Abstract
The 90-kDa heat shock protein (Hsp90) is an essential molecular chaperone in eukaryotic cells, with key roles in the folding and activation of proteins involved in signal transduction and control of the cell cycle. A search for Hsp90 sequences in the Arabidopsis thaliana genome revealed that this family includes 7 members. The AtHsp90-1 through AtHsp90-4 proteins constitute the cytoplasmic subfamily, whereas the AtHsp90-5, AtHsp90-6, and AtHsp90-7 proteins are predicted to be within the plastidial, mitochondrial, and endoplasmic reticulum compartments, respectively. The deduced amino acid sequences of each of the cytoplasmic proteins contains the highly conserved C-terminal pentapeptide MEEVD. All of the AtHsp90 sequences include a conserved adenosine triphosphate–binding domain, whereas only the cytoplasmic and endoplasmic reticulum–resident sequences include an adjacent charged linker domain that is common in mammalian and yeast sequences. The occurrence of multiple AtHsp90 proteins in the cytoplasm and of family members in other subcellular compartments suggests a range of specific functions and target polypeptides.
INTRODUCTION
The 90-kDa heat shock protein (Hsp90) is an abundant and highly conserved molecular chaperone that is essential for eukaryotic cell viability. Studies, primarily in animals, have revealed that Hsp90 has key roles in the folding, activation, and possibly trafficking of proteins involved in signal transduction, such as steroid receptors and a variety of protein kinases, and cell cycle control. The Hsp90-dependent folding and activation of client proteins involve a number of accessory factors, or cochaperones, that participate in multiprotein complexes (reviewed in Toft 1998; Pearl and Prodromou 2000).
The nomenclature of Hsp90 derives from its identification first as belonging to a unique set of proteins that are up-regulated in stressed cells, giving the term heat shock protein of 90 kDa. Further characterization of Hsp90 expression revealed that although some members of the family are stress inducible, others are constitutively expressed. The seminal article by Rutherford and Lindquist (1998) argues that Hsp90 buffers genetic variation in nature by keeping mutant proteins in wild-type conformations. When this buffering is compromised, for example, by temperature stress that diverts Hsp90 from its normal target protein to other partially denatured proteins, variations are exposed, allowing selection to remodel developmental processes. This work elegantly links the dual involvement of Hsp90 in signal transduction with cellular responses to stress.
Hsp90 is present also in prokaryotic cells (reviewed in Parsell and Lindquist 1993; Csermely et al 1998). The prokaryotic ortholog HtpG is 42% identical in amino acid sequence to human Hsp90, and, unlike eukaryotic Hsp90, it is dispensable. In animal cells, the predominantly cytoplasmic Hsp90 has 2 isoforms, Hsp90-α and Hsp90-β, which are 76% identical at the amino acid sequence level. A 94-kDa glucose-regulated protein, grp94, which is 50% identical to cytoplasmic Hsp90, resides in the endoplasmic reticulum (ER). Recently, a distant relative of Hsp90, TRAP1, with approximately 35% identity to human Hsp90-β and a mitochondrial localization, has been reported (Felts et al 2000).
Hsp90 proteins consist of conserved N-terminal and C-terminal domains that are joined by a charged linker region of variable length (Buchner 1999; Pearl and Prodromou 2000). The eukaryotic TRAP1 and the bacterial HtpG proteins resemble one another in that they have only a very short charged region and lack the C-terminal MEEVD sequence, which is characteristic of cytoplasmic Hsp90 (Felts et al 2000). However, based on sequence similarities between different Hsp90 members, it is difficult to say yet if TRAP1 is the eukaryotic homologue of HtpG (Felts et al 2000).
