Abstract
The impact of population bottlenecks is an important factor to consider when assessing species survival. Population declines can considerably limit the evolutionary potential of species and make them more susceptible to stochastic events. New Zealand has a well documented history of decline of endemic avifauna related to human colonization. Here, we investigate the genetic effects of a recent population decline in the endangered kea (Nestor notabilis). Kea have undergone a long-lasting persecution between the late 1800s to 1970s where an estimated 150,000 kea were culled under a governmental bounty scheme. Kea now number 1,000–5,000 individuals in the wild and it is likely that the recent population decline may have reduced the genetic diversity of the species. Comparison of contemporary (n = 410), historical (n = 15) and fossil samples (n = 4) showed a loss of mitochondrial diversity since the end of the last glaciation (Otiran Glacial) but no loss of overall genetic diversity associated with the cull. Microsatellite data indicated a recent bottleneck for only one population and a range-wide decline in Ne dating back some 300 – 6,000 years ago, a period predating European arrival in NZ. These results suggest that despite a recent human persecution, kea might have experienced a large population decline before stabilizing in numbers prior to human settlement of New Zealand in response to Holocene changes in habitat distribution. Our study therefore highlights the need to understand the respective effects of climate change and human activities on endangered species dynamics when proposing conservation guidelines.
Introduction
Genetic diversity is an essential component of both the short and long-term persistence of species. It can be regarded as the basis for evolutionary potential [1] and favors an adaptive response to changing environments [2]. Endangered species with small population sizes often show low genetic diversity [3, 4], which compromises long-term viability and adaptability, and increases chances of inbreeding [4–6]. Even though recent population declines or extinctions are often attributed to human activities (e.g. habitat destruction, over-exploitation) [7–9], humans are not the sole driver of population dynamics of species. For instance, the population dynamics of the musk ox (Ovibos moschatus) were best explained by climate and associated habitat change [10] while the extinction of woolly mammoths (Mammuthus primigenius) appears to have been caused by the concomitant effect of over-hunting and climate change [11]. It is therefore necessary to identify the underlying cause of decline, to determine the appropriate management strategy of a species.
In order to distinguish anthropogenic from natural drivers of population decline, one can refer to pre-human fossil records of the distribution of species. However, such data is often lacking or incomplete, leading to a potential bias in the estimation of population size and oversimplification of population dynamics, as critically addressed by Rawlence et al. [7] and Allentoft et al. [12].
DNA-based approaches using contemporary samples only are routinely used to detect population declines [13–16] but can also have limited resolution. Indeed, a direct comparison of historical and contemporary genetic diversity is often advantageous because it allows the event associated with population fluctuations to be dated with more accuracy and provides a unique opportunity to study genetic variability in pre-bottleneck populations [17–19].
The avifauna of New Zealand evolved in the absence of mammalian predators [20, 21] and multiple lines of evidence support a history of population bottlenecks and extinctions linked to human colonization [8, 22]. The main cause of decline or population extinction is often attributed to introduced predators [23–26] or over-hunting [7–8, 27]. Additionally, some species were historically persecuted. For instance, the kea (Nestor notabilis), an endemic alpine parrot prehistorically present in both the North and South Islands of New Zealand [28–32] and now only found in the South Island was considered a pest [33]. Its inquisitiveness [34, 35], opportunistic feeding behavior [36–38] and more notably attacks on sheep [39–41] prompted a bounty scheme where over 150,000 kea were killed between the late 1800s and 1971 (when they received partial legislative protection). Today the species is mainly found in the alpine areas of the South Island above the tree line [42] while a few populations remain in podocarp forest in Westland [43] but it is still declining due to ongoing predation by introduced mammals [25]. However, accurate estimates of current effective population size are difficult to obtain because kea are often found in remote and rugged habitat. The most commonly accepted and conservative estimates of contemporary population size range between 1,000–5,000 individuals [44]. The estimates of the number of birds culled and current population size therefore suggest that kea went through a significant population bottleneck since European arrival.
Previous results have shown that despite its strong flying capabilities, the kea is subdivided into three distinct genetic clusters and that this population structure is most likely the result of postglacial recolonization processes but not a recent human-induced fragmentation [45]. Kea populations might also have fluctuated through time in response to climate variations during the Pleistocene and Holocene as has been shown in many other New Zealand species [7, 46]. The history of kea thus provides an excellent opportunity to test for genetic signatures of population size changes associated with past climatic events and recent human activities.
Here we test whether kea have experienced pre-human population declines or if the species started to decline only recently because of human persecution. We first assess changes in genetic diversity over time using microsatellite and mitochondrial data comparing contemporary, historical and fossil samples. Second, we look for a signature of ancient and recent bottlenecks using conventional methods for microsatellite data. We also look for a signature of population decline using coalescent-based Bayesian methods and estimate its timing and magnitude. Due to the magnitude of the reported recent population collapse (i.e. loss of approximately 95% of the historical population), we expect to find extinct haplotypes and alleles among historical and fossil samples that are not present in contemporary samples and also a significant signature of a recent bottleneck.
Material and Methods
Ethics Statement
Fieldwork was conducted under the approval of the University of Otago Ethics Committee (No. 74/09) and DoC (Permits No. OT-26311; WC-26101-FAU). No permits were required to sample fossil material from museums. Permission to sample museum specimens was obtained by NJR from individual institutions.
Sampling
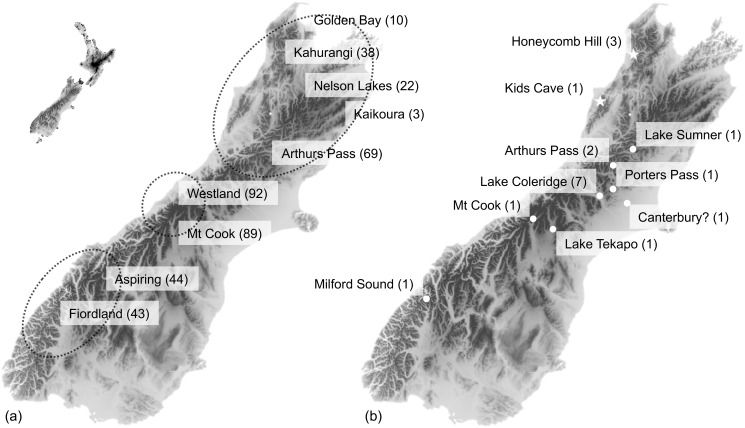
We utilized a contemporary dataset from birds caught between 2010 and 2012 comprising 91 mitochondrial (mtDNA) control region (CR) sequences and 410 nuclear microsatellite genotypes (17 microsatellite loci) as described in Dussex et al. [45]. Additionally, historical skins (n = 15) with robust locality data, and Late Glacial (10–14 kya) and Holocene fossil bones (< 11.6 kya; n = 4) [31, 47] from across the geographic range of kea were obtained from New Zealand and overseas museum collections (Table 1, Fig. 1). To ensure independence of samples, only common elements of the left or right orientation were sampled from an individual deposit, or bones were sampled from different stratigraphic units within a deposit (e.g. layers, squares). While most of the historical skins used in this study come from the Canterbury region, fossil samples from other regions extend the coverage to the whole contemporary range of the species.
Table 1. Historical and fossil samples used to obtain genotypic and sequencing data for the mitochondrial control region.
Information on mitochondrial fragment length, sampling location and the corresponding cluster identified in TESS (See [45]) are given. The four fossil samples were not amplified for microsatellite markers. Abbreviations call on Hum, humerus; Tt, tibiotarsus.
| Sample number | Museum | Tissue type | Sample age/collection date | Region | Sampling location | mtDNA fragment length | Corresponding cluster |
|---|---|---|---|---|---|---|---|
| NMNZ S.43574 | National Museum of New Zealand Te Papa Tangarewa (NMNZ) | fossil bone | 13–17 ky | West Coast | Kids Cave (0.5 cm depth; Hum) | 450 | North |
| NMNZ S.22664.1 | (NMNZ) | fossil bone | 11–14 ky | West Coast | Honeycomb Hill (Terrace edge, Graveyard; Ulna) | 245 | North |
| NMNZ S.22039.1 | (NMNZ) | fossil bone | 11–14 ky | West Coast | Honeycomb Hill (Terrace, Graveyard (L1+ L2 Ex2); Tt) | 245 | North |
| NMNZ S.22039.2 | (NMNZ) | fossil bone | 11–14 ky | West Coast | Honeycomb Hill (Terrace, Graveyard; Tt) | 245 | North |
| AlM LB2467 | Auckland Museum (AlM) | toepad | 1932 | Canterbury | Lake Sumner | 223* | North |
| AlM LB2466 | (AlM) | toepad | 1932 | Canterbury | Canterbury | 616 | North |
| CM AV84 | Canterbury Museum (CM) | toepad | 1908 | Canterbury | Wilberforce River (Lake Coleridge) | 616 | North |
| CM AV86 | (CM) | toepad | 1913 | Canterbury | Wilberforce River (Lake Coleridge) | 616 | North |
| CM AV88 | (CM) | toepad | 1913 | Canterbury | Lake Coleridge | 223* | North |
| CM AV89 | (CM) | toepad | 1907 | Canterbury | Windwhistle (Lake Coleridge) | 616 | North |
| CM AV90 | (CM) | toepad | 1913 | Canterbury | Lake Coleridge | 223* | North |
| CM AV91 | (CM) | toepad | 1913 | Canterbury | Wilberforce River (Lake Coleridge) | 616 | North |
| NMW 12.208 | Museum of Natural History Vienna, Austria (NMW) | toepad | 1877 | Canterbury | Arthur’s Pass | 223* | North |
| NMW 12.209 | (NMW) | toepad | 1877 | Canterbury | Arthur’s Pass | 616 | North |
| NMW 49.760 | (NMW) | toepad | 1877 | Canterbury | Porter’s Pass | 616 | North |
| NMW 12.211 | (NMW) | toepad | 1877 | Canterbury | Wilberforce River (Lake Coleridge) | 616 | North |
| AlM LB2469 | (AlM) | toepad | 1934 | Fiordland | Milford Sound | 616 | South |
| CM AV2039 | (CM) | toepad | 1932 | Mt Cook | Lake Tekapo, Canterbury Land District | 616 | Central |
| NHM 1927.12.18.125 | Natural History Museum London (NHM) | toepad | 1889 | Mt Cook | Mt Cook | 616 | Central |
*toepads with mtDNA fragment shorter than 450 bp were not used for comparison of contemporary and historical genetic diversity
Fig 1. Sampling locations for (a) contemporary samples with circles representing genetic clusters in TESS as described in [45] and (b) the 15 historical skin samples (dots) and four fossil bones (stars) for kea.
Numbers in brackets represent the sample size.
Ancient DNA extraction, amplification and sequencing
All ancient DNA (aDNA) extractions and PCR set up was carried out at the University of Otago in a purpose built aDNA laboratory physically isolated from other molecular laboratories [48]. Strict aDNA procedures were followed to minimize contamination of samples with exogenous DNA [49], including the use of negative extraction and PCR controls to ensure the authentication of sequences. DNA was extracted from historical skins using the Qiagen DNeasy Blood and Tissue Kit following the manufactures instructions, except the lysis step was performed as an overnight incubation at 56°C. Replicate extractions were performed on 10% of samples, chosen randomly. DNA was extracted from up to 250 mg of fossil bone powder following Rohland et al. [50]. PCR amplification and all downstream procedures were carried out in a modern genetics laboratory.
