Abstract
Introduction
HIV-1 transmitted drug resistance (TDR) in treatment-naïve individuals is a well-described phenomenon. Baseline genotypic resistance testing is considered standard of care in most developed areas of the world.
Methods
In the Strategic Timing of AntiRetroviral Treatment (START) trial, baseline genotypic resistance testing results were collected at study entry and analysed centrally to determine the prevalence of TDR in the study population. Resistance was based on a modified 2009 World Health Organization definition to reflect newer resistance mutations.
Results
Baseline resistance testing was available in 1946 study participants. Higher rates of testing occurred in Europe (86.7%), the United States (81.3%), and Australia (89.9%) as compared to Asia (22.2%), South America (1.8%), and Africa (0.1%). The overall prevalence of TDR was 10.1%, most commonly to non-nucleoside reverse transcriptase inhibitors (4.5%) and nucleoside reverse transcriptase inhibitors (4%) compared to protease inhibitors (2.8%). The most frequent TDR mutations observed were M41L, D67N/G/E, T215F/Y/I/S/C/D/E/V/N, 219Q/E/N/R, K103N/S, and G190A/S/E in reverse transcriptase, and M46I/L and L90M in protease. By country, prevalence of TDR was highest in Australia (17.5%), France (16.7%), the United States (12.6%), and Spain (12.6%). No participant characteristics were identified as predictors for the presence of TDR.
Conclusion
START participants enrolled in resource-rich areas of the world were more likely to have baseline resistance testing. In Europe, the United States, and Australia, TDR prevalence rates varied by country.
Keywords: HIV, drug resistance, antiretroviral therapy
INTRODUCTION
Transmitted drug resistance (TDR) results from infection with an HIV-1 strain containing one or more resistance-associated mutations (RAMs). Transmission of a drug-resistant strain usually occurs at the time of initial infection but can also occur with a subsequent exposure, referred to as HIV-1 super-infection. These strains can be transmitted from treatment-naive individuals who may be unaware of their infection or from treatment-experienced persons whose virus may have evolved drug resistance in association with treatment failure. TDR has been associated with an increased risk of suboptimal virologic response to the initial regimen and can also impact future treatment options (1–5). When TDR is detected in a treatment-naïve individual this has implications for selection of the initial antiretroviral regimen, as it is currently recommended that three fully active agents be prescribed.
A number of studies have described the prevalence of TDR in treatment-naïve patients, which varies by geographic region (5–8). Studies from the United States and Europe have shown that the prevalence of transmitted HIV-1 containing one or more RAMs in treatment-naïve individuals typically ranges from 5–15% (5, 6, 9–19). In these resource-rich countries TDR has been most commonly detected to the first generation non-nucleoside reverse transcriptase inhibitors (NNRTIs) and nucleoside reverse transcriptase inhibitors (NRTIs), with a lower prevalence usually reported for TDR to protease inhibitors (PIs) (6, 15). Given the prevalence of TDR, in most resource-rich areas of the world it is recommended that resistance testing be performed at the time of diagnosis or before initiating antiretroviral therapy in the setting of either acute or chronic infection (20–22).
In recent years, TDR has also been described as an emerging health issue in resource-limited countries, although limited surveillance data has been available (6, 23–25). In these regions antiretroviral therapy (ART) has been introduced more recently and has significantly impacted morbidity and mortality, but access to virologic monitoring techniques is often limited when compared to those available in resource-rich regions. In the absence of state-of-the-art laboratory monitoring, there is increased opportunity for the development and transmission of drug-resistant HIV-1 given the prolonged time between the onset of initial virologic failure and subsequent clinical consequences. It is not surprising that TDR prevalence in resource-limited countries is directly correlated with the number of years since ART roll-out programs were initiated (6).
The Strategic Timing of AntiRetroviral Treatment (START) study has recruited participants from both resource-rich and resource-limited regions of the world, including the United States, Europe, Israel, Australia, South America, Mexico, Africa, and Asia. Participants enrolled are required to have a CD4 cell count greater than 500 cells/μL and therefore are anticipated to have relatively recent infection with HIV-1. Baseline resistance testing was not an entry criteria for the study and therefore results were only available if performed according to local practice. Baseline data collected in the study thus provide an opportunity to examine the utilisation of resistance testing and prevalence of TDR in treatment-naïve individuals and assess differences by region. This analysis describes results from locally collected genotypic resistance testing performed on study participants at or before enrolment in START, when available.
METHODS
The design of START has been described elsewhere (26). Baseline resistance testing was not required by the protocol. The study did however collect laboratory results from any locally performed genotypic resistance tests, where available. Specifically, enrolment sites were asked to submit results for the participant’s first ever performed resistance test and the most recent test available. The laboratory reports were collected centrally at the INSIGHT Statistical and Data Management Center, and data were entered into a standard format per the HIV Collaboration Data Exchange Protocol (HICDEP) by staff at the Copenhagen HIV Programme (CHIP) (27).
All resistance tests were conducted at local sites using a variety of genotypic testing systems, based on bulk (Sanger) sequencing of the protease and reverse transcriptase genes. The integrase gene was also sequenced in a small number of samples, but these data are not reported here. Each site submitted the results of local resistance testing in the form of mutations detected and, when available, the assigned HIV-1 subtype (sequences were not centralised). To obtain a standard interpretation across tests, HIV-1 transmitted drug resistance was defined according to the World Health Organization (WHO) 2009 surveillance list, with the addition of T215N (a revertant of T215F/Y omitted from the list) and E138K (a nonpolymorphic rilpivirine-associated mutation) (28). Similarly, susceptibility to antiretroviral drugs was standardised using the Stanford HIVdb algorithm 7.0.1, which defines resistance as: none, potential low level, low level, intermediate, or high (29).
A total of 11 participants had two or more prerandomisation resistance tests that showed different mutational patterns. Seven participants showed evidence of fading of mutations and four acquired new mutations, presumably as a result of super-infection or a lack of initial detection. For the purposes of this analysis mutations have been cumulated across tests, the highest level of predicted resistance (per individual drug) selected, and the date of the most recent test used. Furthermore, for analyses by region Israel was included with European countries and Mexico was included with South America.
