Abstract
Background & Aims
The DNA mismatch repair (MMR) system recognizes certain DNA adducts caused by alkylation damage in addition to its role in recognizing and directing repair of interstrand nucleotide mismatches and slippage mistakes at microsatellite sequences. Because defects in the MMR system can confer tolerance to acquired DNA damage and, by inference, the toxic effects of certain chemotherapeutic agents, we investigated the effect of 5-fluorouracil (5-FU) on colon cancer cell lines.
Methods
We determined growth selection by cell enrichment assay and cloning efficiency after treatment with 5 μmol/L 5-FU, assayed nucleic 3H–5-FU incorporation, and analyzed the cell cycle by flow cytometry.
Results
5-FU treatment provided a growth advantage for MMR-deficient cell lines, indicating a relative degree of tolerance to 5-FU by the MMR-deficient cell lines. Enhanced survival was statistically significant after 5 days of growth, and a 28-fold reduction in survival was noted in the MMR-proficient cells by clonagenic assays after 10 days of growth. Differences in nucleotide uptake of 5-FU did not account for the observed growth differences, and specific cell cycle checkpoint arrest was not detected.
Conclusions
Intact DNA MMR seems to recognize 5-FU incorporated into DNA but may do so in a different manner than other types of alkylation damage. Defective DNA MMR might be one mechanism for tumor resistance to 5-FU.
The DNA mismatch repair (MMR) system consists of a complex of proteins that recognizes and directs repair of nucleotide base mismatches and slippage mistakes at simple repetitive sequences termed microsatellites (reviewed in Marra and Boland1,2). These errors, which can result from mistakes by DNA polymerase, are passed on to progeny cells if the MMR system is defective and may result in insertion or deletion mutations in microsatellites or mutations in key growth-regulatory genes.3–7 In addition, the MMR system participates in DNA repair during transcription,8 meiosis,9 and homeologous recombination events.10 In humans, MMR activity requires the proper functioning of hMSH2,11,12 hMLH1,13,14 hPMS1, and hPMS215 proteins; some overlapping functions occur between the hMSH616–18 and hMSH3 proteins because either one may heterodimerize with hMSH2.19,20 Germline mutations of hMSH2, hMLH1, hPMS1, hPMS2, and hMSH6 have been linked to hereditary nonpolyposis colon cancer,11–15,21 and somatic inactivation of these genes have been implicated in approximately 15% of sporadic colorectal cancers based on the presence of microsatellite instability.22
The human MMR system is also capable of recognizing certain DNA adducts caused by alkylation damage.23–25 The agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) creates O6-methylguanine as a principal adduct. This adduct is recognized by the MMR system and results in a G2/M cell cycle arrest and cell death.23 Similarly, when 6-thioguanine (6-TG) is incorporated into the DNA of MMR-proficient cells, a G2/M cell cycle arrest is induced.24 Both the MNNG adduct and 6-TG are thought to be recognized by the MMR system because of mispairing by T (or A) with the altered nucleotide on the DNA strand. Cell cycle arrest at the G2/M checkpoint prevents mutations (G to A transitions) in daughter cells. Adducts formed by cisplatin and carboplatin intercalate and distort DNA, which are also recognized by the MMR proteins.25 However, substituted amine analogues of cisplatin, namely, oxaliplatin, tetraplatin, transplatin, JM335, and JM216, form different types of adducts that are not recognized by the MMR system.25 Similarly, 8-azaguanine (8-AG) is not recognized in MMR-intact cells.24
It is unclear how the selectivity of recognition of adducts occurs. For instance, 8-AG does not interfere with classical Watson–Crick hydrogen bonding as does 6-TG and O6-methylguanine, and thus it may escape recognition. The MMR proteins can recognize specific intrastrand and interstrand cross-links induced by cisplatin or carboplatin, and not those by its analogues. Because both cisplatin and its substituted amine analogues would be expected to cause distortion of double-stranded DNA, it has been suggested that lack of recognition of the cisplatin analogues by the MMR system might be due to steric hindrance of binding by the hMSH2 protein to the altered DNA.25
Because a defective MMR system has been linked to hereditary nonpolyposis colon cancer and a subset of sporadic colon cancers, we hypothesized that MMR-defective tumors might be resistant to certain types of chemotherapy. Because 5-fluorouracil (5-FU) is the most widely used agent to treat colorectal cancer,26 we investigated the response of MMR-proficient and -deficient colorectal cancer cells to this agent in vitro.
