Significance
To our knowledge, this work describes the first genome-wide annotation of transcriptional start sites in Vibrio cholerae and the discovery and characterization of a regulatory RNA, named VqmR, which controls collective behaviors in this major human pathogen. We show that VqmR is activated by the VqmA transcriptional regulator. VqmR represses expression of multiple mRNA targets including those encoding the Rtx (repeats in toxin) toxin and VpsT, which is required for biofilm formation. Indeed, we show that VqmR controls biofilm formation through repression of vpsT.
Keywords: biofilm, quorum sensing, RNA-seq, sRNA, Vibrio cholerae
Abstract
Quorum sensing (QS) is a process of cell-to-cell communication that enables bacteria to transition between individual and collective lifestyles. QS controls virulence and biofilm formation in Vibrio cholerae, the causative agent of cholera disease. Differential RNA sequencing (RNA-seq) of wild-type V. cholerae and a locked low-cell-density QS-mutant strain identified 7,240 transcriptional start sites with ∼47% initiated in the antisense direction. A total of 107 of the transcripts do not appear to encode proteins, suggesting they specify regulatory RNAs. We focused on one such transcript that we name VqmR. vqmR is located upstream of the vqmA gene encoding a DNA-binding transcription factor. Mutagenesis and microarray analyses demonstrate that VqmA activates vqmR transcription, that vqmR encodes a regulatory RNA, and VqmR directly controls at least eight mRNA targets including the rtx (repeats in toxin) toxin genes and the vpsT transcriptional regulator of biofilm production. We show that VqmR inhibits biofilm formation through repression of vpsT. Together, these data provide to our knowledege the first global annotation of the transcriptional start sites in V. cholerae and highlight the importance of posttranscriptional regulation for collective behaviors in this human pathogen.
Quorum sensing (QS) is a process of bacterial cell-to-cell communication that relies on the production, release, accumulation, and population-wide detection of extracellular signal molecules called autoinducers. Processes controlled by QS are unproductive when undertaken by an individual bacterium but become effective when performed in unison by the group. In the major human pathogen, Vibrio cholerae, QS orchestrates processes including biofilm formation and virulence factor production (1). In the V. cholerae QS circuit, at low cell density (LCD), LuxO∼P activates transcription of genes encoding four small regulatory RNAs (sRNAs), Qrr1–4, which promote translation of the low cell density master QS regulator, AphA, and repress translation of HapR, the high cell density (HCD) master QS regulator (2, 3). AphA directs the gene expression program for V. cholerae cells to act as individuals. At high cell density, in the presence of autoinducers, LuxO is dephosphorylated and inactivated (4). The Qrr sRNAs are not transcribed, AphA is not produced, and by contrast, the hapR mRNA is translated into HapR protein. HapR controls the gene expression program underpinning collective behaviors (5, 6). Thus, the absence or presence of the Qrr sRNAs directs whether or not V. cholerae engages in QS. The Qrr sRNAs belong to the family of Hfq-binding sRNAs that regulate gene expression through base pairing with target mRNAs (7).
An explosion in the discovery of noncoding transcripts, including sRNAs, has occurred due to recently developed transcriptomic technologies such as high-throughput sequencing of cDNAs, e.g., RNA sequencing (RNA-seq). (8). Major advantages of RNA-seq over conventional methods include high sensitivity for low abundance transcripts, independence from species-specific probes, and single-nucleotide resolution mapping of transcript termini (9). Several modifications have further diversified the possible uses of RNA-seq. One such technology is called differential RNA sequencing (dRNA-seq), in which enrichment of primary transcript termini can be combined with strand-specific generation of cDNA libraries to achieve genome-wide identification of transcriptional start sites (TSSs) (10).
In this study, we applied dRNA-seq to the study of QS in V. cholerae. We performed dRNA-seq on wild-type V. cholerae under conditions of low and high cell density. We performed parallel analyses on a V. cholerae luxO D47E mutant that constitutively expresses the qrr sRNA genes. This strategy allowed us to discriminate between QS-specific gene expression changes and gene expression changes resulting from differences in growth rate or, for example, nutrient availability. To our knowledge, our results present the first global annotation of TSSs in V. cholerae with a focus on QS-mediated transcriptome changes. Surprisingly, ∼47% of the TSSs initiate as antisense transcripts in both coding and noncoding regions of the genome. Combining our TSS data with phylogenetic analyses allowed us to reannotate 167 translational start sites in V. cholerae. Our analyses also revealed 107 putative sRNAs and we studied one of them, which we name VqmR, in detail. The vqmR gene is highly conserved among the vibrionaceae. vqmR is located upstream of the vqmA gene encoding the VqmA transcription factor. VqmA activates vqmR transcription. VqmR, in turn, employs two conserved base-pairing domains to repress translation of at least eight trans-encoded genes, including the rtx (repeats in toxin) toxin transcript and the vpsT mRNA, encoding a major transcriptional activator of biofilm formation. Accordingly, overexpression of VqmR strongly inhibits biofilm formation in V. cholerae.
Results
Quorum Sensing and the Global V. cholerae Transcription Profile.
Profiling the transcriptional output of an organism provides a powerful means to gain an understanding of its underlying physiology. Earlier studies have investigated the transcriptome of V. cholerae using RNA-seq, with a concentration on virulence-related functions (11, 12) and discovery of sRNAs (13, 14) or type VI-mediated protein secretion (15). To define the contribution of QS to the transcriptional landscape of V. cholerae, we developed an approach that combines dRNA-seq, bioinformatics, mutagenesis, and phenotypic analyses. First, we isolated total RNA from wild-type V. cholerae C6706 at low (OD600 of 0.1) and high (OD600 of 2.0) cell density and compared the number of mapped cDNA reads corresponding to the 3,693 genes on both chromosomes of V. cholerae N16961. V. cholerae C6706 is a clinical isolate belonging to the O1 El Tor biotype responsible for the current cholera pandemic that is closely related to the sequenced V. cholerae strain N16961 (16). We performed identical analyses with RNA obtained from an isogenic luxO D47E mutant that is locked in the low cell density mode. LuxO D47E mimics LuxO∼P and drives constitutive production of the Qrr sRNAs. Cross-comparisons allowed us to define genes exhibiting differential expression with respect to growth phase (OD600 of 0.1 vs. OD600 of 2.0) and QS status (wild-type vs. luxO D47E). We chose high cell density (OD600 of 2.0) wild type as the reference. A high number of genes (2,544) displayed differential expression (>1.5-fold change) in response to growth status (wild-type OD600 of 0.1 vs. wild-type OD600 2.0, Fig. 1A and SI Appendix, Dataset S1). The majority of these genes (∼94%; 2,402) showed alterations in both wild-type and the luxO D47E strain, suggesting, not surprisingly, that growth phase has a major influence on V. cholerae gene expression. Indeed, canonical stationary phase marker genes, for example, rpoS encoding the alternative sigma factor σS, were up-regulated at high cell density, independently of QS status (SI Appendix, Dataset S1).
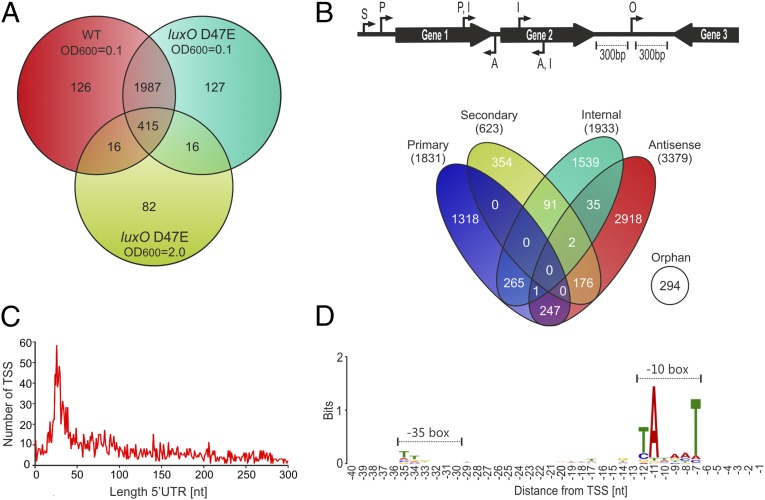
Fig. 1.
