Abstract
One of the temperature-sensitive alleles of CEG1, a guanylyltransferase subunit of the Saccharomyces cerevisiae capping enzyme, showed 6-azauracil (6AU) sensitivity at the permissive growth temperature, which is a phenotype that is correlated with a transcription elongation defect. This temperature-sensitive allele, ceg1-63, has an impaired ability to induce PUR5 in response to 6AU treatment and diminished enzyme-GMP formation activity. However, this cellular and molecular defect is not primarily due to the preferential degradation of the transcript attributed to a lack of cap structure. Our data suggest that the guanylyltransferase subunit of the capping enzyme plays a role in transcription elongation as well as cap formation. First, in addition to the 6AU sensitivity, ceg1-63 is synthetically lethal with elongation-defective mutations in RNA polymerase II. Secondly, it produces a prolonged steady-state level of GAL1 mRNA after glucose shutoff. Third, it decreases the transcription read through a tandem array of promoter-proximal pause sites in an orientation-dependent manner. Taken together, we present direct evidence that suggests a role of capping enzyme in an early transcription. Capping enzyme ensures the early transcription checkpoint by capping of the nascent transcript in time and allowing it to extend further.
The eukaryotic mRNAs produced by RNA polymerase II (Pol II) are capped with an inverted 7-methyl-guanosine (m7G) linked to the first residue of the mRNA (36). This event occurs by a series of three enzymatic reactions; The 5′ triphosphate end of the nascent RNA Pol II transcript is cleaved by 5′ RNA triphosphatase to produce the diphosphate end. RNA guanylyltransferase forms a covalent enzyme-GMP complex and subsequently caps the RNA substrate by adding a guanosine residue in a 5′-5′ triphosphate linkage. The cap is then methylated by RNA (guanine-7) methyltransferase (23, 39). In higher eukaryotes, a bifunctional monomeric polypeptide carries both the RNA triphosphatase and guanylyltransferase activities, while the capping enzyme from yeast is a complex of RNA triphosphatase and guanylyltransferase subunits (40). The polypeptides are encoded by the CET1 and CEG1 genes, respectively in Saccharomyces cerevisiae, and both are essential for the cell viability.
Capping, the first mRNA modification, occurs by the time the transcript is only 25 to 30 nucleotides long in an early transcription phase. Such cotranscriptional capping is mediated by recruitment of capping enzyme machinery to the phosphorylated carboxy-terminal domain (CTD) of the largest subunit of Pol II (7, 15, 22, 51). The CTD of Pol II has an unusual structure with many heptapeptide repeats (YSPTSPS). The capping enzyme binds directly and specifically to the phosphorylated CTD of Pol II via the Ceg1 subunit (yeast) or the guanylyltransferase domain (metazoan). Furthermore, the guanylyltransferase activity of Ceg1 associated with phosphorylated CTD is allosterically regulated by both the Cet1 and phosphorylated CTD to ensure it has a coordinated capping activity (8). The mammalian capping enzyme is also allosterically regulated by an interaction with the phosphorylated CTD (16, 25). The CTD phosphorylated at serine 5 of the heptapeptide repeat appears to be important for the capping reaction because it stimulates the guanylyltransferase activity, although both serine 2 phosphorylation and serine 5 phosphorylation can mediate the protein interaction. An interesting extension to these findings was added by a chromatin immunoprecipitation assay, which provides in vivo evidence that the capping enzyme machinery interacts dynamically with Pol II during a transcription cycle (20, 35). The capping enzyme interacts with Pol II immediately after the serine 5 of CTD is phosphorylated. As serine 5 phosphorylation decreases in an early elongation phase, the capping enzyme dissociates from the transcription complex.
Recently, data from several groups have raised the concept of “checkpoints” in transcription, especially in an early phase (summarized in reference 26). As operated during the cell cycle to ensure that each phase of the cycle is complete before the next one begins, checkpoint in early transcription is suggested to play a role in ensuring that only the properly modified RNA at the 5′ end is extended. The Pol II transcription is thus subjected to checkpoint control for the coordinated transcription with mRNA capping (6, 25, 26). Several transcription factors have been reported to play in this window. DSIF, a human homolog of the yeast transcription factor Spt4/Spt5, increases the pausing of Pol II and thus plays a role as a negative factor (42, 46, 49). Within this temporal and spatial interval, while Pol II with the hypophosphorylated CTD is paused at the promoter-proximal region, many factors are intended to target the capping enzymes to increase their recruitment or to enhance their catalytic activities. In addition to TFIIH, which creates a binding epitope for the capping enzyme on CTD by serine 5 phosphorylation, as described above, Spt5 itself interacts with the triphosphatase and guanylyltransferase components of the capping enzyme (21, 27, 44). In the case of human immunodeficiency virus (HIV), DSIF/Spt5-induced transcription arrest allows HIV-encoded Tat to interact with capping enzymes and to stimulate their catalytic activities (5, 6). Phosphorylation of Pol II CTD is critical for the transition to the elongation phase. At this step, the elongation factor P-TEFb, a DRB-sensitive protein kinase, phosphorylates the Pol II CTD and Spt5 (19, 29, 38). HIV Tat also interacts with P-TEFb in this step (29). In S. cerevisae, Ctk1 kinase complex and the Bur1 kinase complex facilitate the transcription elongation (18, 50). The phosphorylation of Pol II CTD is thus meant to lead to the formation of the processive transcription elongation complexes. According to recent reports, the capping enzyme in Schizosaccharomyces pombe interacts with Cdk9/Pch1, a yeast P-TEFb homolog (28).
How does the capping enzyme fit into the complicated scheme to delineate it in the order of pausing, capping, and the reversion of pausing in the checkpoint model? If the elongation-competent transition does not occur until the RNA is capped and the capping is the major determinant to shift the Pol II status, it could be indicative that a capping enzyme plays a critical role in regulation of an early transcription in addition to its role in simple cap formation.
