Abstract
DNA topoisomerases control the topology of DNA (e.g. the level of supercoiling) in all cells. Type IIA topoisomerases are ATP-dependent enzymes that have been shown to simplify the topology of their DNA substrates to a level beyond that expected at equilibrium (i.e. more relaxed than the product of relaxation by ATP-independent enzymes, such as type I topoisomerases, or a lower than equilibrium level of catenation). The mechanism of this effect is currently unknown, although several models have been suggested. We have analysed the DNA relaxation reactions of type II topoisomerases to further explore this phenomenon. We find that all type IIA topoisomerases tested exhibit the effect to a similar degree and that it is not dependent on the C-terminal domains of the enzymes. As recently reported, the type IIB topoisomerase, topo VI (which is only distantly related to the type IIA enzymes), does not exhibit topology simplification. We find that topology simplification is not significantly dependent on circle size in the range ~2–9 kbp, and is not altered by reducing the free energy available from ATP hydrolysis by varying the ATP:ADP ratio. A direct test of one model (DNA tracking, i.e. sliding of a protein clamp along DNA to trap supercoils) suggests that this is unlikely to be the explanation for the effect. We conclude that geometric selection of DNA segments by the enzymes is likely to be a primary source of the effect but that it is possible that other factors contribute. We also speculate whether topology simplification might simply be an evolutionary relic, with no adaptive significance.
Keywords: topoisomerase, DNA topology, gyrase, energy coupling
Introduction
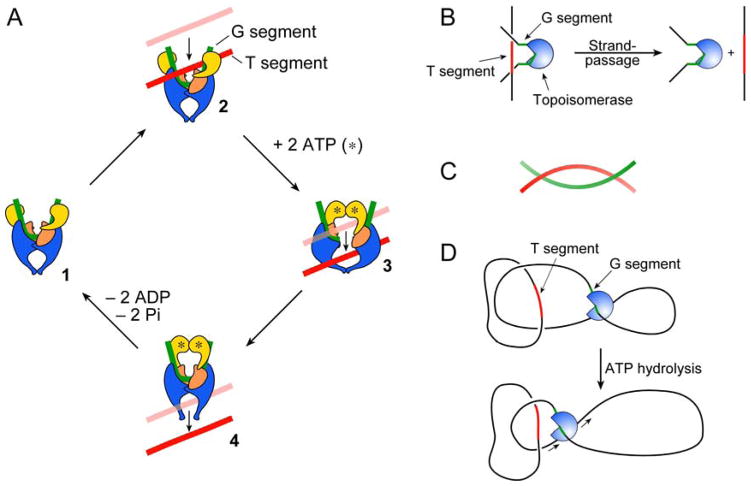
DNA topoisomerases are present in every organism and perform vital roles, controlling the topology of DNA and supporting DNA replication and transcription.1,2 Topoisomerases carry out relaxation and supercoiling of closed-circular DNA, catenation/decatenation of inter-linked DNA, and knotting/unknotting of DNA circles. The enzymes are divided into two types, I and II, corresponding respectively to mechanisms involving breakage of one or both strands of the DNA. Each group is further sub-divided into sub-types A and B (and C3), based on mechanistic and evolutionary considerations.4 Type II enzymes (e.g. topoisomerase (topo) II and DNA gyrase) are known to operate by a strand-passage mechanism5–7 in which a segment of DNA (the gate or G segment) is cleaved in both strands by the enzyme and another double-stranded segment (the transported or T segment) is captured by ATP-dependent dimerisation of the N-terminal ATPase domains (the ‘ATP-operated clamp’). The T segment is passed through the break in the G segment, which is then resealed (Figure 1A).8 When strand passage is intermolecular, i.e. the G and T segments are located on separate circular DNA molecules, the reaction leads to catenation or decatenation of two DNA circles. In an intramolecular reaction, strand passage can lead to a change in linking number of the DNA resulting in the introduction or removal of supercoils, or to knotting or unknotting. All topoisomerases can relax DNA, but DNA gyrase can also introduce negative supercoils into DNA,9 an endergonic reaction coupled to the hydrolysis of ATP.10
Figure 1.
Mechanism of type II DNA topoisomerases. (A) Strand-passage mechanism. (B–D) Models for product simplification: (B) preferential passage of a T segment (red) from the inside to the outside of a bent G segment (green);13 (C) hooked juxtapositions14 (D) tracking of a type II topoisomerase to capture a T segment.11 (Redrawn from ref. 10 with permission.)
DNA relaxation is energetically favourable and can be carried out by ATP-independent type I enzymes, such as eukaryotic topo I, and by gyrase in the absence of ATP. However, all non-supercoiling type II topos require ATP hydrolysis for relaxation, a property that posed something of a puzzle until Rybenkov et al. showed that relaxation by type II topoisomerases simplifies the topology of the products beyond the level expected at equilibrium.11 DNA relaxation by a type I topoisomerase leads to an equilibrated set of products, DNA topoisomers, where each topoisomer is characterised by a different degree of supercoiling [or more technically linking number (Lk)1]. The equilibrated products form a distribution (see Figure 2) around the topoisomer with the highest concentration, which has an integer Lk (= Lkm) closest to that of fully relaxed DNA (with mean Lk = Lk°).1 Rybenkov et al. demonstrated that, in the presence of a type II topoisomerase and ATP, a steady-state distribution was formed that was narrower (less supercoiled) than the equilibrium distribution of topoisomers produced by topo I under the same conditions (e.g. Figure 2A);11 this has been termed the ‘Rybenkov effect’.10 Similarly, the ATP-dependent action of the enzymes resulted in a reduction in the steady-state level of catenation (linking of circular DNAs) and knotting below the equilibrium value. This perturbation of the equilibrium is dominated by a reduction in the entropy of the system. In terms of thermodynamics, it could appear that type II topoisomerases act as a ‘Maxwell’s Demon’, opening an imaginary door to operate only on certain (more supercoiled, catenated or knotted) isomers.12 However, it is well-established that the strand-passage reaction of type II topos is coupled to ATP hydrolysis.10 Thus, there is no thermodynamic conundrum: the free energy of hydrolysis can be used to drive the reaction away from equilibrium, but the question as to how a relatively small enzyme is able to recognise and perturb the global topology of large DNA molecules still remains unanswered.