Expression profiling and TSS mapping of wild-type and luxO D47E V. cholerae. (A) Venn diagram of differentially expressed genes in the wild-type and luxO D47E V. cholerae C6706 strains at LCD (OD600 of 0.1) and HCD (OD600 of 2.0). Numbers of cDNA reads for annotated genes were compared to wild-type cells at HCD (OD600 of 2.0). Statistically significant genes (P < 0.05) that changed >1.5-fold are shown. (B) Identification and classification of TSSs in V. cholerae. A total of 7,240 TSSs were identified by dRNA-seq and classified according to their genomic locations (Top). A, antisense; I, internal; O, orphan; P, primary; S, secondary. (C) Length distribution of 5′ UTRs in V. cholerae. For each primary and secondary TSS, the distance to the cognate translation initiation site was calculated and the frequency of each 5′ UTR length was plotted. (D) Consensus motif for V. cholerae promoters. DNA sequences from −40 to +1 upstream of the 7,240 TSSs were analyzed for conserved sequence elements using the MEME tool.
Comparison of genes differentially expressed in the wild-type and the luxO D47E strain at high cell density revealed 529 transcripts (Fig. 1A, light green circle). These genes are candidates for QS control via direct or indirect regulation by the Qrr sRNAs. Evidence confirming this prediction comes from our identification of altered expression of known Qrr target mRNAs such as aphA (17) and vca0107 (18), as well as genes that are known to be controlled only indirectly by the Qrr sRNAs (SI Appendix, Dataset S1). For example, we detected reduced expression of HapR-activated targets such as the vca0691-0688 operon involved in poly-β-hydroxybutyrate synthesis and type VI secretion genes (6, 18, 19).
Significant Antisense Transcription Occurs in V. cholerae.
To distinguish primary from processed transcripts, we used the recently developed dRNA-seq protocol (10), which hinges on differential sequencing of two cDNA libraries. The first library (−) is generated from total RNA, whereas the second library (+) is treated with terminator exonuclease (TEX), to specifically degrade processed RNAs with 5′-monophosphate ends. We identified 7,240 total TSSs under all conditions, and we classified them with respect to their locations in the genome (Fig. 1B). We define the primary TSS as the strongest promoter of each gene based on the number of reads obtained. We classify secondary TSSs as those using alternative promoters for the same gene. We define internal TSSs as those located within annotated genes on the sense strand. We name antisense TSSs as those that initiate inside genes or within ≤100 bp of a gene on the antisense strand. Finally, orphan TSSs are those that have no clear association with flanking genes (>300 bp from the nearest annotated gene) (Fig. 1B). The data featured in this paper are provided in an easily searchable online site with a visual interface (imib-wuerzburg.de/research/vibrio/).
Only ∼5% of all TSSs were specific to a single condition (SI Appendix, Dataset S2), suggesting that our approach captures the basal expression of most transcription units. We validated our approach by comparing our global TSS annotation with 35 experimentally determined TSSs from published studies exploring V. cholerae biology. In 74% (26/35) of cases, our prediction exactly matched the reported TSS, and in 94% (33/35) of the cases, the TSS matched within a distance of one nucleotide (SI Appendix, Fig. S1). These statistics are in agreement with previous RNA-seq–based TSS predictions from other bacteria (10, 20) and confirm the accuracy of our approach. Approximately 47% of the TSSs were antisense transcripts (Fig. 1B), which is a considerably higher fraction than reported for bacterial species such as Salmonella enterica (∼13%) (20) but is in good agreement with RNA-seq data from Helicobacter pylori (41%) (10), Campylobacter jejuni (44–47%) (21), and Escherichia coli (37%) (22).
The 5′ UTRs of bacterial mRNAs are hotspots for posttranscriptional regulation and commonly involved in base pairing with regulatory RNAs. We analyzed the 5′ UTR length of 2,454 primary and secondary TSSs (Fig. 1B). We found enrichment for 5′ UTRs ranging between 20 and 40 nt (616, Fig. 1C). We also identified 12 transcripts without 5′ UTRs and 57 additional 5′ UTRs shorter than 10 nt (Fig. 1C and SI Appendix, Dataset S2), suggesting translation that is independent of conventional ribosomal recruitment signals (23). An alternative interpretation could be that the ORFs encoded on leaderless transcripts are misannotated in the genome. To test this latter hypothesis, we investigated conservation of all translational start sites of mRNAs carrying 5′ UTRs shorter than 10 nt or TSSs within the first 100 bp downstream of the annotated ORF. In 167 of 356 cases, we predict that, in fact, translation starts downstream of the previously annotated site. In addition, we identify 28 mRNAs with 5′ UTRs shorter than 10 nt (SI Appendix, Dataset S3). We examined the sequences 50 bp upstream of the 7,240 TSSs and used the MEME toolkit (24) to predict conserved transcription control motifs. This analysis revealed a clear −10 box, and only poor conservation of the −35 box (Fig. 1D). Overall, the motif we identified resembles the recognition sequence of the housekeeping sigma factor, σ70. This finding is consistent with reports in other enterobacterial species (20).
Multiple Classes of Noncoding RNAs Exist in V. cholerae.
Most available bacterial genome annotations report ORFs, tRNA genes, and rRNA genes, whereas genes specifying noncoding regulators and small proteins are often absent. This gap in genome annotation has slowed the general understanding of gene regulation and has muddled genetic and bioinformatic screens. Our high-throughput dRNA-sequencing approach allowed us to identify transcripts that are not associated with annotated genes. We consider these transcripts noncoding sRNAs, although in a few cases these sRNAs have been reported to also encode small proteins (25).
We predict 107 new candidate transcripts (Vcr001–Vcr107, SI Appendix, Dataset S4), and we confirmed the expression of 35 previously described sRNAs (SI Appendix, Dataset S4). To garner support for the former, we validated expression of 32 of the newly identified sRNAs on Northern blots (Fig. 2 and SI Appendix, Figs. S2 and S3). Specifically, we collected total RNA from wild-type, luxO D47E, and Δhfq V. cholerae strains at different time points spanning low to high cell density. We categorized the sRNAs according to their genomic localizations, i.e., intergenic, 5′ UTR, 3′ UTR, or overlapping with ORFs.
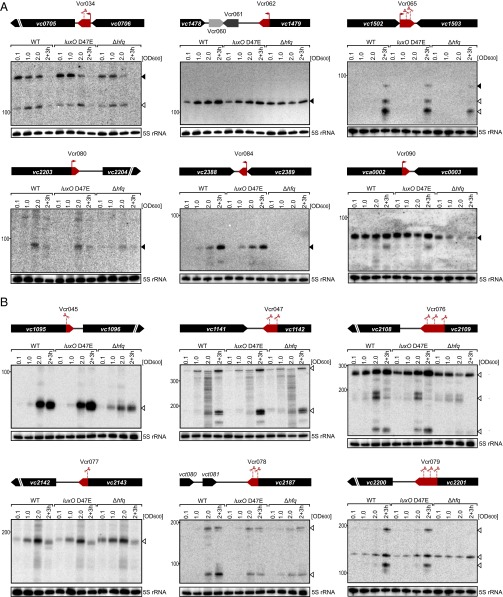
Fig. 2.