To study whether the capping enzyme plays a key role in coordinating mRNA processing and transcription elongation, we used a well-characterized yeast system. Because Ceg1, a capping enzyme subunit, contributes to transcription via its typical cap formation activity, this study examined various ceg1 temperature-sensitive alleles to determine if there is any additional role in transcription elongation. Among them ceg1-63 displayed 6-azauracil (6AU) sensitivity and a defect in PUR5 induction by 6AU treatment. This study shows that transcription through the pause sites artificially inserted at the promoter-proximal region was severely inhibited in ceg1-63. We show that such an elongation defect was coupled to the reduced guanylyltransferase activity. However, it happened independently of the turnover of uncapped transcripts. These results indicate that the transcription ternary complexes are held at the promoter as long as their RNAs have not been properly capped. That is, capping enzyme plays a critical role in the promoter-proximal checkpoint window by reinforcing the checkpoint security circuit and probably by reversing the transcription arrest in time. This finding also strongly supports the transcription checkpoint model, in which an early transcription is tightly regulated.
MATERIALS AND METHODS
Plasmids.
pRS315-CEG1, pRS315-ceg1-12, pRS315-ceg1-34, pRS315-ceg1-63, pRS315-ceg1-237, and pRS315-ceg1-250 are kind gifts from S. Buratowski and are described by Fresco and Buratowski (12). The plasmid pC1023, the GAL1 promoter driving tandem repeats of the Ia pause sequences inserted in the correct orientation, and pC1024 in the reverse orientation were kind gifts from D. Reines (45). pYF1561 (TRP CEN/ARS), pYF1866 (RPO21 on pYF1561), pYF1868 (rpo21-17 on pYF1561), pYF1867 (rpo21-7 on pYF1561), pYF1863 (rpo21-23 on pYF1561), and pYF1864 (rpo21-24 on pYF1561) are described by Archambault et al. (2). YEpPPR2 carries PPR2 and a 2μm origin (provided by S. Buratowski).
Yeast strains.
The yeast strains used in this study are summarized in Table 1. YSB244, YSB228, YSB229, YSB230, YSB231, and YSB232 containing the wild-type or mutant CEG1 alleles have been described previously (12). For the drug sensitivity assay, the above strains were transformed with pRS316, which provides URA3. YC50 was generated by crossing YC49 to AJY892 followed by a random sporulation. Each marker was confirmed on a selective media. This strain represents all the characteristic phenotypes of ceg1-63 in the RPB1 shuffling background. YC9 was constructed as follows: The linearized pUL9 (10) was transformed into YSB516. The LEU2 marker was used to select the homologous recombination into chromosomal TRP1, rendering the cells auxotrophic for tryptophan. Medium preparation, yeast transformation, and other yeast manipulations were performed by the standard methods as described previously (13).
TABLE 1.
Characteristics of the yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| AJY892 | MATaura3-52 leu2,112 ceg1-63 lys2-801 ade2 ade3 | 4 |
| YSB516 | MATaura3-52 or ura3-1 leu2-3,112 his3Δ200 or his3-11, 15 rpb1Δ1::HIS3 ceg1-250 pRP112 (RPB1 URA3 CEN/ARS) | 7 |
| YC50 | MATaura3-52 or ura3-1 leu2-3,112 trp1-1 his3Δ200 or his3-11, 15 rpb1Δ187::HIS3 ceg1-63 pRP112 (RPB1 URA3 CEN/ARS) | This study |
| YC9 | MATaura3-52 or ura3-1 leu2-3,112 trp1Δ::LEU2 his3Δ200 or his3-11, 15 rpb1Δ187::HIS3 ceg1-250 pRP112 (RPB1 URA3 CEN/ARS) | This study |
| DY103 | MATaura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 pRP214 (RPB2 LEU2 CEN/ARS) pRS316 | 45 |
| DY108 | MATaura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 dst1(ppr2)Δ::hisG pRP2-10L (rpb2-10 LEU2 CEN/ARS) pRS316 | 45 |
Growth conditions and analysis.
The yeasts were grown on either YPD or SC containing 2% of glucose. For the 6AU sensitivity assay, the yeasts were transformed with pRS316 and grown in SC lacking uracil (SC−uracil). Yeast strains were freshly grown to an A600 of 1.0 and serially diluted 10-fold. Three microliters from each sample was applied as spots to either YPD or SC−uracil plates containing 6AU (50 μg/ml) or caffeine (10 mM). The plates were incubated for 3 to 4 days at 30°C. For the PUR5 induction assay, the cells grown to an A600 of 0.5 to 0.8 were treated with 6AU and further grown for the indicated time. For the glucose shutoff experiment, yeasts were grown to an A600 of 0.5 to 0.8 in SC containing 2% raffinose. Galactose was added to a final concentration of 2% to allow the GAL1 promoter to be induced. After an hour, glucose was added to a final concentration of 2%, and a sample was drawn at each time point to prepare the RNA.
RNA analysis.
Yeasts grown to the exponential phase (A600 = 0.5 to 1.0) were used for the RNA isolation. The total RNA was prepared by using the Trizol method according to the manufacturer's instructions (Invitrogen). Before reverse transcription-PCR (RT-PCR), the RNA was usually treated with DNase I (Promega) to remove any residual chromosomal DNA that remained in the sample. Double-stranded cDNA was made from 1 μg of total RNA, using reverse transcriptase (Promega). Semiquantitative PCR amplification of the target regions of GAL1, PUR5, SED1, and the reporter construct was performed in a 25-μl reaction mixture by using the following program: 5 min at 94°C followed by 16 to 20 cycles with 40 s at 94°C, 40 s at 50°C, and 50 s at 72°C. The reaction was finished by an extra extension step at 72°C for 10 min. Signals were quantified by Chemi-digital image analysis system using Labworks software (UVP, Upland, Calif.). In some cases, PCR was repeated in the presence of [α-32P]dATP and the signal quantification was confirmed by PhosphorImager analysis with ImageQuant software. The quantified signals were normalized by dividing them with those of SED1 and displayed relative to 100% (or a value of 1) signal for a nominated reference. The following oligonucleotide primers were used in RT-PCR: PUR5f (5′ GGTAGGCTATTGGTTGTTG 3′), PUR5r (5′ TTCCCTGGTGACAACGTTA 3′), SED1f (5′ AACTACATCTTTGCCACCA 3′), SED1r (5′ GTGGTAGTGCCCTTAGATT 3′), GAL1f (5′ GAAGGATCGAGCACGAATA 3′), GAL1r (5′ TTCGTCGGCAGTAAAGCTCG 3′), CYC1TERf (5′ TATATCCATCACACTGGC 3′), and CYC1454 (5′ GACCTAGACTTCAGGTTGT 3′).
