Abstract
We sought to test whether vaccine-induced immune responses could protect rhesus macaques (RMs) against upfront heterologous challenges with an R5 simian-human immunodeficiency virus, SHIV-2873Nip. This SHIV strain exhibits many properties of transmitted HIV-1, such as tier 2 phenotype (relatively difficult to neutralize), exclusive CCR5 tropism, and gradual disease progression in infected RMs. Since no human AIDS vaccine recipient is likely to encounter an HIV-1 strain that exactly matches the immunogens, we immunized the RMs with recombinant Env proteins heterologous to the challenge virus. For induction of immune responses against Gag, Tat, and Nef, we explored a strategy of immunization with overlapping synthetic peptides (OSP). The immune responses against Gag and Tat were finally boosted with recombinant proteins. The vaccinees and a group of ten control animals were given five low-dose intrarectal (i.r.) challenges with SHIV-2873Nip. All controls and seven out of eight vaccinees became systemically infected; there was no significant difference in viremia levels of vaccinees vs. controls. Prevention of viremia was observed in one vaccinee which showed strong boosting of virus-specific cellular immunity during virus exposures. The protected animal showed no challenge virus-specific neutralizing antibodies in the TZM-bl or A3R5 cell-based assays and had low-level ADCC activity after the virus exposures. Microarray data strongly supported a role for cellular immunity in the protected animal. Our study represents a case of protection against heterologous tier 2 SHIV-C by vaccine-induced, virus-specific cellular immune responses.
Keywords: HIV vaccine, SHIV-C, heterologous tier 2 virus, rhesus monkey, cellular immunity, ADCC
1. Introduction
According to UNAIDS global estimates, each year of the last decade added >2.5 million new HIV-1 infections. Considering the recent success of non-vaccine prevention tools, such as microbicides [1], pre-exposure chemoprophylaxis [2], male circumcision (reviewed in [3]), and treatment of infected individuals [4], it may be expected that their increased use will slow the spread of HIV in coming years (reviewed in [5]). However, maintaining control over the HIV epidemic through drug prevention is unconvincing due to suboptimal compliance. Therefore, a prophylactic HIV-1 vaccine is still needed. The pessimistic view regarding prophylactic HIV-1 vaccines has been replaced by cautious optimism due to the RV144 trial outcome [6-8] and recent primate model data indicating prevention of virus acquisition by active immunization against upfront heterologous virus challenges [9-13]. The RV144 vaccine trial showed a moderate efficacy (31.2%) and engendered “no acquisition” as a new ambition for HIV vaccine development [14]. However, reaching this goal is still one of the greatest challenges and requires experimental evaluation of novel vaccine concepts and candidates.
Considering the complexity and challenges involved in testing vaccine candidates in human efficacy trials, biologically relevant animal models that mimic as many aspects of HIV-1 transmission as possible are needed. Due to host restrictions for HIV-1 replication in macaques, the simian immunodeficiency virus (SIV) / rhesus macaque (RM) model has been widely used. However, SIV differs significantly from HIV-1 and therefore cannot be used as a challenge virus to evaluate the efficacy of anti-HIV-1 vaccine candidates. To overcome this difficulty, recombinant simian-human immunodeficiency viruses (SHIVs) that carry HIV-1 envelope have been developed. Such SHIVs can be used as challenge viruses to evaluate the efficacy of HIV-1 envelope-based immunogens in RMs. Of note, envelope is the most important target for antibody-mediated immune defenses, and antibodies are the best correlates of protection in most licensed vaccines (reviewed in [15]). Therefore, SHIVs can significantly contribute to the preclinical evaluation of HIV-1 vaccine candidates. However, earlier SHIV strains were found to be unsuitable due to X4 tropism and overly too rapid disease progression. New SHIV strains that mimic most aspects of HIV-1 transmission and disease have been developed [16-22].
We constructed a series of exclusively CCR5-tropic clade C SHIVs (SHIV-Cs) [18-20], which offer the advantage of reflecting the world's most prevalent HIV-1 subtype. Among our newly developed SHIV strains, SHIV-2873Nip [19] carries a primary pediatric HIV-1 clade C (HIV-C) env isolated from a recently infected Zambian infant who showed rapid disease progression and died within one year of birth. SHIV-2873Nip is a tier 2 virus (less sensitive to neutralizing antibodies), similar to the majority of acutely transmitted HIV-1 strains [23] and causes AIDS in RMs with clinical parameters and disease progression rates similar to those in humans (unpublished data). Hence, we sought to induce immune responses in RMs that would protect against our biologically relevant challenge virus.
In our earlier vaccine efficacy study, simultaneous induction of cellular immunity and challenge virus-specific neutralizing antibodies (after immunization with SIV Gag-Pol particles, HIV-1 Tat and multimeric HIV-1 gp160) were significantly associated with protection against multiple low-dose challenges with the tier 1 SHIV-1157ipEL-p [13, 24]. However, these immune responses were induced only in a fraction of vaccinees. Variable levels of cellular responses may be due to differential protein processing by outbred RMs. To overcome this issue, we immunized a group of RMs with overlapping synthetic peptides (OSP) that were 15 amino acids (aa) in length with an overlap of 11 aa (for Gag, Tat, and Nef proteins). The 15-mer peptides stimulate antigen-specific CD4+ and CD8+ cells in commonly used in vitro assays (ELISPOT assay, intracellular cytokine staining) and represent all potential CD4+ and CD8+ T cell epitopes. These peptides may bind directly to MHC class II molecules of antigen presenting cells (APC) and need only partial processing for binding to MHC class I molecules. In our earlier studies, this approach generated peptide-specific cellular immune responses in all vaccinated outbred mice and also in different strains of inbred mice [25, 26]. The number of peptides made available to MHC molecules after antigen processing is limited [27, 28], but MHC molecules are potentially very promiscuous and can bind to more than million different peptides with significant affinity [29]. Our approach was to make a large number of 15-mer peptides available to APC through direct administration.
For the induction of humoral immune responses against HIV-1 Env, we used our earlier successful strategy of protein-only immunization [13, 30, 31] but used two different (heterologous) Env proteins in a prime-boost strategy. Sequential immunization with different HIV-1 Env versions can lead to more antibody maturation and broadening of neutralizing antibody (nAb) responses [32]. We present immunogenicity and efficacy data of our novel vaccination strategy against a biologically relevant heterologous challenge virus: SHIV-2873Nip [19].
2. Materials and methods
2.1 Immunogens and vaccination
The OSP (15-mers with an 11 aa overlap between sequential peptides) for SIVmne Gag, HIV-1 Tat Oyi [33] and SIVsmE543-3 Nef were commercially synthesized (RS synthesis, Louisville, KY). The peptides represented entire proteins (124, 23 and 63 peptides for Gag, Tat and Nef, respectively). Positively or negatively charged peptides were dissolved in phosphate buffer saline (PBS), whereas neutral peptides were dissolved in DMSO. For Gag peptides, four pools were prepared (pools #1 to #4 consisting of peptides 1-31, 32-62, 63-93, and 94-124, respectively); for Nef peptides, two pools were prepared (pools #1, #2 consisting of peptides 1-32 and 33-63, respectively); for Tat peptides, a single pool was prepared. The concentration of each peptide in the pool was 1.5 mg/ml. Each peptide pool (200 μl, i.e., 300 μg of each peptide) was administered subcutaneously along with an equal volume of Incomplete Freund's adjuvant (IFA). Three doses were given at intervals of five or six weeks (Fig. 1).
