SUMMARY
An interesting concept in the organization of cellular membranes is the proposed existence of lipid rafts. Membranes of eukaryotic cells organize signal transduction proteins into membrane rafts or lipid rafts that are enriched in particular lipids such as cholesterol and are important for the correct functionality of diverse cellular processes. The assembly of lipid rafts in eukaryotes has been considered a fundamental step during the evolution of cellular complexity, suggesting that bacteria and archaea were organisms too simple to require such a sophisticated organization of their cellular membranes. However, it was recently discovered that bacteria organize many signal transduction, protein secretion, and transport processes in functional membrane microdomains, which are equivalent to the lipid rafts of eukaryotic cells. This review contains the most significant advances during the last 4 years in understanding the structural and biological role of lipid rafts in bacteria. Furthermore, this review shows a detailed description of a number of molecular and genetic approaches related to the discovery of bacterial lipid rafts as well as an overview of the group of tentative lipid-protein and protein-protein interactions that give consistency to these sophisticated signaling platforms. Additional data suggesting that lipid rafts are widely distributed in bacteria are presented in this review. Therefore, we discuss the available techniques and optimized protocols for the purification and analysis of raft-associated proteins in various bacterial species to aid in the study of bacterial lipid rafts in other laboratories that could be interested in this topic. Overall, the discovery of lipid rafts in bacteria reveals a new level of sophistication in signal transduction and membrane organization that was unexpected for bacteria and shows that bacteria are more complex than previously appreciated.
INTRODUCTION
Cellular membranes define a dynamic boundary with the environment. Biological membranes are constituted by a specific set of lipids and proteins, and their correct organization influences all cellular processes (1). Consequently, the organization of membrane components has been an important research topic in the past decades (2–4). The pioneering fluid mosaic model (5) proposed by Singer and Nicolson in 1972 suggests that all membrane constituents diffuse freely and, thus, distribute randomly. In this way, all membrane-embedded lipids and proteins are laterally mobile. The fluid mosaic model left open the possibility of the existence of mechanisms to obtain long-range order in a homogeneous fluid system (6), which seeded further investigations to demonstrate that cellular membranes are much more complex organelles than previously thought (7). In particular, it was discovered that biological membranes are constituted by a large variety of different lipid species showing distinct physicochemical properties (8–10). Importantly, the existence of diverse lipid species results in their lateral segregation into membrane microdomains because they tend to coalescence due simply to their physicochemical affinities (11, 12). The heterogeneous organization of membrane lipids into discrete microdomains leads to a diverse distribution of embedded membrane proteins, which appears to be essential for their functionality (12–14). This lateral separation of membrane lipids and proteins is now referred to as membrane domains (15, 16).
Many types of cells contain distinct membrane domains, although their presence has been long recognized in eukaryotic cells only. For instance, polarized epithelial cells show a lateral organization of the membrane to distinguish a basolateral and an apical membrane macrodomain, showing different lipid and protein compositions and being specialized in different roles (17–19). Neurons also have membrane domains with different lipids and proteins, which are catalogued according to their role in synapsis (20, 21). However, the existence of membrane domains is not an exclusive feature of eukaryotic cells. Membrane domains are also evident in bacteria and archaea. In fact, membrane organization is particularly important in unicellular organisms, as it represents the boundary between the organism and the environment and therefore orchestrates many cellular processes that are essential for life, such as cell division or signal transduction (22–25). For instance, the use of specific lipid dyes (e.g., nonyl-acridine orange [NAO]) has demonstrated the presence of cardiolipin-enriched domains at the cell poles and at the division septum in Escherichia coli and Bacillus subtilis bacterial cells (26–29). Although the specificity of this dye for the detection of cardiolipin has been questioned recently (30), the localization pattern of NAO-enriched domains in E. coli and B. subtilis suggests a lateral organization of lipids in bacterial membrane that may be correlated with cell division and morphogenesis (26–29).
An interesting concept in membrane organization is the proposed existence of lipid rafts or membrane rafts (31). Membranes of eukaryotic cells organize a variety of proteins related to signal transduction and membrane trafficking into microdomains or rafts that are enriched in particular lipids such as cholesterol or sphingolipids (31). Lipid rafts also harbor specific proteins. One of the raft-associated proteins is commonly referred to as reggie or flotillin (32–36). Flotillin proteins are membrane-bound chaperones that localize to lipid rafts, where they may recruit the proteins that need to be localized in lipid rafts to be active and facilitate their interaction and oligomerization (32–36). Thus, flotillin proteins are important components of lipid rafts and play a central role in the organization of lipid rafts (33, 35, 36). Because of this, flotillin proteins are considered bona fide markers of the subcellular localization of lipid rafts (32–36). The activity of flotillin is important for the correct functionality of numerous raft-associated cellular processes, including membrane sorting, trafficking, cell polarization, and signal transduction. Consequently, the perturbation of the activity of flotillin causes serious defects in signal transduction and membrane trafficking (32–36), which seems to be related to the occurrence of severe diseases such as Alzheimer's disease and Parkinson's disease (37).
The existence of lipid rafts has been traditionally associated with eukaryotic cells because their assembly depends on the presence of cholesterol, which is absent from the membranes of most bacteria and archaea. Thus, the assembly of lipid rafts in eukaryotes has been considered a fundamental step during the evolution of cellular complexity, suggesting that bacteria and archaea were too simple to require such a sophisticated organization of their signaling networks and membrane-associated protein complexes. However, bacteria also show a variety of membrane-associated sensory complexes, such as the ones involved in bacterial chemotaxis, which organize into large clusters that integrate and amplify stimuli before transmitting the signal to downstream proteins (38, 39). Thus, this demonstrates that bacteria are also able to organize their signal transduction systems into signaling platforms of a certain complexity.
In addition to the above-mentioned findings, it was recently shown that bacteria are also able to organize many signal transduction cascades and protein transport into functional membrane microdomains (FMMs) constituted by specific lipids (40); i.e., bacterial membranes contain lipid rafts similar to those found in eukaryotic cells (31). The assembly of FMMs involves the biosynthesis of polyisoprenoid lipids in the membrane and their colocalization with flotillin-like proteins, which are also present in bacteria (41). Bacterial flotillins seem to play a role similar to the one played by eukaryotic flotillins, acting as protein scaffolds in recruiting proteins that need to be localized in lipid rafts to promote interactions and oligomerization (42, 43). Similarly to eukaryotic flotillin proteins, flotillins in bacteria play an essential role in organizing and maintaining the correct architecture of the FMMs. The discovery of FMMs in bacterial membranes led many laboratories in the last 5 years to experimentally test critical aspects of this discovery. This review compiles the information that is currently available about the existence and biological role of bacterial lipid rafts. Furthermore, current protocols that are important in exploring the existence of bacterial lipid rafts are detailed, as are the possible lines of evolution of this new research field of growing importance.
DISCOVERY OF FUNCTIONAL MEMBRANE MICRODOMAINS IN BACTERIA
FMMs of bacteria were unexpectedly discovered during investigations of biofilm formation using the model organism B. subtilis. The membrane-associated sensor kinase KinC, which triggers biofilm formation in B. subtilis (44, 45), lost its functionality in a ΔyisP mutant that was unable to produce certain membrane-related polyisoprenoid lipids. The activity of KinC and, thus, biofilm formation were recovered in the ΔyisP mutant when polyisoprenoid lipids were added to the cultures at different concentrations (40). Initially, it was assumed that YisP is a squalene synthase. However, it was recently shown that YisP acts as a phosphatase, catalyzing the formation of farnesol from farnesyl diphosphate (46). The ΔyisP mutant is severely affected in the activity of several membrane-associated proteins, including KinC (40), thereby inhibiting biofilm formation. This led to the hypothesis that bacteria could compartmentalize membrane-bound sensor kinases into discrete FMMs differing in lipid composition from the rest of the membrane.
Analysis of bacterial membrane microdomains was performed according to the approaches established for the examination of eukaryotic lipid rafts (47, 48). This procedure is based on the ability of lipid rafts to resist disaggregation by nonionic detergent treatment by taking advantage of their variable lipid composition. Their variable lipid composition makes them more compact and gives them more-hydrophobic regions than the rest of the membrane, and they are thus more resistant to detergent disaggregation (47, 48). The different membrane fragments can be separated according to their size in a sucrose gradient. This treatment resulted in one membrane fraction that is sensitive to detergents (detergent-sensitive membrane [DSM] fraction) and another fraction composed of larger membrane fragments that is more resistant to detergent disruption (detergent-resistant membrane [DRM] fraction). The importance of not equating the DRM fraction with lipid rafts is emphasized several times during this review. Results obtained by using this technique need further validation. Nevertheless, there is strong evidence that the DRM fraction includes proteins thought to be present in lipid rafts, and the DRM fraction is considered a membrane fraction enriched in lipid rafts (47, 48). Purification of DSM and DRM fractions from bacterial membranes and analysis of their protein composition showed a heterogeneous distribution of proteins in B. subtilis. Mass spectrometry analysis of the proteins from the DRM fractions revealed a number of proteins involved in cell signaling, transport, and protein secretion (40). When these membrane regions were purified and their protein contents were analyzed, KinC was found exclusively in association with the DRM fraction, along with many other signaling proteins (40). This result demonstrates that the FMMs of B. subtilis are enriched in proteins that specialize in signal transduction and protein secretion.
It is important to highlight that the above-mentioned membrane fractionation experiments were preceded by other studies that demonstrated membrane fractionation in Bacillus halodurans and Bacillus subtilis (49, 50). These two publications provided evidence for the existence of DSM and DRM fractions in bacterial membranes (49, 50). Importantly, the DRM fraction was enriched in an uncharacterized membrane protein, which is a homologue of the flotillin proteins of eukaryotic cells (50). The YuaG (renamed FloT) flotillin-like protein from B. subtilis showed a heterogeneous distribution in the cytoplasmic membrane, displaying a punctate pattern along the entire cell (49) (Fig. 1). While no exact function was ascribed to these bacterial proteins, a B. subtilis mutant lacking FloT showed a reduced sporulation efficiency as a consequence of a defective activation of the signaling pathway for sporulation, demonstrating that this protein plays an important role in the signaling pathways in B. subtilis (49). Attempts to determine the type of lipid that led to the punctate distribution of the B. subtilis flotillin-like protein were inconclusive, finding only that the FloT-containing DRM fraction was enriched in cardiolipin (49).
FIG 1.
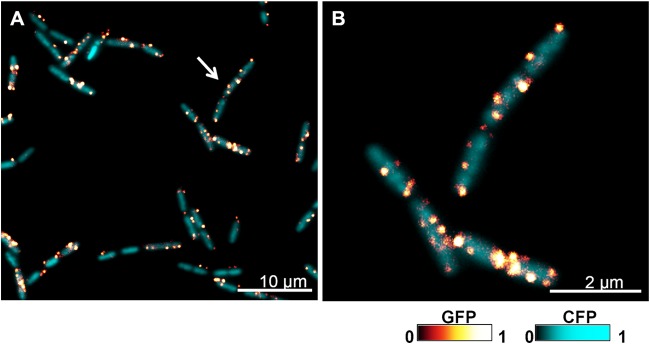
Subcellular localization of flotillin in B. subtilis cells. Shown are fluorescence microscopy images of B. subtilis cells labeled with the translational fusion FloT-GFP (green fluorescent protein). The GFP signal is false colored on a glow-dark logarithmic scale. Cells also constitutively expressed CFP (cyan fluorescent protein) to facilitate the visualization of cells (false colored on a cyan-dark scale). (A) Fluorescence microscopy image of a field of cells labeled with FloT-GFP (glow-dark scale) expressed under the control of its natural promoter and the Pspac-cfp reporter, which constitutively expresses CFP (cyan-dark scale). The arrow indicates the subset of cells that are magnified in panel B. Bar, 10 μm. (B) Fluorescence microscopy detail of cells magnified from panel A. Cells are labeled with FloT-GFP and Pspac-cfp reporters. Bar, 2 μm.
Further protein analysis of the DRM fraction in B. subtilis identified, besides FloT, a number of signaling proteins along with a second flotillin-like protein, which was referred to as YqfA (renamed FloA) (40). Importantly, these two flotillin-like proteins were found to colocalize with KinC in the same membrane regions. B. subtilis mutants lacking flotillin genes showed a noticeable impairment in the activity of KinC, which caused an abrogation of the ability of B. subtilis to form biofilms (40). These results were consistent with the idea that FloA and FloT were two different flotillin-like proteins that act as scaffold proteins in making the activation of the raft-associated signaling pathways more efficient (40, 51).
The colocalization of FloA and FloT with KinC and other signaling-related proteins disappeared when the production of membrane polyisoprenoid lipids was inhibited (40, 52, 53). This effect was achieved by chemically inhibiting YisP using competitive inhibitors of this specific enzyme, such as zaragozic acid. Zaragozic acid can be efficiently incorporated into the active site of YisP to prevent the processing of farnesol (52). As a consequence, nanomolar concentrations of these compounds added to cultures of B. subtilis caused a strong inhibition of the production of the constituent lipids and concomitantly a severe disruption of FMMs, manifested by the dispersion of the protein cargo to the overall cellular membrane (40). The dispersion of protein cargo resulted in a severe reduction of the activity of the associated signal transduction pathways. For instance, nanomolar concentrations of zaragozic acid added to cultures of B. subtilis inhibit the activity of the raft-associated kinase KinC and, therefore, cause a severe inhibition of biofilm formation (40).
