Abstract
Mammalian ATR and ATM checkpoint kinases modulate chromatin structures near DNA breaks by phosphorylating a serine residue in the carboxy-terminal tail SQE motif of histone H2AX. Histone H2A is similarly regulated in Saccharomyces cerevisiae. The phosphorylated forms of H2AX and H2A, known as γ-H2AX and γ-H2A, are thought to be important for DNA repair, although their evolutionarily conserved roles are unknown. Here, we investigate γ-H2A in the fission yeast Schizosaccharomyces pombe. We show that formation of γ-H2A redundantly requires the ATR/ATM-related kinases Rad3 and Tel1. Mutation of the SQE motif to AQE (H2A-AQE) in the two histone H2A genes caused sensitivity to a wide range of genotoxic agents, increased spontaneous DNA damage, and impaired checkpoint maintenance. The H2A-AQE mutations displayed a striking synergistic interaction with rad22Δ (Rad52 homolog) in ionizing radiation (IR) survival. These phenotypes correlated with defective phosphorylation of the checkpoint proteins Crb2 and Chk1 and a failure to recruit large amounts of Crb2 to damaged DNA. Surprisingly, the H2A-AQE mutations substantially suppressed the IR hypersensitivity of crb2Δ cells by a mechanism that required the RecQ-like DNA helicase Rqh1. We propose that γ-H2A modulates checkpoint and DNA repair through large-scale recruitment of Crb2 to damaged DNA. This function correlates with evidence that γ-H2AX regulates recruitment of several BRCA1 carboxyl terminus domain-containing proteins (NBS1, 53BP1, MDC1/NFBD1, and BRCA1) in mammals.
Genomic DNA is assembled into nucleosomes by histones in most eukaryotes (69). DNA-dependent processes, such as replication, repair, and transcription, must therefore be understood in the context of chromatin. Numerous factors that covalently modify the amino-terminal tail (N-tail) or carboxy-terminal tail (C-tail) of histones have been identified (34, 37). These modifications, which include acetylation, methylation, phosphorylation, and ubiquitination, alter the electrostatic charge of histones and may thereby regulate replication, repair, and transcription (34). Additionally, histone modifications can provide specific binding surfaces that recruit protein complexes to DNA. Specific combinations of various histone modifications can create a “histone code” that controls gene expression and higher-order chromosome structure (37).
In mammalian cells, DNA double-strand breaks (DSBs) induce phosphorylation of serine-139 in the C-tail serine-glutamine-glutamate (SQE) motif of the histone variant H2AX (63). This phosphorylation, creating γ-H2AX, is carried out by the phosphatidylinositol 3-kinase-like protein kinases ATM (ataxia-telangiectasia mutated), ATR (ATM- and Rad3-related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit). These kinases are activated in response to DNA DSBs caused by ionizing radiation (IR) and other DNA damaging agents, stalled DNA replication forks, and replication-associated DSBs (63). H2AX is required for efficient accumulation of several BRCA1 carboxyl terminus (BRCT) domain-containing proteins (NBS1, 53BP1, MDC1/NFBD1, and BRCA1) into IR-induced foci (IRIF) that are thought to be sites of DNA damage (7, 16, 23, 29, 60, 73, 74, 76, 89). The initial formation of NBS1, 53BP1, and BRCA1 foci occurs independently of H2AX, but H2AX is required for prolonged maintenance of these foci (15). NBS1, 53BP1, and MDC1 appear to directly bind γ-H2AX (40, 76, 83).
Most studies have used H2AX−/− cell lines to infer the role of γ-H2AX. Specific evidence for the role of γ-H2AX was provided in a recent study that showed that a defect in 53BP1 IRIF formation in H2AX−/− mouse cells was not corrected by transfection of a human H2AX mutant allele in which the serine in the SQE motif was mutated to alanine (83). Another study showed that the ability of NBS1 to form IRIF was lost in mouse cells expressing an H2AX (S136A/S139A) mutant allele, suggesting that phosphorylation events at S136 or S139 are required for NBS1 accumulation at DSBs (15).
Deletion of H2AX in the mouse causes sensitivity to IR at both the cellular and organism level, and H2AX−/− cells experience elevated levels of chromosomal translocations (7, 16). These phenotypes may be associated with a defect in repair of DSBs by homologous recombination (HR), although the nature of this defect has not been defined and repair of DSBs in meiosis does not appear to be defective in H2AX−/− cells (16). Repair of DSBs by nonhomologous end joining (NHEJ) appears to be proficient in H2AX−/− cells (7, 16). H2AX−/− cell lines show defects in G2-M checkpoint responses upon exposure to low doses of IR (16, 23). In addition, condensation and silencing of the sex chromosomes during male meiosis and telomere clustering during meiotic prophase I are defective in H2AX−/− cells (24, 25).
Major H2A species in the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe have an SQE motif in their C-tails (20, 63). Upon DNA damage, the H2A proteins in S. cerevisiae are phosphorylated at serine-129 in the SQE motif (20, 64, 88). This phosphorylation is abolished in a strain that lacks both Mec1 and Tel1, which are the ATR- and ATM-related kinases in budding yeast. It is unknown whether γ-H2A is required for recruitment of proteins to DSBs in budding yeast. Elimination of γ-H2A in S. cerevisiae, either by deletion of the C-tail or by mutating the SQE motif to AQE, imparts sensitivity to drugs that damage DNA, such as methyl methanesulfonate (MMS), phleomycin, bleomycin, and camptothecin (CPT) (20, 64, 88). Unlike mouse H2AX−/− cells, the γ-H2A mutation in S. cerevisiae was originally reported to cause a modest defect in DNA repair by NHEJ, but not HR (20). However, a more recent report found that budding yeast cells carrying the histone H2A-AQE mutation were neither sensitive to bleomycin nor defective in NHEJ (88) and, thus, the role of γ-H2A in DNA repair in S. cerevisiae is uncertain. In addition, checkpoint signaling was reported to be intact in the H2A-AQE mutant yeast cells (20, 64).
The fission yeast S. pombe has been an excellent model system for the investigation of DNA damage response mechanisms. The checkpoint responses in S. pombe require a group of six “checkpoint Rad proteins” (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) that are thought to function as sensors of DNA replication arrest and DNA damage (11). Rad1, Rad9, and Hus1 have weak sequence similarities to proliferating cell nuclear antigen and form a ring-shaped complex (12, 31, 38). Rad17 has sequence similarity to replication factor C (RFC) proteins (32) and associates with other RFC subunits (38). The RFC complex recruits proliferating cell nuclear antigen onto DNA; therefore, it has been proposed that the Rad17 complex loads a Rad1-Rad9-Hus1 complex onto sites of DNA damage. Rad3 is related to mammalian ATR and ATM protein kinases. Rad3 associates with its regulatory subunit Rad26 (22, 87). The protein kinases Chk1 and Cds1 are effectors of the DNA damage and replication checkpoints, respectively (65). These kinases are activated by Rad3-dependent phosphorylation and prevent the onset of mitosis by controlling Cdc25 and Mik1, regulators of the cyclin-dependent kinase Cdc2 (5, 66).
Crb2/Rhp9 is a mediator of the DNA damage checkpoint in fission yeast. It is required for activation of Chk1 by Rad3 (71, 84). We recently reported that yellow fluorescent protein (YFP)-tagged Crb2 forms foci at DSBs (21). Surprisingly, initial Crb2 focus formation was independent of the checkpoint Rad proteins Rad1, Rad3, and Rad17, although retention of Crb2 foci was dependent on these proteins (21). Crb2 has a tandem pair of BRCT domains at its C terminus. This domain structure is shared with Rad9 in S. cerevisiae and 53BP1 in higher eukaryotes, which are thought to be functional homologs of Crb2 (49).
We have investigated γ-H2A and its relationship to DNA repair and checkpoint signaling in fission yeast. We report that γ-H2A is needed for proficient survival of a wide range of DNA damaging agents and for prevention of spontaneous DNA damage. We also report that γ-H2A is required for formation of Crb2 foci, for efficient Rad3-dependent phosphorylation of Crb2 and Chk1, and for prolonged maintenance of a checkpoint arrest. Surprisingly, γ-H2A has negative consequences in survival of crb2Δ cells exposed to IR, suggesting that modification of chromatin structure around a DNA break by γ-H2A modulates a repair mechanism that is facilitated by large-scale accumulation of Crb2.
MATERIALS AND METHODS
Yeast strains.
The fission yeast strains used in this study were constructed by standard techniques (2) and are listed in Table S1 in the supplemental material. Detailed descriptions for construction and origin of various deletion mutations and epitope-tagged strains are described in the Materials and Methods section of the supplemental material. Sequences of DNA primers used for strain construction are listed in supplemental Table S2.
Microscopy.
For visualization of phosphorylated histone H2A by indirect immunofluorescence microscopy, cells were fixed with 3.7% formaldehyde and processed as previously described (42) using anti-phospho-H2AX (serine-139) polyclonal immunoglobulin G (IgG; Upstate Biotechnology) and Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) as primary and secondary antibodies, respectively. Cells were photographed using a Nikon Eclipse E800 microscope equipped with a Photometrix Quantix charge-coupled device camera. For green fluorescent protein (GFP) fluorescence microscopy, images were photographed at eight Z-sections separated by 0.5-μm intervals and projected into one image as previously described (21) using a DeltaVision optical sectioning microscope model 283 equipped with a Photometrix CH350L charge-coupled device camera.