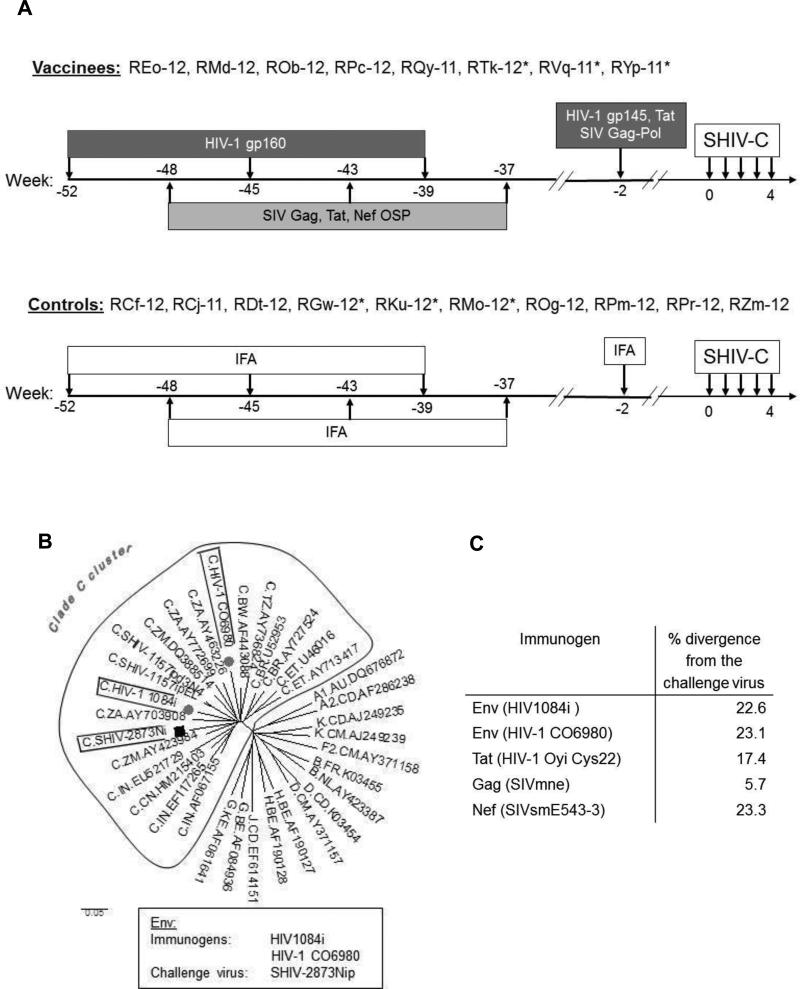
Fig. 1.
Experimental time line and HIV-1 Env phylogenetic analysis. (A) RMs were immunized with 100 μg of recombinant HIV-1 1084i gp160 in IFA at weeks −52, −45 and −37 by the intramuscular (i.m.) route. The animals were also immunized with overlapping synthetic peptides (OSP) representing the entire Gag (SIVmne), Tat (Oyi), and Nef (SIVsmE543-3) at weeks −48, −43 and −37. For OSP immunization, 300 μg of each peptide with IFA was administered by the subcutaneous (s.c.) route. All animals received a final boost with HIV-1 CO6980 gp145, SIVmne Gag-Pol particles and HIV-1 Tat Oyi two weeks before the first SHIV-C exposure; the recombinant protein immunogens were administered with IFA. Control RMs received IFA alone at corresponding time points. All animals were challenged at weeks 0, 1, 2, 3, and 4 with low-dose SHIV-2873Nip by the intrarectal (i.r.) route. *, Mamu A*001+ RMs. (B) Phylogenetic tree of HIV-1 clade C Env sequences of immunogens and challenge virus. Sequences of HIV-1 reference strains were obtained from the Los Alamos HIV-1 sequence database. The evolutionary tree was inferred using the Neighbor-Joining method by MEGA4 software. (C) The percent divergence between amino acid sequences of immunogens and challenge virus.
The multimeric HIV-C gp160 was derived from a recombinant vaccinia virus expressing env of HIV1084i, a recently transmitted HIV-C isolate from a Zambian infant [34]. The same technology was also used for the preparation of SIVmne Gag-Pol particles [35]. The CHO cell-expressed HIV-1 gp145 was produced from the HIV-C06980v0c22 env, cloned from the plasma of an acutely viremic but seronegative individual from a Tanzanian heterosexual cohort. The Tat Oyi protein was synthesized based on the tat gene of HIV-1 Oyi [33, 36]. HIV-1 Oyi is an isolate from a transiently viremic African (Gabon) patient, who was later described as highly exposed, persistently seronegative (HEPS) [37]. Immunization with Tat Oyi induced immune responses in rabbits [38] and also provided partial protection to RMs against SHIV challenge [39]. For each protein immunization (Fig. 1), 100 μg of protein in IFA was administered by the intramuscular (i.m.) route.
2.2 Animals
Indian-origin RMs (Macaca mulatta) were housed at the Yerkes National Primate Research Center (YNPRC), Atlanta, Georgia, USA. YNPRC is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). Approval for all procedures was received from the Institutional Animal and Care and Use Committees of Dana-Farber Cancer Institute (DFCI) and Emory University.
A group of 8 RMs was immunized with OSP and recombinant proteins as shown in Fig. 1. The control group consisted of 10 RMs that received IFA only. All RMs were negative for Mamu- B*008 and B*017 alleles. Six Mamu-A*001+ RMs were equally distributed among vaccinees and controls (Fig. 1).
2.3 Challenge virus
The SHIV-2873Nip stock was grown in concanavalin A (Con A)-stimulated RM peripheral blood mononuclear cells (PBMC) and the 50% tissue culture infectious dose (TCID50) was determined using TZM-bl cells. All RMs were challenged intrarectally (i.r.) with 5,000 TCID50 of the virus (low-dose challenge). The challenge was given once a week up to a maximum of five inoculations. A titration by the i.r. route using low-dose inocula had been performed to verify the virus dose before the challenges of the current experimental groups started. Animals that became systemically infected (>10,000 viral RNA copies/ml) during these multiple challenges were excluded from the subsequent virus inoculations. SHIV-2873Nip is relatively neutralization resistant (tier 2); the 50% inhibitory concentration (IC50) for each of the broadly neutralizing antibodies VRC01, IgG1b12, 2G12, 2F5 and 4E10 was >25 μg/ml [19, 40].
2.4 Measurement of plasma viral RNA (vRNA)
Plasma vRNA was isolated by QiaAmp Viral RNA Mini-Kits (Qiagen, Germantown, MD, USA); vRNA levels were measured by quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) for SIV gag sequences [41]. Additionally, primers/probes according to Lifson were used [42]. Assay sensitivity was 50 vRNA copies/ml [41]. Time to first detection of viremia was analyzed by Kaplan-Meier analysis.