Microsatellite DNA amplification
Historical samples were genotyped for 17 microsatellite loci [45] using cycling conditions outlined in [51] with an increase in the number of cycles from 8 to 12 and from 25 to 30. All samples were independently amplified five times in order to decrease the chances of allelic dropout [52]. Microsatellite markers (> 200 bp) were not amplified for fossil samples, because of the short fragments obtained for mtDNA (∼150 bp) and associated low chance of successful amplification.
Mitochondrial DNA amplification
The mtDNA CR from historical skins was amplified and sequenced using four primer pairs (producing ∼250 bp fragments including primers) (S1 Table, S1 Fig.) and the same PCR conditions described in Dussex et al. [45] but with an increase in cycle number to 60.
Fossil samples were amplified and sequenced using seven primer pairs (∼150 bp fragments including primers) (S1 Table, S1 Fig.) to cover Domain I of the mtDNA CR. Each PCR reaction (20 µL) consisted of: 1 M Betaine (Sigma), 4 mM MgCl2 (Life Technologies), 1 x Gold Buffer II (Life Technologies), 2.5 mM dNTPs (Bioline), 250 nM each primer, 1.25 U of AmpliTaq Gold DNA Polymerase (Life Technologies), and 2 µL DNA. Unsuccessful PCRs were repeated with 2 U AmpliTaq Gold DNA Polymerase and 4 µL DNA or 2 µL 1:10 DNA. PCR thermocycling conditions consisted of 94°C 9 min, 60 cycles 94°C 30 s, 50°C 45 s, 72°C 1 min, followed by a final extension of 72°C 10 min.
PCR products were run on a 2% 1 x TAE agarose gel. PCR products were purified (using ExoSap (1.5 U ExoI, I U SAP; GE Healthcare) by incubation at 37°C for 30 min and 80°C for 15 min), and sequenced bi-directionally from independent PCR products using Big Dye Terminator technology and an ABI 3730xl. When an inconsistency between sequences from an individual was observed due to DNA damage (C-T and G-A transitions), additional PCRs and bi-directional sequencing were conducted, and a majority rule consensus applied to the four independent PCRs [53].
Consensus sequences were obtained from the alignment of fragments made in Geneious 5.6.3 [54].
Data analysis
Genetic diversity
For microsatellite markers, we checked for scoring error due to stuttering, large allele drop-out, and null alleles using MICRO-CHECKER 2.2.3 [55]. Tests for Hardy-Weinberg proportions and genotypic disequilibrium for both historical and contemporary samples were conducted using exact probability tests with 1,000 iterations as implemented in GENEPOP 4.1 [56]. An unbiased estimate of the exact P-value was determined using a Markov chain method following the permutation algorithm of Guo and Thompson [57]. The significance threshold values for multiple statistical tests was adjusted using Bonferroni correction [58].
Indices of genetic diversity were calculated for both contemporary and museum samples. To assess the level of genetic variation in microsatellites, FSTAT was used to calculate means of allelic richness corrected for sample size [59], observed and expected heterozygosity (unbiased H, [60]) for each population. Statistical comparison between contemporary and historical samples for expected and observed heterozygosity, number of alleles and allelic richness were matched by loci and tested using a pairwise Wilcoxon signed-ranked test in R [61].
Mitochondrial diversity indices, such as number of haplotypes (h), nucleotide diversity (π; [62]), number of polymorphic sites (S) and haplotypic diversity (Hd) were computed in ARLEQUIN 3.5 [63] for contemporary, historical and fossil samples focusing on 450 bp of Domain I of the mtDNA CR.
We then constructed a temporal network to look at mitochondrial variation in kea through time using the R script TempNet v1.4. [64].
Historical population structure and assignment of historical samples
In order to compare historical and contemporary population structure, we examined the assignment of historical samples to contemporary populations. The rationale was that if the contemporary structure corresponded to the historical one, museum samples should cluster with samples of the same geographical origin. On the other hand, if the contemporary structure is the result of genetic drift caused by the recent population decline, historical samples should not cluster well with individuals of the same geographical origin or have more admixed membership. We first used STRUCTURE 2.2 [65, 66] to estimate the membership coefficient of each historical sample to one of three genetic clusters previously identified by Dussex et al. [45]. The dataset was analyzed using the LOCPRIOR option and the most probable value of K was estimated from 25 iterations of K values ranging from 2 to 4 (as the most likely K selected was 2 and 3 for contemporary data). The results obtained with STRUCTURE 2.2 were used as input in STRUCTURE HARVESTER [67] to produce the input files used for the following averaging step among runs. For the selected K-value the individuals‚ assignment coefficient (q) for each genetic cluster were averaged over the 10 runs with the lowest posterior probability using CLUMPP software [68]. Finally, the output obtained in CLUMPP was used and visualized in the program DISTRUCT [69].
Demographic history
For all analyses described below, we first considered the three genetic clusters (north, central and south) identified in Dussex et al. [45] (Fig. 1a) as advised by Broquet et al. [70] and Chikhi et al. [71] in order to avoid bias or the generation of a false bottleneck signal. Even though our historical data span nearly 60 years and may not represent a conventional population, we also considered the 14 historical samples (hereafter Historical Canterbury) independently for the analyses described below. Because kea are long-lived and have a long generation time, minimal bias caused by overlapping generations is expected.
Evidence for a genetic bottleneck was first evaluated using three moment-based methods: (i) the heterozygosity excess method implemented in the software BOTTLENECK v. 1.2.02 [72]. This approach assumes that a population that has gone through a recent reduction in effective size will show a high heterozygosity relative to heterozygosity at mutation-drift equilibrium [73]. We used the most appropriate TPM model for microsatellites [74] with 90% stepwise mutations as recommended by [75] as well as 95% of stepwise mutations and SMM as a comparison and chose a variance of 12 to encompass the range of multistep mutations observed in natural populations [74]; (ii) the mode-shift test implemented in the same software; (iii) the M-ratio test [75] using M-P-Val and Critical_M [75], which is more appropriate to detect genetic bottlenecks that occurred over relatively long periods of time (<100 generations). We used TPM mutation models with an amount of single step mutations of ps = 0.9 and ps = 0.78, 3.5 steps for multi-step mutations and pre-bottleneck Ne values ranging from 500 and 5000. The mutation rate remained constant at μ of 5 × 10-4 [75]. The M-ratio was also calculated using only polymorphic loci to account for the upward bias in the M-ratio estimate, as implemented in ARLEQUIN 3.11 [63]. Secondly, in order to estimate variations of Ne through time, we used the coalescent-based Bayesian method of Storz and Beaumont [76] implemented in the program MSVAR 1.3. This approach considers the demographic history of a closed population that increases or decreases exponentially from an historical size NH at T years in the past to the current NC, where the data are allele frequencies for unlinked microsatellites. These loci are assumed to have independent genealogies and to be evolving according to the strict single-step mutation model. Given the demographic parameters (contemporary population size, NC; historical population size NH; and the time interval T) and the mutation rate (μ), the genealogical history of the sampled data can be described using coalescent theory, and the posterior probability distribution of the genealogical and demographic parameters is estimated using Markov chain Monte Carlo (MCMC) simulations. We performed five simulations per dataset (north, central and south clusters and Historical Canterbury) with 2 × 109 iterations with parameter values recorded every 1 × 105 iterations resulting in 20,000 records. Prior distributions were set to the parameters of simulation 1 (S2 Table). After checking for chain convergence, the last 50% of the data from each chain was combined (50,000 sample points) and the mode and 90% highest posterior densities (HPD) were calculated for each parameter using the R-package Locfit 1.5–6 [77]. We then evaluated the strength of evidence for population expansion versus decline by calculating the Bayes factor for each of the models [78, 79] as described by Storz and Beaumont [80]. The Bayes factor indicates the following levels of support for the model; BF, 0.33 = false detection of contraction/expansion, 0.33–3 = no support, 3–10 = substantial support, and >10 = strong support [78]. We ran theses analyses for both exponential and linear models while putting more emphasis on the exponential model, which is more accurate at modeling recent population declines [81].
Finally, two different demographic scenarios were compared within an ABC framework [82] using DIYABC [83] for each dataset (north, central and south clusters and Historical Canterbury). This allowed us to determine whether bottlenecks occurred and also to estimate values for key parameters of interest such as pre- and post bottleneck effective population sizes and time and duration of the bottleneck. The first model described a population bottleneck occurring some 0 to 1,000 generations ago, a period largely encompassing the arrival of humans to New Zealand [84] if assuming a generation time of 7 to 10 years for kea (J. Kemp pers. comm.). The prior for historical Ne (NH) was uniformly distributed between 10 and 2 × 105, the upper bound encompassing 150,000 kea reported being killed between the 1870s and 1970s [44]. The prior for the contemporary Ne (NC) was uniformly distributed between 10 and 104. These values included the current and conservative population estimate of 1,000 to 5,000 birds [44] and also accounted for the difficulty in obtaining accurate estimates of contemporary population size. The duration of the bottleneck was uniformly distributed between 1 and 100 generations. For comparison, a second model of constant population size was also defined, in which Ne was uniformly distributed through time for 0 to 1,000 generations and bounded between 10 and 2 × 105 individuals. For the two models, a generalized stepwise mutation model [85] was implemented with a mean rate uniformly distributed between 1.00 × 10-5 and 1.00 × 10-3 substitutions/generations [86–88]. The posterior probabilities of each scenario were then estimated using both a direct and logistic regression approach providing both point estimated and 95% confidence intervals [89, 90]. The 10,000 datasets (1%) with the smallest Euclidian distances were then retained to build posterior parameter distributions.
Further details on tests of bottleneck detection using moment-based methods, variations of Ne through time and demographic scenarios are available as S1 Supporting information.
Results
Genetic diversity
For museum samples, two microsatellite loci (Nmed31 and Strhab43) failed to amplify. After Bonferonni correction, no significant departure from Hardy-Weinberg equilibrium was observed and no tests for linkage disequilibrium were significant. Signs of null alleles were detected in single loci in the Kahurangi (Strhab43), Arthurs Pass (Nnot8) and Mt Cook (Nnot38) populations.
When comparing microsatellite genetic diversity among our historical and contemporary datasets, no extinct alleles were identified within the 15 skin samples (Table 2, S3 Table). Allelic richness was comparable in both datasets, being slightly lower in historical samples (Table 3). Mean number of alleles and expected heterozygosity were significantly different with historical samples showing lower diversity.
