Abstract
Phosphotyrosine (pTyr) signaling is involved in development and maintenance of metazoans’ multicellular body through cell-to-cell communication. Tyrosine kinases (TKs), tyrosine phosphatases, and other proteins relaying the signal compose the cascade. Domain architectures of the pTyr signaling proteins are diverse in metazoans, reflecting their complex intercellular communication. Previous studies had shown that the metazoan-type TKs, as well as other pTyr signaling proteins, were already diversified in the common ancestor of metazoans, choanoflagellates, and filastereans (which are together included in the clade Holozoa) whereas they are absent in fungi and other nonholozoan lineages. However, the earliest-branching holozoans Ichthyosporea and Corallochytrea, as well as the two fungi-related amoebae Fonticula and Nuclearia, have not been studied. Here, we analyze the complete genome sequences of two ichthyosporeans and Fonticula, and RNAseq data of three additional ichthyosporeans, one corallochytrean, and Nuclearia. Both the ichthyosporean and corallochytrean genomes encode a large variety of receptor TKs (RTKs) and cytoplasmic TKs (CTKs), as well as other pTyr signaling components showing highly complex domain architectures. However, Nuclearia and Fonticula have no TK, and show much less diversity in other pTyr signaling components. The CTK repertoires of both Ichthyosporea and Corallochytrea are similar to those of Metazoa, Choanoflagellida, and Filasterea, but the RTK sets are totally different from each other. The complex pTyr signaling equipped with positive/negative feedback mechanism likely emerged already at an early stage of holozoan evolution, yet keeping a high evolutionary plasticity in extracellular signal reception until the co-option of the system for cell-to-cell communication in metazoans.
Keywords: evolution, tyrosine kinase, ichthyosporeans, corallochytreans, multicellularity
Introduction
The phosphotyrosine-mediated signaling (pTyr signaling) plays a critical role in development and maintenance of multicellular bodies of animals (metazoans) (Hunter 2009). By organizing cell-to-cell communication, pTyr signaling controls concerted movement, differentiation, and proliferation of numerous cells constituting metazoan bodies (Lim and Pawson 2010). Studying the evolution of pTyr signaling is thus essential for understanding the origin of metazoan multicellularity, one of the most fascinating questions in evolutionary biology (Bonner 1998). Tyrosine kinase (TK or PTK), one of the main components of pTyr signaling, initiates the cascade by transducing specific extracellular signals through the cell membrane into the cytoplasm and nucleus (Fantl et al. 1993; van der Geer et al. 1994; Hubbard and Till 2000). All TKs share a conserved catalytic domain (TK domain) for tyrosine phosphorylation, but at the same time the catalytic domain is combined with a vast variety of other protein domains, which characterize the function of TKs within the context of signaling cascade (Fantl et al. 1993; van der Geer et al. 1994; Suga et al. 1999; Hubbard and Till 2000). TKs are roughly divided by structure into two groups: receptor TKs (RTKs) and cytoplasmic or nonreceptor TKs (CTKs). An RTK has a transmembrane (TM) segment, protruding the N-terminus into the extracellular space while the C-terminus with the catalytic domain remains in the cytoplasm. The N-terminal receptor regions of RTKs are very diverse in domain architecture, reflecting the variety of their specific ligands that are secreted into the extracellular space by other cells (van der Geer et al. 1994). The ligand binding triggers a series of conformational changes and multimerization events in RTKs, and eventually initiates the intracellular pTyr signal, which is relayed by various other proteins (Hubbard and Till 2000). TKs are thus regarded as the “writer” of the pTyr signaling (Lim and Pawson 2010). The pTyr signal transduction system is also made up of the “eraser” tyrosine-specific protein phosphatases (PTPs) and a variety of “reader” proteins having Src homology 2 (SH2) domains or pTyr binding (PTB) domains, both of which bind to the pTyr residue. The catalytic domains of writers and erasers, and the two reader domains SH2 and PTB were diversified by gene duplication, and combined as modules with each other, or with other protein domains, giving rise to the complex pTyr signaling cascade (or network) currently seen in metazoans (Hunter 2009; Lim and Pawson 2010). In particular, the combination of the reader domains with the writer or the eraser catalytic domains was crucial for the evolution of efficient signal transduction by positive or negative feedback loops (Lim and Pawson 2010).
The catalytic domains of TKs evolved from eukaryotic protein kinases, which are considered, with some exceptions, to specifically phosphorylate serine and threonine residues, but not tyrosine residues, and thus they are grouped into the serine/threonine kinase (STK) family (Hanks et al. 1988; Manning et al. 2002). Historically, TKs had been considered metazoan specific, due to their absence in plants, fungi, and other analyzed eukaryotes, and their particular importance in metazoan development (Hanks et al. 1988; Hanks and Hunter 1995). Later, putative TKs were found in several single-cellular eukaryotes that are distantly related to metazoans, such as the amoebozoan Dictyostelium discoideum and Entamoeba histolytica, the green alga Chlamydomonas reinhardtii, and the oomycete Phytophthora infestans (Tan and Spudich 1990; Shiu and Li 2004). However, these nonmetazoan TKs do not show diversified domain architectures like those in metazoans, and moreover, a phylogenetic study suggests different evolutionary origins of these two TK groups (Suga et al. 2012). The metazoan-type TKs (referred to as group A TKs) are likely to have evolved monophyletically from the other type of TKs (group B TKs) in nonmetazoan eukaryotes. Group A and group B TKs do not coexist in the genomes of a single species so far studied. It is thus likely that the ancestor of metazoans have lost group B TKs, replacing them with newly evolved group A TKs. Therefore, studies on the evolution of group A TKs are essential for understanding the evolution of metazoan multicellularity.
Metazoan TKs, which are all in group A, are classified into 29 common families on the basis of their protein domain architectures and phylogenetic relationship of the catalytic (kinase) domains (Suga et al. 1997, 2012; Hubbard and Till 2000). Nineteen of the 29 TK families are RTKs. Metazoan RTKs have diverse domain architectures in the ligand-binding regions, which characterize the diversity and specificity of the cell-to-cell signaling. Importantly, many CTK and RTK family are shared among all extant metazoans (Suga et al. 2001; Srivastava et al. 2010), suggesting that the establishment of this basic TK set was critical for the origin of Metazoa.
However, the genomes of two closest metazoan relatives that make up the clade Holozoan together with metazoans, namely choanoflagellates and filastereans (fig. 1), showed the presence of group A TKs in premetazoans, and thus the origin of group A TKs turned out to be much earlier than the metazoan onset itself (Manning et al. 2008; Pincus et al. 2008; Suga, Sasaki, et al. 2008; Suga et al. 2012). The genomes of choanoflagellates and filastereans do not encode any group B TK, similar to the metazoan genomes. These data also showed that the basic repertoire of CTK families had already been established in the last common ancestor of these three holozoan clades, whereas their RTK sets were extensively diversified independently in each clade (Suga et al. 2012).
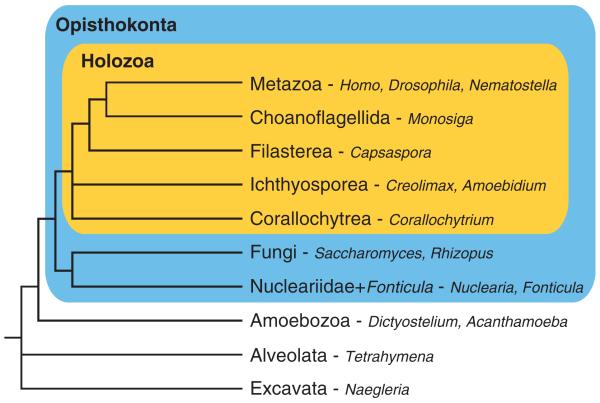
FIG. 1.