Expression analysis of 3′ UTR-derived sRNAs. (A) sRNAs with dedicated promoters. Total RNA was obtained at the designated times during growth from wild-type, luxO D47E, and Δhfq V. cholerae strains. Northern blots were probed for six 3′ UTR-derived sRNAs. The genomic locations and relative orientations are shown above the gels. Genes are shown in black; sRNAs are shown in red or gray. Arrows and scissors indicate TSSs and processing sites, respectively. Filled triangles indicate bands derived from TSSs; open triangles indicate bands derived from processing. The 5S rRNA served as the loading control. (B) sRNAs derived from transcript processing. The designations are the same as in A except that the promoters are shared between the sRNA and the mRNA. The mRNAs undergo ribonucleolytic cleavage to yield the sRNAs.
sRNAs produced from intergenic regions and 5′ UTRs.
Our RNA-seq approach revealed 19 previously unidentified sRNAs in intergenic regions (SI Appendix, Dataset S4). We validated production of a subset of nine of this class of sRNA using Northern blots (SI Appendix, Fig. S2A). Accumulation of Vcr017, Vcr082, Vcr089, and Vcr092 depended on Hfq. Most interesting from our perspective is that Vcr071, Vcr082, Vcr094, and Vcr099 displayed differential expression with respect to QS status. According to our dRNA-seq data, 14 of the 107 putative sRNA candidates could also serve as 5′ UTRs of mRNAs (SI Appendix, Dataset S4). These sRNA candidates are characterized by their proximities to the translational start sites of ORFs and they harbor signatures of rho-independent terminator sequences at their 3′ ends. In most cases, inspection of individual cDNA reads enabled the identification of read-through products extending from the rho-independent terminator into the ORF of the downstream gene. Probing the expression of four of them (Vcr002, Vcr043, Vcr087, and Vcr098) showed stable expression, Vcr043 levels were significantly reduced in the absence of Hfq, and Vcr087 appears to be QS controlled (SI Appendix, Fig. S2B). We do not know whether the V. cholerae 5′-encoded sRNAs act in cis, trans, or both.
3′ UTR-derived sRNAs.
The 3′ UTRs of bacterial mRNAs have recently been recognized as a reservoir for bacterial sRNAs (26). Consistent with this notion, we discovered a significant number (44 of 107) of candidate sRNAs encoded in the 3′ UTRs of functional mRNAs (SI Appendix, Dataset S4). We classified sRNAs as 3′ UTR derived if they shared a rho-independent terminator with the immediate upstream gene but displayed 5′ ends that were independent of those genes. We subdivided the 3′ end-derived RNAs into transcripts that possess their own promoters [20, enriched in the TEX (+) samples] and those derived from processing of longer mRNA transcripts [24, no enrichment in the TEX (+) sample, but significant cDNA reads in the TEX (−) samples]. We tested the expression of a representative 15 of these 3′-derived sRNAs including examples of independently transcribed sRNAs (Fig. 2A and SI Appendix, Fig. S3A) and examples produced by transcript processing (Fig. 2B and SI Appendix, Fig. S3A). The absence of Hfq affected transcript levels for Vcr034, Vcr036, Vcr045, Vcr065, Vcr076, Vcr080, Vcr084, Vcr090, and Vcr105, suggesting that a subset of 3′ UTR encoded sRNAs functions to control trans-encoded mRNA targets (Fig. 2 A and B and SI Appendix, Fig. S3A). In the case of processed 3′ UTR-derived sRNAs, the situation might be more complicated than for sRNAs with dedicated promoters. In the former case, the mRNA itself could be under the control of Hfq, which could change the expression of both the mRNA and the sRNA linked to it. In several instances, the presence of Hfq also affected the processing patterns. For example, Vcr065 failed to accumulate an intermediate processing product in the Δhfq strain (Fig. 2A), whereas Vcr079 required Hfq for cleavage or for stability of two major processing products (Fig. 2B). We have not yet assessed at what point Hfq acts in each case. Hfq could facilitate maturation or stability of the sRNAs through interaction with ribonucleases such as RNase E in the degradosome (27); alternatively, processing could be the consequence of Hfq-assisted base pairing with target mRNAs (7).
sRNAs overlapping with ORFs (cis-encoded sRNAs).
Our analyses also identified sRNAs that overlap with annotated ORFs (SI Appendix, Dataset S4 and Fig. S3B). These so-called cis-encoded sRNAs usually control the expression of the gene with which they overlap (28), although exceptions have been reported (29). The cis-encoded sRNAs are typically expressed antisense to their target genes, and this arrangement promotes extensive base pairing, generating substrates for cleavage by RNase III (28). RNase III degrades both the mRNA and the sRNA partner, thus cis-encoded sRNAs are often difficult to detect by RNA-seq or similar methods (22). Nonetheless, we were able to validate the expression of four cis-encoded sRNAs using Northern blots (SI Appendix, Fig. S3B). Vcr038, Vcr095, and Vcr103 are, as is typical, encoded antisense to the annotated ORFs. Interestingly, Vcr038 is transcribed antisense to the transposase gene vc0870 and therefore might function to silence transposon activity.
VqmR Is a Highly Abundant Hfq-Dependent Regulatory sRNA.
The arrangement of the candidate cis-encoded sRNA Vcr107 struck us as peculiar. Rather than running in the opposite direction of its overlapping gene (vca1078, also known as vqmA; SI Appendix, Fig. S3B), vcr107 is encoded in the same orientation. Interestingly, our dRNA-seq data revealed two transcriptional start sites (Fig. 3A). We identified a proximal TSS (position 1,031,438 on chromosome II) initiating upstream of the annotated vqmA ORF and a distal TSS (position 1,031,656) that is located inside the annotated gene, which would be expected to produce a truncated, nontranslatable vqmA mRNA. An alternative explanation is that the translational start site of vqmA is misannotated, similar to the 167 genes discovered in our above analyses (SI Appendix, Dataset S3). To test this hypothesis, we aligned the 10 closest homologs of VqmA and examined conservation of their start codons (SI Appendix, Fig. S4A). According to the V. cholerae N16961 genome annotation, translation of vqmA starts with the AUG at position 1,031,460 of chromosome two (16). In six other vibrio species, annotation suggests that VqmA translation commences 73 codons downstream of this nucleotide (position 1,031,679 in the V. cholerae N16961 genome; SI Appendix, Fig. S4A). Importantly, V. cholerae N16961 possesses a conserved GTG at this downstream position, which could serve as a translational start site. To test this possibility, we generated a GFP translational fusion to VqmA and individually mutated the two candidate start codons (SI Appendix, Fig. S4B). We changed the annotated start codon from ATG to ATC and no significant change in GFP production occurred (SI Appendix, Fig. S4B). By contrast, when we mutated the alternative GTG start codon to GTC, production of VqmA::GFP was eliminated (SI Appendix, Fig. S4B), suggesting that VqmA translation initiates 73 codons downstream of the site annotated in the genome database and that Vcr107 encodes an independent sRNA. Given its proximity to the vqmA gene, we renamed Vcr107: VqmR.
Fig. 3.
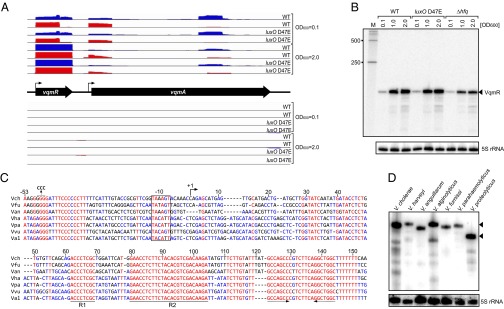
Identification and expression of the VqmR sRNA. (A) dRNA-seq data for the vqmR-vqmA locus. Shown are cDNA reads mapping to the vqmR and vqmA genes in V. cholerae. Blue bars indicate expression in the TEX (−) samples; red bars show expression in the TEX (+) samples. (Upper) cDNA reads mapping to the plus strand of the V. cholerae genome. (Middle) Schematic representation of the vqmR-vqmA genomic arrangement. (Bottom) cDNA reads mapping to the minus strand of the V. cholerae genome. Strains and growth conditions are indicated on the Right. (B) Production of VqmR in V. cholerae. Total RNA obtained at the designated ODs for wild-type, luxO D47E, and Δhfq V. cholerae strains was probed for VqmR on Northern Blots. A DNA-marker (M) is provided on the Left. The 5S rRNA served as the loading control. (C) Sequence alignment of vqmR genes. The sequences, including the promoter regions, of vqmR genes from different vibrio species were aligned. The −10 box, the TSS, and the conserved sequences R1 and R2 are denoted. Dotted arrows indicate the rho-independent terminator element. Mutations introduced into the vqmR promoter (used in EMSA studies; SI Appendix, Fig. S5C) are indicated above the alignment. Vch, V. cholerae; Vfu, Vibrio furnissii; Van, Vibrio anguillarum; Vha, Vibrio harveyi; Vpa, Vibrio parahaemolyticus; Vvu, Vibrio vulnificus; Val, Vibrio alginolyticus. (D) Production of VqmR in different vibrios. VqmR was assessed by Northern blot for the indicated vibrio species using a probe for conserved region R2. Triangles indicate full-length VqmR. The 5S rRNA served as the loading control.