Table 2. Number of alleles sampled and allelic richness for contemporary and historical (1877–1934) samples.
| Contemporary | Historical | |||||||
|---|---|---|---|---|---|---|---|---|
| Locus | A | RS | HO | HE | A | RS | HO | HE |
| Cfor0809‡ | 5 | 1.337 | 0.323 | 0.29 | 3 | 1.325 | 0.111 | 0.121 |
| Nnot8* | 3 | 1.488 | 0.52 | 0.46 | 2 | 1.495 | 0.444 | 0.329 |
| Nnot14* | 11 | 1.759 | 0.758 | 0.76 | 9 | 1.822 | 1 | 0.663 |
| Nnot24* | 4 | 1.492 | 0.408 | 0.40 | 3 | 1.384 | 0.778 | 0.429 |
| Nnot37* | 2 | 1.391 | 0.294 | 0.33 | 2 | 1.389 | 0.652 | 0.409 |
| Nnot38* | 2 | 1.241 | 0.283 | 0.27 | 1 | 1 | 0 | 0.000 |
| Nnot39* | 2 | 1.36 | 0.335 | 0.31 | 2 | 1.254 | 0.061 | 0.099 |
| Nnot49* | 3 | 1.504 | 0.483 | 0.46 | 2 | 1.508 | 0.833 | 0.495 |
| Nnot49* | 2 | 1.202 | 0.193 | 0.20 | 2 | 1.129 | 0.361 | 0.193 |
| Strhab8° | 2 | 1.351 | 0.253 | 0.24 | 2 | 1.349 | 0.182 | 0.132 |
| Strhab13° | 9 | 1.736 | 0.728 | 0.69 | 6 | 1.791 | 0.939 | 0.663 |
| Strhab16° | 2 | 1.336 | 0.258 | 0.27 | 2 | 1.271 | 0.367 | 0.252 |
| Strhab25° | 7 | 1.764 | 0.742 | 0.73 | 5 | 1.709 | 0.97 | 0.608 |
| Strhab33° | 7 | 1.776 | 0.695 | 0.72 | 7 | 1.836 | 0.939 | 0.630 |
| Strhab35° | 8 | 1.811 | 0.719 | 0.75 | 7 | 1.794 | 0.576 | 0.458 |
Table 3. Estimates of genetic diversity of kea microsatellite data within contemporary and historical (1877–1934) samples with the sample size for each region/population (n), observed heterozygosity (HO), expected heterozygosity (HE), allelic richness (RS), mean allele numbers (A) and P-values for HE and RS.
| Sample | n | HO | HE * | RS | A * |
|---|---|---|---|---|---|
| Historical (1877–1934) | 15 | 0.548 | 0.365 | 1.470 | 3.667 |
| Contemporary (2010–2011) | 410 | 0.449 | 0.439 | 1.482 | 4.529 |
*significant difference with P <0.01
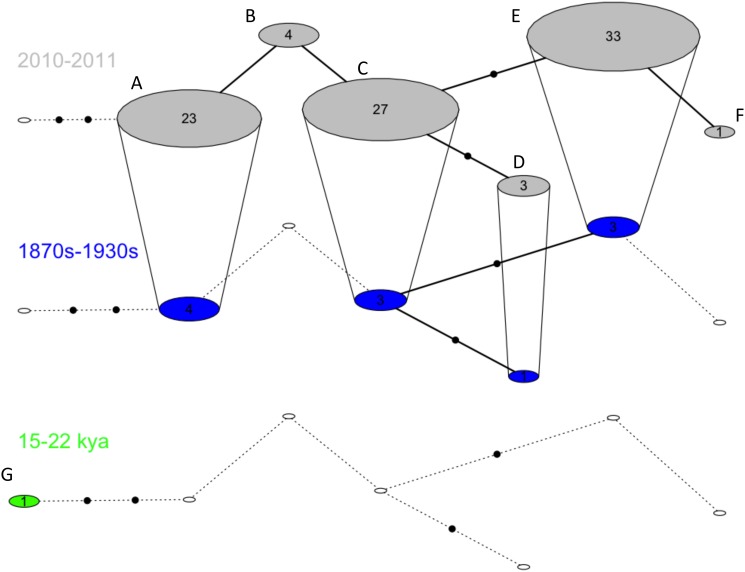
Consensus sequences for the mtDNA CR for historical and fossil samples varied between 245 and 616 bp (Table 4). Three new substitution sites and two new haplotypes were found among fossil samples, while four of the haplotypes and five substitution sites found in contemporary samples were found among historical samples (Table 4). When considering only samples that amplified for 450 bp of the mtDNA CR (11 historical skins and 1 fossil bone), the temporal network was indicative of a loss of haplotypic diversity between the Late Glacial and contemporary period (late 1800s and 2000s) but further sampling of archeological, pre-human Holocene (0–11.6 kya), Late Glacial (11.6–14 kya) and Otiran Glacial (14–70 kya) material is required to confirm this trend. However, there is no recent indication of a loss of genetic diversity for mtDNA CR between the late 1800s to the present (Table 4, 5, Fig. 2).
Table 4. List of haplotypes for control region in kea with changes relative to haplotype A.
Dots represent no change, dash unknown base due to partial sequence and boldened letters represent new haplotypes and mutations.
| Haplotype | n | Sample type | Total length of sequence obtained | Nucleotide site | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 668 | 676 | 677 | 729 | 745 | 754 | 775 | 808 | 969 | ||||
| A | 23 | contemporary | 1026 | A | G | C | A | G | C | G | A | A | A |
| B | 4 | contemporary | 1026 | . | . | . | . | . | . | . | . | G | . |
| C | 27 | contemporary | 1026 | . | . | T | . | . | . | . | . | G | . |
| D | 3 | contemporary | 1026 | G | . | T | G | . | . | . | G | G | . |
| E | 33 | contemporary | 1026 | . | A | T | G | . | . | . | . | G | . |
| F | 1 | contemporary | 1026 | . | A | T | G | . | . | A | . | G | . |
| G | 1 | fossila | 450 | - | . | . | G | A | T | . | . | . | - |
| H | 3 | fossilb | 245 | - | - | - | - | - | - | - | G | . | G |
| A | 4 | historicalc | 616 | - | . | . | . | . | . | . | . | . | - |
| C | 3 | historicald | 616 | - | . | T | . | . | . | . | . | G | - |
| D | 1 | historicale | 616 | - | . | T | G | . | . | . | G | G | - |
| E | 3 | historicalf | 616 | - | A | T | G | . | . | . | . | G | - |
Sample reference numbers:
aNMNZ S.43574;
bNMNZ S.22664.1;
cCM AV84, CM AV89, CM AV91, AlM LB2466;
dAlM LB2469, NMW 12.209, NMW 12.211;
eCM AV89;
fCM AV2039, NHM 1927.12.18.125, NMW 49.760
Table 5. Estimates of genetic diversity (Domain 1 of the mitochondrial control region, 450 bp) within all, historical and fossil, and contemporary kea samples with the sample size for each dataset (n), number of observed haplotypes (h), number of polymorphic sites (S), haplotypic diversity (Hd), nucleotide diversity (π).
| Dataset | n | h | S | Hd | SD | π | SD |
|---|---|---|---|---|---|---|---|
| Contemporary, Historical and Fossil* | 103 | 7 | 8 | 0.728 | 0.018 | 0.00426 | 0.00271 |
| Historical and Fossil* | 12 | 5 | 7 | 0.813 | 0.070 | 0.00556 | 0.00362 |
| Contemporary | 91 | 6 | 6 | 0.721 | 0.020 | 0.00412 | 0.00264 |
*only samples for which 450 bp were amplified were considered here
Fig 2. Temporal haplotype network displaying the relationships of kea haplotypes for 450 bp of the mitochondrial control region through time.
Haplotypes are represented by circles and numbers represent sample size. Empty circles represent absent haplotypes for a given time period. Haplotypes found in multiple time period are connected by vertical lines. Within each time period, black dots represent one mutation.
Historical population structure and assignment of historical samples
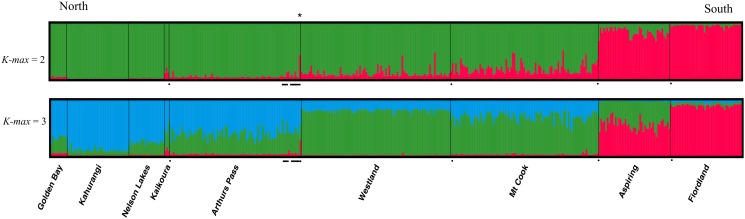
When comparing historical and contemporary population structure using the Bayesian algorithm implemented in STRUCTURE, 14 individuals out of 15 clustered well with contemporary individuals from the same sampling locations. Of the 15 historical samples, 13 had high membership coefficients (q > 0.85) to their putative cluster of origin when considering two distinct genetic clusters (S4 Table). One sample originating from the Mt Cook population (1927.12.18.125) did not assign as strongly to its putative cluster of origin, with a q-value of 0.66 for the north cluster. However this is perhaps not surprising, as contemporary samples from the same location also show admixed ancestry (Fig. 3). Another historical sample originating from the central South Island (NMW 12.211, Lake Coleridge) showed shared membership to one of the two main clusters, with a q-value of 0.56 for the northern cluster and 0.44 for the south cluster (Fig. 3, S4 Table). When considering three genetic clusters, the Mt Cook sample (1927.12.18.125) showed admixed membership to the north and central cluster, but with the highest membership coefficient for its putative central cluster of origin (q = 0.69). Individual NMW 12.211 (Lake Coleridge, north cluster) showed again shared membership with q-values of 0.44, 0.28 and 0.28 for the north, central and south cluster respectively. Samples NMW 12.208 and NMW 49.760, putatively originating from the north cluster, also had admixed origins with q-values of 0.64 and 0.57 to the central cluster. However, those two samples had very high q-values of 0.94 and 0.88 to their cluster of origin when considering a scenario of two clusters. It is worth noting that because the distinction between the north and central cluster is not very strong and as gene flow is marked between these two clusters, shared membership between those two clusters is perhaps not surprising.
Fig 3. Individual clustering assignment for 410 contemporary and 15 historical Kea averaged for the 25 runs with the lowest ln(K) implemented in STRUCTURE for each of a different number of clusters (K-max ∈ [2, 3]) in an admixture model.
Historical samples are shown with a black dot and were grouped with samples of the same geographical location for the analysis. Sample NMW 12.211 referred in the text is shown with an asterisk.
Demographic history
Using the heterozygosity excess approach to bottleneck detection, TPM models with 90% and 95% of stepwise mutations only showed significant excess in heterozygotes for the central cluster (Table 6). The more statistically conservative SMM model [93] did not support a bottleneck in any of the clusters tested. Finally, there was no evidence in any population tested for a significant deviation from the normal L-shaped distribution of allele frequencies that could be expected in a population that is not in mutation-drift equilibrium.
Table 6. Bottleneck results implemented with three different tests for kea including P-values for a signed rank Wilcoxon test for heterozygous excess (one tail) with different models of mutations, the shifted mode test and the M-ratio method.
For the heterozygous excess, TPM and SMM correspond to two-phase mutation model and single-step mutation model respectively. For the M-ratio method, ps stands for the proportion of sing-step mutations, MC for the critical M for a given theta where θ = 1 represents the initial (pre-decline) Ne of 500 and θ = 10 an Ne of 5000. M-ratio values that fall below the MC value are considered statistically significant at α = 0.05.
| Test | Parameter | North n = 142 | Central n = 181 | South n = 87 | Historical Canterbury n = 15 | |
|---|---|---|---|---|---|---|
| He excess | TPM (90% SMM) | 0.09 | 0.02 | 0.08 | 0.60 | |
| TPM (95% SMM) | 0.11 | 0.05 | 0.17 | 0.62 | ||
| SMM | 0.18 | 0.14 | 0.36 | 0.74 | ||
| Shifted Mode | No | No | No | No | ||
| M-Ratio | ps = 0.9 | Δg = 3.5 | ||||
| θ = 1; Ne = 500 | Mc | 0.81 | 0.81 | 0.81 | 0.79 | |
| θ = 10; Ne = 5000 | Mc | 0.75 | 0.76 | 0.74 | 0.65 | |
| M-Ratio | 0.76 | 0.79 | 0.86 | 0.83 | ||
| ps = 0.78 | Δg = 3.5 | |||||
| θ = 1; Ne = 500 | Mc | 0.71 | 0.71 | 0.71 | 0.69 | |
| θ = 10; Ne = 5000 | Mc | 0.70 | 0.71 | 0.68 | 0.56 | |
| M-Ratio | 0.76 | 0.79 | 0.86 | 0.83 | ||
When using the M-ratio approach, we found mixed results for signatures of a more ancient bottleneck (i.e. <100 gen.). Depending on the parameter set used, M-ratio values ranged from 0.76 to 0.86 (Table 6). These values are above or slightly below the critical value (MC) that represents the threshold under which a bottleneck is supported. We only found evidence of a bottleneck when using a θ value of 1, corresponding to a pre-decline Ne of 500 and only for the north and central clusters.
