Abstract
An examination of the Arabidopsis thaliana genome sequence led to the identification of 29 predicted genes with the potential to encode members of the chaperonin family of chaperones (CPN60 and CCT), their associated cochaperonins, and the cytoplasmic chaperonin cofactor prefoldin. These comprise the first complete set of plant chaperonin protein sequences and indicate that the CPN family is more diverse than previously described. In addition to surprising sequence diversity within CPN subclasses, the genomic data also suggest the existence of previously undescribed family members, including a 10-kDa chloroplast cochaperonin. Consideration of the sequence data described in this review prompts questions about the complexities of plant CPN systems and the evolutionary relationships and functions of the component proteins, most of which have not been studied experimentally.
INTRODUCTION
Chaperonins are a diverse family of molecular chaperones that are present in the plastids, mitochondria, and cytoplasm of all eukaryotes and eubacteria. Type I chaperonins include mitochondrial and chloroplast proteins of approximately 60 kDa, which function in a tetradecameric, double-torus–shaped complex consisting of either 1 or 2 subunit types. Type II chaperonins are the cytoplasmic counterparts of the type I chaperonins and consist of double-torus complexes of 16 subunits, which, in eukaryotes, are composed of 8 subunit types. Although type I chaperonins function with a cochaperonin protein, no analogous cochaperonin has been found to be required for type II chaperonin function, although a protein cofactor, prefoldin (PFD), has been identified. Using resources associated with the Arabidopsis Genome Initiative (http://www.arabidopsis.org/info/agi.html), we have examined the Arabidopsis sequence and annotation databases for information relating to members of the chaperonin protein families.
METHODS
Databases and sequence identification
Sequence databases used in the analysis presented herein include GenBank (http://www.ncbi.nlm.nih.gov), TIGR Arabidopsis thaliana Annotation Database (http://www.tigr.org), and The Arabidopsis Information Resource (http://www.arabidopsis.org/). Arabidopsis sequence databases were examined for annotated sequences belonging to the CPN family. Any annotated chaperonin sequences were used as query sequences in BLASTp searches of the sequence databases to identify unannotated sequences of interest. Additionally, other plant, fungi, and animal chaperonin sequences retrieved from GenBank were used as BLASTp query sequences in searches of the Arabidopsis sequence databases.
Target-transit peptide analysis
Putative mitochondrial targeting peptides and chloroplast transit peptide sequences were identified by sequence alignment or by using Predotar v0.5 (http://www.inra.fr/Internet/Produits/Predotar/) and TargetP v1.01 (http://www.cbs.dtu.dk/services/TargetP/) (Nielsen et al 1997; Emanuelsson et al 2000).
Alignments and phylogenetic trees
Protein sequences were aligned with CLUSTALw v1.8, and alignments were viewed with GeneDoc v2.5.006 (K. B. Nicholas 1998). Phylogenetic analysis was done using the PHYLIP software package. Bootstrapping iterations of aligned sequences were generated using seqboot, and distance calculations were performed using the PAM matrix option in protdist. Trees were generated from distance data by neighbor joining. Consensus trees were calculated using consense, and branch lengths were imposed on the consensus trees using fitch. Trees were viewed with TreeView v1.6.1 (R. D. M. Page 2000). All software for alignment and tree generation was accessed through the Canadian Bioinformatics Resource (http://www.cbr.nrc.ca).
TYPE I CHAPERONIN AND COCHAPERONIN PROTEINS
Cpn60
The term chaperonin was originally coined as a group term to refer to homologous, approximately 60-kDa polypeptides that were identified in plant chloroplasts and in Escherichia coli and that function as molecular chaperones (Hemmingsen et al 1988). These proteins (Cpn60) are ubiquitous in eubacteria and in the plastids and mitochondria of eukaryotes. Typically, they function as a pair of stacked rings, each containing 7 subunits (Saibil 2000), although there is evidence that single rings may be functional (Viitanen et al 1992; Nielsen and Cowan 1998; Nielsen et al 1999). Plastid Cpn60 appears to be distinct from homologous proteins in bacteria and mitochondria in that it is composed of 2 distinct subunit types, α and β (Martel et al 1990; Nishio et al 1999).
Eleven Arabidopsis genomic sequences were identified as having potential to encode Cpn60 proteins (Table 1). Six of these appear to encode plastidic Cpn60 subunits, including 2 α subunits and 4 β subunits. Each includes a putative transit peptide sequence (Table 2). A seventh genomic sequence appears to be a plastidic Cpn60 β-subunit pseudogene. The remaining 4 genomic sequences include 3 that appear to encode mitochondrial Cpn60 polypeptides and an apparent mitochondrial protein pseudogene.
Table 1.
Predicted type I and II chaperonins in the Arabidopsis genome
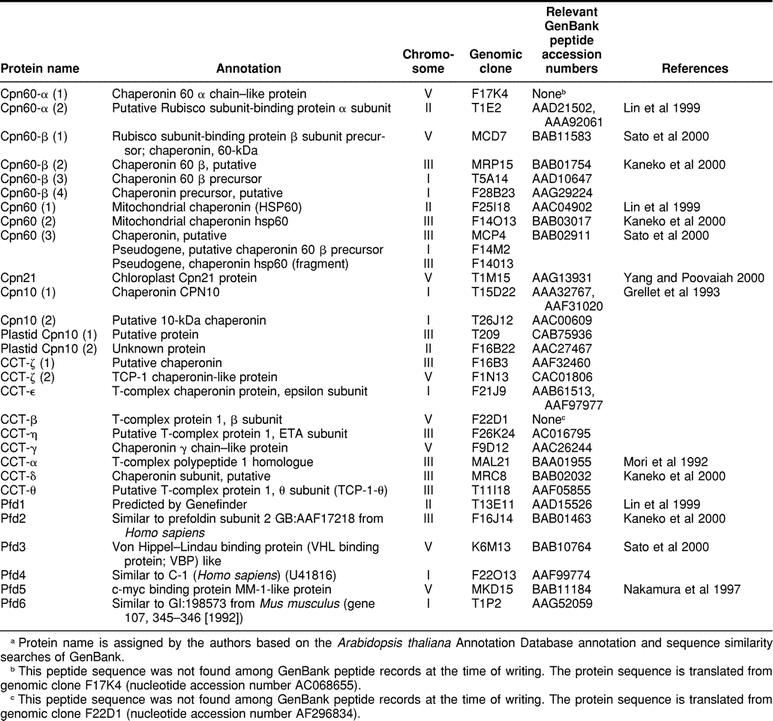
Table 2.