Capping enzyme activity assay.
Yeast cells expressing the various forms of Ceg1p were grown at 30°C to an exponential phase. The preparation of total cell extract was described elsewhere (7). The same amount of total extract (200 μg) was used for an immunoprecipitation assay. The total extract was precipitated by anti-Ceg1 or anti-Cet1 antibody. After vigorous washing, the precipitate was resuspended in guanylyltransferase-GMP (E-GMP) complex formation buffer, which contains 1 μM GTP and 3 μCi of [α-32P]GTP (3,000 Ci/mmol) (7). The labeled E-GMP was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to the membrane, and visualized by an autoradiogram. Immunoblotting assay was performed with anti-Cet1 antibody by using the same blot.
RESULTS
A mutation in the guanylyltransferase subunit gene (CEG1) of the mRNA capping enzyme confers an allele-specific sensitivity to 6AU.
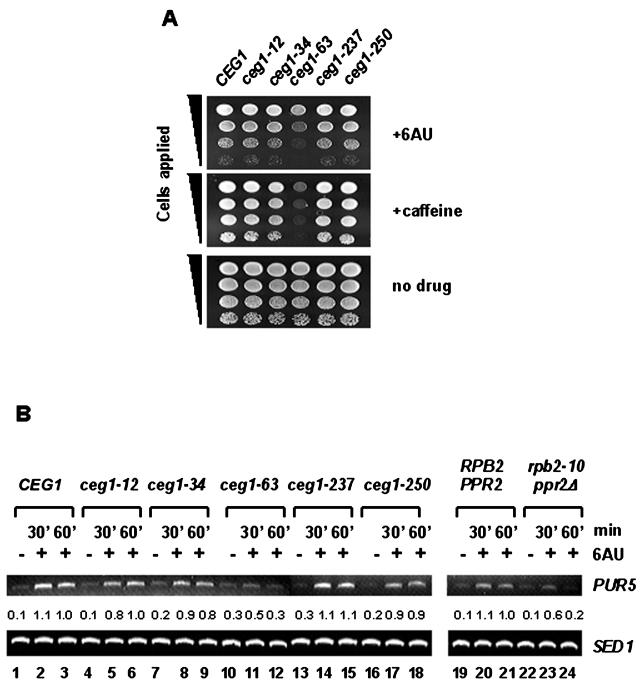
The sensitivity to 6AU and mycophenolic acid (MPA) is a phenotype of yeast with mutations that are broadly related to defects in transcription elongation. 6AU affects the nucleotide metabolism and causes a reduction in the levels of GTP and UTP in the cell (11). Therefore, sensitivity to this drug is often associated with mutations that inactivate transcription elongation (41). In order to test the possibility that a capping enzyme plays an additional role in an early transcription elongation phase in addition to a simple cap formation when a transcription checkpoint is applied, we examined the sensitivity of various alleles of the CEG1 guanylyltransferase subunit gene to 6AU. Yeast strains carrying a variety of the ceg1 temperature-sensitive alleles ceg1-12(C354Y), ceg1-34(P346L), ceg1-63(D152N), ceg1-237(G45A), and ceg1-250(P288S) have been reported previously (12). The growth of each strain carrying the temperature-sensitive allele was tested on the 6AU (50 μg/ml) plates at the permissive temperature (30°C). Among different strains with temperature-sensitive alleles, only the ceg1-63 strain showed reduced growth on the medium containing 6AU (Fig. 1A). MPA (15 μg/ml), a drug structurally different from 6AU but also known to decrease the cellular concentration of the nucleotides, inhibited the growth of the ceg1-63 strain as well (data not shown). In addition, we observed that ceg1-63 repeatedly displayed sensitivity to caffeine (10 mM) and weak sensitivity to 1 M NaCl (Fig. 1A) (data not shown) at the permissive temperature while the other mutant allele strains grew normally compared to their wild-type parent. This shows that the Ceg1 guanylyltransferase capping enzyme subunit might be needed for efficient transcript elongation in addition to its known activity in cap formation.
FIG. 1.
The mutant with mutation in the gene (CEG1) coding for the guanylyltransferase subunit of the capping enzyme shows an elongation-defective phenotype. (A) The ceg1-63 mutant, one of the temperature-sensitive alleles of the gaunylyltransferase subunit, displays an allele-specific sensitivity to 6AU and caffeine. The 10-fold serial dilutions of the cells (A600 = 1.0) from the wild-type CEG1 allele (YSB242) or the indicated strains carrying the ceg1 temperature-sensitive alleles (YSB228, ceg1-12; YSB229, ceg1-34; YSB230, ceg1-63; YSB231, ceg1-237; YSB232, ceg1-250) were applied as spots to SC−uracil medium containing 50-μg/ml 6AU or 10 mM caffeine. As a control, the same dilution was applied as spots to the YPD medium. Photographs were taken after growth at 30°C for 3 days (6AU and no drug) or 4 days (caffeine). The yeasts in the 6AU test are URA3 as provided by pRS316. (B) The ceg1-63 mutant strain was defective in the PUR5 induction. The yeast cells, as described in panel A, were grown at 30°C in the SC−uracil medium containing 2% glucose. When growth reached 0.5 to 0.8 U at A600, the culture was divided into two parts and grown in the presence or absence of 6AU (50 μg/ml). The RNA was purified from each sample, and the induction of PUR5 was analyzed by RT-PCR, as described in Materials and Methods. The level of the SED1 transcript was included as an internal control. Each band was quantified, normalized by SED1 level, and compared to the value of the wild type obtained at an hour of treatment of 6AU (lane 3 for ceg1 mutants or lane 21 for rpb2/ppr2, assigned as 1). The experiments were repeated three times, and a representative experiment is shown. +, samples treated with 6AU. RT-PCR with RNA from DY108 (rpb2-10/ppr2Δ) and its isogenic parent DY103 (RPB2/PPR2) was included as a control.