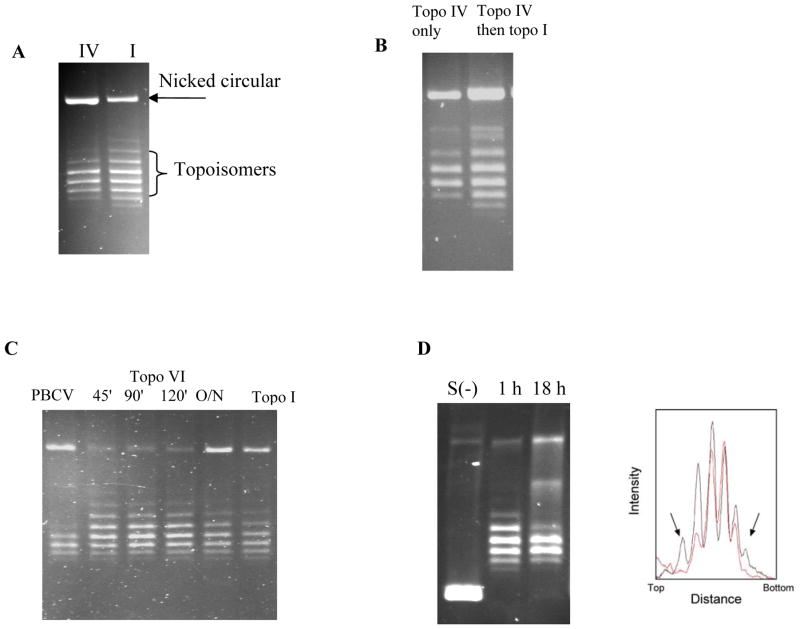
Figure 2.
Topology simplification by type II topoisomerases. A. Plasmid pBR322 relaxed by topo IV (left) and topo I (right) and resolved on a 1% agarose gel in presence of chloroquine, 0.5 μg mL−1. B. Plasmid pBR322 DNA relaxed by topo IV overnight (left lane) and with the subsequent addition of topo I (right lane). C. Time course of relaxation of negatively supercoiled pBR322 by topo VI (times are in minutes; O/N = overnight), compared with PBCV and topo I (overnight incubations). D. Time course of relaxation of negatively supercoiled pBR322 (S(−)) by topo IV; at 1 h the product distribution resembles that generated by topo I. The right-hand panel shows the densitometric traces of the product distributions at 1 h (black) and 18 h (red). Arrows show topoisomers present at 1 h but absent at 18 h.
Several models have been suggested to explain the phenomena of topology simplification in type II topoisomerases.10 All the models are based on increasing the probability of the enzyme transporting specific DNA T segments in a strand-passage reaction; that is, those where strand passage would result in a decrease in topological complexity from an already equilibrated situation. This differential probability of enzyme-T segment interaction could potentially arise via geometric (1) or kinetic (2) selection:
(1). Geometric “bending”, or “kinking” models (Figure 1B)
Vologodskii et al.13 suggested that type II topos bend (or kink) the G segment such that the enzyme adopts a preferred orientation with respect to the global topology of the DNA (indicated by the local curvature of the DNA). If ATP-dependent strand passage then occurs preferentially from inside to outside of the bent DNA hairpin, then a preference for, for example, the capture and strand passage of catenated over uncatenated strands would be expected. Computer simulations confirm that the steady-state fraction of knots could be reduced significantly by this mechanism, although apparently insufficiently to explain the full magnitude of the effect.13 An alternative geometric idea is that ‘hooked juxtaposition’ of DNA double strands (Figure 1C) will be over-represented in supercoiled, knotted or catenated DNA14. It has been reasoned that a reduction of topological complexity could be achieved if the enzyme recognises DNA curvature (hooked juxtapositions) rather than other DNA segments, and simulations support the ability of this mechanism to explain the experimental simplification effect.14–17 This is a geometric selection mechanism viewed from the perspective of the DNA alone, rather than an enzyme-DNA complex. For the enzymes to be operating by this mechanism, it would seem to require them to be sensitive to a bend in both the G and the T segment.
(2). Kinetic “tracking” or “corral effect” models (Figure 1D)
An alternative way to shift the equilibrium was suggested in the original Rybenkov et al. paper11 and envisages that the enzyme binds to a third site on the DNA (in addition to the T and G segments). ATP-dependent translocation of the enzyme along the DNA reduces the apparent DNA length and traps a T segment in a small loop. Such trapped T segments are more likely to correspond to supercoiled, knotted or catenated DNA segments, whose preferential capture would promote reduction of topological complexity. A seemingly related model involving three-segment binding has also been proposed by Trigueros et al.,18 although this model has no kinetic role for ATP hydrolysis, and hence seems not in itself to be able to explain the effect.10 In addition, a kinetic proofreading model has been suggested19,20 that involves two collisions of the enzyme with DNA, and thus ‘proofreading’ of the first reaction by the second. This idea could form a part of a kinetic explanation for bending or tracking models, but it represents a general kinetic scheme rather than a specific mechanism.10
In principle, these models may be able to account for the observations, but there are potential drawbacks and experimental support is scant. Tracking suffers from the obvious problem of requiring three DNA-binding sites on the enzyme, for which there is only indirect evidence.18,21 The potential ability of the enzyme to carry out ATP-dependent translocation on DNA is likewise seemingly inconsistent with the well-established role of ATP binding in DNA clamp closure. Moreover, bioinformatic analysis of topo IIs has failed to reveal obvious sequence similarity with DNA translocating enzymes, such as helicases (data not shown). Evidence for the bending of the G segment by Escherichia coli topo IV has been obtained by measuring cyclisation of small DNA fragments,13 but similar measurement of cyclisation factors for yeast (Saccharomyces cerevisiae) topo II showed the opposite result, i.e. no bending.18 Recently, however, the crystal structure of a complex between yeast topo II and DNA representing a bound G segment has been solved.22 It reveals that the enzyme can bind to a sharply bent DNA. While the exact angle and path of the DNA in the co-crystal structure may be influenced by crystal packing and the use of a doubly-nicked DNA substrate, bending appears to be caused by a highly conserved isoleucine that intercalates into the DNA minor groove. The action of this amino acid in the context of the topo II dimer generates two 75° kinks to give rise to a global 150° bend, a mechanism of deformation reminiscent of that used by other DNA remodeling proteins such as Integration Host Factor (IHF).23 This observation supports the bending model for topology simplification, particularly if combined with the two-gate model for unidirectional DNA transport with type II topoisomerases suggested by Roca and Wang8 and further developed by Vologodskii et al.13 However, the distortion observed may be influenced by the nature of the DNA sequence used in the crystallography work, in particular by nicks in the DNA backbone, and by crystal packing forces, which may contribute to stabilisation of the DNA bend. Therefore the observed G-segment bend must be interpreted cautiously.