DNA damage sensitivity and checkpoint studies.
Exponentially growing cells were irradiated with IR in YES (yeast extract supplemented) medium (2) with a 3.3-Gy/min 137Cs source at room temperature (∼26°C) and then plated onto YES plates. In NHEJ mutant experiments, cells were synchronized in G1 as described elsewhere (27). For UV irradiation, cells were plated onto YES plates and then irradiated at 254 nm with a Stratalinker (Stratagene). For bleomycin treatment, cells were incubated with 5 mU of bleomycin/ml in YES for the indicated times and then plated onto YES plates. For hydroxyurea (HU), MMS, or CPT treatment, cells were plated onto YES or minimal medium (2) plates containing the indicated concentrations of drugs. For transient HU treatment, cells were incubated in YES with 12 mM HU for the indicated times at 32°C and then plated onto YES plates. Plasmids pAL-SK+ and pAL-Cds1 have been described previously (78). For the NHEJ plasmid repair assay, pAL-SK+ was digested with BamHI or SacI, purified with a QIAGEN column, and used for transformation of exponentially growing cells with a MicroPulser electroporator (Bio-Rad).
For cdc25-22 synchronization studies, cells were incubated at 35°C for 2.5 h to arrest in G2, irradiated with IR at room temperature (∼26°C), fixed with 2.5% glutaraldehyde at various time points, stained with calcofluor, and monitored for cell cycle progression at 25°C by microscopically counting septated cells. For bleomycin checkpoint studies, cells were incubated with 5 mU of bleomycin/ml for 2 h at 25°C, shifted to 35°C, fixed with glutaraldehyde, and stained with calcofluor prior to microscopic observation (2).
For the artificial G2 cell cycle arrest experiments involving adh-wee1-50 cells, which overexpress Wee1-50 protein from the adh1 promoter in a wee1+ background, cells were grown overnight at 36°C, arrested for 2 h at 25°C, irradiated with 1,200 Gy, and then plated onto YES plates at 25°C. Plates were then moved to 36°C at the indicated times after irradiation, and colonies were counted after 3 days.
PFGE.
Agarose plugs for pulsed-field gel electrophoresis (PFGE) were prepared as previously described (58). Chromosomal DNA was fractionated in a 0.7% agarose gel with 0.5× Tris-borate-EDTA buffer at 14°C, using the CHEF-DR II system (Bio-Rad) at 2 V/cm (200 V), a 106° angle, and a pulse time of 1,800 s for 48 h, stained with ethidium bromide, and photographed.
Immunoblot analysis.
Whole-cell extracts were prepared from exponentially growing cells and processed for Western blot analysis as described elsewhere (54). For Chk1-myc detection, a mouse anti-myc antibody (9E10; Santa Cruz Biotechnology) was used. For Crb2 detection, a rabbit polyclonal anti-Crb2 antibody (21) was used. For γ-H2A Western blotting, histone-enriched protein extracts were prepared as previously described and the rabbit polyclonal antibody against S. cerevisiae γ-H2A was used (64).
H2A C-tail peptide in vitro binding assay.
For tandem affinity purification of Crb2, cell extracts were clarified by high-speed centrifugation (12,000 rpm for 30 min; JA-20 rotor; Beckman-Coulter) and then purified as previously described (67). Peptide binding experiments were performed by incubating purified Crb2 from S. pombe with Hta1 C-terminal peptide that was either phosphorylated or unphosphorylated at serine-129 [CRTGKP(p)SQEL] and had been immobilized on SulfoLink coupling gel (Pierce). Bound proteins were isolated by incubating the mixture for 3 h at 4°C in 200 μl of binding buffer (50 mM HEPES-NaOH [pH 7.4], 100 mM Na-acetate, 20 mM NaF, 20 mM glycerol phosphate, 2 mM sodium vanadate, 20 μM pepstatin, 1× Complete Mini protease inhibitor cocktail [Roche], 1% NP-40, 0.5 mg of bovine serum albumin/ml, 1 mM dithiothreitol), washing five times with binding buffer, eluting the proteins with Laemmli buffer, separating them by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotting with anti-Crb2 polyclonal antibody (21).
RESULTS
Formation of γ-H2A in response to DNA damage.
Fission yeast has two histone H2A genes that encode proteins that are 98% identical and have C-terminal SQE motifs (Fig. 1A) (18, 48). These motifs were mutated to AQE (hta1-S129A and hta2-S128A) by gene replacement (see the Materials and Methods section of the supplemental material). Cells carrying either single or double H2A-AQE mutations showed no obvious growth defect compared to wild-type cells (see below).
FIG. 1.
Fission yeast histone H2A is phosphorylated in response to IR. (A) S. pombe histone H2A genes and H2A-AQE mutations. (Top) Organization of histone H2A genes (hta1+ and hta2+). hta1+ is paired to the histone H2B gene (htb1+). AQE mutations are indicated (S129A and S128A). (Bottom) Alignment of histone H2A C-tail sequences from human (H2AX), mouse (H2AX), Tetrahymena thermophila (H2A.1), S. cerevisiae (H2A.1 and H2A.2), and S. pombe (H2A.1 and H2A.2). Conserved amino acids are shaded, and the serine in the SQE motif is indicated with an arrow. (B) H2A phosphorylation detected by Western blotting. Histone-enriched cell extracts were prepared from unirradiated (−) and irradiated (+) S. pombe (S.p.) or S. cerevisiae (S.c.) cells and separated by sodium dodecyl sulfate-13.5% polyacrylamide gel electrophoresis. γ-H2A signals were detected using the budding yeast γ-H2A peptide antibody (right). The same protein extracts used in the Western blot assay were loaded on another gel and visualized by Coomassie blue staining (left). (C) H2A phosphorylation detected by immunofluorescence microscopy. Unirradiated (-IR) and irradiated (+IR; 192 Gy) S. pombe cells were fixed with formaldehyde immediately (less than 5 min) after irradiation and processed for indirect immunofluorescence microscopy using mammalian γ-H2AX peptide antibody. (D and E) The percentages of cells with γ-H2A signal were determined from three independent experiments with at least 500 cells counted microscopically for each IR dose or each time after IR. Error bars indicate standard deviations. Strains used were wild type (TMN3289), hta1-S129A hta2-S128A (TMN3291), rad3Δ tel1Δ (TMN2978), rad3Δ (TMN2947), tel1Δ (TMN2967), and rhp51Δ (PS2383).
We tested whether the SQE motifs were phosphorylated in response to DNA damage. Western blot analysis with an antibody that recognizes S. cerevisiae γ-H2A detected an S. pombe protein that migrated at the position expected of H2A (Fig. 1B). As expected for γ-H2A, this signal strongly increased in IR-treated cells. In immunofluorescence studies, an antibody that recognizes mammalian γ-H2AX detected bright nuclear signals in wild-type S. pombe cells immediately after irradiation (Fig. 1C to E). As expected for γ-H2A, these signals were not observed in unirradiated wild-type or irradiated hta1-S129A hta2-S128A cells. The percentage of cells with these signals increased at higher doses of IR (Fig. 1D), and the signals disappeared at a slower rate in rhp51Δ cells that were defective in HR repair (Fig. 1E). These observations indicated that γ-H2A is rapidly generated in response to DNA damage and disappears upon completion of repair.
Rad3 and Tel1 regulate H2A phosphorylation.
In S. cerevisiae, γ-H2A is generated by Mec1 and Tel1, which are ATR- and ATM-related kinases, respectively (20, 64). We tested if Rad3 (ATR/Mec1 homolog) and Tel1 (ATM/Tel1 homolog) were involved in generation of γ-H2A in fission yeast. The γ-H2A signal was reduced in rad3Δ and tel1Δ single mutants, with a greater reduction observed in rad3Δ cells, and it was absent in the rad3Δ tel1Δ double mutant (Fig. 1C and D). These data indicated that both Rad3 and Tel1 generate γ-H2A in response to IR, with Rad3 having the greater role.
Histone H2A-AQE cells are sensitive to DNA damage.
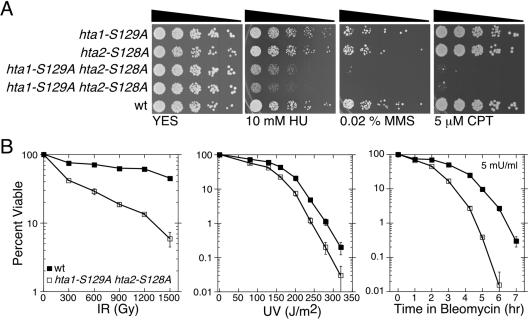
Single-mutant hta1-S129A or hta2-S128A cells were not abnormally sensitive to a variety of DNA damaging agents, but double mutant cells displayed increased sensitivity to the DNA replication inhibitor HU, the DNA alkylating agent MMS, the topoisomerase I inhibitor CPT, IR, UV light, and the radiomimetic DNA-cleaving agent bleomycin (Fig. 2). Therefore, γ-H2A is needed for proficient survival in a wide variety of DNA damaging agents in S. pombe.
FIG. 2.