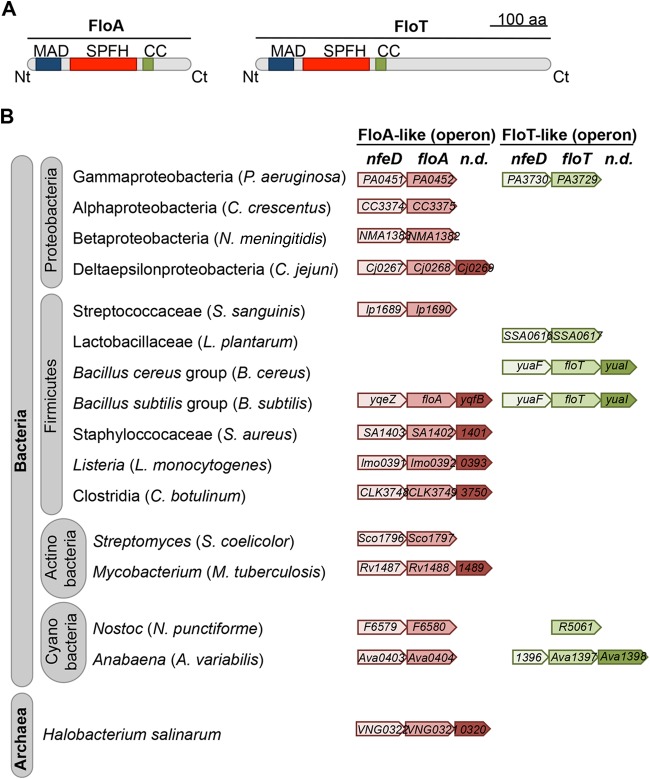
It is possible that the existence of FMMs is universal in bacteria, given that almost all bacterial species harbor at least one flotillin-like protein-encoding gene in their genome. An overview of the presence of flotillin genes in bacteria is shown below in Fig. 3. For instance, the opportunistic pathogen Staphylococcus aureus contains one flotillin gene in its genome, which codes for a protein that is highly similar to FloA of B. subtilis (∼90% identity) (40). Furthermore, when the protein content of the staphylococcal FMMs was isolated and identified, a significant number of proteins related to signal transduction were detected along with FloA (40). Nanomolar concentrations of zaragozic acid added to cultures of S. aureus caused an inhibitory effect similar to the one described for cultures of B. subtilis (40), which is manifested by the inability of cells to produce the golden-colored carotenoid staphyloxanthin that gives the typical yellow coloration to S. aureus (54). In addition to the loss of pigmentation, S. aureus cells showed a strong inhibition of the signal transduction pathways that were detected in association with the staphylococcal FMMs. Consequently, the perturbation of the architecture of FMMs by inhibiting the biosynthetic pathway responsible for the production of the constituent lipids in S. aureus causes a potent and simultaneous inhibition of processes related to the development of infections (40). After the existence of FMMs was reported, the increasing interest of the scientific community in this topic has generated a considerable number of publications, which explore the implications of bacterial lipid rafts in the activation of diverse signaling pathways as well as the aggregation of specific lipids (51, 55–58). The increasing interest in this new field has already consolidated this topic into an emerging area of research in microbiology.
FIG 3.
Taxonomic distribution of FloA and FloT in bacteria. (A) Schematic representation of the molecular structures of two different flotillin proteins, FloA and FloT, from the model organism Bacillus subtilis. MAD represents a membrane-anchoring region (whether this is a transmembrane helix or a hairpin loop has not yet been experimentally addressed). SPFH is a typical protein domain of flotillin proteins, and CC represents a coil-coiled region that localizes at the C-terminal regions of these two proteins. (B) Distribution of the FloA and FloT operons in bacteria. The first gene of the operon codes for an NfeD-like protein. The second gene is the flotillin-encoding gene. The third gene, coding for a protein of unknown function, is less well conserved and is absent from various species. Shown is the architecture of the operons from bacterial species from different phyla as a reference. The operons contain provisional gene names given by genome annotation. S. sanguinis, Streptococcus sanguinis; C. botulinum, Clostridium botulinum; n.d., not determined.
MOLECULAR ARCHITECTURE OF FMMs IN BACTERIA
FMMs are supposed to be membrane platforms highly enriched in polyisoprenoid lipids; thus, their lipid composition differs from that of the surrounding membrane. The constituent polyisoprenoid lipids confer compact and hydrophobic properties to these membrane regions, which probably causes a selective reduction in the diffusion of specific membrane proteins that ultimately tend to coalesce and concentrate in the FMMs (11, 12, 20, 31, 59, 60). The identity of the harbored proteins (here referred to as protein cargo) varies considerably according to the physiological state of the cells. Therefore, the identification of the protein components of the cargo depends on the experimental conditions. In contrast, the FMMs contain two important structural components that provide consistency to these signaling platforms and are present under all experimental conditions tested. These components are the constituent lipids and the flotillin protein. These two structural components can be considered bona fide markers of the presence of FMMs under any experimental condition under study (61).
Constituent Lipids of the FMMs
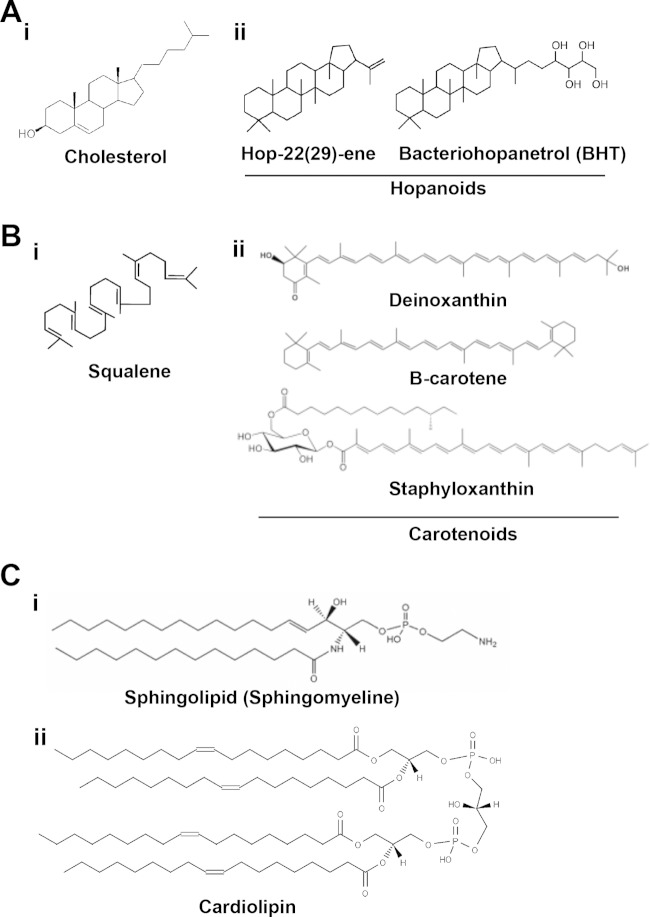
Cellular membranes are composed of a large number of distinct lipid species, which differ in their molecular structures and physicochemical properties (62). These constituent lipids tend to coalesce into microdomains. This phenomenon is known as “lipid ordering” and is the organizing principle of membrane microdomains (11, 63). For instance, the ability of cholesterol and sphingolipids to modulate lipid ordering in cellular membranes is a hallmark of the existence of lipid rafts in eukaryotic cells (31). Bacterial membranes are also composed of distinct lipid species, showing different molecular structures and physicochemical properties. Therefore, it is plausible that lipid ordering also occurs in bacterial membranes, in a fashion similar to that which occurs in eukaryotic cells (11, 12, 31). However, cholesterol is absent from the membranes of most bacteria (with notable exceptions, such as Borrelia burgdorferi, Helicobacter pylori, Mycoplasma spp., Ehrlichia chaffeensis, and Anaplasma phagocytophilum), and therefore, the organization of FMMs should depend on the presence and self-aggregation of sterol surrogates. Alternatively, microorganisms such as Borrelia burgdorferi display specific mechanisms to sequester extracellular cholesterol and chemically modify it to incorporate it into their membranes and generate cholesterol-enriched membrane microdomains (64, 65). It was shown that sterols support lipid raft formation in B. burgdorferi similar to the formation of eukaryotic lipid rafts (66). Membrane domain formation was visualized by using electron microscopy (EM) immunogold labeling of B. burgdorferi with antibodies directed against cholesterol glycolipid. The formation of cholesterol glycolipid-containing membrane domains was retained when cholesterol was replaced by other sterols such as ergosterol and stigmasterol. The loss of sterols from B. burgdorferi membranes prevents the isolation of detergent-resistant membranes (66).
Although the molecular structure of the constituent lipids of the bacterial FMMs is yet to be elucidated, numerous genetic and molecular assays suggest that they are polyisoprenoid lipids similar to cholesterol (40). Self-aggregation of polyisoprenoid lipids confers rigid, compact, and hydrophobic properties to the membrane microdomains (like a floating raft, hence the name lipid raft), similar to the lipid rafts of eukaryotic cells (11, 12, 31). Bacterial membranes contain several lipid species that could coalesce into microdomains and should be considered potential candidates for the assembly of FMMs. For instance, it is possible to detect cyclic polyisoprenoid lipids structurally similar to the cholesterol of eukaryotic cells. These molecules are commonly referred to as hopanoids (from the plant genus Hopea, from which they were isolated as components of the resin) and as sporulenes, in the case of B. subtilis (67, 68) (Fig. 2). Hopanoids are structurally diverse (69–75), although diplopterol or amino-functionalized methylbacteriohopanepolyols may be the most abundant (76). The production of hopanoids requires the cyclization of the polyisoprenoid lipid squalene by the enzymatic action of a squalene-hopene cyclase (SqhC), which releases polycyclic terpenoids called hopanoids (73, 74, 77, 78). There are many unknowns regarding the production of hopanoids in bacteria. For instance, the reaction catalyzed by SqhC seems to occur in the absence of oxygen, but many hopanoids are produced by aerobic bacteria (mostly methanotrophs, heterotrophs, and cyanobacteria), and only a few hopanoid species have been detected in facultative anaerobes and strict anaerobes (68). This could be due to the bias that exists for the cultivation of anaerobic organisms, because hopanoids are typically much more abundant in anoxic environments. In general, there is no clear taxonomic pattern other than that hopanoids seem to be very prevalent in alphaproteobacteria and cyanobacteria, and hopanoids do not occur in archaea and eukaryotes (73, 74, 77). The biological role of hopanoids in bacterial membranes is yet to be elucidated. Nevertheless, it is largely assumed that hopanoids modulate the fluidity of membranes to increase the degree of lipid order or membrane rigidity (57) at high temperatures. It is apparent that hopanoids are important for the correct functionality of numerous cellular processes and membrane-associated signal transduction cascades in bacteria (67, 79–82). For instance, the Gram-negative bacterium Rhodopseudomonas palustris divides asymmetrically into a mother cell and a swarmer cell. Membrane hopanoids seem to play a crucial role in guiding the asymmetrical division of this bacterium. Hopanoid mislocalization causes defective division such that cells remain connected by their cell wall, forming long filaments. From these results, one can hypothesize that the lack of hopanoids severely compromises cell growth in this bacterium, as has been reported (83). Additionally, it has been reported that hopanoids can replace cholesterol in Mycoplasma cells without compromising cell growth (79). In fact, it was recently demonstrated that hopanoid molecules inserted into membranes are able to coalescence and induce phase separation (57), which supports the possibility that hopanoids play a role similar to that of cholesterol in the assembly of lipid rafts.
FIG 2.
Molecular structure of the constituent lipids of eukaryotic and bacterial lipid rafts. (A) Polycyclic terpenoids. (i) Molecular structure of cholesterol, the main constituent lipid of eukaryotic lipid rafts. (ii) Cholesterol is not present in bacterial membranes. Bacterial membranes contain other polycyclic terpenoids, which are structurally similar to cholesterol and referred to as hopanoids. (B) Noncyclic terpenoids. (i) Molecular structure of squalene, the precursor molecule of polycyclic and noncyclic terpenoids. (ii) An example of noncyclic terpenoids is carotenoids, which are widely distributed in bacteria. For instance, the carotenoid lipid staphyloxanthin is responsible for the golden coloration of the pathogen Staphylococcus aureus. (C) Molecular structure of sphingolipids (i) and cardiolipin (ii). Sphingolipids are sphingosine-based membrane lipids known to provide consistency to the lipid rafts of eukaryotic cells.
Although the main constituent lipids of bacterial membranes are glycerol-based phospholipids, other types of lipids in addition to hopanoids can be found at a relatively high frequency in bacterial membranes. Bacterial membranes contain a large variety of noncyclic polyisoprenoid lipids. Despite the structural differences between noncyclic and cyclic polyisoprenoid lipids, their polyisoprenoid nature confers similar physicochemical properties to these lipids. An important example of noncyclic polyisoprenoid lipids that are abundant in bacteria is carotenoids (84) (Fig. 2). The carotenoids are a large family of pigmented membrane-associated polyisoprenoid lipids that exhibit antioxidant properties and are capable of scavenging reactive oxygen species (84–86). They play an important role in regulating the rigidity of the bacterial membrane in a manner similar to that observed for cholesterol in eukaryotic cells (84, 85, 87, 88). However, the high level of structural diversity between members of the large family of carotenoid molecules is worth noting. It is possible that various subfamilies of carotenoids are present in the membrane, exhibiting different physicochemical properties, which may exert distinct effects on the physiology and structure of the cellular membrane. A large number of bacterial species are able to produce carotenoids, including Staphylococcus (89). Specifically, the ability of the genus Bacillus to produce carotenoids was recently discovered (90, 91). This finding is consistent with the hypothesis that carotenoids or similar noncyclic polyisoprenoid molecules are relevant constituent lipids of the FMMs of B. subtilis (40).
The biosynthetic pathway that leads to the production of the constituent polyisoprenoid lipids in B. subtilis requires the activity of YisP (40, 46, 92). Consequently, ΔyisP mutants are unable to produce the membrane-related polyisoprenoid lipids or sesquiterpenes, and the integrity of the FMMs is selectively affected. In S. aureus, a squalene synthase, CrtM, synthesizes squalene by head-to-head condensation of farnesol. CrtM likely converts two molecules of farnesylpyrophosphate into squalene or dehydrosqualene, depending on whether the reaction includes or does not include NADPH as a reducing agent, respectively (92, 93). The B. subtilis YisP enzyme is a structural homologue but likely dephosphorylates farnesylpyrophosphate into farnesol (46, 94). Dehydrosqualene or phytoene is the precursor molecule of carotenoid molecules. Thus, it is reasonable to think that molecules related to carotenoids are part of the constituent lipids of S. aureus FMMs. Moreover, the supplementation of cultures with different carotenoids or farnesol can complement the ΔyisP mutant of B. subtilis and partially recover the loss of functionality of the raft-harbored kinases, leading to a partial recovery of the ability to form a biofilm (40, 46). This result also argues that a lipid factor produced by YisP and CrtM is necessary for the assembly of flotillins in the FMMs of B. subtilis and S. aureus, respectively.