Generally, the hypotheses to explain topology simplification by type II topos that have been formulated since the original Rybenkov et al. paper11 are theoretical; relatively few experiments have been carried out to address this issue. In this work, we further investigate the Rybenkov et al. observations and carry out experiments to test the proposed mechanisms. We have focused on the DNA relaxation reaction of type II topos, which is readily accessible experimentally. In addition, the free energy relations for relaxed topoisomer distributions are well known.1 We assume that these findings will also apply to the catenation/decatenation and knotting/unknotting reactions, which conform to the same general strand-passage mechanism and showed similar results in topology simplification experiments.11
Results
DNA relaxation by all type IIA topoisomerases produces a narrow product distribution
To determine whether topology simplification is a general feature of type IIA topoisomerases, we examined the linking number distribution produced after relaxation of plasmid pBR322 with a range of enzymes: E. coli topo IV, yeast topo II, human topo IIα and human topo IIβ. This serves to both confirm and extend previous data.11 The variance of the topoisomer distributions, <ΔLk2>, was determined,1 and compared to that of a control equilibrated distribution; the results are summarised in terms of the parameter R, which is the ratio of the equilibrium variance, <ΔLk2>eq, to the steady-state variance of the topoisomer distribution11 and measures the degree of topology simplification for each enzyme (Figure 3). (The variance is the mean square linking difference of the topoisomers from the completely relaxed centre of the distribution (Lk°), i.e. it is a measure of the breadth of the distribution).
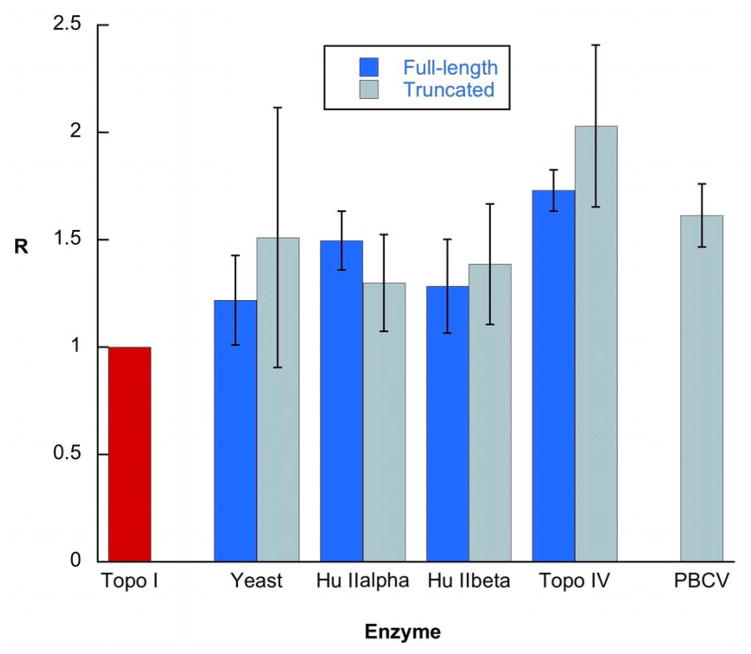
Figure 3.
Degree of product simplification (R) for DNA relaxation by type II topoisomerases. R = <ΔLk2>eq / <ΔLk2>steady state; Hu = human; PBCV = Paramecium bursaria chlorella virus 1; Yeast = Saccharomyces cerevisiae. Solid bars = full-length enzymes; stippled bars = C-terminally truncated enzymes; red bar = wheat germ topo I. Error bars show estimates of the 95% confidence intervals of R based on multiple measurements.
The results confirm that the non-equilibrium product distribution is general and applies to a wide range of type IIA topoisomerases. All four enzymes narrowed the Lk distribution significantly (Figure 3). In the Rybenkov et al. paper, the product distributions varied depending on the enzyme used: E. coli topo IV showed the most extreme effects (R ~1.8) and yeast topo II showed the least (R ~1.35); our results showed similar levels of topology simplification. We found relatively little evidence of significant differences between different enzymes, although the data suggest that topo IV is the most effective enzyme in terms of topology simplification.11 The R-values showed some differences from those reported by Rybenkov et al.,11 although there was no estimate of the precision of the measurements in that case. One potential problem with these measurements is that we found that it can take a considerable time for reactions to reach steady state. To ensure that steady state had been reached, we ran long time courses and verified that there was no further change in product distribution. We also carried out reactions using positively- and negatively-supercoiled DNA as substrates. We found that even where the reaction rate changed significantly with different enzymes or substrates, the final product distributions were always the same (Figure 3).
To further verify that the topo IV relaxation products are at steady-state, we performed sequential incubations of supercoiled DNA with topo IV (which generates a narrow Lk distribution) followed by heating to inactivate the enzyme and incubation with topo I. This was also carried out vice versa, i.e. an equilibrium distribution of DNA isomers produced by topo I was then treated with topo IV. The result was that the topoisomer distribution always corresponded to that of the last enzyme of incubation and the effect of previous enzyme was neutralised (Figure 2B)). In addition, we gel purified the two most prominent topoisomers from the final topo I distribution, to yield a sample with a narrower distribution than that produced by topo IV. This sample was then incubated with topo I and topo IV. The topo I reaction yielded the expected equilibrium distribution and the topo IV reaction yielded the same steady-state distribution (data not shown). These results confirm that the control topo I relaxation products are equilibrated and that the topo IV reaction proceeds in both forward and backward (relaxation and supercoiling) directions to a true topo IV-derived steady-state.