Historical Canterbury and all three contemporary genetic clusters examined showed larger historical (NH) than contemporary (NC) effective population sizes for both demographic models (exponential and linear). Bayes factor values were >10 for all clusters and models indicating strong evidence for a population decline (Table 7). The ratio of the posterior distributions of contemporary and historical population sizes (r = NC/NH) clearly supported a population decline. Using an exponential model of population size change, the 90% highest posterior density (HPD) of the ratio r was of 0.0019–0.052, 0.053–0.068, 0.019–0.031 and of 0.0003–0.017 for Historical Canterbury, the north, central and south cluster samples respectively. These r values indicated that the contemporary Ne is less than 10% of the historical NH.
Table 7. The Bayes Factor (BF) of each model, mode and 90% highest posterior density (in parentheses) for NC, NH and T for the demographic models implemented in MSVAR.
| Cluster | BF | model | NH | NC | Time (T) |
|---|---|---|---|---|---|
| North | 979 | Exp | 4,493 (1,112–19,085) | 279 (76–1,016) | 6,261 (1,208–34,300) |
| 5624 | Linear | 4,676 (1,198–17,124) | 118 (24–521) | 11,766 (2,126–67,321) | |
| Central | inf | Exp | 3,823 (1,111–14,791) | 106 (22–470) | 2,172 (354–12,843) |
| inf | Linear | 3,955 (1,122–14,701) | 65 (4.4–461) | 6,989 (1,122–41,399) | |
| South | inf | Exp | 4,620 (1,359–16,253) | 28 (0.43–290) | 271 (7–3,345) |
| inf | Linear | 4,677 (1,396–15,015) | 11 (0.07–155) | 2,417 (484–12,280) | |
| Historical Canterbury | 276 | Exp | 3,995 (889–20,427) | 131 (1.8–1,067) | 1,302 (20.2–39,058) |
| 624 | Linear | 4,266 (926–20,914) | 103 (1–1,090) | 6,708 (497–104,451) |
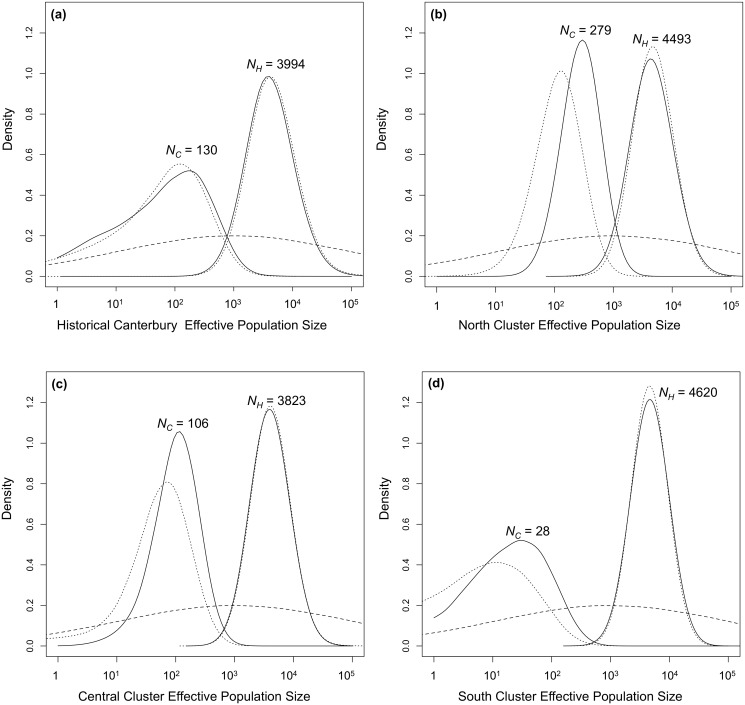
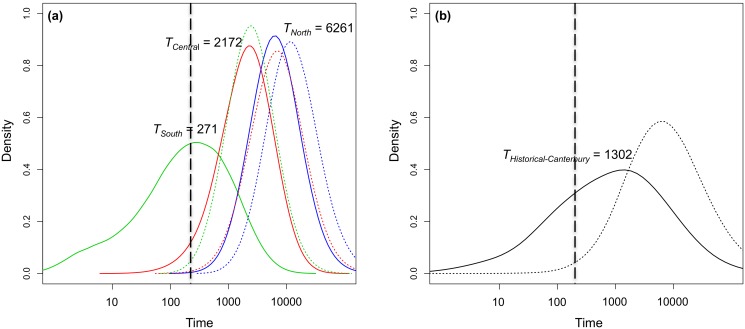
The modes of the 90% HPD of the posterior distributions for historical effective population size (NH) for the exponential model were Nhistorical-Canterbury = 3,995, Nnorth = 4,493, Ncentral = 3,823, and Nsouth = 4,620 compared to modal values for contemporary effective population sizes (NC) of 131, 279, 106, and 28 respectively (Table 7, Fig. 4). Estimates of the time of population decline varied between populations. There was strong support for a population decline occurring well before human arrival in New Zealand in 1280 AD [84] for the Historical Canterbury, north and central clusters with Thistorical-Canterbury = 1,302 years before present [YBP], T north = 6,261 and T central = 2,172 YBP, while a more recent decline was supported for the south cluster with T south = 271 YBP assuming a generation time of seven years (J. Kemp pers. comm.) (Table 7, Fig. 5).
Fig 4. Prior and posterior distributions for the contemporary (NC) and historical (NH) effective population size (a) Historical Canterbury (b) north, (c) central (d) south clusters using both the exponential (full lines) and linear (dotted lines) models.
The dashed line shows the prior distribution for NC and NH.
Fig 5. Posterior distribution of time of decline (T) (in years before present, assuming a generation time of seven years) (a) north, central and south clusters (b) Historical Canterbury using both the exponential (full lines) and linear (dotted lines) models.
The vertical dotted line represents the approximate start of the governmental bounty scheme of the late 1800s.
Estimates for NH and NC for the linear model were similar to the exponential model (Table 7) while estimates for the timing of the decline were longer for the linear model for all genetic clusters (Table 7, Fig. 4). However, we focus here on the results of the exponential model because it is likely more realistic when modeling population dynamics [81].
Evaluation of two historical scenarios indicated that a model of population bottleneck best described the genetic data. This model received a posterior probability of 0.97, 0.99, 0.94 and 0.82 for the north, central, south clusters and Historical Canterbury respectively. Also, Type I and Type II error rates for the selection of the stable population size model ranged between 0.01 to 0.015 and between 0.02 to 0.03 respectively. Posterior estimation of parameters using values drawn from the 10,000 datasets closest to the observed ones for each cluster indicated that the effective population size has declined from 56,300, 10,800, 4,020 and 13,900 to 248, 380, 370 and 579 for the north, central, south clusters and Historical Canterbury respectively (Table 8). Timing of decline was estimated at 1,827, 6,461, 847 and 496.3 YBP assuming a generation time of seven years for the north, central, south clusters and Historical Canterbury respectively.
Table 8. The mode and 95% confidence interval for NC, NH and T (in number of generations) for the demographic models implemented in DIYABC for the scenario with the highest probability (population bottleneck).
| Cluster | Parameter | Prior range | Posterior mode | 5% | 95% |
|---|---|---|---|---|---|
| North | NH | 10 – 2 × 105 | 56,300 | 12,200 | 423,000 |
| NC | 10 – 104 | 248 | 150 | 5,390 | |
| Time (T) | 0 – 1,000 | 261 | 71 | 924 | |
| Bottleneck duration | 1 – 100 | 86 | 6 | 96 | |
| μ rate (microsatellite) | 10-6 – 10-3 | 1 × 10-5 | 1 × 10-5 | 1.53 × 10-5 | |
| μ rate (mtDNA) | 10-8 – 10-7 | 9.93 × 10-8 | 3.49 × 10-8 | 9.94 × 10-8 | |
| Central | NH | 10 – 2 × 105 | 10,800 | 8840 | 378,000 |
| NC | 10 – 104 | 380 | 172 | 5,720 | |
| Time (T) | 0 – 1,000 | 923 | 114 | 963 | |
| Bottleneck duration | 1 – 100 | 78 | 6.1 | 95.7 | |
| μ rate (microsatellite) | 10-6 – 10-3 | 1 × 10-5 | 1 × 10-5 | 1.37 × 10-5 | |
| μ rate (mtDNA) | 10-8 – 10-7 | 9.70 × 10-8 | 2.71 × 10-8 | 9.9 × 10-8 | |
| South | NH | 10 – 2 × 105 | 4,020 | 2,530 | 32,400 |
| NC | 10 – 104 | 370 | 188 | 5,800 | |
| Time (T) | 0 – 1,000 | 121 | 60.8 | 895 | |
| Bottleneck duration | 1 – 100 | 6.83 | 4.2 | 94 | |
| μ rate (microsatellite) | 10-6 – 10-3 | 1 × 10-5 | 1 × 10-5 | 1.79 × 10-5 | |
| μ rate (mtDNA) | 10-8 – 10-7 | 9.79 × 10-8 | 2.51 × 10-8 | 9.9 × 10-8 | |
| Historical Canterbury | NH | 10 – 2 × 105 | 13,900 | 7,860 | 361,000 |
| NC | 10 – 104 | 579 | 214 | 6,820 | |
| Time (T) | 0 – 1,000 | 70.9 | 29.4 | 850 | |
| Bottleneck duration | 1 – 100 | 31.2 | 5.4 | 95.3 | |
| μ rate (microsatellite) | 10-6 – 10-3 | 1 × 10-6 | 1 × 10-6 | 1.17 × 10-5 | |
| μ rate (mtDNA) | 10-8 – 10-7 | 9.93 × 10-8 | 3.13 × 10-8 | 9.93 × 10-8 |
Discussion
Genetic diversity in historical and contemporary samples
No significant loss of genetic diversity was observed in sampled kea over the last 100 to 150 years, as no previously undetected haplotypes or alleles were identified among the historical samples. The higher diversity (mean number of alleles and expected heterozygosity) in contemporary samples was probably due to the paucity of samples available to our study with robust locality data. However, aDNA data shows two haplotypes and three new substitution sites found among fossil samples suggesting that a loss of genetic diversity might have occurred prior to the cull of the late 1800s. A larger sample size representing a wider time period between the Otiran Glacial to European arrival in New Zealand would be required to estimate the timing of such a decline and the extent of genetic diversity loss with more accuracy.
It is possible that our historical samples were collected after the peak of the government cull had already occurred. This is particularly true when considering the nature of the bottleneck. If for instance, the cull was rapid, of great intensity and resulted in populations being extirpated locally, it is quite likely that our historical sampling was not able to capture the effect of the bottleneck. If on the other hand, the cull lasted over a longer time span (> 10 years), we would be more likely to detect a loss of genetic diversity with our samples, as the commonly accepted date for the start of the government cull is 1860 [94]. However, we have little knowledge of the regional intensity and exact onset of this cull and it is possible that it varied among regions due to the regional difference in farming intensity.