2.5 Virus-specific cellular immune responses
Interferon-γ (IFN-γ) ELISPOT assays were performed as described earlier [12] using SIVmne Gag, HIV-1 Tat Oyi or SIV Nef peptides. T-cell proliferative responses were measured by CFSE dilution assay [12]. The responses were measured against pools of SIVmne Gag or HIV-1 Tat Oyi peptides.
2.6 Virus-specific humoral immune responses
Binding antibody titers against viral proteins were determined by ELISA as described earlier [31]. The viral proteins used were SIVmac251 p27 (Immunodiagnostics, Inc., Woburn, MA), HIV-1 Tat and HIV-C96ZM651 gp120 (both obtained from NIH AIDS Research and Reference Reagent Program, ARRRP).
Sera collected on the day of the first virus challenge were tested for nAb titers against three tier 1 (clade C: SHIV-1157ipEL-p, GS015.EC12PV and clade B: SF162PV) viruses and the tier 2 challenge virus (SHIV-2873Nip) using TZM-bl cells [43]. We used challenge virus-related replication-competent molecular clones of HIV-1 expressing Renilla luciferase (NL-LucR.2873Ni and NL-LucR.2878Nipd14) to measure virus-neutralizing activity in A3R5 cell-based assays. These molecular clones were generated by swapping the env ectodomain of NL-LucR.T2A with the corresponding regions of SHIV-2873Nip-related molecular clones as described [44]. A3R5 cells, a human lymphoblastoid cell line engineered to express CCR5, are substantially more sensitive than TZM-bl cells to detect virus neutralization [45]. The nucleotide sequences of HIVC env used to construct NL-LucR.2873Ni and NL-LucR.2878Nipd14 have been deposited in GenBank under accession numbers KJ941152 and KJ941153, respectively. Sera were also tested against SHIV-2873Nip in human PBMC-based neutralization assays as described earlier [12]. Virus neutralization was also tested using Con A-stimulated RM PBMC.
Serum antibody-dependent cellular cytotoxicity (ADCC) was tested using CEM-NKR cells coated with HIV-C gp120 [46]. For antibody-dependent cell-mediated viral inhibition (ADCVI), SHIV-2873Nip-infected CEM-NKR-CCR5 cells were incubated with 1:100 diluted serum + human PBMC effectors (effector: target = 10:1) and virus replication was monitored [47].
2.7 Dissection of antibody responses linked to the protection or non-protection
Plasma samples from the vaccine-protected RMs was screened for antibody responses specific for protection using a peptide phage display-based approach as described earlier [24]. In brief, total IgG was purified from plasma samples of the protected and a non-protected animal, having comparable binding antibody titers against HIV-1 gp120. Equal amounts of purified polyclonal IgGs were coated on paramagnetic protein G beads (Dynabeads Protein G; Life Technologies). The biopanning was performed using different phage display peptide libraries (7-mer, cyclic 7-mer, 12-mer, and 16-mer). For the protection-linked biopanning, IgG from the protected RM was used for positive selection and IgG from a non-protected RM was used for negative selection. For the inverted biopanning, positive selection was done using IgG from non-protected RM and negative selection using IgG from the protected RM.
2.8 Gene expression microarray analysis
Blood was collected before vaccination, on the day of first virus exposure and six weeks after last virus challenge. Lymph node and rectal pinch biopsies were performed before vaccination and six weeks after last virus challenge. Blood was collected in Tempus tubes, processed immediately according to the manufacturer's instructions, and stored at −80°C. The biopsy specimens were cut into small pieces and immediately placed into RNAlater solution (Qiagen, Valencia, CA) and also stored at −80°C. Total RNA from blood, lymph node and rectal biopsies was extracted using RNAeasy extraction kits (Qiagen, Valencia, CA). cDNA labeling, hybridization, staining and scanning were performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA) for rhesus gene expression arrays. Array quality was assessed using the R/Bioconductor package [48]. Affymetrix CEL files were processed and normalized using the robust multiarray average (RMA) algorithm [49]. Results were adjusted for multiple testing using the Benjamini and Hochberg (BH) method [50], and significance was determined using a false-discovery-rate cutoff of <5% and a log fold-change cutoff of >2 as compared to baseline. Gene interaction network analysis and visualization were performed on significant probe sets using the Ingenuity pathway analysis (IPA) software package. Microarray data were deposited into the gene expression omnibus database (Accession number: GSE60368).
2.9 Statistical analysis
Log-rank comparisons were used for time-to-infection analyses between vaccinees vs. controls. The Wilcoxon rank sum test was used to compare continuous variables (number of virus challenges, peak viremia and area-under-the-curve) between groups. The reported P-values are based on two-sided testing.
3. Results
3.1. Vaccine safety and immunogenicity
OSP immunization consisted of subcutaneous administration of 300 μg of each peptide (210 peptides representing SIV Gag, HIV-1 Tat and SIV Nef). Recombinant protein immunization consisted of i.m. administration of 100 μg of each protein (HIV-1 gp160, SIV Gag-Pol particles, HIV-1 Tat, and HIV-1 gp145). Peptides as well as proteins were administered in IFA. None of the vaccinees developed untoward side effects, and no local reactogenicity was noted.
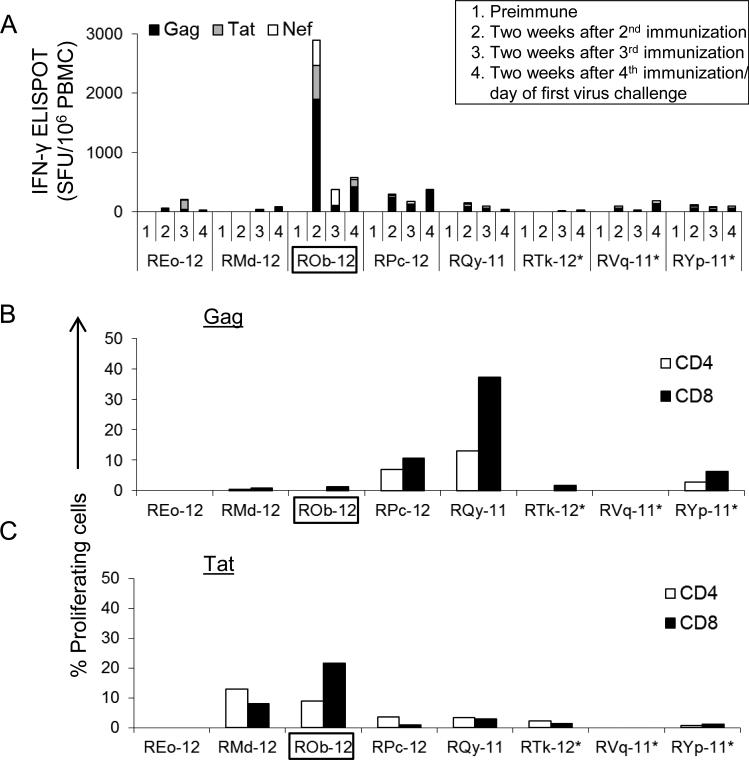
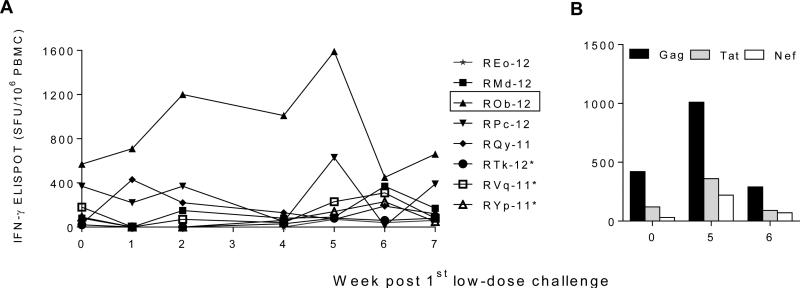
Cellular immune responses were measured at two weeks after the 2nd, 3rd and 4th administrations of Gag, Tat and Nef immunogens by IFN-γ ELISPOT assay (Fig. 2A). On the day of first virus exposure (two weeks after the last immunization, referred to as day or week 0), the total number of virus-specific IFN-γ secreting cells among vaccinees ranged between 20 to 570 Spot Forming Units (SFU)/106 PBMC. We also measured antigen-specific proliferation of CD4+ and CD8+ T cells at day 0. Two RM showed >10% Gag or Tat-specific proliferating T cells (Fig. 2B and 2C).