We have thus confirmed that topology simplification occurs for all type IIA topos tested and is independent of the topological state of the DNA substrate. Interestingly it has been reported that topo VI (a type IIB enzyme) from the archeon Methanosarcina mazei does not exhibit topology simplification, i.e. it produces a topoisomer distribution following DNA relaxation that resembles that of topo I.24 We have repeated this experiment under a range of conditions, including those used in Figure 2A, and confirmed that this enzyme does not simplify topoisomer distributions (Figure 2C). This result suggests that the ability of type IIA enzymes to perform topology simplification may reside in a structural feature that differs between topo IIA and B, e.g. lack of a C-terminal domain (but see below), lack of a third “exit” or “C-gate” gate in topo IIB24, or the ability of type IIA topos to bend a G segment. Current evidence supports the existence of bent G segments in yeast topo II22 and topo IV.13 Structural data shows that topo VI lacks the tower domain and the conserved isoleucine found in topo II, both of which are shown in the DNA-bound structure of yeast topo II to facilitate bending,24–26 consistent with the idea of a correlation between G-segment bending and topology simplification. However, as the current topo VI crystal structures lack DNA this remains to be established.24–26
The role of the C-terminal domains of type IIA topos in topology simplification
It is known that all type IIA topoisomerases have discrete domains, including an ATP-binding domain, a DNA binding and DNA breakage-reunion domain, and a C-terminal domain.2,27 The C-terminal domains of the GyrA and ParC subunits of gyrase and topo IV, and of the eukaryotic enzymes, are very divergent in sequence, and are responsible for the gross differences between the behaviour of the enzymes. In gyrase, this domain is responsible for the wrapping of DNA around the enzyme tetramer, delivering an intramolecular T segment to the ATP-dependent clamp with the correct orientation to result in reduction of linking number.28 The non-supercoiling type IIA enzymes do not wrap DNA in the same way.29 However, in topo IV, Corbett et al. have shown that the C-terminal domain acts in a related way as a sensor for substrate selection, modulating the binding between the enzyme and the T segment, and is responsible for topo IV’s ability to preferentially relax positive over negative supercoils.30 In addition, human topo IIα and β, which differ markedly in their C-terminal domains, show significant differences in their substrate preferences.31 Furthermore, topo IIs from Chlorella virus (PBCV-1 and CVM-1), which naturally lack the C-terminal domain, show no preference for relaxing positively or negatively supercoiled substrates.32 Since models for topology simplification depend on the selection of appropriate T segments, it is plausible that the phenomenon might be influenced by the presence of the C-terminal domain. To investigate this hypothesis, we have analysed the relaxation reactions of enzymes truncated at their C-terminus, including the PBCV-1 enzyme. The artificially truncated enzymes were found to be able to narrow the distribution of DNA topoisomers beyond the range seen at equilibrium, albeit at significantly lower reaction rates (Figure 3). The naturally-truncated Chlorella virus enzyme is an efficient topoisomerase and likewise reaches a similar end-point to the other enzymes (Figures 2C & 3). This finding indicates that the C-terminal domain does not play a significant role in topology simplification by type IIA topoisomerases. Further it has been suggested that the bent G-segment model can also explain the preference of topo IV for relaxing positively supercoiled DNA over negatively supercoiled DNA.33 As it is thought that the C-terminal domain of topo IV is responsible for distinguishing positive from negative supercoiled DNA,30 the finding that the C-terminal domains of type IIA topoisomerases plays no role in topology simplification therefore suggests that topology simplification and chiral sensing employ different mechanisms.
Relaxation by type IIA topoisomerases passes through an intermediate equilibrium distribution of topoisomers
It is clear that DNA relaxation by type IIA topos results in product distributions that are significantly narrower than the equilibrium distributions generated by topo I (Figure 2). In order to further investigate the topology simplification reaction, we have followed the time courses of DNA relaxation of negatively supercoiled DNA by type IIA topoisomerases. Figure 2D shows relaxation of negatively supercoiled DNA by topo IV. The surprising feature of this reaction is that during the course of the topo IV reaction (1 h), slightly positively supercoiled topoisomers appear that are present in a topo I final distribution but not visible in the final topo IV product distribution (Figure 2D), i.e. topo IV apparently generates an equilibrated distribution en route to the final steady-state product; this phenomenon was also shown with other type IIA topos (data not shown).
These data may suggest that the ability of the enzyme to carry out selective strand-passage events capable of narrowing the topoisomer distribution happens at a much lower rate than strand passage in general, i.e. that the level of selection is very low. One possibility is that the early reaction of topo IV is dominated by an essentially ‘non-specific’ strand passage, i.e. a ‘phantom-chain’ process leading to relaxed products,13 which is then only slowly modified by selective relaxation. This finding is consistent with the fact that the reactions take much longer to reach steady state than the time required to equilibrate negatively-supercoiled DNA, and also with the apparently very low transduction of free energy into the topology simplification process (see below).
Topology simplification does not depend on DNA circle size
Trigueros et al.18 investigated the topoisomer-narrowing effect using yeast topo II, and showed that the capacity of yeast topo II to simplify product distributions was correlated with a shift in the centre of the topoisomer distribution in the direction of positive supercoiling. However, in our experiments, we have not detected a significant shift in the centre of the final topoisomer distributions of plasmid pBR322 for the four type II topoisomerases tested. Trigueros et al. also showed that the effect was independent of DNA length between 7 kbp and 3 kbp, but below 3 kbp, topology simplification and the topoisomer shift gradually diminish to disappear altogether by 1.4 kbp, i.e. there is a limiting length (< ~2 kbp) below which both reduction of <ΔLk2> and the shift in the mean position of topoisomers vanish. This result potentially supports a tracking model for DNA relaxation by topo IIs, since in small substrates it would be difficult to bind three DNA segments and achieve any advantage from tracking.
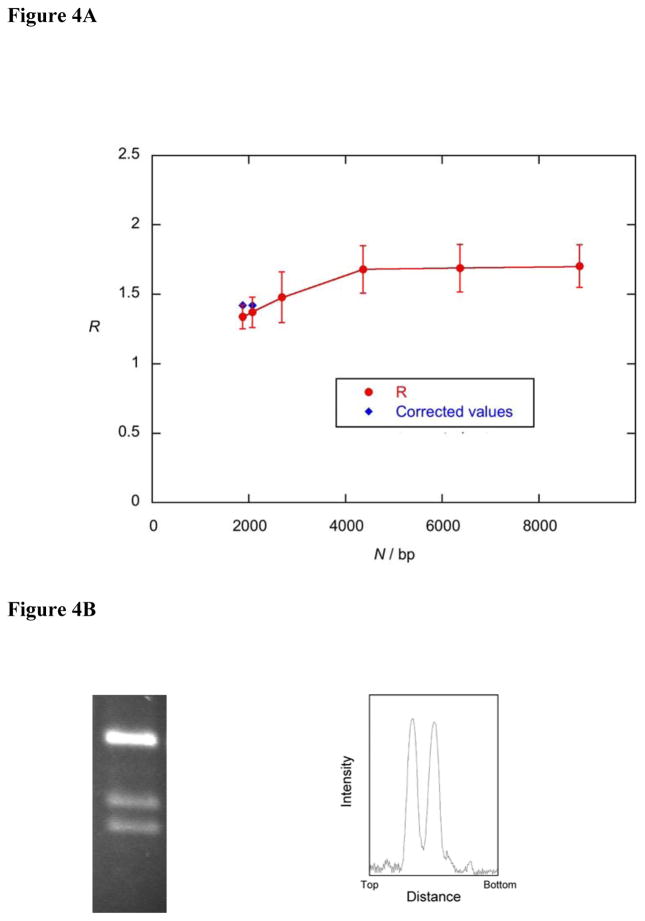
In order to explore these issues further, we performed DNA relaxation experiments with topo IV and DNA circles of different lengths. DNA circles from 1.9 to 8.8 kbp were incubated with catalytic amounts of topo IV or topo I, and the variance ratio R was determined as described above (Figure 4A). These data show that the shift from equilibrium does not depend significantly on DNA length, with only a slight reduction in R for smaller circles. However, a complication arises as circle sizes decrease to about 2 kbp; the narrow topo II distribution may reach a limit corresponding to the presence of two topoisomers of equal intensity, if the distribution is centred between two topoisomers. At smaller sizes, the true R value will be underestimated, as the topo I distribution continues to narrow, while the topo IV distribution cannot. This effect is further complicated by the fact that a type II topo changes Lk in steps of ± 2, and hence cannot interconvert adjacent topoisomers. This leads to a skewing and a further limit to the narrowing of the distribution at smaller circle sizes. This rather complex behaviour is discussed in more detail in the Supplementary Material. These effects are sufficient to explain the apparent reduction in the topology simplification effect previously observed in small DNA circles.18 Figure 4B illustrates this effect; a 2.07 kb circle relaxed with topo IV results in two topoisomers of equal intensity. We have estimated the apparent R values for the 2.07 kbp and a 1.87 kbp circle, and these values are plotted in Figure 4A, along with corrected values from a simulation of these effects discussed in the Supplementary Material.