Hsp90 genes have been isolated from several plant species (Conner et al 1990; Felsheim and Das 1992; Koning et al 1992; Takahashi et al 1992; Marrs et al 1993; Yabe et al 1994; Krishna et al 1995). A comparison of the predicted amino acid sequences of plant Hsp90 with Hsp90 of yeast and animal origin shows identities ranging from 63–71%, and between Hsp90 from different plant species, from 88–93%. In addition to genes encoding cytoplasmic Hsp90, complementary DNAs for the ER- and plastid-localized Hsp90 homologues have also been isolated from plants (Schroder et al 1993; Walther-Larsen et al 1993; Schmitz et al 1996). Notwithstanding the availability of Hsp90 genes from plants for more than a decade, our understanding of the functions and the mechanism of action of plant Hsp90 is limited. The Hsp90 system is of particular interest in plants first because of an additional subcellular compartment, the plastid, and second because plants are subjected to sudden changes in their environment, which invoke rapid molecular responses. Plant Hsp90 gene products, heterocomplexes and Hsp90 mode of action, are likely to encompass unique features. Specific knowledge of these is required to understand how proteins involved in signal transduction pathways and other processes are activated, processed, and trafficked within plant cells. This review focuses on the Hsp90 protein family in Arabidopsis thaliana to provide a comprehensive sequence-based understanding of the different family members in plants and to highlight the similarities and differences between Hsp90 proteins of plant and animal origin. We hope that the data will provide a useful starting point for analysis of the functions of Hsp90 in plants and further consolidate our views on the structure-function relationships of Hsp90 in general.
MATERIALS AND METHODS
Identification of A thaliana Hsp90 sequences
All sequences were obtained from the GenBank server (http://www.ncbi.nlm.nih.gov:80/Entrez/batch.html and saved as a flat FASTA file. Selected Hsp90 sequences were used as search queries, using the SSEARCH implementation of the Smith-Waterman algorithm (Smith and Waterman 1981). Unlike the commonly used heuristic search algorithms, such as BLAST and FASTA, the Smith-Waterman algorithm is guaranteed to identify all sequence similarities within a given database (Pearson 1996). Default values were used for the matrix (BLOSUM50) (Henikoff and Henikoff 1992) and gap opening and extension penalties (−12, −2), and low-complexity regions were not considered in the search (Wootton and Federhen 1996).
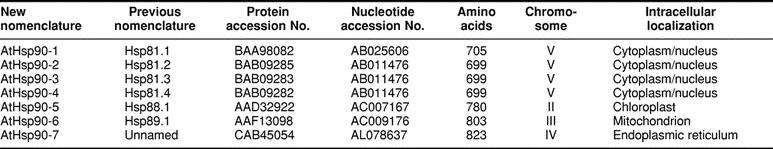
An iterated approach was used to identify all Hsp90 sequences. The AtHsp90-1 (hsp81.1), AtHsp90-2 (hsp81.2), and AtHsp90-3 (hsp81.3) sequences were used as initial query sequences, and 3 classes of sequences were identified. The first class contained sequences more than 99% identical to the query. It was assumed that these are redundant entries for the same sequence, and they were clustered under the protein name of the query (data not shown). We found 8 entries in the database corresponding to AtHsp90-1, 5 corresponding to AtHsp90-2, and 2 corresponding to AtHsp90-3. The second class comprised sequences less than 99% identical to the query but that matched better than would be expected by chance. It was assumed that these sequences are related to the query sequence but are the products of different genes from the same gene family. Each of these was used in turn as a query sequence to identify new members of the Hsp90 family. We found 2 entries for AtHsp90-4 (hsp81.4), 2 entries for AtHsp90-5 (hsp88.1), 2 entries for AtHsp90-6 (hsp89.1), and 4 entries for the AtHsp90-7 (data not shown). The third class of sequences matched the query no better than expected by chance. It was assumed that these sequences are unrelated to the query and they were not further examined. Additional searches were conducted using the Homo sapiens and Drosophila melanogaster mitochondrial (TRAP1) Hsp90 and Secale cereale plastidial Hsp90 sequences. Searching with these heterologous queries failed to reveal any additional members of the A thaliana Hsp90 family. Ultimately, 7 distinct genes or proteins were identified (Table 1).
Table 1.
The Arabidopsis thaliana Hsp90 proteins
Comparison of AtHsp90 sequences
Relatedness values for each member of the AtHsp90 family were calculated after pairwise alignment (Smith and Waterman 1981; Henikoff and Henikoff 1992). The identity values exclude gapped regions. The alignments can be viewed at www.biochem.uwo.ca/fac/gloor/Arab.html. Similar results were obtained using the SSEARCH program plus the BLOSSUM50 or BLOSSUM62 matrices or with the BLAST server plus the BLOSSUM62 matrix. The AtHsp90 sequences were aligned with CLUSTALX, using the default alignment options with the PAM series of matrices. Gap regions in the multiple alignment were ignored, and a correction was made for multiple amino acid substitutions over time. NJPlot was used to generate the distance matrix-based unrooted phylogenetic tree. Multiple alignments and phylogenetic trees constructed from multiple sequence alignments made with either the GONNET or BLOSSUM series of matrices were extremely similar. In no case was the branch order changed, and the branch lengths varied by less than 5%.