5-FU is a fluoropyrimidine that is incorporated into RNA (messenger RNA, ribosomal RNA, and transfer RNA), is an inhibitor of thymidylate synthetase (which catalyzes the conversion of deoxyuridine monophosphate [dUMP] to deoxythymidine monophosphate [dTMP]), and may be incorporated into DNA.27,28 It is an accepted concept that incorporation of 5-FU into RNA is the important mechanism of 5-FU toxicity.27 Under normal conditions, deoxyuridine triphosphatase prevents the incorporation of deoxyuridine triphosphate (dUTP) and 5-fluoro-deoxyuracil triphosphate (FdUTP) into DNA by dephosphorylating the nucleotides to dUMP and 5-fluoro-deoxyuracil monophosphate (FdUMP), respectively.29,30 However, because 5-FU can inhibit thymidylate synthetase, accumulation of dUMP and FdUMP occurs, which exhausts the ability of deoxyuridine triphosphatase to metabolize dUTP and FdUTP. As dUTP and FdUTP levels increase and those of thymidine triphosphate (TTP) decrease, dUTP and FdUTP replace TTP as substrates for DNA polymerases and are incorporated into DNA.27 Nonetheless, uracyl-N-glycosylase, an enzyme that removes uracil bases from DNA after the spontaneous deamination of deoxycytidine, will typically remove the incorporated uracil bases. Despite this, TTP is not available and the DNA strand will be repaired using dUTP or FdUTP as a substrate. 5-FU has been detected in cellular DNA previously,28 but no correlation between 5-FU incorporation into DNA and cytotoxicity has been reported.31–33
In this study, we show that (1) MMR-deficient cells continue to be tumorigenic in nude mice even after DNA MMR has been restored by chromosomal transfer, showing that properly regulated MMR genes are not tumor suppressors per se; (2) MMR-proficient cells treated with 5-FU grow more slowly than MMR-deficient cells; (3) 5-FU is incorporated into DNA of MMR-deficient and MMR-proficient cells at a rate of 7–9-fold less than into RNA and does not correlate with the observed growth effects of 5-FU; and (4) the presence of 5-FU does not lead to a G2/M cell cycle arrest as do those adducts that interfere with Watson–Crick hydrogen binding. These findings suggest that an alternative mechanism of DNA adduct recognition takes place by the MMR system that does not involve cell cycle arrest and that DNA MMR mechanisms contribute to the cytotoxicity of 5-FU in cancer cells.
Materials and Methods
Reagents
5-FU was obtained from Sigma Chemical Co. (St. Louis, MO) and dissolved in Iscove’s modified Dulbecco’s medium at a stock concentration of 1 mmol/L and maintained at 4°C. 3H–5-FU was obtained from Sigma and stored according to the manufacturer’s instructions.
Cell Lines and Cultures
The human colon cancer cell lines HCT116, SW480, and LoVo were obtained from American Type Culture Collection (Rockville, MD) and maintained in growth medium containing 10% fetal bovine serum (FBS). The HCT116 cell lines containing transferred chromosome 2 (HCT116+ch2) and chromosome 3 (HCT116+ch3) were developed as described previously34 and maintained in growth medium containing 10% FBS and 400 μg/mL of G418 (GIBCO BRL, Gaithersburg, MD). The ovarian cell line 2774 was a gift from Kim Orth, Ph.D. (University of Michigan). Of these cell lines, HCT116+ch3 and SW480 cells have been described proficient in and stable at microsatellites.34,35
In Vitro Growth Determinations
Cell lines were grown to 75% confluency in Dulbecco’s modified Eagle medium supplemented with 10% FBS in a 7% CO2 environment. Cultures were harvested by brief trypsinization (0.05% trypsin–0.02% EDTA in Hank’s balanced salt solution without calcium and magnesium), washed several times with calcium- and magnesium-free phosphate-buffered saline (PBS), and plated into 35 × 10–mm tissue culture dishes (Costar, Cambridge, MA) at an initial density of 105 cells/dish. Cells were counted daily using an automated cell counter (Coulter, Hialeah, FL); counts were determined in triplicate. All experiments were duplicated to confirm the results. Doubling times were determined from the slope of the curves.
In Vivo Tumorigenicity of HCT116+ch3 Cells
The tumorigenicity of HCT116, HCT116+ch2, and HCT116+ch3 was compared after subcutaneous injection into athymic nude mice. Cells were grown to 75% confluency in Dulbecco’s modified Eagle medium supplemented with 10% FBS in a 7% CO2 environment. Cultures were harvested by brief trypsinization (0.05% trypsin–0.02% EDTA in Hank’s balanced salt solution without calcium and magnesium), washed several times with calcium- and magnesium-free PBS, and suspended at a final concentration of 5 × 107 cells/mL in serum-free medium. The presence of single-cell suspensions were confirmed by phase-contrast microscopy, and cell viability was determined by trypan blue exclusion. Pathogen-free BALB/c NCR-NU athymic mice (3–5-week-old females obtained from Simonsen Laboratories, Gilroy, CA) were injected with 5 × 106 tumor cells (0.1 mL) subcutaneously into the left posterior flank. Animals were observed daily for tumor growth. Tumors were measured with a caliper at least every other day, and tumor volume was calculated as described previously.36 Five mice were injected per group, and experiments were performed in duplicate. In vivo growth curves were generated from the mean volumes for tumor volume on each day. The difference between groups was analyzed using analysis of variance modified for multiple determinations (Bonferroni and Tuckey-B).