Even if we cannot exclude the possibility of populations being extirpated locally in areas of high conflict with farmers, it is likely that some other populations could have persisted in more remote areas without farming and remained unaffected by the cull, thus maintaining historical levels of genetic diversity. Although this has never been reported, because of the dispersive [95, 96] and cognitive abilities of kea [34, 35], a temporary or permanent range shift might have occurred possibly in response to local persecution.
Historical population structure and assignment of historical samples
Assignments of historical samples scored at 15 microsatellite loci did not suggest a different historical population structure, with most historical samples assigning well to their putative cluster of origin. This gives further support to the contemporary structure not resulting from genetic drift caused by the recent human-led population decline [45].
Bottleneck detection using moment-based methods
The demographic histories of endangered and fluctuating populations is often complex. While conservationists often focus on the effect of human pressures on natural populations, the identification of a single decline event can be difficult, as the genetic variation of natural populations is influenced by multiple events varying in timing and nature. In this respect, kea is a good example as it has presumably been influenced by Pleistocene glaciations, climate change at the onset of the Holocene (11.6 kya), human colonization of New Zealand and by recent European persecution as well. Also, the usual rarity of kea fossils throughout New Zealand contrasting with a high abundance of remains in some sites [28] possibly reflects the effects of non-anthropogenic forces on the distribution of the species [30] while the lack of identified kea bones in middens [97] suggests that kea might not have experienced a range reduction resulting from over-hunting by Polynesians as in other endemic avian species (However the archeological record needs to be reassessed in light of the findings of this study). Kea history is therefore more complicated than previously thought since our results suggest that multiple forces may have influenced kea demography.
Moment-based methods of bottleneck detection yielded contrasting results. The M-ratio method did not detect any bottleneck while a signature of a bottleneck was detected in the central cluster using the heterozygote excess method and the most appropriate TPM model with 90% stepwise mutations [74]. As theory suggests, sign test under the IAM may be prone to (wrongly) detecting heterozygosity excess in non-bottlenecked populations [93]. Decreasing the proportion of stepwise mutations and thus going towards an IAM, therefore increased the significance of bottleneck detection in the samples we considered (Table 6). This became particularly obvious when using less than 90% stepwise mutation models (results not shown). While such a proportion is still likely, it can greatly vary among species, loci and alleles [75, 98]. Moreover, a well-known and recurrent problem with classical bottleneck detection methods is that one needs to make assumptions about microsatellite evolution [99]. Often, parameter values for the species of interest are unknown and must be borrowed from other closely-related species [75]. Thus, precise knowledge about the mutation rate of our markers is essential to be sure of the occurrence of a bottleneck. The lack of resolution of our markers in bottleneck detection might explain in part these contrasting results. Even though we are above the limit of number of loci recommended by the authors of BOTTLENECK (i.e. 8–10 polymorphic loci: [93]), an average of five alleles per loci might limit the power of the analyses.
The detection of a bottleneck signal in the central cluster is consistent with the fact that the McKenzie area was an easily accessible area and one of the first regions where intensive farming occurred [100–102]. There is no clear evidence for a variation in the intensity of the cull among regions, but if such variation in intensity of culling existed, it could explain why a bottleneck has only been detected in this region. The fact that results from both moment-based methods used here are not congruent, casts some doubt on this conclusion. One explanation for the observed pattern might be that the signal obtained with the heterozygote excess method is artificial. Bottleneck tests assume random mating (no population structure) and population closure (no gene flow). Non-random mating can produce genealogies that resemble bottlenecks, whereas gene flow is generally predicted to resemble recent expansion by introducing rare alleles [13, 73, 103]. It is therefore plausible that overlooking kea population substructure, although a rather weak one, could overshadow the effect of the bottleneck through an effect of immigration and replacement of genetic diversity lost locally [104].
Detection of an ancient population decline
The modeling approaches implemented in MSVAR and DIYABC were the only methods that consistently identified population declines in the historical and contemporary samples examined. While mode estimates were slightly different, posterior distribution of parameters were overlapping among the two methods. However, the timing of this decline was not consistent with the reported cull of the late 1800s. Rather, our analysis identified a decline dating back a few hundreds of years ago (south cluster) to up to 6,000 years ago (north and central clusters) and suggests that kea could have experienced a population decline prior to human arrival in the northern part of its range at least. The finding of kea fossils from the Pleistocene in some areas of the South Island where kea are now absent such as central and eastern Otago [105], coastal North Otago [105, 106], Westland [30] and eastern Southland [107], further supports the possibility that kea went through at least one population contraction predating human arrival in New Zealand as suggested by [30]. The timing of this population decline is not unrealistic and although the present data does not allow the identification of the exact driver, a few events could have induced a population decline in kea.
Climate warming at the end of the last glacial period (Otiran Glacial) induced a change in environment in the South Island of New Zealand with the expansion of podocarp forest first [108, 109] and then its replacement by southern beech forest (∼5,000 YBP in the South) [108, 110, 111]. It is therefore possible that kea distribution could have changed with warming temperature and changing habitat. For instance, it has been shown that the extinct crested moa (Pachyornis australis) changed altitudinal, longitudinal and latitudinal ranges through the Late Quaternary in response to alterations in the distribution of suitable habitat [7]. The finding of a kea skeleton of Otiran age in the North Island [29] and apparent absence of North Island kea fossils from the Holocene first suggested that the species probably did not survive post-glacial warming and that the highly-forested North Island was not a suitable habitat for kea during the Holocene (however see below for new evidence). It is therefore possible that kea distribution shifted southwards towards the alpine areas of the South Island as kea were tracking their habitat. Also, the drier climate and more diverse vegetation in the eastern South Island [112, 113] may have provided a more favorable habitat for kea than the tall rain forests of Westland and the North Island. Such a difference in the timing of the decline in the South Island (i.e. ∼300 YBP in South and ∼6,000 YBP the North) can however not be fully be explained by the later southward establishment of forest in the South Island. Rather, fossils from the Holocene in lowland Canterbury [114] and newly discovered Holocene material from the North Island [32] suggest that the present distribution is relictual and reflects where kea survived human-caused extinctions and that they might have naturally occurred at lower altitude. This means that kea might not have been wiped out from the North Island because of climate warming during the Holocene but rather because of direct or indirect human activities such as hunting, predation by introduced mammals or habitat modification. This also suggests that the decline in effective population size in the South Island as shown by genetic data might not represent a sheer decline likely to drive the species to extinction but rather natural population fluctuations and range shifts in response to habitat change during the Holocene. During this period, kea populations could have remained stable with respect to the carrying capacity of their habitat, as has been shown for Australian mega- and meso-fauna [115, 116]. However, the rarity of Holocene specimens does not allow to detect a decline in response to Pleistocene-Holocene climate transition or to estimate its magnitude.
Another important aspect to consider is the possible impact of pre-European Maori on kea population dynamics. Estimates of the timing of decline predate Maori arrival in New Zealand (circa 1280 A.D., [84]) for the north and central clusters, but a much more recent decline for the south cluster (MSVAR, exponential model) seems concordant with Maori being linked with kea decline. While mentions of kea in Maori lore are rare [94, 117, 118], it is well known that Maori impacted on other New Zealand biotas through direct hunting [8, 119–121], habitat modification [122–124] and the introduction of rats [125, 126]. Since kea are scavengers [41, 127, 128], which has also been suggested by beak marks on fossil moa [114], it is possible that the extinction of moa and other birds potentially used as reliable food sources, could have somehow impacted kea population dynamics. The reason why such a recent decline is only observed on the south cluster and not in the other clusters examined remains unclear. The posterior distribution for the decline is rather wide, suggesting a possible methodological artifact and that a mode of ∼300 years should be taken with caution. However, if this estimation was not the result of a methodological artifact, it could be that kea had not been declining in the south prior human arrival and that they only recently declined.
Non-detection of a recent population decline
Our results do not exclude the possibility of a recent (1800s to 1970s) human-induced population decline in kea and are consistent with the conclusion of Girod et al. [129] and the observation by Salmona et al. [130] that recent declines are not likely to be detected by MSVAR. However, a study on the heavily exploited Antarctic fur seal (Arctocephalus gazella) during the 1800’s suggests that ABC approaches are powerful enough to detect recent demographic declines [16]. The question therefore remains why a recent human-induced population decline was not detected with our ABC and moment-based methods. These incongruences among the various methods of bottleneck detection and the general pattern of non-detection of genetic bottleneck in the face of known demographic collapse is however not uncommon [19, 99, 103, 131–136] and a few factors can influence the efficiency of methodologies to detect genetic bottlenecks.
First of all, the detection of a bottleneck is highly dependent on the amount of genetic diversity present before the decline. If kea populations declined before human arrival as supported by the results of MSVAR and DIYABC, it is very likely that the inherent paucity in genetic variation following this ancient decline (300–6,000 years ago) could have precluded further genetic loss [73], making the detection of a recent decline impossible.
Secondly, the nature of the bottleneck is an important factor to consider. Even though classical methods of genetic bottleneck tests have reasonable power [73, 75, 93, 137], only very extreme population declines in Ne (10- to 1000-fold) have been evaluated (e.g. Mexican Wolves, Canis lupus mexicanus [138]). If the extent of the population reduction of kea had been exaggerated and only moderate, it would not have been able to create a detectable signature in the form of heterozygote excess or M-ratio signal. With between 800 to 1,600 birds killed in a single season on some farming estates [139], it is possible that the bottleneck was short, rapid and intense, with most of the cull occurring in the first few years. Interestingly, large numbers of kea were still killed in the late 1940s [140], with nearly 7,000 birds killed over three years, suggesting kea might have in part recovered or still be maintained at high densities, probably thanks to new food sources such as wild ungulates or sheep [41, 128, 141]. Such a brief and extreme population decline and a possible quick population recovery may therefore not have left a signature of genetic bottleneck [15, 142, 143].
Thirdly, it is plausible that the cull was not homogeneous over the whole range of the species and that population density could have been significantly reduced mainly close to sheep stations, while populations in remote areas might have survived the cull [41]. Also, because of their cognitive behavior [34, 35] and high mobility [95, 96, 144], local populations could have easily shifted their range. It has been observed that the availability of new food sources might have attracted kea in numbers [41] and that groups of birds would move through the landscape, stopping to attack sheep and then disappear from the area for days or even weeks [95, 96]. Range shift could have also been induced by human persecution, as has been shown in other hunted species like wild boars [145]. It is thus quite likely that a combination of range-shift and immigration might have insured the maintenance of genetic diversity despite a localized population declines.
Finally, as has been suggested for long-lived species such as white-tailed eagle [146], Hawaiian petrel [19] or white-tailed black cockatoo [147], long generation time could lead to retention or buffering of genetic diversity because of a shorter effective time of exposure to bottleneck compared to a species with short generation time. Kea being able to reach up to 40 years of age in captivity [148] and with a generation time of 7 to 10 years (J. Kemp pers. comm.) could well fall in this category.
Conservation implications
Although genetic signatures of historical contraction were detected, no recent bottleneck consistent with the report of an important cull was noted. This suggests that the cull was not strong enough to leave a genetic signature or that a combination of the factors discussed above reduced the probability of bottleneck detection. The discovery of additional mitochondrial diversity among fossil samples supports an ancient and possibly non-anthropogenic decline in kea, but further study of fossil samples is required to determine with more certainty the importance of habitat change during the Holocene on kea population dynamics. In a context of global climate change, our findings suggest that kea abundance could be further reduced by increasing temperatures either through associated habitat change, or through the effects of increased predator pressure in alpine habitats. While we cannot rule out the possibility that kea was once present in the lowlands and that it is now an alpine relict species because of habitat loss, persecution and predation, our study highlights the need to examine histories of endangered species and try and understand the respective anthropogenic and non-anthropogenic effects on population dynamics when proposing conservation guidelines.
