Significance
The human lysosomal polypeptide transporter TAPL belongs to the ATP-binding cassette (ABC) superfamily. Transport studies of ABC exporters are rare, and, most often, function is inferred from ATP hydrolysis rather than translocation measurements. For mechanistic understanding of solute transport, the proteins have to be purified and incorporated into lipid vesicles. Macroscopic measurements with ensembles of molecules are often problematic due to inhomogeneities in reconstitution. Therefore, we developed a method called dual-color fluorescence-burst analysis to analyze peptide transport into single liposomes with, on average, one to three TAPL complexes per vesicle. We show that the turnover number is in a physiologically meaningful range and that TAPL is indeed an active transporter accumulating peptides against a concentration gradient.
Keywords: ABCB9, DCFBA, lysosome, trans-inhibition, uphill transport
Abstract
ATP-binding cassette (ABC) transporters use ATP to drive solute transport across biological membranes. Members of this superfamily have crucial roles in cell physiology, and some of the transporters are linked to severe diseases. However, understanding of the transport mechanism, especially of human ABC exporters, is scarce. We reconstituted the human lysosomal polypeptide ABC transporter TAPL, expressed in Pichia pastoris, into lipid vesicles (liposomes) and performed explicit transport measurements. We analyzed solute transport at the single liposome level by monitoring the coincident fluorescence of solutes and proteoliposomes in the focal volume of a confocal microscope. We determined a turnover number of eight peptides per minute, which is two orders of magnitude higher than previously estimated from macroscopic measurements. Moreover, we show that TAPL translocates peptides against a large concentration gradient. Maximal filling is not limited by an electrochemical gradient but by trans-inhibition. Countertransport and reversibility studies demonstrate that peptide translocation is a strictly unidirectional process. Altogether, these data are included in a refined model of solute transport by ABC exporters.
The transporter associated with antigen processing-like (TAPL, ABCB9) belongs to the superfamily of ATP-binding cassette (ABC) proteins, which is one of the largest membrane protein families present in all kingdoms of life (1, 2). Most members of this superfamily are transporters (3–5). ABC transporters play crucial roles in many cellular processes; defects in some of these proteins generate diseases, such as cystic fibrosis and immune deficiency, whereas other members of this family cause antibiotic or multidrug resistance (6, 7). Based on the transport direction, ABC transporters are subdivided in importers mediating solute uptake into the cytosol and exporters extruding substrates out of the cytosol. During transport cycle, the transmembrane domain interconverts between inward- and outward-facing conformations, which is driven by binding and hydrolysis of ATP in the cytosolic nucleotide-binding domains (NBDs).
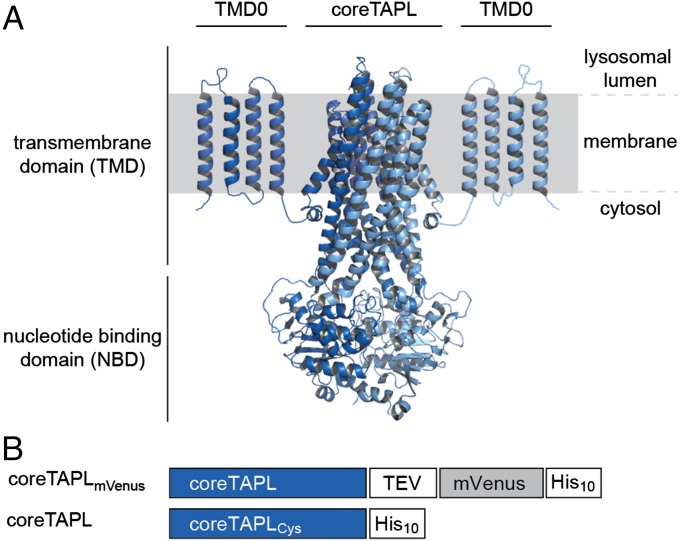
TAPL is a so-called exporter, which shuttles 6- up to 59-mer peptides from the cytosol into the lysosome in a process that is coupled to ATP hydrolysis. Peptide selectivity is restricted to the N- and C-terminal residues, where positive-charged and large hydrophobic residues are favored (8). Orthologs of TAPL are not only found in vertebrates but also in invertebrates and even in plants (9, 10). TAPL expression shows a broad tissue distribution with high expression in dendritic cells and macrophages (11, 12). TAPL is a homodimeric protein complex with a core domain for peptide transport and an accessory N-terminal membrane-embedded domain, called TMD0 (transmembrane domain 0), responsible for correct lysosomal targeting and interaction with the lysosomal associated membrane proteins LAMP-1/2 (8, 13, 14) (Fig. 1A).
Fig. 1.
Architecture and constructs of the homodimeric TAPL complex. (A) CoreTAPL was modeled on the X-ray structure of Sav1866 (Protein Data Bank ID: 2ONJ) (4). The core complex consists of 2 × 6 transmembrane helices and two nucleotide-binding domains. The accessory N-terminal membrane-embedded domain TMD0 is schematically depicted by four predicted transmembrane helices connected by short loops. (B) A fusion construct of human coreTAPL (637 amino acids, 69.7 kDa) linked to the YFP derivative mVenus via a tobacco etch virus (TEV) cleavage site and a single-cysteine coreTAPL mutant were designed for expression in Pichia pastoris.