Chloroplast transit peptides (cTPs) and mitochondrial targeting peptides (mTPs) in Arabidopsis chaperonins
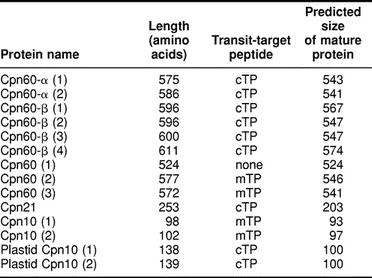
Of the 3 potential mitochondrial Cpn60 proteins shown in Table 1, only 2 contain predicted mitochondrial targeting peptides (Table 2). Cpn60(1) is only 524 amino acids in length, and if aligned with Cpn60(2) and Cpn60(3), it appears that the shorter length is accounted for by an N-terminal truncation of 60 residues. An examination of the genomic DNA sequence upstream of the predicted start of the Cpn60(1) coding sequence shows the potential for additional N-terminal sequence very similar to the corresponding sequences of Cpn60(2) and Cpn60(3), so the possibility exists for an alternative start for Cpn60(1) that would contain the expected mitochondrial targeting peptide. The expression of a complementary DNA (cDNA) corresponding to Cpn60(2) was described as being developmentally regulated and induced by heat shock (Prasad and Stewart 1992). However, the cDNA probe the authors used to identify other mitochondrial Cpn60 sequences in the Arabidopsis genome failed to recognize the other mitochondrial Cpn60 genes apparently present in the genome.
Zabaleta et al (1992) identified 2 closely related Cpn60-β transcripts and suggested, based on Southern analysis of the genome, that the Cpn60-β family of Arabidopsis likely contained at least 3 very similar Cpn60-β proteins. However, the authors did not recognize the apparent fourth member of this family, perhaps not surprisingly, considering that although the first 3 Cpn60-β proteins are 90–95% identical to each other, the outlier Cpn60-β (Cpn60-β[4] in Table 1 and Fig 1) is only on the order of 63% identical to any of the others. The protein identified in Table 1 and Figure 1 as Cpn60-β(3) has been further characterized with respect to its expression patterns and its possible roles in the chloroplast (Zabaleta et al 1994a, 1994b).
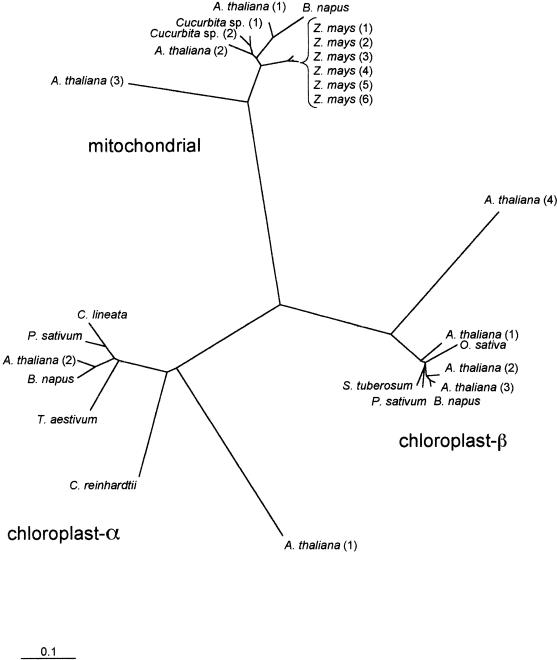
Fig 1.
Phylogenetic relationships of plant mitochondrial and chloroplast α and β Cpn60 proteins. Sequences included in the tree were predicted mature peptides (transit peptides removed). Scale bar indicates 0.1 substitution per site. Bootstrap values for the α, β, and mitochondrial clusters were all greater than 80%, indicating that the clusters are robust. Mitochondrial Cpn60 sequences included in this tree are from A thaliana (GenBank accession numbers AAC04902, BAB03017, BAB02911), Cucurbita sp. (CAA50217, CAA50218), B napus (CAA81689), and Zea mays (AAA44350, AAA33451, AAA33452, CAA77645, CAA78100, CAA78101). Cpn60-α sequences are from Canavalia lineata (AAC68501), Chlamydomonas reinhardtii (AAA98642), Triticum aestivum (HHWTBA), B napus (CAA81736), Pisum sativum (AAA87731), and A thaliana (TIGR locus 68105.t01491, AAD21502). Cpn60-β sequences are from Solanum tuberosum (AAB39827), P sativum (AAA66365), B napus (AAA32980), O sativa (BAA92724), and A thaliana (BAB11583, BAB01754, AAD10647, C079829_21)
The 9 Arabidopsis plastidic and mitochondrial Cpn60 sequences were included in a multiple sequence alignment that also included the available full-length sequences from other plant and algal species. A phylogenetic tree derived from this alignment shows the 3 expected clusters of sequences for the 2 plastidic subunits and for the mitochondrial sequences (Fig 1).
Genes encoding the distinct plastidic subunit types are assumed to have arisen from an ancient gene duplication event (Wastl et al 1999; Archibald et al 2000) and thus α and β genes are considered to be paralogous (Fitch 1970). Genes encoding plastidic and mitochondrial homologues are believed to be derived from independent endosymbiotic events and as such the terms paralogous and orthologous do not apply.