Moreover, B. subtilis flotillins can be copurified with an additional type of lipid known as cardiolipin (49) (Fig. 2). Cardiolipin is a specific type of diphosphatidylglycerol lipid, so it shows a dimeric structure in which two phosphatidyl moieties are linked by a glycerol molecule. Cardiolipin is found exclusively in the inner mitochondrial membrane and in most bacterial membranes. Here, cardiolipin represents ∼30% of the total membrane lipids during the stationary growth phase (95). In mitochondria, cardiolipin is essential for the functionality of numerous membrane-bound proteins that are involved in energy metabolism (96). In bacteria, mutants lacking cardiolipin are viable, but they show severe growth defects when exposed to high-salt conditions, indicating the importance of cardiolipin in modulating the membrane composition to adapt to stress conditions and adjust membrane fluidity (26, 27, 97, 98). Recent advances in the development of new fluorescent probes and visualization techniques allowed the identification of cardiolipin domains in bacterial membranes (26, 27). For instance, the lipid dye NAO allows the visualization of the subcellular distribution of cardiolipin domains in bacterial membranes (26, 27), although the specificity of this dye for cardiolipin was recently questioned (30). Nevertheless, this dye organizes in membrane microdomains, preferentially positioned at the poles and septal regions of E. coli and B. subtilis cells, which are enriched in cardiolipin (26, 27). The subcellular distribution of cardiolipin influences many membrane-associated proteins and serves as a cue for the localization of proteins at specific sites in the cell, more specifically the poles and the septation site (99). As it has already been demonstrated that cardiolipin participates in the lipid ordering of bacterial membranes, it is possible that cardiolipin participates as one of the constituent lipids of FMMs and serves as a cue for the recruitment of other proteins that need to be in the FMMs to be active.
Bacterial Flotillin
Flotillin is an abundant protein of lipid rafts and therefore is considered an important protein marker for the detection and visualization of lipid rafts (32, 35, 43, 100–102). Flotillins are evolutionarily well conserved from bacteria to humans. Flotillin proteins (also called reggie proteins) (103) are considered scaffold proteins that favor the recruitment of proteins that need to be in the lipid rafts to be functional, organizing signaling complexes and promoting the interactions of raft-associated proteins (42, 43, 104). Thus, the scaffold activity of flotillins is expressed in tethering protein or lipid components to facilitate their efficient interaction and oligomerization and to mediate the efficient activation of the signal transduction pathways (42, 43, 104). Consequently, alterations in the functionality of flotillins significantly influence the large number of raft-associated cellular processes (35, 37, 105, 106). Flotillin proteins were found in the insoluble fraction of Triton X-100, which floated after sucrose centrifugation and hence were named flotillins (100). Flotillins are also highly upregulated in regenerating axons after lesion formation and thus are called reggies (101). Eukaryotic lipid rafts contain two homologous members of the flotillin or reggie proteins, referred to as flotillin-1 and flotillin-2 (reggie-2 and reggie-1, respectively). For simplicity, we do not use the reggie terminology in this review. Both flotillins are ubiquitously present in mammalian tissues and associated with each other in hetero-oligomeric complexes (33, 34, 36, 106) and seem to have a strong regulatory correlation (107–109).
Flotillin-1 and flotillin-2 are anchored to the cytoplasmic membrane by their N-terminal regions via myristoyl and palmitoyl moieties, respectively, like many other proteins that are known to localize in lipid rafts (110, 111). Flotillins belong to a large family of proteins termed the SPFH (stomatin, prohibitin, flotillin, and HflK/C) family (41, 112). This protein family was described based on sequence comparisons and homology searches (40, 41, 49, 50, 113–116). All members of the SPFH family present the so-called PHB domain (PHB stands for prohibitin; SPFH/PHB and Band7 are synonyms of these protein modules). It is believed that the PHB protein domain is important for the functionality of the SPFH proteins, but the definitive role of the PHB domain is unknown (41, 112). Bacterial flotillins also belong to the family of SPFH proteins, as they contain a PHB domain in their molecular structure. However, the N-terminal region of the bacterial flotillins is not decorated with lipidic moieties. Instead, the association of these proteins with the bacterial membrane occurs via their transmembrane regions or a hairpin loop inserting into the lipid bilayer. Moreover, flotillins usually harbor a coiled-coil region downstream of the SPFH domain, which seems to be involved in specific protein-protein interactions and oligomerization (117).
The discovery and identification of flotillin proteins in bacteria were reported in 1999 by Tavernarakis et al., with the description of a large of number of bacterial proteins that belonged to the SPFH family (41). Among these proteins, the YuaG protein of B. subtilis was detected and showed a high level of homology to eukaryotic flotillins (41). Experimental evidence of the existence of bacterial flotillins was reported in 2005 by Zhang et al., who showed that the flotillin-like protein BH3500 of B. halodurans was partially purified in association with the DRM fraction (50) by using an experimental methodology similar to the one used for the purification of eukaryotic flotillins (50). This work also showed that the flotillin protein BH3500 was induced under alkaline conditions (50), which is consistent with additional studies of gene expression showing an induction of the yuaG gene in B. subtilis under certain membrane stress conditions (113, 118). Rivera-Milla et al. performed structural and functional comparative studies of diverse flotillin proteins in 2006 and provided evidence for the similarities between the putative flotillin-like protein YuaG and eukaryotic flotillins (112).
In 2009, Donovan and Bramkamp showed the subcellular localization of YuaG, which is heterogeneously distributed in discrete puncta across the bacterial membrane (49). Attempts to determine the type of lipid that led to the punctate distribution of YuaG were inconclusive, finding only that its localization was not strictly dependent on lipids containing phosphatidylglycerol and cardiolipin (49). However, this report presented the first evidence for the role of YuaG in specific cellular processes. YuaG was found to influence sporulation in B. subtilis, as cells lacking YuaG were delayed in the onset of sporulation in comparison to wild-type cells (49), suggesting that one or more kinases involved in the signal cascade at the onset of sporulation are affected. In 2010, YuaG was found to colocalize with a second flotillin-like protein, YqfA, in discrete membrane puncta, which also clustered with other proteins related to signal transduction and cell-cell communication (40). The integrity of these regions depended on the production of the flotillins YuaG and YqfA (here referred to as FloT and FloA, respectively), such that the absence of flotillins caused a severe misfunctionality of the associated signaling processes (40). Remarkably, alterations in the production of membrane-associated polyisoprenoid lipids impaired the localization and functionality of the proteins harbored within FMMs and resulted in a general disruption of the related cellular processes, including biofilm formation, sporulation, and protease secretion (40), in B. subtilis but also in other bacterial species such as S. aureus (40).
In 2012, Dempwolff et al. reported a battery of physiological assays in which a flotillin-defective B. subtilis strain showed severe impairments in sporulation, the activation of natural competence, and motility (51). In that same year, the AAA membrane-bound protease FtsH was reported to associate with FloT and FloA of B. subtilis, and there was a subsequent defect in the functionality of FtsH in a flotillin-defective mutant (58). Cells lacking flotillins showed alterations in cell shape and cell division (119). In 2013, Bach and Bramkamp demonstrated that flotillins play an important role in regulating membrane fluidity and scaffolding raft-associated cellular processes related to protein secretion and transport (120). Furthermore, this work used biochemical and molecular approaches to show that the loss of flotillins in bacterial membranes reduces membrane heterogeneity and leads to a coalescence of ordered lipid regions (120). This finding is in agreement with previous work by Lee et al., who showed that the expression of FloT reduces membrane fluidity under membrane stress conditions (56). Altogether, data from these two publications connect the functionality of flotillins to membrane organization in bacteria.
Presence of flotillins in various bacterial species.
The study of lipid rafts in bacteria is a field of growing importance, which aims to expand investigations to other bacterial species to ultimately determine whether the organization of FMMs is a universal feature of bacteria and whether there are fundamental differences between different species. The use of bioinformatic tools is currently the most robust preliminary approach to screen for the existence of FMMs in diverse bacterial species. Bioinformatic analysis is based on genome-wide searches for genes that code for flotillin-like proteins or constituent lipids, as these are the most important structural components of FMMs in bacteria. It is important to consider that the molecular structure of the constituent lipids may vary from one bacterial species to another, and therefore, the biosynthetic pathways that lead to the production of these constituent lipids may vary as well. Therefore, the best initial approach to ascertain whether a given bacterial species is able to organize FMMs is probably via detection of a flotillin-like protein-encoding gene(s) within its genome. Detection of flotillin-encoding genes is a relatively straightforward task. The first step in the identification of a flotillin protein in any given bacterial species is the evaluation of the number of membrane-associated proteins that contain the PHB domain characteristic of flotillins. This step can be performed, for instance, by using the bioinformatic tool SMART (http://smart.embl-heidelberg.de/) (121). The pool of tentative protein candidates should be further investigated, as other flotillin-related proteins that belong to the large SPFH family of proteins also contain PHB domains (41, 112). Verification can be performed at the genetic level. For instance, flotillin-encoding genes are generally found as part of an operon in bacterial genomes (122). The flotillin-encoding gene is usually the second gene of the operon, while the first gene generally codes for a homologue of the NfeD protein (122), a protease that is confined to bacteria and archaea and proven to interact directly with flotillin-like proteins. The third gene of the operon shows no homologies and is absent from the operons of some species.
(i) Presence of flotillin in Gram-positive bacteria.
Most work regarding the existence of flotillin proteins in bacteria has been performed on Gram-positive organisms. The bacterial model B. subtilis is currently the most consolidated model for the study of bacterial flotillins. B. subtilis produces two different flotillin-like proteins, which are referred to as FloA and FloT (40, 51). FloA and FloT share a similar molecular structure, with a predicted transmembrane region adjacent to a PHB domain, which is typically present in flotillin proteins. FloT (509 amino acids [aa]) is a larger protein than FloA (331 aa) because its C-terminal region extends 178 amino acids farther. Bacillus species that are closely related to B. subtilis, such as B. amyloliquefaciens, B. megaterium, B. licheniformis, and B. pumilus, contain the genes necessary to express both the FloA and FloT flotillin-like proteins, similarly to B. subtilis (Fig. 3). However, members of the most distant group of Bacillus species, called the Bacillus cereus group, which includes the human pathogens B. cereus and B. anthracis and the insect pathogen B. thuringiensis, harbor the gene that codes for a FloT-like flotillin, but they do not express a FloA-like flotillin. The floT gene is annotated BCE_0618, BA_0557, and BALH_0497 in B. cereus, B. anthracis, and B. thuringiensis, respectively (Fig. 3). FloT shows an identical amino acid sequence among the B. cereus group, and it shares 63% amino acid identity (319/509 aa) and 81% positive amino acids (413/509 aa) with FloT of B. subtilis. The reason why the B. cereus group lacks the floA gene is unknown, as it is not clear what causes the expression of two different types of flotillin in one species or one single FloT-like flotillin in another species.
The Firmicutes contain the genus Bacillus along with other genera such as Listeria and Staphylococcus. The non-spore-forming, motile bacterium Listeria monocytogenes is probably the most representative species of the genus Listeria and is responsible for causing severe infections in humans, including listeriosis (123). Similarly to the B. cereus group, Listeria species harbor only one flotillin-encoding gene in their genomes, but in contrast to the B. cereus group, the gene that is present in the genome of L. monocytogenes codes for a FloA-like flotillin (lmo0392 gene) (Fig. 3). Therefore, L. monocytogenes does not contain FloT. FloA of L. monocytogenes shares 62% identity (187/301 aa) and 76% positive amino acids (229/301 aa) with FloA of B. subtilis (Fig. 3). Similarly, the genus Staphylococcus has in S. aureus its most significant example, which generally causes severe disease, including hospital- and community-associated infections (124). Staphylococcus aureus contains one floA-like flotillin gene in its genome (SA1402 gene), similar to what is observed for L. monocytogenes (Fig. 3). Staphylococcal FloA shares 84% identity (249/301 aa) and 90% positive amino acids (277/301 aa) with FloA of B. subtilis.
The class Actinobacteria contains Mycobacterium and Streptomyces as the most relevant genera, and its members harbor one flotillin-encoding gene. The life-threatening pathogen Mycobacterium tuberculosis has one flotillin-encoding gene (Rv1488 gene) (Fig. 3). In this case, the taxonomic distance between Mycobacterium and Bacillus prevented a significant overlap in the sequence alignment of the flotillins. However, the putative flotillin shares 76% identity (277/365 aa) and 87% positive amino acids (320/368 aa) with flotillin of Streptomyces coelicolor (SCO1796 gene) (Fig. 3). Corynebacterium glutamicum contains four genes that encode proteins with the PHB domain typically found in flotillin-like proteins (cgl0650, cgl1533, cgl2836, and cgl3067). However, their functionality as flotillin proteins has not been experimentally addressed.
(ii) Presence of flotillin in Gram-negative bacteria.
The role of flotillin proteins and the study of lipid raft organization in Gram-negative bacteria have not been addressed. Escherichia coli belongs to the class Gammaproteobacteria, along with Salmonella and Pseudomonas. Escherichia coli possesses one flotillin-like gene in its genome, termed yqiK. YqiK presents an N-terminal transmembrane region that is located next to the PHB domain that is typically found in flotillin proteins. YqiK possesses a large C-terminal region that makes this protein a flotillin of 553 amino acids. In this respect, the molecular structure of YqiK resembles that of FloT of B. subtilis, despite sharing only 26% amino acid identity and 45% similarity to FloT of B. subtilis. There are already reports of the presence of YqiK in E. coli cells and its role in membrane quality maintenance (125). There is additional evidence for a role of YqiK in lipid metabolism. In their review, Hinderhofer et al. commented: “Overexpression of YqiK shows a marked effect on cell morphology. Bacteria that overexpress YqiK are larger than wild-type cells and contain opaque cellular inclusions suggesting that they might contain overproduced lipids” (126). However, these data are unpublished, and thus, further experiments should be performed in the future to confirm this claim.