Phylogeny of eukaryotes. Schema of a widely accepted eukaryotic phylogeny. Genera described in this study are indicated after the hyphens. Blue and orange boxes represent the Opisthokonta and the Holozoa, respectively.
These surveys however still contain a large taxonomic gap, in which whole-genome scale information has been critically lacking. For example, ichthyosporeans (or mesomycetozoans; [Mendoza et al. 2002; Glockling et al. 2013; for review]), which are a highly diverse group of animal symbionts and are the sister group to filastereans, choanoflagellates, and metazoans (Shalchian-Tabrizi et al. 2008; Torruella et al. 2012; Paps et al. 2013) (fig. 1), have not been analyzed. Moreover, there has been no information from the enigmatic Corallochytrium limacisporum (Raghu-kumar 1987; Cavalier-Smith and Allsopp 1996), whose phylogenetic position within the Holozoa remains unsettled (Ruiz-Trillo et al. 2006; Steenkamp et al. 2006; Paps et al. 2013). In addition, no data have been available from Nuclearia or Fonticula alba, sister lineages to the Fungi (Brown et al. 2009; Liu et al. 2009). For a deeper understanding of the origin and evolution of TKs, these earlier-branching lineages of the Opisthokonta (metazoans, fungi, and their closest-relative protists [Cavalier-Smith 1998]; fig. 1) should be explored.
In this study, we search for TKs, PTPs, and proteins containing SH2 domains and PTB domains in all of the aforementioned opisthokont lineages. The origin of complex pTyr signaling system seen now in metazoans is here pinpointed with unprecedented detail. Furthermore, we examine numerical and architectural diversification of these proteins, and discuss how eventually the metazoans’ cell-to-cell communication through pTyr signaling evolved in the course of holozoan evolution.
Results
Early Expansion of pTyr Signal Transduction Components in Holozoan Evolution
We produced three ichthyosporean data: the draft genome sequence of Creolimax fragrantissima and RNAseq of Abeoforma whisleri and Pirum gemmata. Moreover, we generated RNAseq data of the free-living marine holozoan Corallochytrium limacisporum, and the nucleariid amoeba Nuclearia sp. (ATCC50694), a sister lineage of fungi. We also analyzed the draft genome sequences of the ichthyosporean Sphaeroforma arctica and the amoeba Fonticula alba, another fungi-relative, and RNAseq data of the ichthyosporean Amoebidium parasticium, generated in the context of UNICORN project (Ruiz-Trillo et al. 2007).
By combining multiple homology searches for TK catalytic domains, phylogenetic analyses, and importantly a careful manual inspection of gene prediction, we identified 173 group A TKs in the analyzed holozoan data, but no group B TKs. Notably, neither group A nor group B TKs were identified in Nuclearia and Fonticula. The number of TK catalytic domains identified in ichthyosporeans and Corallochytrium ranges from 8 to 67, which are much smaller than those of the filasterean Capsaspora owczarzaki (103) (Suga et al. 2012) and the choanoflagellate Monosiga brevicollis (136) (Manning et al. 2008; Pincus et al. 2008), but still comparable to that of Drosophila melanogaster (34) (table 1).
Table 1.
Numbers of pTyr Signaling Domains.
| Species | Group A TK | PTP | SH2 | PTB |
|---|---|---|---|---|
| Nonopisthokonta | ||||
| Naegleria | 0 | 3 | 5 | 0 |
| Tetrahymena a | 0 | 3 | 1 | 0 |
| Acanthamoeba b | 0 | 6 | 51 | 0 |
| Dictyostelium a | 0 | 3 | 14 | 0 |
|
| ||||
| Fungi and their relatives | ||||
| Rhizopus | 0 | 6 | 1 | 0 |
| Saccharomyces a | 0 | 7 | 1 | 0 |
| Fonticula | 0 | 1 | 3 | 0 |
| Nucleariac | 0 | 1 | 5 | 0 |
|
| ||||
| Corallochytrea | ||||
| Corallochytrium c | 8 (5) | 7 | 18 | 1 |
|
| ||||
| Ichthyosporea | ||||
| Creolimax | 31 (30) | 7 | 22 | 1 |
| Sphaeroforma | 38 (37) | 7 | 22 | 1 |
| Abeoforma c | 18 (8) | 13 | 56 | 0 |
| Pirum c | 11 (3) | 16 | 45 | 0 |
| Amoebidium c | 67 (56) | 9 | 55 | 0 |
|
| ||||
| Filasterea | ||||
| Capsaspora | 103 (92) | 8 | 39 | 7 |
|
| ||||
| Choanoflagellida | ||||
| Monosiga a | 136 (93) | 40 | 143 | 31 |
|
| ||||
| Metazoa | ||||
| Nematostella | 85 (—)d | 66 | 29 | 27 |
| Drosophila a | 34 (21) | 23 | 34 | 10 |
| Homo a | 94 (58) | 50 | 120 | 51 |
Note.—Sequence data reported in this study are underlined. Number of protein domains, but not number of proteins (or genes) is shown. Numbers of kinase domains occurring in RTKs are in parentheses.
Numbers from the previous publication (Manning et al. 2008).
Numbers from the previous publication (Clarke et al. 2013).
RNAseq data.
Uncountable because the predicted Nematostella TK sequences in the current assembly are very fragmented.
We then searched for the classical (i.e., tyrosine-specific [Tonks 2006]) PTP catalytic domains, SH2 domains, and PTB domains. We identified between 7 to 16 PTPs in the whole genomes and RNAseq data of the five ichthyosporeans, as well as 7 PTPs in the RNAseq data in Corallochytrium (table 1). In contrast, only one PTP was identified both in Fonticula and in Nuclearia. The SH2 domain repertoire is also highly expanded in the Ichthyosporea (22-56 SH2 domains) and Corallochytrium (18 domains), whereas those of Fonticula and Nuclearia are poorly diversified (3 and 5 domains, respectively). PTB domain was also identified in the Ichthyosporea and Corallochytrium, but not in Fonticula and Nuclearia. Among the studied ichthyosporeans, only one PTB domain was identified in each of the two genome-sequenced lineages, Creolimax and Sphaeroforma, but none in the RNAseq data of the other three species, which may underrepresent their whole genomic information.
Overall, our data show that the repertoires of pTyr signaling components greatly increased at the onset of Holozoa. Group A TK and PTB domain appear to be an earliest holozoan innovation. Although a few flowering plant genomes encode a short motif with a partial homology to the PTB domain (e.g., GenBank entries XP_003545025, XP_002301956, and BAJ53102), this motif seems to be a product of either convergent evolution or lateral gene transfer event because it is not found in other plants. The origins of classical PTP and SH2 domain, which show a paneukaryotic distribution, are much more ancient than those of group A TK and PTB domain. However, the numbers of PTPs and SH2 domains appear to have leaped at the holozoan onset (3–4 times more on average in ichthyosporeans and Corallochytrium than in the nonholozoan taxa analyzed) (table 1).