The transcription pattern of each ceg1 allele was analyzed at the molecular level too. Reines and colleagues have reported that PUR5, which encodes IMP dehydrogenase, is transcriptionally induced by 6AU (37). Therefore, it is generally believed that the 6AU-sensitive phenotype coupled with the inability to induce PUR5 in response to 6AU is indicative of a defect in transcriptional elongation.
The induction of PUR5 by 6AU was monitored in the different ceg1 alleles. Following a treatment with 6AU, the total RNA was purified at two different time points from an equal amount of each cells and subjected to RT-PCR to analyze the PUR5 mRNA level (Fig. 1B). As a control, SED1 mRNA was amplified from the same batch of RNA samples. SED1 encodes a cell wall protein whose expression is unaffected by 6AU and remains constant throughout the growth condition (37). The amplified DNA products were quantified, normalized by SED1 level, and compared between samples. As shown in Fig. 1B, a large induction of PUR5 mRNA was observed in the wild type. The PUR5 induction in the ceg1-63 strain was greatly affected compared to that in the others, while SED1 mRNA was detected at levels that were relatively unchanged. RT-PCR was performed properly, since it reproduced the characteristic behavior of the transcription elongation mutants when they were challenged with 6AU (Fig. 1B, lanes 19 to 24). The yeast double mutant, which carries rpb2-10 and ppr2Δ, is very sensitive to 6AU, and it was reported that PUR5 induction was severely affected by an hour of treatment (37). RPB2 and PPR2 code for the second largest subunit of Pol II and the transcription elongation factor TFIIS, respectively. This result opens a possibility that Ceg1 may play a role in the transcription elongation as observed for the typical elongation factors.
The ceg1-63 mutant shows reduced guanylyltransferase activity.
The sensitivity to 6AU is often but not always indicative of a defect in transcription elongation. Theoretically, any step that depends on the optimum concentration of nucleotides would be affected by this kind of drugs. In this regard, the sensitive phenotype of the ceg1-63 mutant might have arisen from defective catalytic activity because this drug consumes GTP, which is a substrate of guanylytransferase. In fact, the ceg1-63 mutant carries a mutation at position 152 with Asp changed to Apn within the N-terminal nucleotidyltransferase domain (14, 33). Although this residue is not in direct contact with GTP, its substitution in mammalian capping enzyme is lethal (33).
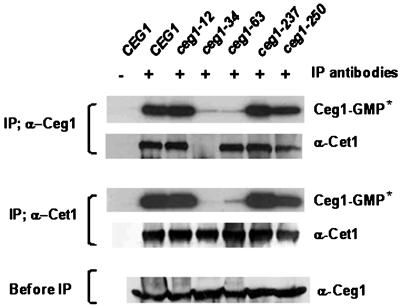
To test whether the guanylyltransferase activity of ceg1-63 was affected or not, we attempted to measure its catalytic activity directly. During the capping reaction, Ceg1 forms a covalent intermediate with GMP. Therefore, it is possible to quantitate the enzyme activity by measuring the formation of the enzyme-GMP (Ceg1-GMP) intermediate. The whole-cell extract was prepared from the culture grown to an exponential phase at 30°C. Since it was very difficult to achieve an assay condition under the quantitative control with total extract, capping enzyme was partially purified from whole extract with anti-Ceg1 or anti-Cet1 antibodies. Immunoprecipitation was able to remove any reverse or substrate-depleting activity. After an extensive washing, immunoprecipitate was incubated with [α-32P]GTP followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and an autoradiogram to analyze the Ceg1-GMP complex formation or an immunoblot assay to detect the amount of proteins (Fig. 2). As shown in Fig. 2, the Ceg1-GMP formation activity of ceg1-34 and ceg1-63 mutants was greatly diminished compared to the wild type or other alleles. The immunoblot with anti-Cet1 antibody showed that Ceg1-63p still retained the subunit interaction with Cet1p (top panel). The reciprocal precipitation with anti-Cet1 antibody indicated that the compromised catalytic activity was not caused by simple interference by antibodies (middle panel). In contrast to Ceg1-63p, the loss of activity of Ceg1-34p was accompanied by the loss of Cet1p interaction. The gene ceg1-34 (P346L) carries the mutation in the CTD that is meant to mediate Cet1 subunit interaction. The temperature-sensitive phenotype of this mutant has been suppressed by an extra copy of CET1 (8). Although in vitro immunoprecipitation showed that the capping enzyme formation was disturbed in this particular mutant, it is not yet clear whether the lack of Ceg1-GMP formation activity was directly attributed from the loss of subunit interaction. However, the cellular phenotype shown at the permissive temperature described in previous figure implies that the diminished ability of complex formation and the Ceg1-GMP formation of the ceg1-34 mutant suffice for yeast cell growth and PUR5 induction in vivo, in contrast to the ceg1-63 mutant. Unfortunately, Ceg1p migrates while overlapped with immunoglobulin heavy chain, so it was not possible to detect the amount of Ceg1p coprecipitated directly. However, according to careful analysis among autoradiographies and immunoblottings, at least for the ceg1-63 mutant, it seemed to be able to produce a capping enzyme complex but with a diminished catalytic activity.
FIG. 2.
The in vitro Ceg1-GMP formation assay shows that the ceg1-63 allele has reduced catalytic activity. Immunoprecipitation (IP) was performed with Ceg1 polyclonal antibody (top panel) or Cet1 antibody (middle panel) from the total extract (200 μg) of each strain grown at 30°C. The beads were pelleted and washed extensively. Capping enzyme in the pellet was detected by both an autoradiogram of enzyme-GMP formation (Ceg1-GMP*) and immunoblotting with anti-Cet1 antibody (α-Cet1). −, immunoprecipitation without antibody. Whole-cell extracts (60 μg) were probed with Ceg1 antibody (bottom panel).
The Ceg1-GMP formation activity from total extract of each different temperature-sensitive allele has been reported previously (12). In this report, there was variable amount of reduction in the enzymatic activity of guanylyltransferase in various mutant alleles. This showed decreased Ceg1-GMP formation activity in the ceg1-12 and ceg1-237 mutants too. We assume that it is due to the different reaction condition; total extract was used for enzyme activity in the presence of 0.1 μM GTP. The location of the mutation in ceg1-12 is close to the motif that exerted a strong dependency on GTP concentration (33). Anyway, although there are variable changes in enzymatic activities in Ceg1 mutants, only the ceg1-63 mutant seems to be well below the critical threshold expressed as a cellular phenotype, such as 6AU sensitivity and lack of PUR5 induction at the permissive temperature.