Primary structures of plastid α and β subunits and mitochondrial subunits are highly divergent, a fact illustrated by the phylogenetic tree in Figure 1. For example, the α and β subunits of Arabidopsis Cpn60 are approximately 51% identical to each other and approximately 45% identical to the mitochondrial Cpn60 protein. These intersubunit identities are similar to those found between prokaryotic Cpn60 homologues and any of the eukaryotic subunits: the E coli Cpn60 homologue is 48%, 52%, and 57% identical to the α, β, and mitochondrial Cpn60 proteins of Arabidopsis, respectively.
Diversity of primary structure is also apparent within each subunit type, particularly within the α subunits. One Arabidopsis Cpn60-α polypeptide (Cpn60-α[2]) appears to have orthologues in other dicots and monocots and an apparent paralogue (Cpn60-α[1]) that is only 57% identical in peptide sequence (60%, excluding the putative transit peptide) and for which, to date, orthologues have not been described in other species. The 3 distinct α subunit peptide sequences reported from Brassica napus chloroplast stroma (Cloney et al 1994) are each much closer in sequence to Cpn60-α(2) than to Cpn60-α(1). The 2 Arabidopsis Cpn60-α proteins are similar in length (575 and 586 amino acids, Table 2), and sequence differences are evenly distributed along the length of the proteins. It seems likely that orthologues of Cpn60-α(1) exist in other plant species but have not been identified to date, perhaps in part because of the divergence in sequence. An intriguing possibility is that this outlier sequence somehow relates to the observations made by Schlicher and Soll (1996), which suggest a 1-component Cpn60 complex in the thylakoid lumen of pea chloroplasts composed of a Cpn60-α–like protein. As suggested by the authors, this thylakoid Cpn60 could be identical to one of the stromal Cpn60-α proteins, transported across the thylakoid membrane into the lumen, or an independent gene product exclusively targeted to the thylakoid lumen.
There is also sequence diversity within plastid Cpn60-β subunits. Arabidopsis has 3 Cpn60-β subunit paralogues that are similar to each other (pairwise sequence identities are 92%, 86%, and 85%) and a fourth that is only 60% identical to each of the other 3. In this case, examination of the sequence predicted for this fourth Cpn60-β subunit reveals that relative to the other paralogues it has a C-terminal extension of about 27 residues, raising the possibility that most of the differences between this sequence and other Cpn60-β sequences could be accounted for by this region and that it may constitute an error in the available sequence data or its interpretation. However, an examination of the nucleotide sequence data encoding Cpn60-β(4) did not reveal a likely alternative translation product, and if the C-terminal 27 residues of Cpn60-β(4) are ignored in the pairwise comparison of Arabidopsis Cpn60-β sequences, Cpn60-β(4) is still only 67–68% identical to the other 3 Cpn60-β proteins.
Cochaperonins Cpn10 and Cpn21
Cpn60 proteins act in concert with cochaperonin proteins, typically 10-kDa polypeptides that function as a ring containing 7 subunits (Saibil 1996). Plastids contain a cochaperonin subunit that is approximately double this size, which appears to have evolved as an endoduplication (Bertsch et al 1992). Plant plastid and mitochondrial cochaperonin proteins have been referred to by a number of names, including Cpn21 and Cpn10, the latter referring to both the mitochondrial protein and the larger, 21-kDa plastid homologue (Viitanen et al 1998).
Five genomic sequences were identified as potentially encoding cochaperonin proteins (Table 1). One of these sequences potentially encodes the 21-kDa chloroplast cochaperonin protein, 2 encode putative 10-kDa mitochondrial cochaperonins, and the other 2 may correspond to the 10-kDa chloroplast thylakoid luminal cochaperonin observed by Schlicher and Soll (1996) in pea chloroplasts.
The chloroplast Cpn21 is a functional homologue of the mitochondrial Cpn10 and Cpn21 proteins that have been identified in Arabidopsis and pea (Schlicher and Soll 1996) and spinach (Bertsch et al 1992; Baneyx et al 1995; Bertsch and Soll 1995). Expressed sequence tag data for a putative Cpn21 homologue from Lycopersicon esculentum are also available (Fig 3). Cpn21 proteins consist of 2 Cpn10-like domains following a transit peptide. The 2 Cpn10-like domains are generally less than 50% identical to each other in sequence.
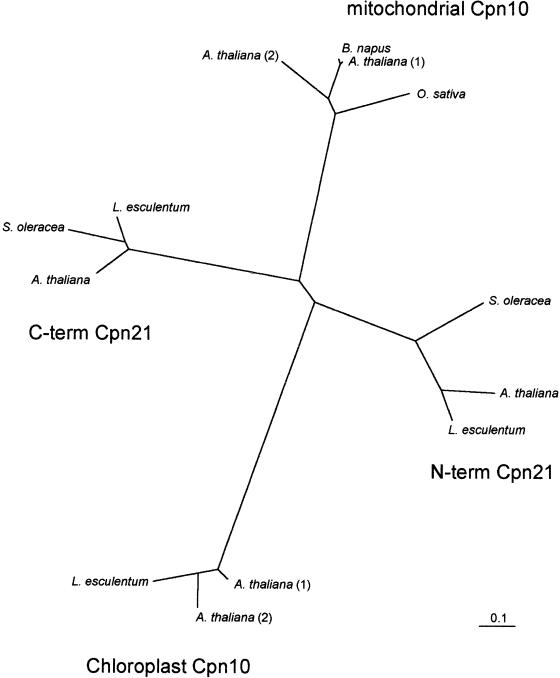
Fig 3.