Contrary to ABC importers, there is no compelling evidence that ABC exporters translocate solutes against their concentration gradient, and trans-inhibitory effects are poorly understood. Generally, the activity of ABC exporters is determined by measuring the rate of ATP hydrolysis, because sensitive direct translocation assays are not available. Moreover, stimulation of basal ATPase activity by substrates does not correlate with transport. For Pdr5 from Saccharomyces cerevisiae, no ATPase stimulation by substrates was detected, whereas human ABCC3 (multidrug resistance-associated protein 3) shows a weak coupling between ATP hydrolysis and substrate transport for leukotriene C4 but not for carboxydichlorofluorescein (15, 16). The lysosomal transporter TAPL is an ideal model system to address these outstanding questions because it transports hydrophilic peptides that do not partition in the membrane and can be labeled by fluorophores.
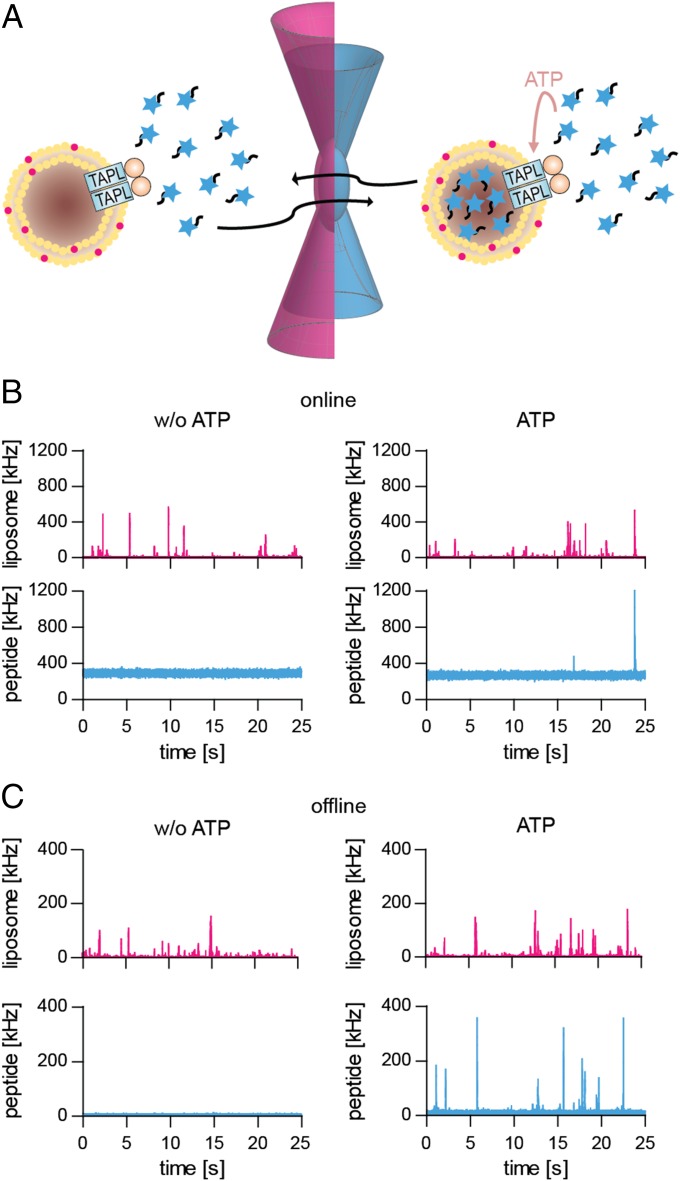
Detailed mechanistic studies of membrane transport are typically performed by macroscopic measurements. With small vesicles and high protein-to-lipid ratios, required to obtain a good signal-to-noise ratio, it is difficult to estimate true initial transport rates and to dissect variations in accumulation ratios from trans-inhibitory effects. We applied dual-color fluorescence-burst analysis (DCFBA) to determine the activity of single transporters in individual liposomes (17, 18). DCFBA relies on the coincidence of fluorescence bursts in the confocal volume of a microscope, which originate from spectrally well-separated fluorophores that report the number of TAPL molecules per vesicle and the number of peptides translocated (Fig. 2A). DCFBA is related to confocal fluorescence coincidence analysis and two-color coincidence detection, but the analysis method differs (19, 20). Up to now, this technique has been used to follow high-affinity binding of ligands to membrane-embedded proteins (21) or efflux of substrates out of liposomes, where the fluorescently labeled substrates are infinitely diluted and do not interfere with the measurements (17, 18).
Fig. 2.
Experimental setup and fluorescence traces of DCFBA. (A) Coincident fluorescent bursts caused by the diffusion of proteoliposomes and peptides through the confocal volume. Liposomes are labeled with DiD (633-nm He−Ne laser excitation) or reconstituted coreTAPLmVenus (488-nm argon laser excitation); peptides are conjugated with either ATTO488 (488 nm) or ATTO655 (633 nm). (B and C) Peptide transport using DiD-labeled proteoliposomes (2.5 µg coreTAPL, 50 µg lipid), incubated with 5 µM RRYCATTO488KSTEL at 37 °C for 30 min in the absence (w/o ATP) or presence of 3 mM Mg-ATP, was directly analyzed at the microscope (B) or after removal of external peptide (C). Laser powers were kept constant: 488, 1.5 µW (B) and 3 µW (C); 633, 5 µW (B) and 1 µW (C). For direct measurements, the last 25 s of a 30-min transport period are depicted.
By adjusting DCFBA to probe solute import, we determined the kinetics of peptide transport via TAPL on the single liposome level. Moreover, we could evaluate uphill transport and trans-inhibition not biased by inhomogeneity in reconstitution.
Results and Discussion
Expression, Purification, and Macroscopic Transport Studies.