Fig. 2.
Cellular immune responses after vaccination. (A) SIVmne Gag, HIV-1 Tat Oyi and SIVsmE543-3 Nef-specific, IFN-γ-secreting cells (spot-forming units (SFU) / 106 PBMC) as determined by ELISPOT assay at different time points. (B & C) Ex vivo proliferation of CD4+ and CD8+ T cells after stimulation with SIVmne Gag, or HIV-1 Tat Oyi peptides. The assay was performed after the final immunization, i.e., on the day of the first SHIV-C challenge. The RM ROb-12 (boxed) remained aviremic after all heterologous SHIV-C challenges. *, Mamu A*001+ RMs.
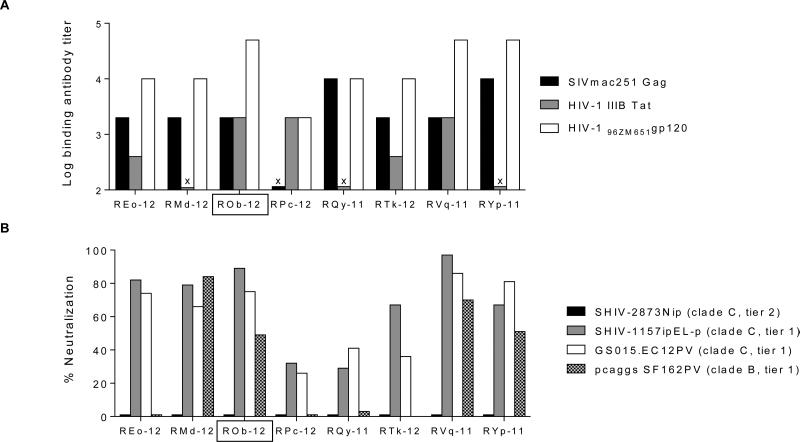
Sera collected on day 0 were tested by ELISA to determine binding antibody titers against SIVmac251 Gag, HIV-1 IIIB Tat and heterologous clade C HIV96ZM651 gp120 (Fig. 3A). The median binding antibody titers (highest reciprocal serum dilution showing binding by ELISA) against Gag and Tat were 2000 and 400, respectively. As expected, binding antibody titers induced by peptide immunizations against Gag and Tat were substantially lower than those in our earlier study [30] where Gag-Pol particles and Tat protein had been used for immunization. The Env binding titers ranged from 2,000 to 50,000 (Fig. 3A).
Fig. 3.
Humoral immune responses after vaccination (on the day of first virus exposure). (A) Reciprocal serum antibody ELISA titers against SIVmac251 Gag, HIV-1 IIIB Tat and heterologous clade C HIV96ZM651 gp120. (B) NAb activity against tier 1 (two clade C and one clade B) strains and against the tier 2 challenge virus (SHIV-2873Nip), as measured by TZM-bl assay. The % virus neutralization by sera collected on the day of first virus challenge (at 1:20 dilution with regard to autologous pre-immune serum) is presented. X, binding titer < 200. The RM ROb-12 (boxed) remained aviremic after all heterologous SHIV-C challenges. None of the sera showed neutralizing activity against SHIV-2873Nip.
Sera collected on day 0 were tested for neutralizing activity against SHIV-2873Nip (tier 2 challenge virus), SHIV-1157ipEL-p (tier 1, SHIV-C), GS015.EC12PV (tier 1, HIV-C) and SF162PV (tier 1, HIV-1 subtype B) by TZM-bl assay (Fig. 3B). None of the vaccines sera showed neutralization of challenge virus (SHIV-2873Nip), but four out of eight vaccinees showed ≥50% neutralization of all three tier 1 viruses (Fig. 3B). The A3R5 cell-based assay is more sensitive to detect virus neutralization [45]. In this assay, nAb responses against challenge virus-related molecular clones (NL-LucR.2873Ni and NL-LucR.2878Nipd14) were measured. Similar to the TZM-bl assay results, virus inhibition was not shown by sera of any vaccinee in the A3R5 cell-based assay (data not shown). ADCC responses were also undetectable against gp120-coated target cells on day 0 (tested using serum as well as purified IgG). In the ADCVI assay, four out of eight vaccinees (REo-12, RMd-12, ROb-12 and RVq-11) showed >50% inhibition of SHIV-2873Nip replication at 1:100 serum dilution.
3.2 Vaccine efficacy against multiple, low-dose challenges with heterologous tier 2 SHIV-C
To recapitulate HIV-1 transmission as closely as possible, the animals were challenged with heterologous SHIV-2873Nip, a virus with exclusive CCR5 tropism, tier 2 neutralization profile, and gradual pathogenicity [19]. Repeated (5x, one dose/week), low-dose (5000 TCID50) challenges were given by the mucosal (i.r.) route.
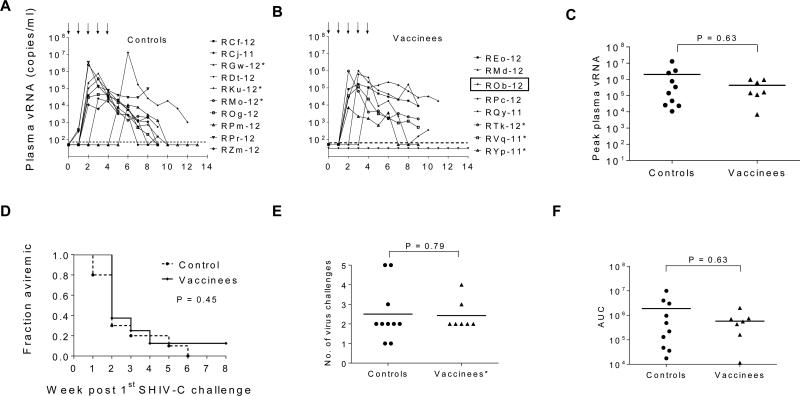
The animals were monitored weekly for plasma vRNA levels (Fig. 4A-B). All controls became viremic and showed peak viremia ranging from 1.1 x 104 to 1.3 x 107 (median: 2 x 105) vRNA copies/ml (Fig. 4C). However, one vaccinee (ROb-12) showed complete protection (no detectable plasma viremia). Among vaccinees with breakthrough infection, peak plasma vRNA ranged from 6.9 x 103 to 9 x 105 (median: 2 x 105) copies/ml (Fig. 4C). There was no difference in the time for onset of viremia (Fig. 4D) or the number of virus challenges before detection of viremia (Fig. 4E) among the vaccinees compared to the controls. All animals were monitored for seven weeks after first detection of viremia to determine AUC; no significant difference was noted among controls and vaccinees (Fig. 4F).
