Array-based comparative genomic hybridization is widely used within precision medicine programs to match molecular alterations with therapeutic orientations. In an analysis of cell lines and two large precision medicine trials, we show that different centralization strategies may have a profound impact on alteration calls, and consequently, on therapeutic recommendations.
Keywords: precision medicine, comparative genomic hybridization, aCGH, targeted therapy
Abstract
Background
Comparative genomic hybridization (CGH) arrays are increasingly used in personalized medicine programs to identify gene copy number aberrations (CNAs) that may be used to guide clinical decisions made during molecular tumor boards. However, analytical processes such as the centralization step may profoundly affect CGH array results and therefore may adversely affect outcomes in the precision medicine context.
Patients and methods
The effect of three different centralization methods: median, maximum peak, alternative peak, were evaluated on three datasets: (i) the NCI60 cell lines panel, (ii) the Cancer Cell Line Encyclopedia (CCLE) panel, and (iii) the patients enrolled in prospective molecular screening trials (SAFIR-01 n = 283, MOSCATO-01 n = 309), and compared with karyotyping, drug sensitivity, and patient-drug matching, respectively.
Results
Using the NCI60 cell lines panel, the profiles generated by the alternative peak method were significantly closer to the cell karyotypes than those generated by the other centralization strategies (P < 0.05). Using the CCLE dataset, selected genes (ERBB2, EGFR) were better or equally correlated to the IC50 of their companion drug (lapatinib, erlotinib), when applying the alternative centralization. Finally, focusing on 24 actionable genes, we observed as many as 7.1% (SAFIR-01) and 6.8% (MOSCATO-01) of patients originally not oriented to a specific treatment, but who could have been proposed a treatment based on the alternative peak centralization method.
Conclusion
The centralization method substantially affects the call detection of CGH profiles and may thus impact precision medicine approaches. Among the three methods described, the alternative peak method addresses limitations associated with existing approaches.
introduction
The detection of gene copy number aberrations (CNAs) by array-based comparative genomic hybridization (aCGH) is extensively used to decipher the molecular landscape of tumors in modern scientific programs [e.g. The Cancer Genome Atlas, Cancer Cell Line Encyclopedia (CCLE), the Cancer Genome Project] [1–3]. Combined with the identification of gene mutations, aCGH profiling is also part of precision medicine programs (e.g. SAFIR-01, MOSCATO-01, WINTHER) [4–6], guiding the prescription of a number of molecular targeted agents [7–9]. However, the rules to define amplifications from aCGH profiles are still unclear. Particularly, amplifications are related to signal magnitudes from a baseline, considered as a neutral (or 2n) copy numbers (CNs). Therefore, this baseline appears to be fundamental in genomic analysis of CN alterations, and may have profound consequences on the use of aCGH in precision medicine programs.
Regarding the aCGH analysis framework itself (supplementary Figure S1, available at Annals of Oncology online), most attention has been focused on the development of highly efficient segmentation algorithms, such as the circular binary segmentation (CBS) or hidden Markov models [10, 11], and on identifying significant regions of interest, such as GISTIC [12]. However, the importance of the centralization step is often underestimated. Its aim is to adjust the entire profile on a zero value, which is to facilitate comparisons across samples based using a neutral level (a normal 2 copies count), from which DNA fragments will be defined as gained or lost. Therefore, it is a crucial step that may affect decision-making criteria in the matching of genomic aberrations with targeted therapies.
A commonly used strategy consists in centralizing the LogR on their mean or their median [13]. Several more elaborated methods have been proposed, and are included in global analysis pipelines. CGHcall [14] uses a supplementary post-segmentation centralization. CGHnormaliter [15] performs an iterative normalization method, where the centralization and imbalances are optimized by a repeated two-step procedure. The popLowess algorithm suggests an efficient alternative for adjusting bias due to cyanines, when two-channel hybridizations are used [16]. Two other algorithms, PAIR and genome alteration print, dramatically differ from the others since they use snp probes signals to infer the tumor ploidy [17, 18]. Unfortunately, this latter information is not available on all the platforms (e.g. Agilent platforms), which precludes the use of these methods on all aCGH arrays. A detailed discussion of these methods is beyond the scope of this article and has been addressed comprehensively elsewhere [14–18].