CoreTAPL C-terminally coupled with the fluorescent protein mVenus (coreTAPLmVenus) or coreTAPL containing a single cysteine (coreTAPL) were expressed in P. pastoris and purified with a yield of ∼45 mg per liter of fermenter culture (Fig. 1B and Figs. S1 and S2). Both homodimeric complexes were reconstituted into liposomes composed of Escherichia coli polar lipids and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) (7:3 weight ratio). The reconstitution efficiency was 22% as evaluated by sodium carbonate treatment followed by a sucrose gradient centrifugation to isolate membrane-inserted TAPL (Fig. S3). As determined by nanoparticle tracking analysis, the proteoliposomes had a mean particle diameter of 143 ± 49 nm (Fig. S4) (22). On average, nine TAPL complexes were inserted per liposome, and one TAPL in vesicles with a diameter of ∼50 nm. As derived from macroscopic filter assays, coreTAPL variants followed Michaelis−Menten kinetics for the import of the peptide RRYCATTO488KSTEL, a modified naturally occurring degradation product of histone H3 (23). The Michaelis−Menten constant is similar to that of wild-type TAPL in membranes derived from Sf9 insect cells (8, 24), indicating that neither cysteine mutation nor fusion with mVenus or deletion of TMD0 influence TAPL activity (Fig. 3A and Table 1). However, all former and present studies of reconstituted TAPL show a maximal transport rate of 0.1 peptides per minute, which is slow in comparison with the activity in membrane vesicles isolated from TAPL expressing Sf9 insect cells (8). The reasons for low activity are loss of vesicles in the filter assay (Table S1), the inactivation of TAPL during reconstitution, and the dual orientation of TAPL in the proteoliposomes (Fig. S5).
Fig. 3.
Michaelis−Menten kinetics of transport by coreTAPL. Proteoliposomes (50 µg lipid) containing coreTAPL (0.5 µg) were incubated for 20 min at 37 °C in the presence of 3 mM Mg-ATP, and the amount of transported RRYCATTO488KSTEL was quantified by a filter assay (A) or DCFBA (B). Peptide transport analyzed by DCFBA in Cav per minute was normalized at the highest substrate concentration. Laser powers were kept constant: 488, 3 µW; 633, 1 µW. Data were fitted by the Michaelis−Menten equation, and KM values are given in Table 1.
Table 1.
Kinetic parameters of peptide transport by TAPL
| Macroscopic | DCFBA | |
| KM | 2.8 ± 0.5 µM | 0.9 ± 0.3 µM |
| kcat | 0.064 ± 0.004 min−1 | 8 ± 2 min−1 |
DCFBA for Single-Liposome Import.
To characterize the reconstituted vesicles, DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine)-labeled liposomes (DiD:lipid molar ratio 1:4,000) containing coreTAPLmVenus were studied by DCFBA. From the DiD-derived fluorescence bursts (5−15 ms per vesicle), a mean vesicle diameter of 91 ± 38 nm (n = 115 liposomes) was calculated by taking the DiD fluorescence in detergent as reference (Figs. S4B and S6B). The DCFBA-derived vesicle sizes are somewhat smaller than those estimated by nanoparticle tracking analysis, which relates the diffusion coefficient to particle size. We found that 61% of all liposomes (n = 17,527) with a lipid-to-protein mass ratio of 20 (corresponding to a molar ratio of 6,000) contain coreTAPLmVenus. By evaluating the coincident fluorescent bursts from ATTO488-labeled peptide (RRYCATTO488KSTEL, 3 µM), accumulated for 60 min in the presence of 3 mM ATP and coreTAPL reconstituted in DiD-labeled liposomes, we found that ∼8% of the liposomes (n = 9,738) are active in peptide transport. Importantly, DCFBA allows dissection of the vesicle heterogeneity typical for reconstitutions of membrane proteins.
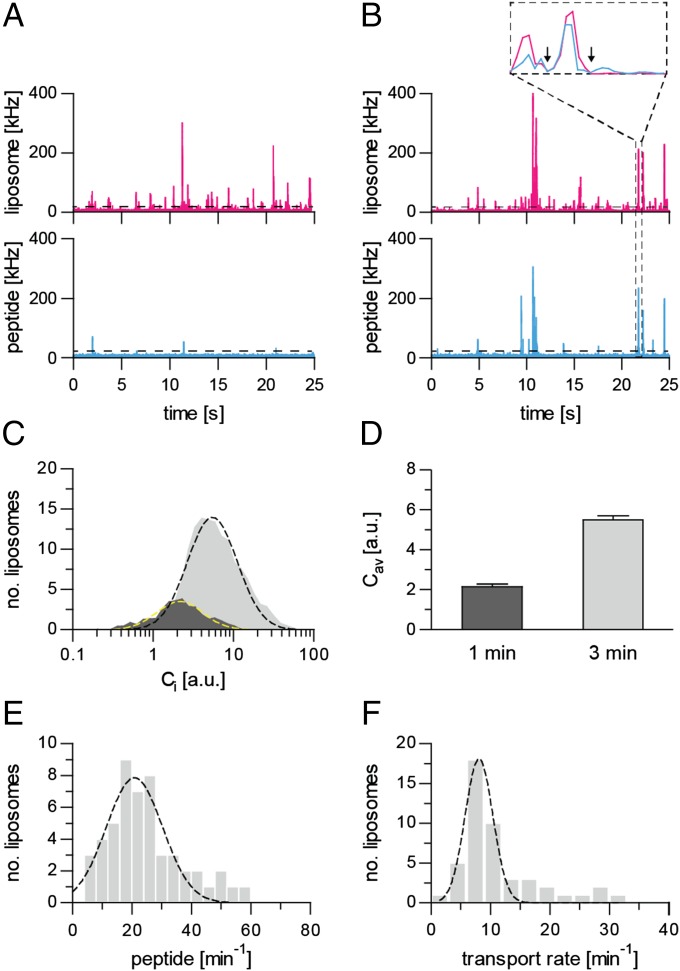
We adopted the DCFBA method to determine the import of peptides into individual liposomes with one or a few functional TAPL complexes per vesicle. The accumulation of the peptide RRYCATTO488KSTEL in TAPL-containing proteoliposomes (0.5 mg/mL lipids), labeled with DiD, could not be detected by laser-scanning confocal microscopy because of a too-high background fluorescence of external peptide, even in the presence of 100 mM of the collisional quencher iodide (100 mM KI). We thus removed external fluorescent peptide by ultracentrifugation. The coincidence of fluorescence bursts from labeled peptide and vesicles was observed in the presence but not in the absence of ATP (Fig. 2). For quantification, all coincident fluorescent bursts above a certain offset were used to calculate the