Histone H2A C-tail SQE motif is important for survival in response to DNA damage. (A) Fivefold serial dilutions of wild-type and various histone H2A-AQE mutant S. pombe cells (hta1-S129A and/or hta2-S128A) were plated onto YES medium with the indicated concentrations of HU, MMS, or CPT. Pictures were taken after 4 days at 32°C. Strains used were hta1-S129A (TMN3294), hta2-S128A (TMN3295), hta1-S129A hta2-S128A (TMN3292 and TMN3293), and wild type (TMN2665). (B) Survival of wild-type and H2A-AQE mutant cells after exposure to the indicated doses of IR, UV, or incubation with 5 mU of bleomycin/ml for the indicated lengths of time. Strains used were wild type (TMN3296) and hta1-S129A hta2-S128A (TMN3293). Error bars indicate standard deviations from at least three experiments.
Epistasis analysis involving H2A-AQE and DNA repair mutations.
Genetic epistasis studies were performed to investigate why H2A-AQE cells are sensitive to DNA damage. Deletion mutations of genes involved in DNA repair by HR (rad22Δ, rhp51Δ, rad32Δ, and rqh1Δ) or NHEJ (pku70Δ and lig4Δ) were combined with the H2A-AQE mutations and tested for IR sensitivity. Rad22 and Rhp51 are homologs of Rad52 and Rad51, respectively (55, 77, 82). Rad32 is an Mre11 homolog that forms a complex with Rad50 and Nbs1 (17, 80). Rqh1 is a RecQ-like DNA helicase whose homologs are Sgs1 in S. cerevisiae and Werner's syndrome helicase (WRN) and Bloom's syndrome helicase (BLM) in mammalian cells (56, 75). Pku70 and Lig4 are homologs of KU70 and DNA ligase 4, respectively (46). The sensitivity of NHEJ mutants was tested with G1-synchronized cultures, because pku70Δ and lig4Δ cells show sensitivity to IR only in G1 phase (46).
H2A-AQE mutations made rad22Δ, rhp51Δ, rad32Δ, rqh1Δ, pku70Δ, lig4Δ, and rhp51Δ pku70Δ cells all more sensitive to IR (Fig. 3A to D). These data showed that γ-H2A is not specifically involved in only HR or NHEJ. These findings were generally consistent with studies in S. cerevisiae that showed that histone H2A C-tail SQEL truncation mutations displayed additive sensitivity with mutations that cause defects in HR (RAD52) or NHEJ (YKU70) for DNA damage caused by continuous exposure to MMS (20). Interestingly, rad22Δ showed a very strong synergistic genetic interaction with H2A-AQE for IR sensitivity. The rad22Δ hta1-S129A hta2-S128A strain was substantially more sensitive to IR than rhp51Δ cells or even the rhp51Δ hta1-S129A hta2-S128A strain (Fig. 3A). While both Rhp51 (Rad51) and Rad22 (Rad52) are important for HR repair, a recent study in S. cerevisiae has shown that Rad52 has roles in three distinct stages during HR: (i) a presynaptic role prior to Rad51 filament assembly on single-stranded DNA, (ii) a synaptic role together with Rad51 filament, and (iii) a postsynaptic role after Rad51 filament disassembly (51). Histone H2A-AQE mutations might affect any of these steps, but further studies are necessary to understand the synergistic interaction between rad22Δ and H2A-AQE.
FIG. 3.
Genetic interactions of histone H2A-AQE mutations with mutations in DNA repair genes. (A to D) Survival of cells carrying various combinations of mutations in H2A (hta1-S129A and hta2-S128A, indicated as hta1 hta2) and DNA repair (rad22Δ, rhp51Δ, rad32Δ, rqh1Δ, pku70Δ, and lig4Δ) genes after exposure to the indicated doses of IR. For data in panels C and D, G1 indicates that cells were arrested in G1 by nitrogen starvation for 48 h prior to IR treatment (see Materials and Methods), while G2 indicates exponentially growing cells. Strains used were rad22Δ (TMN3317), rad22Δ hta1 hta2 (TMN3318), rhp51Δ (TMN3319), rhp51Δ hta1 hta2 (TMN3320), rqh1Δ (TMN3301), rqh1Δ hta1 hta2 (TMN3302), rad32Δ (TMN2800), rad32Δ hta1 hta2 (TMN3321), wild type (TMN2665), hta1 hta2 (TMN3293), pku70Δ (TMN3001), pku70Δ hta1 hta2 (TMN3322), lig4Δ (PS2818), lig4Δ hta1 hta2 (TMN3323), rhp51Δ pku70Δ (TMN3419), and rhp51Δ pku70Δ hta1 hta2 (TMN3420). Error bars indicate standard deviations from at least three experiments. (E) A plasmid repair assay to measure NHEJ efficiency was carried out for the indicated strains. Relative colony numbers obtained from transformation with cut versus uncut plasmid are plotted. Strains used were wild type (TMN2663), hta1 hta2 (TMN3292), pku70Δ (TMN3001), pku70Δ hta1 hta2 (TMN3322), lig4Δ (PS2818), and lig4Δ hta1 hta2 (TMN3323). Error bars indicate standard deviations from at least three experiments. (F) Generation times for indicated strains, determined from two independent experiments. Error bars indicate differences between two experiments. Strains used were the same as those used for panels A and B, except for wild-type (TMN3296), hta1 hta2 (TMN3291), rad22Δ (TMN3460), and rad32Δ (TMN3461) strains.
We also tested NHEJ efficiency using a standard assay that compares the number of colonies obtained from transformation of linear versus circular plasmid DNA. Efficient transformation of the linear plasmid requires ligation of DNA by NHEJ and, therefore, a defect in NHEJ can be detected as a reduction in the relative linear and circular plasmid transformation efficiencies. As shown in Fig. 3E, there was no reduction in the relative linear plasmid transformation efficiency in H2A-AQE cells, suggesting that there is no major NHEJ defect for H2A-AQE mutants in S. pombe. In fact, ligation efficiency was increased relative to wild type with BamHI-digested plasmid (Fig. 3E). This effect was also observed in pku70Δ cells, but not in lig4Δ cells. These results suggested that the loss of γ-H2A stimulates Ku-independent and DNA ligase 4-dependent religation of the linear plasmid.
H2AX−/− mouse embryo fibroblast cells exhibited reduced repair of IR-induced DSBs, as assayed by PFGE of DNA (16). We carried out a similar analysis of H2A-AQE cells by monitoring the reappearance of intact chromosomes after IR. No obvious defect in DNA repair rate was observed in H2A-AQE cells (Fig. 4A), although small differences in repair rate or quality cannot be detected by PFGE. The rate of DNA repair, as monitored by the time-dependent disappearance of IR-induced nuclear foci of the HR protein Rad22 tagged with YFP (Fig. 4B), also showed no obvious difference between wild-type and H2A-AQE cells. However, H2A-AQE cells showed a twofold increase in the frequency of spontaneous Rad22-YFP foci (Fig. 4B). A large proportion of these foci were brighter than those seen in wild-type cells (Fig. 4B), suggesting recruitment of a greater number of Rad22 molecules to DNA breaks in H2A-AQE cells.
FIG. 4.
DNA repair rate for H2A-AQE mutant cells is essentially the same as for wild-type cells. (A) DNA repair rates of wild-type and hta1-S129A hta2-S128A cells after exposure to IR (192 Gy) were monitored by visualizing reappearance of intact chromosomes after the indicated times by PFGE. Strains used were wild type (TMN2665) and hta1 hta2 (TMN3293). (B) IRIF formation of Rad22 is not affected by H2A-AQE mutations. Time-dependent changes in Rad22-YFP foci after 36-Gy IR for YFP-rad22 (TMN3333) and YFP-rad22 hta1-S129 hta2-S128A (TMN3334) cells are plotted on the right. Representative changes in rad22-YFP localization patterns in response to IR (45 min past IR) are shown in the left panels.
The increase in the number of spontaneous Rad22-YFP foci in H2A-AQE cells was indicative of a higher frequency of DNA replication errors or genomic instability. Consistent with such possibilities, we found that H2A-AQE mutations substantially reduced the growth rate in rad22Δ, rhp51Δ, rad32Δ, and rqh1Δ genetic backgrounds (Fig. 3F). These genetic interactions presumably reflect a need for DNA repair to cope with elevated spontaneous DNA damage in H2A-AQE cells.
Formation of Crb2 foci at DSBs requires γ-H2A.
In view of evidence that H2AX is required for efficient accumulation of 53BP1 and several other BRCT domain-containing proteins at IRIF in mouse cells (7, 15, 16, 23, 29, 60, 73, 74, 76, 89), we decided to test if focus formation by the BRCT domain protein Crb2 required γ-H2A in S. pombe. We observed that IR-induced Crb2 foci were almost completely lost in H2A-AQE cells (Fig. 5A and B). These observations suggested that Rad3- and Tel1-dependent generation of γ-H2A is necessary for formation of Crb2 foci. Indeed, IR-induced Crb2 foci were abolished in rad3Δ tel1Δ cells but remained in the single mutants (Fig. 5A). It is worth noting that rad3Δ, which had a greater effect on formation of γ-H2A signals (Fig. 1D), also had a greater impact on Crb2 foci retention after IR treatment (Fig. 5B).
FIG. 5.