The ceg1-63 mutant shows a reduced transcription elongation activity.
The above results so far seem to indicate that the ceg1-63 mutant allele mimicked the typical phenotype of the elongation factor mutations. In other words, it began to develop the phenotype when the GTP level was decreased, since the mutant with the ceg1-63 mutation had an impaired ability of Ceg1 to transfer GMP to a diphosphate end of the nascent RNA. As a result, the RNA transcript without the 5′ cap might be subjected to preferential degradation. To test whether the low induction of PUR5 in the ceg1-63 mutant is attributed to a lack of mRNA stability or from a possible impaired ability of transcription elongation, expression of the GAL1 gene was examined.
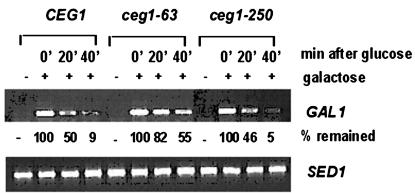
Yeasts with a transcription elongation mutation often have a defect in the induction of GAL1 gene by galactose. In addition to the delayed induction, the steady-state level of GAL1 mRNA after the glucose shutoff is usually prolonged in those mutants (37). Therefore, this study examined the steady-state level of the GAL1 transcripts, particularly the kinetics of RNA disappearance. The wild-type CEG1 and ceg1-63 and ceg1-250 yeasts were grown at 30°C in raffinose medium followed by galactose induction for an hour. During this period, the wild type and both mutants reached the maximum level of GAL1. Glucose was then added to repress further de novo initiation of transcription from the GAL1 promoter. RNA was collected at different time points thereafter, and the decrease in the GAL1 transcript was analyzed by RT-PCR. Figure 3 shows that wild-type CEG1 and mutant ceg1-250 yeasts have decreased levels of the GAL1 in the time course once the glucose was added. Glucose rapidly repressed further formation of the initiation complex at the GAL1 promoter. The half-life of the GAL1 transcript already synthesized in the presence of galactose was found to be approximately 20 min in both the wild type and the ceg1-250 mutant. However, surprisingly, the GAL1 transcript lived longer after the glucose shutoff in the ceg1-63 mutant. It required more than 40 min to drop to 50% of the starting level. This result shows that the ceg1-63 mutant generated more long-lived RNA than either the wild type or the ceg1-250 mutant. In fact, the half-life of the GAL1 transcript in the ceg1-63 mutant must be similar to those of others. In other words, as observed in typical elongation factor mutants, the decreased RNA level must be counterbalanced by the slowly elongating Pol II, which continuously synthesizes RNA as it moves to the 3′ end of the GAL1 gene even after the de novo initiation has been shut off. Consequently, it is unlikely that lower level of the transcript from PUR5 (Fig. 1B) is solely due to an increase in the degradation of mRNA from the lack of a guanosyl cap. It is rather likely that, as reported in other transcription elongation-defective mutants, the ceg1-63 mutant has a lower rate of RNA synthesis. The nascent RNAs were capped at reduced efficiency in the ceg1-63 mutant. This caused reduced release of elongating complex at the promoter region and in turn led to a slow extension of transcript. This result indicates that the guanylyltransferase subunit of the capping enzyme has a distinctive role in transcription elongation.
FIG. 3.
ceg1-63 produces the GAL1 transcript with a higher steady-state level following the glucose shutoff. Each of the wild type CEG1 and mutant ceg1-63 and ceg1-250 strains was grown in SC medium supplemented with raffinose at 30°C. The culture was induced with galactose for an hour prior to the glucose treatment. The steady-state level of the GAL1 transcript at the indicated time points after glucose addition was analyzed by RT-PCR. −, GAL1 level from the cells grown in raffinose prior to galactose induction. The level of the SED1 transcript was included as described for Fig. 1B. The signal was quantified as in the legend to Fig. 1, normalized with SED1, and compared to that of the wild type obtained by an hour of treatment of galactose (lane 2 was considered as 100%).
An allele-specific synthetic lethal assay shows a role of Ceg1 in the transcription elongation.
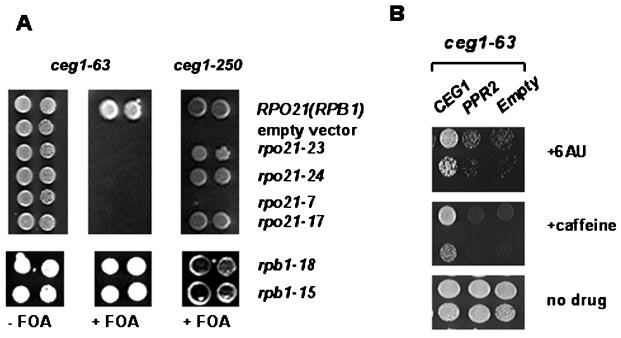
To further determine if Ceg1 plays a role in transcription elongation and to examine whether they behave allele specifically, we constructed an rpo21(rpb1) deletion strain in the background of the ceg1-250 or ceg1-63 strain suitable for plasmid shuffling. The defects in the transcription elongation activity often display synthetic interactions with defects in the other known elongation factors. Several RPO21 (RPB1) alleles, which code for the largest subunit of RNA Pol II, are well characterized to display 6AU sensitivity and confer compromised elongation. These mutations have been mapped to the region between G and H, which are two of the regions highly conserved among the largest subunits of prokaryotic and eukaryotic RNA polymerases, where the elongation factor TFIIS (PPR2 in yeast) is supposed to interact (2, 47). The above yeast strains were shuffled to introduce the elongation-defective rpo21-7, rpo21-17, rpo21-23, and rpo21-24 alleles as well as the wild-type RPO21 (RPB1) allele in the ceg1-63 or ceg1-250 background. When the ceg1-63 allele was combined with each of these alleles, it resulted in synthetic lethality, while the ceg1-250 mutant did not exhibit an additive growth defect in combination, except for the rpo21-7 strain, which has a stringent temperature-sensitive phenotype and a slow growth rate even at the permissive temperature (Fig. 4A, top panel). It was reported that the ceg1-250 strain is sensitive to rpb1 CTD truncation and is growth defective in combination with the TFIIH kin28 CTD kinase mutation (7, 31). In contrast to the ceg1-250 strain, the ceg1-63 mutant was dependent on the presence of the elongation-competent polymerase for growth at the permissive temperature in the absence of 6AU. We also tested the other temperature-sensitive alleles, rpb1-18, and rpb1-15, whose mutations were mapped to the region between A/B and near H, respectively (34). Both the ceg1-63 and ceg1-250 mutants did not show any synergistic effect with them (Fig. 4A, bottom panel). This result shows an allele-specific functional link between Ceg1 and Rpb1, in further support of the notion that the capping enzyme Ceg1 subunit has a distinctive role in the transcription elongation.