Dramatic Increase in the Architectural Diversity of Ichthyosporean and Corallochytrean TKs
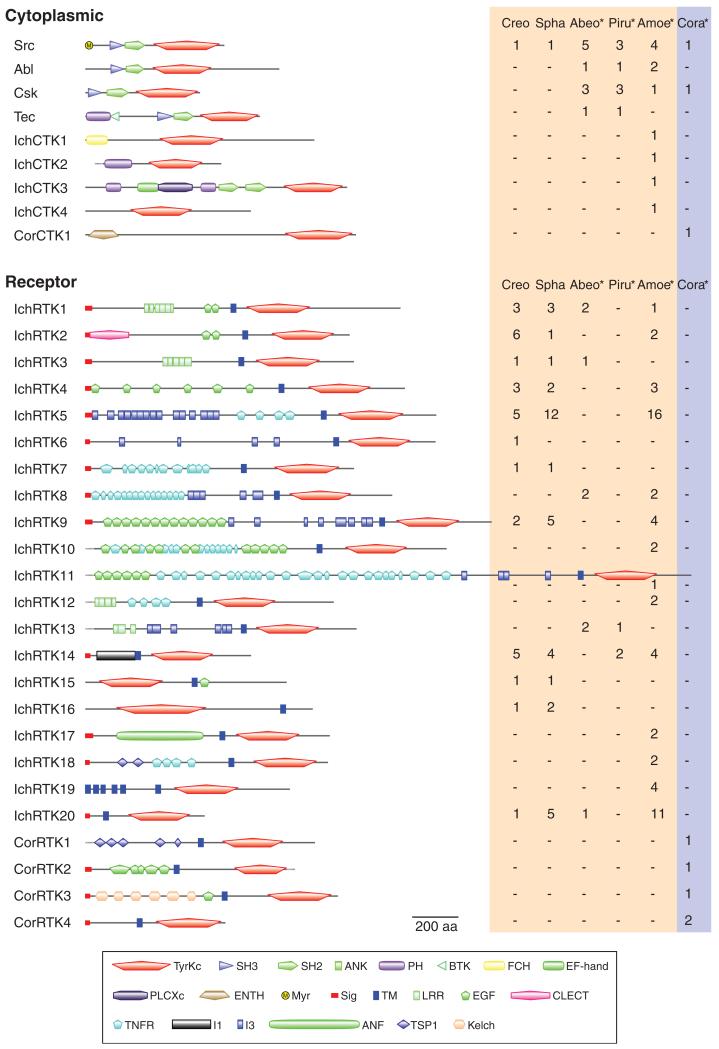
We analyzed the domain architecture of each identified TK by a search of Pfam and SMART databases. The search was complemented with manual inspection, which recovered many domains with low statistical significance. As a result, we found a large architectural variety in both ichthyosporean and corallochytrean TKs (fig. 2; supplementary fig. S1, Supplementary Material online). The identified holozoan TKs were classified into 9 CTK families (34 genes) and 24 RTK families (139 genes) according to their protein domain architectures and the similarity in their catalytic domain sequences (see also the phylogenetic analyses discussed later).
FIG. 2.
Ichthyosporean and corallochytrean TKs. TKs found in the whole genome data of two ichthyosporeans (Creo, Creolimax; Spha, Sphaeroforma) and the RNAseq data (asterisks) from three other ichthyosporeans (Abeo, Abeoforma; Piru, Pirum; Amoe, Amoebidium), and a corallochytrean (Cora, Corallochytrium) are classified according to their domain architectures and phylogenetic positions. CTKs are classified into ichthyosporean-specific families (IchCTK1-4) or corallochytrean-specific families (CorCTK1) unless they are homologous to metazoan CTKs. The domain architecture of a family member is schematically shown. The number of genes within each family is shown on the right (red and blue shades for ichthyosporeans and corallochytreans, respectively). See supplementary figure S1, Supplementary Material online, for the full list of annotated TKs. ANF, atrial natriuretic factor receptor-like ligand binding region; ANK, ankyrin repeats; BTK, Bruton’s tyrosine kinase Cys-rich motif; CLECT, C-type lectin (CTL) or carbohydrate-recognition domain (CRD); EF-hand, EF-hand-like domain; EGF, epidermal growth factor-like domain; ENTH, epsin N-terminal homology domain; FCH, Fes/CIP4 homology domain; I1, I1 domain; I3, I3 domain; Kelch, Kelch motif; LRR, leucine-rich repeat; Myr, predicted myristoylation site; PH, Pleckstrin homology domain; PLCXc, phospholipase C catalytic domain X; SH2, Src homology 2 domain; SH3, Src homology 3 domain; Sig, signal peptide; TM, transmembrane segment; TNFR, tumor necrosis factor receptor/nerve growth factor receptor repeat; TSP1, thrombospondin type 1 repeats; TyrKc, tyrosine kinase catalytic domain. Incomplete sequences are shown by dotted lines.
Twenty-nine group A TKs from ichthyosporeans and Corallochytrium belong to the four CTK families (Src, Abl, Csk, and Tec; fig. 2) that are shared among metazoans, choanoflagellates, and filastereans (Suga et al. 2012). Although the two complete genomes of Creolimax and Sphaeroforma encode only a single CTK (Src), the other three ichthyosporean species have a much diverse CTK repertoire, even shown by their RNAseq data. This suggests an earliest holozoan diversification of the CTK family repertoire, which already resembled that in extant metazoans, followed by its secondary reduction in the ichthyosporean lineage leading to Creolimax and Sphaeroforma.
Both ichthyosporeans and Corallochytrium show a much more expanded repertoire of RTK families than that of CTK families. Their RTKs have unique architectures that are not homologous to any RTK family of metazoans, choanoflagellates, or filastereans (fig. 2; supplementary fig. S1, Supplementary Material online). Moreover, the RTK repertoires of ichthyosporeans and Corallochytrium show little, if any, overlap among them, suggesting that they diversified independently. In this context, the architectural homology between IchRTK4 and CorRTK2 (fig. 2; supplementary fig. S1, Supplementary Material online) is likely a product of convergent evolution, as also shown by a phylogenetic analysis (described in the following section) of the catalytic domain sequence.
We identified two novel protein domains (named as I1 and I3 domains) in the ichthyosporean RTKs. The I1 domain of around 170 amino acids in length (supplementary fig. S2A, Supplementary Material online) appears as a single domain only in the extracellular regions of the ichthyosporean IchRTK14 family proteins. In most cases, the I1 domain contains four conserved cysteine residues, suggesting formation of disulfide bonding in its tertiary structure. The I3 domain, which has the conserved sequence motif GGA(V/I)(Y/F) XXXX(V/L)X(V/I)X(N/D)XXFXXNXA (supplementary fig. S2B, Supplementary Material online), is much smaller, mostly composed of 25 amino acids, but occurs repeatedly in a single protein (fig. 2). In contrast to the I1 domain, which is unique to ichthyosporeans, the I3 domain is identified in many eubacterial or archaeal proteins of unknown function (supplementary fig. S2C, Supplementary Material online). Interestingly, there is an overall structural similarity between these prokaryotic I3-containing proteins and other bacterial proteins designated adhesins, which are involved in host infection of bacteria (Klemm and Schembri 2000; Coutte et al. 2003); both proteins always have numerous internal repeats, and often a signal peptide and a TM segment, suggesting their localization at the outer membrane, or secretion onto the cell surface. Similar to many bacterial adhesins, the prokaryotic I3-containing proteins often have as well the hem-agglutination activity domain (supplementary fig. S2C, Supplementary Material online), which is considered to be involved in the host cell attachment (Inatsuka et al. 2005). The I3 domain is also found in some protists such as Trichomonas but not in any examined metazoan or nonichthyosporean holozoan. In ichthyosporeans, this domain always occurs in the receptor regions of RTKs, suggesting their potential function of interacting with extracellular molecules.
