Abstract
Phylogenomic analyses have revealed several important metazoan clades, such as the Ecdysozoa and the Lophotrochozoa. However, the phylogenetic position of a few taxa, such as ctenophores, chaetognaths, acoelomorphs, and Xenoturbella remain contentious. Thus, the finding of qualitative markers or “Rare Genomic Changes” seem ideal to independently test previous phylogenetic hypotheses. We here describe a rare genomic change, the presence of the gene UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase (GNE). We show that GNE is encoded in the genomes of deuterostomes, acoelomorphs and Xenoturbella while it is absent in protostomes and non-bilaterians. Moreover, the GNE has a complex evolutionary origin involving unique lateral gene transfer events and/or extensive hidden paralogy for each protein domain. However, rather than using GNE as a phylogenetic character, we argue that rare genomic changes such as the one presented here should be used with caution.
Keywords: molecular markers, metazoan phylogeny, lateral gene transfer, Xenoturbella, UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase, Acoels
Molecular signatures that characterize a clade are an important tool to create or corroborate phylogenetic groupings (reviewed in Telford and Copley 2011). Examples of such molecular synapomorphies include the presence of Hox/ParaHox genes in the ParaHoxozoa (Placozoa, Cnidaria and Bilateria) (Ryan et al. 2010), an indel in the gene elongation-factor 1-alpha as a character of opisthokonts (Steenkamp, Wright and Baldauf 2006) or the the NAD5 mithocondrial gene that is exclusive to protostomes (Papillon et al 2004).
The monophyly of deuterostomes (the clade that comprises chordates, echinoderms, and hemichordates) has been consistently recovered on phylogenetic and phylogenomic studies (Hejnol et al. 2009; Paps, Baguñà and Riutort 2009). However, it remains contentious whether Xenoturbella and/or the acoelomorphs (acoels and nemertodermatids) are members of the deuterostomes or basal bilaterians (Ruiz-Trillo et al. 1999; Ruiz-Trillo et al. 2002; Bourlat et al. 2003; Bourlat et al. 2006; Philippe et al. 2007; Dunn et al. 2008; Hejnol et al. 2009; Paps, Baguñà and Riutort 2009; Mwinyi et al. 2010; Philippe et al. 2011). Thus the identification of diagnostic molecular synapomorphies for deuterostomes is important to both corroborate previous molecular analyses and independently test the putative deuterostome affiliation of acoels and Xenoturbella.
We here show that the bi-functional enzyme UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase (GNE), is exclusive to deuterostomes, acoels and Xenoturbella, being absent from all sequenced protostomes and non-bilaterian taxa. Our data show that GNE is encoded in the genomes of all sequenced deuterostomes except for the urochordates Ciona savignyi, C. intestinalis and Oikopleura dioica, most likely an effect of secondary gene loss (D’Aniello et al. 2008; Churcher and Taylor 2009). Moreover, a small fragment of the gene is present in the expressed sequence tags (EST) of Xenoturbella bocki and we have amplified the gene GNE from the acoel Symsagittifera roscoffensis (GenBank JF826132). We searched publicly available EST data as well as unpublished transcriptome data from nemertodermatids and did not get any hit. However, since a complete genome of a nemertodermatid is not available, we can not discard the presence of GNE in this group. Interestingly, the GNE encoded by chordates, echinoderms and hemichordates all share the same 9 introns (both in position and phase), while the GNE of the acoel S. roscoffensis does not share any intron with deuterostomes (Figure S1). Unfortunately, we could not elucidate the intronexon structure of the GNE encoded by X. bocki since only a small cDNA fragment is available.