To bolster our hypothesis that vqmR is a trans-acting sRNA, we tested its dependency on Hfq. There was approximately threefold less VqmR sRNA in the Δhfq strain compared with the wild type (Fig. 3B and SI Appendix, Fig. S3B), and VqmR stability declined from >32 min in the wild-type strain to ∼12 min in the Δhfq strain (SI Appendix, Fig. S5A). Further, we aligned the DNA sequences of putative vqmR genes from different vibrio species and scanned them for signature sequences indicative of Hfq-dependent sRNAs (Fig. 3C) (30, 31). In all cases, there exist −10 boxes, one or more strongly conserved regions required for base pairing with target mRNAs, and rho-independent transcription termination sites. Two regions of VqmR (which we name R1 and R2) show the highest sequence conservation among vibrios. We exploited the extended sequence conservation in region R2 to design a specific oligonucleotide probe to assess whether VqmR is produced in vibrios other than V. cholerae. VqmR was indeed present as a stable transcript in all seven species tested (Fig. 3D). A shorter transcript was produce by Vibrio proteolyticus than was made by other vibrios. This result is supported by sequence alignment showing that full conservation of the R2 domain exists but the R1 domain is absent in V. proteolyticus vqmR (SI Appendix, Fig. S5B). In summary, our data suggest that VqmR is an abundant stationary phase Hfq-dependent sRNA and it is highly conserved among the vibrionaceae.
VqmA Activates the vqmR Promoter.
To define the biological role of VqmR, we first investigated how vqmR itself is controlled by performing a genetic screen to identify vqmR regulators. We constructed a vqmR::lacZ transcriptional fusion and incorporated it at the lacZ locus in the V. cholerae chromosome. We mutagenized this strain using Tn5 to generate a library of ∼100,000 random mutants, and we screened them for altered β-galactosidase activity. We did not identify mutants with increased β-galactosidase levels. We identified 16 mutants with reduced β-galactosidase activity. We used PCR to eliminate mutants harboring insertions in lacZ. Six mutants remained, and all possessed insertions in vqmA. The VqmA protein is a transcriptional regulator that harbors a LuxR-like DNA-binding domain at the C terminus and a conserved N-terminal PAS (Per-Arnt-Sim) fold, which often interact with ligands (32). To verify our results, we generated an in-frame deletion of vqmA and compared production of VqmR between the ΔvqmA mutant and the wild type (Fig. 4A). No VqmR could be detected in the ΔvqmA strain under any condition. Expression of vqmA from a plasmid restored VqmR production to the ΔvqmA strain (Fig. 4A), suggesting that VqmA is a transcriptional activator of VqmR.
Fig. 4.
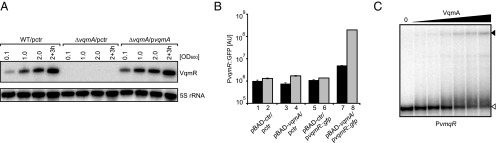
VqmA activates vqmR transcription. (A) Total RNA was obtained for wild-type/pctr, ΔvqmA/pctr, and ΔvqmA/pvqmA V. cholerae strains at the indicated times during growth. The Northern blot was probed for VqmR. The 5S rRNA served as loading control. (B) GFP production from a vqmR transcriptional reporter was measured in E. coli carrying the indicated plasmids following 12 h growth in LB with 0.2% (final concentration) glucose (black bars) or 0.2% (final concentration) l-arabinose (gray bars). Error bars represent SD of three replicates. (C) Electrophoretic mobility shift assay (EMSA) showing that VqmA protein binds the vqmR promoter sequence. Migration of the [32P] end-labeled vqmR promoter fragment in the absence and presence of increasing concentrations of purified VqmA::3XFLAG protein (indicated by the black triangle above the gel) was determined by native polyacrylamide gel electrophoresis and autoradiography. The open triangle indicates free DNA; filled triangle indicates DNA in complex with VqmA::3XFLAG. A negative control consisting of a mutated version of the vqmR promoter is shown in SI Appendix, Fig. S5C.
To test if VqmA functions directly at the vqmR promoter, we performed two experiments. First, we introduced a plasmid harboring a vqmR::gfp transcriptional fusion into E. coli and provided either a control plasmid (pBAD-ctr, Fig. 4B, bars 1, 2, 5, and 6) or a plasmid carrying vqmA under the control of the inducible pBAD promoter (bars 2, 3, 7, and 8). Fig. 4B shows that basal GFP production occurred in the reporter strain carrying pBAD-ctr (compare bars 1 and 2 to bars 5 and 6). The absence (black bars) or presence of arabinose (gray bars) made no difference. Introduction of pBAD-vqmA increased GFP production ∼5-fold above the control level (black bar 5 vs. black bar 7). Addition of the inducer l-arabinose to increase VqmA production elevated vqmR::gfp expression by over 140-fold (gray bar 6 vs. gray bar 8). In a second analysis, we purified VqmA::3XFLAG protein and performed gel-mobility shift assays with the 32P-labeled vmqR promoter sequence. VqmA protein readily bound and shifted the vqmR promoter DNA (Fig. 4C). By contrast, when we mutated the conserved residues −47 to −49 upstream of the vqmR TSS from GGG to CCC (Fig. 3C), VqmA binding was eliminated (SI Appendix, Fig. S5C). These data show that VqmA is a direct transcriptional activator of the vqmR promoter and that interaction with a conserved upstream region in the vqmR promoter is required for binding.
VqmR Posttranscriptionally Regulates Multiple Target mRNAs.
Our next goal was to link VqmR to downstream mRNA targets to obtain clues to the biology underlying the QS-VqmA-VqmR circuit. We used microarrays to assess changes in mRNA levels following transient (10 min) overexpression of vqmR in a ΔvqmRA V. cholerae strain. In total, 17 genes organized in 10 transcription units displayed significant changes (SI Appendix, Table S1). These genes encoded the biofilm regulator, VpsT (vca0952), the Rtx-toxin (vc1449-1450), and a homolog of the siderophore receptor FhuA (vc0200). To validate our findings, we used quantitative real-time PCR (qPCR) to compare target gene expression in the wild type, the ΔvqmR mutant, and the ΔvqmR mutant carrying vqmR on a plasmid. mRNAs that showed decreases in the pulse-expression experiment increased in the ΔvqmR mutant, whereas expression of the activated vc1188 gene decreased (Fig. 5A and SI Appendix, Table S1). Introduction of the plasmid carrying vqmR increased VqmR ∼4-fold, and in all cases, restored candidate target mRNA production in the ΔvqmR stain to the level of the wild type (Fig. 5A). There was one discrepancy, vc0676, which was repressed 3.6-fold by VqmR in the pulse-expression experiment, remained repressed in the absence of vqmR (Fig. 5A and SI Appendix, Table S1). We do not understand this pattern and have therefore excluded vc0676 from further analyses.