Phylogenetic relationships of plant mitochondrial and putative chloroplast Cpn10 proteins and N- and C-terminal portions of plant Cpn21 proteins derived from predicted mature peptide sequences (transit peptides removed). The tree was generated with distance data calculated using the PAM matrix option of protdist, followed by neighbor joining. Scale bar indicates 0.1 substitution per site. Bootstrap values for the clusters (chloroplast Cpn10, mitochondrial Cpn10, N-terminus of Cpn21, and C-terminus of Cpn21) were all greater than 80%, indicating the robustness of the clusters. Mitochondrial Cpn10 sequences included in this tree are from O sativa (GenBank accession number AAB63591), B napus (AAB07452), and A thaliana (AC012189_2, AAC00609). Chloroplast Cpn10 sequences are from L esculentum (AW649119) and A thaliana (CAB75936, AAC27467). Cpn21 proteins sequences are from L esculentum (AAF60293), Spinacia oleracea (AAB59307), and A thaliana (AF268068_1)
Tetramers of Cpn21 were detected when a cDNA corresponding to Arabidopsis Cpn21 was expressed in E coli (Hirohashi et al 1999; Koumoto et al 1999). It has also been reported that Arabidopsis Cpn21 forms tetramers in vivo and that these tetramers interact with the Cpn60 tetradecamer (Koumoto et al 1999). The predicted chloroplast localization of this Cpn21 protein has been confirmed experimentally (Koumoto et al 1999). It has been recently observed that Arabidopsis Cpn21 is a calmodulin-binding protein and that the calcium-calmodulin messenger system may be involved in regulating Rubisco assembly in the chloroplast (Yang and Poovaiah 2000).
Experimental evidence for the existence of the protein designated Cpn10(1) in Table 1 has been provided in a study that demonstrated that this Arabidopsis cochaperonin is localized to the mitochondria of transgenic tobacco and that it can functionally complement E coli GroES (Koumoto et al 1996). Our analysis of the Arabidopsis genome suggests the existence of a second mitochondrial Cpn10, which is 75% identical in sequence to Cpn10(1).
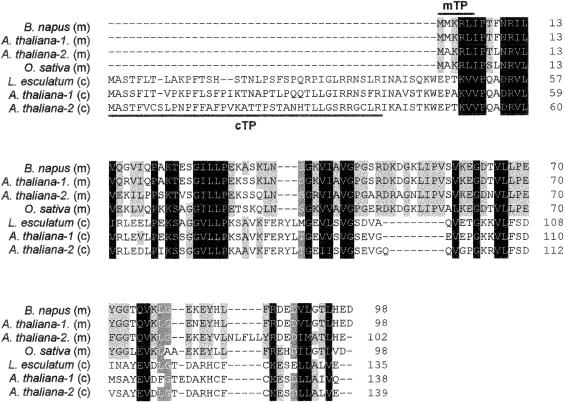
The most interesting finding in the Arabidopsis genome sequence regarding potential Arabidopsis cochaperonins was the identification of 2 potential chloroplastidic, 10-kDa cochaperonins (Table 1). Both of these protein sequences include a predicted chloroplast transit peptide that when cleaved would result in a mature protein size of 100 amino acids for both proteins (Table 2, Fig 2). Neither of these sequences has been reported in the literature, but sequence database searches resulted in the identification of a potential homologue from L esculentum (Fig 2). Also, a report by Schlicher and Soll (1996) described the presence of a 10-kDa cochaperonin in the thylakoid lumen of pea chloroplasts, which could be a homologue of the predicted Arabidopsis sequences. This putative 10-kDa cochaperonin may have been observed in other studies of pea chloroplasts (Bertsch et al 1992) and spinach chloroplasts (Ryan et al 1995).
Fig 2.
CLUSTALw alignment of plant Cpn10 sequences. Localization of the mature peptide is predicted to be either mitochondrial (m) or chloroplastidic (c) based on an analysis of predicted transit peptide sequences. Mitochondrial Cpn10 sequences are from B napus (GenBank accession number AAB07452), A thaliana (AAA32767, AAC00609), and O sativa (AAB63591). Chloroplast Cpn10 sequences are from L esculentum (AW649119) and A thaliana (CAB75936, AAC27467). The predicted chloroplast transit peptides (cTP) are underlined. The predicted mitochondrial targeting peptides (mTP) are overlined
Figure 3 shows the phylogenetic relationships of available cochaperonin protein sequences from plants, including mitochondrial Cpn10, the N- and C-terminal halves of the “double-Cpn10,” chloroplast Cpn21, and the putative chloroplast Cpn10. It is clear from this dendrogram that the sequences cluster according to protein family and that members of the same Cpn10 subtype from different species are more closely related to each other than different Cpn10 types within the same species. For example, considering only the mature forms of the proteins, Arabidopsis mitochondrial Cpn10(1) is only 31% identical (42% similar) to Arabidopsis chloroplast Cpn10(1), whereas it is 67% identical (78% similar) to Oryza sativa mitochondrial Cpn10. As with the different organellar forms of Cpn60, it seems likely that the mitochondrial and chloroplastidic forms of Cpn10 arose from independent endocytic events during the evolution of plants. It is also interesting that the 2 halves of Cpn21 are as different from each other (42% identical, 55% similar) as they are from either of the other forms of Cpn10.
TYPE II CHAPERONIN AND PFD PROTEINS
The cytoplasmic chaperonin CCT
Like the organellar Cpn60 chaperonin, the eukaryotic cytosolic chaperonin CCT (for chaperonin containing TCP1) (also called TRiC for TCP1 ring complex) is a double toroidal protein complex. However, rather than the 1- or 2-subunit composition of Cpn60, the CCT complex is composed of 8 related but distinct subunits (CCT-α, β, χ, δ, ε, η, θ, and ζ) (reviewed in Gutsche et al 1999). Based on sequence similarities, CCT is thought to be related to the archaeal thermosome. Although there are thousands of possible combinations of the 8 subunit types, it is likely that each ring of the double torus consists of 1 of each of the 8 subunit types in a particular arrangement (Liou and Willison 1997). Sequence is available for many CCT subunits from a variety of eukaryotic organisms, and complete sets of 8 subunit sequences are available for several species, including mouse, human, Caenorhabditis elegans, and Saccharomyces cerevisiae. Before the completion of the Arabidopsis genome, there has been little information available for plant CCT proteins: only CCT-ε subunit sequences and partial characterization for Cucumis sativus (Ahnert et al 1996) and Avena sativa (Ehmann et al 1993; Moser et al 2000).