The lack of detection of a recent genetic bottleneck associated with a documented human-induced demographic decline further highlights the need to interpret results of tests for population bottleneck with great caution. As failure to detect a genetic bottleneck does not exclude the possibility of a recent population contraction, our results further highlight the need to consider historical and fossil genetic diversity and effective population size estimates to develop efficient conservation recommendations for species of conservation concern.
While the species has been fully protected since 1986 [44], the introduction of mammalian predators in New Zealand has been an important driver of population decline in kea [25, 141]. It is also likely that the low genetic diversity in declining kea populations and ensuing inbreeding might contribute to the risk of extinction of the species [149]. We therefore recommend that further studies be done on the current levels of inbreeding and on future predicted loss of genetic diversity in extant kea populations.
Supporting Information
(PDF)
The left hand side of the figure represents the 5‚ end of the sequence.
(PDF)
(PDF)
Columns 3–6 show the starting values for the mean and variance of the prior distributions. Columns 7–10 show the means and variances (and their means and variances) of the hyperprior distributions. Parameters listed are generation interval (g), contemporary Ne (NC), historical Ne (NH), mutation rate scaled in terms of contemporary population size (θ), and time (T). scale.
(PDF)
(PDF)
Individuals with unusual assignments and referred to in the text are in bold.
(PDF)
(TXT)
(TXT)
Acknowledgments
We thank Josh Kemp, Tamsin Orr-Walker, Raoul Schwing, Jacinda Amey, Joseph Fraser, Lucy Rossiter, Francesca Cunningham, Brent Barrett, Phil Bradfield for their major contribution in kea sampling. We thank Trevor Worthy, Alan Tennyson and Paul Scofield for helpful comments on the Discussion.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by University of Otago funding to Bruce C. Robertson. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Conner JK, Hartl DL (2004) A Primer of Ecological Genetics. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 2. Hedrick PW, Kalinowski ST (2000) Inbreeding depression in conservation biology. Annual Review of Ecology and Systematics 31: 139–162. 10.1146/annurev.ecolsys.31.1.139 [DOI] [Google Scholar]
- 3. Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends in Ecology & Evolution 17: 230–241. 10.1016/S0169-5347(02)02489-8 [DOI] [Google Scholar]
- 4. Frankham R, Ballou JD, Briscoe DA (2010) Introduction to Conservation Genetics. Introduction to Conservation Genetics: 617. [Google Scholar]
- 5. Lacy RC (1997) Importance of genetic variation to the viability of mammalian populations. Journal of Mammalogy 78: 320–335. 10.2307/1382885 [DOI] [Google Scholar]
- 6. Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conservation Biology 17: 230–237. 10.1046/j.1523-1739.2003.01236.x [DOI] [Google Scholar]
- 7. Rawlence NJ, Metcalf JL, Wood JR, Worthy TH, Austin JJ, Cooper A (2012) The effect of climate and environmental change 36 on the megafaunal moa of New Zealand in the absence of humans. Quaternary Science Reviews 50: 141–153. 10.1016/j.quascirev.2012.07.004 [DOI] [Google Scholar]
- 8. Allentoft M, Heller R, Oskam CL, Lorenzen ED, Hale ML, Gilbert MT et al. (2014) Extinct New Zealand megafauna were not in decline before human colonization. Proceedings of the National Academy of Sciences of the United States of America 11: 4922–4927. 10.1073/pnas.1314972111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandom C, Faurby SR, Sandel B, Svenning JC (2014) Global late Quaternary megafauna extinctions linked to humans, not climate change Global late Quaternary megafauna extinctions linked to humans, not climate change. Proceedings of the Royal Society B Biological Sciences 281: 1–9. 10.1098/rspb.2013.3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campos PF, Willerslev E, Sher A, Orlando L, Axelsson E, Tikhonov A et al. (2010) Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. Proceedings of the National Academy of Sciences of the United States of America 107: 5675–80. 10.1073/pnas.0907189107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. MacDonald GM, Beilman DW, Kuzmin YV, Orlova La, Kremenetski KV, Shapiro B et al. (2012) Pattern of extinction of the woolly mammoth in Beringia. Nature communications 3: 893 10.1038/ncomms1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allentoft ME, Bunce M, Scofield RP, Hale ML, Holdaway RN (2010) Highly skewed sex ratios and biased fossil deposition of moa: ancient DNA provides new insight on New Zealand’s extinct megafauna. Quaternary Science Reviews 29: 753–762. 10.1016/j.quascirev.2009.11.022 [DOI] [Google Scholar]
- 13. Goossens B, Chikhi L, Ancrenaz M, Lackman-Ancrenaz I, Andau P, Bruford MW et al. (2006) Genetic signature of anthropogenic population collapse in orang-utans. Plos Biology 4: 285–291. 10.1371/journal.pbio.0040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alter SE, Rynes E, Palumbi SR (2007) DNA evidence for historic population size and past ecosystem impacts of gray whales. Proceedings of the National Academy of Sciences of the United States of America 104: 15162–15167. 10.1073/pnas.0706056104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okello JBA, Wittemyer G, Rasmussen HB, Arctander P, Nyakaana S, Douglas-Hamilton I et al. (2008) Effective population size dynamics reveal impacts of historic climatic events and recent anthropogenic pressure in African elephants. Molecular Ecology 17: 3788–3799. 10.1111/j.1365-294X.2008.03871.x [DOI] [PubMed] [Google Scholar]
- 16. Hoffman JI, Grant SM, Phillips CD (2011) Bayesian inference of a historical bottleneck in a heavily exploited marine mammal. Molecular Ecology 20: 3989–4008. 10.1111/j.1365-294X.2011.05248.x [DOI] [PubMed] [Google Scholar]
- 17. Schwartz MK, Aubry KB, Mckelvey KS, Pilgrim KL, Copeland JP, Squires JR et al. (2007) Inferring geographic isolation of wolverines in California using historical DNA. Journal of Wildlife Management 71: 2170–2179. 10.2193/2007-026 [DOI] [Google Scholar]
- 18. Wandeler P, Hoeck PEA, Keller LF (2007) Back to the future: museum specimens in population genetics. Trends in Ecology & Evolution 22: 634–642. 10.1016/j.tree.2007.08.017 [DOI] [PubMed] [Google Scholar]
- 19. Welch AJ, Wiley AE, James HF, Ostrom PH, Stafford TW, Fleischer RC et al. (2012) Ancient DNA Reveals Genetic Stability Despite Demographic Decline: 3,000 Years of Population History in the Endemic Hawaiian Petrel. Molecular Biology and Evolution 29: 3729–3740. 10.1093/molbev/mss185 [DOI] [PubMed] [Google Scholar]
- 20. Gibbs G (2006) Ghosts of Gondwana. Nelson, New Zealand: Craig Potton Publishing. [Google Scholar]
- 21. Tennyson AJD (2010) The origin and history of New Zealand’s terrestrial vertebrates. New Zealand Journal of Ecology: 6–27. [Google Scholar]
- 22. Holdaway R (1999) Introduced predators and avifaunal extinction in New Zealand. Kluwer Academic & Plenum Publishers, 189–238 pp. [Google Scholar]
- 23. Holdaway RN, Worthy TH, Tennyson AJD (2001) A working list of breeding bird species of the New Zealand region at first human contact. New Zealand Journal of Zoology 28: 119–187. 10.1080/03014223.2001.9518262 [DOI] [Google Scholar]
- 24. Duncan RP, Blackburn TM (2004) Extinction and endemism in the New Zealand avifauna. Global Ecology and Biogeography 13: 509–517. 10.1111/j.1466-822X.2004.00132.x [DOI] [Google Scholar]
- 25. Elliott G, Kemp J (2004) Effect of hunting and predation on kea, and a method of monitoring kea populations. Results of kea research on the St. Arnaud range. DOC Science Internal Series 181: 1–17. [Google Scholar]
- 26. Innes J, Kelly D, Overton JM, Gillies C (2010) Predation and other factors currently limiting New Zealand forest birds. New Zealand Journal of Ecology 34: 86–114. [Google Scholar]
- 27. Holdaway RN, Jacomb C (2000) Rapid Extinction of the Moas (Aves: Dinornithiformes): Model, Test, and Implications. Science 287: 2250–2254. 10.1126/science.287.5461.2250 [DOI] [PubMed] [Google Scholar]
- 28. Worthy TH, Mildenhall DC (1989) A late Otiran-Holocene paleoenvironment reconstruction based on cave excavations in northwest Nelson, New Zealand. New Zealand Journal of Geology and Geophysics 32: 243–253. 10.1080/00288306.1989.10427586 [DOI] [Google Scholar]
- 29. Holdaway RN, Worthy TH (1993) First North Island fossil record of Kea, and 37 morphological and morphometric comparison of Kea and Kaka. Notornis 40: 95–108. [Google Scholar]
- 30. Worthy TH, Holdaway RN (1993) Quaternary fossil faunas from caves in the Punakaiki area, West Coast, South Island, New Zealand. Journal of the Royal Society of New Zealand 23: 147–254. 10.1080/03036758.1993.10721222 [DOI] [Google Scholar]
- 31. Worthy TH, Holdaway RN (1994) Quaternary Fossil Faunas from Caves in Takaka Valley and on Takaka Hill, Northwest Nelson, South-Island, New-Zealand. Journal of the Royal Society of New Zealand 24: 297–391. 10.1080/03014223.1994.9517474 [DOI] [Google Scholar]
- 32. Tennyson AJD, Easton LJ, Wood JR (2014) Kea (Nestor notabilis)—another North Island human-caused extinction. Notornis 61: 174–176. [Google Scholar]
- 33. Myers JG (1924) The realtion of birds to agriculture in New Zealand, 10: The kea or mountain parrot. New Zealand Journal of Agriculture 29: 25. [Google Scholar]
- 34. Gajdon GK, Fijn N, Huber L (2004) Testing social learning in a wild mountain parrot, the kea (Nestor notabilis). Learning & Behavior 32: 62–71. 10.3758/BF03196007 [DOI] [PubMed] [Google Scholar]
- 35. Huber L, Gajdon GK (2006) Technical intelligence in animals: the kea model. Animal Cognition 9: 295–305. 10.1007/s10071-006-0033-8 [DOI] [PubMed] [Google Scholar]
- 36. Clarke CMH (1970) Observations on population, movements and food of the kea (Nestor notabilis). Notornis 17: 105–114. [Google Scholar]
- 37. Bond AB, Diamond J (1992) Population estimates of Kea in Arthur’s Pass National Park. Notornis 39: 151–160. [Google Scholar]
- 38. Diamond J, Bond AB (1999) Kea, bird of paradox: the evolution and behavior of a New Zealand parrot. Kea, bird of paradox: the evolution and behavior of a New Zealand parrot: 230. [Google Scholar]
- 39. White T (1895) The Kea (Nestor notabilis), a sheep-eating Parrot. Tr N Zealand Inst xxvii: pp. 273–280. [Google Scholar]
- 40. Benham WB (1907) Notes on the flesh-eating propensity of the Kea (Nestor notabilis). Wellington, Trans N Zeal Inst 39: (71–89). [Google Scholar]
- 41. Marriner GR (1908) The Kea: a New Zealand Problem, including a full description of this very interesting bird, its habitat and ways, together with a discussion of the theories advanced to explain its sheep-killing propensities. 152 pp. [Google Scholar]
- 42. Falla RA, Sibson RB, Turbott EG (1979) The new guide to the birds of New Zealand. The new guide to the birds of New Zealand: 1–247. [Google Scholar]
- 43.O’Donnell CF, Dilks PJ (1986) Forest birds in South Westland: status and habitat use. Technical report, NZ Wildlife Service, Department of Internal Affairs, Wellington.