Statistical methods
Confidence intervals for proportions were derived using the method of Clopper-Pearson. Multivariate logistic regression was used to examine participant-level predictors of receiving a resistance test and, among participants whose virus was successfully sequenced, the likelihood of harbouring a TDR mutation. The former analysis also included a random effect for clinical site to allow for potential localised variation in the availability of resistance testing; in the latter analysis, geographical variation was adjusted using a fixed-effects approach (see footnote to Table 3). To maximise the available sample size, cases with missing data were included by creating an unknown/missing category for the variable in question, which is approximately valid when there is a small fraction of missing data, as in the present analysis. Significance tests in the logistic regression models are based on Wald tests, ignoring other and unknown/missing categories for the variable in question. Chi-squared and Fisher’s exact tests were used to compare categorical variables. All statistical analyses were conducted using Stata, version 13 (StataCorp, Houston, Texas, United States).
Table 3.
Predictors of transmitted drug resistance (any class)
| No. participants | No. (%) with TDR | Unadjusted OR | Adjusted OR* (95% CI) | p-value | |
|---|---|---|---|---|---|
| 1869 | 188 (10.1) | ||||
|
| |||||
| Age | 0.62 | ||||
| 18–29 | 463 | 49 (10.6) | 1.23 | 1.18 (0.78, 1.80) | |
| 30–39 | 617 | 54 (8.8) | 1.00 | 1.00 | |
| 40–49 | 553 | 58 (10.5) | 1.22 | 1.20 (0.80, 1.79) | |
| ≥50 | 236 | 27 (11.4) | 1.35 | 1.39 (0.83, 2.32) | |
|
| |||||
| Gender | 0.29 | ||||
| Male | 1699 | 168 (9.9) | 1.00 | 1.00 | |
| Female | 170 | 20 (11.8) | 1.22 | 1.48 (0.72, 3.07) | |
|
| |||||
| Race | 0.24 | ||||
| White | 1352 | 131 (9.7) | 1.00 | 1.00 | |
| Black | 255 | 28 (11.0) | 1.15 | 0.86 (0.50, 1.48) | |
| Hispanic | 123 | 15 (12.2) | 1.29 | 0.92 (0.49, 1.73) | |
| Asian | 100 | 11 (11.0) | 1.15 | 2.78 (0.98, 7.89) | |
| Other | 39 | 3 (7.7) | 0.78 | 1.04 (0.30, 3.56) | |
|
| |||||
| HIV mode | 0.43 | ||||
| MSM | 1440 | 148 (10.3) | 1.00 | 1.00 | |
| Heterosexual | 292 | 30 (10.3) | 1.00 | 0.79 (0.43, 1.47) | |
| IDU | 43 | 2 (4.7) | 0.43 | 0.40 (0.08, 1.91) | |
| Other/unknown | 94 | 8 (8.5) | 0.81 | 0.75 (0.35, 1.63) | |
|
| |||||
| CD4 count (cells/μL) | 0.64 | ||||
| 500–599 | 609 | 59 (9.7) | 1.00 | 1.00 | |
| 600–699 | 623 | 67 (10.8) | 1.12 | 1.15 (0.78, 1.68) | |
| ≥700 | 637 | 62 (9.7) | 1.01 | 0.97 (0.66, 1.44) | |
|
| |||||
| HIV RNA (copies/mL) | 0.75 | ||||
| 0–399 | 50 | 5 (10.0) | 1.00 | 1.19 (0.45, 3.17) | |
| 400–9,999 | 643 | 71 (11.0) | 1.11 | 1.09 (0.77, 1.53) | |
| 10,000–99,999 | 936 | 94 (10.0) | 1.00 | 1.00 | |
| ≥100,000 | 235 | 17 (7.2) | 0.70 | 0.80 (0.46, 1.39) | |
|
| |||||
| anti-HCV | 0.34 | ||||
| No | 1708 | 179 (10.5) | 1.00 | 1.00 | |
| Yes | 102 | 7 (6.9) | 0.63 | 0.66 (0.28, 1.57) | |
|
| |||||
| Year of resistance test† | 0.29 | ||||
| 2000–2007 | 127 | 16 (12.6) | 1.25 | 1.33 (0.73, 2.45) | |
| 2008 | 126 | 17 (13.5) | 1.35 | 1.25 (0.70, 2.21) | |
| 2009 | 248 | 21 (8.5) | 0.80 | 0.81 (0.48, 1.36) | |
| 2010 | 452 | 39 (8.6) | 0.82 | 0.75 (0.50, 1.12) | |
| 2011–2013 | 916 | 95 (10.4) | 1.00 | 1.00 | |
|
| |||||
| Interval HIV-resistance test‡ | 0.08 | ||||
| <1 year | 1381 | 129 (9.3) | 1.00 | 1.00 | |
| ≥1 year | 488 | 59 (12.1) | 1.33 | 1.38 (0.97, 1.95) | |
Analysis based on data from Europe, United States, Australia and Asia.
Adjusted for all other variables listed and for region (United States, Asia, Australia, United Kingdom, Germany, Spain, France, Belgium, Greece, Other European)
Year of the most recent resistance test
Time between diagnosis of HIV and most recent resistance test
MSM=men who have sex with men; IDU=injecting drug use; HCV=hepatitis C.
RESULTS
Baseline resistance testing by region
The results of one or more prerandomisation resistance tests were available on 1946 (41.5%) of the total 4685 HIV-positive participants. The median interval between the date of the resistance test and date of randomisation was 19 weeks. Testing was infrequent at sites in South America (1.8%; 21/1,174), Asia (22.2%; 79/356), and Africa (0.1%; 1/1,000). Table 1 examines predictors of having received a resistance test (whether or not this was successful) in sites in the remaining regions of Europe, the United States, and Australia. The overall frequency of testing was 85.6%, with no significant variation between these three regions after accounting for participant-level demographic and clinical variables. Testing was more frequent among participants with a later date of HIV diagnosis. Participants with a viral load of <400 copies/ml at their screening visit were less likely to have a genotype performed, presumably due to concern about insufficient virus to amplify. White ethnicity (versus black or Asian, but not versus Hispanic) and male-to-male sexual transmission were independently associated with a greater likelihood of testing. In subanalyses, these effects were apparent in each of the three geographical regions (data not shown).
Table 1.