YqiK shares >90% identity to the flotillins of Pseudomonas aeruginosa. The lung-associated pathogen P. aeruginosa contains two different flotillin-encoding genes in its genome. The genes are annotated PA3729 and PA0452 (Fig. 3). PA3729 encodes a 688-amino-acid flotillin-like protein that presents a long C-terminal region. According to the size of the protein and the extension of the C-terminal region, this protein is structurally more similar to FloT of B. subtilis. In contrast, PA0452 encodes a 264-amino-acid flotillin-like protein that shows a small C-terminal region, and thus, this protein seems structurally more similar to FloA of B. subtilis (Fig. 3).
Alphaproteobacteria are represented by the freshwater bacterium Caulobacter crescentus. C. crescentus has a gene in its genome, annotated CC3375, that encodes a flotillin that is structurally similar to FloA of B. subtilis (Fig. 3). CC3375 codes for a 310-amino-acid protein. The FloA-like flotillin of C. crescentus is also similar to the FloA-like flotillin of the betaproteobacterium Neisseria meningitidis (annotated NM1220), a 315-amino-acid protein with a small C-terminal region (Fig. 3). The class of delta-/epsilonproteobacteria also harbors flotillin-encoding genes in their genomes. Campylobacter and Helicobacter are within this class. Campylobacter jejuni and Helicobacter pylori both contain tentative flotillin-encoding genes, Cj0268c and HP0248, respectively. These two proteins seem to be structurally more similar to FloA than to FloT (Fig. 3).
We also found several bacterial species in which the flotillin-encoding genes seemed to be absent from their genomes. One example is the bacterium Mycoplasma pneumoniae, which has the smallest bacterial genome (816 kb) (127). Its reduced genome atypically provides this bacterium with no signal transduction systems (127). Hence, it is probably not surprising that cells lacking signal transduction systems do not show any evidence for the organization of functional membrane microdomains in their membranes and therefore contain no flotillin-encoding genes.
Finally, FMMs may also exist in the membranes of archaea, since it is possible to detect flotillin-like proteins in several species of archaea by using SMART software (http://smart.embl-heidelberg.de/) (121). For instance, Halobacterium species possesses a flotillin-like protein (VNG_0321G). This protein is 392 amino acids long and possesses a N-terminal membrane-anchoring region along with a PHB domain. Importantly, it was recently reported that the membrane of Halobacterium exhibits domains that are constituted by the lipid squalene. The energy-transducing purple membrane in Halobacterium species. was shown to accumulate high concentrations of squalene (128). This is indeed an interesting connection between the production of polyisoprenoid lipids and flotillin in archaea (129).
Protein Cargo of the FMMs
One of the most relevant features of lipid rafts is the capacity of these discrete membrane regions to organize a specific subset of proteins in space and time, to ultimately achieve a fine-tuned specificity in the organization of signaling networks and transport machineries. Thus, a key question regarding the organization of lipid rafts, and, by extension, the FMMs in bacteria, is the identities of the proteins tethered in the microdomains and the signaling networks in which these proteins participate. It is possible to isolate and identify the membrane proteins that are associated with the functional microdomains based on their ability to resist membrane disaggregation when treated with a mixture of nonionic detergents (47). This allows a physical separation of two membrane fractions by zonal centrifugation: one is the DSM fraction, which is sensitive to detergents, and the other is the DRM fraction, which is composed of larger membrane fragments and is more resistant to detergent disruption (47). Whereas it is extremely important not to equate the pool of proteins present in the DRM fraction with raft-associated proteins (60), it is known that the DRM fraction is highly enriched in proteins associated with lipid rafts (Table 1). Therefore, this technique represents an excellent starting point to investigate whether a protein of interest is harbored in the functional membrane microdomains (47).
TABLE 1.
List of proteins identified by mass spectrometry in association with the DRM fraction of B. subtilis membranesa
| Protein | Function | Functional category | Reference(s) |
|---|---|---|---|
| FloA | Flotillin | Scaffold protein | 40 |
| FloT | Flotillin | Scaffold protein | 40, 49 |
| YqeZ | Unknown | Unknown | 58 |
| YuaF | Unknown | Unknown | 58 |
| KinC | Sensor kinase | Signaling | 40 |
| McpABC | Sensor protein | Signaling | 120 |
| OppA | Peptide transporter | Signaling | 40, 58, 120 |
| OpuAC | Peptide transporter | Signaling | 40, 58, 120 |
| FtsH | AAA protease | Cell wall metabolism | 40, 58, 120 |
| FtsX | Cell division protein | Cell wall metabolism | 120 |
| Pbp5 | Penicillin-binding protein | Cell wall metabolism | 58, 120 |
| TagU | Phosphotransferase | Cell wall metabolism | 120 |
| GtaB | Biosynthesis of teichoic acid | Cell wall metabolism | 120 |
| MreC | Cell shape-determining protein | Cell wall metabolism | 58 |
| ErzA | Cell division protein | Cell wall metabolism | 58 |
| FhuD | Siderophore uptake | Iron uptake | 58, 120 |
| YclQ | Petrobactin uptake | Iron uptake | 58, 120 |
| YfiY | Athrobactin uptake | Iron uptake | 58 |
| YhfQ | Iron/citrate uptake | Iron uptake | 58 |
| FeuA | Bacillibactin uptake | Iron uptake | 40, 58, 120 |
| FeuB | Bacillibactin uptake | Iron uptake | 40, 58 |
| SecY | Translocase protein | Protein secretion | 120 |
| SppA | Signal peptidase | Protein secretion | 120 |
| PrsA | Translocase protein | Protein secretion | 40, 58 |
| YxeM | ABC transporter | Membrane transport | 58, 120 |
| RbsB | ABC transporter | 58 | |
| YxeB | ABC transporter | 58 | |
| YwjA | ABC transporter | 58 | |
| YknZ | ABC transporter | 58 | |
| GltT | Glutamate uptake | Uptake of metabolites | 120 |
| MntA | Manganese uptake | 120 | |
| RbsB | Ribose uptake | 120 | |
| AcsA | Acetyl-CoA synthetase | Energy metabolism | 120 |
| AtpD | ATP synthase | 120 | |
| AtpG | ATP synthase | 120 | |
| NagA | NAG utilization | 120 | |
| PtsI | Sugar transport | 120 | |
| QoxA | Quinol oxidase | 40, 58, 120 | |
| ResC | Cytochrome c biogenesis | 120 | |
| SdhA | Succinate dehydrogenase | 120 | |
| BdbD | Thiol-disulfide oxidoreductase | 40, 58, 120 |
NAG, N-acetylglucosamine.
It is crucial to perform further experiments to validate whether any potential protein candidate identified in the DRM fraction is actually part of the cargo of the FMMs. For instance, one can assay protein-protein interactions with the protein candidate and the scaffold protein flotillin, using pulldown assays or a bacterial two-hybrid system. Also, whether mutants lacking the constituent lipids and/or flotillin show any effect on the functionality of the protein of interest can be evaluated. It is important at this point to emphasize that raft separation by detergent disaggregation should not be the main criterion to classify a protein as part of the protein cargo of lipid rafts (12, 20, 60). The unrigorous use of this approach in the past generated results that were difficult to reconcile, which led to questioning of the existence of lipid rafts and whether lipid rafts were artifacts generated during the preparation of samples (130, 131). The solubilization of membranes by detergent treatment can render biased results depending on the concentration and type of detergent or the temperature, and an alternative methodology is required to validate any protein of interest as part of the cargo of the FMMs. Table 1 shows a list of proteins that were identified in the DRM fraction of B. subtilis. Below, some examples of protein cargo that are already characterized are detailed.
In order to generate a biofilm, cells switch from a planktonic to a sessile state by downregulating the expression of flagellar genes and inducing the expression of genes required for matrix production (132). This self-produced extracellular matrix contains exopolysaccharides (EPSs) and proteins (133) and provides the rigidity that is required for the formation of robust biofilms (134, 135). The genes responsible for the production of EPSs are part of an 11-gene operon termed the epsA-O operon (or the eps operon) (133, 136, 137). Two secreted proteins provide structural integrity to the matrix. These proteins are TasA and TapA, which are encoded by the three-gene operon tapA-sipW-tasA (tapA operon) (133). TasA is a functional amyloid protein (138) that is anchored to the cell wall by TapA (139). The activation of the master regulator Spo0A is necessary to trigger the activation of the eps and tasA genes, which leads to biofilm formation (140–145). The activation of Spo0A requires the phosphorylative action of five different kinases (KinA to KinE [KinA-E]) that are responsible for transferring a phosphoryl group to Spo0A via a Spo0F/Spo0B phosphorelay system (45, 146). The activation of the KinA-E kinases is driven by the action of specific signals of an unknown nature (147, 148). It was recently discovered that a signaling molecule termed surfactin, produced by B. subtilis itself, activates histidine kinase C (KinC) and in turn phosphorylates Spo0A, with subsequent activation of the signaling pathway (44, 149).
An association of the sensor kinase KinC with the protein cargo or the FMMs of B. subtilis occurred when KinC was detected in the DRM fraction (40) (Fig. 4). Validation of this result was obtained by colocalization of the protein KinC and the flotillin-like protein FloT. The localization of KinC to the FMMs of B. subtilis requires FloT and FloA (40). Deletion of floT and floA results in a mislocalization of KinC and also abrogates KinC activity, which prevents the flotillin-defective strain from expressing matrix genes and forming a biofilm in response to the signal surfactin, similarly to a mutant defective in kinC (40). Similarly, the ΔyisP mutant that is unable to produce constituent lipids of FMMs was unable to form a biofilm in a Spo0A-dependent manner. It was not possible to detect KinC in the DRM fraction of the ΔyisP mutant by Western blotting. It was possible to recover biofilm formation in the ΔyisP mutant with expression of the Sad67 allele (150), which codes for a variant of Spo0A that is constitutively active and does not need the phosphorylative action of KinC to induce biofilm formation (40). This provided evidence that KinC is one component of the protein cargo of the FMMs (Fig. 4). Alterations in the lipid or proteinic architecture of the FMMs severely affected the functionality of KinC and, thus, the activation of the signaling cascade for biofilm formation (40).
FIG 4.
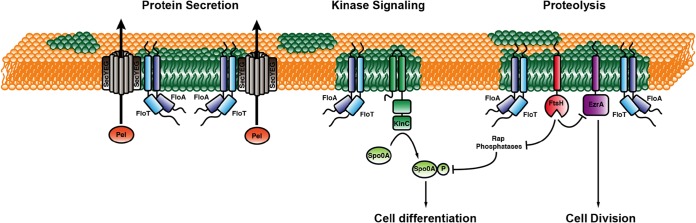
Cellular processes influenced by flotillin in B. subtilis. Shown is a schematic representation of a B. subtilis cell membrane, with FMMs represented in green. The pathways in transport/secretion, signaling, and proteolysis that are scaffolded by flotillins in lipid rafts are indicated.
However, a ΔfloT ΔfloA mutant displayed a more severe defect in biofilm formation and sporulation than did a ΔkinC mutant (49, 58). Both biofilm formation and sporulation are Spo0A-regulated processes (142, 151), suggesting that the ΔfloT ΔfloA mutant had other defects that further inhibited the activation of the Spo0A genetic cascade. Supporting this hypothesis, the direct interaction of FloA and FloT with the protein cargo protease FtsH was important for the protease activity of FtsH (58) (Fig. 4). FtsH was found to be associated with the DRM fraction and to colocalize with FloA and FloT (58). FtsH has been reported to indirectly affect the levels of phosphorylated Spo0A (Spo0A∼P) by degrading four regulatory phosphatases, RapA, RapB, RapE, and Spo0E, which feed into the Spo0A phosphorelay to decrease the levels of Spo0A∼P (152). Accordingly, previous studies have shown that the ΔftsH mutant has a severe defect in sporulation, consistent with a decrease in the levels of Spo0A∼P (153, 154). Further analysis has shown that FtsH may have a degradation effect on cell division-related proteins to ultimately regulate cell division in B. subtilis (119, 120). Thus, it is possible that the FMMs of bacteria indirectly affect cell division via the regulation of FtsH (Fig. 4).
In E. coli, the FtsH protease is associated with the HflC/K proteins. HflC and HflK are members of the SPFH protein family that are essential for the correct oligomerization of FtsH in E. coli (155). B. subtilis lacks HflC/K proteins, and hence, it was speculated that FloT/A might substitute for HflC/K in B. subtilis to allow FtsH oligomerization into a functional membrane-integral protease hexamer (58). FtsH is not only localized into mobile foci in B. subtilis but also enriched at the division septum. In accordance with a FloT/A interaction with FtsH, FloT and FloA are less dynamic when they are localized to the septum than in lateral foci (58). FtsH is a membrane-embedded proteolytic machinery that is involved in several essential cellular processes. Although FtsH is not absolutely essential in B. subtilis, mutants have a severe, pleiotropic phenotype. FtsH is required for biofilm formation, likely because cells do not differentiate efficiently into matrix producers (58). An increase in FtsH activity has a dual effect on B. subtilis cells. First, the increased degradation of Rap phosphatases leads to an elevated level of Spo0A∼P and an increase in biofilm formation. The overproduction of FloT/A and FtsH activities in B. subtilis leads to cell length reduction and an increase in the number of FtsZ rings (119). Because EzrA, a negative regulator of FtsZ, was found in the DRM fraction, molecular concentrations of EzrA were analyzed under flotillin overexpression conditions. Indeed, EzrA levels were downregulated when flotillins were overexpressed, and a similar overexpression of FtsH led to a reduction in EzrA levels. These data support the notions that EzrA is a target of the FtsH protease and that the activity of FtsH can be triggered by flotillins (119). In contrast, however, the activity of the E. coli AAA protease FtsH is negatively regulated by HflC/K (156, 157). Thus, both flotillins and HflC/K have chaperoning roles but regulate the activity of the protease complex differently. Interestingly, an interaction between a eukaryotic homologue of the FtsH protease and prohibitin, a PHB domain protein, has been described. The prohibitins Phb1b and Phb2b regulate the turnover of membrane proteins by modulating the activity of the m-AAA protease, a conserved ATP-dependent protease in the inner membrane of Saccharomyces cerevisiae mitochondria that is homologous to the bacterial FtsH protein. The m-AAA protease of S. cerevisiae and other yeasts is a hetero-oligomer that is composed of the subunits Yta10P (Agf3p) and Yta12p (Rca1p) (158). However, in the yeast system, a lack of Phb1/2 leads to an increase in the level of the m-AAA protease (158) and thus to an effect opposite of that observed for the flotillin-FtsH interaction in B. subtilis but similar to that observed for the E. coli HflC/K-FtsH complex.