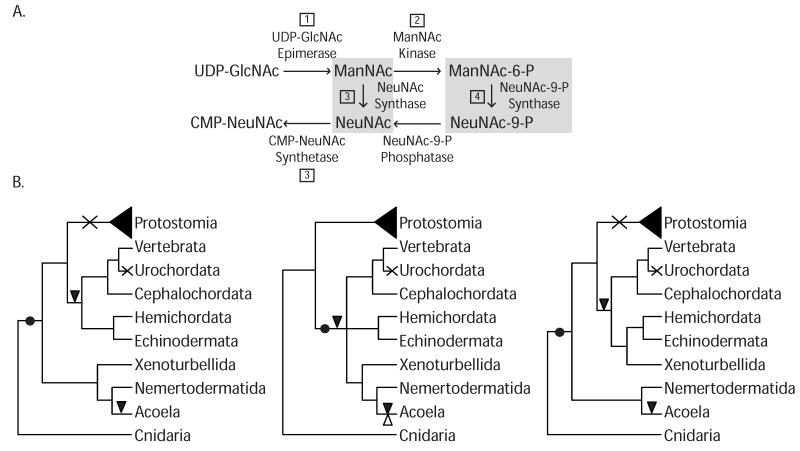
GNE is known to play an important role in the biosynthesis of sialic acids, which are monosaccharides that act in a wide range of biological and pathological events, such as cellular adhesion, recognition determinants, tumorigenesis and stem cells (Effertz, Hinderlich and Reutter 1999; Tanner 2005; Weidemann et al. 2010). In mammals, the metabolic precursor of sialic acids is the N-acetylneuraminic (Neu5Ac) acid, which derives from UDP-N-acetylglucosamine (UDP-GlcNAc). The first two steps of this reaction are catalyzed by the bi-functional enzyme UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase (GNE) (Figure 1). The bi-functional activity of GNE comes from two different protein domains: the UDP-N-acetylglucosamine 2-epimerase domain (PF02350) (from herein the “epimerase-2 domain”) and a kinase domain known as ROK (Repressor, ORF, Kinase PF00480) (Tanner 2005). The epimerase-2 domain converts UDP-GlcNAc to N-acetylmannosamine (ManNAc), which is consecutively phosphorylated to ManNAc-6-P by the ROK domain (Figure 1). Interestingly, while in prokaryotes the epimerase and kinase functions are carried out by two separate enzymes, in vertebrates those two domains have been fused, allowing an allosteric site to appear, and thus conferring the potential for new functions to arise.
Figure 1.
A) Schematic representation of the biosynthesis pathway of CMP-NeuNAc both in bacteria and mammals (adapted from (Tanner 2005). Steps 1 and 2 are performed by GNE in deuterostomes. Step 3 is exclusive of Bacteria; while the additional step 4 has so-far only been described in mammals. B) Cladogram representing GNE evolution among the Bilateria according to different current phylogenetic scenarios. Black dot indicates acquisition of the GNE, cross shows secondary loss, black triangle shows intron gain and white triangle indicates intron loss.
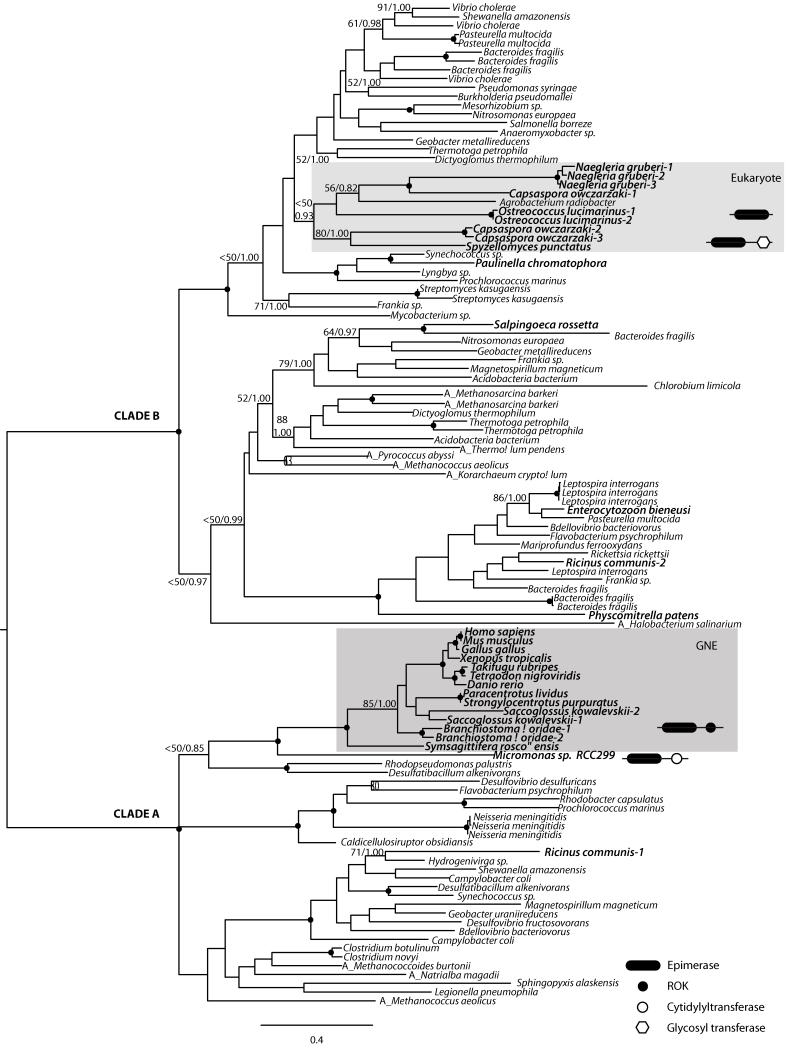
Thus, to gain further insights into the evolutionary origin of GNE, we performed both exhaustive searches across the public databases and phylogenetic analyses of the two domains independently. Our data show that both domains have a patchy distribution across eukaryotes. The Epimerase-2 domain is encoded in a few eukaryotic genomes, being, in contrast, ubiquitous among Archaea and Eubacteria (Figure 2). The