A search of the Arabidopsis genome yielded 9 predicted coding regions distributed among chromosomes I, III, and V, which encode proteins similar to CCT proteins (Table 1). The annotations available in the Arabidopsis thaliana Annotation Database assigned β, γ, ε, η, and θ subunits, whereas the remaining 4 putative translation products were annotated as “chaperonin” or “T-complex protein” with no subunit designation. A comparison of these semiannotated proteins to GenBank proteins permitted their identification as CCT-α, δ, and ζ, with 2 of them corresponding to CCT-ζ. Using these identified proteins as sequence similarity probes in the Arabidopsis genome did not lead to the identification of any additional, unannotated CCTs.
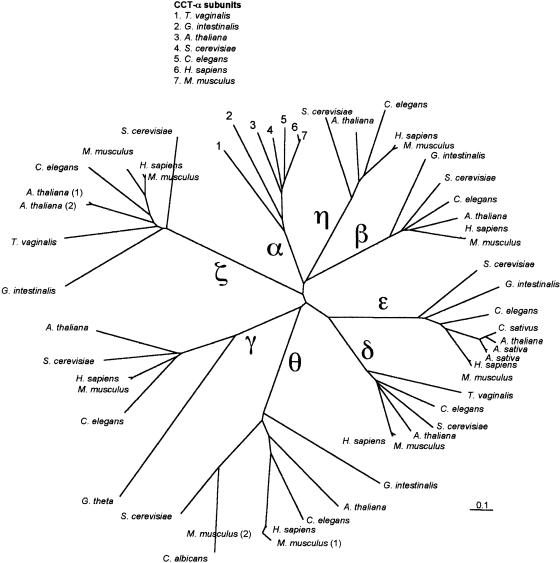
The 2 predicted CCT-ζ coding regions are found on chromosomes III and V, and the predicted protein sequences are 96% identical. The presence of multiple copies of CCT subunits is not unprecedented, since a similar arrangement is found in human and Kubota et al (1997) demonstrated that a second CCT-ζ subunit is facultatively expressed in mouse testis. Figure 4 indicates the phylogenetic relationships of a number of eukaryotic CCT subunits. The tree clearly illustrates that homologous subunits from different eukaryotic species are more similar to each other than are the different subunits from the same species. For example, Arabidopsis CCT-α and CCT-θ are 31% identical (42% similar), whereas Arabidopsis CCT-α and C elegans CCT-α are 63% identical (73% similar). The intrasubunit similarities are even greater for more closely related taxa: Arabidopsis CCT-ε is 91% identical (96% similar) to C sativus CCT-ε. There is currently no published characterization of any predicted Arabidopsis CCT proteins.
Fig 4.
Phylogenetic relationships of eukaryotic cytoplasmic chaperonin (CCT) subunits. The subunit clusters are indicated by labels on the major branches (α, β, γ, δ, ε, η, θ, and ζ). The tree was generated with distance data calculated using the PAM matrix option of protdist, followed by neighbor joining. Scale bar indicates 0.1 substitution per site. Bootstrap values for the 8 subunit clusters were all greater than 90%, indicating the robustness of these clusters. CCT-α subunit proteins included in the tree are from Trichomonas vaginalis (GenBank accession number AAG18494), Giardia intestinalis (AAG18500), A thaliana (BAA01955), C elegans (AAB05072), S cerevisiae (NP_010498), Homo sapiens (P17987), and Mus musculus (228954). CCT-β subunit proteins are from G intestinalis (AAG18501), C elegans (AAA93233), S cerevisiae (S48232), H sapiens (P78371), M musculus (CAA83428), and A thaliana (TIGR locus 68077.t00006). CCT-γ subunit proteins are from Guillardia theta (CAB40401), S cerevisiae (NP_01250), C elegans (AAF35963), A thaliana (AAC26244), H sapiens (NP_005989), and M musculus (CAA83431). CCT-δ subunit proteins are from M musculus (BAA81875), H sapiens (AAC96010), A thaliana (BAB02032), S cerevisiae (CAA98716), C elegans (AAA92842), and T vaginalis (AAG18497). CCT-ε subunit proteins are from Avena sativa (CAA53396, CAA53397), A thaliana (AAB61513), C sativus (1587206), M musculus (CAA83430), S cerevisiae (S57083), C elegans (AAA92843), and G intestinalis (AAG18504). CCT-η subunit proteins are from S cerevisiae (CAA98716), A thaliana (C016795_12), C elegans (AAC19229), M musculus (CAA83274), and H sapiens (AAC96011). CCT-θ subunits are from Candida albicans (P47828), G intestinalis (AAG18505), A thaliana (AC011698_6), H sapiens (XP_009716), C elegans (AAF60806), S cerevisiae (P47079), and M musculus (CAA85521, BAA81879). TriC-ζ subunit proteins are from S cerevisiae (P39079), C elegans (P46550), G intestinalis (AAG18506), T vaginalis (AAG18498), A thaliana (AAF32460, CAC01806), H sapiens (P40227), and M musculus (CAA83432, BAA81891)
Prefoldin
Given the evolutionary relationship of group I and group II chaperonins (Kubota et al 1995), the lack of a cytosolic homologue of the organellar cochaperonins, Cpn10 and Cpn21, is somewhat surprising, although it has been suggested that the α-helical extensions of the apical domain of the CCT subunits may be able to form a cap structure functionally analogous to that provided for Cpn60 by its cochaperonin (Saibil 2000). Recent developments in the search for a CCT cofactor have led to the identification of the PFD complex of proteins (Vainberg et al 1998) (also called GimC, for genes involved in microtubule biogenesis complex (Geissler et al 1998)).