average (Cav) of the internal peptide concentration (Ci) over all liposomes active in peptide import (Fig. 4). Offsets for the peptide channel were derived from DCFBA traces in the absence of ATP, whereas the offsets for the liposome channel were obtained from DCFBA traces with nonlabeled liposomes. An increase in peptide or ATP concentration as well as elongation of transport increased the number of peptide-enriched liposomes as well as the concentration of accumulated peptide (Fig. 4). A plot of the mean of the log-normal Gaussian distributed histograms (Cav) against peptide concentration shows a hyperbolic behavior with Michaelis−Menten constant KM of 0.9 ± 0.3 µM, which is in good agreement with that measured by conventional macroscopic methods (Fig. 3 and Table 1).
Fig. 4.
DCFBA of TAPL-dependent peptide transport. (A and B) Labeled proteoliposomes (2.5 µg coreTAPL, 50 µg lipid) were incubated at 37 °C for 1 min (A) or 3 min (B) with 5 µM RRYCATTO488KSTEL and 3 mM Mg-ATP. DCFBA was performed with intensity traces for DiD-labeled liposomes (red) and luminal peptide RRYCATTO488KSTEL (blue). The relative amount of transported peptide was calculated from the ratio of fluorescence bursts in both channels above a certain offset (22 kHz for liposomes; 25 kHz for peptides). Only 25 s of the whole traces (5 min) are presented. Inset depicts single burst events of approximately 8 ms (arrows indicate the duration of the event). The laser power was kept constant: 488, 3 µW; 633, 1 µW. (C and D) Fitting the concentration distribution of transported RRYCATTO488KSTEL with a log Gaussian equation (black and yellow dashed lines) resulted in a mean internal peptide concentration Cav. (E) Accumulation of peptides in single liposomes. The actual number of transported peptides per liposome was quantified using standards as depicted in Fig. S6. A Gaussian fit of the transport rate distribution of liposomes (n = 51) results in a mean transport rate of 21 ± 9 peptides per liposome per minute. (F) Turnover number of TAPL. Proteoliposomes (50 µg lipid) containing coreTAPLmVenus (2.5 µg) were incubated with 10 µM RRYCATTO655KSTEL for 1 min at 37 °C in the presence of 3 mM Mg-ATP. The mean transport rate per TAPL complex was determined from individual liposomes (n = 47) containing one to three TAPL complexes. The transport rate distribution was fitted by a Gaussian equation resulting in a mean turnover number of 8 ± 2 peptides per minute. Laser powers were kept constant: 488, 4.5 µW; 633, 2 µW.
We used DCFBA not strictly as a single-molecule technique, because liposomes were filled with 20–300 peptide molecules. We estimated the absolute number of peptides accumulated in liposomes from intensity calibration curves, using free peptide and DiD as standards (Fig. S6 A and B). A transport rate of 21 ± 9 peptides per liposome per minute was obtained under saturating ATP and peptide concentrations (Fig. 4E). The variation in transport rates results from heterogeneity in TAPL distribution and liposome size. In conclusion, DCFBA is a valuable method to analyze solute accumulation in individual vesicles.
Turnover Number of Peptide Translocation.
Next, we determined the turnover number of TAPL by DCFBA. Because we could apply only two laser lines simultaneously, we used proteoliposomes containing coreTAPLmVenus and followed the accumulation of RRYCATTO655KSTEL. The number of TAPL complexes per liposome was estimated from the mVenus fluorescence (Fig. S6 C and D). Transport was performed for 1 min under saturating ATP (3 mM) and peptide (10 µM) concentrations, and only liposomes containing one to three TAPL complexes were considered for initial rate measurements. By analyzing 47 liposomes, a turnover number of 8 ± 2 transported peptides per minute was obtained (Fig. 4F), which is close to the value obtained with crude membranes (8). Because purified coreTAPLmVenus is very stable (Fig. S5), it is unlikely that we overestimated the transport rate by proteolytic cleavage of mVenus from TAPL. In addition, peptide fluorescence bursts that are not coinciding with mVenus fluorescence were identical with and without ATP and may reflect some aggregation of peptides. Our microscopic transport rate correlates with ATP hydrolysis rates of other ABC exporters like MsbA reconstituted in nanodiscs (19 ATP per min) (25) or ABCC3 reconstituted in liposomes (36 ATP per min) (26).
Peptide Accumulation Is Restricted by Trans-Inhibition.