The ichthyosporean RTK families IchRTK15, IchRTK16, and IchRTK19 show unique membrane topologies that no other RTK exhibits. The IchRTK15 and IchRTK16 families were predicted to be “inverted,” having their TM segments on the C-termini of the TK catalytic domains and lacking signal peptides (fig. 2). The epidermal growth factor- (EGF-) like domains, which are normally located in the N-terminus of the TM segment (i.e., ligand-binding regions), reside in the C-terminus. We confirmed that these genes are expressed as predicted by RT-PCR. Interestingly, the electrostatic properties on the edges of TM segments of these families are inverted as well; they carry positively charged amino acid stretches adjacently on the N-termini and moderately negatively charged ones on the C-termini, which are oppositely aligned in canonical RTKs (supplementary fig. S3, Supplementary Material online). These RTKs are most likely inversely inserted into the membrane so that the catalytic domains are exposed to the cytoplasm like in canonical RTKs. Certain membrane-bound proteins such as the asialoglycoprotein and transferrin receptors are proposed to be similarly inserted in the membrane (von Heijne 1990; Lodish et al. 2000). Such receptor proteins usually lack a signal peptide, but instead the proteins use a TM segment for the membrane localization signal (Lodish et al. 2000). The same seems to apply to the ichthyosporean RTK10 and RTK11 proteins that also lack a signal peptide (fig. 2). The Amoebidium-specific IchRTK19 family, which has six putative TM segments, is another unique example demonstrating the plasticity in membrane topology of ichthyosporean RTKs.
Domain Architectures of Readers and Erasers in pTyr Signaling
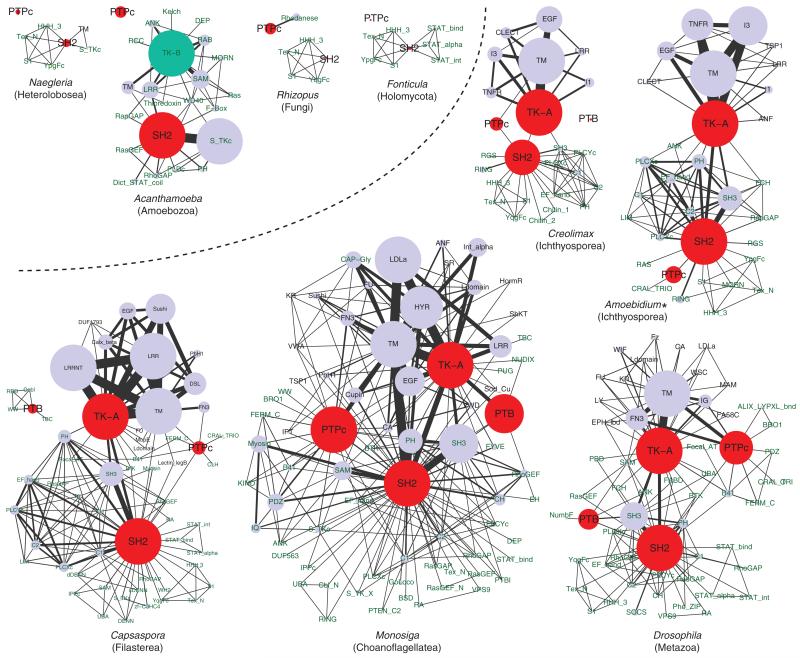
We examined the detailed domain architectures of classical PTPs and SH2- and PTB-domain containing proteins. Figure 3 shows the domain combinations occurring in single proteins. The network roughly illustrates the architectural complexity of pTyr signaling proteins enhanced in the Holozoa, compared with nonholozoans. Within the Holozoa, the choanoflagellate Monosiga recruits exceptionally diverse domain combinations in the pTyr signaling proteins.
FIG. 3.
Complex architecture of pTyr signaling proteins in holozoans. Domain is represented by a circle, whose size is proportional to the domain count (more than 25 counts are shown in identical size). Each line directly connecting two domains represents their co-occurrence in single proteins, and its width is proportional to the frequency of co-occurrence (more than 25 connections are shown in identical width). Duplicated domains in a protein were not counted. The four core pTyr signaling modules are in red, while the group B TK catalytic domain is in turquoise. SMART or Pfam domain names are shown, except for TK-A (group A TK catalytic domain), TK-B (group B TK catalytic domain), and TM (transmembrane segment). Names of extracellular and intracellular domains are shown in violet and green, respectively. Holozoan and nonholozoan data are divided by a dotted line. Asterisk, RNAseq data.
Contrary to TKs, PTPs of ichthyosporeans and Corallochytrium show much less diversity than those of metazoans. Nevertheless, some of the cytoplasmic PTP in ichthyosporeans and Corallochytrium are clearly homologous to certain metazoan PTPs: PTPN6/11 family genes (Tonks 2006) with two SH2 domains, PTPN9 with the CRAL-TRIO/SEC14 domain, PTPN21 with the B41 (FERM) domain, and PTPN23 with the BRO1 domain (supplementary figs. S4 and S5, Supplementary Material online). On the other hand, clear homologs of metazoan PTPs, or even a complex domain combination usually seen in the holozoan PTPs, are absent in Fonticula, Nuclearia, and any other nonholozoan taxa analyzed (fig. 3; Lim and Pawson 2010; Clarke et al. 2013). Contrary to the cytoplasmic PTPs, none of the receptor PTPs in ichthyosporeans and Corallochytrium is clearly homologous to any metazoan PTP (supplementary fig. S5, Supplementary Material online; Tonks 2006). These data suggest, similar to TKs, an early diversification of major cytoplasmic PTP families in extant metazoans at the onset of Holozoa, as well as a simplification of the PTP repertoires in the line-age leading to Creolimax and Sphaeroforma, which have less PTPs with little architectural diversity than other ichthyosporeans (supplementary fig. S4, Supplementary Material online).
The SH2 domain-containing proteins exhibit highly diversified architectures in ichthyosporeans and Corallochytrium. At least 34 different domains in total are involved in building the SH2 domain-containing protein sets of the six holozoan taxa (supplementary fig. S5, Supplementary Material online). This architectural variation in basal holozoans clearly outnumbers those of the fungi S. cerevisiae and R. oryzae (only one [Lim and Pawson 2010] and four domains combined with SH2, respectively) and other nonholozoan eukaryotes such as N. gruberi (six domains combined; fig. 3). The amoebozoan A. castellanii, which has the most expanded SH2 domain-containing protein repertoire among nonholozoan lineages known to date (Clarke et al. 2013; table 1 and fig. 3), has a unique set of SH2-containing proteins that may have evolved independently—their SH2 domains are frequently combined with STK catalytic domains (S_TKc), but not with TK or PTP catalytic domains (fig. 3). The network in figure 3 also illustrates the exceptionally highly diversified architectures of SH2-containing proteins in M. brevicollis (123 proteins containing 143 SH2 domains, which are combined with 53 other domains), suggesting the presence of a highly diversified pTyr signaling uniquely evolved in choanoflagellates.
The presence of the combination of SH2 domains with TK or PTP domains in ichthyosporeans and Corallochytrium is especially important, because the evolution of this combination likely contributed to building a complex and efficient pTyr signaling circuit by creating the positive or negative feedback loops (Lim and Pawson 2010). Such recruitment of the reader module into the writer/eraser proteins of pTyr signaling appears to be exclusive to the Holozoa (fig. 3).