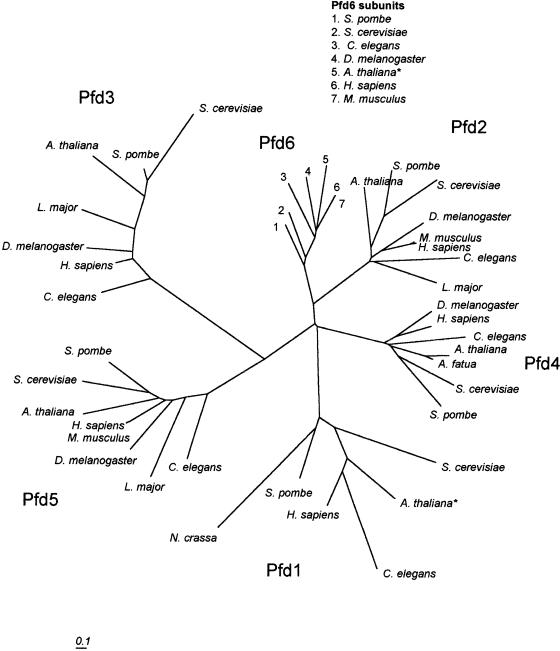
The PFD complex consists of 6 sequence-related subunits (Pfd1–6) and appears to function in binding nascent proteins (especially actin and tubulin) and transferring them to CCT for folding into native protein structure (Geissler et al 1998; Vainberg et al 1998; Hansen et al 1999). The PFD subunits of eukaryotes and of the archaeal PFD homologue fall into 2 classes, α and β, based on their size and predicted structure (Leroux et al 1999). Although archaea generally have only 1 of each subunit type, eukaryotes possess 2 subunits of the α type (Pfd2 and Pfd5) and 4 subunits of the β type (Pfd1, Pfd3, Pfd4, Pfd6). Both subunit types consist of 2 long coiled-coil domains separated by either 2 (in β class subunits) or 4 (in α class subunits) β strands. The archaeal PFD structure has been solved and shows that the overall structure is reminiscent of a jellyfish, with the coiled-coils forming 6 double tentacles hanging from the body of β strands (Siegert et al 2000). In addition to chaperoning nascent cytoskeletal proteins and escorting them to CCT for folding, PFD may play a role in protecting unfolded proteins during CCT “cycling” (the repeated capture and release of proteins during the folding process). Although complete sets of 6 PFD subunit sequences are available for several organisms, including human, S cerevisiae, Schizosaccharomyces pombe, and C elegans, the only plant sequence data available to date have been for Pfd4 of Avena fatua.
Representatives of all 6 PFD subunits were found in the Arabidopsis genome distributed on chromosomes I, II, III, and V; however, only 1 of these (Pfd2) was annotated as a PFD (Table 1). The remaining 5 PFD subunits were identified by sequence similarity searching within the Arabidopsis genome using yeast and mammalian PFD subunit sequences as probes. In 2 cases (Pfd1 and Pfd6), exons not included in the Arabidopsis thaliana Annotation Database predicted translation products were included to produce a full-length PFD subunit sequence.
Figure 5 illustrates the phylogenetic relationships of PFD subunits, including those encoded by Arabidopsis. Bootstrap analysis of the tree shown in Figure 5 showed that although the subunits consistently grouped into 6 clusters, the arrangement of branches connecting the clusters were inconsistent and received low bootstrap values. This is similar to the situation observed in a previous analysis of fewer PFD sequences, including archaea, eubacteria, and eukaryotes (Leroux et al 1999). Similarly to CCT subunits, PFD subunits are more similar to their homologues from other species than to other subunits within the same organism. Arabidopsis Pfd4 is only 19% identical (27% similar) to Arabidopsis Pfd1, but it is 34% identical (46% similar) to C elegans Pfd4 and 71% identical (80% similar) to A fatua Pfd4. In general, PFD subunit sequences are less conserved than CCT subunits both within subunit types and between species. One possible explanation for this is that a large portion of the PFD sequence is accounted for by 2 coiled-coil domains, and beyond the required coiled-coil heptad repeat, there would be little selective pressure to conserve primary structure in these regions. Currently, there is no published characterization of any of the putative Arabidopsis PFD subunit proteins.
Fig 5.
Phylogenetic relationships of eukaryotic PFD subunits Pfd1–6. The tree was generated with distance data calculated using the PAM matrix option of protdist, followed by neighbor joining. Scale bar indicates 0.1 substitution per site. Pfd1 sequences included in the tree are from S cerevisiae (Genbank accession number P46988), A thaliana (AAD15526*), C elegans (Q17827), H sapiens (O60925), S pombe (O14334), and Neurospora crassa (T50987). Pfd2 sequences are from A thaliana (BAB01463), S pombe (Q9UTC9), S cerevisiae (P40005), Drosophila melanogaster (Q9VTE5), Mus musculus (O70591), Homo sapiens (AAF17218), C elegans (Q9N5M2), and Leishmania major (CAB71282). Pfd3 sequences are from S cerevisiae (P48363), S pombe (Q10143), A thaliana (BAB10764), L major (CAB98114), D melanogaster (Q9VTE5), H sapiens (CAA76761), and C elegans (O18054). Pfd4 sequences are from D melanogaster (Q9VRL3), H sapiens (CAB98782), C elegans (Q17435), A thaliana (AAF99774), A fatua (Q9M4C4), S cerevisiae (P53900), and S pombe (Q9UTD4). Pfd5 sequences are from S pombe (Q94307), S cerevisiae (Q04493), A thaliana (BAB11184), H sapiens (Q99471), M musculus (NP_064415), D melanogaster (Q9VCZ8), L major (AAF01572), and C elegans (Q21993). Pfd6 sequences are from S pombe (O14450), S cerevisiae (P52553), C elegans (P52554), D melanogaster (Q9VW56), A thaliana (AAG52059*), H sapiens (O15212), and M musculus (Q03958). A thaliana sequences marked with asterisks are modified by the authors to include exons adjacent to those annotated by TIGR Arabidopsis thaliana Annotation Database (see text for details)
CONCLUSIONS
The availability of the complete genomic sequence of Arabidopsis provides an unprecedented opportunity to examine and consider what may constitute the entire chaperonin family of proteins from a plant. Perhaps most importantly, this analysis points to many questions to be explored regarding the members of this family in plants and other organisms.