FIG. 4.
The ceg1-63 allele is synthetically lethal with rpo21 mutations that are specifically compromised in transcription elongation. (A) The ceg1-250 and ceg1-63 mutants were analyzed in combination with the rpo21 elongation-defective mutant upon shuffling pYF1561 (empty vector), pYF1866 (RPO21), pYF1868 (rpo21-17), pYF1867 (rpo21-7), pYF1863 (rpo21-23), and pYF1864 (rpo21-24) into YC9 and YC50, respectively. For the genetic interaction assay with rpb1-18 and rpb1-15, the YSB516 shuffling strain was used for the ceg1-250 strain (31). Growth of the transformants carrying a double mutation were assayed by applying spots of a 10- or 100-fold dilution of an A600 of 0.5 culture onto plates containing SC plus fluoroorotic acid (FOA) and incubating for 3 days at 30°C. (B) The 6AU- and caffeine-sensitive phenotype was not suppressed by overexpression of PPR2. CEG1, pRS316-CEG1; PPR2, YEpPPR2; empty, pRS426.
The rpo21 elongation-defective alleles confer a 6AU-sensitive phenotype, which is suppressed by the overexpression of PPR2 (2). Since PPR2 is a typical elongation factor that facilitates the polymerase read through the intrinsic arrest sites in the transcribed region, this study also examined the genetic interaction with PPR2. No significant augmentation of the ceg1-63 phenotype was observed in the presence of the ppr2Δ (data not shown). In addition, its 6AU-sensitive phenotype was not suppressed by overexpression of PPR2 (Fig. 4B). This suggests that Ceg1 might have a role that differs from that of Ppr2 in transcription elongation.
ceg1-63 cells are unable to transcribe passing through the artificial pause sites in vivo.
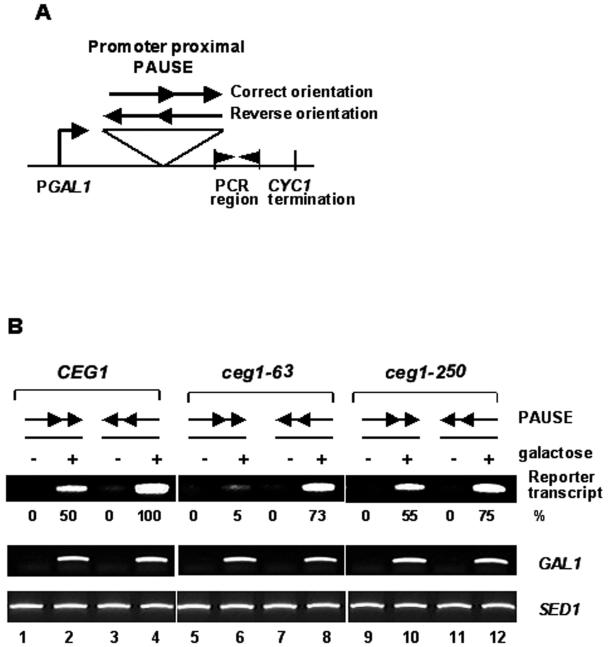
Since the ceg1-63 mutant has defects in transcription elongation in vivo as well as guanylyltransferase activity, we attempted to determine its effect on the transcription of the reporter gene with a strong pause site. One of the well-characterized pause sequences, which was originally derived from the human histone gene, H3.3 (Ia site), has been studied both in vitro and in vivo (9, 24, 45). At least in vitro, this Ia site efficiently attenuated transcription by eukaryotic polymerase as well as by prokaryotic polymerase (24). Approximately 50% of the RNA Pol II stops elongation at the Ia site in the presence of a high concentration of the nucleoside triphosphate substrates. Tandem repeats of the Ia sequences increase the attenuation efficiency up to 75%. In addition, its effect is known to depend on the orientation (30). To our knowledge, there is no report that Ia transcriptional pause sites interact with capping enzyme or affect the capping activity. Therefore, if there is any transcriptional synergistic defect that is dependent on the orientation of pausing sequence, it must be attributed to the transcription elongation defect independently of its direct effect on mRNA capping.
The high-copy-number plasmid carrying two repeats of the Ia pause under the GAL1 promoter (a kind gift from D. Reines) was used for this study (Fig. 5A). Although it was reported that these constructs failed to attenuate the mRNA production differentially in vivo (45), it is worth examining whether other factors that are abrogated in transcription elongation cause an arrest on the pause site. Plasmids with a correct (pC1023) or reverse (pC1024) orientation of the pause sites were transformed into the wild-type CEG1, ceg1-63, and ceg1-250 strains separately. The yeast cells were treated with galactose for an hour to induce transcription from the GAL1 promoter. Total RNA before or after galactose induction was purified, and the specific transcripts were analyzed by RT-PCR. The primers used for PCR were designed to amplify the region immediately downstream of the tandem repeats of the Ia pause. This study used RT-PCR instead of Northern analysis. All of the RNA levels were normalized by the one produced from the genomic SED1 and compared to that of wild type that was produced with a reverse pause in the presence of galactose (considered as 100%). In this application, the standard deviation of genomic GAL1 RNA between even lanes was within 5%. By semiquantitative analysis by RT-PCR, we were able to observe the pause effect in vivo (Fig. 5B). The Ia sequence in an arresting orientation usually produced RNA with 47 to ∼83% of the one in a nonarresting orientation. In ceg1 mutants, transcription from the nonarresting Ia was reduced to 73 or 75% of that of wild type (Fig. 5B, compare lanes 4, 8, and 12). However, surprisingly, the yeasts with the ceg1-63 allele have dramatically decreased the level of transcript from the reporter with a pause in an arresting orientation (Fig. 5B, compare lanes 6 and 8). This decreased the RNA level below 6.8% compared to the reverse orientation. In contrast to ceg1-63, ceg1-250 was able to transcribe reporters with a similar ratio as shown in the wild type (Fig. 5B compare lanes 10 and 12). Although, ceg1 mutants were turned out to be less efficient in pause transcription even with a nonarresting orientation, ceg1-63 but not ceg1-250 showed a strong pausing effect in an orientation-dependent manner. The mRNA capping by capping enzyme and mRNA synthesis by Pol II are not interdependent in vitro. However, our in vivo analysis showed that they are tightly coupled, such that reduced capping activity affected transcription dramatically in ceg1-63.