An interesting approach models the LogR as a mixture of several Gaussian distributions: after estimating the parameters of the mixture, the mean of the highest peak, ∼95% of the main density peak, is used as a centralization value [19]. Exploring the LogR densities gives a good overview of the different levels of imbalances. However, using this method for choosing the right profile centralization value appears frequently not trivial, since there is not always only one clear and unambiguous peak density choice (supplementary Figure S1, available at Annals of Oncology online). Moreover, the main density peak corresponds to the region of the most commonly observed values. In case of predominantly aneuploid samples, this region would not represent a neutral 2/2 copies ratio, but rather a higher ratio, relative to the main sample ploidy. For this reason, we are introducing a new central value estimator, which we called the alternative centralization. This approach, based on the LogR density analysis, uses a more flexible rule in order to capture the remaining 2-copies population, when exists, and use it as a possibly more accurate adjustment value.
By comparing this rule to standard approaches, we investigated in this work how different centralization methods influence on the genomic profiles, and thus impact the decision making in precision medicine programs. We first applied different centralization strategies on the NCI60 cell lines panel and evaluated each approach by comparing the corresponding genomic profiles with the expected values deduced from the karyotypes. Next, we processed a large panel of cell lines, labeled for drug sensitivity, and correlated gene CNs with drug sensitivities for their recognized companion actionable genes. Finally, we described the impact of these centralization methods on the identification of actionable genes using patients' data from two prospective molecular screening trials (SAFIR-01 and MOSCATO-01, NCT01414933, and NCT01566019, respectively).
patients and methods
karyotypes
The NCI panel karyotypes information was obtained from SKY/M-FISH and CGH Database [20, 21]. We generated genomic-like profiles from the karyotypic annotations as follows: for each cell line, each fully annotated segment count was used as an estimate for the corresponding region CN. Not fully annotated segments (missing start and/or end cytoband) were not considered. Data were then transformed into Log2(CN/2). The python script is available at http://nbviewer.ipython.org/gist/fredcommo/9334224. Among the NCI60 cell lines, 57 of 60 had both aCGH data and karyotype with sufficient information to reconstruct a profile. In case of replicates, only the best aCGH profile was considered (lowest derivative Log2 Ratio spread).
cell lines panels and patients datasets
The NCI60 NimbleGen Whole Genome 385K microarray data were downloaded from Gene Expression Omnibus (id: GSE30291). These data represented 71 aCGH experiments carried out on 60 individual cell lines.
The aCGH SAFIR01 Affymetrix-snp6 CEL files (n = 125) and Agilent-4 × 180K FE files (n = 158) were downloaded from Synapse (https://www.synapse.org/#!Synapse:syn2286494) [4].
The MOSCATO-01 Agilent-4 × 180K FE files (n = 309) were downloaded from Gene Expression Omnibus (GEO id under process).
The CCLE Affymetrix snp6 CEL files and drug responses were downloaded from http://www.broadinstitute.org/ccle/home. We only considered the 487 cell lines for which responses for the 24 explored compounds were available.
processing the aCGH profiles
To estimate the NCI60 aCGH-based genomic profiles, Log2(Cy3/Cy5) were computed from the provided paired files, after cyanine bias correction and GC% adjustment.
For the MOSCATO-01 Agilent FE files, LogR were computed using the two-channel intensities, after cyanine bias correction and GC% adjustment.
The SAFIR01 and the CCLE Affymetrix snp6 CEL files were preprocessed using the Affymetrix Genotyping Console, version 4.1.4.840.