Figure 4.
Dependence of topology simplification upon circle size. (A) Degree of product simplification (R) vs DNA length (N) for relaxation by topo IV for circles of various sizes. Blue diamonds show corrected R values for small circles (see Supplementary Material). Error bars show estimates of the 95% confidence intervals of R based on multiple measurements. (B) Relaxation reaction with plasmid pQR722 (2.07 kbp) showing the final topoisomers on a gel and densitometric scan; R = 1.37.
These data suggest that, in the size range 2–9 kbp, the topology simplification effect is largely unaffected, i.e. circle size does not appreciably affect the ability of type II topos to simplify product distributions. Below 2 kbp, R cannot be reliably determined. One might expect the models involving a bent G segment and detection of hooked juxtapositions to predict enhanced effects with small circles and reduced effects with larger ones, since the tendency for the bend to orient the enzyme to capture a T segment from within the contour of the DNA circle or the prevalence of hooked juxtapositions would be decreased in highly flexible large circles. Although we do not see this (Figure 4A), it is not clear whether the range of circles we have examined would exhibit any length dependence. In relation to tracking, we would have expected to see some change in the effect as a function of circle size, i.e. due to the difficulty in binding 3 segments of DNA in small circles; the fact that we see no significant differences disfavours tracking.
Topology simplification is not sensitive to the free energy of ATP hydrolysis
DNA gyrase performs an efficient transduction of the free energy of ATP hydrolysis into negative supercoiling, carrying out topoisomer conversions for which the positive ΔG0 is of the same order as the free energy available from the hydrolysis of two ATP molecules.34–36 The enzyme can maintain a level of supercoiling corresponding to a free energy difference from relaxed DNA of more than 500 kJ mol−1 (see Supplementary Material). Consistent with this behaviour, it has previously been shown for DNA gyrase that the degree of supercoiling depends on the free energy available from ATP hydrolysis,35 which can be changed by varying the ATP:ADP ratio.36 In contrast, the non-supercoiling type II enzymes maintain a non-equilibrium distribution of topoisomers with a ΔG of <0.2 kJ mol−1, more than 1000 times lower than that maintained by gyrase. The corresponding values for the unknotting and decatenation reactions investigated by Rybenkov et al. are even lower: <10 J.mol−1 in the case of the decatenation reaction (see Supplementary Material).
To examine this issue further, we carried out experiments with topo IV using varying ATP concentrations and ATP:ADP ratios. We found that the same narrow range of DNA topoisomers is produced independent of the concentration of ATP and the amount of ADP, up to a ADP:ATP ratios of 5:1 (Figure 5) and 10:1 (data not shown), and also using an ATP-regeneration system that maximises the ATP:ADP ratio, and hence the free energy of hydrolysis (data not shown). The addition of inorganic phosphate (up to 4.4 mM) to these reactions, further reducing the free energy of hydrolysis, also had no effect on the reaction products. The reaction proceeds very slowly above a certain ADP:ATP ratio; in some cases, at the higher ratios, ~20 h was required for the reaction to reach steady-state. However, the same product distribution was always seen (Figure 5). Complete inhibition of relaxation occurs at low [ATP] (0.2 mM) when the ADP:ATP ratio is 2.5:1.
Figure 5.
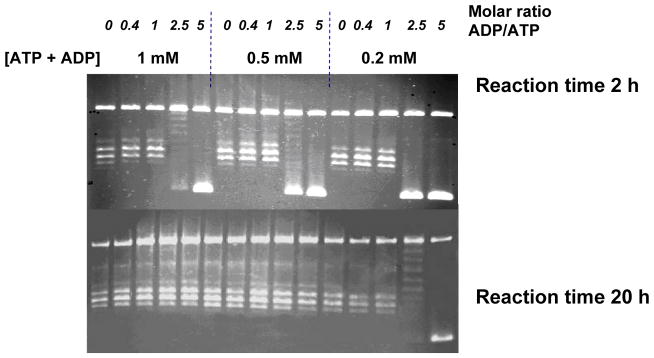
Effect of nucleotides on topology simplification. DNA relaxation by topo IV in the presence of varying concentrations of ATP and ADP, showing products at 2 h and 20 h. Gels were run under the same conditions as in Figure 2.
These results suggest that, within the limits of these experiments, the product distribution of the relaxation reaction of topo IV does not vary significantly with ATP concentration or the ADP:ATP ratio. The free energy available from ATP hydrolysis (assuming 2 ATPs are consumed per reaction cycle) varies between approximately 100 and 150 kJ mol−1 in Figure 5 (see Supplementary Material); this difference does not have any impact on topology simplification. These results would appear to rule out a mechanism in which most of the free energy available is transduced into changes in the free energy of the DNA, or in which a constant proportion of that energy contributes to the free energy change. The data are, however, consistent with a scheme in which the ATP hydrolysis facilitates a geometric or possibly kinetic selection of T segments by determining the unidirectionality of the strand-passage reaction. At some level, the free energy of ATP hydrolysis must provide the free energy to perturb the DNA equilibrium, but we cannot reduce the free energy of hydrolysis to a low enough level to modulate the effect.