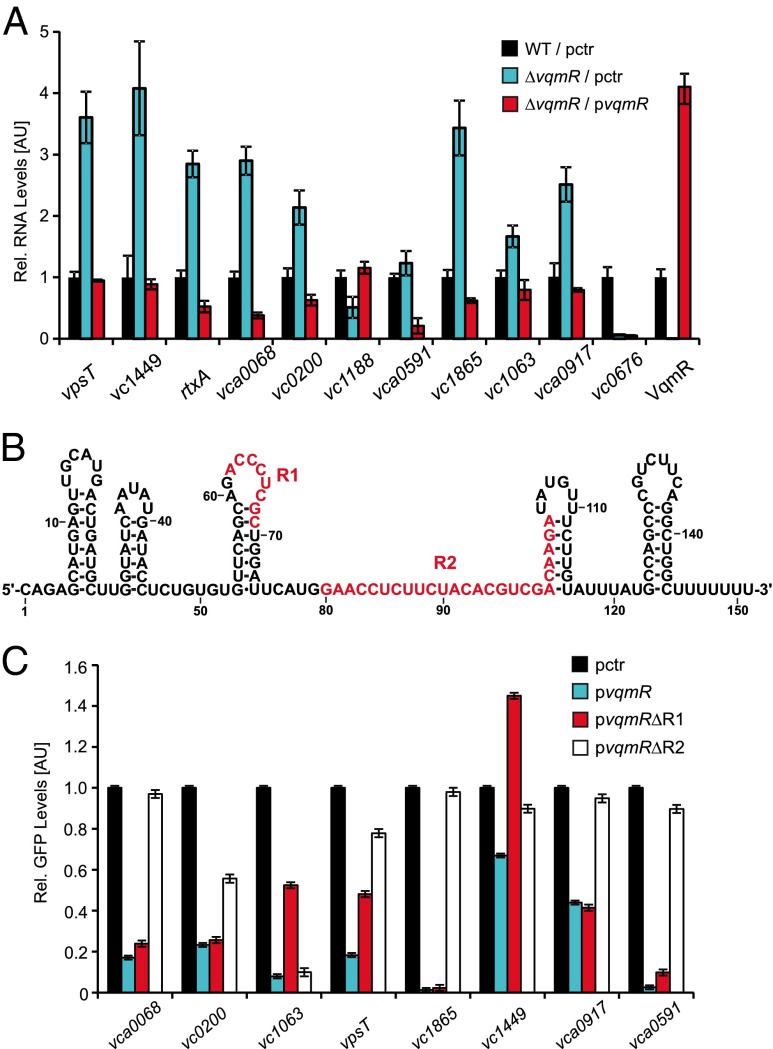
Fig. 5.
The VqmR mRNA targets. (A) Expression of VqmR-controlled target mRNAs was measured in wild-type and ΔvqmR V. cholerae strains. The strains carried either a control plasmid (pctr) or a plasmid harboring the vqmR gene (pvqmR). RNA was monitored using qRT-PCR. Expression in the wild-type strain was set to 1. Error bars represent SD of three replicates. (B) Secondary structure of VqmR. The secondary structure of VqmR was derived from SI Appendix, Fig. S6. Conserved regions R1 and R2 are indicated in red. (C) VqmR regulation of target mRNAs. E. coli harboring plasmids carrying the eight genes denoted on the x axis each fused to gfp were cotransformed with a control plasmid (pctr) or the indicated VqmR-expressing plasmids. gfp and vqmR transcription were driven by constitutive promoters. Strains were grown in LB for 8 h and GFP production was measured. GFP levels in strains carrying the control plasmid (pctr) were set to 1. Error bars represent SD of three replicates.
To explore the mechanism underpinning VqmR-mediated regulation of the above target mRNAs, we first probed the secondary structure of VqmR using the sequence/structure-specific ribonucleases RNase T1, RNase V1, and RNase A (SI Appendix, Fig. S6). These analyses revealed five stem loops in VqmR with the most distal stem loop serving as a rho-independent terminator (Fig. 5B). The conserved R1 and R2 regions are primarily single stranded. Region R1 is exposed in the loop of stem-loop 3 (Fig. 5B). Region R2 represents the major single-stranded sequence of VqmR and extends into stem-loop 4. This result suggests that the single-stranded regions R1 and R2 participate in base pairing with target mRNAs (33). To test this hypothesis, we cloned eight of the repressed target mRNAs into a posttranscriptional reporter system (34). The 5′ UTR and the nucleotides encoding the first 20 amino acids of each target gene were fused to gfp under the pTetO promoter. These plasmids were introduced into E. coli along with a second plasmid carrying the vqmR gene under pLac. In all cases, overexpression of VqmR caused decreased target-GFP production (Fig. 5C). We also performed these experiments in an E. coli Δhfq strain, and no repression of the targets occurred (SI Appendix, Fig. S7A). To assess the roles of the R1 and R2 regions, we individually deleted the R1 and R2 sequences in vqmR and monitored the targets using the same assay. Importantly, deletion of either domain did not affect VqmR production (SI Appendix, Fig. S7B). No repression occurred when region R2 was absent except in the case of vc1063, which required region R1 for full repression (Fig. 5C). Region R1 was dispensable for repression of the other targets. Of note, both the R1 and R2 mutants were less capable of repressing vc1449 than was wild-type VqmR, suggesting that additional sequence elements could be relevant for base pairing. In summary, VqmR is a direct repressor of at least eight trans-encoded mRNAs and region R2 is critical for regulation of the majority of the targets.
VqmR Represses Target mRNAs Using Conserved Regions R1 and R2.
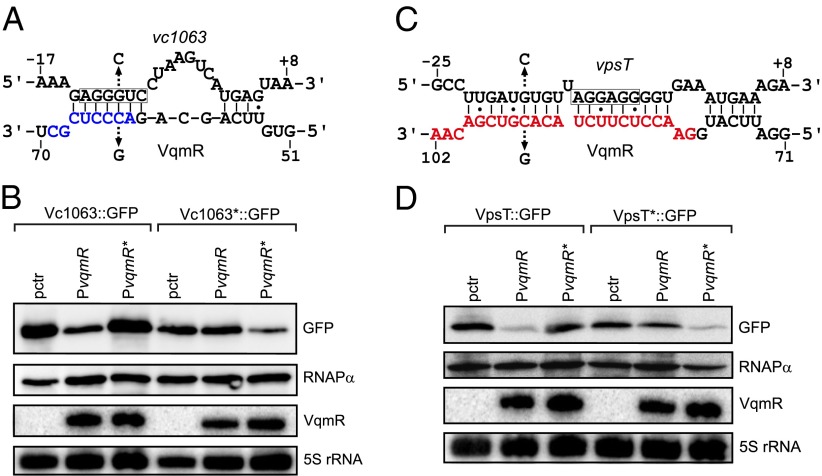
To understand how regions R1 and R2 engage in base pairing with target mRNAs, we used the RNA hybrid algorithm (35) to predict interactions between region R1 and vc1063 and region R2 and the seven other target mRNAs. In all cases, RNA hybrid predicted base pairing between R1 or R2 and the ribosome binding sites of the target genes (SI Appendix, Fig. S7C). To validate these predictions, we chose two representative examples to test: vc1063 for region R1 and vpsT for region R2. We altered cytosine to guanine at position 63 of VqmR and tested GFP production from Vc1063::GFP (Fig. 6 A and B). The VqmR C63G mutant was incapable of repression of vc1063 (Fig. 6B, lanes 1–3). We made a compensatory mutation by changing guanosine at −10 to cytosine in the reporter (Vc1063*::GFP) and this restored repression by VqmR C63G, whereas wild-type VqmR lost its repression capability (Fig. 6 A and B). We performed analogous experiments to test region R2 using vpsT as the target. First, we changed cytosine 94 of VqmR to guanosine and found that VqmR C94G was incapable of repression of vpsT (Fig. 6 C and D). Introduction of a compensatory mutation in vpsT (guanosine −17 to cytosine, VpsT*::GFP) abolished repression by wild-type VqmR and restored repression by VqmR C94G (Fig. 6 C and D). Thus, VqmR indeed uses region R1 to bind to vc1063 and region R2 to bind the vpsT mRNA, presumably inhibiting ribosome binding.