5-FU Treatment
Determination of the linear portion of a 5-FU dose-response curve was done by treating cell lines with increasing doses of 5-FU (0–20 μmol/L) and counting the viable cells remaining (data not shown). The linear range of the dose response curve for the various cell lines was between 1 and 5 μmol/L 5-FU, and thus doses from this range were used. This range corresponds to concentrations reported previously for the HCT116 cells37 and is identical to the median inhibitory concentration range (1–5 μmol/L 5-FU) for five colorectal cancer cell lines, including LoVo cells, reported by Sugimoto et al.38 Our mean median inhibitory concentration was 4.3 μmol/L 5-FU for the HCT116 cells. This also was consistent with reported tissue concentrations after intravenous injection of 5-FU for colon tumor chemotherapy in which levels of 2–5 μmol/L were sustained in the tissue for 48 hours after the injection.39
Determination of 5-FU Incorporation
The procedure used to determine 5-FU incorporation into RNA and DNA has been described previously.28 Exponentially growing cells (107) were plated on 100 × 20–mm dishes (Costar). After 24 hours to ensure attachment, the cells were washed twice with PBS, and 4 mL of complete media were added to each sample, containing 50 μCi of 3H–5-FU (1μmol/L 5-FU) and 10 μCi/mL of H3-32PO4 in Iscove’s modified Dulbecco’s medium with 10% FBS, and incubated at 37°C for 24 hours. The media was then removed, and the cells washed twice with PBS. Cells were then harvested with a cell scraper (Costar) into a 2-mL cryotube (Nalge, Rochester, NY), washed three times with PBS, and resuspended in 0.5 mL PBS. To isolate RNA and DNA, the cell suspension was incubated for 1 hour at 60°C with a proteinase K solution (10 mg/mL proteinase K, 0.01 mol/L Tris, 0.01 mol/L EDTA, 0.5% sodium dodecyl sulfate), phase extracted with phenol: chloroform:isoamyl alcohol (25:24:1), and precipitated with 2 volumes of ethanol. The mixture was centrifuged at 14,000 rpm for 30 minutes, and the precipitate was resuspended in 0.005 mol/L EDTA and formamide. After heating to 80°C for 5 minutes, the sample was placed in cesium chloride (starting density, 1.52 g/mL) and centrifuged at 36,000 rpm for 60 hours. The resulting gradient was fractionated, and portions of each fraction were weighed and counted for 3H and 32P radioactivity in a scintillation counter (Beckman, Fullerton, CA).
Enrichment Assay
HCT116-GFP cells were developed as previously described.40 GFP was expressed in high levels in 90%–95% of these cells. Cells were subjected to flow cytometric analysis on a FACScan (Becton Dickinson, Bedford, MA) using an argon ion laser tuned to 488 nm to identify GFP-positive cells. Fluorescence was observed with a 515/545 nm/bandpass filter.
HCT116+ch3 and HCT116-GFP cells were mixed in a 50:50 ratio and analyzed by flow cytometry to document that the population contained 50% GFP-expressing cells. Enrichment assays were performed by plating 200,000 cells into 100-mm tissue culture dishes. After 24 hours, 0 or 5 μmol/L of 5-FU was added to the dishes as a continuous exposure. Flow cytometric analysis was performed 5 days later. Each experiment was performed three separate times. The HCT116 and HCT116-GFP cells, as well as the HCT116 cells transfected with an empty pCL vector, were tested by clonogenic assay, and no difference in sensitivity to 5-FU was found.
Cloning Efficiency
Exponentially growing cells (106) were trypsinized and washed twice with PBS. Cells were then plated on 60 × 15–mm plates (Costar) in Iscove’s modified Dulbecco’s medium supplemented with 10% FBS and containing various concentrations of 5-FU (0, 1, 2.5, and 5 μmol/L), then incubated at 37°C and 5% CO2. Fresh medium containing 10% FBS, and the appropriate concentration of 5-FU was exchanged every 3 days. After at least 10 days of growth, the culture plates were washed with PBS, fixed with methanol for 15 minutes, then rewashed with PBS. The colonies were then stained with 3% Giemsa (Sigma) for 15 minutes and rinsed with water. Previously, viable clonal colonies of at least 50 cells were counted using a low-power objective (15×). The relative surviving fraction for each cell line was expressed as a ratio of the plating efficiency in treated cultures to that observed in the controls.
Cell Cycle Analysis
Cell cycle analysis was performed by flow cytometric measurement of the DNA content of isolated nuclei from the cells. The methods used for nuclear isolation have been described previously.41 Cells were trypsinized at 24-hour intervals for 5 days, washed twice with PBS, and centrifuged at 250g for 10 minutes. The cells were resuspended at a density of 106 cells/mL in a lysis buffer containing 10 mmol/L Tris-HCl (pH 7.5), 0.32 mol/L sucrose, 3 mmol/L MgCl2, 2 mmol/L CaCl2, and 0.2% (vol/vol) Nonidet P-40 (N-P40; Sigma) and incubated on ice for 10 minutes. The suspension was centrifuged at 2000g for 10 minutes and resuspended in the above lysis buffer, except without N-P40, at a density of 4 × 106 nuclei/mL. The nuclei were pelleted by centrifugation at 2000g for 10 minutes and resuspended in a buffer containing 0.1 mol/L Tris-HCl (pH 7.4), 0.15 mol/L NaCl, 1 mmol/L CaCl2, 0.5 mmol/L MgCl2, 0.01% N-P40, 0.1% ribonuclease A (RNase A; Sigma), and 2 μg/mL propidium iodide (Sigma). Subsequently, nuclei were analyzed for DNA content by the Coulter Epics C Clinical flow cytometer (Coulter Corp.), using excitation at 488 nm with detection at 620 nm for red fluorescence. Analysis of cell cycle data was made with Multicycle Software Autofit Version 2.50 (Phoenix Flow Systems, San Diego, CA).