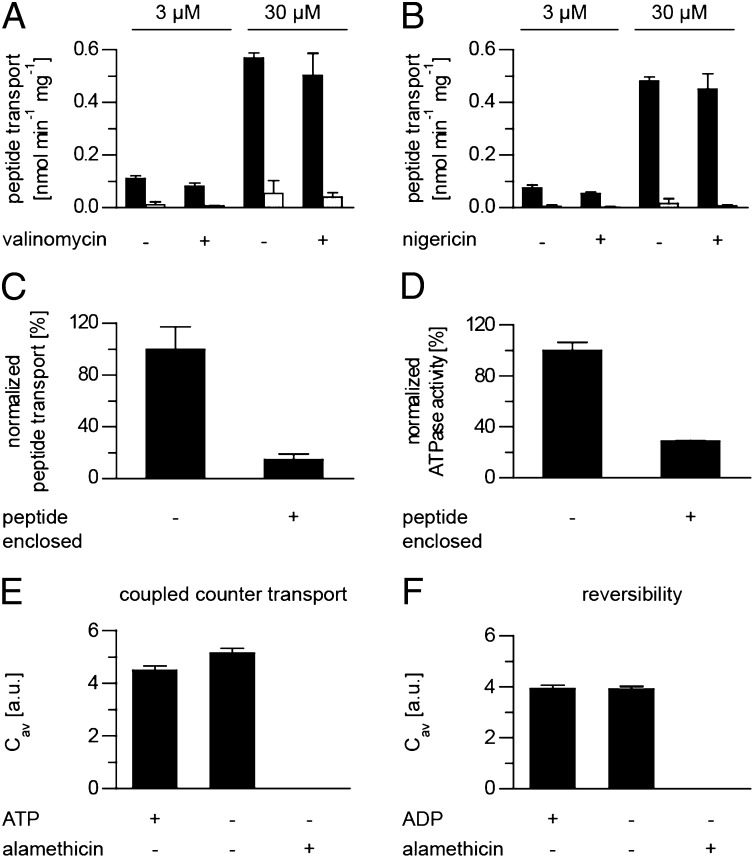
Although the free energy of ATP is potentially sufficient to establish large electrochemical gradients, the evidence that ABC exporters pump solutes uphill is scarce (27, 28). The classical macroscopic studies average accumulation levels over all liposomes and do not consider vesicles with and without active transporters, as well as possible trans-inhibitory effects in relation to volume, and thus neglect concentration differences. In contrast, DCFBA allows the quantification of transport into individual liposomes. For maximal filling, we incubated proteoliposomes with 0.3–30 µM of RRYCATTO488KSTEL. After 60 min of transport, luminal peptide concentrations of 0.3–0.9 mM were obtained, which corresponds to an approximate 1,000-fold and 30-fold accumulation, respectively (Table 2). The highest level of peptide accumulation was observed in small liposomes with a diameter of 50 nm. Peptide accumulation was not inhibited by an electrochemical gradient, resulting from the import of net positively charged peptide, because peptide transport was not affected by the ionophores valinomycin and nigericin (Fig. 5 A and B). Hence peptide transport in proteoliposomes, prefilled with 3 mM unlabeled peptide (RRYQKSTEL), is limited by trans-inhibition (Fig. 5 C and D). Such product inhibition has been reported for ion (29) and amino acid transporters (30–32), including ABC importers like MetNI from E. coli (33), the glycine/betaine importer from Lactobacillus plantarum and Listeria monocytogenes (34, 35), and ModBC from Methanosarcina acetivorans (36). However, compelling evidence for trans-inhibition in ABC exporters is not available, except for the heterodimeric ABC-transporter TAP. The trans-inhibitory peptide concentration for TAP is 50-fold lower than for TAPL (27). In summary, TAPL is an active pump whose activity is limited by threshold levels of luminal peptide.
Table 2.
Peptide accumulation via TAPL over 60 min
| peptideout, µM | peptidein, µM | peptidein/ peptideout | Lipososome diameter, nm | Liposome, number |
| 0.3 | 252 ± 88 | 840 | 48 ± 12 | 27 |
| 3 | 556 ± 216 | 185 | 54 ± 18 | 21 |
| 30 | 901 ± 539 | 30 | 52 ± 8 | 22 |
Fig. 5.
Peptide accumulation by TAPL is limited by trans-inhibition and is unidirectional. (A and B) Proteoliposomes (50 µg lipid, 2.5 µg coreTAPL) were incubated in 140 mM KPi, pH 7.5, containing peptide RRYCATTO488KSTEL (3 or 30 µM) with (black) or without (white) Mg-ATP (3 mM) at 37 °C for 60 min in the presence or absence of either 1 µM valinomycin (A) or nigericin (B). (C) Proteoliposomes (2.5 µg coreTAPL, 50 µg lipid), empty or prefilled with 3 mM unlabeled peptide (RRYQKSTEL), were incubated with 3 mM Mg-ATP and 3 µM RRYCATTO488KSTEL at 37 °C for 15 min. Background fluorescence measured in the absence of Mg-ATP was subtracted. The transport assay was performed in triplicate, with error bars indicating SD. (D) ATPase activity of proteoliposomes (2 µg coreTAPL, 40 µg lipid), empty or prefilled with 3 mM unlabeled peptide (RRYQKSTEL), was measured for 15 min at 37 °C in the presence of 3 mM Mg−ATP. The ATPase assay was performed in triplicate, with error bars indicating SD. (E and F) TAPL acts as strictly unidirectional ABC exporter. Proteoliposomes prefilled for 20 min at 37 °C in the presence of 3 µM RRYCATTO488KSTEL were incubated with either 60 µM unlabeled peptide RRYQKSTEL in the presence or absence of Mg-ATP (E) or were incubated in phosphate buffer (20 mM) containing 3 mM Mg−ADP (F), and peptide efflux was analyzed by DCFBA over 10 min at 37 °C. As control, addition of alamethicin (9.8 µM) induced instantaneous peptide efflux. Laser powers were kept constant: 488, 3 µW; 633, 1 µW.