In all cases, genomic profiles were generated from LogR, using the same pipeline.
expectation–maximization optimization
In each case, the LogR distribution was modeled as a mixture of several Gaussian variables, with potentially different mean and standard deviation. The parameters of the Gaussian mixture were estimated using an expectation–maximization (EM) algorithm [22] using the R package mclust [23]. Then, the centralization values were chosen using two different strategies, as follow: (i) maximal centralization: the centralization value was defined as the mean of the major density peak, (ii) alternative centralization: the centralization value was defined as the mean of the major-left peak, if its maximum density was at least 50% of the major peak, and at a distance of at least 0.14, in LogR. Our strategy was to increase the tolerance for choosing a minor population, when compared with the 95% threshold suggested in Chen et al. [19]: a lower threshold would catch neutral ratios related to 2-copies DNA segments, in case of a predominantly aneuploid sample. A distance of at least 0.14 from the maximum peak, was added as a supplementary criterion, and was deduced from preliminary tests more extensively described is supplementary Methods, available at Annals of Oncology online.
In parallel, LogR medians were considered as the centralization values.
In order to increase the efficacy of the EM algorithm on large sets of values, we applied a resampling strategy, as described in the supplementary Material, available at Annals of Oncology online, section (supplementary Methods, available at Annals of Oncology online, and https://github.com/fredcommo/EMnormalize for R code).
For each sample, LogR were adjusted by subtracting each centralization values, separately, and segmented using the CBS algorithm, with the appropriate parameters.
In order to minimize differences with karyotype-based profiles, and because of their low resolution compared with aCGH, segments with lengths lower than 200 markers were deliberately merged with the closest segment, considering the previous and next segment LogR value.
distances from karyotypes
As changing the correction value leads to a simple translation of the entire vector of values, comparing the centralization approaches using correlations between array-based genomic profiles and their corresponding karyotypes would not be an appropriate comparison. Instead, we computed gene-by-gene squared distances with the reconstructed karyotype profile, considered as a reference profile. Mean squared distances from karyotypes were then compared across the different centralization methods by using paired t-tests, after log-transformation.
correlations with drug responses
Spearman correlations between CNs and drug responses (active area scores) were computed for a selected panel of four actionable genes (ERBB2, EGFR, FGFR1, and MET), and their related inhibitor (lapatinib, erlotinib, TKI258, and PHA665752, respectively). Significance of differences between correlations was evaluated after Fisher Z-transformation of the correlation values.
decisions in patient cohorts
For the SAFIR-01 and the MOSCATO-01 data, we focused on the 24 actionable genes used in André et al. [4]. Since nearly all the actionable CN alterations today are amplifications, only amplifications were depicted here, and calls were defined according to the criteria previously published in this same paper.
results
comparison of the CGH profiled centralization methods using a panel of cell lines with known karyotypes (NCI60)
To investigate how the centralization impacts on the profiles accuracy, we first analyzed a panel of cell lines for which the karyotypes were available. When using LogR densities, an alternative choice for centralizing the profile occurred in 18/57 cases (31.6%), mainly on the 3n (57.9%) and the 4n (38.5%) cell lines. Conversely, an alternative adjustment was detected for only 2 of the 22 cell lines with 2n− to 2n+ ploidies (8.3%). No other choice than the maximum peak was observed for the unique 5n−/+ SF-295 cell line (supplementary Figure S2, available at Annals of Oncology online).