Predictors of receiving a resistance test
| No. participants | No. (%) with resistance test | Unadjusted OR | Adjusted OR* (95% CI) | p-value | |
|---|---|---|---|---|---|
| 2155 | 1845 (85.6) | ||||
|
| |||||
| Age | 0.36 | ||||
| 18–29 | 523 | 439 (83.9) | 0.72 | 0.81 (0.52, 1.25) | |
| 30–39 | 688 | 605 (87.9) | 1.00 | 1.00 | |
| 40–49 | 643 | 561 (87.2) | 0.94 | 0.96 (0.63, 1.47) | |
| ≥50 | 301 | 240 (79.7) | 0.54 | 0.67 (0.40, 1.11) | |
|
| |||||
| Gender | 0.26 | ||||
| Male | 1900 | 1673 (88.1) | 1.00 | 1.00 | |
| Female | 255 | 172 (67.5) | 0.28 | 0.72 (0.41, 1.27) | |
|
| |||||
| Race | 0.01 | ||||
| White | 1579 | 1388 (87.9) | 1.00 | 1.00 | |
| Black | 340 | 259 (76.2) | 0.44 | 0.57 (0.34, 0.97) | |
| Hispanic | 161 | 132 (82.0) | 0.63 | 0.53 (0.26, 1.06) | |
| Asian | 36 | 27 (75.0) | 0.41 | 0.26 (0.09, 0.73) | |
| Other | 39 | 39 (100.0) | ∞ | ∞ | |
|
| |||||
| HIV mode | 0.005 | ||||
| MSM | 1583 | 1412 (89.2) | 1.00 | 1.00 | |
| Heterosexual | 402 | 296 (73.6) | 0.34 | 0.51 (0.30, 0.86) | |
| IDU | 60 | 44 (73.3) | 0.33 | 0.26 (0.09, 0.74) | |
| Other/unknown | 110 | 93 (84.5) | 0.66 | 1.10 (0.51, 2.35) | |
|
| |||||
| Year diagnosed | 0.001 | ||||
| 1982–2006 | 357 | 273 (76.5) | 0.42 | 0.39 (0.24, 0.64) | |
| 2007–2008 | 341 | 293 (85.9) | 0.78 | 0.75 (0.46, 1.24) | |
| 2009–2010 | 728 | 633 (87.0) | 0.86 | 0.89 (0.59, 1.35) | |
| 2011–2013 | 729 | 646 (88.6) | 1.00 | 1.00 | |
|
| |||||
| CD4 count (cells/μL) | 0.29 | ||||
| 500–599 | 680 | 582 (85.6) | 1.00 | 1.00 | |
| 600–699 | 709 | 620 (87.4) | 1.17 | 0.93 (0.62, 1.40) | |
| ≥700 | 766 | 643 (83.9) | 0.88 | 0.74 (0.50, 1.10) | |
|
| |||||
| HIV RNA (copies/mL) | <0.001 | ||||
| 0–399 | 104 | 56 (53.8) | 0.15 | 0.13 (0.07, 0.24) | |
| 400–9,999 | 775 | 650 (83.9) | 0.65 | 0.75 (0.52, 1.07) | |
| 10,000–99,999 | 1022 | 908 (88.8) | 1.00 | 1.00 | |
| ≥100,000 | 249 | 226 (90.8) | 1.23 | 1.06 (0.60, 1.90) | |
|
| |||||
| Anti-HCV | 0.47 | ||||
| No | 1956 | 1683 (86.0) | 1.00 | 1.00 | |
| Yes | 129 | 103 (79.8) | 0.64 | 1.32 (0.62, 2.78) | |
|
| |||||
| Region | 0.97 | ||||
| Europe | 1539 | 1335 (86.7) | 1.00 | 1.00 | |
| United States | 507 | 412 (81.3) | 0.66 | 1.09 (0.46, 2.58) | |
| Australia | 109 | 98 (89.9) | 1.36 | 0.93 (0.21, 4.23) | |
Analysis based on data from Europe, the United States and Australia.
Random effects logistic regression analysis (random effect for study site), adjusted for all other variables listed
MSM=men who have sex with men; IDU=injecting drug use; HCV=hepatitis C.
All further analyses, which are based on the results of genotyping, consider sites in Europe, the United States, Australia, and Asia.
HIV-1 subtype distribution
A valid genotype result was available for 1869 (97.1%) of 1924 participants from Europe, the United States, Australia, and Asia who were tested at least once. Of these, HIV subtype was reported for 1228 participants: 961 (78.3%) were subtype B, 86 (7.0%) were CRF01(AE), 52 (4.2%) subtype C, 37 (3.0%) CRF02(AG), and 92 (7.5%) were other subtypes. Subtype CRF01(AE) was observed mainly in Asia (n=58), and subtypes C and CRF02(AG) mainly in Europe (n=48 and n=36, respectively).
Prevalence of transmitted drug resistance
Overall, the prevalence of NRTI, NNRTI, and PI TDR mutations was 4.0%, 4.5%, and 2.8%, respectively; 10.1% of participants had resistance-associated mutations to one or more of these drug classes (Table 2). Cross-class resistance (involving two or three classes) was observed in only 22 (11.7%) of 188 participants with any form of resistance. Major and statistically significant geographical variation in patterns of TDR was observed. Most strikingly, the overall prevalence of TDR was markedly higher in the United States (12.6%) than Europe (8.8%), driven by higher rates of NNRTI-related mutations (8.4% versus 3.4%, p<0.001). Highly significant heterogeneity was also observed between different European countries, with relatively high rates of TDR mutations in France and Spain and relatively low rates in the United Kingdom. Sites in Australia had the highest level of TDR and sites in Asia the lowest, although the estimates in these regions are imprecise due to smaller sample sizes. There were no clearly significant individual-level predictors of the presence of TDR mutations (Table 3). The large difference between the adjusted and unadjusted odds ratios for Asian participants is due to a strong association with geographical region, i.e., there were few Asian participants other than at Asian sites.
Table 2.