γ-H2A mediates accumulation of Crb2 at sites of DNA DSBs. (A) YFP-Crb2 focus formation is dependent on γ-H2A generated by Rad3 and Tel1 kinases. YFP-Crb2 localization was observed immediately after irradiation with IR (36 Gy). Strains used were wild type (LLD3260), hta1 hta2 (LLD3340), rad3Δ tel1Δ (LLD3341), rad3Δ (LLD3342), and tel1Δ (LLD3343). (B) Time-dependent change in Crb2 foci after exposure to IR (36 Gy). The number of cells with Crb2 foci at various times after IR treatment was counted and plotted as a percentage of cells with foci. (C) In vitro binding of partially purified Crb2 protein (from LLD3344) to the phosphorylated H2A C-tail peptide. Phosphorylated or unphosphorylated H2A C-tail peptide was conjugated to agarose beads and incubated with Crb2 protein prepared by two-step purification with IgG and calmodulin beads. Peptide-bound Crb2 was then visualized by Western blotting using polyclonal anti-Crb2 antibody. (D) IRIF formation by Rad26 and Rad32 is not affected by H2A-AQE mutations. Cells were irradiated with 180 Gy for YFP-Rad26 cells (TMN3337 and TMN3338) and 120 Gy for Rad32-GFP cells (LLD3335 and TMN3336) and observed immediately after irradiation.
Loss of IRIF in the H2A-AQE mutant was specific to Crb2, as Rad26 (homolog of mammalian ATRIP and S. cerevisiae Lcd1), Rad22 (Rad52 homolog), and Rad32 (Mre11 homolog) formed IRIF in H2A-AQE cells (Fig. 4B and 5D). Direct interaction between the phosphorylated H2A C-tail and Crb2 might be responsible for Crb2 recruitment, as we observed the phosphorylation-dependent interaction of partially purified Crb2 protein and the H2A C-tail peptide in vitro (Fig. 5C). Similar studies of 53BP1 and MDC1 have indicated that these proteins might directly bind to γ-H2AX (76, 83).
Histone H2A-AQE mutations impair checkpoint maintenance.
Our finding that focus formation by the checkpoint adaptor Crb2 is defective in H2A-AQE cells prompted an examination of checkpoint signaling in the histone H2A-AQE mutants. We observed a reproducible reduction in Chk1 phosphorylation and an even greater reduction in Crb2 phosphorylation at lower doses of IR (Fig. 6A). These results correlated with evidence of inefficient phosphorylation of 53BP1 in H2AX−/− mouse cells at lower doses of IR (23).
FIG. 6.
Histone H2A-AQE mutant cells show defects in checkpoint signaling. (A) H2A-AQE mutations cause reduced Rad3-dependent phosphorylation of Chk1 and Crb2 in response to IR. Extracts were prepared from cells exposed to the indicated doses of IR immediately after irradiation and visualized by Western blotting. Arrows indicate phosphorylated forms of Chk1 and Crb2. Strains used were chk1+ (TMN2665), chk1-myc (TMN3309), chk1-myc hta1 hta2 (TMN3310), chk1-myc rad3Δ (NR2648), crb2Δ (LLD3339), crb2+ (KS1599), crb2+ hta1 hta2 (TMN3290), and crb2+ rad3Δ (TMN2938). (B) H2A-AQE mutant cells prematurely reentered the cell cycle after irradiation with high-dose IR. Wild-type (cdc25-22; TMN3311) or H2A-AQE mutant (cdc25-22 hta1-S129A hta2-S128A; TMN3312) cells were synchronized at 35°C for 2.5 h, exposed to the indicated doses of IR at room temperature, fixed with glutaraldehyde, and stained with calcoflour, and the percentage of septated cells was counted for each time point. A schematic drawing of the S. pombe cell cycle with the arrest points for cdc25-22 and adh-wee1-50 cells is indicated on the right. (C) Artificial cell cycle arrest of the H2-AQE mutant can rescue only a small fraction of cells from IR-induced DNA damage. The experimental scheme is drawn on the right. Cells expressing the constitutive temperature-sensitive allele (wee1-50) of Cdc2 inhibitor Wee1 were arrested at 25°C 2 h prior to IR (1,200 Gy) and held arrested during irradiation. After completion of IR treatment, cells were plated at 25°C and then shifted to permissive temperature (36°C) at the indicated time. Wild-type (adh-wee1-50; TMN2713) and hta1 hta2 adh-wee1-50 (TMN3421) cells were used.
We also tested if checkpoint arrest in response to IR is defective in H2A-AQE cells. The temperature-sensitive cdc25-22 mutation was used to synchronize cells at the G2-M boundary. These cells were irradiated with IR and then shifted to permissive temperature. We found no difference in the duration of the IR-induced cell cycle arrest after exposure to 96-Gy IR (Fig. 6B). However, histone H2A-AQE mutant cells showed premature release from cell cycle arrest after exposure to 300- and 600-Gy IR. Therefore, γ-H2A is important for prolonged checkpoint arrest in response to IR. However, artificial cell cycle arrest after IR treatment, enforced by overexpression of the Cdc2 inhibitor Wee1 kinase, only partially rescued the IR sensitivity of H2A-AQE cells (Fig. 6C). Therefore, the IR sensitivity of H2A-AQE cells cannot be attributed solely to premature reentry into the cell cycle.
Since cdc25-22 experiments suggested that H2A-AQE cells are less effective in maintaining prolonged cell cycle arrest, we decided to monitor checkpoint arrest in the presence of persistent DNA damage, generated by continuous treatment of cells with bleomycin. As expected, the H2A-AQE cells prematurely resumed division in the presence of bleomycin (Fig. 7A, top panels). This result is consistent with the idea that H2A-AQE cells are defective in checkpoint maintenance.
FIG. 7.
H2A-AQE mutant cells show a defect in checkpoint maintenance under continuous exposure to bleomycin. (A) Wild-type, H2A-AQE (hta1-S129A hta2-S128A), or tel1Δ cells in either a rad3+ or rad3ts (rad3ts-V2217A) background were incubated with bleomycin for 2 h at 25°C, shifted to 35°C, fixed with glutaraldehyde, and stained with calcofluor, and the percentage of septated cells was counted for each time point. The micrographic pictures on the right are representative cells from the experiments graphed on the left. Cells were fixed and stained with 4,6-diamidino-2-phenylindole (DAPI) to visualize DNA and septa. Strains used were wild type (TMN2665), hta1 hta2 (TMN3293), rad3ts (TMN3313), and rad3ts hta1 hta2 (TMN3314). (B) Cells with integrated chk1-myc in either wild-type (TMN3309), hta1-S129A hta2-S128A (TMN3310), rad3ts (TMN3315), or rad3ts hta1-S129A hta2-S128A (TMN3316) backgrounds were incubated with bleomycin for 2 h at 25°C and then shifted to 35°C. Aliquots were taken at the indicated time points after the temperature shift, and cell extracts were prepared. Chk1-myc was visualized by Western blotting. Arrows indicate phosphorylated forms of Chk1.
We also tested how cell cycle arrest maintenance was affected by inactivation of Rad3 kinase after a checkpoint arrest had been established. A previous study has shown that cells can maintain G2 arrest if Rad3 is inactivated after DNA damage, suggesting that Rad3 is required for activation of checkpoint but is dispensable for maintenance of checkpoint (47). Temperature-sensitive rad3 cells (rad3-A2217Vts, hereafter abbreviated as rad3ts) (47) were first incubated with bleomycin at permissive temperature (25°C) and then shifted to nonpermissive temperature (35°C). We found that rad3ts cells were able to maintain the cell cycle arrest for 90 min after inactivation of Rad3 (Fig. 7A). In contrast, rad3ts hta1-S129A hta2-S128A cells immediately resumed division after the shift to 35°C (Fig. 7A, middle panels). This effect correlated with premature loss of Chk1 phosphorylation in rad3ts hta1-S129A hta2-S128A cells compared to rad3ts cells (Fig. 7B), although division occurred before complete dephosphorylation of Chk1, as was also observed in previous studies (47). These data underscored the importance of γ-H2A in checkpoint arrest maintenance in the absence of Rad3 activity.
We also tested the role of Tel1 in maintaining a checkpoint arrest after Rad3 inactivation in the presence of bleomycin (Fig. 7A, bottom panels). Lack of Tel1 accelerated cell cycle reentry after Rad3 inactivation, but its effect was smaller relative to the effects of H2A-AQE mutations. To our knowledge, this is the first demonstration that Tel1 can contribute to cell cycle arrest in S. pombe.
In S. pombe, Mrc1 and Cds1 define a checkpoint pathway that responds to replication fork arrest (1, 78). Phosphorylation of Mrc1 by Rad3 is required for Rad3-dependent activation of Cds1 (91). HU sensitivity assays in mrc1Δ or cds1Δ genetic backgrounds can reveal defects in the DNA damage checkpoint. Cells defective in both DNA replication checkpoint (Mrc1-Cds1) and DNA damage checkpoint (Crb2-Chk1) pathways show greater sensitivity to HU and exhibit much greater occurrences of premature mitosis in the presence of HU (“cut” phenotype) than cells with only one of these two pathways inactivated (78). We found that H2A-AQE mutations enhanced the HU-sensitive phenotype in mrc1Δ and cds1Δ backgrounds (Fig. 8A and C). These cells also displayed a much higher occurrence of the cut phenotype than wild-type, hta1-S129A hta2-S128A, cds1Δ, or mrc1Δ cells (Fig. 8B and data not shown). These data further supported the idea that the H2A-AQE mutations caused a defect in the DNA damage checkpoint. Overexpression of Cds1 rescued HU sensitivity in both mrc1Δ and mrc1Δ hta1-S129A hta2-S128A cells (Fig. 8C). These data were consistent with the notion that γ-H2A participates in the Crb2-Chk1 checkpoint pathway and has little or no role in the Mrc1-Cds1 pathway.