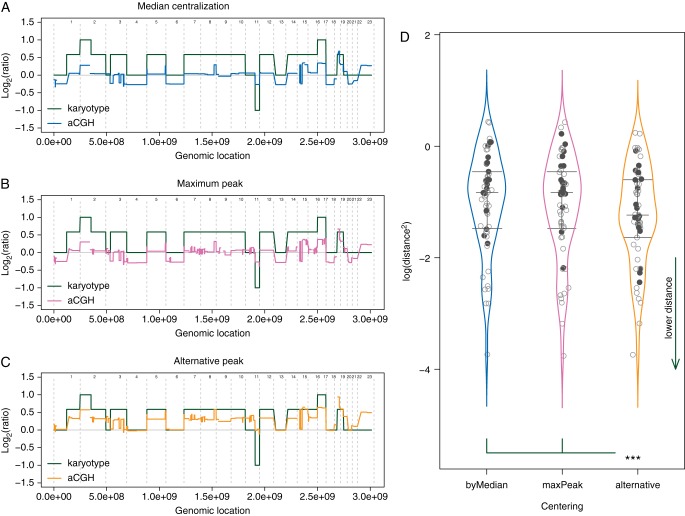
Focusing on the 18 cases where an alternative centralization was available, mostly the 3n and 4n cell lines, we observed that using the maximum peak or the LogR median for adjusting the profiles was unable to detect imbalances revealed by the karyotypes: in case of aneuploidy, entire chromosomes, or chromosome arms, in numbers corresponding to the main cell line ploidy on karyotypes, appeared as in neutral counts, i.e. 2 copies, on the genomic profiles. In these cases, 2-copy DNA regions on karyotypes appeared as lost on the same genomic profiles (supplementary Figure S3, available at Annals of Oncology online). In such cases, the alternative centralization resulted in more consistent profiles with the karyotypes, for 17/18 and 18/18 cell lines compared with the maximum peak and the median centralization, respectively. These results were confirmed by paired t-tests on mean squared distances between profiles and their corresponding reconstructed karyotype (P = 1.13e−4 and 6.35e−5 for the same comparisons, respectively) (Figure 1). Interestingly, the 5n−/+ SF-295 cell line did not show any alternative, as previously defined, for adjusting the profile. That said, none of the possible choices would have lead to a genomic profile consistent with this cell line karyotype (supplementary Figure S4, available at Annals of Oncology online).
Figure 1.
Distance from karyotypes. (A–C) In order to estimate the effect of the centralization methods, squared distances between genomic profiles (colored line) and karyotypes (green line) have been calculated. Distances are symbolized by the colored areas. (D) An alternative peak was available for 18 of the 57 analyzed cell lines (bold black points). Choosing the alternative peak for adjusting the genomic profiles significantly reduce the discrepancies with the corresponding karyotypes, compared with the other methods. P = 1.13e−4 and 6.35e−5, compared with the maximum peak and the LogR median, respectively. Vertical colored curves represent the densities, and horizontal gray segments are the Q25, Q50, and Q75 quantiles of each distribution.
comparison of the outputs of different centralization methods on large panels of cell lines labeled for drug sensitivity (CCLE)
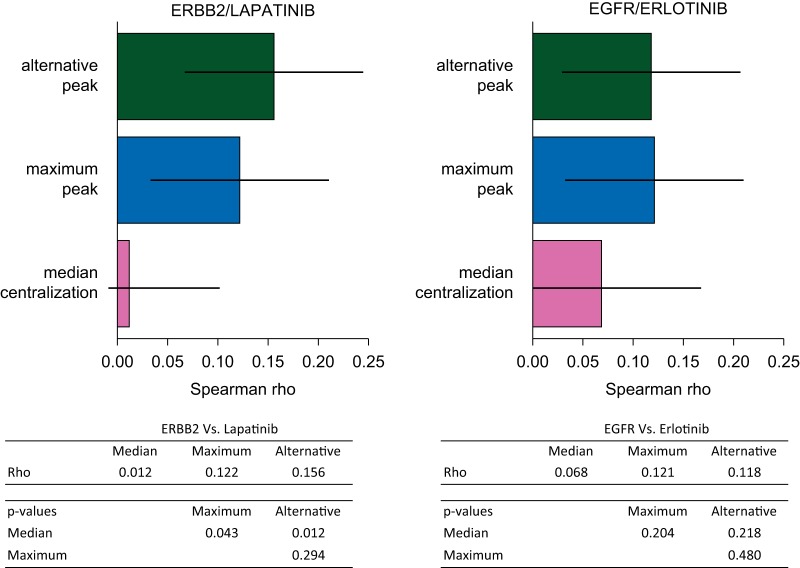
Applying similar comparisons on the CCLE data, we first noted that of 995 cell lines' profiles, an alternative peak was detected in 160 cases (16%), and in 92 of the 487 sub-panel cell lines (18.9%) tested for drug sensitivities. To assess the impact of different centralization methods, we selected this latest sub-panel, and computed Spearman's correlations between centralized CN values and drug sensitivities. We focused on four genes for which the amplification is known to be associated with an increased sensitivity to the related inhibitor and are currently used in the clinic. For ERBB2 and lapatinib, the alternative and the maximum peak centralization both increased significantly the correlation when compared with the median method (P = 0.012 and P = 0.043, respectively). Further, regarding EGFR and erlotinib, both the maximum peak and the alternative peak tended to be associated with higher corrrelations when compared with the median centralization approach (Figure 2). No significant improvement was observed in correlations between FGFR1 and MET, and their respective inhibitors: all ρ values where lower than 0.1 for FGFR1, and close to 0 for MET, and P values were at least >0.27 in all centralization comparisons (supplementary Figure S5, available at Annals of Oncology online).