phylogenetic analyses show two major clades, one comprising the bilaterian-specific GNE genes within a mostly prokaryotic clade (clade A in Figure 2), the other comprising most other (non-metazoan) eukaryotes branching also within a prokaryotic clade (clade B in Figure 2). Both clades are divided with high nodal support (Bootstrap Value (BV) = 100%, and Bayesian Posterior Probability (PP) = 1.00), and both have specific indel characters (see Figure S2). The general topology of the epimerase-2 domain is probably due to either i) an extreme case of hidden paralogy (i.e, this protein domain was present at the origin of eukaryotes and lost in all lineages except in deuterostomes, Xenoturbella, Acoela and a few other eukaryotes), ii) domain convergence, or iii) the consequence of several independent Lateral Gene Transfer (LGT) events to the different eukaryotic lineages, one being a LGT event to the last common bilaterian ancestor (and then subsequently lost in protostomes and in urochordates), or to the last common ancestor of deuterostomes (and lost in urochordates), if xenacoelomorphs (Xenoturbella + acoelomorphs) are indeed deuterostomes as recently suggested (Philippe et al. 2011). Interestingly, the sequence from Micromonas sp. falls as sister group to bilaterians. However, the Micromonas gene, in contrast to the deuterostome sequences, has no introns. Moreover, neither M. pusilla (the other congeneric species with its complete genome sequenced), or any other sequenced clorophyte (except Ostreococcus lucimarinus, whose epimerase gene is far related; see Figure 2) encode this domain. Thus, Micromonas epimerase-2 sequence, as well as Ricinus homolog which branches within another independent bacterial clade (Figure 2), most probably come from independent LGT events (see Supplementary Material).
Figure 2.
Maximum likelihood (ML) phylogenetic tree of the Epimerase-2 domain as obtained by RAxML (WAG + Γ + I). The tree is rooted using the midpoint-rooted tree option. Nodal support was obtained by RAxML 100-bootstrap replicates (BV) and Bayesian Posterior Probabilities (PP). Both values are shown on key branches. A black dot in the node indicates BV >95% and PP > 0.95. Eukaryotes are shown in bold. “A_” before species name indicates archaeal sequences. Characteristic domain architecture shown in key taxa or lineages.