Fig. 6.
Base pairing of VqmR with vc1063 and vpsT. (A) Predicted base pairing between VqmR region R1 and vc1063 mRNA. VqmR region R1 is indicated in blue and the vc1063 Shine Dalgarno sequence is boxed. Mutations introduced in B are indicated with arrows. (B) VqmR repression of vc1063 requires region R1. Vc1063::GFP and Vc1063*::GFP (G-10C) were measured using Western blot. RNAPα served as the loading control. VqmR (PvqmR) and VqmR C63G (PvqmR*) were probed on Northern blot. The 5S rRNA served as loading control. (C) Predicted base pairing between VqmR region R2 and vpsT mRNA. Region R2 is indicated in red and the vpsT Shine Dalgarno sequence is boxed. Mutations introduced in D are indicated with arrows. (D) VqmR repression of vpsT requires region R2. VpsT::GFP and VpsT*::GFP (G-17C) were measured using Western blot. RNAPα served as the loading control. VqmR (PvqmR) and VqmR C94G (PvqmR*) were probed on Northern blot. The 5S rRNA served as loading control.
VqmR Inhibits V. cholerae Biofilm Formation via Repression of vpsT.
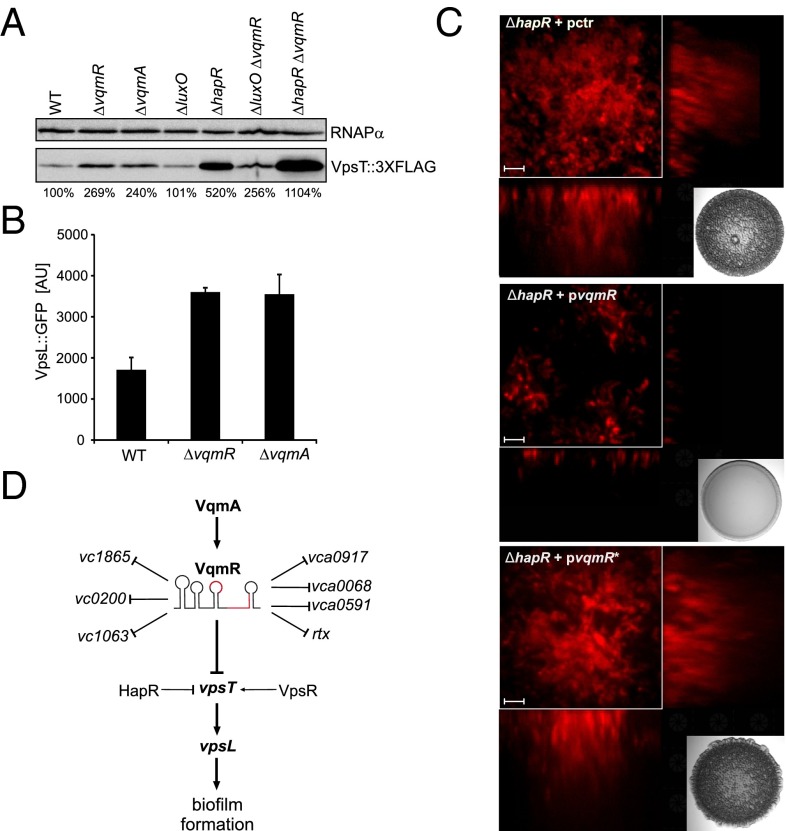
Our ultimate aim is to understand the VqmA–VqmR regulatory network and how it influences the physiology of V. cholerae. We were intrigued by our identification of vpsT as a VqmR target because vpsT encodes a major transcriptional regulator of biofilm formation. At low cell density, VpsT activates the expression of genes required for production of vibrio polysaccharide (vps), and, thus, mutation of vpsT strongly inhibits biofilm formation (36). Transcription of vpsT is itself activated by another biofilm regulator, VpsR (37). Finally, vpsT is repressed at high cell density by the master QS regulator, HapR (38). Our results (Fig. 5A) show that increased vpsT mRNA is present in the ΔvqmR strain. To examine whether there is a corresponding increase in VpsT protein, we engineered a vpsT::3XFLAG construct, introduced it onto the V. cholerae chromosome at the vpsT locus, and measured protein production. Both the ΔvqmR and ΔvqmA strains possessed ∼2.5-fold more VpsT protein than wild-type V. cholerae (Fig. 7A, lanes 1–3). We also investigated VpsT levels in QS mutants. Consistent with our understanding of the QS circuitry, mutation of luxO did not affect VpsT protein levels (Fig. 7A, lane 4), whereas deletion of hapR strongly activated VpsT production (∼fivefold, Fig. 7A, lane 5). A ΔluxO, ΔvqmR double mutant had the same level of VpsT as the single ΔvqmR mutant (lane 6), whereas mutation of both the vqmR and hapR repressors caused an additive effect and increased VpsT levels ∼11-fold (lane 7). These results indicate that both HapR and VqmR repress vpsT expression. We suggest that HapR operates at the level of transcription (38) whereas VqmR acts at the level of translation.
Fig. 7.
VqmR represses biofilm formation in V. cholerae. (A) Western blot analysis of VpsT::3XFLAG. Wild-type, ΔvqmR, ΔvqmA, ΔluxO, and ΔhapR single mutants and ΔluxO vqmR and ΔhapR vqmR double mutant V. cholerae strains were grown to OD600 of 1.0 and examined for VpsT::3XFLAG production using an anti-FLAG antibody. RNAPα served as the loading control. Changes in VpsT::3XFLAG protein production are indicated below the figure (WT was set to 100%). (B) Production of VpsL::GFP. Wild-type, ΔvqmR, and ΔvqmA V. cholerae strains carrying a vpsL::gfp transcriptional reporter on a plasmid were grown for 6 h and GFP production was measured. Error bars represent SD of three replicates. (C) Biofilm production of vqmR-expressing strains. Biofilm formation of the ΔhapR V. cholerae strain constitutively expressing the fluorescent mKate2 protein was assayed in the presence of a control plasmid (Top, pctr), a plasmid carrying vqmR (Middle, pvqmR), or a plasmid carrying VqmR C94G (pvqmR*) that does not base pair with vpsT (Bottom). Biofilms were imaged following a 24-h incubation at 30 °C. (Scale bar: 5 µm.) The inlays show the colony morphologies of the indicated strains grown on agar plates and incubated for 24 h at 30 °C. (D) Model of VqmR activity in V. cholerae. Transcription of vqmR is activated by VqmA. VqmR directly regulates at least eight target mRNAs including vpsT. VpsT, in turn, acts as an activator of VpsL production and biofilm formation.
Binding of VpsT to the second messenger cyclic di-GMP alters VpsT oligomerization and activates its activity (39). To test if the increased VpsT protein present in the ΔvqmR and ΔvqmA mutants correlated with increased VpsT activity, we measured expression of the VpsT target vpsL (Fig. 7B). Indeed, at high cell density, vpsL::gfp was approximately twofold higher in both the ΔvqmR and ΔvqmA strains than in the wild type. To verify that this regulatory architecture indeed affects V. cholerae biofilm formation, we overexpressed vqmR in the hyperbiofilm producing (i.e., rugose) ΔhapR V. cholerae strain and measured biofilms. The ΔhapR strain displayed the characteristic rugose phenotype caused by vps overproduction [Fig. 7C, Top (40, 41)]. Overexpression of vqmR restored the nonrugose, wild-type colony morphology (Fig. 7C Middle). Likewise, biofilm formation on glass surfaces was strongly inhibited when VqmR was overexpressed (Fig. 7C, compare Top and Middle). No alteration in colony morphology or biofilm formation capability occurred in either assay when the mutated vqmR C94G (pvqmR*) allele was expressed (Fig. 7C Bottom). These results suggest that VqmR affects biofilm formation through posttranscriptional repression of vpsT, which in turn, regulates biofilm-associated genes, such as vpsL (Fig. 7D).