In exploring the hypothesis that FloA and FloT facilitate the interaction and oligomerization of the protein cargo, pulldown experiments showed an additional number of FloT-interacting proteins, which include a number of oligomeric proteins related to signal transduction and protein secretion (120). Accordingly, protein secretion was significantly reduced in cells lacking flotillins, providing evidence for the existence of a functional link between the protein secretory machinery and the flotillin proteins (120). Among the proteins that coeluted with FloT, the channel subunit SecY, the central component of the Sec system, as well as the signal peptidase SppA were identified (120) (Fig. 4). Localization of the secretory machinery in puncta across the bacterial membrane was described and was dependent on flotillin membrane domains. The localization and functionality of the Sec system are dependent on the presence of negatively charged phospholipids (e.g., cardiolipin) in the bacterial membrane (159). In an in vitro nanodisc assay of the function of the Sec system, it was shown that negatively charged phospholipids (e.g., phosphatidylglycerol) are essential for activity, while nanodiscs with a high percentage of uncharged lipids such as phosphatidylethanolamine led to inactive sec channels (160). These in vitro experiments support the idea that protein secretion needs a highly specialized membrane environment for correct functioning. Thus, it is probably not surprising that both FloT and Sec proteins colocalize in foci within the membrane (Fig. 4).
Moreover, the concentration of secreted proteins was reduced up to 40% in the flotillin-defective mutant compared to the wild type (120), suggesting that secretion in cells lacking the FloA and FloT proteins is less effective, affecting particularly Sec-dependent secreted proteins. This experimental approach suggested that the functionality of the protein secretory machinery and probably its oligomerization requires the scaffolding activity of FloA and FloT in B. subtilis cells (120). Several transport proteins were isolated in coelution experiments with FloT (120). Among these proteins was OppA, a peptide transporter linked to quorum sensing. Opp proteins are also conserved in Gram-negative bacteria, where they are involved in the recycling of cell wall peptides released from the growing cell wall (161). In Gram-positive species such as B. subtilis, Opp proteins play a major role in the uptake of peptide pheromones such as Phr peptides (162). However, there is no experimental evidence showing a decreased activity of OppA in B. subtilis in relation to flotillins, but the colocalization of OppA and FloT supports a functional interaction (120).
Various proteins involved in cell wall synthesis/metabolism were identified in FloT pulldown experiments. Pbp5, TagU, GtaB, and FtsX were found to interact with FloT. FtsX was also shown to colocalize with FloT (120). FtsX is the membrane-integral part of the FtsEX complex that has similarities to an ABC transporter. Although a transported substrate has not yet been identified, recent studies point toward a role of FtsEX in the regulation of peptidoglycan hydrolases (163, 164). The conserved hydrolase CwlO is involved in cell wall elongation in B. subtilis, and its activity is controlled by FtsEX. In accordance with this function, FtsEX localizes in a punctate manner along the lateral wall in B. subtilis, where it colocalizes with FloT (120). TagU is involved in the final step of transferring teichoic acid polymers from the lipid-linked precursor to peptidoglycan and localizes in foci and bands along the lateral axis of B. subtilis (165). Although individual knockouts of floA and floT have no apparent cell shape phenotype, cell shape and morphology defects have been described for floA floT double mutants (51). The lack of both PHB domain proteins led to twisted, irregular-shaped cells. Membrane staining of these double mutants showed severe membrane distortions. Furthermore, double mutants have been described to be impaired in competence, likely because the DNA uptake apparatus is misplaced under these conditions (51). Indeed, FloT and FloA have been copurified with the minor competence pilin ComGG (166). However, in this study, the lack of either FloT or FloA led to an increase in competence (166) Thus, the influence of bacterial flotillins on competence development is therefore still unclear.
DISASSEMBLY OF BACTERIAL FMMs
Targeting of FMMs provides an interesting strategy to simultaneously inhibit a myriad of physiological processes that are related to microbial development. There are a number of small molecules that severely perturb the architecture of FMMs by inhibiting the biosynthetic pathway responsible for the production of the constituent lipids (40). Importantly, most of these “antiraft” compounds are nontoxic to humans and commercially available as cholesterol-lowering drugs to treat patients with hypercholesterolemia (167–169). When bacteria are exposed to nanomolar concentrations of these compounds, FMMs disperse, and raft-harbored proteins, including flotillin and part of the protein cargo, diffuse across the membrane, losing their functionality (40). While the activity of raft-localized signaling pathways is severely compromised in the presence of antiraft molecules, these molecules do not represent a serious threat to cell viability and show lower pressure on the development of spontaneous mutation-acquired resistance to these compounds (40), which makes them excellent candidates for the development of antimicrobial therapies.
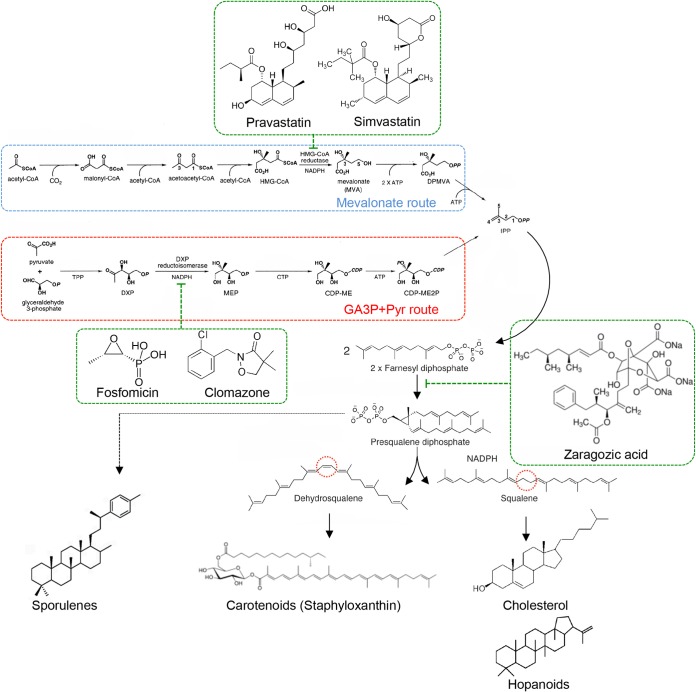
The metabolic pathway that leads to the production of isoprene and other polyisoprenoid lipids in bacteria is relatively well known, as are the enzymes that catalyze the reactions that constitute the biosynthetic pathway. The production of isoprene in bacteria follows two different biosynthetic routes. One of them is called the mevalonate route and is the pathway that is found in species such as S. aureus, Streptococcus pneumoniae, and Enterococcus faecalis (170, 171) (Fig. 5). This is also the same pathway that human cells use to synthesize cholesterol. Consequently, cholesterol-lowering drugs that are designed to inhibit the mevalonate pathway in patients with hypercholesterolemia also show a potent inhibitory activity toward the biosynthetic pathway of the constituent lipids in S. aureus, S. pneumoniae, and E. faecalis, blocking most of the signal transduction pathways that are known to localize in FMMs (167–169).
FIG 5.
Inhibition of isoprenoid biosynthesis in bacteria. Isoprenoid biosynthesis in bacteria follows the mevalonate route or the GA3P-pyruvate (Pyr) route. Statin molecules such as simvastatin or pravastatin are potent competitive inhibitors of the HMG-CoA reductase (hydroxymethylglutaryl-CoA-reductase) enzyme that participates in the mevalonate route. Furthermore, compounds such as fosfomycin or clomazone are conventionally used to inhibit the GA3P-pyruvate route, as they are potent inhibitors of DXP reductoisomerase (1-deoxy-d-xylulose 5-phosphate reductoisomerase). The condensation of several isoprenoid molecules renders polyisoprenoid molecules such as squalene, which are precursors of polycyclic and noncyclic terpenoids. Zaragozic acids are competitive inhibitors of the enzyme squalene synthase and cause an inhibition of the production of squalene in bacteria that use the mevalonate route and the GA3P-pyruvate route. The inhibitor molecules are shown in dashed green frames.
However, there is another isoprenoid biosynthesis pathway in bacteria, which is referred to as the glyceraldehyde-3-phosphate (GA3P)–pyruvate route (172) (Fig. 5). This is the route that is found in species such as B. subtilis, Mycobacterium tuberculosis, and Escherichia coli, and it is the same route that plants use to synthesize phytosterols. This route is not present in human cells, and consequently, the enzymes that participate in this biosynthetic route are not present in human cells. Because of this, a number of compounds that target the functionality of these enzymes have been developed for many diverse treatments of plants. Since human cells do not produce the target protein(s), the inhibitory compounds do not represent any harm to humans. For instance, the herbicide clomazone can inhibit biofilm formation in B. subtilis by targeting the production of constituent lipids (40) (Fig. 5). The inhibition of constituent lipids causes a dispersion of the protein cargo of the membrane microdomains across the whole bacterial membrane, losing their functionality and therefore causing a strong inhibition of the related physiological features, including biofilm formation (40).
The distribution of the GA3P-pyruvate and mevalonate biosynthetic pathways seems to not follow any particular pattern along the bacterial phylogeny, except that the GA3P-pyruvate route is more represented among bacteria (archaea use only the mevalonate pathway) (173). A remarkable exception is the Gram-positive pathogen Listeria monocytogenes, which seems to contain the genes for both the mevalonate route and the GA3P-pyruvate route. It is not yet known whether these biosynthetic routes are redundant in L. monocytogenes or if they produce a different type of isoprenoid lipid. The existence of different biosynthetic pathways for the production of the lipids of FMMs implies that the collection of inhibitory compounds that target these pathways have a restricted spectrum of action. There is, however, a class of small molecules that can target both pathways indistinctively. These small molecules are the squalestatins, which are represented by the family of compounds called zaragozic acids (52) (Fig. 5). Zaragozic acids are a family of fungal natural products that are very potent inhibitors of squalene synthase, which inhibit cholesterol synthesis and lower plasma cholesterol levels in humans (52). They also inhibit fungal ergosterol and plant phytosterol synthesis and are potent fungicidal compounds. These potent natural product-based inhibitors of squalene synthase are universal inhibitors of sterol synthesis because they act upon a later stage in the biosynthetic pathway, once acetyl coenzyme A (acetyl-CoA) is converted to mevalonate and is processed to become squalene. Consequently, zaragozic acids represent an example of a family of natural products that can inhibit the production of constituent lipids in all kinds of bacterial species and therefore are likely to cause a dispersion of FMMs in bacteria, regardless of the species under consideration (40).
ANALYSIS OF BACTERIAL LIPID DOMAINS BY USING ADVANCED MICROSCOPY TECHNIQUES
Biochemical analysis of cellular structures has provided fundamental insights into the cellular organization and function of cellular macromolecules, such as proteins and lipids. However, visualization of cellular structures is seen as prime evidence for their existence and is often more convincing to the community. Visualization of membrane domains has been particularly difficult since these domains are in the nanometer range below the resolution limit of conventional light microscopy. However, recent advances in technology now allow us to visualize lipid rafts with great precision. For instance, a recent study used secondary ion mass spectrometry (NanoSIMS) to directly image lipid microdomains in bacteria by using stable-isotope labeling and avoiding localization artifacts (174). Furthermore, the past decade has seen a dramatic change in microscopy techniques. The use of fluorescent labels has helped to address the subcellular positioning of proteins, lipids, and DNA. Furthermore, new microscopic techniques that allow resolution below the diffraction limit of visible light (200 to 300 nm) have been developed. These techniques include stimulated emission depletion (STED), structured illumination microscopy (SIM), as well as techniques using stochastic fluorescence detection, such as photoactivated localization microscopy (PALM) and direct stochastic optical reconstruction microscopy (dSTORM). These superresolution techniques can provide resolution down to single molecules. These different superresolution microscopy techniques have different strengths and bottlenecks (175–177). For imaging of bacterial cells, PALM/STORM seems to us to be the most useful approach because it can exploit total internal-reflection (TIRF) microscopy setups and is technically less demanding than STED or SIM. Furthermore, the theoretical resolution limit in the lateral and axial directions is highest for PALM/STORM. For stochastic fluorescence detection, photoswitchable fluorophores are used. These fluorophores are capable of efficient photoconversion, changing their excitation and emission spectra in response to light irradiation or any other environmental perturbation. An elegant way to use these photoswitchable fluorophores to track lipid domains with high resolution was described recently (178). Those authors used a fusion between the C-terminal D4 domain of the theta-toxin, a known cholesterol binder, and the photoactivatable fluorescent protein Dronpa. The fusion protein localized to distinct membrane domains in a cholesterol-dependent manner when the isolated protein was incubated with HeLa cells. Due to the use of photoswitchable Dronpa, PALM revealed a high-resolution localization of cholesterol domains in plasma membranes. Examples of dual-color experiments have also been described for eukaryotic plasma membranes. By using the N-terminal domain of the T-cell receptor pathway kinase Lck and the N-terminal domain of the Src kinase fused to different photoswitchable fluorophores, a discrete distribution of these two proteins into defined membrane areas was shown (15). These data are in nice agreement with data from yeast showing that proteins cluster together in distinct domains based on their membrane-anchoring domain. This technique should in principle work ideally for bacterial membrane proteins as well (15, 16).