- 44. Anderson R (1986) Keas for keeps. Forest and Bird 17: 2–5. [Google Scholar]
- 45. Dussex N, Wegmann D, Robertson BC (2014) Postglacial expansion and not human influence best explains the population structure in the endangered kea (Nestor notabilis). Molecular Ecology 23: 2193–2209. 10.1111/mec.12729 [DOI] [PubMed] [Google Scholar]
- 46. Wallis GP, Trewick SA (2009) New Zealand phylogeography: evolution on a small continent. Molecular Ecology 18: 3548–3580. 10.1111/j.1365-294X.2009.04294.x [DOI] [PubMed] [Google Scholar]
- 47. Worthy TH, Zhao JX (2006) A late Pleistocene predator-accumulated avifauna from Kids Cave, West Coast, South Island, New Zealand. Alcheringa: An Australasian Journal of Palaeontology 30: 389–408. 10.1080/03115510609506874 [DOI] [Google Scholar]
- 48. Knapp M, Clarke AC, Horsburgh KA, Matisoo-smith EA (2012) Setting the stage—Building and working in an ancient DNA laboratory. Annals of Anatomy 194: 3–6. 10.1016/j.aanat.2011.03.008 [DOI] [PubMed] [Google Scholar]
- 49. Cooper A, Poinar HN (2000) Ancient DNA: Do it right or not at all. Science 289: 1139 10.1126/science.289.5482.1139b [DOI] [PubMed] [Google Scholar]
- 50. Rohland N, Siedel H, Hofreiter M (2010) A rapid column-based ancient DNA extraction method for increased sample throughput. Molecular Ecology Resources 10: 677–683. 10.1111/j.1755-0998.2009.02824.x [DOI] [PubMed] [Google Scholar]
- 51. Dussex N, Robertson BC (2013) Isolation and characterization of 8 mi-crosatellites for the Kea Nestor notabilis . Conservation Genetics Resources 5: 1–3. 10.1007/s12686-013-9975-8 [DOI] [Google Scholar]
- 52. Allentoft ME, Oskam C, Houston J, Hale ML, Gilbert MTP, Rasmussen M et al. (2011) Profiling the dead: generating microsatellite data from fossil bones of extinct megafauna–protocols, problems, and prospects. PloS one 6: e16670 10.1371/journal.pone.0016670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J et al. (2007) Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Research 35: 5717–5728. 10.1093/nar/gkm588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al. (2012) 40 Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538. 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- 56. Rousset F (2008) GENEPOP’ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources 8: 103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 57. Guo SW, Thompson EA (1992) Performing the Exact Test of Hardy-Weinberg Proportion for Multiple Alleles. Biometrics 48: 361–372. 10.2307/2532296 [DOI] [PubMed] [Google Scholar]
- 58. Rice WE (1989) Analyzing tables of statistical tests. Evolution 1: 223–225. 10.2307/2409177 [DOI] [PubMed] [Google Scholar]
- 59. ElMousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L) Skeels) endemic to Morocco. Theoretical and Applied Genetics 92: 832–839. 10.1007/BF00221895 [DOI] [PubMed] [Google Scholar]
- 60. Nei M (2000) Molecular evolution and phylogenetics. Oxford University Press, New York. [Google Scholar]
- 61. R Development Team (2005) No Title. R: A Language and Environment for Statistical Computing: R Foundation for Statistical Computing, Vienna, Au. [Google Scholar]
- 62.Nei M (1987) Molecular evolutionary genetics.
- 63. Excoffier L, Estoup A, Cornuet JM (2005) Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics 169: 1727–1738. 10.1534/genetics.104.036236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prost S, Anderson CNK (2011) TempNet: a method to display statistical parsimony networks for heterochronous DNA sequence data. Methods in Ecology and Evolution 2: 663–667. 10.1111/j.2041-210X.2011.00129.x [DOI] [Google Scholar]
- 65. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Earl DA, Vonholdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- 68. Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- 69. Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4: 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- 70. Broquet T, Angelone S, Jaquiery J, Joly P, Lena JP, Lengagne T et al. (2010) Genetic Bottlenecks Driven by Population Disconnection. Conservation Biology 24: 1596–1605. 10.1111/j.1523-1739.2010.01556.x [DOI] [PubMed] [Google Scholar]
- 71. Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA (2010) The Confounding Effects of Population Structure, Genetic Diversity and the Sampling Scheme on the Detection and Quantification of Population Size Changes. Genetics 186: 983–995. 10.1534/genetics.110.118661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity 90: 502–503. 10.1093/jhered/90.4.502 [DOI] [Google Scholar]
- 73. Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dirienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB et al. (1994) Mutational Processes of Simple-Sequence Repeat Loci in Human-Populations. Proceedings of the National Academy of Sciences of the United States of America 91: 3166–3170. 10.1073/pnas.91.8.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318. 10.1046/j.1365-294x.2001.01190.x [DOI] [PubMed] [Google Scholar]
- 76. Storz JF, Beaumont MA (2002) Testing for genetic evidence of population expansion and contraction: An empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56: 154–166. 10.1554/0014-3820(2002)056[0154:TFGEOP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 77. Loader C (2007) Locfit: Local regression, likelihood and density estimation. R Package Version: 1.5–4. [Google Scholar]
- 78. Jeffreys H (1961) Theory of probability. Oxford, UK: Oxford University Press; 447 p. [Google Scholar]
- 79. Kass RE, Raftery AE (1995) Bayes Factors. Journal of the American Statistical Association 90: 773–795. 10.1080/01621459.1995.10476572 [DOI] [Google Scholar]
- 80. Storz JF, Beaumont MA, Alberts SC (2002) Genetic evidence for long-term population decline in a savannah-dwelling primate: Inferences from a hierarchical Bayesian model. Molecular Biology and Evolution 19: 1981–1990. 10.1093/oxfordjournals.molbev.a004022 [DOI] [PubMed] [Google Scholar]
- 81. Beaumont MA (1999) Detecting population expansion and decline using microsatellites. Genetics 153: 2013–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beaumont MA, Zhang WY, Balding DJ (2002) Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cornuet JM, Ravigne V, Estoup A (2010) Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinformatics 11 10.1186/1471-2105-11-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH (2008) Dating the late prehistoric dispersal of polynesians to New Zealand using the commensal Pacific rat. Proceedings of the National Academy of Sciences of the United States of America 105: 7676–7680. 10.1073/pnas.0801507105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Estoup A, Jarne P, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology 11: 1591–1604. 10.1046/j.1365-294X.2002.01576.x [DOI] [PubMed] [Google Scholar]
- 86. Weber JL, Wong C (1993) Mutation of Human Short Tandem Repeats. Human Molecular Genetics 2: 1123–1128. 10.1093/hmg/2.8.1123 [DOI] [PubMed] [Google Scholar]
- 87. Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Molecular Ecology 11: 2453–2465. 10.1046/j.1365-294X.2002.01643.x [DOI] [PubMed] [Google Scholar]
- 88. Anmarkrud JA, Kleven O, Bachmann L, Lifjeld JT (2008) Microsatellite evolution: Mutations, sequence variation, and homoplasy in the hypervariable avian microsatellite locus HrU10. BMC Evolutionary Biology Evolutionary Biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fagundes NJR, Ray N, Beaumont M, Neuenschwander S, Salzano FM, Bonatto SL et al. (2007) Statistical evaluation of alternative models of human evolution. Proceedings of the National Academy of Sciences of the United States of America 104: 17614–17619. 10.1073/pnas.0708280104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cornuet JM, Santos F, Beaumont MA, Robert CP, Marin JM, Balding DJ et al. (2008) Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24: 2713–2719. 10.1093/bioinformatics/btn514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chan CH, Ballantyne KN, Lambert DM, Chambers GK (2005) Characterization of variable microsatellite loci in Forbes’ parakeet (Cyanoramphus forbesi) and their use in other parrots. Conservation Genetics 6: 651–654. 10.1007/s10592-005-9021-9 [DOI] [Google Scholar]
- 92. Robertson BC, Frauenfelder N, Eason DK, Elliott G, Moorhouse R (2009) Thirty polymorphic microsatellite loci from the critically endangered kakapo (Strigops habroptilus). Molecular Ecology Resources 9: 664–666. 10.1111/j.1755-0998.2008.02506.x [DOI] [PubMed] [Google Scholar]
- 93. Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology 12: 228–237. 10.1046/j.1523-1739.1998.96388.x [DOI] [Google Scholar]
- 94. Temple P (1996) Book of the kea. Auckland: Hodder Moa Beckett. [Google Scholar]
- 95. Menzies MLC (1878) Memorandum of the kea. Transactions of the New Zealand Institute 11: 376–377. [Google Scholar]
- 96. Kirk T (1895) The Displacement of species in New Zealand. Transactions of the New Zealand Institute 28: 1–27. [Google Scholar]
- 97. Smith I, James-Lee T (2010) Data for an Archaeozoological Analysis of Marine Resource Use in Two New Zealand Study Areas. Otago Archaeological Laboratory Report No. 7. Technical Report 6, Anthropology Department, University of Otago, Dunedin. [Google Scholar]
- 98. Ellegren H (2004) Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics 5: 435–445. 10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- 99. Peery MZ, Kirby R, Reid BN, Stoelting R, Doucet-Bëer E, Vásquez-Carrillo C et al. (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Molecular Ecology 21: 3403–18. 10.1111/j.1365-294X.2012.05635.x [DOI] [PubMed] [Google Scholar]
- 100. O’Connor KF, Kerr IGC (1978) The history and present pattern of pastoral range production in New Zealand In: Proceedings fo the 1st International Congress. Denver, pp. 104–107. [Google Scholar]
- 101. O’Connor KF (1982) The implications of past exploitation and current de-46 velopments to the conservation of South Island tussock grasslands. New Zealand Journal of Ecology 5: 97–107. [Google Scholar]
- 102. Phillips J (2004) Te Ara: the online encyclopedia of New Zealand. Cultural Geographies 11: 342–346. 10.1191/1474474004eu310xx [DOI] [Google Scholar]
- 103. Busch JD, Waser PM, DeWoody JA (2007) Recent demographic bottlenecks are not accompanied by a genetic signature in banner-tailed kangaroo rats (Dipodomys spectabilis). Molecular Ecology 16: 2450–2462. 10.1111/j.1365-294X.2007.03283.x [DOI] [PubMed] [Google Scholar]
- 104. Keller LF, Jeffery KJ, Arcese P, Beaumont MA, Hochachka WM, Smith JNM et al. (2001) Immigration and the ephemerality of a natural population bottleneck: evidence from molecular markers. Proceedings of the Royal Society of London Series B-Biological Sciences 268: 1387–1394. 10.1098/rspb.2001.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Worthy TH (1998) Quaternary fossil faunas of Otago, South Island, New Zealand. Journal of the Royal Society of New Zealand 28: 421–521. 10.1080/03014223.1998.9517575 [DOI] [Google Scholar]