Results
MMR-Corrected HCT116+ch3 Cells Are Tumorigenic in Nude Mice
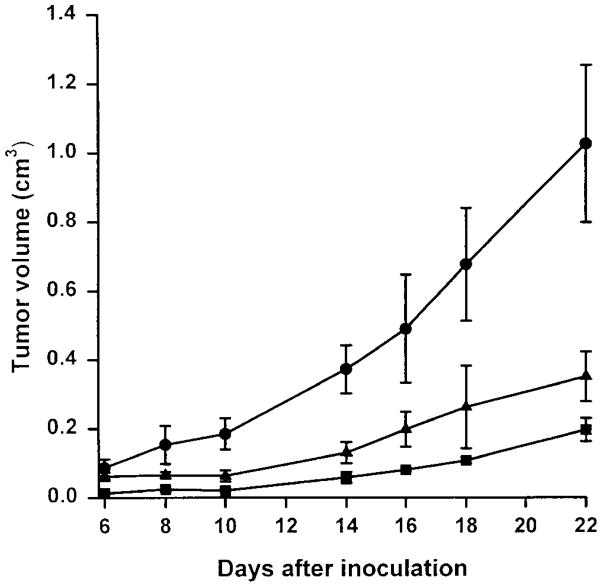
To assess the tumorigenicity of the chromosome 3–transferred cell lines before the in vitro treatment with 5-FU, HCT116+ch3 cells were injected into nude mice. HCT116+ch3 cells do not exhibit microsatellite instability,34 possess a full-length hMLH1 protein by in vitro transcription/translation assay, and have reduced tolerance to alkylation damage compared with HCT116 cells.23 Despite a prolongation in the in vitro doubling time for both the HCT116+ch2 and HCT116+ch3 cell lines (20.7 and 25.3 hours, respectively) compared with parental HCT116 cells (18.6 hours), both cell lines produced tumors in the nude mice. Measurable tumors were detected 6 to 7 days after injection of the parental HCT116 cell line, and 8–10 days after injection of the cell lines HCT116+ch2 and HCT116+ch3. Tumorigenicity was high for all cell lines, and tumors were detected in all animals. However, HCT116+ch2 and HCT116+ch3 cells produced tumors that grew at a significantly lower rate than the parental cell line HCT116 (P < 0.005) (Figure 1). Despite a correction of the MMR-deficient phenotypes and the restoration of DNA MMR activity,34 HCT116+ch3 cells remain tumorigenic in nude mice.
Figure 1.
Tumorigenicity of HCT116 (●), HCT116+ch2 (■), and HCT116+ch3 (▲) cell lines. Each cell line was grown in Dulbecco’s modified Eagle medium with 10% FBS in 7% CO2 and injected into the left flank of 5 BALB/c NCR-NU athymic mice, performed in duplicate. Tumor volume was calculated at each time point by the method in Kwan et al.36 Each point represents the mean ± SD.
MMR-Proficient Cell Lines Are Sensitive to 5-FU
We determined the linear portion of a 5-FU dose response curve on HCT116 cells using concentrations of 5-FU ranging from 0 to 20 μmol/L. The linear range for the various cell lines was consistently between 1 and 5 μmol/L 5-FU (data not shown) and is identical to ranges previously reported for several colorectal cancer cell lines.38 This was consistent with reported tissue concentrations after intravenous injection of 5-FU for colon tumor chemotherapy, in which levels 2–5 μmol/L are sustained in the tissue for 48 hours after intravenous injection.39
To determine any difference in sensitivity with 5-FU treatment, we first performed an enrichment assay of HCT116 and HCT116+ch3 cell lines in the presence of 0 or 5 μmol/L 5-FU. A 50% population of HCT116 cells that were infected with a retrovirus encoding the GFP gene driven by a cytomegalovirus promoter was mixed with a 50% population of HCT116+ch3 cells and subjected to continuous treatment with 5-FU. Five days later, the population was analyzed by flow cytometry to document the proportion of GFP-expressing MMR-deficient cells within the mixture. The mean percent of HCT116-GFP cells in the mixture after three independent experiments was 50.6% on day 0, 53.1% on day 5 for 0 μmol/L 5-FU, and 64.2% on day 5 for 5 μmol/L 5-FU. Thus, the 5-FU–treated population contained 21% more GFP-expressing MMR-deficient HCT116 cells than the control, untreated population after 5 days (P < 0.05 in a two-sided t test).
To assess the growth response of cells to 5-FU, we treated six cell lines in which the MMR phenotype or MMR gene status had been previously determined. HCT116+ch2 cells, like parental HCT116 cells, are MMR-deficient because of inactivation of the hMLH1 gene.34 SW480 is a colon cancer cell line proficient in MMR and has no recognized defects in any of the known MMR genes.35 LoVo is a colon cancer cell line with homozygous partial deletions of the hMSH2 gene,42 and the ovarian cell line 2774 has an hMSH2 mutation in one allele coupled with loss of the other allele and exhibits microsatellite instability.43
The ability of these cancer cells to form colonies with 5-FU treatment was evaluated. HCT116+ch3 cells exhibited a 51-fold reduction in clonal surviving fraction with 5 μmol/L 5-FU compared with HCT116 cells, indicating enhanced sensitivity to the substituted pyrimidine (Figure 2). The MMR-proficient SW480 cell line showed a cloning efficiency similar to HCT116+ch3 cells, showing enhanced sensitivity to 5-FU compared with MMR-deficient cell lines at 2.5 and 5 μmol/L 5-FU concentrations. Overall colony-forming ability, determined 10 days after treatment with 5-FU, was reduced an average of 28-fold at 5 μmol/L 5-FU in MMR-proficient cells compared with MMR-deficient cell lines.
Figure 2.