The surprising degree of sequence diversity found among members of the Cpn60 family suggests that orthologues of the outlier Cpn60-α and β remain to be discovered in other plant species and that the chloroplast Cpn60 family may in fact be larger and more diverse than currently available sequence data would suggest. It is also possible that one or both of the outlier chloroplast Cpn60 sequences encodes the thylakoid luminal Cpn60 observed in pea chloroplasts (Schlicher and Soll 1996).
Another important observation resulting from the analysis presented herein is the possibility of a 10-kDa chloroplast cochaperonin, which supports the findings of Schlicher and Soll (1996), who recognized the possibility of the coexistence of 2 chaperonin systems in the stroma and thylakoid lumen of the chloroplast. The potential presence of a 10-kDa chloroplast cochaperonin in addition to the previously recognized Cpn21 is intriguing. What is the role of this protein in the chloroplast? Do both Cpn21 and 10-kDa cochaperonin proteins interact with the same Cpn60 complexes or are chaperonin duties in the chloroplast performed between 2 different systems, divided between the stroma and the thylakoid lumen? The answers to these and other questions will naturally depend on continued efforts to identify and characterize plant chaperonin proteins, but the availability of sequence data for all the potential candidate proteins from a model organism such as Arabidopsis will certainly narrow and focus the search.
REFERENCES
- Ahnert V, May C, Gerke R, Kindl H. Cucumber T-complex protein: molecular cloning, bacterial expression and characterization within a 22-S cytosolic complex in cotyledons and hypocotyls. Eur J Biochem. 1996;235:114–119. doi: 10.1111/j.1432-1033.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Logsdon JM, Doolittle WF. Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol. 2000;17:1456–1466. doi: 10.1093/oxfordjournals.molbev.a026246. [DOI] [PubMed] [Google Scholar]
- Baneyx F, Bertsch U, Kalbach CE, van der Vies SM, Soll J, Gatenby AA. Spinach chloroplast cpn21 co-chaperonin possesses two functional domains fused together in a toroidal structure and exhibits nucleotide-dependent binding to plastid chaperonin 60. J Biol Chem. 1995;270:10695–10702. doi: 10.1074/jbc.270.18.10695. [DOI] [PubMed] [Google Scholar]
- Bertsch U, Soll J. Functional analysis of isolated cpn10 domains and conserved amino acid residues in spinach chloroplast co-chaperonin by site-directed mutagenesis. Plant Mol Biol. 1995;29:1039–1055. doi: 10.1007/BF00014976. [DOI] [PubMed] [Google Scholar]
- Bertsch U, Soll J, Seetharam R, Viitanen PV. Identification, characterization, and DNA sequence of a functional “double” groES-like chaperonin from chloroplasts of higher plants. Proc Natl Acad Sci U S A. 1992;89:8696–8700. doi: 10.1073/pnas.89.18.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloney LP, Bekkaoui DR, Feist GL, Lane WS, Hemmingsen SM. Brassica napus plastid and mitochondrial chaperonin-60 proteins contain multiple distinct polypeptides. Plant Physiol. 1994;105:233–241. doi: 10.1104/pp.105.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann B, Krenz M, Mummert E, Schafer E. Two Tcp-1-related but highly divergent gene families exist in oat encoding proteins of assumed chaperone function. FEBS Lett. 1993;336:313–316. doi: 10.1016/0014-5793(93)80827-h. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Von Heijne G. Predicting subcellular localization of proteins based on their N- terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grellet F, Raynal M, Laudie M, Cooke R, Giraudat J, Delseny M. cDNA encoding a putative 10-kilodalton chaperonin from Arabidopsis thaliana. Plant Physiol. 1993;102:685. doi: 10.1104/pp.102.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche I, Essen LO, Baumeister W. Group II chaperonins: new TRiC(k)s and turns of a protein folding machine. J Mol Biol. 1999;293:295–312. doi: 10.1006/jmbi.1999.3008. [DOI] [PubMed] [Google Scholar]
- Hansen WJ, Cowan NJ, Welch WJ. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J Cell Biol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hirohashi T, Nishio K, Nakai M. cDNA sequence and overexpression of chloroplast chaperonin 21 from Arabidopsis thaliana. Biochim Biophys Acta. 1999;1429:512–515. doi: 10.1016/s0167-4838(98)00268-4. [DOI] [PubMed] [Google Scholar]
- Koumoto Y, Shimada T, Kondo M, Takao T, Shimonishi Y, Hara-Nishimura I, Nishimura M. Chloroplast Cpn20 forms a tetrameric structure in Arabidopsis thaliana. Plant J. 1999;17:467–477. doi: 10.1046/j.1365-313x.1999.00388.x. [DOI] [PubMed] [Google Scholar]
- Koumoto Y, Tsugeki R, Shimada T, Mori H, Kondo M, Hara-Nishimura I, Nishimura M. Isolation and characterization of a cDNA encoding mitochondrial chaperonin 10 from Arabidopsis thaliana by functional complementation of an Escherichia coli groES mutant. Plant J. 1996;10:1119–1125. doi: 10.1046/j.1365-313x.1996.10061119.x. [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes G, Willison K. The chaperonin containing t-complex polypeptide 1 (TCP-1)—multisubunit machinery assisting in protein folding and assembly in the eukaryotic cytosol. Eur J Biochem. 1995;230:3–16. doi: 10.1111/j.1432-1033.1995.tb20527.x. [DOI] [PubMed] [Google Scholar]
- Kubota H, Hynes GM, Kerr SM, Willison KR. Tissue-specific subunit of the mouse cytosolic chaperonin-containing TCP-1. FEBS Lett. 1997;402:53–56. doi: 10.1016/s0014-5793(96)01501-3. [DOI] [PubMed] [Google Scholar]