Prevalence of transmitted drug resistance by geographical region
| Region | No. participants | Transmitted Drug Resistance | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Any | NRTI | NNRTI | PI | ||||||
| N | % (95% CI)† | N | % (95% CI) | N | % (95% CI) | N | % (95% CI) | ||
| Total | 1,869 | 188 | 10.1 (8.7, 11.5) | 75 | 4.0 (3.2, 5.0) | 85 | 4.5 (3.6, 5.6) | 52 | 2.8 (2.1, 3.6) |
| United States | 405 | 51 | 12.6 (9.5, 16.2) | 15 | 3.7 (2.1, 6.0) | 34 | 8.4 (5.9, 11.5) | 8 | 2.0 (0.9, 3.9) |
| Australia | 97 | 17 | 17.5 (10.6, 26.6) | 9 | 9.3 (4.3, 16.9) | 4 | 4.1 (1.1, 10.2) | 5 | 5.2 (1.7, 11.6) |
| Asia | 75 | 6 | 8.0 (3.0, 16.6) | 2 | 2.7 (0.3, 9.3) | 3 | 4.0 (0.8, 11.2) | 2 | 2.7 (0.3, 9.3) |
| Europe | 1,292 | 114 | 8.8 (7.3, 10.5) | 49 | 3.8 (2.8, 5.0) | 44 | 3.4 (2.5, 4.5) | 37 | 2.9 (2.0, 3.9) |
| United Kingdom | 320 | 15 | 4.7 (2.6, 7.6) | 6 | 1.9 (0.7, 4.0) | 3 | 0.9 (0.2, 2.7) | 6 | 1.9 (0.7, 4.0) |
| Germany | 271 | 29 | 10.7 (7.3, 15.0) | 12 | 4.4 (2.3, 7.6) | 10 | 3.7 (1.8, 6.7) | 11 | 4.1 (2.0, 7.1) |
| Spain | 191 | 24 | 12.6 (8.2, 18.1) | 6 | 3.1 (1.1, 6.7) | 11 | 5.8 (2.9, 10.1) | 10 | 5.2 (2.5, 9.4) |
| France | 96 | 16 | 16.7 (9.8, 25.7) | 12 | 12.5 (6.6, 20.8) | 4 | 4.2 (1.1, 10.3) | 3 | 3.1 (0.6, 8.9) |
| Belgium | 88 | 5 | 5.7 (1.9, 12.8) | 3 | 3.4 (0.7, 9.6) | 2 | 2.3 (0.3, 8.0) | 0 | 0 (0, 4.1) |
| Greece | 92 | 8 | 8.7 (3.8, 16.4) | 3 | 3.3 (0.7, 9.2) | 8 | 8.7 (3.8, 16.4) | 0 | 0 (0, 3.9) |
| Other | 234 | 17 | 7.3 (4.3, 11.4) | 7 | 3.0 (1.2, 6.1) | 6 | 2.6 (0.9, 5.5) | 7 | 3.0 (1.2, 6.1) |
| p-value | Transmitted Drug Resistance | |||
|---|---|---|---|---|
| Any | NRTI | NNRTI | PI | |
| Comparing regions* | 0.01 | 0.09 | 0.001 | 0.35 |
| Comparing countries within Europe | 0.002 | 0.004 | 0.004 | 0.06 |
Confidence intervals defined using the Clopper-Pearson Method
TDR defined according to WHO classification (2009) plus E138K and T215N
Other countries in Europe with <50 patients: Austria, Czech Republic, Denmark, Estonia, Finland, Ireland, Israel, Italy, Luxembourg, Norway, Poland, Portugal, Sweden, Switzerland.
Comparing regions=United States, Europe, Australia and Asia
Specific TDR mutations are shown in Table 4. NRTI mutations were overwhelmingly thymidine analogue mutations, particularly M41L and T215 revertants. The M184VI (associated with lamivudine and emtricitabine exposure) and K65R (associated with didanosine, abacavir, and tenofovir exposure) mutations were not detected in any samples. The most prevalent NNRTI mutation was K103NS, observed in 62% (53/85) of samples that harboured virus with resistance to this class. Similarly, M46IL (42%; 22/52) and L90M (25%; 13/52) were the most common PI mutations. There was no evidence in variation of specific mutations (within drug class) by geographical region (data not shown).
Table 4.
Individual TDR mutations
| NRTI | N (%) | NNRTI | N (%) | PI | N(%) | |||
|---|---|---|---|---|---|---|---|---|
| Any | 75 (4.0) | Any | 85 (4.5) | Any | 52 (2.8) | |||
| M41 | L | 28 (1.5) | L100 | I | 0 (0.0) | L23 | I | 0 (0.0) |
| K65 | R | 0 (0.0) | K101 | EP | 4 (0.2) | L24 | I | 1 (0.1) |
| D67 | NGE | 12 (0.6) | K103 | NS*** | 53 (2.8) | D30 | N | 3 (0.2) |
| T69 | D, Ins* | 3 (0.2) | V106 | MA | 2 (0.1) | V32 | I | 0 (0.0) |
| K70 | RE | 6 (0.3) | E138 | K | 5 (0.3) | M46 | IL | 22 (1.2) |
| L74 | VI | 0 (0.0) | V179 | F | 0 (0.0) | I47 | VA | 2 (0.1) |
| V75 | MTAS | 0 (0.0) | Y181 | CIV | 9 (0.5) | G48 | VM | 0 (0.0) |
| F77 | L | 4 (0.2) | Y188 | LHC | 5 (0.3) | I50 | VL | 1 (0.1) |
| Y115 | F | 0 (0.0) | G190 | ASE | 13 (0.7) | F53 | LY | 4 (0.2) |
| F116 | Y | 0 (0.0) | P225 | H | 1 (0.1) | I54 | VLMATS | 6 (0.3) |
| Q151 | M | 0 (0.0) | M230 | L | 1 (0.1) | G73 | STCA | 5 (0.3) |
| M184 | VI | 0 (0.0) | L76 | V | 1 (0.1) | |||
| L210 | W | 8 (0.4) | V82 | ATFSCML | 7 (0.4) | |||
| T215 | YFISCDVEN** | 51 (2.7) | N83 | D | 0 (0.0) | |||
| K219 | QENR | 16 (0.9) | I84 | VAC | 2 (0.1) | |||
| I85 | V | 3 (0.2) | ||||||
| N88 | DS | 2 (0.1) | ||||||
| L90 | M | 13 (0.7) | ||||||
Participants may have more than 1 mutation within each class, similarly they may have mutations for more than 1 class.
Based on 1869 participants from United States, Europe, Australia and Asia.
TDR defined according to WHO classification (2009) & E138K and T215N
1 participant had an insertion at position 69
2 participants had T215F, and 3 patients with T215Y mutations, 0 participants with T215N
7 participants had a K103S mutation.
TDR=transmitted drug resistance; NRTI=nucleoside reverse transcriptase inhibitor; NNRTI=non- nucleoside reverse transcriptase inhibitor; PI=protease inhibitor.