Colony-forming ability of MMR-deficient and -proficient cell lines in response to 5-FU. Cells were plated in media containing 0, 1, 2.5, or 5 μmol/L 5-FU and allowed to form colonies over a 10-day period. The plates were then fixed with methanol and stained with 3% Giemsa, and previously viable colonies were counted. The mean number of colonies obtained after 10 days for untreated cells were (plating factor of 103): HCT116, 390; HCT116+ch2, 438; HCT116+ch3, 602.5; SW480, 610; LoVo, 176; and 2774, 385. Results are expressed as the mean ± SE of the relative surviving fraction. Data are from 5 independent experiments. MMR-proficient cell lines are noted with the asterisk.
Differences in 5-FU Incorporation Into the Nucleic Acid of the HCT116 Cell Lines Does Not Account for Differences in Cell Viability
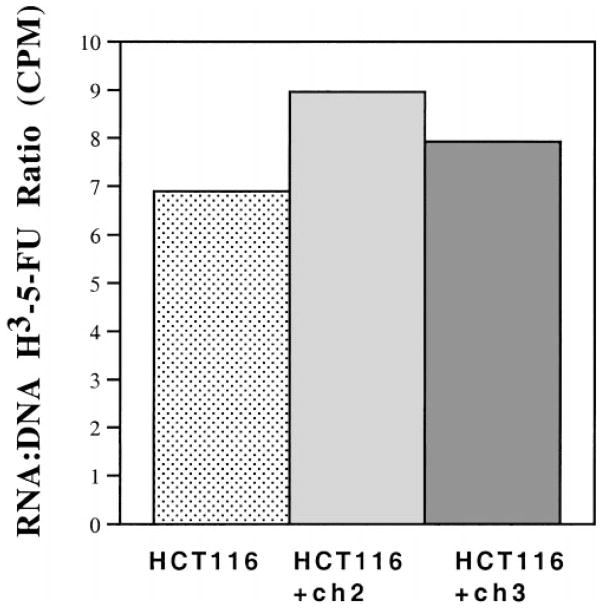
To determine the extent that the 5-FU content of DNA might be responsible for differences in sensitivity among the HCT116, HCT116+ch2, and HCT116+ch3 cell lines, we examined the amount of 3H–5-FU incorporated into each. Cells were treated with H3-32PO4 and 1 μmol/L 3H–5-FU for 24 hours, after which the nucleic acids were extracted. The nucleic acid extract was separated into RNA and DNA fractions by centrifugation in cesium chloride. As shown in Figure 3, there was a 7–9-fold difference in the 3H–5-FU incorporation into RNA vs. DNA among the HCT116 cell lines. The highest ratio (indicating relatively more incorporation into RNA) and the lowest ratio (indicating relatively more incorporation into DNA) corresponded to HCT116 and HCT116+ch2 cells, respectively. Thus the ratio of incorporation into the nucleic acids does not seem to correlate with survival among the cell lines and suggests that differences in incorporation of 3H–5-FU was an unlikely cause for the difference in cell viability.
Figure 3.
Incorporation of 3H–5-FU into nucleic acids of HCT116, HCT116+ch2, and HCT116+ch3 cell lines. Cells were coincubated with 1 μmol/L 3H–5-FU and 10 μCi/mL of H3-32PO4 for 24 hours. The nucleic acids were separated by a cesium chloride gradient and fractionated. Linear regression was determined by cesium chloride density for each fraction. The 3H–5-FU and H3-32PO4 counts banding in the RNA region (between density 1.62 and 1.68 g/mL) and DNA region (between density 1.42 and 1.48 g/mL) of the gradient were determined and used as a measure of the incorporation of 5-FU and phosphate.28 The RNA to DNA ratio is the total tritium counts per minute from the RNA fraction divided by the total counts per minute from the DNA fraction. Results are from 2 independent experiments.
There Is No G2/M Arrest After Treatment With 5-FU in MMR-Proficient Cell Lines
Previously we have reported that the alkylating agents MNNG23 and 6-TG24 induce a G2/M cell cycle arrest in MMR-proficient cell lines. To determine if the response to 5-FU in MMR-proficient cell lines included a G2/M arrest, we analyzed the cell cycle of the two MMR-proficient cell lines, HCT116+ch3 and SW480, after treatment with 5 μmol/L 5-FU. We detected no difference between cells treated with 5-FU and untreated cells through 120 hours (data not shown). Unlike treatment with MNNG or 6-TG, 5-FU incorporation into nucleic acids does not trigger G2/M cell cycle arrest.
Discussion
The DNA MMR system repairs strand-to-strand base mispairs, slippage mistakes at microsatellite sequences,1,2 and recognizes certain DNA adducts caused by alkylation damage.23–25 It has been suggested that the MMR system may be the afferent limb for the detection of certain types of DNA damage.44 We have found that the widely used agent 5-FU is selectively more cytotoxic for MMR-proficient cells than for MMR-deficient cells. Cytotoxicity with 5-FU was shown with a cell enrichment assay in a family of cells derived from a parental cell line and confirmed with clonagenic assays of 6 cell lines. This selective cytotoxicity occurs in the tumorigenic MMR-proficient HCT116+ch3 cells and was specific for MMR-proficient cells among the six cell lines tested.