Although there is as yet no report that describes an analysis of bacterial membrane domains using PALM/STORM, there is clearly the potential for PALM/STORM to increase our knowledge about the organization of bacterial membrane domains. Some of these new microscopy techniques, such as three-dimensional structured illumination microscopy (3D-SIM), PALM, and STORM, may even allow live-cell imaging, and hence, these light microscopy techniques are more versatile than electron microscopy. Transmission electron microscopy (TEM) has been used intensively to study biological membrane systems and, together with immune-labeling techniques, was also successfully employed to visualize membrane rafts in bacteria (65). Electron microscopy tomography and cryofixation have allowed enhancement of the preservation of biological structures in comparison to chemical fixation. Tomography has been used to analyze caveola structures in eukaryotes (179, 180) but has also proven tremendously useful in studies of bacterial protein complexes and organelles (181, 182).
A new method uses genetically encoded probes, such as mini-singlet-oxygen generator (miniSOG), for determination of protein localization by EM. MiniSOG is a fluorescent flavoprotein (106 amino acids) that can be used for conventional light microscopy but also for TEM. Singlet-oxygen production is triggered by blue light illumination and can be used to polymerize diaminobenzidine into a polymer. This method works effectively on fixed cells and can be used after staining with osmium for high-resolution EM (183). Although these new imaging techniques have been widely used to study lipid and protein domains in eukaryotic cell membranes, there are only a few examples for bacterial cells. However, in view of the great impact of cryo-electron tomography on bacterial cell biology, these techniques are promising tools that can be used to understand the ultrastructure of the bacterial membrane in a new dimension.
Despite new high-resolution imaging techniques, membrane research has long been facilitated by the use of fluorescent dyes that integrate into the membrane bilayer and provide a specific readout that allows the drawing of conclusions about the membrane composition and physical state of the lipids. Laurdan is one of the polarity-sensitive fluorescent probes that has been intensively used by the community to study membrane fluidity and heterogeneity in biological membranes. A prerequisite for the use of Laurdan for membrane analysis is that Laurdan distributes evenly into the membrane, independent of its physicochemical properties. Furthermore, insertion of proteins into the lipid bilayer does not interfere with Laurdan intercalation (184). The dye inserts into the lipid bilayer and displays a phase-dependent shift of the emission spectrum. Tightly ordered lipids in membranes (liquid-ordered [Lo] regions) exclude water molecules from penetrating deeply between the lipid head groups. Laurdan that has intercalated into Lo regions exhibits a blue shift of its emission spectrum. In less-ordered membrane regions (liquid-disordered [Ld] regions) the emission spectrum is red shifted. This allows one to obtain direct spatial information about the physical state of the membrane by using fluorescence microscopy. The ratio between the emission maxima in the blue- and red-shifted peaks (420 to 470 nm and 500 to 570 nm, respectively) provides a relative value for membrane order, called generalized polarization (GP). Due to bleaching effects, Laurdan is most often visualized by using 2-photon microscopy (185). However, there are examples of the use of conventional wide-field microscopy for eukaryotes (186) and also for bacteria (120, 187). The use of Laurdan confirmed phase separation and lipid domain coexistence in model membranes (188) and plasma membranes (120, 189).
Laurdan can be used in combination with fluorophores emitting red light fluorescence. The membrane dye rhodamine-DPPE [1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-n-(lissamine rhodamine B sulfonyl)] is an example of a dye that enriches in Ld regions and can be used together with Laurdan. In bacterial cells, the fluorescent dye Nile Red has been used together with Laurdan to visualize highly fluid membrane regions. Interestingly, these fluid membrane domains depend on MreB, the bacterial action homologue (187). Proteins that were reported to be excluded from these fluid membrane domains are, for example, succinate dehydrogenase (SdhA) and FoF1 ATPase (AtpA). Both proteins have been identified in bacterial lipid rafts (40, 120). Together, these data suggest that the bacterial cell controls membrane domain formation via at least two distinct pathways, with MreB organizing the fluid regions and flotillins organizing the more ordered membrane domains.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
The assembly of functional microdomains in the membranes of diverse bacteria, similar in structure and function to the lipid rafts of eukaryotic cells, is interesting due to different aspects. First, it reveals a remarkable level of sophistication in signal transduction and membrane organization that is unprecedented in bacteria. Second, the presence of these signaling platforms in many very distant domains of life strongly argues in favor of their critical role in regulating important cellular processes. Third, bacteria are genetically tractable model organisms that will help to solve questions related to the architecture and functionality of lipid rafts, which have been traditionally difficult to address in eukaryotic models due to complications with genetic manipulation in these systems. Fourth, the perturbation of the functional membrane microdomains impairs key developmental processes in bacteria and opens new avenues to use FMMs as targets to control, for instance, biofilm-related bacterial infections.
The discovery of the existence of functional membrane microdomains in the membranes of bacteria is recent, and therefore, the number of questions that need to be answered to acquire a better understanding of raft-like microdomains in bacteria is still overwhelming. Nevertheless, there are two main research aspects that need to be addressed in the near future in order to make progress in this field. One aspect is the structural components of these signaling platforms, to precisely determine the molecular structure of the constituent lipids, the role of the scaffold protein flotillin, and the molecular mechanism that cells activate to assemble these platforms. Moreover, a functional component needs to be addressed to fully understand the biological role of these signaling hubs in bacterial membranes and whether there is functional variation between different bacterial species. When these two important and challenging experimental aspects of the FMMs are addressed, the field will be more established and consolidated within the overall aspects to take into consideration in microbiology.
APPENDIX
Protocol for the Purification and Analysis of Detergent-Resistant Membranes from B. subtilis Cells
This section described a detailed methodology for isolating and analyzing FMMs by detergent insolubility using B. subtilis cells as a model system. FMMs can be purified based on the selective advantage of these microdomains in resisting membrane disaggregation by detergent treatment. We detail below the crucial role of this separation technique in understanding the composition of lipid rafts and, by extension, the FMMs of bacteria. The technical procedure for the purification of the detergent-resistant membrane (DRM) fraction can be achieved simply by using this methodology. Briefly, the DRM is isolated by flotation on sucrose density gradients after detergent disruption. DRM proteins can be analyzed by SDS-PAGE and immunoblotting. Alternatively, the technical procedure for the purification of the DRM fraction can be achieved simply by using a purification kit that has been developed by Sigma-Aldrich, the CelLytic MEM protein extraction kit (catalogue number CE0050). This kit provides a mixture of nonionic detergents optimized to achieve high-performance purification of the DRM fraction and a gel that separates the membrane fraction in the DSM and DRM phases after detergent treatment. We have compared the performance of this kit to that of the traditional purification method and detected minimal variations between both approaches. When using either of these two technical approaches, it is important that all steps involved in the manipulation of cellular membranes, including detergent disruption, are performed at 4°C. Low temperatures favor the separation of the different lipid species and the stabilization of lipid ordering in bacterial membranes.
Step 1: preparation of cell membranes for detergent disruption.
For the isolation of cell membranes, the respective strains of B. subtilis were grown in 100 ml of tryptic soy broth (TSB) medium at 37°C for 24 h. Subsequently, cells were pelleted (10 min at 4,000 rpm at 4°C), and the supernatant was removed. The dry pellet was either stored at −20°C until further use or resuspended in 10 ml of buffer H (20 mM HEPES [pH 8], 20 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]). To lyse the cells, 200 μl of lysozyme (1 mg/ml), 100 μl of PMSF (100 μM), and 5 μl of DNase I were added to the suspension before disruption by using a French press (10,000 lb/in2 and 4 passes). Centrifugation was performed to eliminate cell debris, and the membrane fraction was further precipitated by ultracentrifugation (100,000 × g for 1 h at 4°C). The supernatant was discarded, and the membrane fraction was dissolved in buffer H supplemented with 10% glycerol.
Step 2: separation of DSMs and DRMs by centrifugation in a sucrose gradient.
Disruption of the membranes was performed by using the CelLytic MEM protein extraction kit (Sigma-Aldrich). This kit provides a mixture of nonionic detergent and is adapted for high efficiency. Alternatively, other detergents, such as Triton X-100, Lubrol, Brij 96, Nonidet, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and octyl-β-d-glucopyranoside, have been largely used at concentrations from 1 to 4% for the disruption of cell membranes. Optimization of the detergent treatment needs to be determined empirically. Incubation of the detergents with the membranes was performed at 4°C for 30 min. The total volume was mixed 1:1 with 80% sucrose in 0.2 M sodium carbonate, transferred into ultracentrifugation tubes, and carefully overlaid with a 5× volume of 20% sucrose in buffer H. Separation of sucrose gradient fractions was performed by ultracentrifugation (15 h at 100,000 × g at 4°C). Membrane fractions were collected by aspiration and kept on ice for protein precipitation.
Step 3: analysis of proteins associated with DSMs and DRMs.
For protein precipitation, 4 volumes of ice-cold acetone were added to the sample. After 2 h of incubation at −20°C, denatured proteins were precipitated by centrifugation (20 min at 15,000 rpm at 4°C). Afterwards, the supernatant was removed, and pellets were dried at room temperature for up to 2 h. Proteins were resuspended in 1× protein loading buffer and stored at −20°C. Protein analysis of the DRM and DSM fractions was performed by resolving both fractions independently in SDS-PAGE gels. For this purpose, samples were boiled for 5 min before loading into a 12% acrylamide-bisacrylamide gel (37:1:1).
ACKNOWLEDGMENTS
Research related to flotillins by the group of Marc Bramkamp is supported by funds from the Deutsche Forschungsgemeinschaft (INST 216/498-SFB 635; BR 2915/4-1). Research related to flotillins by the group of Daniel Lopez is supported by funds from the European Research Council (ERC-335568).
We thank our coworkers for constructive criticism on the manuscript and figure preparation.
Biographies

Marc Bramkamp obtained his Ph.D. in microbiology at the University of Osnabrück (Germany) in 2003. He worked in the laboratory of Prof. Dr. Karlheinz Altendorf on the structure and function of the potassium transport P-type ATPase KdpFABC. With a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft, he joined the group of Prof. Dr. Jeff Errington at the Sir William Dunn School of Pathology in Oxford (United Kingdom), where he started to work on cell division in Bacillus subtilis. In 2006, he moved to the University of Cologne and joined the laboratory of Prof. Dr. Reinhard Krämer, where he started a junior group working on bacterial cell biology in B. subtilis and Corynebacterium glutamicum. Dr. Bramkamp received his venia lengendi in Biochemistry in 2011 and was appointed Professor of Microbiology at the Ludwig Maximilians University Munich (Germany) in 2012. His research focuses on membrane-associated processes in bacterial cell division and development.

Daniel Lopez received his Ph.D. in Biochemistry at the University of Murcia (Spain). His Ph.D. project used a number of molecular approaches to understand the role of the production of melanin in a novel bacterium isolated from the Mediterranean Sea. After receiving his doctorate, he trained as a postdoctoral fellow with Dr. Roberto Kolter at Harvard Medical School, where he became interested in cell-cell communication in bacteria and the number of small molecules that are involved in this process, now using the soil-dwelling bacterium B. subtilis as a model organism. In 2011, he joined the Research Centre for Infectious Diseases (ZINF) at the University of Wuerzburg (Germany) as a Junior Research Group Leader. He built a research group that focuses on exploring the molecular organization of signaling networks in pathogenic bacteria and their role during the infection process.