Figure 2.
Correlation between copy number variation and sensitivity to related inhibitors. The Spearman correlation between ERBB2 and lapatinib increased significantly when applying the maximum peak or the alternative peak centralizations, compared with the median value adjustment (ρ = 0.122, 0.156, and 0.012, respectively. P = 0.043 and 0.012, respectively). The alternative centralization even improved the correlation, but not significantly (P = 0.294). The same trend was observed for EGFR and erlotinib, even though none of the improvements appeared significant.
comparison of the outputs of different centralization methods on aCGH profiles from patients prospectively enrolled in precision medicine programs (SAFIR01 and MOSCATO-01)
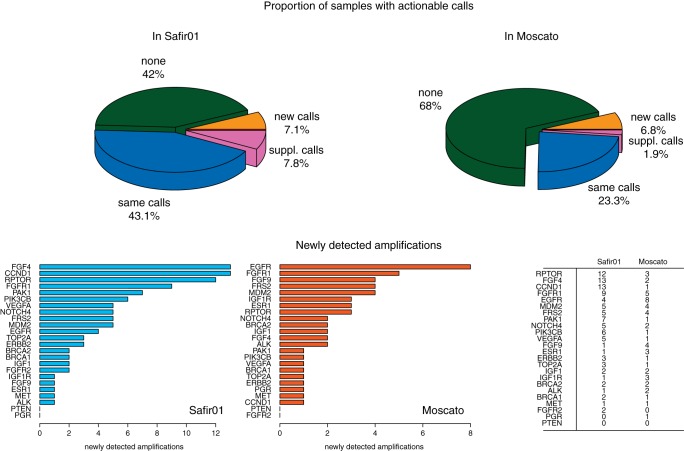
Due to lower performances of the median centralization on cell lines, only the maximum peak and the alternative centralization were considered. The alternative peak detection method was applied on 283 breast metastasis samples from SAFIR01, and on 309 MOSCATO-01 tumor samples. In the SAFIR01 cohort, we observed an alternative centralization peak in 76 of the 283 profiles (26.9%), with similar proportions on both platforms: 31/125 profiles generated using Affymetrix (24.8%), and 45/158 generated using Agilent 4 × 180K (28.5%). Importantly, when applying the alternative centralization, an actionable amplification was detected in 20 of the 283 patients (7.07%), for whom no actionable trait was previously identified with the maximum peak method. Further, supplementary amplifications, not seen with the maximum peak centralization, were detected in 22 patients (7.8%).
Similar results were obtained using the MOSCATO-01 cohort: an alternative peak was detected in 79 of 309 samples (25.6%). As a major consequence, an actionable amplification was detected using the alternative centralization in 21 patients (6.8%), while no aberration was previously identified using the maximum peak method, and supplementary amplifications were found in six patients (1.9%).