Selective resistance to alkylation toxicity in MMR-deficient cell lines has been reported with several compounds. The adduct O6-methylguanine, induced by MNNG, MNU, procarbazine, and temozolomide, causes mispairing with T on the opposing DNA strand, leading to a G to A transversion.45 However, the MMR system recognizes this mispair and attempts to correct the mispair by replacing the “correct” nucleotide on the newly synthesized strand before mitosis.46 Without a perfect complementary match for O6-methylguanine, the MMR proteins undertake a futile attempt to replace the nucleotide, which locks the cell cycle in the G2/M phase or triggers a cell death program.23,43,46,47 Similarly, 6-TG, when incorporated into DNA, is recognized by the MMR system and triggers a G2/M arrest in HCT116+ch3 cells.24 These experiments indicate that toxic cellular response to these compounds requires an intact MMR system and that alkylation damage to DNA by these compounds in MMR-deficient tumors is well tolerated.
Other chemotherapeutic agents such as cisplatin and carboplatin form bulky adducts, which distort DNA and may account for recognition by the MMR complex and subsequent cytotoxicity.25 The specificity of this recognition is not currently understood because amine-substituted analogues of cisplatin form different adducts that are not recognized by the MMR system.25 Recognition of cisplatin-damaged DNA by the MMR system has been recently shown to activate an intracellular signaling cascade involving c-Jun-N–terminal kinase 1 and c-abl kinase, a response not found in MMR-deficient cells.48 Thus, detection of damaged DNA, if recognized by an intact MMR system, signals events that may lead to programmed cell death.
It is not known whether the MMR system can recognize and respond to 5-FU incorporated into DNA. Unlike bulky intercalating adducts such as cisplatin, or incorporated purines such as 6-TG and O6-methylgua-nine adducts formed by treatment with MNNG, 5-FU incorporation may not physically distort the DNA double helix. In addition, we have shown that 8-AG is not recognized by the MMR system.24 Both 5-FU and 8-AG are incorporated into DNA but do not directly interfere with interstrand hydrogen bonding. Recognition of 5-FU may be caused by the position of attachment of chemical groups on the altered nucleotide or by the physical nature of the compound itself. We speculate that because the fluoropyrimidine is highly charged, this may deform the DNA double strand enough to be recognized by MMR proteins. Another possibility is that the MMR system recognizes strand distortion produced when uracil-N-glycosylase removes incorporated FdUTP or dUTP. These pyrimidines would eventually be reincorporated in place of TTP because thymidylate synthetase is inactivated.
We found no cell cycle alterations in the MMR-proficient cells after 5-FU treatment. Previously, we reported that treatment with MNNG or incorporation of 6-TG caused a G2/M cell cycle arrest within the first 24 hours after treatment, and this was sustained for more than 5 days.23,24 Both of these agents interfere with classical Watson–Crick hydrogen bonding (positions N1, C2, and C6 of the purine rings). Incorporation of 5-FU into DNA does not interfere with hydrogen bonding (positions N3 and C4 of the pyrimidine rings). We suggest that distortion of hydrogen bonding between double-stranded DNA recruits the MMR proteins and signals cell cycle arrest in the 6-TG and MNNG models. Lack of cell cycle perturbations after 5-FU treatment suggests that 5-FU cytotoxicity induces an alternative pathway of response by the MMR system that does not trigger G2/M cycle arrest. Elucidation of the intracellular signaling pathways that control these processes should give insight into control mechanisms that differ between the MNNG and 5-FU models.
Although our findings are the results of in vitro studies, these data may have important clinical implications. Selection of patients with colorectal cancer for 5-FU treatment is based on the stage of tumor rather than the biology of the tumor.49 A recent meta-analysis of seven randomized trials indicated that the tumor response rate to 5-FU treatment is 22%.50 Our study suggests that the presence of microsatellite instability in a colorectal tumor might be one mechanism of tumor resistance to 5-FU.
Certain colorectal cancer patients whose tumor has a defective MMR system (~15% of sporadic tumors as judged by the presence of microsatellite instability) might not benefit from 5-FU chemotherapy if these data are confirmed in clinical trials. This would be particularly important in patients with colorectal cancer <35 years of age because most exhibit microsatellite instability in their tumors.51 In this study, we used in vitro concentrations of 5-FU that are similar to tissue concentrations achieved in vivo39 and include the effects of hepatic and mononuclear dihydropyrimidine dehydrogenase, the initial rate-limiting enzymes in the catabolism of 5-FU.52 These concentrations used clinically would select for resistant, MMR-deficient tumor cells.
Incorporation of 5-FU into DNA has been reported,28 but we show that incorporation into DNA is 7–9-fold less than RNA. The resultant incorporation of 5-FU into DNA is thought to be facilitated by exhaustion of deoxyuridine triphosphatase when thymidylate synthetase is inhibited and the lack of TTP as the conventional substrate for incorporation even when uracyl-N-glycosylase excises 5-FU from DNA. It is not known what effect the incorporation of 5-FU into DNA might have on subsequent cell divisions. Furthermore, unlike O6-methylguanine adducts or 6-TG incorporation, it is not apparent that 5-FU would induce point mutations.
In summary, a competent MMR system participates in the cytotoxicity of 5-FU–treated cells but does not induce the cell cycle alterations that have been reported in other types of DNA-damaging cytotoxic agents. Because 5-FU is widely used clinically for treatment of advanced colonic adenocarcinomas and other solid epithelial tumors, its efficacy in treating tumors with microsatellite instability should be evaluated.
Acknowledgments
Supported by National Institutes of Health grants DK02433 and CA72851, the Robert Wood Johnson Foundation, and the Veterans Affairs Research Service.