Discussion
Whole-transcriptome RNA sequencing analyses have enabled a quantitative understanding of the gene expression patterns underlying the basic biology of organisms. Nevertheless, it has been difficult to distinguish transcription initiation from posttranscriptional regulation because the output of most RNA-seq technologies is the sum of both processes. In this study, we used dRNA-seq to investigate the transcriptional and posttranscriptional landscape of the human pathogen V. cholerae. We annotated a total of 7,240 TSSs, 3,379 (47%) of which initiated antisense transcripts (Fig. 1B). Pervasive antisense transcription has now been reported for bacterial species from diverse taxa (42), suggesting this class of transcripts could be the norm in the bacterial world.
We detected 107 previously unannotated sRNA candidates, 44 of which have the potential to also function as 3′ UTRs of mRNAs (SI Appendix, Dataset S4). This finding, together with earlier reports, suggests that mRNA UTRs can serve as reservoirs for bacterial sRNAs (26, 43). Our data indicate that there exist two general pathways to generate sRNAs from the 3′ ends of mRNAs. In the first case, sRNA genes can be driven by dedicated promoters. This mechanism has the advantage of enabling transcriptional regulation of the sRNA independent from that controlling the mRNA from which the sRNA is derived (Fig. 2A). In the second case, sRNA production is strictly linked to the upstream promoter of the overlapping gene and requires ribonucleolytic cleavage to separate the sRNA from the mRNA (Fig. 2B). Production of this second class of sRNAs might come at a cost: processing of the mRNA could render the transcript inactive for additional rounds of translation. For both types of sRNAs, rho-independent terminator elements at the 3′ ends could provide the basis for Hfq binding and potential trans-regulatory functions (44).
Our dRNA-seq analysis revealed the VqmR sRNA encoded upstream of vqmA. To explore the relationship between vqmA and vqmR, we performed a gene synteny analysis to identify homologs of vqmR. In all cases, the presence of vqmA coincided with an upstream vqmR gene (SI Appendix, Fig. S8A). Similar approaches have identified homologs of the SgrS (45) and GcvB (46) sRNAs, which, like VqmR, are located adjacent to the genes encoding the cognate transcriptional regulators. We do not yet know the functions of VqmR in other vibrios, but the high degree of sequence conservation in VqmR regions R1 and R2 (Fig. 3C) strongly suggests that regulation of trans-encoded target mRNAs is a conserved function.
The current set of confirmed VqmR targets in V. cholerae includes eight directly regulated mRNAs (Fig. 5C). Repression of seven of the targets depends on conserved region R2, whereas vc1063 requires region R1 (Figs. 5C and 6 A–D and SI Appendix, Fig. S7C). One of the main findings of our study is that VqmR regulates biofilm formation through repression of vpsT (Fig. 7C). We already know that vpsT is also controlled by QS via HapR (38). Thus, our work reveals that VqmR represents a previously unidentified link between biofilm formation and QS. VpsT is a homolog of CsgD in enterobacterial species. CsgD controls biofilm formation and multicellular development in E. coli (47) and regulation of csgD depends on six Hfq-dependent sRNAs (48). Our finding that vpsT is under the control of the unrelated VqmR sRNA in V. cholerae could indicate evolution of parallel regulatory pathways. Other examples like this exist: the V. cholerae VrrA sRNA and the E. coli MicA sRNA are not related to one another but both are controlled by the alternative sigma factor σE and both base pair with the ompA mRNA (49, 50). Likewise, in numerous bacterial species, sRNAs that are not related to one another are nonetheless required for responses to iron limitation, and all function by repressing mRNAs encoding iron-binding proteins (51).
Why certain pathways preferentially use regulatory RNAs rather than proteins has been the subject of intense speculation (52). One major difference is that sRNAs accumulate to significantly higher copy numbers than transcriptional regulators. Indeed, we found that VqmR accumulates to >500 molecules per cell in stationary phase (SI Appendix, Fig. S8B), whereas copy numbers of transcription factors are typically <100 (53). The higher copy numbers of sRNAs can provide an effective buffer against noise from transcriptional bursting. This feature of sRNAs is proposed to make signaling through biological networks robust to fluctuations in mRNA production (54). Given that Vps production comes at a high metabolic cost (55), VqmR-mediated repression of vpsT might further enhance transcriptional (37, 38) and posttranslational (39) mechanisms to curtail noisy gene expression. This hypothesis is supported by the notion that QS phenotypes and biofilm formation are traits that require group-wide coordination and thus the regulatory circuits underpinning them must possess mechanisms that minimize fluctuations (56). Another possible explanation for the necessity of VqmR to act between VqmA and the downstream targets could be because VqmA is strictly an activator. Recruiting the VqmR sRNA, which we show is capable of activation and repression, could expand the regulatory capability of VqmA.
VqmA was previously reported to be connected to QS via activation of transcription of hapR (32). We do not observe this regulatory link. We wonder if the constructs used in the previous analyses unknowingly included vqmR, which caused indirect effects. Recent V. cholerae infection studies also report that vqmA expression is regulated by the interspecies autoinducer AI-2 and that AI-2 regulation of vqmA is independent of HapR (57). Indeed, our results show that neither vqmA nor vqmR display differential expression in the luxO D47E QS mutant (SI Appendix, Dataset S1 and Fig. 3B). It is possible that binding of AI-2 (or an AI-2–controlled factor) alters VqmA activity to activate VqmR transcription, and VqmR, in turn, represses biofilm formation and toxin production. Identifying components of this putative QS-mediated but HapR-independent pathway in V. cholerae biofilm formation would be fascinating given that, in nature, hapR mutations commonly occur in pandemic V. cholerae strains to produce hyperbiofilm formers (58).
Materials and Methods
Strains, Plasmids, and Growth Conditions.
Strains are listed in SI Appendix, Table S2. V. cholerae and E. coli were grown aerobically in LB or M9 medium (0.4% glucose, 0.4% casamino acids) at 37 °C or at 30 °C for biofilm assays. Antibiotics were used at the following concentrations: 50 units⋅ml−1 polymyxin B, 200 μg⋅ml−1 ampicillin, 100 μg⋅ml−1 kanamycin, 5,000 μg⋅ml−1 streptomycin, and 20 μg⋅ml−1 chloramphenicol.
Sample Collection for dRNA-Seq Analysis.
V. cholerae wild-type and luxO D47E strains were grown with shaking in LB medium at 37 °C and whole cell samples (biological duplicates) were collected at OD600 of 0.1 and 2.0. Transcription and translation were terminated by the addition of STOP Mix [95% (vol/vol) EtOH and 5% (vol/vol) phenol] and cells were frozen in liquid nitrogen. Samples were stored at −80 °C until RNA preparation.
Oligonucleotides and Plasmids.
Plasmids and DNA oligonucleotides are listed in SI Appendix, Tables S3 and S4, respectively. Details on plasmid construction are provided in SI Appendix, SI Materials and Methods.
Northern Blot Analyses.
Total RNA was prepared and blotted as described (59). Membranes were hybridized in Rapid-Hyb buffer (GE Healthcare) at 42 °C with [32P] end-labeled DNA oligonucleotides. Signals were visualized using a Typhoon phosphorimager (GE Healthcare) and band intensities were quantified using GelQuant software. Oligonucleotides for sRNA probing are listed in SI Appendix, Dataset S4 and Table S4.
Purification of VqmA::3XFLAG and EMSA.