In both cohorts, the alternative centralization never missed any amplification detected using the maximum centralization method. In the two studies, the new (or possibly supplementary) actionable amplifications included the same genes, but with different frequencies. This can be due to the differences between the two cohorts: metastasis of breast tumors and metastasis from all type of tumors, in SAFIR01 and MOSCATO-01, respectively (Figure 3 and supplementary Table S6, available at Annals of Oncology online).
Figure 3.
Effect of different genomic profiles centralization methods on possible therapeutic orientations. Top panel: Centralizing on an alternative peak leaded to identify actionable amplifications (new calls) in 20 more samples (7.1%) in the SAFIR01 data, and in 21 supplementary samples (6.8%) in MOSCATO, for whom no amplification was found by using the maximum peak for centralizing the genomic profiles. In 22 (7.8%) and 6 (1.9%) cases in SAFIR01 and MOSCATO, respectively, supplementary amplifications (sup. calls) were also identified, leading to supplementary options for a therapeutic decision making. For 42% and 68%, in Safir01 and Moscato, respectively, no therapeutic option appeared, and in 43.1% and 23.3%, the same actionable genes were identified with both methods. Bottom panel: FGF4, CCND1, and RPTOR were the most frequently newly detected amplified genes in Safir01, while EGFR and FGFR1, principally, were impacted by the centralization strategy in Moscato data (frequencies are summarized in the table, on right).
discussion
Array-based genomic profiling is widely used to estimate gains and losses of DNA segments and ultimately to guide the therapeutic decision in personalized medicine programs. Herein, we demonstrated the importance of the centralization step to determinate the LogR of the array signal intensities by comparing the effect of three different methods on several panels of cell lines and patients cohorts. To our knowledge, this is the first time that such a comparison between various centralization methods is carried out.
Using the NCI60 panel of cell lines for which the related karyotypes are known allowed us to prove that some centralization rules can lead to erroneous profiles. To note, his effect appears prominent in the aneuploidy setting, which is frequent in cancer. For instance, in cell lines with high ploidy, centering on the LogR median or on the highest density peak led to inappropriate values. In the latter setting normal 2-copy regions are estimated as losses; thus, amplifications are likely to be underestimated and deletions are likely to be overestimated. Though, even after the centralization adjustment, we did observe remaining discrepancies between the genomic profiles and karyotypes. Several reasons could be advocated to explain this such as technical issues like the adjustment step of DNA samples to a fixed quantity before being used could reduce ploidy differences between the sample and the reference used in the CGH array. Similar effects have been observed on gene expression analysis [24].
Second, using a large panel of cell lines (CCLE), we showed that applying the alternative centralization peak method led to a significantly improved correlation between ERBB2 CN and the sensitivity to lapatinib when compared with the median and the maximum peak methods. However, this had little impact on the correlation between EGFR and the sensitivity to erlotinib. To note, none of the centralization procedures tested lead to significant differences in correlations between FGFR1, and MET, with their respective known inhibitors. This latter result may be secondary to the fact that the drugs tested were relatively weak inhibitors of FGFR1 (TKI258) and MET (PHA665752), thus rendering the correlation hazardous. Further, we cannot formally exclude issues in the estimation of the drug sensitivity in these large panels of cell lines, as this was previously noted before [25].
Finally, applying similar centralization comparisons on the SAFIR01 and the MOSCATO-01 data also unveiled ambiguities for determining the CGH calls in tumor samples, and thus the therapeutic decisions. Applying the alternative centralization method would have changed the decision for 20/283 (7.07%) and 21/309 (6.8%) patients, in SAFIR01 and MOSCATO-01, respectively, for whom a standard approach did not reveal any actionable gene amplification. Since array-based genomic profilings give a global overview of a diversity of events that occur in a tumor, and may provide possible misinterpretations, fluorescent in situ hybridizations may be considered as a necessary validation step. However, such verification is rarely performed because of evident cost and time issues, and thus reinforces the importance of the centralization step in the CGH profiling.