A direct test of DNA tracking
To directly address the tracking model (Figure 1B), we used a mutant of EcoRI (E111Q) known to bind tightly to EcoRI recognition sites but with a much reduced cleavage rate.37 In previous work this mutant has been utilised as a ‘roadblock’ to enzymes translocating along DNA.38,39 We constructed plasmids containing 4 and 7 EcoRI sites, and carried out relaxation reactions with topo IV and topo I in the absence and presence of the EcoRI mutant (Figure 6). For both topo I and topo IV the rate of relaxation in the presence of the mutant EcoRI was greatly reduced, but we found no significant differences in the final product distributions: for topo IV, the R values were 1.8 in the presence and absence of the EcoRI mutant (Figure 6). As a control, we showed that under the conditions of these experiments, the ability of wild-type EcoRI to cleave the DNA in the presence of the mutant was greatly diminished, i.e. the recognition sites were largely inaccessible (not shown).
Figure 6.
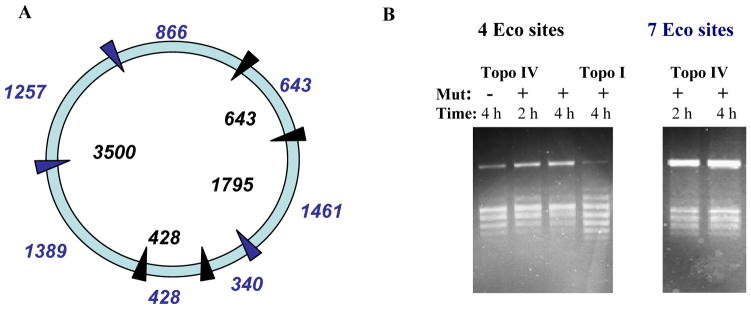
DNA relaxation in the presence of a ‘roadblock’. Relaxation of plasmid pKER3 (~6.5 kbp) containing 4 EcoRI sites (blue arrows) and 7 sites (blue and black arrows). A. Plasmid map. Numbers in black inside circle indicate distances (in bp) between sites in the 4-site plasmid; numbers in blue outside circle indicate distances (in bp) between sites in the 7-site plasmid. B. The gels show relaxation by topo I and IV of the 4-site and 7-site plasmids in the presence and absence of the EcoRI mutant protein (Mut). Gels were run under the same conditions as in Figure 2.
These results suggest that tracking is not a significant feature of the ability of type II topos to simplify their reaction products. We note that, even with the 7-Eco-site plasmid, the largest distance between adjacent sites is ~1.4 kbp. Thus, it is feasible that this separation is sufficient for the enzyme to track; nonetheless, we would expect to detect some global difference in topology simplification as the shorter tracking might be expected to lead to a reduced topology simplification effect.
Discussion
We have shown, using DNA relaxation as a model reaction, that topology simplification is a ubiquitous feature of the type IIA topoisomerases tested, both eukaryotic topo IIs and prokaryotic topo IV (Figure 3). Measurement of time courses of relaxation reactions with topo IV suggests that the reaction may proceed in two kinetic phases, with a relatively rapid acquisition of the equilibrium distribution, followed by slow narrowing to the steady-state level (Figure 2D). The level of the effect is similar in all enzymes tested, and is essentially a property of the core type IIA topoisomerase with no obvious dependence on the presence of the divergent C-terminal domain of ParC or topo II. The effect is also largely independent of the length of the circular DNA substrate used, from 2–9 kbp (Figure 4). Type II topoisomerases require free energy from ATP hydrolysis to maintain non-equilibrium product distributions, but as the free energy requirements are very small, it is perhaps unsurprising that we found the level of the effect to be independent of the free energy available from ATP hydrolysis, in experiments where the ATP concentration and [ATP]:[ADP] ratio is varied (Figure 5). We found that it is not possible to reduce the free energy available to very low levels without the reaction becoming too slow to measure.
It is not straightforward to directly test each of the proposed models for topology simplification; however we have tested the ability of a type IIA topoisomerase to track using up to seven tightly-bound mutant EcoRI complexes on a ~6.5 kbp plasmid (Figure 6). The steady-state distribution achieved in these experiments was independent of the presence of the protein roadblocks, suggesting that tracking is unlikely to be a dominant mode of operation. The evidence to date is therefore most consistent with geometric models, such as the hypothesis based on bending of the G segment,13 recently found to be a feature of the yeast topo II core DNA complex.22 Monte-Carlo simulations of the unknotting reaction using DNA with an intrinsically bent hairpin have demonstrated that such a bend might be a source of the selectivity for topology simplification,13 although the magnitude of the effect observed by Rybenkov et al.,11 has not been reproduced in simulations. The selection of hooked juxtapositions by the enzyme might also be the source of the selectivity, and simulations again indicate that this type of mechanism might explain the experimental observations.15–17 On the other hand, it is unclear whether a mechanism involving these types of geometrical selections is consistent with the absence of any observable dependence on circle-size (Figure 4A). In addition, the lack of dependence on the C-terminal domain of the enzymes casts doubt on the most obvious physical location of potential bent T segment selection, which would seem to be a pre-requisite of the hooked juxtaposition model.
Alternatively, it is possible that that the mechanism of topology simplification by type II topoisomerases might involve more than a simple geometric selection step. Kinetic proof-reading has been proposed to feature in the mechanism of action of type II topoisomerases.19,20 A prerequisite for proofreading would be a degree of uncoupling between ATPase and strand passage. In the case of DNA gyrase, it is well-established that ATP hydrolysis can occur in the absence of strand passage and it is likely that T segments are captured and released without being transported through the enzyme.40–42 In the case of yeast topo II, the rate of ATP hydrolysis can significantly exceed the rate of the topoisomerase reaction.43 Under these conditions it is possible that a proofreading process is occurring that could amplify the selection of T segments through geometric selection and contribute to the observed topology simplification.10 Further work is needed to explore this possibility.
Discussions of topology simplification to date have suggested that the effect should be a useful adaptation for organisms, since it allows them to decatenate and unknot their DNA with high efficiency.10,11 It is certainly true that decatenation of daughter chromosomes is likely to be the primary activity of the non-supercoiling type II topos,44 and that complete decatenation is clearly crucial for successful DNA partitioning. However, it has been suggested that the forces of partition tending to pull apart the daughter replicons will lower the effective concentration of decatenated DNA and allow a phantom-chain type II enzyme to complete decatenation without the requirement for the topology simplification effect.45,46 Furthermore, it is unlikely that the hyper-relaxation reaction that we have studied here has much biological significance, since the level of supercoiling in DNA in vivo is dynamically regulated, and DNA may be only transiently relaxed. Even then, an equilibrium mixture of topoisomers is unlikely to cause a problem that organisms need to solve.