The VqmA::3XFLAG protein was expressed from a pET15b expression vector in BL21(DE3) cells (Agilent). Protein expression was induced by addition of IPTG (1 mM final) for 4 h before harvesting. Cells were resuspended in lysis buffer (Sigma) and lysed by sonication. Protein purification was performed using the FLAG Immunoprecipitation Kit (Sigma) following the manufacturer’s instructions. Protein purification was verified by SDS/PAGE analysis. For EMSA studies, the vqmR promoter sequence (∼100 bp upstream of the TSS) or a mutagenized variant were PCR amplified and [32P] end-labeled using PNK enzyme (Fermentas). The labeled DNA fragments (∼0.2 pmol) were incubated with increasing amounts of VqmA::3XFLAG protein (0, 23, 46, 93, 187, 345, and 750 ng) in binding buffer (10 mM Tris⋅HCl pH 8.0, 1 mM EDTA, 1 mM DTT, 50 mM KCl, 1.5 mg/mL poly IC, 50 µg/mL BSA, 10% glycerol) for 30 min at 37 °C. DNA–protein complexes were separated on 6% native polyacrylamide gels. Protein signal was visualized using a Typhoon phosphorimager (GE Healthcare).
Western Blot Analysis.
Western blot analyses of GFP and FLAG fusion proteins followed previously published protocols (60). Signals were visualized using an ImageQuant LAS-4000 imager (GE Healthcare) and band intensities were quantified using GelQuant software.
qRT-PCR and Microarray Analysis.
Quantitative real-time PCR was described previously (59). Briefly, RNA was isolated using the SV40 Total RNA Isolation kit (Promega) according to manufacturer instructions. Expression of VqmR-controlled target genes was assessed by qRT-PCR in a 7900HT instrument (Applied Biosystems), with hfq as reference. Oligonucleotides used in qRT-PCR experiments are listed in SI Appendix, Table S4. For microarray analyses, V. cholerae strain C6706 ΔvqmR/ΔvqmA (ZLV101) carrying either the control plasmid pCMW-1 (pctr) or pBAD-vqmR was grown in LB at 37 °C to OD600 of 1.0. Expression from the pBAD promoter was induced by addition of 0.2% (final concentration) l-arabinose. Duplicate samples were collected 10 min after arabinose treatment. Four arrays were performed comparing two independent cultures as well as a dye‐swap control. Data analyses were performed using the Princeton University Microarray Database and are publicly available at puma.princeton.edu/.
Construction of cDNA Libraries and Illumina Sequencing.
cDNA libraries were constructed as described (21) by Vertis Biotechnology and sequenced using a HiSEq. 2000 machine (Illumina) in single-read mode for 100 cycles. The raw, demultiplexed reads and coverage files have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus with accession code GSE62084. Statistics on cDNA library sequencing are provided in SI Appendix, Table S5. Detailed descriptions of procedures used for read mapping, expression graph construction, and normalization of expression graphs are provided in SI Appendix, SI Materials and Methods.
Biofilm Assays.
The V. cholerae ΔhapR strain was grown for 12 h in M9 medium at 37 °C and 200-µL aliquots were deposited into 96-well plates. Following incubation for 1 h at 30 °C, cells were washed three times with M9 medium and incubation at 30 °C was continued for 12 h. Cells were washed three times, and images of biofilms were acquired using a Nikon Ti-Eclipse microscope and a TCS SP5, Leica confocal laser-scanning microscope.
Supplementary Material
Acknowledgments
We thank Jun Zhu for strain ZLV101, Kathrin Fröhlich for help with structure probing experiments, Carey Nadell for support with biofilm assays, Yi Shao for help with the microarrays, Richard Reinhardt for help with deep sequencing analyses, and all members of the B.L.B. laboratory for insightful discussions and suggestions. K.P. is supported by a postdoctoral fellowship from the Human Frontiers in Science program. C.M.S. is supported by the ZINF Young Investigator program and the young academy program of the Bavarian Academy of Sciences and Humanities. B.L.B. is supported by the Howard Hughes Medical Institute, National Institutes of Health Grant 5R01GM065859, and National Science Foundation Grant MCB-0343821.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE62084).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500203112/-/DCSupplemental.
References
- 1.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11) doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118(1):69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25(4):397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31(2):665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 5.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol. 2013;195(3):436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99(5):3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9(8):578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma CM, Vogel J. Differential RNA-seq: The approach behind and the biological insight gained. Curr Opin Microbiol. 2014;19:97–105. doi: 10.1016/j.mib.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Sorek R, Cossart P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat Rev Genet. 2010;11(1):9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 10.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 11.Bradley ES, Bodi K, Ismail AM, Camilli A. A genome-wide approach to discovery of small RNAs involved in regulation of virulence in Vibrio cholerae. PLoS Pathog. 2011;7(7):e1002126. doi: 10.1371/journal.ppat.1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandlik A, et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10(2):165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JM, et al. Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing. Nucleic Acids Res. 2009;37(6):e46. doi: 10.1093/nar/gkp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raabe CA, et al. The rocks and shallows of deep RNA sequencing: Examples in the Vibrio cholerae RNome. RNA. 2011;17(7):1357–1366. doi: 10.1261/rna.2682311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012;40(16):7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Y, Bassler BL. Quorum-sensing non-coding small RNAs use unique pairing regions to differentially control mRNA targets. Mol Microbiol. 2012;83(3):599–611. doi: 10.1111/j.1365-2958.2011.07959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao Y, Bassler BL. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol Microbiol. 2014;92(5):921–930. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol. 2009;191(1):169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kröger C, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2012;109(20):E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dugar G, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9(5):e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomason MK, et al. Global transcriptional start site mapping using dRNA-seq reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2014;197(1):18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malys N, McCarthy JE. Translation initiation: Variations in the mechanism can be anticipated. Cell Mol Life Sci. 2011;68(6):991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93(11):1943–1949. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31(20):4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandyra KJ, Bouvier M, Carpousis AJ, Luisi BF. The social fabric of the RNA degradosome. Biochim Biophys Acta. 2013;1829(6-7):514–522. doi: 10.1016/j.bbagrm.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75(2):286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayed N, Jousselin A, Felden B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol. 2012;19(1):105–112. doi: 10.1038/nsmb.2193. [DOI] [PubMed] [Google Scholar]
- 30.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12) doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Hsiao A, Joelsson A, Zhu J. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J Bacteriol. 2006;188(7):2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papenfort K, et al. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68(4):890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 34.Corcoran CP, et al. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol. 2012;84(3):428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 35.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186(5):1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193(22):6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190(7):2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jobling MG, Holmes RK. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26(5):1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 41.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53(2):497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 42.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: A new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol. 2013;11(2):75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 43.Guo MS, et al. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28(14):1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc Natl Acad Sci USA. 2011;108(32):13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horler RS, Vanderpool CK. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 2009;37(16):5465–5476. doi: 10.1093/nar/gkp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21(21):2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18(4):661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 48.Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014;11(5):494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song T, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70(1):100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udekwu KI, Wagner EG. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007;35(4):1279–1288. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salvail H, Massé E. Regulating iron storage and metabolism with RNA: An overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip Rev RNA. 2012;3(1):26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 52.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol Rev. 2010;34(5):866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehta P, Goyal S, Wingreen NS. A quantitative comparison of sRNA-based and protein-based gene regulation. Mol Syst Biol. 2008;4:221. doi: 10.1038/msb.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA. 2011;108(34):14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long T, et al. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 2009;7(3):e68. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515(7527):423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talyzina NM, Ingvarsson PK, Zhu J, Wai SN, Andersson A. Molecular diversification in the quorum-sensing system of Vibrio cholerae: Role of natural selection in the emergence of pandemic strains. Appl Environ Microbiol. 2009;75(11):3808–3812. doi: 10.1128/AEM.02496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papenfort K, et al. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol. 2006;62(6):1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell. 2013;153(2):426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.