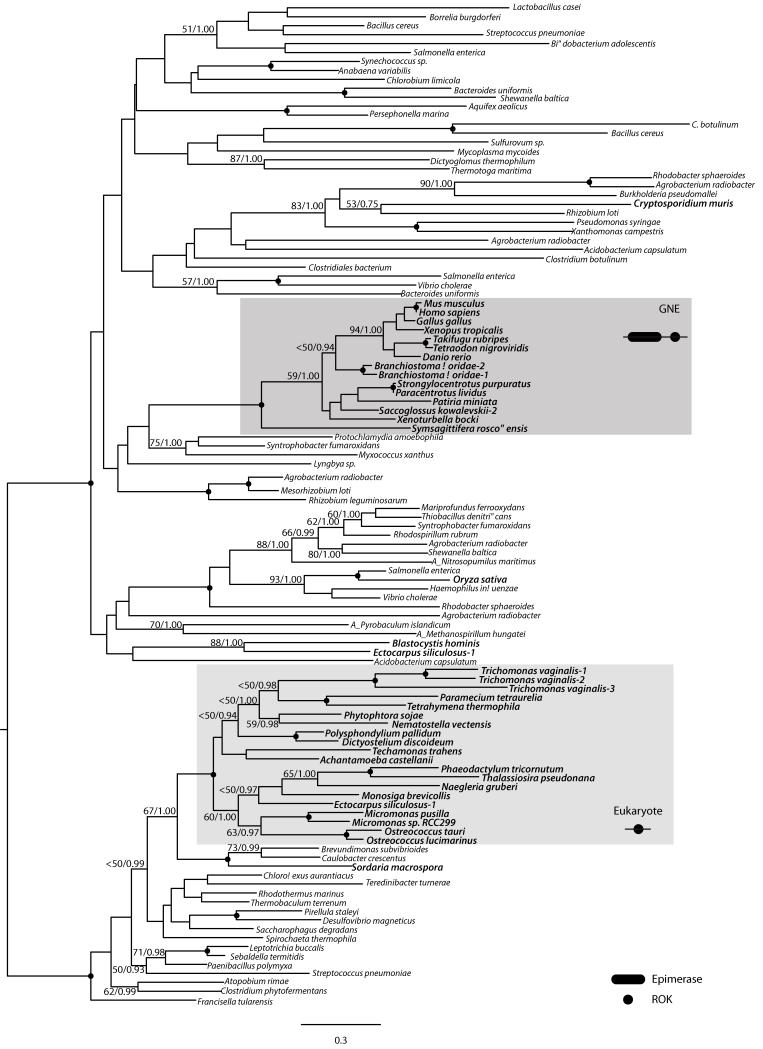
The ROK domain also bears a complex evolutionary history (Figure 3). The phylogenetic analysis shows most eukaryotic sequences in a single monophyletic group that also includes Eubacteria. The bilaterian GNE genes are monophyletic and unrelated to the other eukaryotic sequences (also supported by indel characters, see Figure S2B). Again, this topology can either be explained by LGT or by hidden paralogy.
Figure 3.
Maximum likelihood (ML) phylogenetic tree of the ROK domain as obtained by RAxML (WAG + Γ + I). The tree is rooted using the eukaryotes as out-group. RAxML 100-bootstrap replicates (BV) and Bayesian Posterior Probabilities (PP) values are shown on key branches. A black dot in the node indicates BV > 95% and BPP > 0.95. Eukaryotes are shown in bold. “A_” before species name indicates archaeal sequences. Characteristic domain architecture are shown in key lineages.
Our data show that GNE (the derived gene fusion of epimerase-2 and ROK domains) is exclusive to most deuterostomes, acoelomorphs and Xenoturbella. How this presence/absence scheme is interpreted remains unclear. One could easily propose the presence of the gene GNE as a molecular synapomorphy of deuterostomes (Figure 1B). This would corroborate the recent, although lowly supported, proposal that xenacoelomorphs are deuterostomes (Philippe, H. et al 2011). However if Xenacoelomorpha or just the Acoelomorpha are not deuterostomes but basal bilaterians, as most phylogenetic analyses suggest (Ruiz-Trillo et al. 1999; Ruiz-Trillo et al. 2002; Hejnol et al. 2009; Paps, Baguñà and Riutort 2009; Mwinyi et al. 2010), then GNE was secondarily lost in the last common protostome ancestor. This is indeed not a difficult scenario, specially considering that gene loss must already be hypothesized for urochordates (Figure 1B). The analysis of intron composition shows the GNE encoded by acoels and deuterostomes independently evolved their own introns (Figure S1), somehow supporting acoels are not deuterostomes. However, independent intron evolution within acoels could easily be argued as well; as has been described, for example, in the urochordate Oikopleura dioica (Edvardsen et al. 2004). To sum up, although rare genomic changes can be important phylogenetic markers, they should be used with caution, since gene loss has been shown to play an important role in eukaryotic evolution (see for example Sebé-Pedrós et al. 2011 and Zmasek and Godzik 2011). Additional data, especially from phylogenomics analyses, should be taken into account. Finally, the two domains that make up the GNE gene have complex evolutionary histories, most likely involving LGT events or extreme hidden paralogy.
Supplementary Material
Acknowledgments
We thank Albert Poustka and Pere Martínez for allowing us to use the genomic raw data from Symsagittifera roscoffensis. We thank Andreas Hejnol for accession to Meara stichopi data and helpful discussion. We thank Marta Chiodin for providing the cDNA from S. roscoffensis. We thank Jordi Paps, Lora L. Shadwick, Marta Riutort, Jaume Baguñà and Pere Martínez for helpful insights and motivation. This work was supported by an ICREA contract, an European Research Council Starting Grant (ERC-2007-StG-206883), and a grant (BFU2008-02839/BMC) from Ministerio de Ciencia e Innovación (MICINN) to I.R.-T. A.d.M.’s salary was supported by a pregraduate FPI grant from MICINN.