FIG. 5.
The ceg1-63 cell is unable to transcribe passing through the pause sites. (A) The reporter constructs containing a tandem array of the pause Ia sequence under the galactose-inducible promoter (GAL1) is shown. The arrowhead represents the orientation of the cloned Ia pause. The region flanked by two flags was amplified by RT-PCR. (B) The reporter assay was performed with the CEG1 wild type (YSB242) and the ceg1-250 (YSB232) and ceg1-63 (YSB230) mutants. Each strain was grown in SC−uracil medium containing raffinose. The reporter was induced by 2% galactose for an hour. RNA from each cell culture was purified and analyzed by RT-PCR with the primers CYC1TERf and CYC1454, which were annealed immediately downstream of the pause. The RNA level was quantitated as in Fig. 1, normalized with SED1, and compared to that of wild type obtained in lane 4 (considered as 100%).
DISCUSSION
This study showed that one of the temperature-sensitive alleles of the CEG1 capping enzyme guanylyltransferase subunit gene displayed the transcription elongation-defective mutant phenotypes in addition to the compromised catalytic activity in the permissive temperature. The ceg1-63 mutant showed 6AU sensitivity and had a low PUR5 induction in response to 6AU. It also displayed a genetic interaction specifically with the elongation-defective Pol II mutants. Interestingly, the ceg1-63 mutant failed to pass through the artificial pause sites inserted at the promoter-proximal region in an orientation-dependent manner. The reduced guanylyltransferase activity of the ceg1-63 mutant was coupled to a reduced pass of Pol II through the artificial pause sites. Overall, these results demonstrate that the capping enzyme plays a role in the early transcription elongation. The temporal arrest is released only when the nascent RNA is successfully capped. In other words, our observation proves that early transcription is tightly regulated by checkpoint systems that target capping enzyme for efficient coupling of transcription and mRNA capping.
The capping enzyme associates with the Pol II transcription complexes, apparently during the transition from initiation to elongation (20, 35). At the same time and in the same place, the transcription checkpoint is suggested to occur. During this time, elongation is prevented by elongation factors such as DSIF (Spt4/5 complex in yeast) in conjugation with the NELF (negative elongation factor) (42). This early transcriptional arrest is supposed to ensure a temporal window so a timely modification of the 5′ end of the mRNA can be obtained (26). The temporal arrest must be resumed in concert with the subsequent recruitment of positive elongation factors, such as P-TEFb (43). P-TEFb (CTDK-I and/or Bur1/Bur2 complex in yeasts) adds phosphates onto CTD of Pol II and allows it to proceed into a productive elongation. Therefore, repression and reactivation seem to be precisely coordinated with the transfer of the cap onto the nascent RNA.
The time and location when and where the RNA transcripts are capped and Pol II escapes from the promoter-proximal arrest overlap with those of the capping enzymes appear on the transcription complex. This observation raises an interesting possibility that the capping enzyme might play a direct role to ensure that Pol II does not extend uncapped RNA (25, 26). In this regard, we directly showed that a reduced capping event resulted in diminished progression of Pol II transcription independent of transcript turnover.
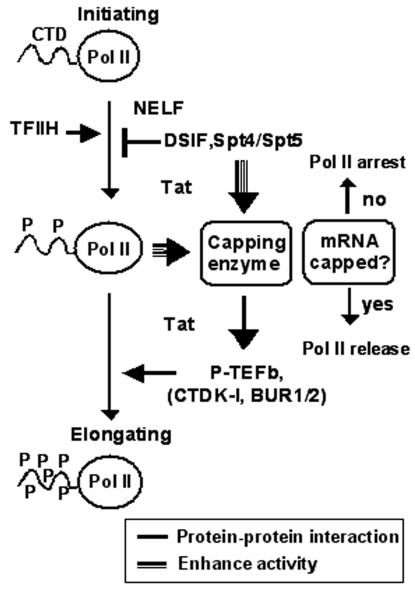
A checkpoint is supposed to adopt a timely mechanism for reactivating transcription once a transcript has been appropriately capped. In our scheme, the capping enzyme might be able to tell when to transit the transcription status in concert with the RNA capping (Fig. 6). HIV-encoded Tat protein must support the role of capping enzyme to overcome the rate-limiting step in the mammalian system (5). It has been independently shown that it interacts with P-TEFb to override the Spt5-induced promoter-proximal arrest (6, 25). However it must be coupled to capping in an ordered fashion. In this regard, the capping of RNA at the 5′ end must be the most critical determinant of the Pol II status. The reversion of the arrested transcription is directly coupled to the cap formation. Indeed, none of the proteins in this early checkpoint window is known to interact with the RNA transcript or differentiate the capped messages from those without. Accordingly, the capping enzyme might be an excellent candidate to determine the time when to reverse the repression. Interestingly, the capping enzyme undergoes a large conformational change during transfer of the guanylyl moiety (14). The conformational change might allow a microenvironmental reconfiguration of the transcription complex to signal that the situation is to be reversed.
FIG. 6.
Schematic model of how capping enzyme plays a role in an early transcription.