Compared with TK, PTP, and SH2, PTB domain-containing proteins show a much poorer diversity regarding the combination with other domains. None of the three PTB-containing proteins of ichthyosporeans and Corallochytrium is combined with other recognizable domains, whereas in filastereans, choanoflagellates, or metazoans, PTB-containing proteins show much complex architectures (fig. 3). The filasterean Capsaspora, in which seven PTB domains are found but not combined with TK or PTP catalytic domains, exhibits a kind of intermediate stage of PTB domain evolution between ichthyosporeans and choanoflagellates + metazoans (fig. 3).
Evolution of the Diverse TKs of Holozoans
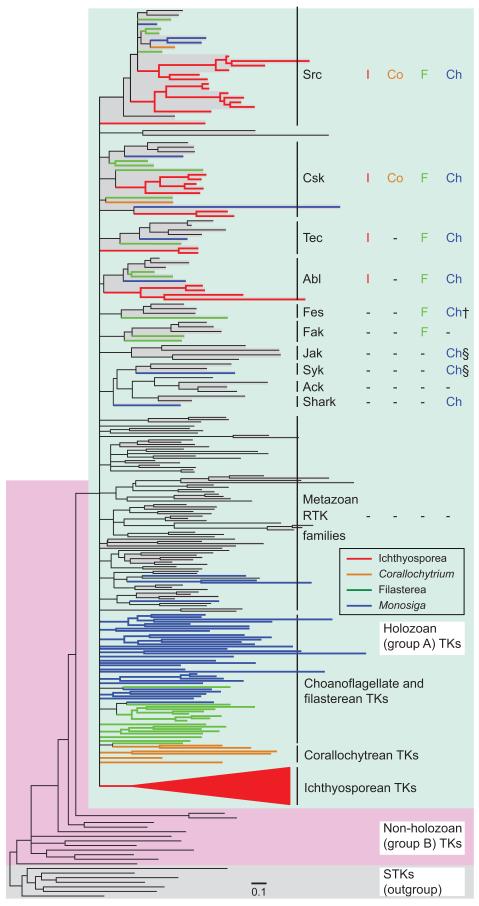
To elucidate the evolutionary history of TKs, we performed a Maximum Likelihood (ML) phylogenetic analysis (fig. 4; the details are in supplementary fig. S6, Supplementary Material online) based on the common catalytic domain sequences. All the 173 TKs identified in ichthyosporeans and Corallochytrium form a monophyletic cluster (the group A TK cluster; fig. 4 green shade) together with all the other holozoan TKs, supporting the single evolutionary origin of group A TKs from a nonholozoan group B TK (Suga et al. 2012). There is however no support for a branching close to the root of the group A TK subtree, suggesting a rapid diversification process of group A TK families.
FIG. 4.
Phylogenetic tree of the TK family. Branches with low support (bootstrap values <10%) are collapsed and represented by multifurcated nodes. The full tree showing each gene name and bootstrap value is in the supplementary figure S6, Supplementary Material online. Seven STKs are used as an outgroup. TK families are highlighted by grey shading. Group A and group B TKs are shaded in green and red, respectively. The subtree containing all RTKs and a few CTKs of ichthyosporeans is represented by a red triangle, which is fully shown in supplementary figure S8, Supplementary Material online. Presence of orthologs of 10 common metazoan CTK families in ichthyosporeans (I), a corallochytrean (Co), filastereans (F), and choanoflagellates (Ch) are shown on the right. Orthologs of metazoan RTK were not identified in premetazoans, as shown in the abbreviated row “Metazoan RTK families”. †Ortholog not found in the Monosiga brevicollis genome, but identified in Codonosiga gracilis (Suga, Sasaki, et al. 2008). §A CTK with domain architecture similar to Jak is present in M. brevicollis, but its TK domain is phylogentically close to that of Syk (Suga et al. 2012).
Our phylogenetic tree analysis confirmed the classification of the 29 ichthyosporean and corallochytrean CTKs into the four metazoan CTK families (Src, Csk, Tec, and Abl; designated as Src-related CTKs), which are also present in choanoflagellates and filastereans (fig. 4; the detailed CTK tree in supplementary fig. S7, Supplementary Material online). Frequent gene duplications in the ichthyosporean Src and Csk families are also apparent (supplementary fig. S7, Supplementary Material online). The other five CTKs of ichthyosporeans and Corallochytrium do not associate with metazoan CTK families (fig. 2; supplementary fig. S6, Supplementary Material online), indicating their independent origins.
As RNAseq data do not represent the whole transcriptome, it remains unclear whether the evolution of the other major CTK families (Fes, Fak, Jak, Ack, Syk, and Shark; fig. 4), which are common in metazoans but missing in the studied ichthyosporeans, antedates that split. The possibility that these six CTK families emerged in the last common ancestor of filastereans, choanoflagellates, and metazoans after its divergence from ichthyosporeans cannot be excluded. However, the dramatically reduced CTK repertoires in Creolimax and Sphaeroforma suggest a massive loss of CTK families that occurred as a product of evolutionary adaptation in the Ichthyosporea.
All the ichthyosporean TKs except the Src-related CTKs fall into a monophyletic cluster (red triangle in fig. 4), including all the ichthyosporean RTKs and a few non-Src-related CTKs of Amoebidium (supplementary fig. S8, Supplementary Material online). This clustering suggests an extensive RTK expansion in the ichthyosporean clade, as well as several gene duplication and domain shuffling events that transformed certain Amoebidium RTKs into CTKs. Yet, the phylogenetic tree does not provide support for monophyly of the TK family from choanoflagellates, filastereans, or corallochytreans. This suggests either their paraphyletic origins or obscured phylogenetic signal due to deep divergence.
Overall, in the basal holozoans analyzed, RTKs show contrasting diversification patterns from CTKs during holozoan evolution, similar to the results obtained from the filasterean and choanoflagellate data (Suga et al. 2012). Although the basic repertoire of CTKs was already established very early in holozoan evolution, probably at the onset of Holozoa, RTKs diversified much later independently in each of the Ichthyosporea, Corallochytrea, Filasterea, Choanoflagelliida, and Metazoa.
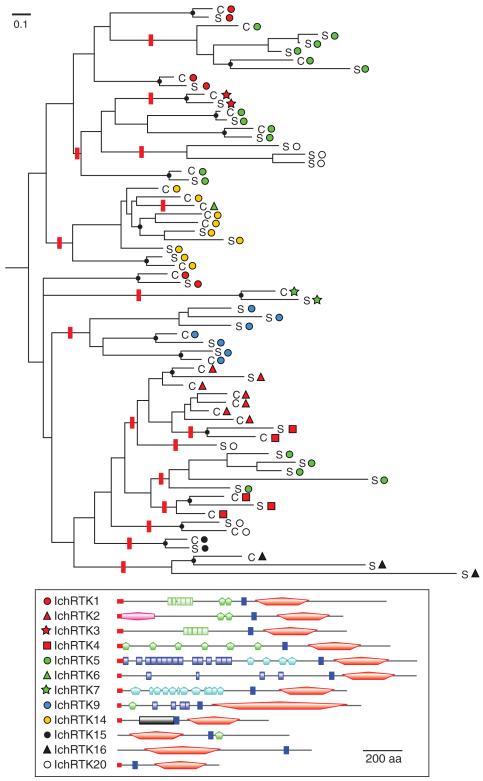
Rapid RTK Diversification within the Ichthyosporean Clade
We examined in detail the diversification history of the ichthyosporean RTKs using the whole genomic information from Creolimax and Sphaeroforma (fig. 5). These two species are very close, with their divergence time inferred as 87–163 Ma (the Mesozoic Era) from molecular dating by Bayesian analysis (fig. 6). Consistent with this dating, Creolimax and Sphaeroforma have similar RTK repertoires; 40 out of 67 RTKs are most likely orthologous to their counterparts (fig. 5; supplementary fig. S9, Supplementary Material online), sharing identical domain architectures. However, we could not identify orthologs among the remaining 27 RTKs. It is likely that these RTKs were either produced by species-specific gene duplication or were orphaned by loss of the counterparts.