Abbreviations used in this paper
- 8-AG
8-azaguanine
- dTMP
deoxythymidine monophosphate
- dUMP
deoxyuridine monophosphate
- dUTP
deoxyuridine triphosphate
- FBS
fetal bovine serum
- FdUMP
5-fluoro-deoxyuracil monophosphate
- FdUTP
5-fluoro-deoxyuracil triphosphate
- 5-FU
5-fluorouracil
- MMR
mismatch repair
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- 6-TG
6-thioguanine
- TTP
thymidine triphosphate
References
- 1.Marra G, Boland CR. DNA repair and colorectal cancer. Gastroenterol Clin North Am. 1996;25:755–772. doi: 10.1016/s0889-8553(05)70273-9. [DOI] [PubMed] [Google Scholar]
- 2.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome the genes and historical perspectives. J Natl Cancer Inst. 1996;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz S, Wang J, Myeroff L, Parsons R, Sun LZ, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Willson JKV. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 5.Souza RF, Appel R, Yin J, Wang S, Smolinski KN, Abraham JM, Zou TT, Shi YQ, Lei J, Cottrell J, Cymes K, Biden K, Simms L, Leggett B, Lynch PM, Frazier M, Powell SM, Harpaz N, Sugimura H, Young J, Meltzer SJ. Microsatellite instability in the insulin-like growth factor receptor gene in gastrointestinal tumours. Nat Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 6.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 7.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN. Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science. 1996;272:557–560. doi: 10.1126/science.272.5261.557. [DOI] [PubMed] [Google Scholar]
- 9.Arnheim N, Shibata D. DNA mismatch repair in mammals: role in disease and meiosis. Curr Opin Genet Dev. 1997;7:364–370. doi: 10.1016/s0959-437x(97)80150-5. [DOI] [PubMed] [Google Scholar]
- 10.Selva EM, New L, Crouse GF, Lahue RS. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, Guan X-Y, Zhang J, Meltzer PS, Yu J-W, Kao F-T, Chen DJ, Cerosaletti KM, Fournier REK, Todd S, Lewis T, Leach RJ, Naylor SL, Weissenbach J, Mecklin J-P, Jarvinen H, Petersen GM, Hamilton SR, Green J, Jass J, Watson P, Lynch HT, Trent JM, de la Chapelle A, Kinzler KW, Volgelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 12.Fishel R, Lescoe MK, Rao MRS, Copeland NG, Jenkins NA, Garber J, Kane MR. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulos N, Nicolaides NC, Wei Y-F, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Hamilton SR, Petersen GM, Watson P, Lynch HT, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler KW, Volgelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 14.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag RJ, Godwin AR, Ward D, Nordenskjold M, Fishel R, Kolodner R, Liskay RM. Mutation in the DNA mismatch repair homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaides NC, Papadopoulos N, Liu B, Wei Y-F, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Dunlop MG, Hamilton SR, Peteresen GM, de la Chapelle A, Vogelstein B, Kinzler KW. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 16.Drummond JT, Li G-M, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 17.Palombo F, Gallinari P, Iaccarino I, Lettieri TM, D’Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 18.Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson JKV, Kinzler KW, Jiricny J, Vogelstein B. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 19.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 20.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 22.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR, Petersen GM, Kinzler KW, Vogelstein B, de la Chapelle A. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 23.Carethers JM, Hawn MT, Chauhan DP, Luce MC, Marra G, Koi M, Boland CR. Competency in mismatch repair prohibits clonal expansion of cancer cells treated with N-methyl-N′-nitro-N-nitrosoguanidine. J Clin Invest. 1996;98:199–206. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawn MT, Umar A, Carethers JM, Marra G, Kunkel TA, Boland CR, Koi M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 25.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 26.Moertel CG. Chemotherapy for colorectal cancer. N Engl J Med. 1994;330:1136–1142. doi: 10.1056/NEJM199404213301608. [DOI] [PubMed] [Google Scholar]
- 27.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 28.Major PP, Egan E, Herrick D, Kufe DW. 5-Fluorouracil incorporation in DNA of human breast carcinoma cells. Cancer Res. 1982;42:3005–3009. [PubMed] [Google Scholar]
- 29.Caradonna SJ, Cheng YC. The role of deoxyuridine triphosphate nucleotidohydrolase, uracil-DNA glycosylase, and DNA polymerase alpha in the metabolism of FUdR in human tumor cells. Mol Pharmacol. 1980;18:513–520. [PubMed] [Google Scholar]
- 30.Ingraham HA, Tseng BY, Goulian M. Mechanism for exclusion of 5-fluorouracil from DNA. Cancer Res. 1980;40:998–1001. [PubMed] [Google Scholar]
- 31.Danenberg PV, Lockshin A. Fluorinated pyrimidines as tight-binding inhibitors of thymidylate synthetase. Pharmacol Ther. 1981;13:69–90. doi: 10.1016/0163-7258(81)90068-1. [DOI] [PubMed] [Google Scholar]
- 32.Lonn U, Lonn S. DNA lesions in human neoplastic cells and cytotoxicity of 5-fluoropyrimidines. Cancer Res. 1986;46:3866–3870. [PubMed] [Google Scholar]