Footnotes
Supplementary material file 1 includes all sequences and alignments used in this study. Alignments can also be downloaded from the webpage www.multicellgenome.com.
Supplementary material file 2 includes the Methods as well as Figure S1.
Supplementary material file 3 includes Figure S2 and Supplementary material file 4 includes Figure S3.
Literature cited
- Bourlat SJ, Nielsen C, Lockyer AE, Littlewood DTJ, Telford MJ. Xenoturbella is a deuterostome that eats molluscs. Nature. 2003;424:925–928. doi: 10.1038/nature01851. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–8. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Rota-Stabelli O, Lanfear R, Telford MJ. The mitochondrial genome structure of Xenoturbella bocki (phylum Xenoturbellida) is ancestral within the deuterostomes. BMC Evol Biol. 2009;9:107. doi: 10.1186/1471-2148-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher AM, Taylor JS. Amphioxus (Branchiostoma floridae) has orthologs of vertebrate odorant receptors. BMC Evol Biol. 2009;9:242. doi: 10.1186/1471-2148-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jimenez-Delgado S, Bertrand S, García-Fernàndez J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol. Biol. Evol. 2008;25:1841–1854. doi: 10.1093/molbev/msn132. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. 18 co-authors. [DOI] [PubMed] [Google Scholar]
- Edvardsen RB, Lerat E, Maeland AD, Flat M, Tewari R, Jensen MF, Lehrach H, Reinhardt R, Seo HC, Chourrout D. Hypervariable and highly divergent intron-exon organizations in the chordate Oikopleura dioica. J. Mol Evol. 2004;59:448–57. doi: 10.1007/s00239-004-2636-5. [DOI] [PubMed] [Google Scholar]
- Effertz K, Hinderlich S, Reutter W. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J Biol Chem. 1999;274:28771–28778. doi: 10.1074/jbc.274.40.28771. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. 17 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwinyi A, Bailly X, Bourlat SJ, Jondelius U, Littlewood DT, Podsiadlowski L. The phylogenetic position of Acoela as revealed by the complete mitochondrial genome of Symsagittifera roscoffensis. BMC Evol Biol. 2010;10:309. doi: 10.1186/1471-2148-10-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon D, Perez Y, Caubit X, Le Parco Y. Identification of chaetognaths as protostomes is supported by the analysis of their mitochondrial genome. Mol. Biol. Evol. 2004;21:2122–2129. doi: 10.1093/molbev/msh229. [DOI] [PubMed] [Google Scholar]
- Paps J, Baguñà J, Riutort M. Bilaterian phylogeny: a broad sampling of 13 nuclear genes provides a new Lophotrochozoa phylogeny and supports a paraphyletic basal acoelomorpha. Mol. Biol. Evol. 2009;26:2397–2406. doi: 10.1093/molbev/msp150. [DOI] [PubMed] [Google Scholar]
- Philipp H, Brinkmann H, Martinez P, Riutort M, Baguñà J. Acoel flatworms are not platyhelminthes: evidence from phylogenomics. PLoS ONE. 2007;2:e717. doi: 10.1371/journal.pone.0000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DT, Herniou EA, Baguñá J. Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Paps J, Loukota M, Ribera C, Jondelius U, Baguñá J, Riutort M. A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proc Natl Acad Sci U S A. 2002;99:11246–11251. doi: 10.1073/pnas.172390199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo. 2010;1:9. doi: 10.1186/2041-9139-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol. Biol. Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol. Biol. Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Tanner ME. The enzymes of sialic acid biosynthesis. Bioorg. Chem. 2005;33:216–228. doi: 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Copley RR. Improving animal phylogenies with genomic data. Trends in genetics : Trends Genet. 2011;27:186–195. doi: 10.1016/j.tig.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Weidemann W, Klukas C, Klein A, Simm A, Schreiber F, Horstkorte R. Lessons from GNEdeficient embryonic stem cells: sialic acid biosynthesis is involved in proliferation and gene expression. Glycobiology. 2010;20:107–117. doi: 10.1093/glycob/cwp153. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 2011;12:R4. doi: 10.1186/gb-2011-12-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.