Fig. 4.
Post challenge viremia data. (A–B) Five low-dose i.r. challenges of SHIV-2873Nip were given at weeks 0, 1, 2, 3, & 4 (as shown by black arrows) and plasma vRNA levels were measured. (C) Comparison of peak plasma vRNA levels, (D) Kaplan–Meier plots depicting the fraction of RMs remaining aviremic, (E) comparison of the number of virus challenges before detection of viremia, and (F) comparison of area-under-the-curve (AUC) for viremic RMs. RM ROb-12 (boxed) remained aviremic after all heterologous SHIV-C challenges. Horizontal dashed line in A-B, the lower limit of plasma vRNA detection (50 vRNA copies/ml); horizontal solid lines in C, E, & F, mean peak plasma vRNA, the mean number of virus challenges before detection of viremia, & mean AUC, respectively. *, Mamu A*001+ RMs.
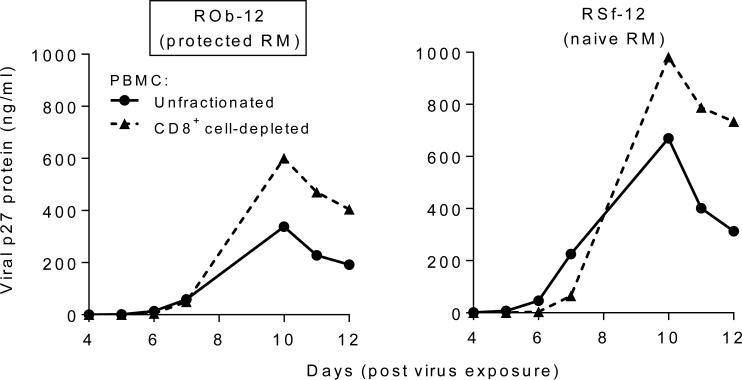
To confirm complete protection of ROb-12, RT-PCR analysis was performed to detect vRNA in a peripheral lymph node as well as in rectal biopsies taken 6 weeks after the last virus challenge; no vRNA copies were detected. Importantly, PBMC of ROb-12 supported SHIV-2873Nip replication in vitro (Fig. 5), which indicates that the cells of ROb-12 were not intrinsically resistant to SHIV-C replication.
Fig. 5.
Ex vivo replication of SHIV-2873Nip in cultured PBMC of ROb-12, the protected RM (boxed). PBMC were stimulated with Con A in the presence of IL-2. Unfractionated PBMC or CD8+ cell-depleted PBMC were exposed to SHIV-2873Nip, and replication was monitored by p27 ELISA of culture supernatants. As control, virus replication in PBMC (unfractionated or CD8+ cell-depleted) of one naïve RM (RSf-12) was also measured.
3.3. Immune responses shown by the completely protected vaccinee
The one protected, persistently aviremic vaccinee, ROb-12, had developed the highest level of virus-specific cellular responses during immunization (two weeks after second OSP dose; Fig. 2A & Fig. 6) and also showed the highest number of virus-specific ELISPOT responses (570 SFU/106 PBMC) on the day of first virus exposure (Fig. 2A & Fig. 6). ROb-12 also exhibited the highest level of HIV-1 Tat-specific proliferating CD8+ T cells on the day of first virus challenge (Fig. 2B). This RM maintained virus-specific cellular responses throughout all virus challenges and indeed the response increased from 570 (week 0) to 1590 SFU/106 PBMC during virus exposures (Fig. 6) suggesting antigenic stimulation delivered by the challenges. Among the remaining seven unprotected vaccinees, such anamnestic cellular responses were not observed. It is important to note that the protected RM (ROb-12) was negative for Mamu-A*001, B*008, and B*17 alleles, the MHC genotypes associated with improved control of SIV replication in RMs with these alleles compared to those without.
Fig. 6.
Virus-specific cellular immune responses during and after virus challenges. (A) The sum of Gag, Tat and Nef-specific, IFN-γ-producing cells measured by ELISPOT assay is shown for the vaccinees. The RM ROb-12 (boxed) remained aviremic after all heterologous SHIV-C challenges. (B) Anamnestic cellular responses against individual vaccine component shown by ROb-12.
On day 0, the protected RM (ROb-12), did not show nAb responses against the challenge virus in the TZM-bl as well as A3R5 cell-based assays. The animal did not show lysis of the HIV-C gp120-coated cells in the ADCC assay. Although ROb-12 showed >50% challenge virus inhibition in the ADCVI assay, similar virus inhibition was also shown by three unprotected animals (data not shown). The serum samples of this protected animal collected during virus exposures and two weeks after last virus exposure were also analyzed for nAb, ADCVI, ADCC and gp120 binding Ab titers. Unlike cellular responses, strong boosting of any humoral responses was not observed during virus exposures (data not shown). The findings are summarized in the Table 1. Mucosal samples (rectal washes or biopsies) could not be collected from any vaccinees just prior to virus exposure as this would have breached mucosal integrity.
Table 1.
Virus-specific humoral responses.
| Week 0 (for all vaccinated RM) | Week 6 (for protected RM, ROb-12) | |
|---|---|---|
| nAb response (TZM-bl assay) | Not detected | Not detected |
| nAb response (A3R5 assay) | Not detected | Not detected |
| Binding Ab titer against HIV-C Env | Did not correlate with peak viremia | Boost (~2 fold) observed |
| ADCC | Not detected | Low level |
| ADCVI titer | Did not correlate with peak viremia | No boost observed |
The differential biopanning strategy designed to identify antibody responses that are present in the protected animal (ROb-12), but absent in the non-protected animal (RVq-11) [24] did not reveal any protection-linked antibody responses in ROb-12. Similarly, the biopanning strategy designed to identify antibody responses that might be present in the non-protected animal (RVq-11), but absent in the protected animal (ROb-12) did not reveal any specific non-protection-linked antibody responses in the RVq-11.