FIG. 5.
Clade-specific RTK expansion in the Ichthyosporea. The phylogenetic tree was inferred from the TK sequences of Creolimax (C) and Sphaeroforma (S). The family classification is represented by symbols. A typical structure of each family is shown at the bottom. Likely orthologous divergences of Creolimax and Sphaeroforma are labeled by black dots. One of the most parsimonious scenarios of domain architecture alteration was manually inferred based on the assumption that the ancestral gene belongs to the most dispersed RTK1 family. Red bars indicate branches where an architectural alteration is assumed. The full figure with bootstrap values and domain architectures is in supplementary figure S9, Supplementary Material online.
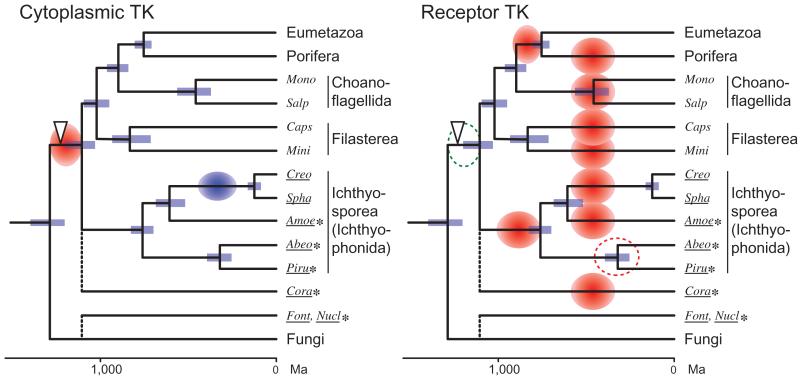
FIG. 6.
Evolution of the holozoan TK diversity. The history of family diversification of the metazoan-type (group A) TK is schematically represented. The tree is based on the time-calibrated phylogeny by Bayesian inference. Blue bars represent the 95% range of highest probability density (Drummond et al. 2012) of the node age. Red and blue circles represent extensive TK diversification and massive reduction of TK diversity, respectively. The period of RTK diversification in Abeoforma whisleri and Pirum gemmata remains unclear (red dotted circle), because RNAseq data may represent a minor fraction of TK diversity. The open arrowhead represents the evolution of the complex pTyr-mediated signaling system with the diversified pTyr signaling modules, as well as their enhanced combination. Note that we do not exclude the possibility of basal holozoan expansion of group A RTKs (green dotted circle), which may be obscured today by their rapid turnover during the holozoan evolution. Amoe, Amoebidium; Abeo, Abeoforma; Creo, Creolimax; Cora, Corallochytrium; Caps, Capsaspora; Font, Fonticula; Mini, Ministeria; Mono, Monosiga brevicollis; Nucl., Nuclearia; Piru, Pirum; Spha, Sphaeroforma; Salp, Salpingoeca. Species that were newly analyzed in this study are underlined, with the RNAseq data labeled with asterisks.
According to this tree, we parsimoniously inferred that the domain architectures of the RTK extracellular regions changed 16 times (red bars) in the course of ichthyosporean evolution. We also found that RTKs in the same family are not always monophyletic, as typically shown by the IchRTK5 family (fig. 5). These data suggest a dynamic evolution and optimization of the ichthyosporean RTK diversity by frequent gene duplications, domain shuffling, and gene conversions, as well as gene disruption and gene loss, which continued after the Mesozoic split between Creolimax and Sphaeroforma.
Discussion
Evolutionary History of pTyr Signaling
The evolution of group A TK diversity is illustrated in figure 6. Concurrently with the numerical expansions of group A TK and other pTyr signaling modules, the earliest holozoans increased the architectural complexity of pTyr signaling proteins through the combination with other protein domains. The positive or negative feedback loops in pTyr signaling cascade, which is proposed to be a critical innovation toward the efficient and complex signal transduction seen in extant metazoans (Lim and Pawson 2010), was already present in the earliest stage of holozoan evolution.
Our study has reinforced, by the use of the data from two holozoan clades (ichthyosporeans and corallochytrean) that were not analyzed previously, the scenario of contrasting diversification patterns between CTKs and RTKs, which was previously proposed from a study of filastereans and choanoflagellates (Suga et al. 2012). Holozoans have been diversifying the RTK repertoire extensively, generating the clade-specific RTK sets that are totally different from each other. Our data covering the five ichthyosporean species further demonstrated that the rapid turnover of RTK repertoire continued within the ichthyosporean clade after the divergence from other holozoan clades.
It has been hypothesized that the pTyr signaling system of premetazoan lineages was originally assembled to deal with the various extracellular information, and later co-opted for the intercellular communication at the unicell–multicell transition (King et al. 2003; Suga et al. 2012). This hypothesis is supported by the observation that the basic set of RTK did not drastically change in the Metazoa, in which RTK generally confronts the internal fluid, whereas in the premetazoan lineages, which are constantly exposed to environment, the RTK repertoire has been rapidly changing.
Possible Function of Ichthyosporean RTKs
The protein domain I3 is present, mostly as tandem repeats, in one-third of ichthyosporean RTKs. It is tempting to speculate that the I3 domains in the ichthyosporean RTKs are involved in sensing the extracellular information such as nutrient condition, or possibly the host tissue for symbiosis, similar to prokaryotic adhesins. The large variety of the I3-containing RTKs of ichthyosporeans may then contribute to the diversity of extracellular information that they are able to detect. The highly repetitive and plastic structure in the receptor regions of these RTKs may have allowed the ichthyosporeans’ rapid adaptation to various environments, by changing the receptor sequence through recombination. It is noteworthy that the hosts of ichthyosporeans are highly diverse, ranging from fish’s gill to digestive tracts of marine crustaceans (Mendoza et al. 2002). Host diversification occurs even within a single ichthyosporean species (Marshall and Berbee 2013).
As previously shown, the RTK repertoire of the filasterean Capsaspora is also highly diverse (Suga et al. 2012). In particular, one specific RTK family of Capsaspora includes 40 genes (39% of the whole TK repertoire) of similar structure, having a long receptor region that contains numerous leucine-rich repeat (LRR; fig. 3). Similar to filastereans, many RTKs of choanoflagellates contain hyalin repeats (Manning et al. 2008; Suga, Sasaki, et al. 2008), which are considered to be involved in cellular adhesion in metazoan tissues (Callebaut et al. 2000). Thus, three unicellular holozoan clades independently use different protein domains to increase the diversity of receptor region structures of RTKs. As seen in animal lens proteins, such flexible recruitment of proteins or protein domains represents an important evolutionary driving force (Piatigorsky 2003).
Conclusions
Our analyses have elucidated the history of pTyr signaling development through the course of holozoan evolution with unprecedented detail. Further molecular-level studies on the function of premetazoan pTyr signaling using available tools (Suga and Ruiz-Trillo 2013) will allow better understanding of the co-option process of this signaling system.
Materials and Methods
Cultures
The live culture of C. fragrantissima, P. gemmata, and A. whisleri were kindly provided by Dr. Wyth Marshall. The live culture of C. limacisporumI was kindly provided by Stuart Donachie (University of Hawaii, USA). The live culture of Nuclearia sp. was purchased from ATCC (ATCC50694).