DNA knotting, on the other hand, has been the subject of some recent interest and may be more problematic for cells. Knotting has been shown to occur in E. coli generally at low frequencies (≤ 1% in plasmid DNA47,48), although increased levels can occur in cells harbouring gyrase and/or topo I mutations, or in cells treated with gyrase inhibitors.48,49 A recent study by Diebler et al.,47 where knotting in E. coli has been artificially increased to high levels, has suggested that knotting can promote genetic rearrangements, implying that efficient removal of knots through topology simplification could be adaptive. On the other hand, a recent theoretical paper50 has suggested that negatively supercoiled DNA decreases the equilibrium level of knotting produced by a phantom-chain topo II to very low levels, again suggesting that topology simplification may not be required for efficient unknotting, at least where the DNA is maintained in a negatively supercoiled state.
An alternative hypothesis is that topology simplification by type II topoisomerases is an evolutionary relic of no adaptive significance. Phylogenetic analysis30,51 suggests that topo IV evolved from gyrase by loss of the full wrapping function of the GyrA C-terminal domain. On the other hand, whether eukaryotic topo IIAs also evolved directly from gyrase or from a simpler, ancestral type IIA topoisomerase is less clear. These enzymes have a C-terminus with no detectable homology to that of the bacterial enzymes, although the strong sequence similarity of the enzyme cores, along with experimental results using topo IV,13 make it likely that all the type IIA enzymes exhibit a G-segment bend similar to that identified in the S. cerevisiae enzyme.22 This bending may plausibly have evolved as part of the DNA wrapping inherent in the gyrase mechanism, and now remains as a feature of the non-supercoiling enzymes after loss of wrapping, along with the ATP-driven conformational changes, giving rise to topology simplification as an accident of evolution.
It could be argued that topology simplification by type II topoisomerases is rather inefficient, as judged by the level of energy transduced into free energy changes of the DNA; we estimate that <0.2 kJ mol−1 is required to maintain the non-equilibrium distribution of topoisomers compared with up to 150 kJ mol−1 from the hydrolysis of two ATPs (see Supplementary Material). In contrast, as we have seen, gyrase can in principle convert most of the free energy of hydrolysis of two ATPs into supercoiling free energy.34–36 A well-designed topology simplification enzyme might be expected to do better than the topo II enzymes in free energy transduction terms, perhaps using a translocation mechanism such as that described by Rybenkov et al.,11 in which the topoisomerase is coupled to a DNA translocase to drive the requisite T segments into a small loop to facilitate capture. Such a mechanism is perhaps not so unlikely in evolutionary terms; it is well established that type IA enzymes have recruited helicase domains or subunits for specialist activities such as in archaeal reverse gyrase,52 or in the interaction between the Sgs1 helicase and topo III in yeast.53
Materials and Methods
Enzymes and DNA
Escherichia coli DNA topo IV, yeast topo II and the C-terminally truncated ParC and yeast topo II were prepared as described previously;30,54 yeast topo II was also provided by John Nitiss. Human topo IIα and IIβ, and their C-terminally truncated derivatives, were gifts from Neil Osheroff and Caroline Austin; PBCV-1 was a gift from Neil Osheroff. Topo VI from Methanosarcina mazei was prepared as described by Corbett et al.24 Wheat-germ topo I was purchased from Promega. The EcoRI mutant E111Q was prepared37 from a clone provided by Paul Modrich; wild-type EcoRI was purchased from New England Biolabs.
Negatively supercoiled and relaxed forms of plasmid pBR322 were purchased from Inspiralis Ltd; positively supercoiled pBR322 was prepared as described previously41 or purchased from Inspiralis Ltd. Plasmid pAL-F (8840 bp) was obtained from LGC Promochem; plasmid pKER3 was a gift from Katy Evans-Roberts. Plasmid pUC19 was purchased from Invitrogen. Plasmids pQR792 (2072 bp) and PQR499 (1873 bp) were gifts from John Ward. Plasmid pKER3, which contains 4 EcoRI sites, was engineered to introduce a further 3 EcoRI sites by cloning appropriate oligonucleotides into the following restriction sites: MfeI (1306), BclI (4055) DraIII (5487).
Topoisomerase assays
Topoisomerase assays (30 μL) were carried out under the following conditions: 5–22 nM enzyme, 5.8 nM DNA (plasmid pBR322) in 40 mM Tris·HCl (pH 7.5), 20 mM KCl, 1 mM dithiothreitol, 6 mM MgCl2, 50 μg mL−1 BSA and 1 mM ATP, and were incubated for up to 20 h at 37 °C, then terminated by addition of 0.5 volume of 20% (w/v) sucrose, 50 mM Tris·HCl (pH 7.5), 50 mM EDTA, 50 μg mL−1 bromophenol blue) and one volume of chloroform:isoamyl alcohol (24:1). For some enzymes, the reactions were stopped by the addition of SDS (to 0.2 %) and proteinase K (to 0.2 mg mL−1) and incubated for 30 min at 37 °C. Extracted DNA was analysed by electrophoresis on 1% agarose gels in 90 mM Tris, 90 mM acetic acid, 1 mM EDTA; where appropriate, gels contained chloroquine to resolve topoisomers. Gels were stained with ethidium bromide or SYBR gold (Invitrogen) and photographed using a Syngene system. Linking number distributions were measured using densitometry to produce the relative intensities of the topoisomer bands. Topoisomer intensities were analysed as Gaussian distributions according to the methods described in detail by Bates and Maxwell1 to produce the variance, <ΔLk2>, of the distributions for both topo I and topo II experiments. Only ethidium-stained gels were quantitated, and control experiments showed a linear dependence of fluorescence intensity and DNA concentration over the appropriate range. The R value was computed as the ratio of variances of topo I to topo II (R = <ΔLk2>eq / <ΔLk2>steady state). For ease of comparability, the topo II variances were also compared to previous estimates of the variance of the equilibrium distributions of topoisomers, using NK = 1150RT bp as a best estimate for the solution conditions, based on Rybenkov et al.,55 and corrected for circle sizes below 2.5 kbp based on the data of Horowitz and Wang56.
DNA relaxation assays were also carried out on plasmid pKER3 (4 EcoRI sites) and on a derivative containing 7 EcoRI sites, in the presence of the E111Q mutant EcoRI enzyme. Reactions contained 1.3 nM of DNA and were incubated on ice (30 min), under the conditions described above, and subsequently at 37°C (5 min) with 43 nM of the E111Q mutant; then topo IV was added. Samples were taken every hour and analysed by electrophoresis.
Supplementary Material
Acknowledgments
We thank Caroline Austin, Katy Evans-Roberts, John Nitiss, Neil Osheroff and John Ward for gifts of enzymes and DNA, and Steph Bornemann and Alex Vologodskii for helpful discussions. This work is supported by BBSRC grant BB/C517376/1 (to AM and ADB) and by the National Cancer Institute grant CA077373 (to JMB).