In addition, capping enzyme interacts physically with many positive elongation factors. It interacts with SPT5 and P-TEFb in human and yeast cells (21, 27, 28, 44). The positive factor, P-TEFb, thus recruited might reverse transcriptional delay by phosphorylating CTD and/or Spt5. Spt4/5, in addition to playing a negative role in the early transcription, is closely associated with a positive elongation (1, 3, 17, 32, 48). Capping enzyme might either directly recruit the positive factors to form an elongation complex or stimulate their activities. However, the mechanism of this activation remains to be elucidated. Also a study should be followed to see whether building an elongation complex is dependent on RNA capping.
Future studies of the cross talk between the capping enzyme and the transcription elongation machineries should provide an insight into understanding the exact role of the early transcription checkpoint and help us determine if a similar mechanism can be used to monitor the later steps of elongation.
Acknowledgments
We are extremely grateful to S. Buratowski, Harvard Medical School, for providing useful comments, plasmids, yeasts, and antibodies; D. Reines, Emory University, for providing plasmids and yeasts; and J. Friesen, A. Johnson, R. Young, and F. Cross for providing the yeast strains and plasmids used for strain construction.
This work was supported by Korea Research Foundation grants (2002-070-C0070) to E.-J. Cho.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. T., X. Yang, and A. W. Johnson. 2000. Inhibition of mRNA turnover in yeast by an xrn1 mutation enhances the requirement for elF4E binding to elF4G and for proper capping of transcripts by Ceg1p. Genetics 55:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., E. Coronel, C. K. Ho., S. Shuman, and T. M. Rana. 2001. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J. Biol. Chem. 276:12959-12966. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, Y. L., C. K. Ho, N. Saha, B. Schwer, S. Shuman, and T. M. Rana. 2002. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell 10:585-597. [DOI] [PubMed] [Google Scholar]
- 7.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, E. J., C. R. Rodriguez, T. Takagi, and S. Buratowski. 1998. Allosteric interactions between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 12:3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, K. R., D. E. Awrey, A. M. Edwards, and C. M. Kane. 1994. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, p37. J. Biol. Chem. 269:936-943. [PubMed] [Google Scholar]
- 10.Cross, F. R. 1997. ′Marker swap' plasmid: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 11.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 12.Fresco, L. D., and S. Buratowski. 1996. Conditional mutants in the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA 2:584-596. [PMC free article] [PubMed] [Google Scholar]
- 13.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-863. [PubMed] [Google Scholar]
- 14.Hakansson, K., A. J. Doherty, S. Shuman, and D. B. Wigley. 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89:545-553. [DOI] [PubMed] [Google Scholar]
- 15.Ho, C. K., V. Sriskanda, S. McCracken, D. Bentley, B. Schwer, and S. Shuman. 1998. The gaunylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxy-terminal domain of RNA polymerase II. J. Biol. Chem. 273:9577-9585. [DOI] [PubMed] [Google Scholar]
- 16.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD ser-2 and ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, C. D., J. R. Morris, C. T. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keogh, M.-C., V. Podolny, and S. Buratowski. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor b phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 20.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual role for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, Amgen EST Program, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizumoto, K., and Y. Kaziro. 1987. Messenger RNA capping enzyme from eukaryotic cells. Prog. Nucleic Acid Res. Mol. Biol. 34:1-28. [DOI] [PubMed] [Google Scholar]
- 24.Mote, J., Jr., and D. Reines. 1998. Recognition of a human arrests site is conserved between RNA polymerase II and prokaryotic RNA polymerases. J. Biol. Chem. 273:16843-16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moteki, S., and D. Price. 2002. Functional coupling of capping and transcription of mRNA. Mol. Cell 10:599-609. [DOI] [PubMed] [Google Scholar]
- 26.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 27.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639-19648. [DOI] [PubMed] [Google Scholar]
- 28.Pei, Y., B. Schwer, and S. Shuman. 2003. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggests an elongation checkpoint for mRNA quality control. J. Biol. Chem. 278:7180-7188. [DOI] [PubMed] [Google Scholar]
- 29.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase ll. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reines, D., P. Ghanouni, W. Gu, J. Mote, Jr., and W. Powell. 1993. Transcription elongation by RNA polymerase II: mechanism of SII activation. Cell. Mol. Biol. Res. 39:331-338. [PubMed] [Google Scholar]
- 31.Rodriguez, C. R., E.-J. Cho, M.-C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rondon, A. G., M. Garcia-Rubio, S. Gonzalez-Barrera, and A. Aguilera. 2003. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO. J. 22:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawaya, R., and S. Shuman. 2003. Mutational analysis of the guanylyltransferase component of mammalian mRNA capping enzyme. Biochemistry 42:8240-8249. [DOI] [PubMed] [Google Scholar]
- 34.Scafe, C., C. Martin, M. Nonet, S. Podos, S. Okamura, and R. A. Young. 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shatkin, A. J. 1976. Capping of eukaryotic mRNAs. Cell 9:645-653. [DOI] [PubMed] [Google Scholar]
- 37.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shim, E. Y., A. K. Walker, Y. Shi, and K. T. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shuman, S. 1995. Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 50:101-129. [DOI] [PubMed] [Google Scholar]
- 40.Takagi, T., E. J. Cho, R. T. K. Janoo, V. Polodny, Y. Takase, M. C. Keogh, S. A. Woo, L. D. Fresco-Cohen, C. S. Hoffman, and S. Buratowski. 2002. Divergent subunit interactions among fungal mRNA 5′-capping machineries. Eukaryot. Cell 1:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcription elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 42.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effects of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen, Y., and A. J. Shatkin. 1999. Transcriptional elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 13:1774-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wind-Rotolo, M., and D. Reines. 2001. Analysis of gene induction and arrest site transcription in yeast with mutations in the transcription elongation machinery. J. Biol. Chem. 276:11531-11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Honda, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, J., D. E. Awrey, A. M. Edwards, J. Archambault, and J. D. Frisen. 1996. In vitro characterization of mutant RNA polymerase II with reduced binding for elongation factor TFIIS. Proc. Natl. Acad. Sci. USA 93:11552-11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-Tat-activation. J. Mol. Biol. 277:179-197. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao, S., A. Neiman, and G. Prelich. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]