- Leroux MR, Fandrich M, and Klunker D. et al. . 1999 MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 18:6730–6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, and Rounsley S. et al. . 1999 Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 402:761–768. [DOI] [PubMed] [Google Scholar]
- Liou AK, Willison KR. Elucidation of the subunit orientation in CCT, chaperonin containing TCP1, from the subunit composition of CCT micro-complexes. EMBO J. 1997;16:4311–4316. doi: 10.1093/emboj/16.14.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel R, Cloney LP, Pelcher LE, Hemmingsen SM. Unique composition of plastid chaperonin-60:α and β polypeptide-encoding genes are highly divergent. Gene. 1990;94:181–187. doi: 10.1016/0378-1119(90)90385-5. [DOI] [PubMed] [Google Scholar]
- Mori M, Murata K, Kubota H, Yamamoto A, Matsushiro A, Morita T. Cloning of a cDNA encoding the Tcp-1, t complex polypeptide 1, homologue of Arabidopsis thaliana. Gene. 1992;122:381–382. doi: 10.1016/0378-1119(92)90231-d. [DOI] [PubMed] [Google Scholar]
- Moser M, Schafer E, Ehmann B. Characterization of protein and transcript levels of the chaperonin containing tailless complex protein-1 and tubulin during light-regulated growth of oat seedlings. Plant Physiol. 2000;124:313–320. doi: 10.1104/pp.124.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Sato S, Kaneko T, Kotani H, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5, III: sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 1997;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen KL, Cowan NJ. A single ring is sufficient for productive chaperonin-mediated folding in vivo. Mol Cell. 1998;2:93–99. doi: 10.1016/s1097-2765(00)80117-3. [DOI] [PubMed] [Google Scholar]
- Nielsen KL, McLennan N, Masters M, Cowan NJ. A single-ring mitochondrial chaperonin, Hsp60-Hsp10, can substitute for GroEL-GroES in vivo. J Bacteriol. 1999;181:5871–5875. doi: 10.1128/jb.181.18.5871-5875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio K, Hirohashi T, Nakai M. Chloroplast chaperonins: evidence for heterogeneous assembly of alpha and beta Cpn60 polypeptides into a chaperonin oligomer. Biochem Biophys Res Commun. 1999;266:584–587. doi: 10.1006/bbrc.1999.1868. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Stewart CR. cDNA clones encoding Arabidopsis thaliana and Zea mays mitochondrial chaperonin HSP60 and gene expression during seed germination and heat shock. Plant Mol Biol. 1992;18:873–885. doi: 10.1007/BF00019202. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Naylor DJ, Hoogenraad NJ, Hoj PB. Affinity purification, overexpression, and characterization of chaperonin 10 homologues synthesized with and without N-terminal acetylation. J Biol Chem. 1995;270:22037–22043. doi: 10.1074/jbc.270.37.22037. [DOI] [PubMed] [Google Scholar]
- Saibil H. The lid that shapes the pot: structure and function of the chaperonin GroES. Structure. 1996;4:1–4. doi: 10.1016/s0969-2126(96)00002-0. [DOI] [PubMed] [Google Scholar]
- Saibil H. Molecular chaperones: containers and surfaces for folding, stabilising or unfolding proteins. Curr Opin Struct Biol. 2000;10:251–258. doi: 10.1016/s0959-440x(00)00074-9. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Katoh T, Asamizu E, Kotani H, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5, X: sequence features of the regions of 3,076,755 bp covered by sixty P1 and TAC clones. DNA Res. 2000;7:31–63. doi: 10.1093/dnares/7.1.31. [DOI] [PubMed] [Google Scholar]
- Schlicher T, Soll J. Molecular chaperones are present in the thylakoid lumen of pea chloroplasts. FEBS Lett. 1996;379:302–304. doi: 10.1016/0014-5793(95)01534-5. [DOI] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–632. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Bacot K, Dickson R, Webb T. Purification of recombinant plant and animal GroES homologs: chloroplast and mitochondrial chaperonin 10. Methods Enzymol. 1998;290:218–230. doi: 10.1016/s0076-6879(98)90021-0. [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Lorimer GH, Seetharam R, Gupta RS, Oppenheim J, Thomas JO, Cowan NJ. Mammalian mitochondrial chaperonin 60 functions as a single toroidal ring. J Biol Chem. 1992;267:695–698. [PubMed] [Google Scholar]
- Wastl J, Fraunholz M, Zauner S, Douglas S, Maier UG. Ancient gene duplication and differential gene flow in plastid lineages: the GroEL/Cpn60 example. J Mol Evol. 1999;48:112–117. doi: 10.1007/pl00006438. [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. Arabidopsis chloroplast chaperonin 10 is a calmodulin-binding protein. Biochem Biophys Res Commun. 2000;275:601–607. doi: 10.1006/bbrc.2000.3335. [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Assad N, Oropeza A, Salerno G, Herrera-Estrella L. Expression of one of the members of the Arabidopsis chaperonin 60β gene family is developmentally regulated and wound-repressible. Plant Mol Biol. 1994a;24:195–202. doi: 10.1007/BF00040585. [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Oropeza A, Assad N, Mandel A, Salerno G, Herrera-Estrella L. Antisense expression of chaperonin 60β in transgenic tobacco plants leads to abnormal phenotypes and altered distribution of photoassimilates. Plant J. 1994b;6:425–432. [Google Scholar]
- Zabaleta E, Oropeza A, Jiménez B, Salerno G, Crespi M, Herrera-Estrella L. Isolation and characterization of genes encoding chaperonin 60β from. Arabidopsis thaliana. Gene. 1992;111:175–181. doi: 10.1016/0378-1119(92)90685-i. [DOI] [PubMed] [Google Scholar]