Drug susceptibility
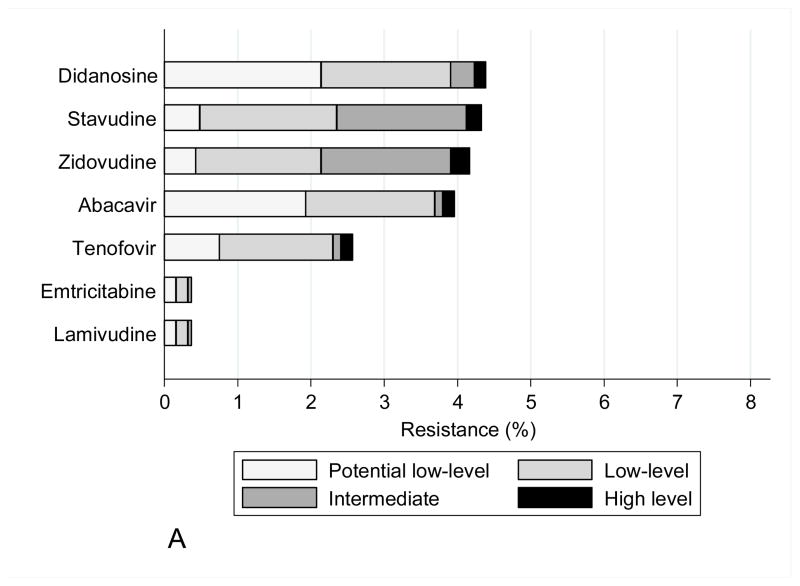
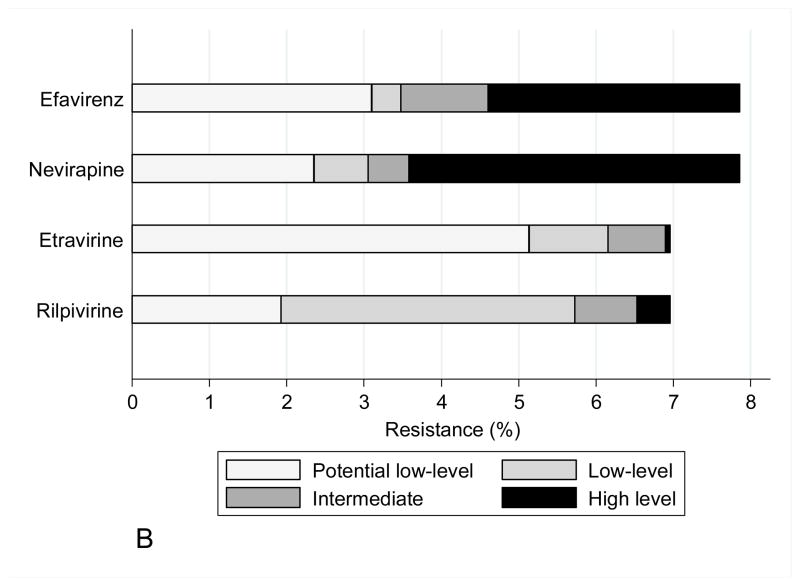
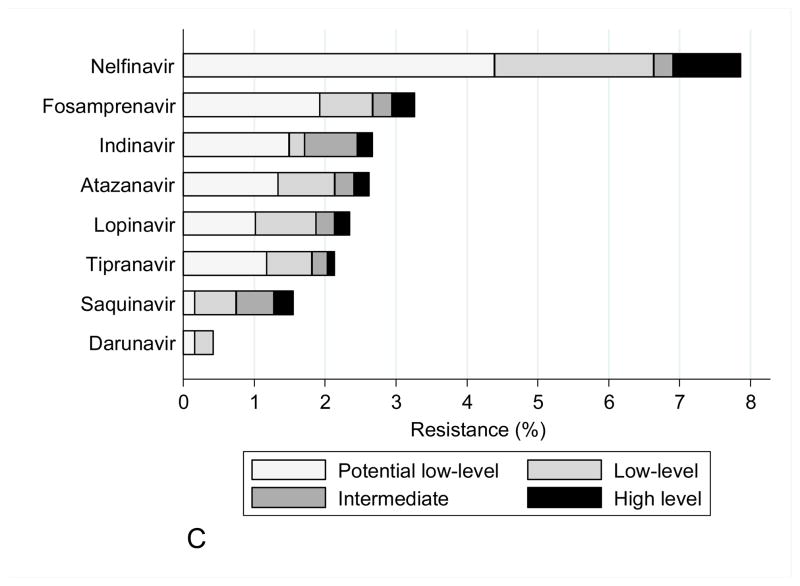
The mutations observed in START study participants did not have a significant impact on predicted drug susceptibility to most agents, other than to zidovudine and stavudine within the NRTI class, efavirenz and nevirapine within the NNRTI class, and nelfinavir within the PI class (Figure 1). Zidovudine resistance was mainly low or intermediate level. NNRTI resistance was mainly high level to efavirenz and nevirapine (at a frequency of 3–4%), largely reflecting the contribution of the K103N mutation. In isolation, this mutation is not associated with etravirine or rilpivirine resistance, and most viruses containing NNRTI mutations remained susceptible to these drugs.
Figure 1.
Figure 1A: Drug susceptibility for NRTI (Stanford v7.0.1)
Figure 1B : Drug susceptibility for NNRTI (Stanford v7.0.1)
Figure 1C : Drug susceptibility for PI (Stanford v7.0.1)
DISCUSSION
HIV-1 TDR has been well documented in resource-rich countries ever since the introduction of antiretroviral therapy more than two decades ago (5–19). Initial reports from these regions involved TDR most commonly to the NRTI class, as these agents were initially widely available for the treatment of HIV infection. Subsequently, there were increasing rates of NNRTI and PI TDR reported, although this has varied by geographic region (6). The source for TDR is from infected individuals in a population with uncontrolled viraemia, who may be treatment naïve or experienced. It has been demonstrated that individuals infected with drug-resistant strains may experience viral reversion to wild type over time after initial infection (17, 30, 31). This occurs when the major HIV population in an infected individual shifts from a predominantly drug-resistant variant to wild-type susceptible virus with improved fitness. However, TDR has been shown to persist in some individuals for many years after initial infection (3, 4, 13, 16, 32). Given that the population in START had CD4 cell counts above 500 cells/μL at study entry, most of these individuals were early in infection and less likely to have experienced reversion to wild-type virus.