In this study, we showed that the centralization step is critical in the evaluation of gene copy number using CGH arrays and is susceptible to substantial effects on the decision-making criteria for patient treatment. Among the three different centralization methods tested, the alternative peak approach appears promising. Since centralization problems are linked with the tumor polidy variation, they are not likely to be restricted to only hybridization-based technologies and may also impact sequencing-based pipelines. Though, dedicated methods remain to be developed for the latter technologies.
funding
FC, CF, SF and JG were supported by the grant U54CA149237 from the Integrative Cancer Biology Program of the National Cancer Institute.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
references
- 1.Cline MS, Craft B, Swatloski T, et al. Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.André F, Bachelot T, Commo F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. doi: 10.1016/S1470-2045(13)70611-9. [DOI] [PubMed] [Google Scholar]
- 5.Hollebecque A, Massard C, De Baere T, et al. Molecular screening for cancer treatment optimization (MOSCATO 01): a prospective molecular triage trial—interim results. ASCO Annual Meeting. 2013 Abstract 2512. [Google Scholar]
- 6.WIN Consortium. http://www.winconsortium.org/ (30 October 2014, date last accessed)
- 7.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353(16):1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 8.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 9.André F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19(13):3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 10.Marioni JC, Thorne NP, Tavaré S. BioHMM: a heterogeneous hidden Markov model for segmenting array CGH data. Bioinformatics. 2006;22(9):1144–1146. doi: 10.1093/bioinformatics/btl089. [DOI] [PubMed] [Google Scholar]
- 11.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23(6):657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 12.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picard F, Robin S, Lavielle M, Vaisse C, Daudin J-J. A statistical approach for array CGH data analysis. BMC Bioinformatics. 2005;6(1):27. doi: 10.1186/1471-2105-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van De Wiel MA, Kim KI, Vosse SJ, et al. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23(7):892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 15.Van Houte BPP, Binsl TW, Hettling H, Pirovano W, Heringa J. CGHnormaliter: an iterative strategy to enhance normalization of array CGH data with imbalanced aberrations. BMC Genomics. 2009;10(1):401. doi: 10.1186/1471-2164-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staaf J, Jönsson G, Ringnér M, Vallon-Christersson J. Normalization of array-CGH data: influence of copy number imbalances. BMC Genomics. 2007;8(1):382. doi: 10.1186/1471-2164-8-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, Pounds S, Zhang K, Fang Z. PAIR: paired allelic log-intensity-ratio-based normalization method for SNP-CGH arrays. Bioinformatics. 2013;29(3):299–307. doi: 10.1093/bioinformatics/bts683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popova T, Manié E, Stoppa-Lyonnet D, et al. Genome Alteration Print (GAP): a tool to visualize and mine complex cancer genomic profiles obtained by SNP arrays. Genome Biol. 2009;10(11):R128. doi: 10.1186/gb-2009-10-11-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HI, Hsu FH, Jiang Y, et al. A probe-density-based analysis method for array CGH data: simulation, normalization and centralization. Bioinformatics. 2008;24(16):1749–1756. doi: 10.1093/bioinformatics/btn321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knutsen T, Gobu V, Knaus R, et al. The interactive online SKY/M-FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosomes Cancer. 2005;44(1):52–64. doi: 10.1002/gcc.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NCI60 cell line panel Genetics. ftp://ftp.ncbi.nih.gov/sky-cgh/ESI/NCI60_cell_line_panel_Genetics_Branch_I.R.Kirsch.esi. (1 October 2014, date last accessed)
- 22.Celeux G, Govaert G. Gaussian parsimonious clustering models. Pattern Recognit. 1995;28(5):781–793. [Google Scholar]
- 23.Fraley C, Raftery AE. Model-based methods of classification: using the mclust Software in Chemometrics. J Stat Softw. 2007;18(6):1–13. [Google Scholar]
- 24.Lovén J, Orlando DA, Sigova AA, et al. Revisiting global gene expression analysis. Cell. 2012;151(3):476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haibe-Kains B, El-Hachem N, Birkbak NJ, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.