3.4. Gene expression microarray analysis for the completely protected vaccinee
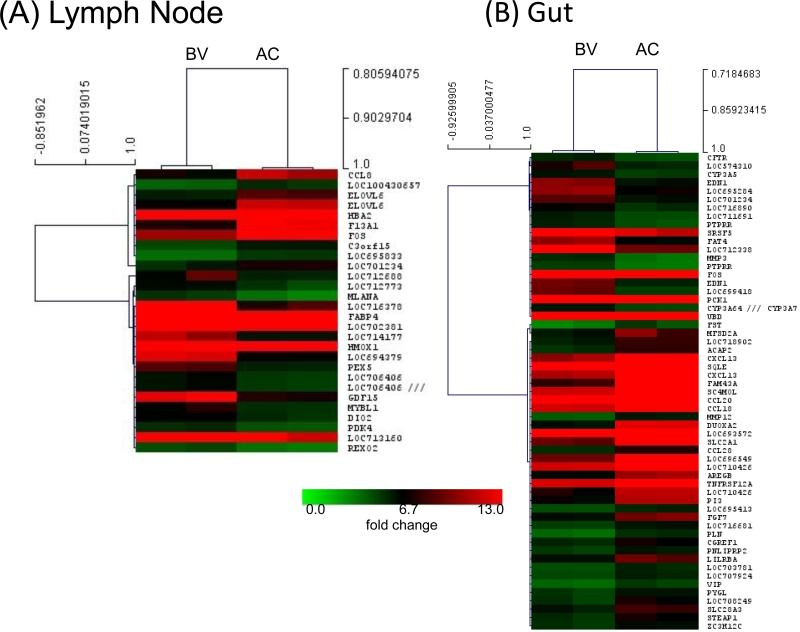
To understand differentially expressed genes in ROb-12, we performed gene expression microarray analysis for the total blood, lymph node (LN) and rectal pinch biopsies collected before and after 5X low dose challenges (Fig. 7). Supervised analysis identified 353, 28 and 57 individual genes in blood, LN and gut biopsies, respectively, that were differentially regulated. We noted a clear upregulation of several genes involved in cell-mediated immunity, cell signaling, immune cell trafficking and cell cycle pathways. Most notably, CCL8 (C-C motif ligand 8; also known as monocyte chemoattractant protein 2 (MCP-2)) was upregulated in the LN. MCP-2 binds to CCR5 with high affinity and blocks HIV-1 replication [51].
Fig. 7.
Microarray analysis for ROb-12, a RM protected against multiple low-dose challenges with a heterologous R5 SHIV-C. Hierarchical clustering and heat map analysis of differentially expressed genes in the lymph node (A) and gut (B). There were 28 and 57 genes in the lymph node (LN) and gut biopsies, respectively, that were differentially expressed after vaccination and virus challenges (AC) compared to the before vaccination (BV). The analysis uses a significance threshold of false discovery rate <5% and log2 fold-change of >2.
4. Discussion
Here, we report a case of complete protection of a vaccinated RM against repeated mucosal challenges with a biologically relevant immunodeficiency virus. The protected animal showed the highest level of vaccine-induced virus-specific cellular immune responses among the group of vaccinees. Although no vRNA was detected in blood, lymph node and rectal biopsies, the animal showed strong anamnestic immune responses after repeated virus exposures. The role of anamnestic cellular responses in controlling virus replication has been documented [13, 52, 53]. We believe that the anamnestic cellular responses were generated due to a cryptic infection and these responses rapidly cleared infected cells before establishment of systemic infection could occur. Although all vaccinees, including the protected RM ROb-12, carried TRIM5α genotypes that were most resistant to SIVsmE543-3 (TRIM TFP/TRIM TFP or TRIM TFP/ TRIM CYPA or TRIM CYPA/ TRIM CYPA), the PBMC of the protected animal supported robust virus replication ex vivo. Thus, this animal was not intrinsically resistant to replication of the challenge virus but presents a case of prevention of viremia by vaccine-induced immune responses.
We have previously shown prevention of SHIV-C acquisition in vaccinated animals [12, 13, 31]. Unlike in the present study, the challenge virus used in all our earlier studies was relatively neutralization sensitive (tier 1). Prevention of virus acquisition was seen in multiple vaccinees that had developed the highest levels of challenge virus-specific cellular as well as nAb responses (Table 2). Due to the bimodal nature of the protective immune responses, the relative contribution of nAb and cellular immune responses could not be assessed. In the present study, challenge virus-specific nAbs did not develop in any vaccinated animal, which was not surprising considering the tier 2 nature of our challenge virus, SHIV-2873Nip. Despite the absence of nAb as well as ADCC responses on day 0, one vaccinee was protected. The ADCVI mechanism could not be linked to the protection. Moreover, no difference was detected in the epitope specificity of the humoral responses of the protected versus unprotected animal by our extensive biopanning efforts using day 0 polyclonal IgG samples. This suggests that cellular responses played a major role in the protection of this animal. Furthermore, microarray data of ROb-12 showed upregulation of several genes involved in cell-mediated immunity, cell signaling, immune cell trafficking, and cell cycle pathways. Most notably, CCL8 (also known as MCP-2) was upregulated in the LN. It is known that MCP-2 is a potent inhibitor of HIV-1, since it binds to CCR5 with high affinity [51]. This mechanism may also have played a role in preventing the establishment of persistent systemic infection in our protected animal.
Table 2.
The immune responses of vaccine-protected (aviremic) vaccinees from present and our earlier published studies.
| Vaccine-protected RM (Undetectable viremia) | Challenge virus | Virus-specific immune responses (week 0) | Ref | |
|---|---|---|---|---|
| Cellular IFN-γ ELISPOT (Total SFU/106 PBMC) | Humoral Challenge virus-specific nAb titer (IC50, TZM-bl assay) | |||
| RAt-9 | SHIV-1157ip (tier 1) | 3830 | 130 | 32 |
| RRi-11 | SHIV-1157ipEL-p (tier 1) | 3560 | 3157 | 13 |
| RTr-11 | SHIV-1157ipEL-p (tier 1) | 920 | 4382 | 13 |
| RQe-10 | SHIV-1157ipEL-p (tier 1) | 520 | 48 | 12 |
| ROb-12 | SHIV-2873Nip (tier 2) | 570 | <20 | Present study |
In one of our earlier vaccine studies [12], strong virus-specific cellular responses were detected in some animals immediately after vaccination but these animals did not show anamnestic cellular responses after repeated virus exposures. The animals became viremic and also did not control virus replication. In contrast, the protected animal in this study showed strong anamnestic cellular responses. This highlights the importance of anamnestic cellular responses in preventing systemic spread of cryptic infection. Other vaccinees from this study did not show anamnestic cellular responses and also lacked nAb responses. This is in agreement with the finding that no significant differences were seen in the peak viremia and AUC of viral load of vaccinees and controls. The protected RM showed a low level of ADCC activity after the virus exposures but was repeatedly negative before the first virus challenge. We previously reported an alteration in the antibody repertoire even in the absence of viremia in vaccine-protected RMs [54]. It is possible that ADCC mediating antibodies were induced after virus exposures and contributed to the prevention of viremia.
In our earlier mouse studies, OSP were able to induce immune responses in different strains of inbred mice as well as in the outbred mice [25, 26]; the strategy was found to be only modestly successful in RMs. This is not a stand-alone case where a mouse-tested vaccine concept could not be translated with equal success into primates/humans. The discrepancy in immunogenicity and efficacy in mice versus primates has been observed for DNA vaccination (reviewed in [55]) and in our Listeria-vector prime, adenovirus boost strategy [12]. Administration of autologous PBMC pulsed with 15-mer viral peptides to SIV-infected macaques [56-58] and administration of 15-mer HIV-1 p24-like peptides to infected patients [59] boosted virus-specific cellular responses. However, 15-mer peptides administered to the naïve animals in our study, did not effectively induce immune responses.