Our study findings are reflective of the routine use of screening resistance testing to detect TDR in resource-rich compared to resource-limited countries. While the majority of study participants had locally performed resistance testing as standard of care in Europe, the United States, and Australia, less than 2% of participants enrolled in clinics in Africa and South America, and only 22% of those enrolled in Asian clinics had testing. In Europe, the United States, and Australia, testing was performed more often in participants diagnosed with HIV closer to study entry, likely due to test availability and updated guidelines over time in these countries. Of note, white participants and men who had sex with men were more likely to have resistance testing in these three geographic regions. This may reflect socioeconomic differences in these populations and access to resistance testing at some sites. Of note, selective use of resistance testing in resource-rich countries is not in line with current guidelines for care (20, 21, 33–35). We did not find a significant gender difference in screening resistance testing, although a large antiretroviral trial in the United States recently reported that women were significantly less likely to have locally performed baseline genotyping (36).
The overall prevalence of TDR in study participants was 10.1%, which is consistent with other reports of TDR prevalence in resource-rich countries (5, 6, 9–19). Australia and France had the highest rates of TDR, although the sample sizes were smaller from these countries. The lowest rate of TDR was observed in the United Kingdom. Although data are limited, one prior study from Australia reported higher rates of TDR (23%) particularly to the NRTI class, which was also seen in our data (6). Interestingly, there have been reports of declining or stable rates of TDR over time in the United Kingdom (6, 37, 38). There were a limited number of available tests from Asia, however we found a 8.0% prevalence rate of TDR, which is higher than prior average prevalence rates (4.2%) reported from pooled Asian data (6). Given that only 22% of Asian participants in START had resistance testing, it is possible that physician selection of those with elevated risk for TDR may have occurred.
When examining the presence of resistance by drug class, we found that overall TDR to NNRTIs and NRTIs was more common that resistance to PIs, which has also been observed in other prevalence studies (6, 9–19). Only a minority of participants had TDR to more than one drug class. The highest rate of transmitted NNRTI resistance (8.4%) was observed in participants enrolled from the United States. A review of global TDR prevalence showed that transmitted NNRTI resistance has historically been higher in North America compared to Europe and that rates increased in North America after the year 2003(6). A large surv eillance study conducted in the United States revealed transmitted NNRTI resistance in 7.8% of newly diagnosed individuals (13).
Among European countries, higher rates of transmitted NNRTI resistance were seen in START participants from Greece and Spain. Higher prevalence of transmitted NNRTI resistance in some countries may reflect the increased use of these agents in initial therapy. In addition, the long half-life of these agents has been associated with the selection of NNRTI RAMs in individuals with treatment interruptions (39, 40). Treatment interruptions have also been associated with a higher risk of detecting drug resistant HIV-1 in female genital tract secretions, potentially increasing the risk for TDR (41). It is unclear why the highest rates of transmitted NRTI resistance were observed in subjects from France and Australia, but this likely reflects higher prior exposure to this class in viruses from the pool of individuals transmitting HIV in these countries.
Of the individual TDR RAMs identified, the two most common positions were T215 and K103 in reverse transcriptase. Most mutations at the T215 position were revertants of the T215Y or T215F mutations, which are associated with thymidine analogue (zidovudine or stavudine) exposure. The next most frequent transmitted NRTI resistance mutations were also thymidine analogue associated positions M41, D67, and K219. Of note, in this patient population we did not observe transmitted NRTI resistance with M184VI. This mutation is often detected in treatment-experienced patients failing therapy but has been less commonly reported in TDR prevalence studies using standard genotyping (3, 9–17, 42). However, the M184VI can be transmitted and may be linked with other RAMs, but tends to wane over time due to overgrowth of more fit wild-type virus (42). Studies employing more sensitive methods of detecting RAMs such as ultra-deep sequencing have found the M184VI in treatment-naive chronically infected individuals with TDR (3, 43, 44).
As a group, the NNRTI mutations were the most frequently identified TDR RAMs. The K103N mutation was the single most frequently identified mutation and has been commonly reported in transmitted NNRTI drug resistance in North America and Europe. This mutation is typically associated with viruses exposed primarily to efavirenz and also to nevirapine. The next most commonly identified NNRTI mutations were at position G190, followed by Y181. The higher frequency of NNRTI mutations along with their effect on the first generation NNRTIs resulted in significantly reduced predicted susceptibility to these agents in approximately 5% of the study population.
Overall, transmitted protease resistance mutations were identified less frequently, with M46IL and L90M being the most common. The M46IL is considered a primary mutation for indinavir, but is also a common secondary mutation associated with the other PIs, except for saquinavir and darunavir (45). The L90M mutation has been associated with exposure to a number of PIs, but is classically associated with saquinavir and is considered a primary mutation for both saquinavir and nelfinavir (45). Given the transmitted protease mutations identified, darunavir was found to be the PI with the lowest predicted resistance.
Our analysis did not identify any significant individual-level predictors for the presence of TDR mutations in this patient population. Other prevalence studies from resource-rich countries have found predictors of TDR, such as age or gender, but others have not found any association with these factors, including socioeconomic status, duration of infection, race, risk group, homelessness, incarceration, or level of education (46). One recent Centers for Disease Control and Prevention (CDC) study of TDR in recently infected individuals in the United States reported higher rates of RAMs in younger individuals (13–19 years), whites and blacks compared to Hispanics/Latinos. (43)
This analysis was based on laboratory results from standard genotypic testing using Sanger sequencing; more sensitive assays examining low-frequency drug-resistance mutations were not available. Some studies have demonstrated higher prevalence of TDR in treatment-naïve populations using more sensitive resistance detection assays, such as ultra-deep sequencing and allele-specific PCR (3, 43, 44, 47, 48). Given the limitations of genotype assays used in this analysis, the actual prevalence of TDR in the START study population is likely greater than what has been reported here. Other limitations of this analysis are that the genotypes were performed locally in different laboratories, which could contribute to variability in reporting of mutations; also, data are lacking from resource-limited regions. Additional analysis using more sensitive assays in a broader sample of the study population will be helpful in further defining the prevalence of TDR by region and would enrich the dataset by testing samples from participants without access to local resistance testing, which was particularly common in resource-limited countries.