FIG. 8.
H2A-AQE cells show defects in HU-induced cell cycle arrest in the absence of S-phase checkpoint proteins. (A) H2A-AQE mutations make cells with mutations in the S-phase checkpoint (mrc1Δ and cds1Δ), but not those with mutations in the DNA damage checkpoint (crb2Δ and chk1Δ), more sensitive to HU. Viability of cells with indicated genotypes was measured by counting colony formation on YES plates after treatment with 12 mM HU for the indicated lengths of time. Strains used were wild type (TMN2665), hta1 hta2 (TMN3293), crb2Δ (TMN2941), crb2Δ hta1 hta2 (TMN3297), chk1Δ (TMN2943), chk1Δ hta1 hta2 (TMN3298), mrc1Δ (TMN3326), mrc1Δ hta1 hta2 (TMN3327), cds1Δ (TMN2945), cds1Δ hta1 hta2 (TMN3324), mrc1Δ chk1Δ (TMN3328), and cds1Δ chk1Δ (TMN3325). (B) Cells of the indicated genotypes (the same strains as in panel A) were mock treated or treated with 12 mM HU for 6 h and then stained with DAPI. Increased occurrence of the cut phenotype in mrc1Δ hta1-S129A hta2-S128A and cds1Δ hta1-S129A hta2-S128A cells, but not in chk1Δ hta1-S129A hta2-S128A cells, is consistent with a defect in DNA damage checkpoint enforced by Chk1. (C) Fivefold serial dilutions of mrc1Δ and mrc1Δ hta1-S129A hta2-S128A cells carrying either the control pAL-SK+ plasmid or the Cds1 overexpression plasmid pAL-Cds1 (TMN3329, TMN3330, TMN3331, and TMN3332) were plated on minimal medium with the indicated concentrations of HU. Pictures were taken after 6 days at 32°C. (D) Fivefold serial dilutions of cells with the indicated genotypes (the same strains as used in panel A) were plated onto YES with the indicated concentrations of CPT. Pictures were taken after 4 days at 32°C.
The H2A-AQE mutations also showed strong synergistic reduction in cell viability in response to CPT treatment for mrc1Δ compared to that in cds1Δ or chk1Δ cells (Fig. 8D). This observation suggests that Mrc1 may have additional Cds1-independent functions in response to DNA replication stress and DNA damage.
γ-H2A and Crb2 are important modulators for Rqh1-dependent DNA repair.
Evidence for the role of γ-H2A in checkpoint signaling prompted epistasis studies involving checkpoint mutations (rad3Δ, crb2Δ, and chk1Δ) and the H2A-AQE mutations. The H2A-AQE mutations did not increase IR sensitivity in a rad3Δ background (Fig. 9A). These observations contrasted with the additive interactions involving the H2A-AQE mutations in rhp51Δ and rad32Δ cells (Fig. 3A and B), whose IR hypersensitivity is comparable to that of rad3Δ cells. These findings indicated that γ-H2A functions in a Rad3-dependent pathway. The rad3Δ mutation reduced but did not eliminate γ-H2A (Fig. 1C and D). Therefore, the epistatic relationship involving rad3Δ and H2A-AQE mutations with respect to IR sensitivity cannot simply be explained by loss of γ-H2A in rad3Δ cells.
FIG. 9.
Genetic interactions of histone H2A-AQE mutations with mutations in DNA damage checkpoint genes. (A to D) Survival of cells carrying various combinations of mutations in H2A (hta1-S129A and hta2-S128A, indicated as hta1 hta2), checkpoint (rad3Δ, crb2Δ, and chk1Δ), and DNA repair (rqh1Δ) genes after exposure to the indicated dose of IR. Strains used were rad3Δ (TMN2947), rad3Δ hta1 hta2 (TMN3299), rad3Δ crb2Δ (TMN3300), crb2Δ (TMN2941), crb2Δ hta1 hta2 (TMN3297), chk1Δ (TMN2943), chk1Δ hta1 hta2 (TMN3298), crb2Δ chk1Δ (TMN3305), crb2Δ chk1Δ hta1 hta2 (TMN3308), rqh1Δ (TMN3301), rqh1Δ hta1 hta2 (TMN3302), crb2Δ rqh1Δ (TMN3303), crb2Δ rqh1Δ hta1 hta2 (TMN3307), chk1Δ rqh1Δ (TMN3304), chk1Δ rqh1Δ hta1 hta2 (TMN3422), and crb2Δ chk1Δ rqh1Δ (TMN3306). Error bars indicate standard deviations from at least three experiments. (E) Model depicting the γ-H2A-dependent modulation of Crb2 functions involving Chk1 and Rqh1, based on genetic interactions of IR sensitivity. (F) Model for the mammalian γ-H2AX-dependent DNA damage response. A speculative negative role of the γ-H2AX in DNA repair is indicated as a gray inhibitory sign covered with question marks.
Analysis of mutant combinations of crb2Δ and chk1Δ with histone H2A-AQE mutations yielded unexpected results. In IR sensitivity assays, crb2Δ cells were more sensitive than chk1Δ cells, consistent with earlier UV sensitivity studies (84). Surprisingly, the H2A-AQE mutations ameliorated the IR sensitivity of crb2Δ cells (Fig. 9B). The crb2Δ hta1-S129A hta2-S128A cells were similar to chk1Δ or chk1Δ hta1-S129A hta2-S128A cells in IR survival assays (Fig. 9B). Analysis of chk1Δ crb2Δ hta1-S129A hta2-S128A cells demonstrated that the rescue of crb2Δ IR sensitivity by H2A-AQE mutations did not require Chk1 (Fig. 9B). These results indicated that C-tail phosphorylation of histone H2A has negative consequences in the absence of Crb2. Given that H2A-AQE mutations eliminate IRIF formation by Crb2 (Fig. 5A and B) and H2A-AQE mutations show strong synergistic interaction with rad22Δ (Fig. 3A), these observations suggest that Crb2 accumulation at DNA damage sites controls how the DNA damage is repaired.
Crb2 has been implicated in regulation of Rqh1 DNA helicase (13). Therefore, we tested how the rqh1Δ mutation affects IR sensitivity of crb2Δ and H2A-AQE mutant cells. Based on IR sensitivity, Rqh1 and Crb2 appeared to work in the same pathway, as double mutant rhq1Δ crb2Δ cells showed no additional sensitivity (Fig. 9C). In addition, crb2Δ rqh1Δ hta1-S129A hta2-S128A cells were as IR sensitive as crb2Δ cells. Thus, the rescue of crb2Δ IR sensitivity by the H2A-AQE mutations requires Rqh1. Moreover, crb2Δ, chk1Δ rqh1Δ, chk1Δ rqh1Δ crb2Δ, and chk1Δ rqh1Δ hta1-S129A hta2-S128A cells were approximately equally sensitive to IR (Fig. 9C and D). These findings indicated that Crb2's function in response to IR can largely and perhaps entirely be explained by regulation of two pathways represented by Rqh1 and Chk1. Furthermore, in the absence of Crb2 protein, γ-H2A appears to interfere with a DNA repair pathway that is dependent on Rqh1 (Fig. 9E).
DISCUSSION
Prior to these studies, it was known that C-terminal phosphorylation of histone H2AX and H2A occurred in mammals and budding yeast, but it was unclear whether the functional consequences of this phosphorylation were conserved. We have shown that C-terminal phosphorylation of histone H2A is conserved in fission yeast, is induced by DNA damage, and is controlled by Rad3 and Tel1. The functional importance of this phosphorylation is evident from the sensitivity of H2A-AQE mutants to a broad range of genotoxic agents, the increased rate of spontaneous DNA damage in these cells, and the genetic interactions with mutations in DNA repair genes. The inability of H2A-AQE cells to form Crb2 foci in response to DNA damage is strikingly similar to the behavior of certain BRCT domain checkpoint proteins in mouse H2AX−/− cells, including 53BP1, the apparent Crb2 functional homolog. These findings establish that γ-H2A in fission yeast and γ-H2AX in mammals have substantially conserved functions and validate yeasts as useful model systems for investigating γ-H2AX.
Generalized role for γ-H2A in survival of DNA damage.
Previous studies of γ-H2AX and γ-H2A used different types of H2AX/H2A mutations that deleted the entire gene (in the case of mouse H2AX), truncated the C terminus of the protein, or mutated one or more phosphorylation sites (7, 15, 16, 20, 64, 88). The use of different mutations and strain backgrounds has complicated the analysis of γ-H2A in budding yeast. For example, γ-H2A has been variously reported to be important or unimportant for resistance to the radiomimetic DNA-cleaving agent bleomycin and for repair of broken DNA by NHEJ (20, 88). Budding yeast H2A-AQE mutants were also reported to be sensitive to MMS and CPT but insensitive to ethyl methanesulfonate and UV (20, 64). Mouse embryonic fibroblast H2AX−/− cells were reported to be sensitive to IR, bleomycin, MMS, and CPT but not UV (7, 14, 16, 28, 61), although the specific requirement for γ-H2A has only been tested for IR survival (14).