Unidirectional Peptide Translocation.
Next, we determined directionality of peptide translocation by TAPL. TAPL-containing proteoliposomes were prefilled with RRYCATTO488KSTEL. Export of fluorescently labeled peptide was studied directly at the microscope in the presence of external unlabeled peptide RRYQKSTEL (60 µM). We found that the fluorescence intensity of prefilled liposomes stayed constant in the presence or absence of external ATP, whereas the addition of the pore-forming peptide antibiotic alamethicin induced the instantaneous release of luminal peptide (Fig. 5E). The transport reaction was also not reversed in the presence of ADP (3 mM) and inorganic phosphate (20 mM) (Fig. 5F). Therefore, the peptide transporter TAPL functions strictly unidirectionally and does not exchange internal for external peptide.
To our knowledge, this is the first report where solute import by an ABC transporter into individual liposomes is monitored. The high sensitivity of the method allowed us to determine true turnover numbers, peptide accumulation, and unidirectionality of transport.
Based on this data in combination with biochemical and structural information of other ABC exporters (37–39), we propose the following model of peptide transport by TAPL. Upon binding of ATP and peptide at the cytosolic site, the NBDs dimerize, which induces a conformational switch of the transmembrane domain from inward to outward facing. After dissociation of the peptide from the low-affinity binding site, ATP is hydrolyzed, which induces dissociation of the NBDs and resetting of the transmembrane domain to the inward-facing conformation. For P-glycoprotein, the low-affinity site can be stabilized by the nonhydrolyzable ATP analog adenosine 5′-(β,γ-imido)triphosphate (40). In the trans-inhibited state, peptide association to the low-affinity binding site is faster than the structural rearrangement from the outward to the inward open conformation, which inhibits the transport.
Materials and Methods
Cloning and Expression of coreTAPL.
A single cysteine variant of human coreTAPL (Q9NP78, UniProt, amino acids 143–766) and a coreTAPL fusion construct were designed for large-scale expression in P. pastoris (SI Materials and Methods).
Purification and Membrane Reconstitution of coreTAPL.
After membrane preparation using the FastPrep-24 system (MP Biomedicals) and glass beads, both coreTAPL constructs were purified via the His10-tag, using Ni-nitrilotriacetic acid agarose (Qiagen). Purified coreTAPL was reconstituted into liposomes composed of E. coli polar lipids and DOPC (Avanti Polar Lipids) in a 7:3 (wt/wt) ratio at a protein-to-lipid ratio of either 1:20 or 1:100 (wt/wt) as described (24). To detect liposomes by DCFBA, the lipid-mimicking dye DiD (λex/em = 648/670 nm; Invitrogen) was added in a molar DiD:lipid ratio of 1:4,000 during liposome formation. See SI Materials and Methods for additional information.
Peptide Transport.
For peptide transport, 50 µL of proteoliposomes (1 mg/mL lipid) were incubated at 37 °C in transport buffer (20 mM Na-Hepes, 5% glycerol, pH 7.5) containing 42.5 mM NaCl and peptides (RRYCKSTEL) labeled with ATTO488- (λex/em = 501/523 nm) or ATTO655-maleimide (λex/em = 663/684 nm) (ATTO-TEC) containing an ATP-regenerating system (41). For macroscopic analysis, proteoliposomes were collected on a filter membrane and peptide transport was quantified by a fluorescence plate reader. For DCFBA, proteoliposomes were pelleted at 270,000 × g for 20 min, washed with 3 mL ice-cold stop buffer (PBS, 10 mM EDTA, pH 7.5) (20 min, 270,000 × g), and suspended in reconstitution buffer (20 mM Na-Hepes, 140 mM NaCl, 5% glycerol, pH 7.5) to a final lipid concentration of 0.5 mg/mL (for details, see SI Materials and Methods).
Dual-Color Fluorescence Burst Analysis.
A droplet of 50 µL of proteoliposomes (0.5 mg/mL lipid) containing TAPL was applied to a coverslip, resulting in a vesicle density of ∼0.1 per confocal volume taking a vesicle diameter of 143 nm and a mean lipid surface of 0.6 nm2. Fluorescence bursts were recorded on a commercial laser-scanning confocal microscope, LSM 710 and ConfoCor3 (Carl Zeiss MicroImaging), equipped with an incubation chamber for temperature control. An argon ion laser (488 nm) and a He−Ne laser (633 nm) focused by an objective C-Apochromat 40×/1.2NA were used for excitation. Fluorescence was collected through the same objective, separated from the excitation beams by a dichroic mirror (MBS 488/543/633) and split into two channels by a dichroic beam splitter (NFT 635 VIS). For 488 excitation, light was directed through a band-pass emission filter 505–610 IR, and for 633 excitation, light was directed through a band-pass emission filter 655–710 IR and detected by two avalanche photodiodes. For both channels, pinholes of 40 µm were used (17, 18). Acquisition was performed with constant laser powers (1−5 µW) at 20 °C for 5 min with a bin width of 1 ms, and data were analyzed by ZEN2010B software (Carl Zeiss MicroImaging). The confocal volumes of 0.20 femtoliter (fL) and 0.45 fL for 488 nm and 633 nm excitation, respectively, were determined from the diffusion time of Alexa fluor 488 and 633 in water (42). The background fluorescence was, for both channels, ∼400 counts per second.