Another limitation is that START study participants were selected for a randomised clinical trial and hence may not be entirely representative of recently diagnosed individuals at these sites, although the simple and broad eligibility criteria for the study should have minimised this potential bias. Furthermore, no participant characteristics were found to be predictive of TDR. Although different resistance test technologies were used at the sites, these assays have become routine in resource-rich regions of the world and have undergone standardisation in last decade. Furthermore, our approach to data capture of genotype information has been used extensively in the EuroSIDA study and proven very reliable compared to centrally performed resistance tests done on stored biological material. Importantly, this data capture approach of information on individual mutations from many study participants allows for central validation and interpretation.
Because this analysis was confined to prerandomisation baseline data, we are unable to assess the impact of TDR on response to the initial regimens prescribed in study participants. Given the lack of availability of baseline resistance testing for a significant portion of the START trial population, those participants who had TDR that was not detected through resistance testing may be at greater risk of suboptimal responses to initial therapy. Comparing baseline TDR in the entire study population, antiretroviral regimens prescribed, and virologic responses will be important in future study analyses. It may also be useful to examine the virologic impact of TDR and the timing of initiating ART. It is possible that over the course of the study initial regimens prescribed, particularly in the deferred treatment group, may be less vulnerable to the effects of TDR.
The general goals of ART include maximal sustained suppression of HIV replication and enhanced quality of life for persons living with HIV (20–22). The selection of the initial antiretroviral regimen provides the greatest chance for long-term success in optimally suppressing viral replication. The practical importance of identifying TDR in an individual is for clinicians to avoid prescribing inactive agents as initial therapy, which may result in a higher likelihood of treatment failure. On a global level, data from TDR surveillance surveys can help guide the need for public health initiatives, such as improving antiretroviral treatment program effectiveness and potentially altering initial regimen selection, particularly in resource-limited countries.
Acknowledgments
Funding
The START study is primarily funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1-AI068641, the Department of Bioethics at the NIH Clinical Center and five NIH institutes: the National Cancer Institute, the National Heart, Lung, and Blood Institute, the National Institute of Mental Health, the National Institute of Neurological Disorders and Stroke and the National Institute of Arthritis and Musculoskeletal disorders. Financial support is also provided by the French Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS), the German Ministry of Education and Research, the European AIDS Treatment Network (NEAT), the Australian National Health and Medical Research Council, and the UK Medical Research Council and National Institute for Heath Research. Six pharmaceutical companies (AbbVie, Inc., Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline/ViiV Healthcare, Janssen Scientific Affairs, LLC, and Merck Sharp and Dohme Corp.) donate antiretroviral drugs to START.
We would like to thank the START participants without whom this work would not be possible. See INSIGHT START Study Group, 2015, this supplement for a complete list of START investigators.
Footnotes
The START study is registered at clinicaltrials.gov (NCT00867048).
Disclosures
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The University of Minnesota, the sponsor of START, receives royalties from the use of abacavir, one of the HIV medicines that can be used in START.
References
- 1.Pillay D, Bhaskaran K, Jurriaans S, et al. The impact of transmitted drug resistance on the natural history of HIV infection and response to first-line therapy. AIDS. 2006;20(1):21–28. doi: 10.1097/01.aids.0000196172.35056.b7. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5(7):e158. doi: 10.1371/journal.pmed.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199(5):693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 4.Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201(5):662–671. doi: 10.1086/650543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 6.Frentz D, Boucher CA, van de Vijver DA. Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS Rev. 2012;14(1):17–27. [PubMed] [Google Scholar]
- 7.Daar ES, Richman DD. Confronting the emergence of drug-resistant HIV type 1: impact of antiretroviral therapy on individual and population resistance. AIDS Res Hum Retroviruses. 2005;21(5):343–357. doi: 10.1089/aid.2005.21.343. [DOI] [PubMed] [Google Scholar]
- 8.Pillay D. Current patterns in the epidemiology of primary HIV drug resistance in North America and Europe. Antivir Ther. 2004;9(5):695–702. [PubMed] [Google Scholar]
- 9.Geretti AM. Epidemiology of antiretroviral drug resistance in drug-naive persons. Curr Opin Infect Dis. 2007;20(1):22–32. doi: 10.1097/QCO.0b013e328013caff. [DOI] [PubMed] [Google Scholar]
- 10.Weinstock HS, Zaidi I, Heneine W, et al. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1-infected persons in 10 US cities. J Infect Dis. 2004;189(12):2174–2180. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- 11.Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192(6):958–966. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 12.Cane P, Chrystie I, Dunn D, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: multicentre observational study. BMJ. 2005;331(7529):1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler WH, Ziebell RA, Zabina H, et al. Prevalence of transmitted drug resistance associated mutations and HIV-1 subtypes in new HIV-1 diagnoses, U.S-2006. AIDS. 2010;24(8):1203–1212. doi: 10.1097/QAD.0b013e3283388742. [DOI] [PubMed] [Google Scholar]
- 14.Ross L, Lim ML, Liao Q, et al. Prevalence of antiretroviral drug resistance and resistance-associated mutations in antiretroviral therapy-naive HIV-infected individuals from 40 United States cities. HIV Clin Trials. 2007;8(1):1–8. doi: 10.1310/hct0801-1. [DOI] [PubMed] [Google Scholar]
- 15.Novak RM, Chen L, MacArthur RD, et al. Prevalence of antiretroviral drug resistance mutations in chronically HIV-infected, treatment-naive patients: implications for routine resistance screening before initiation of antiretroviral therapy. Clin Infect Dis. 2005;40(3):468–474. doi: 10.1086/427212. [DOI] [PubMed] [Google Scholar]
- 16.Little SJ, Frost SD, Wong JK, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virology. 2008;82(11):5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanik EL, Napravnik S, Hurt CB, et al. Prevalence of transmitted antiretroviral drug resistance differs between acutely and chronically HIV-infected patients. J Acquir Immune Defic Syndr. 2012;61(2):258–262. doi: 10.1097/QAI.0b013e3182618f05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agwu AL, Bethel J, Hightow-Weidman LB, et al. Substantial multiclass transmitted drug resistance and drug-relevant polymorphisms among treatment-naive behaviorally HIV-infected youth. AIDS Patient Care STDs. 2012;26(4):193–196. doi: 10.1089/apc.2011.0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castor D, Low A, Evering T, et al. Transmitted drug resistance and phylogenetic relationships among acute and early HIV-1-infected individuals in New York City. J Acquir Immune Defic Syndr. 2012;61(1):1–8. doi: 10.1097/QAI.0b013e31825a289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson MA, Aberg JA, Hoy JF, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA. 2012;308(4):387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 21.Panel in Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agent in HIV-1 infected adults and adolescents. Department of Health and Human Services; [Accessed September 24, 2014]. Available at http://aidsinfo.nih.gov/contentfiles/lvguidelines/AdultandAdolescentGL.pdf. [Google Scholar]
- 22.European AIDS Clinical Society Guidelines Version 7.0. 2013 Oct; Available from: www.eacsociety.org.