To avoid these problems, we precisely mutated the C-terminal SQE motif to AQE in the genomic copies of hta1+ and hta2+ in S. pombe. Unlike studies in other organisms, we have found that these mutant cells are sensitive to a wide variety of genotoxic agents (Fig. 2). This broad-spectrum effect suggests that γ-H2A has a general role in DNA repair and checkpoint signaling. This conclusion is consistent with the evidence that fission yeast mutants defective in checkpoint signaling and HR repair are sensitive to many different types of DNA damage (10).
It should be understood that while the DNA damage survival effects of H2A-AQE mutations are both broad and highly reproducible, they are nevertheless rather modest when compared to the profound sensitivities of some checkpoint and DNA repair mutants (Fig. 3 and 9). This fact demonstrates that there is a strong selective pressure for optimum operation of genome maintenance mechanisms. Even a modest reduction in these mechanisms can severely degrade organism fitness. Thus, it should come as no surprise that there has been strong selective pressure for conservation of γ-H2A even though it makes only a relatively modest contribution in acute DNA damage survival assays.
Genomic instability in H2A-AQE mutants.
There was an approximately twofold increase in spontaneous Rad22-YFP foci in H2A-AQE cells (Fig. 4B). These findings suggest that γ-H2A promotes genomic stability under normal growth conditions. This hypothesis is consistent with the observation that H2A-AQE mutations cause a synergistic growth rate reduction in various DNA repair mutant backgrounds (Fig. 3F). This function of γ-H2A may be evolutionarily conserved, because H2AX−/− mouse cells display increased spontaneous DNA breaks (7, 16). Moreover, elimination of the tumor suppressor p53 in H2AX−/− cells caused a synergistic enhancement of genomic instability and tumor susceptibility (8, 14). Whether γ-H2A is required to prevent DNA damage, or is only needed for efficient repair of spontaneous DNA damage, is a question that cannot be answered from our studies. However, the unusual intensity of the Rad22-YFP foci in H2A-AQE cells suggests that the DNA damage is persistent and may involve formation of long single-stranded DNA regions that are bound by Rad22.
Genetic interactions involving rad22Δ and pku70Δ.
Most DNA repair mutations that impair HR and NHEJ showed additive interactions with H2A-AQE mutations in IR sensitivity assays (Fig. 3 and 9), indicating that γ-H2A does not fit neatly into a single pathway of DNA repair. One interesting exception to this pattern was rad22Δ, which showed a strong synergistic interaction with H2A-AQE mutations (Fig. 3A). The reason for this interaction is unknown, but one possibility is that γ-H2A participates in an HR repair pathway that involves Rti1/Rad22B, a second Rad52 homolog in fission yeast (77, 82). In budding yeast, the H2A C-tail SQEL truncation mutations showed strong synergistic hypersensitivities with rad52 with respect to continued exposure to MMS or HO endonuclease (20). It will be interesting to determine whether rad51Δ and rad52Δ mutations display different genetic interactions with H2A-AQE mutations in S. cerevisiae.
The plasmid-based ligation assay to monitor NHEJ in H2A-AQE cells also yielded interesting results (Fig. 3E). As noted above, there are conflicting observations regarding the role of γ-H2A in NHEJ in S. cerevisiae (20, 88). Our assays resemble those of Wyatt et al. (88), in that we repeatedly observed that ligation of BamHI-digested plasmid was more efficient in H2A-AQE cells (Fig. 3E). More surprising was the observation that the H2A-AQE mutations substantially rescued the plasmid ligation defect in pku70Δ cells but not in lig4Δ cells. These results suggest that Ku-independent ligation of the linear plasmid was enhanced in the absence of γ-H2A. These observations might relate to evidence in S. cerevisiae of a Ku-independent and DNA ligase 4-dependent mechanism for DNA DSB repair, termed microhomology-mediated end joining (44). Our results are intriguing in light of previous findings that plasmid ligation is increased in crb2Δ, rad22Δ, and rqh1Δ cells but not in rhp51Δ or rad32Δ cells (86). Given that H2A-AQE mutations interact synergistically with rad22Δ and rescue crb2Δ IR sensitivity in a Rqh1-dependent manner (Fig. 3A and 9C), the increase in linear plasmid transformation efficiency in H2A-AQE cells suggests that regulation of Crb2 accumulation at DSBs by γ-H2A might modulate utilization of HR and NHEJ pathways. Crb2 was previously proposed to influence repair pathway choice and to regulate Rqh1 (13).
The relationship among Crb2, Rqh1, and γ-H2A.
Our genetic interaction studies involving H2A-AQE mutations and mutations in DNA damage checkpoint proteins revealed a surprising negative role for γ-H2A in crb2Δ cell survival after IR treatment (Fig. 9). We also established that the rescue of crb2Δ IR sensitivity by H2A-AQE mutations require Rqh1 DNA helicase. The data support the idea that in wild-type cells, the functions of Rqh1 in IR survival require Crb2. In the absence of Crb2, γ-H2A in some ways interferes with an activity of Rqh1 that contributes to IR survival (Fig. 9E). Mutations in mammalian homologs of Rqh1 helicase, such as Werner's helicase (WRN) and Bloom's helicase (BLM), are linked to cancer-prone Werner's syndrome and Bloom's syndrome, respectively (33). Therefore, further characterizations of the complex interplay among Crb2, Rqh1, and γ-H2A might lead to better understanding of how WRN and BLM functions might be regulated by γ-H2AX in mammalian cells.
Rqh1 has been recently shown to function both upstream and downstream of the HR DNA repair protein Rhp51 (Rad51 homolog) in S. pombe (41). S. cerevisiae Rad52 (Rad22) has also been suggested to function both upstream and downstream of Rad51 (51). It would be of particular interest to determine if synergistic genetic interaction with rad22Δ and the rescue of the relative linear plasmid transformation efficiency observed in H2A-AQE mutation might be related to Rqh1 regulation by γ-H2A. It may also be interesting to test in mammalian cells if the loss of γ-H2AX rescues DNA damage sensitivity of cells deficient in 53BP1 or MDC1. Such observation would suggest that mammalian γ-H2AX also contributes negatively to the DNA repair process and that this negative contribution of γ-H2AX needs be to overcome by accumulation of BRCT domain proteins to DNA damage sites (Fig. 9F).
γ-H2A contributes to both checkpoint maintenance and DNA repair.
Phosphorylation of Crb2 and Chk1 is defective in H2A-AQE mutants (Fig. 6A), as is cell cycle arrest maintenance in response to DNA damage (Fig. 6B, 7A, and 8B). These results suggest that a defect in checkpoint arrest maintenance might account for the DNA damage-sensitive phenotype of H2A-AQE mutant cells. However, an artificial cell cycle arrest, imposed by overexpression of Wee1, only weakly rescued the IR sensitivity of H2A-AQE cells (Fig. 6C). Therefore, an inability to maintain a long-term checkpoint cannot fully explain why H2A-AQE cells are sensitive to DNA damage.
We found that rad3Δ is epistatic to H2A-AQE mutations in an IR survival assay, suggesting that Rad3 and γ-H2A function in the same DNA damage response pathway (Fig. 9A). In addition to arresting the cell cycle, checkpoint Rad proteins have additional roles in DNA repair, stabilization of the replication fork, and telomere maintenance (10, 58). In fact, Wee1 overexpression after IR treatment only partially rescues DNA damage sensitivity in rad3Δ and crb2Δ cells, and these mutants exhibit slower repair of IR-induced DNA damage when the DNA repair rate is monitored by PFGE or YFP-Rad22 foci disappearance (T. M. Nakamura, L.-L. Du, and P. Russell, unpublished data). Rad3 and Crb2 are not unique among checkpoint proteins in having roles other than the cell cycle arrest in response to DNA damage. There have been reports of mutations in other checkpoint proteins that maintain proficiency in cell cycle arrest yet cause DNA damage sensitivity (3, 22, 32). In S. cerevisiae, the checkpoint proteins Rad24 (S. pombe Rad17) and Rad9 (S. pombe Crb2) affect the rate of DSB processing (4, 43). In common with these mutants, we believe that the DNA damage sensitivity of H2A-AQE mutant cells arises from defects in both cell cycle arrest maintenance and DNA damage repair.
Rad3 and Tel1 act redundantly through γ-H2A to regulate Crb2 accumulation at DSBs.
We previously showed that Crb2 is recruited to DSBs in rad1Δ and rad3Δ mutants, but Crb2 foci prematurely disappear in these mutants (21). These findings suggested that Crb2 is an independent sensor of DNA damage, but its maintenance at DSB sites requires Rad1-Rad9-Hus1 and Rad3-Rad26 complexes. Our present study demonstrates that Crb2 focus formation is regulated by γ-H2A (Fig. 5A and B). Because Rad3 and Tel1 are both able to generate γ-H2A, these findings explain why Crb2 foci form in rad3Δ or tel1Δ single mutants but not the rad3Δ tel1Δ double mutant. Therefore, large-scale recruitment of Crb2 to DSBs is a secondary checkpoint event that occurs after recognition of DNA damage by Rad3 and Tel1.