REFERENCES
- 1.Grecco HE, Schmick M, Bastiaens PI. 2011. Signaling from the living plasma membrane. Cell 144:897–909. doi: 10.1016/j.cell.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Lindner R, Naim HY. 2009. Domains in biological membranes. Exp Cell Res 315:2871–2878. doi: 10.1016/j.yexcr.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 3.Schroeder F, Woodford JK, Kavecansky J, Wood WG, Joiner C. 1995. Cholesterol domains in biological membranes. Mol Membr Biol 12:113–119. doi: 10.3109/09687689509038505. [DOI] [PubMed] [Google Scholar]
- 4.Silvius JR. 2012. Membranes: first steps to rafts? Nat Chem Biol 8:743–744. doi: 10.1038/nchembio.1045. [DOI] [PubMed] [Google Scholar]
- 5.Singer SJ, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 6.Hyman AA, Simons K. 2012. Cell biology. Beyond oil and water-phase transitions in cells. Science 337:1047–1049. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- 7.Engelman DM. 2005. Membranes are more mosaic than fluid. Nature 438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 8.Coskun U, Simons K. 2011. Cell membranes: the lipid perspective. Structure 19:1543–1548. doi: 10.1016/j.str.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Cronan JE. 2003. Bacterial membrane lipids: where do we stand? Annu Rev Microbiol 57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 10.van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 12.Simons K, Sampaio JL. 2011. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraft ML. 2013. Plasma membrane organization and function: moving past lipid rafts. Mol Biol Cell 24:2765–2768. doi: 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann AK, Itano MS, Jacobson K. 2010. Understanding lipid rafts and other related membrane domains. F1000 Biol Rep 2:31. doi: 10.3410/B2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen DM, Rentero C, Rossy J, Magenau A, Williamson D, Rodriguez M, Gaus K. 2010. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics 3:446–454. doi: 10.1002/jbio.200900089. [DOI] [PubMed] [Google Scholar]
- 16.Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Soldner R. 2012. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 17.Schuck S, Simons K. 2004. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci 117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 18.Simons K, van Meer G. 1988. Lipid sorting in epithelial cells. Biochemistry 27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 19.van Meer G, Simons K. 1988. Lipid polarity and sorting in epithelial cells. J Cell Biochem 36:51–58. doi: 10.1002/jcb.240360106. [DOI] [PubMed] [Google Scholar]
- 20.Simons K, Toomre D. 2000. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 21.Tsui-Pierchala BA, Encinas M, Milbrandt J, Johnson EM Jr. 2002. Lipid rafts in neuronal signaling and function. Trends Neurosci 25:412–417. doi: 10.1016/S0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- 22.Barak I, Muchova K. 2013. The role of lipid domains in bacterial cell processes. Int J Mol Sci 14:4050–4065. doi: 10.3390/ijms14024050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barak I, Muchova K, Wilkinson AJ, O'Toole PJ, Pavlendova N. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol 68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto K, Kusaka J, Nishibori A, Hara H. 2006. Lipid domains in bacterial membranes. Mol Microbiol 61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 25.Muchova K, Jamroskovic J, Barak I. 2010. Lipid domains in Bacillus subtilis anucleate cells. Res Microbiol 161:783–790. doi: 10.1016/j.resmic.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. 2004. Cardiolipin domains in Bacillus subtilis Marburg membranes. J Bacteriol 186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mileykovskaya E, Dowhan W. 2000. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol 182:1172–1175. doi: 10.1128/JB.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mileykovskaya E, Dowhan W. 2009. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta 1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosch JW, Hsu FF, Caparon MG. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol 189:801–806. doi: 10.1128/JB.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver PM, Crooks JA, Leidl M, Yoon EJ, Saghatelian A, Weibel DB. 2014. Localization of anionic phospholipids in Escherichia coli cells. J Bacteriol 196:3386–3398. doi: 10.1128/JB.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 32.Morrow IC, Parton RG. 2005. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic 6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 33.Babuke T, Tikkanen R. 2007. Dissecting the molecular function of reggie/flotillin proteins. Eur J Cell Biol 86:525–532. doi: 10.1016/j.ejcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Otto GP, Nichols BJ. 2011. The roles of flotillin microdomains—endocytosis and beyond. J Cell Sci 124:3933–3940. doi: 10.1242/jcs.092015. [DOI] [PubMed] [Google Scholar]
- 35.Stuermer CA. 2011. Reggie/flotillin and the targeted delivery of cargo. J Neurochem 116:708–713. doi: 10.1111/j.1471-4159.2010.07007.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhao F, Zhang J, Liu YS, Li L, He YL. 2011. Research advances on flotillins. Virol J 8:479. doi: 10.1186/1743-422X-8-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel V, Bakovic M. 2007. Lipid rafts in health and disease. Biol Cell 99:129–140. doi: 10.1042/BC20060051. [DOI] [PubMed] [Google Scholar]
- 38.Sourjik V. 2004. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol 12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Bray D, Levin MD, Morton-Firth CJ. 1998. Receptor clustering as a cellular mechanism to control sensitivity. Nature 393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 40.Lopez D, Kolter R. 2010. Functional microdomains in bacterial membranes. Genes Dev 24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tavernarakis N, Driscoll M, Kyrpides NC. 1999. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem Sci 24:425–427. doi: 10.1016/S0968-0004(99)01467-X. [DOI] [PubMed] [Google Scholar]
- 42.Good MC, Zalatan JG, Lim WA. 2011. Scaffold proteins: hubs for controlling the flow of cellular information. Science 332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langhorst MF, Reuter A, Stuermer CA. 2005. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci 62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LeDeaux JR, Yu N, Grossman AD. 1995. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol 177:861–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng X, Hu Y, Zheng Y, Zhu W, Li K, Huang CH, Ko TP, Ren F, Chan HC, Nega M, Bogue S, Lopez D, Kolter R, Gotz F, Guo RT, Oldfield E. 2014. Structural and functional analysis of Bacillus subtilis YisP reveals a role of its product in biofilm production. Chem Biol 21:1557–1563. doi: 10.1016/j.chembiol.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown DA. 2002. Isolation and use of rafts. Curr Protoc Immunol Chapter 11:Unit 11.10. doi: 10.1002/0471142735.im1110s51. [DOI] [PubMed] [Google Scholar]
- 48.Shah MB, Sehgal PB. 2007. Nondetergent isolation of rafts. Methods Mol Biol 398:21–28. doi: 10.1007/978-1-59745-513-8_3. [DOI] [PubMed] [Google Scholar]
- 49.Donovan C, Bramkamp M. 2009. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology 155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang HM, Li Z, Tsudome M, Ito S, Takami H, Horikoshi K. 2005. An alkali-inducible flotillin-like protein from Bacillus halodurans C-125. Protein J 24:125–131. doi: 10.1007/s10930-004-1519-3. [DOI] [PubMed] [Google Scholar]
- 51.Dempwolff F, Moller HM, Graumann PL. 2012. Synthetic motility and cell shape defects for deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay with NfeD proteins. J Bacteriol 194:4652–4661. doi: 10.1128/JB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergstrom JD, Kurtz MM, Rew DJ, Amend AM, Karkas JD, Bostedor RG, Bansal VS, Dufresne C, VanMiddlesworth FL, Hensens OD, Liesch JM, Zink DL, Wilson KE, Onishi J, Milligan JA, Bills G, Kaplan L, Omstead MN, Jenkins RG, Huang L, Meinz MS, Quinn L, Burg RW, Kong YL, Mochales S, Mojena M, Martin I, Pelaez F, Diez MT, Alberts AW. 1993. Zaragozic acids: a family of fungal metabolites that are picomolar competitive inhibitors of squalene synthase. Proc Natl Acad Sci U S A 90:80–84. doi: 10.1073/pnas.90.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Endo A, Kuroda M, Tanzawa K. 1976. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme A reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett 72:323–326. doi: 10.1016/0014-5793(76)80996-9. [DOI] [PubMed] [Google Scholar]
- 54.Pelz A, Wieland KP, Putzbach K, Hentschel P, Albert K, Gotz F. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem 280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 55.Kaiser HJ, Surma MA, Mayer F, Levental I, Grzybek M, Klemm RW, Da Cruz S, Meisinger C, Muller V, Simons K, Lingwood D. 2011. Molecular convergence of bacterial and eukaryotic surface order. J Biol Chem 286:40631–40637. doi: 10.1074/jbc.M111.276444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YH, Kingston AW, Helmann JD. 2012. Glutamate dehydrogenase affects resistance to cell wall antibiotics in Bacillus subtilis. J Bacteriol 194:993–1001. doi: 10.1128/JB.06547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saenz JP, Sezgin E, Schwille P, Simons K. 2012. Functional convergence of hopanoids and sterols in membrane ordering. Proc Natl Acad Sci U S A 109:14236–14240. doi: 10.1073/pnas.1212141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yepes A, Schneider J, Mielich B, Koch G, Garcia-Betancur JC, Ramamurthi KS, Vlamakis H, Lopez D. 2012. The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH. Mol Microbiol 86:457–471. doi: 10.1111/j.1365-2958.2012.08205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajendran L, Simons K. 2005. Lipid rafts and membrane dynamics. J Cell Sci 118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 60.Simons K, Gerl MJ. 2010. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol 11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 61.Browman DT, Hoegg MB, Robbins SM. 2007. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Parsons JB, Rock CO. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaiser HJ, Lingwood D, Levental I, Sampaio JL, Kalvodova L, Rajendran L, Simons K. 2009. Order of lipid phases in model and plasma membranes. Proc Natl Acad Sci U S A 106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ben-Menachem G, Kubler-Kielb J, Coxon B, Yergey A, Schneerson R. 2003. A newly discovered cholesteryl galactoside from Borrelia burgdorferi. Proc Natl Acad Sci U S A 100:7913–7918. doi: 10.1073/pnas.1232451100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LaRocca TJ, Crowley JT, Cusack BJ, Pathak P, Benach J, London E, Garcia-Monco JC, Benach JL. 2010. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe 8:331–342. doi: 10.1016/j.chom.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LaRocca TJ, Pathak P, Chiantia S, Toledo A, Silvius JR, Benach JL, London E. 2013. Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog 9:e1003353. doi: 10.1371/journal.ppat.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bosak T, Losick RM, Pearson A. 2008. A polycyclic terpenoid that alleviates oxidative stress. Proc Natl Acad Sci U S A 105:6725–6729. doi: 10.1073/pnas.0800199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ourisson G, Rohmer M, Poralla K. 1987. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu Rev Microbiol 41:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 69.Cvejic JH, Putra SR, El-Beltagy A, Hattori R, Hattori T, Rohmer M. 2000. Bacterial triterpenoids of the hopane series as biomarkers for the chemotaxonomy of Burkholderia, Pseudomonas and Ralstonia spp. FEMS Microbiol Lett 183:295–299. doi: 10.1111/j.1574-6968.2000.tb08974.x. [DOI] [PubMed] [Google Scholar]
- 70.Dobritsa SV, Potter D, Gookin TE, Berry AM. 2001. Hopanoid lipids in Frankia: identification of squalene-hopene cyclase gene sequences. Can J Microbiol 47:535–540. doi: 10.1139/w01-045. [DOI] [PubMed] [Google Scholar]
- 71.Doughty DM, Hunter RC, Summons RE, Newman DK. 2009. 2-Methylhopanoids are maximally produced in akinetes of Nostoc punctiforme: geobiological implications. Geobiology 7:524–532. doi: 10.1111/j.1472-4669.2009.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hartner T, Straub KL, Kannenberg E. 2005. Occurrence of hopanoid lipids in anaerobic Geobacter species. FEMS Microbiol Lett 243:59–64. doi: 10.1016/j.femsle.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 73.Pearson A, Leavitt WD, Saenz JP, Summons RE, Tam MC, Close HG. 2009. Diversity of hopanoids and squalene-hopene cyclases across a tropical land-sea gradient. Environ Microbiol 11:1208–1223. doi: 10.1111/j.1462-2920.2008.01817.x. [DOI] [PubMed] [Google Scholar]
- 74.Pearson A, Rusch DB. 2009. Distribution of microbial terpenoid lipid cyclases in the global ocean metagenome. ISME J 3:352–363. doi: 10.1038/ismej.2008.116. [DOI] [PubMed] [Google Scholar]
- 75.Schmerk CL, Bernards MA, Valvano MA. 2011. Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J Bacteriol 193:6712–6723. doi: 10.1128/JB.05979-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Y, Cooke MP, Talbot HM, Simpson MJ. 2009. Bacteriohopanepolyol signatures of bacterial populations in Western Canadian soils. Org Geochem 40:79–86. doi: 10.1016/j.orggeochem.2008.09.003. [DOI] [Google Scholar]
- 77.Pearson A, Flood Page SR, Jorgenson TL, Fischer WW, Higgins MB. 2007. Novel hopanoid cyclases from the environment. Environ Microbiol 9:2175–2188. doi: 10.1111/j.1462-2920.2007.01331.x. [DOI] [PubMed] [Google Scholar]
- 78.Siedenburg G, Jendrossek D. 2011. Squalene-hopene cyclases. Appl Environ Microbiol 77:3905–3915. doi: 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kannenberg E, Poralla K. 1982. The influence of hopanoids on growth of Mycoplasma mycoides. Arch Microbiol 133:100–102. doi: 10.1007/BF00413519. [DOI] [PubMed] [Google Scholar]
- 80.Moreau RA, Agnew J, Hicks KB, Powell MJ. 1997. Modulation of lipoxygenase activity by bacterial hopanoids. J Nat Prod 60:397–398. doi: 10.1021/np960611y. [DOI] [PubMed] [Google Scholar]
- 81.Poralla K, Muth G, Hartner T. 2000. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol Lett 189:93–95. doi: 10.1111/j.1574-6968.2000.tb09212.x. [DOI] [PubMed] [Google Scholar]
- 82.Seipke RF, Loria R. 2009. Hopanoids are not essential for growth of Streptomyces scabies 87-22. J Bacteriol 191:5216–5223. doi: 10.1128/JB.00390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doughty DM, Coleman ML, Hunter RC, Sessions AL, Summons RE, Newman DK. 2011. The RND-family transporter, HpnN, is required for hopanoid localization to the outer membrane of Rhodopseudomonas palustris TIE-1. Proc Natl Acad Sci U S A 108:E1045–E1051. doi: 10.1073/pnas.1104209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor RF. 1984. Bacterial triterpenoids. Microbiol Rev 48:181–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dworkin M. 1959. Function of carotenoids in photosynthetic bacteria. Nature 184:1891–1892. doi: 10.1038/1841891b0. [DOI] [PubMed] [Google Scholar]
- 86.Vershinin A. 1999. Biological functions of carotenoids—diversity and evolution. Biofactors 10:99–104. doi: 10.1002/biof.5520100203. [DOI] [PubMed] [Google Scholar]
- 87.Cvetkovic D, Fiedor L, Wisniewska-Becker A, Markovic D. 2013. Organization of carotenoids in models of biological membranes: current status of knowledge and research. Curr Anal Chem 9:1573–4110. doi: 10.2174/1573411011309010086. [DOI] [Google Scholar]
- 88.Subczynski WK, Wisniewska-Becker A, Widomska J. 2012. Can macular xanthophylls replace cholesterol in formation of the liquid-ordered phase in lipid-bilayer membranes? Acta Biochim Pol 59:109–114. [PMC free article] [PubMed] [Google Scholar]
- 89.Hammond RK, White DC. 1970. Carotenoid formation by Staphylococcus aureus. J Bacteriol 103:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duc LH, Fraser PD, Tam NK, Cutting SM. 2006. Carotenoids present in halotolerant Bacillus spore formers. FEMS Microbiol Lett 255:215–224. doi: 10.1111/j.1574-6968.2005.00091.x. [DOI] [PubMed] [Google Scholar]
- 91.Khaneja R, Perez-Fons L, Fakhry S, Baccigalupi L, Steiger S, To E, Sandmann G, Dong TC, Ricca E, Fraser PD, Cutting SM. 2010. Carotenoids found in Bacillus. J Appl Microbiol 108:1889–1902. doi: 10.1111/j.1365-2672.2009.04590.x. [DOI] [PubMed] [Google Scholar]