Abbreviations used
- ADPNP
5′-adenylyl-β,γ-imidodiphosphate
- GyrA
DNA gyrase A protein
- ParC
topoisomerase IV subunit homologous to GyrA
- PBCV
Paramecium bursaria chlorella virus
- topo
topoisomerase
- Lk
linking number
- Lkm
linking number of most concentrated topoisomer in a relaxed distribution
Footnotes
Supplementary Material associated with this article can be found, in the online version, at doi: XXXXXX.
References
- 1.Bates AD, Maxwell A. DNA Topology. Oxford University Press; Oxford: 2005. [Google Scholar]
- 2.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Taneja B, Patel A, Slesarev A, Mondragon A. Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases. EMBO J. 2006;25:398–408. doi: 10.1038/sj.emboj.7600922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 5.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 6.Mizuuchi K, Fisher M, O’Dea M, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JC, Gumport RI, Javaherian K, Kirkegaard K, Klevan L, Kotewicz ML, Tse Y-C. DNA topoisomerases. In: Alberts B, Fox CF, editors. Mechanistic studies of DNA replication and genetic recombination. Academic Press; New York: 1980. pp. 769–784. [Google Scholar]
- 8.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 9.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates AD, Maxwell A. Energy Coupling in Type II Topoisomerases: Why Do They Hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- 11.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 12.Pulleyblank DE. Of topo and Maxwell’s dream. Science. 1997;277:648–9. doi: 10.1126/science.277.5326.648. [DOI] [PubMed] [Google Scholar]
- 13.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc Natl Acad Sci U S A. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J Mol Biol. 2004;340:933–9. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Mann JK, Zechiedrich EL, Chan HS. Topological information embodied in local juxtaposition geometry provides a statistical mechanical basis for unknotting by type-2 DNA topoisomerases. J Mol Biol. 2006;361:268–85. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Zechiedrich EL, Chan HS. Inferring global topology from local juxtaposition geometry: interlinking polymer rings and ramifications for topoisomerase action. Biophys J. 2006;90:2344–55. doi: 10.1529/biophysj.105.076778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randall GL, Pettitt BM, Buck GR, Zechiedrich EL. Electrostatics of DNA-DNA juxtapositions: consequences for type II topoisomerase function. J Phys Condens Matter. 2006;18:S173–S85. doi: 10.1088/0953-8984/18/14/S03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J Mol Biol. 2004;335:723–31. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Magnasco MO, Marko JF. Kinetic proofreading can explain the supression of supercoiling of circular DNA molecules by type-II topoisomerases. Phys Rev E. 2001;63:031909. doi: 10.1103/PhysRevE.63.031909. [DOI] [PubMed] [Google Scholar]
- 21.Roca J. Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA. J Mol Biol. 2001;305:441–50. doi: 10.1006/jmbi.2000.4307. [DOI] [PubMed] [Google Scholar]
- 22.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–5. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 23.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 24.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol. 2007;14:611–9. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 25.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo 11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graille M, Cladiere L, Durand D, Lecointe F, Gadelle D, Quevillon-Cheruel S, Vachette P, Forterre P, van Tilbeurgh H. Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms. Structure. 2008;16:360–70. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem Soc Trans. 2005;33:1465–70. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 28.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc Natl Acad Sci USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng H, Marians KJ. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 30.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J Mol Biol. 2005;351:545–61. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 31.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J Biol Chem. 2005;280:39337–45. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 32.McClendon AK, Dickey JS, Osheroff N. Ability of viral topoisomerase II to discern the handedness of supercoiled DNA: bimodal recognition of DNA geometry by type II enzymes. Biochemistry. 2006;45:11674–80. doi: 10.1021/bi0520838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klenin K, Langowski J, Vologodskii A. Computational Analysis of the Chiral Action of Type II DNA Topoisomerases. J Mol Biol. 2002;320:359–67. doi: 10.1016/S0022-2836(02)00447-3. [DOI] [PubMed] [Google Scholar]
- 34.Bates AD, Maxwell A. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 1989;8:1861–6. doi: 10.1002/j.1460-2075.1989.tb03582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullis PM, Maxwell A, Weiner DP. Energy coupling in DNA gyrase: a thermodynamic limit to the extent of DNA supercoiling. Biochemistry. 1992;31:9642–9646. doi: 10.1021/bi00155a017. [DOI] [PubMed] [Google Scholar]
- 36.Westerhoff HV, O’Dea MH, Maxwell A, Gellert M. DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
- 37.Wright DJ, King K, Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989;264:11816–21. [PubMed] [Google Scholar]
- 38.Pavco PA, Steege DA. Characterization of elongating T7 and SP6 RNA polymerases and their response to a roadblock generated by a site-specific DNA binding protein. Nucleic Acids Res. 1991;19:4639–4646. doi: 10.1093/nar/19.17.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- 40.Bates AD, O’Dea MH, Gellert M. Energy coupling in Escherichia coli DNA gyrase: the relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry. 1996;35:1408–1416. doi: 10.1021/bi952433y. [DOI] [PubMed] [Google Scholar]
- 41.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc Natl Acad Sci USA. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugino A, Cozzarelli NR. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980;255:6299–6306. [PubMed] [Google Scholar]
- 43.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J Biol Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 44.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 45.Holm C. Coming undone: how to untangle a chromosome. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 46.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deibler RW, Mann JK, Sumners de WL, Zechiedrich L. Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation. BMC Mol Biol. 2007;8:44. doi: 10.1186/1471-2199-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shishido K, Komiyama N, Ikawa S. Increased production of a knotted form of plasmid pBR322 DNA in Escherichia coli DNA topoisomerase mutants. J Mol Biol. 1987;195:215–218. doi: 10.1016/0022-2836(87)90338-x. [DOI] [PubMed] [Google Scholar]
- 49.Ishii S, Murakami T, Shishido K. Gyrase inhibitors increase the content of knotted DNA species of plasmid pBR322 in Escherichia coli. J Bacteriol. 1991;173:5551–5553. doi: 10.1128/jb.173.17.5551-5553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–63. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–46. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Nadal M. Reverse gyrase: an insight into the role of DNA-topoisomerases. Biochimie. 2007;89:447–55. doi: 10.1016/j.biochi.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng H, Marians KJ. Escherichia coli topoisomerase IV. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 55.Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res. 1997;25:1412–8. doi: 10.1093/nar/25.7.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horowitz DS, Wang JC. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984;173:75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.