Our data establish that formation of Crb2 foci is not required for checkpoint activation, but it is needed for long-term maintenance of cell cycle arrest (Fig. 6 to 8). How is a checkpoint established without the formation of Crb2 foci? It is possible that a small amount of Crb2, too little to be observed as nuclear foci, might be recruited to DSBs independently of γ-H2A. This recruitment might involve direct physical interactions between Crb2 and Rad3 that do not require γ-H2A (52). The impaired recruitment of Crb2 in H2A-AQE cells may still be sufficient for substantial checkpoint signaling to Chk1. Our data illustrate that focus formation and protein function do not always equate in checkpoint responses.
The role of γ-H2A in efficient accumulation of Crb2 at sites of DNA damage is interesting in light of observations in mammalian cells that γ-H2AX is needed for maintenance but not initial formation of foci of DNA repair and checkpoint proteins (15). It will also be interesting to test if IRIF formation by S. cerevisiae Rad9 (a presumptive Crb2/53BP1 homolog) (50) is regulated by γ-H2A. Previous studies in S. cerevisiae indicated that γ-H2A is not involved in checkpoint signaling (20, 64), but a premature checkpoint adaptation phenotype might have been missed in those studies. In fact, a subtle checkpoint defect in H2AX−/− mouse cells (23) was initially not recognized (16).
IRIF formation of Rad22 (Rad52 homolog), Rad32 (Mre11 homolog), and Rad26 (ATRIP/Lcd1 homolog) is intact in H2A-AQE cells (Fig. 4B and 5D). Rad32 forms a stable complex with Rad50 and Nbs1 in S. pombe (17, 80). This complex participates in DNA damage repair and telomere maintenance. The lack of an effect on Rad32 foci by H2A-AQE was surprising, as sustained IRIF formation of NBS1 appears to require γ-H2A in mouse cells (15) and NBS1 has been shown to bind directly to γ-H2AX (40). Rad32 (Mre11) IRIF formation might be differently regulated in mouse and S. pombe cells. It is also worth noting that S. pombe Rad32 is phosphorylated in response to DNA damage (85), but this phosphorylation is independent of Rad3 and Tel1 (58).
Studies of mammalian NBS1 and MDC1 indicated that BRCT domains are required for IRIF formation (40, 74), while the BRCT domains of 53BP1 were not required for IRIF formation (35, 53, 83). We are currently dissecting domains of Crb2 that might be required for focus formation and binding to γ-H2A. Our group's early studies have shown that a C-terminal truncation that eliminates amino acids 521 to 778, which includes the BRCT domains, prevents focus formation by Crb2 (L.-L. Du and P. Russell, unpublished data). A recent study has shown that Crb2 BRCT domains are required for the DNA checkpoint response (52). It will be interesting to test if the BRCT domains mediate direct binding to γ-H2A, given recent evidence that BRCT domains are phosphopeptide-binding modules (45, 70, 90).
Rad3 and Tel1 contribute to checkpoint maintenance.
A previous study showed that inactivation of Rad3ts just before exposure to IR abolished the Chk1-dependent DNA damage checkpoint, while Rad3ts inactivation just after irradiation allowed cells to arrest almost as well as wild type (47). These studies suggested that Rad3 activity is required for DNA damage checkpoint activation but not for its maintenance (47). Different findings were observed with the replication checkpoint induced by HU, in which it was found that Rad3 was required for both checkpoint activation and maintenance (47). How cells maintain a DNA damage checkpoint in the absence of Rad3 activity remains unknown.
Our studies of checkpoint maintenance in rad3ts cells have identified γ-H2A as a key molecular marker that allows Rad3-independent maintenance of a checkpoint arrest (Fig. 7A). The onset of mitosis was substantially advanced following inactivation of Rad3ts in the H2A-AQE background. Our studies with HU-treated cells indicated that H2A-AQE mutations preferentially impaired the Crb2-Chk1 pathway compared to effects on the Mrc1-Cds1 pathway (Fig. 8). The differential requirements for Rad3 activity in maintenance of DNA damage checkpoint arrest (Crb2-Chk1 pathway) compared to a DNA replication checkpoint arrest (Mrc1-Cds1 pathway) might be explained by a differential dependence on γ-H2A for checkpoint arrest maintenance.
The rad3ts experiment also allowed us to explore the involvement of Tel1 kinase in the DNA damage checkpoint response (Fig. 7A). Tel1 contributes to checkpoint responses in S. cerevisiae (19, 57, 72, 81), but a checkpoint arrest defect has not been reported for S. pombe tel1Δ cells. In S. pombe, Tel1 does play a role in HU-induced phosphorylation of Mrc1 (91), and it also functions in telomere maintenance with the Rad32 (Mre11)-Rad50-Nbs1 complex (17, 58). The Mre11 complex in S. cerevisiae participates in checkpoint responses (19, 30, 81), and it works in the Tel1-dependent pathway of telomere maintenance (68, 79).
While we did not observe a defect for bleomycin-induced cell cycle arrest in tel1Δ cells, the tel1Δ mutations allowed a faster release from the cell cycle arrest upon inactivation of Rad3ts (Fig. 7A). This release was delayed relative to that seen in rad3ts H2A-AQE cells. Therefore, Tel1 kinase does contribute to the cell cycle arrest after inactivation of Rad3, and this function might require γ-H2A. Additional experiments will be required to address this possibility. These observations suggest that Rad3 and Tel1 activities maintain H2A phosphorylation after checkpoint activation. Indeed, the disappearance of the γ-H2A signal was delayed in rhp51Δ cells, suggesting that H2A SQE dephosphorylation is influenced by the rate of DNA repair (Fig. 1E).
Conserved roles for γ-H2AX/γ-H2A: toward understanding histone codes for DNA damage responses.
Increasing attention is being placed on the role of chromatin modifications in DNA repair and checkpoint signaling (26, 34). C-tail phosphorylation of H2AX/H2A is the most-well-characterized DNA damage-dependent histone modification. Based on findings in mammalian cells that H2AX is necessary for IRIF formation for various checkpoint and DNA repair protein complexes, it was suggested that γ-H2AX/γ-H2A might serve as a docking site for these complexes (6). However, no γ-H2AX/γ-H2A-dependent IRIF formation by checkpoint and DNA repair proteins has been demonstrated in nonmammalian cells. Our study provides evidence that a docking function for γ-H2A is evolutionarily conserved. The parallels involving focus formation of the BRCT domain protein Crb2 in fission yeast and mammalian BRCT domain checkpoint proteins, such as 53BP1 and MDC1, are truly remarkable (Fig. 9E and F). The highly conserved nature of this mechanism underscores the importance of the DNA damage response involving γ-H2AX/γ-H2A.
In addition to γ-H2AX/γ-H2A, specific histone acetylation and deacetylation patterns, generated in response to DNA damage, appear to contribute to effective DNA repair (9, 36, 62). There is a potential connection between γ-H2AX/γ-H2A and histone acetylation and deacetylation. For example, histone acetylation has been shown to enhance H2AX phosphorylation in the context of nucleosomes by DNA-Pkcs in vitro (59). An interaction between 53BP1 and the histone deacetylase HDAC4 has also been reported (39). Therefore, many histone modifications might take place in a highly regulated manner to promote efficient DNA damage checkpoint signaling and DNA repair. The challenge for the future will be to understand how various histone modifications work together to generate a histone code for the DNA damage response.
Supplementary Material
Acknowledgments
We thank T. Carr, T. Enoch, S. Forsberg, and A. Pastink for S. pombe strains, K. Tanaka for the purified Crb2 preparation, S. Saitoh and H. Zhao for the Rad26 construct, the Yeast Resource Center at the University of Washington for the YFP vector, and J. Bailis for tips on γ-H2AX immunofluorescence microscopy. We also thank C. McGowan, B. Moser, M. Rodríguez, and P.-H. Gaillard for critical reading of the manuscript and members of the Scripps Cell Cycle groups for discussions.