- 106. Worthy TH, Grant-Mackie JA (2003) Pleistocene avifaunas from Cape Wan-brow, Otago, South Island, New Zealand. Journal of the Royal Society of New Zealand 33: 427–485. 10.1080/03014223.2003.9517738 [DOI] [Google Scholar]
- 107. Worthy TH (1998) The Quaternary fossil avifauna of Southland, South Island, New Zealand. Journal of the Royal Society of New Zealand 28: 537–589. 10.1080/03014223.1998.9517575 [DOI] [Google Scholar]
- 108. Alloway BV, Lowe DJ, Barrell DJA, Newnham RM, Almond PC, Augustinus PC et al. (2007) Towards a climate event stratigraphy for New Zealand over the past 47 30 000 years (NZ-INTIMATE project). Journal of Quaternary Science 22: 9–35. 10.1002/jqs.1079 [DOI] [Google Scholar]
- 109. McGlone M (1995) Lateglacial landscape and vegetation change and the younger dryas climatic oscillation in New Zealand. Quaternary Science Reviews 14: 867–881. 10.1016/0277-3791(95)00068-2 [DOI] [Google Scholar]
- 110. Wardle P (1963) Evolution and distribution of the New Zealand flora, as affected by Quaternary climates. New Zealand Journal of Botany: 3–17. 10.1080/0028825X.1963.10429318 [DOI] [Google Scholar]
- 111. McGlone MS, Newnham RM, Moar NT (2010) The vegetation cover of New Zealand during the Last Glacial Maximum: Do pollen records under-represent woody vegetation? Terra Australis 32: 49–68. 112. [Google Scholar]
- 112. Burrows C (1989) Moa browsing: evidence from the Pyramid Valley mire. New Zealand journal of ecology 12: 51–56. [Google Scholar]
- 113. Rawlence N, Scofield R, Wood J, Wilmshurst J, Moar N, Worthy T et al. (2011) New palaeontological data from the excavation of the Late Glacial Glencrieff miring bone deposit, North Canterbury, South Island, New Zealand. Journal of the Royal Society of New Zealand 41: 217–236. 10.1080/03036758.2011.559663 [DOI] [Google Scholar]
- 114. Holdaway RN, Worthy TH (1997) A reappraisal of the late Quaternary fossil vertebrates of Pyramid Valley Swamp, North Canterbury, New Zealand. New Zealand Journal of Zoology 24: 69–121. 10.1080/03014223.1997.9518107 [DOI] [Google Scholar]
- 115. Prideaux GJ, Roberts RG, Megirian D, Westaway KE, Hellstrom JC, Olley JM et al. (2007) Mammalian responses to Pleistocene climate change in southeastern Australia. Geology 35: 33 10.1130/G23070A.1 [DOI] [Google Scholar]
- 116. Macken AC, Prideaux GJ, Reed EH (2012) Variation and pattern in the responses of mammal faunas to Late Pleistocene climatic change in south-eastern South Australia. Journal of Quaternary Science 27: 415–424. 10.1002/jqs.1563 [DOI] [Google Scholar]
- 117. Best E (1977) Forest Lore of the Maori. Wellington: Keating, E. C. [Google Scholar]
- 118. Beattie J (1994) Traditional lifeways of the southern Maori. Dunedin, N.Z.: University of Otago Press. [Google Scholar]
- 119. Holdaway R (1989) New Zealand’s pre-human avifauna and its vulnerability. New Zealand Journal of Ecology 12: 11–25. [Google Scholar]
- 120. Boessenkool S, Austin JJ, Worthy TH, Scofield P, Cooper A, Seddon PJ et al. (2009) Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proceedings of the Royal Society B-Biological Sciences 276: 815–821. 10.1098/rspb.2008.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Collins CJ, Rawlence NJ, Prost S, Anderson CNK, Knapp M, Scofield P et al. (2014) Extinction and recolonization of coastal megafauna following human arrival in New Zealand Extinction and recolonization of coastal megafauna following human arrival in New Zealand. Proceedings of the Royal Society B Biological Sciences 281 10.1098/rspb.2014.0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. McWethy DB, Whitlock C, Wilmshurst JM, McGlone MS, Li X (2009) Rapid deforestation of South Islands, New Zealands, by early Polynesian fires. Holocene 19: 883–897. 10.1177/0959683609336563 [DOI] [Google Scholar]
- 123. McWethy DB, Whitlock C, Wilmshurst JM, McGlone MS, Fromont M, Li X et al. (2010) Rapid landscape transformation in 52 South Island, New Zealand, following initial Polynesian settlement. Proceedings of the National Academy of Sciences of the United States of America 107: 21343–21348. 10.1073/pnas.1011801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Perry GLW, Wilmshurst JM, McGlone MS, McWethy DB, Whitlock C (2012) Explaining fire-driven landscape transformation during the Initial Burning Period of New Zealand’s prehistory. Global Change Biology 18: 1609–1621. 10.1111/j.1365-2486.2011.02631.x [DOI] [Google Scholar]
- 125. King C (1984) Immigrant killers: introduced predators and the conservation of birds in New Zealand. Auckland, New Zealand: Oxford University Press, 224 pp. [Google Scholar]
- 126. Towns DR, Daugherty CH (1994) Patterns of range contractions and extinctions in the New Zealand herpetofauna following human colonisation. New Zealand Journal of Zoology 21: 325–339. 10.1080/03014223.1994.9518003 [DOI] [Google Scholar]
- 127. Diamond J, Bond AB (1991) Social behavior and the Ontogeny of Foraging behavior in the Kea (Nestor notabilis). Ethology 88: 128–144. 10.1111/j.1439-0310.1991.tb00268.x [DOI] [Google Scholar]
- 128. Schwing R (2010) Scavenging behaviour of kea (Nestor notabilis). Notornis 57: 98–99. [Google Scholar]
- 129. Girod C, Vitalis R, Leblois R, Freville H (2011) Inferring Population Decline and Expansion From Microsatellite Data: A Simulation-Based Evaluation of the Msvar Method. Genetics 188: 165–179. 10.1534/genetics.110.121764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Salmona J, Salamolard M, Fouillot D, Ghestemme T, Larose J, Centon JF et al. (2012) Signature of a pre-human population decline in the critically endangered Reunion Island endemic forest bird Coracina newtoni . PLoS One 7: e43524 10.1371/journal.pone.0043524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Spear SF, Peterson CR, Matocq MD, Storfer A (2006) Molecular evidence for historical and recent population size reductions of tiger salamanders (Ambystoma tigrinum) in Yellowstone National Park. Conservation Genetics 7: 605–611. 10.1007/s10592-005-9095-4 [DOI] [Google Scholar]
- 132. Larsson JK, Jansman HAH, Segelbacher G, Hoglund J, Koelewijn HP (2008) Genetic impoverishment of the last black grouse (Tetrao tetrix) population in the Netherlands: detectable only with a reference from the past. Molecular Ecology 17: 1897–1904. 10.1111/j.1365-294X.2008.03717.x [DOI] [PubMed] [Google Scholar]
- 133. De Oliveira LR, Meyer D, Hoffman J, Majluf P, Morgante JS (2009) Evidence of a genetic bottleneck in an El Nino affected population of South American fur seals, Arctocephalus australis . Journal of the Marine Biological Association of the United Kingdom 89: 1717–1725. 10.1017/S0025315409000162 [DOI] [Google Scholar]
- 134. Henry P, Miquelle D, Sugimoto T, McCullough DR, Caccone A, Russello MA et al. (2009) In situ population structure and ex situ representation of the endangered Amur tiger. Molecular Ecology 18: 3173–3184. 10.1111/j.1365-294X.2009.04266.x [DOI] [PubMed] [Google Scholar]
- 135. Hundertmark KJ, Van Daele LJ (2010) Founder effect and bottleneck signatures in an introduced, insular population of elk. Conservation Genetics 11: 139–147. 10.1007/s10592-009-0013-z [DOI] [Google Scholar]
- 136. Hu Y, Guo Y, Qi D, Zhan X, Wu H, Bruford MW et al. (2011) Genetic structuring and recent demographic history of red pandas (Ailurus fulgens) inferred from microsatellite and mitochondrial DNA. Molecular Ecology 20: 2662–2675. 10.1111/j.1365-294X.2011.05126.x [DOI] [PubMed] [Google Scholar]
- 137. Williamson-Natesan EG (2005) Comparison of methods for detecting bottlenecks from microsatellite loci. Conservation Genetics 6: 551–562. 10.1007/s10592-005-9009-5 [DOI] [Google Scholar]
- 138. USFWS (2010) Mexican Wolf Conservation Assessment Technical Report Region 2, US Fish and Wildlife Service Region 2, Albuquerque, New Mexico. [Google Scholar]
- 139. Buller WL (1888) A History of the Birds of New Zealand. London, 2nd edition. [Google Scholar]
- 140. Cunningham JM (1948) Numbers of Kea. Notornis 2: 154. [Google Scholar]
- 141. Elliott G, Kemp J (1999) Conservation ecology of kea (Nestor notabilis): 49. [Google Scholar]
- 142. Brown JW, de Groot PJV, Birt TP, Seutin G, Boag PT, Friesen VL (2007) Appraisal of the consequences of the DDT-induced bottleneck on the level and geographic distribution of neutral genetic variation in Canadian peregrine falcons, Falco peregrinus . Molecular Ecology 16: 327–343. 10.1111/j.1365-294X.2007.03151.x [DOI] [PubMed] [Google Scholar]
- 143. Yao XH, Ye QG, Kang M, Huang HW (2007) Microsatellite analysis reveals interpopulation differentiation and gene flow in the endangered tree Changiostyrax dolichocarpa (Styracaceae) with fragmented distribution in central China. New Phytologist 176: 472–480. 10.1111/j.1469-8137.2007.02175.x [DOI] [PubMed] [Google Scholar]
- 144. Jackson JR (1960) Keas at Arthurs Pass. Notornis 9: 39–58. [Google Scholar]
- 145. Keuling O, Stier N, Roth M (2008) How does hunting influence activity and spatial usage in wild boar Sus scrofa L.? European Journal of Wildlife Research 54: 729–737. 10.1007/s10344-008-0204-9 [DOI] [Google Scholar]
- 146. Hailer F, Helander B, Folkestad AO, Ganusevich Sa, Garstad S, Hauff P et al. (2006) Bottlenecked but long-lived: high genetic diversity retained in white-tailed eagles upon recovery from population decline. Biology letters 2: 316–9. 10.1098/rsbl.2006.0453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. White NE, Bunce M, Mawson PR, Dawson R, Saunders Da, Allentoft ME (2014) Identifying conservation units after large-scale land clearing: a spatio-temporal molecular survey of endangered white-tailed black cockatoos (Calyptorhynchus spp.). Diversity and Distributions 20: 1208–1220. 10.1111/ddi.12202 [DOI] [Google Scholar]
- 148. Brouwer K, Jones M (2000) Longevity records for Psittaciformes in captivity. International Zoo Yearbook 37: 299–316. 10.1111/j.1748-1090.2000.tb00735.x [DOI] [Google Scholar]
- 149. Frankham R (2005) Genetics and extinction. Biological Conservation 126: 131–140. 10.1016/j.biocon.2005.05.002 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
The left hand side of the figure represents the 5‚ end of the sequence.
(PDF)
(PDF)
Columns 3–6 show the starting values for the mean and variance of the prior distributions. Columns 7–10 show the means and variances (and their means and variances) of the hyperprior distributions. Parameters listed are generation interval (g), contemporary Ne (NC), historical Ne (NH), mutation rate scaled in terms of contemporary population size (θ), and time (T). scale.
(PDF)
(PDF)
Individuals with unusual assignments and referred to in the text are in bold.
(PDF)
(TXT)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.