- 33.Parker WB, Kennedy KA, Klubes P. Dissociation of 5-fluorouracil–induced DNA fragmentation from either its incorporation into DNA or its cytotoxicity in murine T-lymphoma (S-49) cells. Cancer Res. 1987;47:979–982. [PubMed] [Google Scholar]
- 34.Koi M, Umar A, Chauhan DP, Cherian SJ, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 35.Umar A, Boyer JC, Thomas DC, Nguyen DC, Risinger JI, Boyd J, Ionov Y, Perucho M, Kunke TA. Defective mismatch repair in extracts of colorectal and endometrial cancer cell lines exhibiting microsatellite instability. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 36.Kwan SF, Byrd JC, Basbaum C, Kim YS. Characterization of quantitative mucin variants from a human colon cancer cell line. Cancer Res. 1987;47:5715–5724. [PubMed] [Google Scholar]
- 37.Grem JL, Fischer PH, Mulcahy RT. Interaction of deoxyuridine (dUrd) with fluorouracil (FUra) and dipyridamole (DP) in HCT116 cells (abstr) Proc Am Assoc Cancer Res. 1987;28:321, A1271. [Google Scholar]
- 38.Sugimoto Y, Ohe Y, Nishio K, Ohmori T, Fujiwara Y, Saijo N. In vitro enhancement of fluoropyrimidine-induced cytotoxicity by leukovorin in colorectal and gastric carcinoma cell lines but not in non–small-cell lung carcinoma cell lines. Cancer Chemother Pharmacol. 1992;30:417–422. doi: 10.1007/BF00685591. [DOI] [PubMed] [Google Scholar]
- 39.Peters GJ, Lankelma J, Kok RM, Noordhuis P, van Groeningen CJ, van der Wilt CL, Meyer S, Pinedo HM. Prolonged retention of high concentrations of 5-fluorouracil in human and murine tumors as compared with plasma. Cancer Chemother Pharmacol. 1993;31:269–276. doi: 10.1007/BF00685670. [DOI] [PubMed] [Google Scholar]
- 40.Fink D, Nebel S, Norris PS, Aebi S, Kim HK, Haas M, Howell SB. The effect of different chemotherapeutic agents on the enrichment of DNA mismatch repair–deficient tumour cells. Br J Cancer. 1998;77:703–708. doi: 10.1038/bjc.1998.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Black KA, McFarland RD, Grisham JW, Smith GJ. Cell cycle perturbation and cell death after exposure of a human lymphoblastoid cell strain to N-methyl-N′-nitro-N-nitrosoguanidine. Am J Pathol. 1989;134:53–61. [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B, Nicolaides NC, Markowitz S, Willson JKV, Parsons RE, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton SR, Kinzler KW, Vogelstein B. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 43.Orth K, Hung J, Gazdar A, Bowcock A, Mathis JM, Sambrook J. Genetic instability in human ovarian cancer cell lines. Proc Natl Acad Sci USA. 1994;91:9495–9499. doi: 10.1073/pnas.91.20.9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kat A, Thilly WG, Fang W-H, Longley MJ, Li G-M, Modrich P. An alkylation-tolerant mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffin S, Branch P, Xu Y-Z, Karran P. DNA mismatch binding and incision at modified guanine bases by extracts of mammalian cells: implications for tolerance to DNA methylation damage. Biochemistry. 1994;33:4787–4793. doi: 10.1021/bi00182a006. [DOI] [PubMed] [Google Scholar]
- 46.Karran P, Bignami M. Self-destruction and tolerance in resistance of mammalian cells to alkylation damage. Nucleic Acids Res. 1992;20:2933–2940. doi: 10.1093/nar/20.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldmacher VS, Cuzick RA, Jr, Thilly WG. Isolation and partial characterization of human cell mutants differing in sensitivity to killing and mutation by methylnitrosurea and N-methyl-N′-nitro-N-nitrosoguanidine. J Biol Chem. 1986;261:12462–12471. [PubMed] [Google Scholar]
- 48.Nehme A, Baskaran R, Aebi S, Fink D, Nebel S, Cenni B, Wang JYJ, Howell SB, Christen RD. Differential induction of c-Jun NH2-terminal kinase and c-Abl kinase in DNA mismatch repair–proficient and –deficient cells exposed to cisplatin. Cancer Res. 1997;57:3253–3257. [PubMed] [Google Scholar]
- 49.Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Tangen CM, Ungerleider JS, Emerson WA, Tormey DC, Glick JH, Veeder MH, Mailliard JA. Fluorouracil plus levamisole as effective adjuvant therapy after resection of stage III colon carcinoma: a final report. Ann Intern Med. 1995;122:321–326. doi: 10.7326/0003-4819-122-5-199503010-00001. [DOI] [PubMed] [Google Scholar]
- 50.Meta-analysis Group in Cancer. Efficacy of intraveneous continuous infusion of fluorouracil compared with bolus administration in advanced colorectal cancer. J Clin Oncol. 1998;16:301–308. doi: 10.1200/JCO.1998.16.1.301. [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Farrington SM, Petersen GM, Hamilton SR, Parsons R, Papadopoulos N, Fujiwara T, Jen J, Kinzler KW, Wyllie AH, Vogelstein B, Dunlop MG. Genetic instability occurs in the majority of young patients with colorectal cancer. Nat Med. 1995;1:348–352. doi: 10.1038/nm0495-348. [DOI] [PubMed] [Google Scholar]
- 52.McLeod HL, Sludden J, Murray GI, Keenan RA, Davidson AI, Park K, Koruth M, Cassidy J. Characterization of dihydropyrimidine dehydrogenase in human colorectal tumours. Br J Cancer. 1998;77:461–465. doi: 10.1038/bjc.1998.73. [DOI] [PMC free article] [PubMed] [Google Scholar]