- 92.Lee S, Poulter CD. 2008. Cloning, solubilization, and characterization of squalene synthase from Thermosynechococcus elongatus BP-1. J Bacteriol 190:3808–3816. doi: 10.1128/JB.01939-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu CI, Liu GY, Song Y, Yin F, Hensler ME, Jeng WY, Nizet V, Wang AH, Oldfield E. 2008. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu Y, Jia S, Ren F, Huang CH, Ko TP, Mitchell DA, Guo RT, Zheng Y. 2013. Crystallization and preliminary X-ray diffraction analysis of YisP protein from Bacillus subtilis subsp. subtilis strain 168. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:77–79. doi: 10.1107/S1744309112049330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schlame M. 2008. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res 49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mileykovskaya E, Zhang M, Dowhan W. 2005. Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- 97.Mileykovskaya E. 2007. Subcellular localization of Escherichia coli osmosensory transporter ProP: focus on cardiolipin membrane domains. Mol Microbiol 64:1419–1422. doi: 10.1111/j.1365-2958.2007.05766.x. [DOI] [PubMed] [Google Scholar]
- 98.Tsai M, Ohniwa RL, Kato Y, Takeshita SL, Ohta T, Saito S, Hayashi H, Morikawa K. 2011. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol 11:13. doi: 10.1186/1471-2180-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. 2001. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett 507:187–190. doi: 10.1016/S0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- 100.Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. 1997. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem 272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 101.Lang DM, Lommel S, Jung M, Ankerhold R, Petrausch B, Laessing U, Wiechers MF, Plattner H, Stuermer CA. 1998. Identification of reggie-1 and reggie-2 as plasmamembrane-associated proteins which cocluster with activated GPI-anchored cell adhesion molecules in non-caveolar micropatches in neurons. J Neurobiol 37:502–523. doi:. [DOI] [PubMed] [Google Scholar]
- 102.Morrow IC, Rea S, Martin S, Prior IA, Prohaska R, Hancock JF, James DE, Parton RG. 2002. Flotillin-1/reggie-2 traffics to surface raft domains via a novel Golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J Biol Chem 277:48834–48841. doi: 10.1074/jbc.M209082200. [DOI] [PubMed] [Google Scholar]
- 103.Schulte T, Paschke KA, Laessing U, Lottspeich F, Stuermer CA. 1997. Reggie-1 and reggie-2, two cell surface proteins expressed by retinal ganglion cells during axon regeneration. Development 124:577–587. [DOI] [PubMed] [Google Scholar]
- 104.Stuermer CA, Plattner H. 2005. The ‘lipid raft’ microdomain proteins reggie-1 and reggie-2 (flotillins) are scaffolds for protein interaction and signalling. Biochem Soc Symp 72:109–118. [DOI] [PubMed] [Google Scholar]
- 105.Allen JA, Halverson-Tamboli RA, Rasenick MM. 2007. Lipid raft microdomains and neurotransmitter signalling. Nat Rev Neurosci 8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- 106.Stuermer CA. 2010. The reggie/flotillin connection to growth. Trends Cell Biol 20:6–13. doi: 10.1016/j.tcb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 107.Babuke T, Ruonala M, Meister M, Amaddii M, Genzler C, Esposito A, Tikkanen R. 2009. Hetero-oligomerization of reggie-1/flotillin-2 and reggie-2/flotillin-1 is required for their endocytosis. Cell Signal 21:1287–1297. doi: 10.1016/j.cellsig.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 108.Banning A, Tomasovic A, Tikkanen R. 2011. Functional aspects of membrane association of reggie/flotillin proteins. Curr Protein Pept Sci 12:725–735. doi: 10.2174/138920311798841708. [DOI] [PubMed] [Google Scholar]
- 109.Amaddii M, Meister M, Banning A, Tomasovic A, Mooz J, Rajalingam K, Tikkanen R. 2012. Flotillin-1/reggie-2 protein plays dual role in activation of receptor-tyrosine kinase/mitogen-activated protein kinase signaling. J Biol Chem 287:7265–7278. doi: 10.1074/jbc.M111.287599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. 2010. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A 107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neumann-Giesen C, Falkenbach B, Beicht P, Claasen S, Luers G, Stuermer CA, Herzog V, Tikkanen R. 2004. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem J 378:509–518. doi: 10.1042/BJ20031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rivera-Milla E, Stuermer CA, Malaga-Trillo E. 2006. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci 63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cao M, Kobel PA, Morshedi MM, Wu MF, Paddon C, Helmann JD. 2002. Defining the Bacillus subtilis sigma(W) regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J Mol Biol 316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- 114.Malaga-Trillo E, Laessing U, Lang DM, Meyer A, Stuermer CA. 2002. Evolution of duplicated reggie genes in zebrafish and goldfish. J Mol Evol 54:235–245. doi: 10.1007/s00239-001-0005-1. [DOI] [PubMed] [Google Scholar]
- 115.Moszer I, Jones LM, Moreira S, Fabry C, Danchin A. 2002. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res 30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walker CA, Hinderhofer M, Witte DJ, Boos W, Moller HM. 2008. Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis. J Biomol NMR 42:69–76. doi: 10.1007/s10858-008-9261-3. [DOI] [PubMed] [Google Scholar]
- 117.Solis GP, Hoegg M, Munderloh C, Schrock Y, Malaga-Trillo E, Rivera-Milla E, Stuermer CA. 2007. Reggie/flotillin proteins are organized into stable tetramers in membrane microdomains. Biochem J 403:313–322. doi: 10.1042/BJ20061686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wiegert T, Homuth G, Versteeg S, Schumann W. 2001. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol Microbiol 41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- 119.Mielich-Suss B, Schneider J, Lopez D. 2013. Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis. mBio 4(6):e00719-13. doi: 10.1128/mBio.00719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bach JN, Bramkamp M. 2013. Flotillins functionally organize the bacterial membrane. Mol Microbiol 88:1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- 121.Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Green JB, Lower RP, Young JP. 2009. The NfeD protein family and its conserved gene neighbours throughout prokaryotes: functional implications for stomatin-like proteins. J Mol Evol 69:657–667. doi: 10.1007/s00239-009-9304-8. [DOI] [PubMed] [Google Scholar]
- 123.Pizarro-Cerda J, Kuhbacher A, Cossart P. 2012. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med 2:a010009. doi: 10.1101/cshperspect.a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chiba S, Ito K, Akiyama Y. 2006. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol Microbiol 60:448–457. doi: 10.1111/j.1365-2958.2006.05104.x. [DOI] [PubMed] [Google Scholar]
- 126.Hinderhofer M, Walker CA, Friemel A, Stuermer CA, Moller HM, Reuter A. 2009. Evolution of prokaryotic SPFH proteins. BMC Evol Biol 9:10. doi: 10.1186/1471-2148-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kenri T, Horino A, Matsui M, Sasaki Y, Suzuki S, Narita M, Ohya H, Okazaki N, Shibayama K. 2012. Complete genome sequence of Mycoplasma pneumoniae type 2a strain 309, isolated in Japan. J Bacteriol 194:1253–1254. doi: 10.1128/JB.06553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corcelli A, Lattanzio VM, Mascolo G, Papadia P, Fanizzi F. 2002. Lipid-protein stoichiometries in a crystalline biological membrane: NMR quantitative analysis of the lipid extract of the purple membrane. J Lipid Res 43:132–140. [PubMed] [Google Scholar]
- 129.Gilmore SF, Yao AI, Tietel Z, Kind T, Facciotti MT, Parikh AN. 2013. Role of squalene in the organization of monolayers derived from lipid extracts of Halobacterium salinarum. Langmuir 29:7922–7930. doi: 10.1021/la401412t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Munro S. 2003. Lipid rafts: elusive or illusive? Cell 115:377–388. doi: 10.1016/S0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 131.Shaw AS. 2006. Lipid rafts: now you see them, now you don't. Nat Immunol 7:1139–1142. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- 132.Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb Perspect Biol 2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 134.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 135.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol 59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 136.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 138.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Romero D, Vlamakis H, Losick R, Kolter R. 2011. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol 80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 142.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- 143.Hahn J, Roggiani M, Dubnau D. 1995. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J Bacteriol 177:3601–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Grossman AD. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet 29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 145.Zuber P, Losick R. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol 169:2223–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jiang M, Shao W, Perego M, Hoch JA. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol 38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 147.Shemesh M, Chai Y. 2013. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J Bacteriol 195:2747–2754. doi: 10.1128/JB.00028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lopez D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev 34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 149.Lopez D, Gontang EA, Kolter R. 2010. Potassium sensing histidine kinase in Bacillus subtilis. Methods Enzymol 471:229–251. doi: 10.1016/S0076-6879(10)71013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev 7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- 151.Lopez D, Vlamakis H, Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev 33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 152.Le AT, Schumann W. 2009. The Spo0E phosphatase of Bacillus subtilis is a substrate of the FtsH metalloprotease. Microbiology 155:1122–1132. doi: 10.1099/mic.0.024182-0. [DOI] [PubMed] [Google Scholar]
- 153.Lysenko E, Ogura T, Cutting SM. 1997. Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143:971–978. doi: 10.1099/00221287-143-3-971. [DOI] [PubMed] [Google Scholar]
- 154.Zellmeier S, Zuber U, Schumann W, Wiegert T. 2003. The absence of FtsH metalloprotease activity causes overexpression of the sigmaW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis. J Bacteriol 185:973–982. doi: 10.1128/JB.185.3.973-982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ito K, Akiyama Y. 2005. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 156.Kihara A, Akiyama Y, Ito K. 1996. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J 15:6122–6131. [PMC free article] [PubMed] [Google Scholar]
- 157.Kihara A, Akiyama Y, Ito K. 1997. Host regulation of lysogenic decision in bacteriophage lambda: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc Natl Acad Sci U S A 94:5544–5549. doi: 10.1073/pnas.94.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Steglich G, Neupert W, Langer T. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol 19:3435–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hendrick JP, Wickner W. 1991. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem 266:24596–24600. [PubMed] [Google Scholar]
- 160.Dalal K, Chan CS, Sligar SG, Duong F. 2012. Two copies of the SecY channel and acidic lipids are necessary to activate the SecA translocation ATPase. Proc Natl Acad Sci U S A 109:4104–4109. doi: 10.1073/pnas.1117783109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Perego M, Higgins CF, Pearce SR, Gallagher MP, Hoch JA. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol 5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 162.Lazazzera BA. 2001. The intracellular function of extracellular signaling peptides. Peptides 22:1519–1527. doi: 10.1016/S0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- 163.Dominguez-Cuevas P, Porcelli I, Daniel RA, Errington J. 2013. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol 89:1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meisner J, Montero Llopis P, Sham LT, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J 30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mann JM, Carabetta VJ, Cristea IM, Dubnau D. 2013. Complex formation and processing of the minor transformation pilins of Bacillus subtilis. Mol Microbiol 90:1201–1215. doi: 10.1111/mmi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Falagas ME, Makris GC, Matthaiou DK, Rafailidis PI. 2008. Statins for infection and sepsis: a systematic review of the clinical evidence. J Antimicrob Chemother 61:774–785. doi: 10.1093/jac/dkn019. [DOI] [PubMed] [Google Scholar]
- 168.Kopterides P, Falagas ME. 2009. Statins for sepsis: a critical and updated review. Clin Microbiol Infect 15:325–334. doi: 10.1111/j.1469-0691.2009.02750.x. [DOI] [PubMed] [Google Scholar]
- 169.Liappis AP, Kan VL, Rochester CG, Simon GL. 2001. The effect of statins on mortality in patients with bacteremia. Clin Infect Dis 33:1352–1357. doi: 10.1086/323334. [DOI] [PubMed] [Google Scholar]
- 170.Balibar CJ, Shen X, Tao J. 2009. The mevalonate pathway of Staphylococcus aureus. J Bacteriol 191:851–861. doi: 10.1128/JB.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol 182:4319–4327. doi: 10.1128/JB.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Takahashi S, Kuzuyama T, Watanabe H, Seto H. 1998. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci U S A 95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hunter WN. 2007. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J Biol Chem 282:21573–21577. doi: 10.1074/jbc.R700005200. [DOI] [PubMed] [Google Scholar]
- 174.Doughty DM, Dieterle M, Sessions AL, Fischer WW, Newman DK. 2014. Probing the subcellular localization of hopanoid lipids in bacteria using NanoSIMS. PLoS One 9:e84455. doi: 10.1371/journal.pone.0084455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gahlmann A, Moerner WE. 2014. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat Rev Microbiol 12:9–22. doi: 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Owen DM, Gaus K. 2013. Imaging lipid domains in cell membranes: the advent of super-resolution fluorescence microscopy. Front Plant Sci 4:503. doi: 10.3389/fpls.2013.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schermelleh L, Heintzmann R, Leonhardt H. 2010. A guide to super-resolution fluorescence microscopy. J Cell Biol 190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mizuno H, Abe M, Dedecker P, Makino M, Rocha S, Ohno-Iwashita Y, Hofkens J, Kobayashi T, Miyawaki A. 2011. Fluorescent probes for superresolution imaging of lipid domains on the plasma membrane. Chem Sci 2:1548–1553. doi: 10.1039/c1sc00169h. [DOI] [Google Scholar]
- 179.Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, Usukura J, Kusumi A. 2006. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol 174:851–862. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Richter T, Floetenmeyer M, Ferguson C, Galea J, Goh J, Lindsay MR, Morgan GP, Marsh BJ, Parton RG. 2008. High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic 9:893–909. doi: 10.1111/j.1600-0854.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- 181.Tocheva EI, Li Z, Jensen GJ. 2010. Electron cryotomography. Cold Spring Harb Perspect Biol 2:a003442. doi: 10.1101/cshperspect.a003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP. 2013. Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol 11:e1001565. doi: 10.1371/journal.pbio.1001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. 2011. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol 9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dinic J, Biverstahl H, Maler L, Parmryd I. 2011. Laurdan and di-4-ANEPPDHQ do not respond to membrane-inserted peptides and are good probes for lipid packing. Biochim Biophys Acta 1808:298–306. doi: 10.1016/j.bbamem.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 185.Gaus K, Zech T, Harder T. 2006. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol Membr Biol 23:41–48. doi: 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- 186.Wheeler G, Tyler KM. 2011. Widefield microscopy for live imaging of lipid domains and membrane dynamics. Biochim Biophys Acta 1808:634–641. doi: 10.1016/j.bbamem.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Strahl H, Burmann F, Hamoen LW. 2014. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun 5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.de Almeida RF, Fedorov A, Prieto M. 2003. Sphingomyelin phosphatidylcholine cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys J 85:2406–2416. doi: 10.1016/S0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Parasassi T, Gratton E, Yu WM, Wilson P, Levi M. 1997. Two-photon fluorescence microscopy of laurdan generalized polarization domains in model and natural membranes. Biophys J 72:2413–2429. doi: 10.1016/S0006-3495(97)78887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]