Data Mining
Creolimax TKs were searched for in the draft genome sequence, which was preliminary constructed from raw reads of an approximately 70× coverage. The Sphaeroforma genome and the Fonticula genome were sequenced as a part of UNICORN project (Ruiz-Trillo et al. 2007); their version 1 assemblies were retrieved from http://www.broadinstitute.org/annotation/genome/multicellularity_project/ (last accessed December 13, 2013). The RNAseq data of Nuclearia, Corallochytrium, Pirum, and Abeoforma were produced from high quality Illumina paired-end reads of approximately 4 GB (quality score >20 in 40% bases). The RNAseq of Amoebidium (from 22 GB Illumina paired-end reads) was performed by Broad Institute within UNICORN project (data available under the NCBI BioProject PRJNA189477). The predicted proteomes were searched for TKs by the use of BLAST and HMMER (Eddy 2009) programs. For the HMMER search, both the SMART entry SM00219 and a model build from a previously published alignment (Suga et al. 2012) that include choanoflagellates and filasterean TKs were used. To discriminate between TK and STK, we used the ML phylogenetic tree analyses (discussed later), as well as the conservation of the highly characteristic motif in the catalytic loop (typically HRDLAARN/HRDLRAAN [Manning et al. 2008]). For the downstream analyses, we used only those with kinase domain sequences that have less than 50% of missing sequence (i.e., those having kinase domains with more than 112 amino acid sites aligned with other protein kinases). All the discovered ichthyosporean and corallochytrean TKs were then subjected to a manual inspection; in each protein, we examined the integrities of protein domains and the presence of signal peptide (in case of an RTK), and manually amended the prediction. In this process, we took into account also the suboptimum predictions of Augustus (Stanke and Waack 2003), which was specifically trained for each organism. Finally, we compared the domain architectures of the discovered sequences with those of other organisms to confirm the prediction adequacy. Some predicted Sphaeroforma sequences were split into pieces, due to its severely fragmented scaffolds of the current draft assembly. We manually combined these pieces as much as possible by comparing their sequences with those of Creolimax putative orthologs. We assigned the same letter to the name of each putatively orthologous pair of Creolimax and Sphaeroforma TKs (e.g., CfrTK-a and SarTK-a). As the architectures of IchRTK15 and IchRTK16 families are very atypical, we further confirmed the prediction validity of CfrTK-s, one of the IchRTK15 family genes, by RT-PCR and sequencing the obtained fragments. Gene-specific primers used for the RT-PCR are as follows: for the 5′ amplification, CfrTK-s-S0.2 (sense strand), 5′-TACTAAAGCTCAAAATGCCACG-3′; CfrTK-s-S0.3 (sense), 5′-TCTACCGACGCTGCCACC-3′; CfrTK-s-A3 (antisense), 5′-TGTTCTGCGTCCCTATCCAC-3′; for the 3′RACE, CfrTK-s-S1 (sense), 5′-CCTTCGCCATCTTACAATTC-3′; CfrTK-s-S2 (sense), 5′-AACGAGGCATATGAGGTAATG-3′.
The PTP catalytic domain, the SH2 domain, and the PTB domain were similarly searched for by the use of HMMER program, followed by the manual inspection. The used probabilistic models are SM00194 and PF00102 for PTP (for comparison, the other models SM00012 and SM00404 with a lower specificity were also used), SM00252 and PF00017 for SH2, and SM00462, PF08416, and PF00640 for PTB domain. For the PTP-domain containing proteins, only the members of the classical tyrosine-specific phosphatase family (Tonks 2006) were considered. The dual specificity phosphatases were thus excluded.
The pTyr signaling modules were further searched across eukaryotes. In addition to the previously published data (Manning et al. 2008; Suga et al. 2012; Clarke et al. 2013), we searched the whole genome data of the heterolobosean Naegleria gruberi (version 1; http://genome.jgi-psf.org/Naegr1, last accessed December 13, 2013), the zygomycete fungus Rhizopus oryzae (version 3; http://www.broadinstitute.org/annotation/genome/rhizopus_oryzae, last accessed December 13, 2013), the filasterean Capsaspora owczarzaki (version 2; http://www.broadinstitute.org/annotation/genome/multicellularity_project/, last accessed December 13, 2013), and the sea anemone Nematostella vectensis (version 1; http://genome.jgi-psf.org/Nemve1/, last accessed December 13, 2013).
Protein Domain Architecture and TK Classification
Protein domains composing the ichthyosporean and corallochytrean TKs were annotated by the search of SMART (http://smart.embl-heidelberg.de, last accessed December 13, 2013) and Pfam (http://pfam.sanger.ac.uk/, last accessed December 13, 2013) databases, which were then subjected to manual inspections of the result alignments. Sequences that were not mapped to any known domain were further searched for in the public databases and in other ichthyosporean TKs, and two domains (I1 and I3 domains) were newly annotated. TM segments and signal peptides of RTKs were manually predicted. For the detailed classification of CTK families, their N-terminal myristoylation sites were predicted by the Myristoylator program (Bologna et al. 2004), which provides the prediction confidence by the score S. We classified TKs into families based on their domain architectures and phylogenetic positions as in (Suga et al. 2012). TKs without clear domain architectural information or an apparent affiliation to a specific family supported by the phylogeny were provisionally classified into IchRTK20 (for ichthyosporean genes) or CorRTK4 (for corallochytrean genes) families.
Network Analysis of Domain Architecture
Domain architectures of proteins containing the four pTyr signaling domains (TK catalytic, PTP catalytic, SH2, and PTB domains) were manually examined. The domain combinations occurring in these proteins were represented by a network drawn by Cytoscape (http://www.cytoscape.org/, last accessed December 13, 2013). In the network, co-occurring two domains are represented by two circles directly connected by a line. Duplicated domains in a protein were not counted.
Phylogenetic Tree Inference
Phylogenetic trees were inferred from the kinase (catalytic) domain sequences (225 amino acid positions) by the Maximum Likelihood (ML) method, using the LG-Γ model with four rate categories implemented in RAxML version 7.2.8 (Stamatakis 2006). The alignment is in supplementary fig. S10, Supplementary Material online. The obtained topology was further optimized by the genetic algorithm-based heuristic approach (Katoh et al. 2001; Suga, Schmid, et al. 2008; Suga et al. 2012), in which all the possible topologies generated both by crossing over all trees in the population and by the subtree pruning and regrafting (SPR) and tree bisection and reconnection (TBR) algorithms started from the best tree were evaluated. To avoid the long-branch attraction artifact, Awh_RS39176 of Abeoforma and Apa_RS28315.0.1 of Amoebidium were excluded from phylogenetic analyses.
Molecular Dating by Bayesian Inferences
We used the methodology from the previous publication (Parfrey et al. 2011). Data from Salpingoeca rosetta, Ministeria, Creolimax, Corallochytrium, Pirum, and Abeoforma were added to the original alignment constituted by 15 proteins (5,695 amino acid sites in total). The Sphaeroforma and Amoebidium data, which had already been included in the original alignment, were additionally supplemented with the new genome and RNAseq data of this study. The alignment was then manually inspected and amended, and then used for Bayesian inferences by the Beast program (Drummond et al. 2012). We analyzed the data using the setting A (Parfrey et al. 2011), where the root was constrained to the Opisthokonta and all calibration points were used.
Protein Charge Plot
We analyzed the 45 amino acids around the boundary between the TM segment and the non-TM sequence (30 amino acids outside the TM segment and 15 amino acids within the TM segment). As a sliding window of five amino acids moved along the sequence from the TM toward the non-TM region, the averaged charge of five amino acids in the window was calculated. This value was considered as the charge at the first position of the window. In the scanning process, the residue of aspartic acid and glutamic acid gave a charge of − 1, lysine and arginine a charge of + 1, and histidine a charge of + 0.5. The values were plotted along the sequence.