Fluorescent bursts resulting from DiD-labeled proteoliposomes or coreTAPLmVenus and imported fluorescent peptide were evaluated, using DCFBA software available online at bogeert.com/DCFBA/publish.htm. This software recognizes coincident signals in both channels above a given offset that can be set for both channels. The relative concentration Ci of peptide inside the vesicle for each burst i is calculated by
| [1] |
Iliposome and Ipeptide are the fluorescence intensities from the liposomes and peptides above an offset between t1 and t2. Because the fluorescent lipid analogs are associated with the surface of the liposome (2D) whereas the peptides are encapsulated (3D), a scaling factor of 3/2 is introduced.
Ci, a multiplicative parameter, is transformed to log scale, and the Ci distribution is fitted by a log Gaussian function (43),
| [2] |
Cav depicts the mean internal peptide concentration, the amplitude A reflects the height at Cav, and the width is a measure of the geometric SD. Transport rates in Cav per time were normalized and fitted by the Michaelis−Menten equation,
| [3] |
To investigate the turnover number kcat for peptide transport and the accumulation of peptide, the fluorescence intensity per molecule for ATTO488- and ATTO655-labeled peptide, DiD, and coreTAPLmVenus was determined by measuring the fluorescence as a function of concentration in the confocal microscope. The calibration measurements resulted in count rates of 0.4–1.4 kHz per fluorophore (Isingle molecule) (Fig. S6). The number of molecules N in liposomes was determined from individual intensity bursts by
| [4] |
I is the fluorescence of accumulated peptide, DiD, or coreTAPLmVenus between t1 and t2, and bckg is the background fluorescence (0.4 kHz). The diameter (d) and volume (V) of single proteoliposomes were calculated from the DiD fluorescence, taking into account a molar ratio of DiD:lipid of 1:4,000 and assuming a mean lipid surface area of 0.6 nm2,
where NL displays the number of lipids per liposome. Details for investigation of transport directionality are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. Le Gal for critical reading of the manuscript. This work was supported by the German Research Foundation via SFB807—Transport and Communication across Membranes (R.T. and R.A.), Fond der Chemischen Industrie (T.Z. and R.A.), the Netherlands Organisation for Scientific Research (NWO, Top-Subsidy Grant 700.56.302 to B.P.), and the NWO Bonus Incentive Scheme of the Zernike Institute (G.M. and B.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418100112/-/DCSupplemental.
References
- 1.Schmitt L, Tampé R. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2002;12(6):754–760. doi: 10.1016/s0959-440x(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 2.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10(3):218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JY, Yang JG, Zhitnitsky D, Lewinson O, Rees DC. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science. 2014;343(6175):1133–1136. doi: 10.1126/science.1246489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443(7108):180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 5.Mishra S, et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife. 2014;3:e02740. doi: 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 7.Parcej D, Tampé R. ABC proteins in antigen translocation and viral inhibition. Nat Chem Biol. 2010;6(8):572–580. doi: 10.1038/nchembio.410. [DOI] [PubMed] [Google Scholar]
- 8.Wolters JC, Abele R, Tampé R. Selective and ATP-dependent translocation of peptides by the homodimeric ATP binding cassette transporter TAP-like (ABCB9) J Biol Chem. 2005;280(25):23631–23636. doi: 10.1074/jbc.M503231200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao C, Tampé R, Abele R. TAP and TAP-like—Brothers in arms? Naunyn Schmiedebergs Arch Pharmacol. 2006;372(6):444–450. doi: 10.1007/s00210-005-0028-z. [DOI] [PubMed] [Google Scholar]
- 10.Ramos MS, et al. Characterization of a transport activity for long-chain peptides in barley mesophyll vacuoles. J Exp Bot. 2011;62(7):2403–2410. doi: 10.1093/jxb/erq397. [DOI] [PubMed] [Google Scholar]
- 11.Zhang F, et al. Characterization of ABCB9, an ATP binding cassette protein associated with lysosomes. J Biol Chem. 2000;275(30):23287–23294. doi: 10.1074/jbc.M001819200. [DOI] [PubMed] [Google Scholar]
- 12.Demirel Ö, et al. Identification of a lysosomal peptide transport system induced during dendritic cell development. J Biol Chem. 2007;282(52):37836–37843. doi: 10.1074/jbc.M708139200. [DOI] [PubMed] [Google Scholar]
- 13.Demirel Ö, Bangert I, Tampé R, Abele R. Tuning the cellular trafficking of the lysosomal peptide transporter TAPL by its N-terminal domain. Traffic. 2010;11(3):383–393. doi: 10.1111/j.1600-0854.2009.01021.x. [DOI] [PubMed] [Google Scholar]
- 14.Demirel Ö, et al. The lysosomal polypeptide transporter TAPL is stabilized by interaction with LAMP-1 and LAMP-2. J Cell Sci. 2012;125(Pt 18):4230–4240. doi: 10.1242/jcs.087346. [DOI] [PubMed] [Google Scholar]