- 23.Aghokeng AF, Monleau M, Eymard-Duvernay S, et al. Virological outcome and frequency of drug resistance mutations in HIV-infected patients receiving first-line ARV regimen and monitored with the public health approach in Southeast Asia and sub-Saharan Africa. Antivir Ther. 2012;17:A122. [Google Scholar]
- 24.Hassan AS, Mwaringa SM, Obonyo CA, et al. HIV-1 drug resistance amongst adults in a routine rural HIV clinic in Kenya. Antivir Ther. 2012;17:A126. [Google Scholar]
- 25.Nankya I, Mehta S, Akao J, et al. Trends of HIV-1 drug resistance during the past 11 years of ARV treatment in Uganda. Antivir Ther. 2012;17:A127. [Google Scholar]
- 26.Babiker AG, Emery S, Fatkenheuer G, et al. Considerations in the rationale, design and methods of the Strategic Timing of AntiRetroviral Treatment (START) study. Clin Trials. 2013;10(1 Suppl):S5–S36. doi: 10.1177/1740774512440342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.HIV Collaboration Data Exchange Protocol. Available from: www.hidep.org.
- 28.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clinical Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodes B, Garcia F, Gutierrez C, et al. Impact of drug resistance genotypes on CD4+ counts and plasma viremia in heavily antiretroviral-experienced HIV-infected patients. J Med Virol. 2005;77(1):23–28. doi: 10.1002/jmv.20395. [DOI] [PubMed] [Google Scholar]
- 31.Gandhi RT, Wurcel A, Rosenberg ES, et al. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin Infect Dis. 2003;37(12):1693–1698. doi: 10.1086/379773. [DOI] [PubMed] [Google Scholar]
- 32.Castro H, Pillay D, Cane P, et al. Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis. 2013;208(9):1459–1463. doi: 10.1093/infdis/jit345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV Protease Inhibitors: raising the barrier to resistance. Antivir Res. 2010;85(1):59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme AM, Camacho RJ, Ceccherini-Silberstein F, et al. European recommendations for the clinical use of HIV drug resistance testing: 2011 update. AIDS Rev. 2011;13(2):77–108. [PubMed] [Google Scholar]
- 35.Asboe D, Aitken C, Boffito M, et al. British HIV Association guidelines for the routine investigation and monitoring of adult HIV-1-infected individuals 2011. HIV Med. 2012;13(1):1–44. doi: 10.1111/j.1468-1293.2011.00971.x. [DOI] [PubMed] [Google Scholar]
- 36.Smith KY, Tierney C, Mollan K, et al. Outcomes by sex following treatment initiation with atazanavir plus ritonavir or efavirenz with abacavir/lamivudine or tenofovir/emtricitabine. Clin Infect Dis. 2014;58(4):555–563. doi: 10.1093/cid/cit747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dolling D, Sabin C, Delpech V, et al. Time trends in drug resistant HIV-1 infections in the United Kingdom up to 2009: multicentre observational study. BMJ. 2012;345:e5253. doi: 10.1136/bmj.e5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UK Collaborative Group on HIV Drug Resistance, UK Collaborative HIV Cohort Study, and UK Register of HIV Seroconverters.. Evidence of a decline in transmitted HIV-1 drug resistance in the United Kingdom. AIDS. 2007;21(8):1035–1039. doi: 10.1097/QAD.0b013e3280b07761. [DOI] [PubMed] [Google Scholar]
- 39.Hare CB, Mellors J, Krambrink A, et al. Detection of nonnucleoside reverse-transcriptase inhibitor-resistant HIV-1 after discontinuation of virologically suppressive antiretroviral therapy. Clin Infect Dis. 2008;47(3):421–424. doi: 10.1086/589867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox Z, Phillips A, Cohen C, et al. Viral resuppression and detection of drug resistance following interruption of a suppressive non-nucleoside reverse transcriptase inhibitor-based regimen. AIDS. 2008;22(17):2279–2289. doi: 10.1097/QAD.0b013e328311d16f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graham SM, Jalalian-Lechak Z, Shafi J, et al. Antiretroviral treatment interruptions predict female genital shedding of genotypically resistant HIV-1 RNA. J Acquir Immune Defic Syndr. 2012;60(5):511–518. doi: 10.1097/QAI.0b013e31825bd703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wainberg MA, Moisi D, Oliveira M, Toni TD, Brenner BG. Transmission dynamics of the M184V drug resistance mutation in primary HIV infection. J Antimicrob Chemother. 2011;66(10):2346–2349. doi: 10.1093/jac/dkr291. [DOI] [PubMed] [Google Scholar]
- 43.Li JF, Kim D, Linley R, et al. Sensitive screening reveals widespread underestimation of transmitted HIV drug resistance. Conference of Retroviral and Opportunistic Infections; Boston, MA. March 2014; p. Abstract 87. [Google Scholar]
- 44.Lataillade M, Chiarella J, Yang R, et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PloS One. 2010;5(6):e10952. doi: 10.1371/journal.pone.0010952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim R, Baxter JD. Protease inhibitor resistance update: where are we now? AIDS Patient Care STDs. 2008;22(4):267–277. doi: 10.1089/apc.2007.0099. [DOI] [PubMed] [Google Scholar]
- 46.Snedecor SJ, Khachatryan A, Nedrow K, et al. The prevalence of transmitted resistance to first-generation non-nucleoside reverse transcriptase inhibitors and its potential economic impact in HIV-infected patients. PloS One. 2013;8(8):e72784. doi: 10.1371/journal.pone.0072784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305(13):1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geretti AM, Conibear T, Hill A, et al. Sensitive testing of plasma HIV-1 RNA and Sanger sequencing of cellular HIV-1 DNA for the detection of drug resistance prior to starting first-line antiretroviral therapy with etravirine or efavirenz. J Antimicrob Chemother. 2014;69(4):1090–1097. doi: 10.1093/jac/dkt474. [DOI] [PubMed] [Google Scholar]