Brightling CE, Chung KF, et al. Chromosome 17q21 SNP and severe asthma. J Hum Genet. 2011;56:97–98. doi: 10.1038/jhg.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19:458–464. doi: 10.1038/ejhg.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota T, Harada M, Sakashita M, Doi S, Miyatake A, Fujita K, et al. Genetic polymorphism regulating ORM1-like 3 (Saccharomyces cerevisiae) expression is associated with childhood atopic asthma in a Japanese population. J Allergy Clin Immunol. 2008;121:769–770. doi: 10.1016/j.jaci.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 22.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Larivière M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Deeks JJ, Altman DG, Bradburn MJ. Chapter 15. Statistical methods for examining heterogeneity and combining results from several studies in meta-analysis. In: Egger M, Smith GD, Altman DG, editors. Systematic reviews in health care: meta-analysis in context. 2nd ed. London: BMJ; 2001. pp. 285–312. [Google Scholar]
- 27.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Ann Hum Genet. 2005;69:329–335. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 29.Nicodemus KK. Catmap: case-control and TDT meta-analysis package. BMC Bioinformatics. 2008;9:130. doi: 10.1186/1471-2105-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, et al. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179:1084–1090. doi: 10.1164/rccm.200812-1860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madore AM, Tremblay K, Hudson TJ, Laprise C. Replication of an association between 17q21 SNPs and asthma in a French-Canadian familial collection. Hum Genet. 2008;123:93–95. doi: 10.1007/s00439-007-0444-x. [DOI] [PubMed] [Google Scholar]
- 33.Marinho S, Custovic A, Marsden P, Smith JA, Simpson A. 17q12-21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann Allergy Asthma Immunol. 2012;108:402–411.e9. doi: 10.1016/j.anai.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, et al. 17q12-21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–607. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 35.Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, et al. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 36.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–768. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 39.Kang MJ, Yu HS, Seo JH, Kim HY, Jung YH, Kim YJ, et al. GSDMB/ORMDL3 variants contribute to asthma susceptibility and eosinophil-mediated bronchial hyperresponsiveness. Hum Immunol. 2012;73:954–959. doi: 10.1016/j.humimm.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Sleiman PM, Annaiah K, Imielinski M, Bradfield JP, Kim CE, Frackelton EC, et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008;122:1225–1227. doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 41.Sterne JA, Gavaghan D, Egger M. Publication and related bias in meta-analysis: power of statistical tests and prevalence in the literature. J Clin Epidemiol. 2000;53:1119–1129. doi: 10.1016/s0895-4356(00)00242-0. [DOI] [PubMed] [Google Scholar]
- 42.Willemsen G, van Beijsterveldt TC, van Baal CG, Postma D, Boomsma DI. Heritability of self-reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res Hum Genet. 2008;11:132–142. doi: 10.1375/twin.11.2.132. [DOI] [PubMed] [Google Scholar]
- 43.Fagnani C, Annesi-Maesano I, Brescianini S, D'Ippolito C, Medda E, Nisticò L, et al. Heritability and shared genetic effects of asthma and hay fever: an Italian study of young twins. Twin Res Hum Genet. 2008;11:121–131. doi: 10.1375/twin.11.2.121. [DOI] [PubMed] [Google Scholar]
- 44.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 45.Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (Gsdm) localizing to mouse Chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells. Mamm Genome. 2000;11:718–724. doi: 10.1007/s003350010138. [DOI] [PubMed] [Google Scholar]
- 46.Saeki N, Usui T, Aoyagi K, Kim DH, Sato M, Mabuchi T, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium. Genes Chromosomes Cancer. 2009;48:261–271. doi: 10.1002/gcc.20636. [DOI] [PubMed] [Google Scholar]
- 47.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzàlez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002;3:RESEARCH0027. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: e-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 50.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- 51.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 52.Tomita K, Sakashita M, Hirota T, Tanaka S, Masuyama K, Yamada T, et al. Variants in the 17q21 asthma susceptibility locus are associated with allergic rhinitis in the Japanese population. Allergy. 2013;68:92–100. doi: 10.1111/all.12066. [DOI] [PubMed] [Google Scholar]
- 53.Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci U S A. 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP, et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol. 2014;192:3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.CalXMLLink_XYZkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]