To induce anti-Env responses, multimeric HIV-C gp160 protein derived from a recently transmitted HIV1084i was administered three times at intervals of six to seven weeks. To broaden anti-Env nAb specificities, we decided to boost the animals with a different HIV-C Env. The animals were given HIV-1 CO6980 gp145 that was 20.9% and 23.1% divergent compared to Env of HIV1084i and the challenge virus, respectively. The strategy of sequential immunizations with heterologous Env has been reported to mimic in vivo development of broad nAb responses [32, 60]. However, this strategy was not successful in the present study to induce nAbs against our tier 2 challenge virus.
Nonetheless, this study reports a case of prevention of viremia by vaccine-induced immune responses. Considering the challenges associated with the induction of nAbs and the recent recognition of the contribution of vaccine-induced cellular immunity to protection [61-64], it may be important to harness cellular immune defenses as well as ADCC mechanisms for HIV vaccine development.
Highlights.
Vaccine-induced protection against heterologous tier 2 SHIV-C challenge
Protection was associated with anamnestic cellular and ADCC responses
Protection was achieved in the absence of measurable challenge virus-specific neutralizing antibodies
Acknowledgements
We thank Stephanie Ehnert, Chris Souder and Kalpana Patel from the YNPRC for assistance with the primate studies. We acknowledge ABL, Inc. for the production of the clade C gp145 protein. This work was supported by NIH grants R01 AI100703 to RMR and P01 AI048240 to RMR, SLH, RAR and JLE. This project was also funded in part by the Intramural Research Program of the NIH, National Cancer Institute and by the Office of Research Infrastructure Programs/OD P51 OD011132 to the YNPRC and OD P51 OD011133 (previously NCRR grant P51 RR13986) to the Southwest National Primate Research Center. The work conducted by WEJ was supported by NIH grant AI095092.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329:1168–74. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant RM, Lama JR, Anderson PL, McMahan V, Liu AY, Vargas L, et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010;363:2587–99. doi: 10.1056/NEJMoa1011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wamai RG, Morris BJ, Bailis SA, Sokal D, Klausner JD, Appleton R, et al. Male circumcision for HIV prevention: current evidence and implementation in sub-Saharan Africa. J Int AIDS Soc. 2011;14:49. doi: 10.1186/1758-2652-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padian NS, McCoy SI, Karim SS, Hasen N, Kim J, Bartos M, et al. HIV prevention transformed: the new prevention research agenda. Lancet. 2011;378:269–78. doi: 10.1016/S0140-6736(11)60877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 7.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–86. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMichael AJ, Haynes BF. Lessons learned from HIV-1 vaccine trials: new priorities and directions. Nat Immunol. 2012;13:423–7. doi: 10.1038/ni.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, et al. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci Transl Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomsel M, Tudor D, Drillet AS, Alfsen A, Ganor Y, Roger MG, et al. Immunization with HIV-1 gp41 subunit virosomes induces mucosal antibodies protecting nonhuman primates against vaginal SHIV challenges. Immunity. 2011;34:269–80. doi: 10.1016/j.immuni.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Lai L, Kwa S, Kozlowski PA, Montefiori DC, Ferrari G, Johnson WE, et al. Prevention of Infection by a Granulocyte-Macrophage Colony-Stimulating Factor Co-Expressing DNA/Modified Vaccinia Ankara Simian Immunodeficiency Virus Vaccine. J Infect Dis. 2011;204:164–73. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lakhashe SK, Velu V, Sciaranghella G, Siddappa NB, Dipasquale JM, Hemashettar G, et al. Prime-boost vaccination with heterologous live vectors encoding SIV gag and multimeric HIV-1 gp160 protein: efficacy against repeated mucosal R5 clade C SHIV challenges. Vaccine. 2011;29:5611–22. doi: 10.1016/j.vaccine.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakhashe SK, Wang W, Siddappa NB, Hemashettar G, Polacino P, Hu SL, et al. Vaccination against heterologous R5 clade C SHIV: prevention of infection and correlates of protection. PLoS One. 2011;6:e22010. doi: 10.1371/journal.pone.0022010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakhashe SK, Silvestri G, Ruprecht RM. No acquisition: a new ambition for HIV vaccine development? Curr Opin Virol. 2011;1:246–53. doi: 10.1016/j.coviro.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17:1055–65. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harouse JM, Gettie A, Eshetu T, Tan RC, Bohm R, Blanchard J, et al. Mucosal transmission and induction of simian AIDS by CCR5-specific simian/human immunodeficiency virus SHIV(SF162P3). J Virol. 2001;75:1990–5. doi: 10.1128/JVI.75.4.1990-1995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pal R, Taylor B, Foulke JS, Woodward R, Merges M, Praschunus R, et al. Characterization of a simian human immunodeficiency virus encoding the envelope gene from the CCR5-tropic HIV-1 Ba-L. Journal of acquired immune deficiency syndromes. 2003;33:300–7. doi: 10.1097/00126334-200307010-00003. [DOI] [PubMed] [Google Scholar]
- 18.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, et al. Molecularly cloned SHIV-1157ipd3N4: a highly replication- competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol. 2006;80:8729–38. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddappa NB, Song R, Kramer VG, Chenine AL, Velu V, Ong H, et al. Neutralization- sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J Virol. 2009;83:1422–32. doi: 10.1128/JVI.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddappa NB, Watkins JD, Wassermann KJ, Song R, Wang W, Kramer VG, et al. R5 clade C SHIV strains with tier 1 or 2 neutralization sensitivity: tools to dissect env evolution and to develop AIDS vaccines in primate models. PLoS One. 2010;5:e11689. doi: 10.1371/journal.pone.0011689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautam R, Nishimura Y, Lee WR, Donau O, Buckler-White A, Shingai M, et al. Pathogenicity and mucosal transmissibility of the R5-tropic simian/human immunodeficiency virus SHIV(AD8) in rhesus macaques: implications for use in vaccine studies. J Virol. 2012;86:8516–26. doi: 10.1128/JVI.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren W, Mumbauer A, Zhuang K, Harbison C, Knight H, Westmoreland S, et al. Mucosal transmissibility, disease induction and coreceptor switching of R5 SHIVSF162P3N molecular clones in rhesus macaques. Retrovirology. 2013;10:9. doi: 10.1186/1742-4690-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–52. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachler BC, Humbert M, Palikuqi B, Siddappa NB, Lakhashe SK, Rasmussen RA, et al. Novel biopanning strategy to identify epitopes associated with vaccine protection. J Virol. 2013;87:4403–16. doi: 10.1128/JVI.02888-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S, Song R, Popov S, Mirshahidi S, Ruprecht RM. Overlapping synthetic peptides as vaccines. Vaccine. 2006;24:6356–65. doi: 10.1016/j.vaccine.2006.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirshahidi S, Kramer VG, Whitney JB, Essono S, Lee S, Dranoff G, et al. Overlapping synthetic peptides encoding TPD52 as breast cancer vaccine in mice: prolonged survival. Vaccine. 2009;27:1825–33. doi: 10.1016/j.vaccine.2009.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–75. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, et al. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J Clin Invest. 2011;121:2480–92. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisen HN, Hou XH, Shen C, Wang K, Tanguturi VK, Smith C, et al. Promiscuous binding of extracellular peptides to cell surface class I MHC protein. Proc Natl Acad Sci U S A. 2012;109:4580–5. doi: 10.1073/pnas.1201586109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen RA, Lakhashe SK, Ruprecht RM. Bimodal AIDS vaccine approach: induction of cellular as well as humoral immunity can protect from systemic infection. Vaccine. 2010;28(Suppl 2):B25–31. doi: 10.1016/j.vaccine.2009.10.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasmussen RA, Ong H, Song R, Chenine AL, Ayash-Rashkovsky M, Hu SL, et al. Efficacy of a multigenic protein vaccine containing multimeric HIV gp160 against heterologous SHIV clade C challenges. AIDS. 2007;21:1841–8. doi: 10.1097/QAD.0b013e32828684ea. [DOI] [PubMed] [Google Scholar]