- 15.Ernst R, et al. A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc Natl Acad Sci USA. 2008;105(13):5069–5074. doi: 10.1073/pnas.0800191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seelheim P, Wüllner A, Galla HJ. Substrate translocation and stimulated ATP hydrolysis of human ABC transporter MRP3 show positive cooperativity and are half-coupled. Biophys Chem. 2013;171:31–37. doi: 10.1016/j.bpc.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 17.van den Bogaart G, Krasnikov V, Poolman B. Dual-color fluorescence-burst analysis to probe protein efflux through the mechanosensitive channel MscL. Biophys J. 2007;92(4):1233–1240. doi: 10.1529/biophysj.106.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Bogaart G, et al. Dual-color fluorescence-burst analysis to study pore formation and protein-protein interactions. Methods. 2008;46(2):123–130. doi: 10.1016/j.ymeth.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Winkler T, Kettling U, Koltermann A, Eigen M. Confocal fluorescence coincidence analysis: An approach to ultra high-throughput screening. Proc Natl Acad Sci USA. 1999;96(4):1375–1378. doi: 10.1073/pnas.96.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ying L, Green JJ, Balasubramanian S, Klenerman D. Ultrasensitive coincidence fluorescence detection of single DNA molecules. Anal Chem. 2003;75(7):1664–1670. doi: 10.1021/ac026367z. [DOI] [PubMed] [Google Scholar]
- 21.Kusters I, et al. Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure. 2011;19(3):430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jardetzky TS, Lane WS, Robinson RA, Madden DR, Wiley DC. Identification of self peptides bound to purified HLA-B27. Nature. 1991;353(6342):326–329. doi: 10.1038/353326a0. [DOI] [PubMed] [Google Scholar]
- 24.Zhao C, Haase W, Tampé R, Abele R. Peptide specificity and lipid activation of the lysosomal transport complex ABCB9 (TAPL) J Biol Chem. 2008;283(25):17083–17091. doi: 10.1074/jbc.M801794200. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T, Caaveiro JM, Abe R, Katagiri T, Tsumoto K. Catalytic activity of MsbA reconstituted in nanodisc particles is modulated by remote interactions with the bilayer. FEBS Lett. 2011;585(22):3533–3537. doi: 10.1016/j.febslet.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Zehnpfennig B, Urbatsch IL, Galla HJ. Functional reconstitution of human ABCC3 into proteoliposomes reveals a transport mechanism with positive cooperativity. Biochemistry. 2009;48(20):4423–4430. doi: 10.1021/bi9001908. [DOI] [PubMed] [Google Scholar]
- 27.Grossmann N, et al. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat Commun. 2014;5:5419. doi: 10.1038/ncomms6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omote H, Al-Shawi MK. A novel electron paramagnetic resonance approach to determine the mechanism of drug transport by P-glycoprotein. J Biol Chem. 2002;277(47):45688–45694. doi: 10.1074/jbc.M206479200. [DOI] [PubMed] [Google Scholar]
- 29.Marzluf GA. Regulation of sulfate transport in neurospora by transinhibition and by inositol depletion. Arch Biochem Biophys. 1973;156(1):244–254. doi: 10.1016/0003-9861(73)90362-7. [DOI] [PubMed] [Google Scholar]
- 30.Hunter DR, Segel IH. Control of the general amino acid permease of Penicillium chrysogenum by transinhibition and turnover. Arch Biochem Biophys. 1973;154(1):387–399. doi: 10.1016/0003-9861(73)90071-4. [DOI] [PubMed] [Google Scholar]
- 31.Heaton JH, Gelehrter TD. Derepression of amino acid transport by amino acid starvation in rat hepatoma cells. J Biol Chem. 1977;252(9):2900–2907. [PubMed] [Google Scholar]
- 32.Kelley DS, Potter VR. Repression, derepression, transinhibition, and trans-stimulation of amino acid transport in rat hepatocytes and four rat hepatoma cell lines in culture. J Biol Chem. 1979;254(14):6691–6697. [PubMed] [Google Scholar]
- 33.Johnson E, Nguyen PT, Yeates TO, Rees DC. Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition. Protein Sci. 2012;21(1):84–96. doi: 10.1002/pro.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verheul A, Glaasker E, Poolman B, Abee T. Betaine and L-carnitine transport by Listeria monocytogenes Scott A in response to osmotic signals. J Bacteriol. 1997;179(22):6979–6985. doi: 10.1128/jb.179.22.6979-6985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaasker E, Tjan FS, Ter Steeg PF, Konings WN, Poolman B. Physiological response of Lactobacillus plantarum to salt and nonelectrolyte stress. J Bacteriol. 1998;180(17):4718–4723. doi: 10.1128/jb.180.17.4718-4723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science. 2008;321(5886):246–250. doi: 10.1126/science.1156213. [DOI] [PubMed] [Google Scholar]
- 37.Zou P, McHaourab HS. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J Mol Biol. 2009;393(3):574–585. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci USA. 2007;104(48):19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geng J, Sivaramakrishnan S, Raghavan M. Analyses of conformational states of the transporter associated with antigen processing (TAP) protein in a native cellular membrane environment. J Biol Chem. 2013;288(52):37039–37047. doi: 10.1074/jbc.M113.504696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin C, Higgins CF, Callaghan R. The vinblastine binding site adopts high- and low-affinity conformations during a transport cycle of P-glycoprotein. Biochemistry. 2001;40(51):15733–15742. doi: 10.1021/bi011211z. [DOI] [PubMed] [Google Scholar]
- 41.Neumann L, Tampé R. Kinetic analysis of peptide binding to the TAP transport complex: Evidence for structural rearrangements induced by substrate binding. J Mol Biol. 1999;294(5):1203–1213. doi: 10.1006/jmbi.1999.3329. [DOI] [PubMed] [Google Scholar]
- 42.Bacia K, Schwille P. A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods. 2003;29(1):74–85. doi: 10.1016/s1046-2023(02)00291-8. [DOI] [PubMed] [Google Scholar]
- 43.Limpert E, Stahel WA, Abbt M. Log-normal distributions across the sciences: Keys and clues. Bioscience. 2001;51(5):341–352. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.