Supplementary Material
Acknowledgments
The authors thank UNICORN and Broad Institute for sequencing the S. arctica genome and generating the A. parasiticum RNAseq data, Dr Wyth Marshall for providing the cultures of C. fragrantissima, P. gemmata, and A. whisleri, Dr Stuart Donachie (University of Hawaii, USA) for providing the culture of C. limacisporum, and Dr Laura Wegener Parfrey (University of Colorado, Colorado, USA) for providing the alignment used for the molecular dating. This work was supported by the European Research Council Starting grant ERC-2007-StG-206883 to I.R.-T; Ministerio de Ciencia e Innovación grant BFU2008-02839/BMC to I.R.-T.; and the Marie Curie Intra-European Fellowship (MMEMA) within the 7th European Community Framework Programme to H.S.
Footnotes
Supplementary Material Supplementary figures S1–S10 are available at Molecular Biology and Evolution online (http://http://mbe.oxfordjournals.org/).
References
- Bologna G, Yvon C, Duvaud S, Veuthey AL. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics. 2004;4:1626–1632. doi: 10.1002/pmic.200300783. [DOI] [PubMed] [Google Scholar]
- Bonner J. The origins of multicellularity. Integr Biol. 1998;1:27–36. [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Gilges D, Vigon I, Mornon JP. HYR, an extracellular module involved in cellular adhesion and related to the immunoglobulin-like fold. Protein Sci. 2000;9:1382–1390. doi: 10.1110/ps.9.7.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. A revised six-kingdom system of life. Biol Rev. 1998;73:203–266. doi: 10.1017/s0006323198005167. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Allsopp MTEP. Corallochytrium, an enigmatic non-flagellate protozoan related to choanoflagel-lates. Eur J Protistol. 1996;32:306–310. [Google Scholar]
- Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Alonso S, Reveneau N, Willery E, Quatannens B, Locht C, Jacob-Dubuisson F. Role of adhesin release for mucosal colonization by a bacterial pathogen. J Exp Med. 2003;197:735–742. doi: 10.1084/jem.20021153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Fantl WJ, Johnson DE, Williams LT. Signalling by receptor tyrosine kinases. Annu Rev Biochem. 1993;62:453–481. doi: 10.1146/annurev.bi.62.070193.002321. [DOI] [PubMed] [Google Scholar]
- Glockling SL, Wyth LM, Gleason FH. Phylogenetic interpretations and ecological potentials of the Mesomycetozoea (Ichthyosporea) Fungal Ecol. 2013;6:237–247. [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu Rev Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatsuka CS, Julio SM, Cotter PA. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc Natl Acad Sci U S A. 2005;102:18578–18583. doi: 10.1073/pnas.0507910102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Miyata T. Genetic algorithm-based maximum-likelihood analysis for molecular phylogeny. J Mol Evol. 2001;53:477–484. doi: 10.1007/s002390010238. [DOI] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- Klemm P, Schembri MA. Bacterial adhesins: function and structure. Int J Med Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- Lim WA, Pawson T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell. 2010;142:661–667. doi: 10.1016/j.cell.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Steenkamp ET, Brinkmann H, Forget L, Philippe H, Lang BF. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular cell biology, Section 17.5 Insertion of membrane proteins into the ER membrane. W.H. Freeman; New York: [cited 2013 Dec 13]. 2000. Available from: http://www.ncbi.nlm.nih.gov/books/NBK21731/ [Google Scholar]
- Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WL, Berbee ML. Comparative morphology and genealogical delimitation of cryptic species of sympatric isolates of Sphaeroforma (Ichthyosporea, Opisthokonta) Protist. 2013;164:287–311. doi: 10.1016/j.protis.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Mendoza L, Taylor JW, Ajello L. The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu Rev Microbiol. 2002;56:315–344. doi: 10.1146/annurev.micro.56.012302.160950. [DOI] [PubMed] [Google Scholar]
- Paps J, Medina-Chacón LA, Marshall WL, Suga H, Ruiz-Trillo I. Molecular phylogeny of Unikonts: new insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist. 2013;164:2–12. doi: 10.1016/j.protis.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J. Crystallin genes: specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phosphotyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci U S A. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu-kumar S. Occurrence of the thraustochytrid, Corallochytrium limacisporum gen. et sp. nov. in the coral reef lagoons of the Lakshadweep Islands in the Arabian Sea. Botanica Marina. 1987;30:83–89. [Google Scholar]
- Ruiz-Trillo I, Burger G, Holland PW, King N, Lang BF, Roger AJ, Gray MW. The origins of multicellularity: a multi-taxon genome initiative. Trends Genet. 2007;23:113–118. doi: 10.1016/j.tig.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Lane CE, Archibald JM, Roger AJ. Insights into the evolutionary origin and genome architecture of the unicellular opisthokonts Capsaspora owczarzaki and Sphaeroforma arctica. J Eukaryot Microbiol. 2006;53:379–384. doi: 10.1111/j.1550-7408.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS One. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol Biol Evol. 2004;21:828–840. doi: 10.1093/molbev/msh077. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylo-genetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Suga H, Dacre M, de Mendoza A, Shalchian-Tabrizi K, Manning G, Ruiz-Trillo I. Genomic survey of pre-metazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signal. 2012;5:ra35. doi: 10.1126/scisignal.2002733. [DOI] [PubMed] [Google Scholar]
- Suga H, Katoh K, Miyata T. Sponge homologs of vertebrate protein tyrosine kinases and frequent domain shufflings in the early evolution of animals before the parazoan-eumetazoan split. Gene. 2001;280:195–201. doi: 10.1016/s0378-1119(01)00784-3. [DOI] [PubMed] [Google Scholar]
- Suga H, Koyanagi M, Hoshiyama D, Ono K, Iwabe N, Kuma K, Miyata T. Extensive gene duplication in the early evolution of animals before the parazoan-eumetazoan split demonstrated by G proteins and protein tyrosine kinases from sponge and hydra. J Mol Evol. 1999;48:646–653. doi: 10.1007/pl00006508. [DOI] [PubMed] [Google Scholar]
- Suga H, Kuma K, Iwabe N, Nikoh N, Ono K, Koyanagi M, Hoshiyama D, Miyata T. Intermittent divergence of the protein tyrosine kinase family during animal evolution. FEBS Lett. 1997;412:540–546. doi: 10.1016/s0014-5793(97)00639-x. [DOI] [PubMed] [Google Scholar]
- Suga H, Ruiz-Trillo I. Development of ichthyosporeans sheds light on the origin of metazoan multicellularity. Dev Biol. 2013;377:284–292. doi: 10.1016/j.ydbio.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Sasaki G, Kuma K, Nishiyori H, Hirose N, Su ZH, Iwabe N, Miyata T. Ancient divergence of animal protein tyrosine kinase genes demonstrated by a gene family tree including choanoflagellate genes. FEBS Lett. 2008;582:815–818. doi: 10.1016/j.febslet.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. Curr Biol. 2008;18:51–55. doi: 10.1016/j.cub.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Tan JL, Spudich JA. Developmentally regulated protein-tyrosine kinase genes in Dictyostelium discoideum. Mol Cell Biol. 1990;10:3578–3583. doi: 10.1128/mcb.10.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol Biol Evol. 2012;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Geer P, Hunter T, Lindberg RA. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.