T.M.N. was supported in part by the Damon Runyon Cancer Research Foundation (DRG-1565). L.-L.D. is supported by the Leukemia and Lymphoma Society. This work was funded by National Institutes of Health grants CA77325 and GM59447 awarded to P.R.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, C., P. Fantes, J. Hyams, M. McLoed, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.al-Khodairy, F., E. Fotou, K. S. Sheldrick, D. J. Griffiths, A. R. Lehmann, and A. M. Carr. 1994. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell 5:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aylon, Y., and M. Kupiec. 2003. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol. Cell. Biol. 23:6585-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baber-Furnari, B. A., N. Rhind, M. N. Boddy, P. Shanahan, A. Lopez-Girona, and P. Russell. 2000. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol. Biol. Cell 11:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassing, C. H., and F. W. Alt. 2004. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle 3:149-153. [DOI] [PubMed] [Google Scholar]
- 7.Bassing, C. H., K. F. Chua, J. Sekiguchi, H. Suh, S. R. Whitlow, J. C. Fleming, B. C. Monroe, D. N. Ciccone, C. Yan, K. Vlasakova, D. M. Livingston, D. O. Ferguson, R. Scully, and F. W. Alt. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 99:8173-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359-370. [DOI] [PubMed] [Google Scholar]
- 9.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 10.Carr, A. M. 2002. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amsterdam) 1:983-994. [DOI] [PubMed] [Google Scholar]
- 11.Caspari, T., and A. M. Carr. 1999. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie 81:173-181. [DOI] [PubMed] [Google Scholar]
- 12.Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann, K. Papadimitriou, P. Sunnerhagen, and A. M. Carr. 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20:1254-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspari, T., J. M. Murray, and A. M. Carr. 2002. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 16.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell, and N. Rhind. 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 23:6564-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe, J., T. Schuster, and M. Grunstein. 1985. Organization, primary structure, and evolution of histone H2A and H2B genes of the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 5:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 21.Du, L.-L., T. M. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Capetillo, O., H. T. Chen, A. Celeste, I. Ward, P. J. Romanienko, J. C. Morales, K. Naka, Z. Xia, R. D. Camerini-Otero, N. Motoyama, P. B. Carpenter, W. M. Bonner, J. Chen, and A. Nussenzweig. 2002. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 4:993-997. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Capetillo, O., B. Liebe, H. Scherthan, and A. Nussenzweig. 2003. H2AX regulates meiotic telomere clustering. J. Cell Biol. 163:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Capetillo, O., S. K. Mahadevaiah, A. Celeste, P. J. Romanienko, R. D. Camerini-Otero, W. M. Bonner, K. Manova, P. Burgoyne, and A. Nussenzweig. 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4:497-508. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Capetillo, O., and A. Nussenzweig. 2004. Linking histone deacetylation with the repair of DNA breaks. Proc. Natl. Acad. Sci. USA 101:1427-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira, M. G., and J. P. Cooper. 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7:55-63. [DOI] [PubMed] [Google Scholar]
- 28.Furuta, T., H. Takemura, Z. Y. Liao, G. J. Aune, C. Redon, O. A. Sedelnikova, D. R. Pilch, E. P. Rogakou, A. Celeste, H. T. Chen, A. Nussenzweig, M. I. Aladjem, W. M. Bonner, and Y. Pommier. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303-20312. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg, M., M. Stucki, J. Falck, D. D'Amours, D. Rahman, D. Pappin, J. Bartek, and S. P. Jackson. 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952-956. [DOI] [PubMed] [Google Scholar]
- 30.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 31.Griffith, J. D., L. A. Lindsey-Boltz, and A. Sancar. 2002. Structures of the human rad17-replication factor C and checkpoint rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 277:15233-15236. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths, D. J., N. C. Barbet, S. McCready, A. R. Lehmann, and A. M. Carr. 1995. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 14:5812-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 34.Iizuka, M., and M. M. Smith. 2003. Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 13:154-160. [DOI] [PubMed] [Google Scholar]
- 35.Iwabuchi, K., B. P. Basu, B. Kysela, T. Kurihara, M. Shibata, D. Guan, Y. Cao, T. Hamada, K. Imamura, P. A. Jeggo, T. Date, and A. J. Doherty. 2003. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278:36487-36495. [DOI] [PubMed] [Google Scholar]
- 36.Jazayeri, A., A. D. McAinsh, and S. P. Jackson. 2004. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 101:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 38.Kai, M., H. Tanaka, and T. S. Wang. 2001. Fission yeast Rad17 associates with chromatin in response to aberrant genomic structures. Mol. Cell. Biol. 21:3289-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao, G. D., W. G. McKenna, M. G. Guenther, R. J. Muschel, M. A. Lazar, and T. J. Yen. 2003. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J. Cell Biol. 160:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12:1846-1851. [DOI] [PubMed] [Google Scholar]
- 41.Laursen, L. V., E. Ampatzidou, A. H. Andersen, and J. M. Murray. 2003. Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 23:3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 43.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 44.Ma, J. L., E. M. Kim, J. E. Haber, and S. E. Lee. 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23:8820-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manke, I. A., D. M. Lowery, A. Nguyen, and M. B. Yaffe. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302:636-639. [DOI] [PubMed] [Google Scholar]
- 46.Manolis, K. G., E. R. Nimmo, E. Hartsuiker, A. M. Carr, P. A. Jeggo, and R. C. Allshire. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J. 20:210-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsumoto, S., and M. Yanagida. 1985. Histone gene organization of fission yeast: a common upstream sequence. EMBO J. 4:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 50.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyazaki, T., D. A. Bressan, M. Shinohara, J. E. Haber, and A. Shinohara. 2004. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 23:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mochida, S., F. Esashi, N. Aono, K. Tamai, M. J. O'Connell, and M. Yanagida. 2004. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 23:418-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales, J. C., Z. Xia, T. Lu, M. B. Aldrich, B. Wang, C. Rosales, R. E. Kellems, W. N. Hittelman, S. J. Elledge, and P. B. Carpenter. 2003. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 278:14971-14977. [DOI] [PubMed] [Google Scholar]
- 54.Moser, B. A., J. M. Brondello, B. Baber-Furnari, and P. Russell. 2000. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol. Cell. Biol. 20:4288-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray, J. M., H. D. Lindsay, C. A. Munday, and A. M. Carr. 1997. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol. Cell. Biol. 17:6868-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakada, D., T. Shimomura, K. Matsumoto, and K. Sugimoto. 2003. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura, T. M., B. A. Moser, and P. Russell. 2002. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park, E. J., D. W. Chan, J. H. Park, M. A. Oettinger, and J. Kwon. 2003. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31:6819-6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 61.Pilch, D. R., O. A. Sedelnikova, C. Redon, A. Celeste, A. Nussenzweig, and W. M. Bonner. 2003. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem. Cell. Biol. 81:123-129. [DOI] [PubMed] [Google Scholar]
- 62.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 64.Redon, C., D. R. Pilch, E. P. Rogakou, A. H. Orr, N. F. Lowndes, and W. M. Bonner. 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhind, N., and P. Russell. 1998. Mitotic DNA damage and replication checkpoints in yeast. Curr. Opin. Cell Biol. 10:749-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhind, N., and P. Russell. 2001. Roles of the mitotic inhibitors Wee1 and Mik1 in the G2 DNA damage and replication checkpoints. Mol. Cell. Biol. 21:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 68.Ritchie, K. B., and T. D. Petes. 2000. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics 155:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rizzo, P. J. 2003. Those amazing dinoflagellate chromosomes. Cell Res. 13:215-217. [DOI] [PubMed] [Google Scholar]
- 70.Rodriguez, M., X. Yu, J. Chen, and Z. Songyang. 2003. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J. Biol. Chem. 278:52914-52918. [DOI] [PubMed] [Google Scholar]
- 71.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 73.Schultz, L. B., N. H. Chehab, A. Malikzay, and T. D. Halazonetis. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151:1381-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shang, Y. L., A. J. Bodero, and P. L. Chen. 2003. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41-amino acid repeat sequence, is an early participant in DNA damage response. J. Biol. Chem. 278:6323-6329. [DOI] [PubMed] [Google Scholar]
- 75.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stewart, G. S., B. Wang, C. R. Bignell, A. M. Taylor, and S. J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961-966. [DOI] [PubMed] [Google Scholar]
- 77.Suto, K., A. Nagata, H. Murakami, and H. Okayama. 1999. A double-strand break repair component is essential for S phase completion in fission yeast cell cycling. Mol. Biol. Cell 10:3331-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 79.Tsukamoto, Y., A. K. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 80.Ueno, M., T. Nakazaki, Y. Akamatsu, K. Watanabe, K. Tomita, H. D. Lindsay, H. Shinagawa, and H. Iwasaki. 2003. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol. Cell. Biol. 23:6553-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 82.van den Bosch, M., K. Vreeken, J. B. Zonneveld, J. A. Brandsma, M. Lombaerts, J. M. Murray, P. H. Lohman, and A. Pastink. 2001. Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461:311-323. [DOI] [PubMed] [Google Scholar]
- 83.Ward, I. M., K. Minn, K. G. Jorda, and J. Chen. 2003. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278:19579-19582. [DOI] [PubMed] [Google Scholar]
- 84.Willson, J., S. Wilson, N. Warr, and F. Z. Watts. 1997. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 25:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilson, S., M. Tavassoli, and F. Z. Watts. 1998. Schizosaccharomyces pombe rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res. 26:5261-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson, S., N. Warr, D. L. Taylor, and F. Z. Watts. 1999. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 27:2655-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wolkow, T. D., and T. Enoch. 2002. Fission yeast rad26 is a regulatory subunit of the rad3 checkpoint kinase. Mol. Biol. Cell 13:480-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wyatt, H. R., H. Liaw, G. R. Green, and A. J. Lustig. 2003. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu, X., and D. F. Stern. 2003. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 278:8795-8803. [DOI] [PubMed] [Google Scholar]
- 90.Yu, X., C. C. Chini, M. He, G. Mer, and J. Chen. 2003. The BRCT domain is a phospho-protein binding domain. Science 302:639-642. [DOI] [PubMed] [Google Scholar]
- 91.Zhao, H., K. Tanaka, E. Noguchi, C. Noguchi, and P. Russell. 2003. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol. Cell. Biol. 23:8395-8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.