- 32.Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85:5262–74. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peloponese JM, Jr., Collette Y, Gregoire C, Bailly C, Campese D, Meurs EF, et al. Full peptide synthesis, purification, and characterization of six Tat variants. Differences observed between HIV-1 isolates from Africa and other continents. J Biol Chem. 1999;274:11473–8. doi: 10.1074/jbc.274.17.11473. [DOI] [PubMed] [Google Scholar]
- 34.Grisson RD, Chenine AL, Yeh LY, He J, Wood C, Bhat GJ, et al. Infectious molecular clone of a recently transmitted pediatric human immunodeficiency virus clade C isolate from Africa: evidence of intraclade recombination. J Virol. 2004;78:14066–9. doi: 10.1128/JVI.78.24.14066-14069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polacino PS, Stallard V, Klaniecki JE, Pennathur S, Montefiori DC, Langlois AJ, et al. Role of immune responses against the envelope and the core antigens of simian immunodeficiency virus SIVmne in protection against homologous cloned and uncloned virus challenge in Macaques. J Virol. 1999;73:8201–15. doi: 10.1128/jvi.73.10.8201-8215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mediouni S, Darque A, Ravaux I, Baillat G, Devaux C, Loret EP. Identification of a highly conserved surface on Tat variants. J Biol Chem. 2013;288:19072–80. doi: 10.1074/jbc.M113.466011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huet T, Dazza MC, Brun-Vezinet F, Roelants GE, Wain-Hobson S. A highly defective HIV-1 strain isolated from a healthy Gabonese individual presenting an atypical western blot. AIDS. 1989;3:707–15. doi: 10.1097/00002030-198911000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Opi S, Peloponese JM, Jr., Esquieu D, Campbell G, de Mareuil J, Walburger A, et al. Tat HIV-1 primary and tertiary structures critical to immune response against non-homologous variants. J Biol Chem. 2002;277:35915–9. doi: 10.1074/jbc.M204393200. [DOI] [PubMed] [Google Scholar]
- 39.Watkins JD, Lancelot S, Campbell GR, Esquieu D, de Mareuil J, Opi S, et al. Reservoir cells no longer detectable after a heterologous SHIV challenge with the synthetic HIV-1 Tat Oyi vaccine. Retrovirology. 2006;3:8. doi: 10.1186/1742-4690-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sholukh AM, Byrareddy SN, Shanmuganathan V, Hemashettar G, Lakhashe SK, Rasmussen RA, et al. Passive immunization of macaques with polyclonal anti-SHIV IgG against a heterologous tier 2 SHIV: outcome depends on IgG dose. Retrovirology. 2014;11:8. doi: 10.1186/1742-4690-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, et al. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res Hum Retroviruses. 2000;16:1247–57. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 42.Cline AN, Bess JW, Piatak M, Jr., Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–12. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 43.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80:11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408:1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, et al. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis. 2012;206:431–41. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Roman VR, Patterson LJ, Venzon D, Liewehr D, Aldrich K, Florese R, et al. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. Journal of immunology. 2005;174:2185–9. doi: 10.4049/jimmunol.174.4.2185. [DOI] [PubMed] [Google Scholar]
- 47.Forthal DN, Landucci G, Cole KS, Marthas M, Becerra JC, Van Rompay K. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol. 2006;80:9217–25. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carey VJ, Gentry J, Whalen E, Gentleman R. Network structures and algorithms in Bioconductor. Bioinformatics. 2005;21:135–6. doi: 10.1093/bioinformatics/bth458. [DOI] [PubMed] [Google Scholar]
- 49.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clevert DA, Mitterecker A, Mayr A, Klambauer G, Tuefferd M, De Bondt A, et al. cn.FARMS: a latent variable model to detect copy number variations in microarray data with a low false discovery rate. Nucleic Acids Research. 2011;39:e79. doi: 10.1093/nar/gkr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong W, Howard OM, Turpin JA, Grimm MC, Ueda H, Gray PW, et al. Monocyte chemotactic protein-2 activates CCR5 and blocks CD4/CCR5-mediated HIV-1 entry/replication. J Biol Chem. 1998;273:4289–92. doi: 10.1074/jbc.273.8.4289. [DOI] [PubMed] [Google Scholar]
- 52.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–33. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, et al. Vaccine- induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–21. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachler BC, Humbert M, Lakhashe SK, Rasmussen RA, Ruprecht RM. Live-virus exposure of vaccine-protected macaques alters the anti-HIV-1 antibody repertoire in the absence of viremia. Retrovirology. 2013;10:63. doi: 10.1186/1742-4690-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–88. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rose R, Mason RD, Loh L, Peut V, Smith MZ, Fernandez CS, et al. Safety, immunogenicity and efficacy of peptide-pulsed cellular immunotherapy in macaques. J Med Primatol. 2008;37(Suppl 2):69–78. doi: 10.1111/j.1600-0684.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- 57.De Rose R, Fernandez CS, Smith MZ, Batten CJ, Alcantara S, Peut V, et al. Control of viremia and prevention of AIDS following immunotherapy of SIV-infected macaques with peptide-pulsed blood. PLoS Pathog. 2008;4:e1000055. doi: 10.1371/journal.ppat.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chea S, Dale CJ, De Rose R, Ramshaw IA, Kent SJ. Enhanced cellular immunity in macaques following a novel peptide immunotherapy. J Virol. 2005;79:3748–57. doi: 10.1128/JVI.79.6.3748-3757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lind A, Sommerfelt M, Holmberg JO, Baksaas I, Sorensen B, Kvale D. Intradermal vaccination of HIV-infected patients with short HIV Gag p24-like peptides induces CD4 + and CD8 + T cell responses lasting more than seven years. Scand J Infect Dis. 2012;44:566–72. doi: 10.3109/00365548.2011.653581. [DOI] [PubMed] [Google Scholar]
- 60.Klinman DM, Higgins KW, Conover J. Sequential immunizations with rgp120s from independent isolates of human immunodeficiency virus type 1 induce the preferential expansion of broadly crossreactive B cells. J Exp Med. 1991;173:881–7. doi: 10.1084/jem.173.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen SG, Piatak M, Jr., Ventura AB, Hughes CM, Gilbride RM, Ford JC, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–4. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–7. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, et al. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nature medicine. 2012;18:1673–81. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu W, Chen S, Lai C, Guo W, Fu L, Andrieu JM. Induction of CD8+ regulatory T cells protects macaques against SIV challenge. Cell reports. 2012;2:1736–46. doi: 10.1016/j.celrep.2012.11.016. [DOI] [PubMed] [Google Scholar]