Analysis of AtHsp90 sequences for functional domains and localization signals
Predictions of the subcellular location of A thaliana Hsp90 proteins used multiple algorithms for sequence analysis: PSORT (http://psort.nibb.ac.jp/form.html) (Nakai and Horton 1999), TargetP (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al 2000), Predotar (http://www.inra.fr/Internet/Produits/Predotar/index.html), and MITOPROT (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter) (Claros and Vincens 1996).
The AtHsp90 sequences were analyzed for functional motifs or domains using the Prosite (http://ca.expasy.org/prosite/) and Conserved Domain (www.ncbi.nlm.nih.gov/Structure/) databases.
RESULTS AND DISCUSSION
The A thaliana Hsp90 protein family
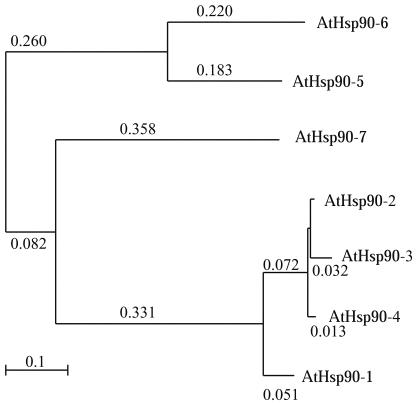
Seven members belonging to the Hsp90 family of proteins were identified in A thaliana. Aiming to be consistent with the nomenclature scheme of members of molecular chaperone families, the Hsp90 proteins are referred to herein as AtHsp90-1 through AtHsp90-7 (Table 1). The 7 members of the A thaliana Hsp90 family share at least 45% sequence identity extending along their entire lengths (Table 2). The AtHsp90-2, AtHsp90-3, and AtHsp90-4 protein sequences are highly similar; all are at least 96% identical to each other. This suggests that they may be functionally redundant. The close relatedness of AtHsp90-2, AtHsp90-3, and AtHsp90-4 is also obvious in Figure 1. Milioni and Hatzopoulos (1997) have previously reported that these 3 genes are clustered within a 15-kb genomic region, with AtHsp90-2 and AtHsp90-4 lying 1.5 kb apart in a head-to-head orientation and AtHsp90-3 having the same orientation as AtHsp90-2.
Table 2.
Percentage of identity among the Arabidopsis thaliana Hsp90 proteins
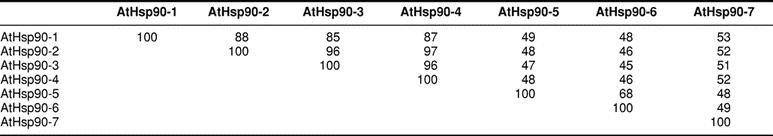
Fig 1.
Molecular phylogeny of the AtHsp90 family of genes. This diagram shows the number of amino acid substitutions per amino acid position that is observed between a given protein and the inferred ancestral protein sequence represented as the node to which the protein is joined. For example, it is estimated that an average of 0.22 amino acid substitution occurred at each position in the sequence of the AtHsp90-6 protein since it diverged from the common ancestor, leading to both it and the AtHsp90-5 protein. Branches without an associated substitution number have fewer than 0.01 substitution per position
Eukaryotic Hsp90 proteins contain 2 highly conserved domains: the adenosine triphosphate (ATP)–binding domain at the N-terminus and the highly charged (glutamic acid–rich) linker region. Crystal structures of the N-terminal domains of human and yeast Hsp90s alone or complexed with nucleotides have been determined. There exists homologous structures between the ATP-binding domains of Hsp90, DNA gyrase, the MutL DNA mismatch repair protein, and bacterial histidine kinase CheA (reviewed in Pearl and Prodromou 2000). Geldanamycin, a benzoquinoid ansamycin, binds to the ATP-binding site, displacing ATP and inhibiting Hsp90 function (Stebbins et al 1997). The recent demonstration of ATP binding by Hsp90 in vitro, together with the finding that ATP binding and hydrolysis is essential for the in vivo functions of Hsp90 (Panaretou et al 1998), has redefined our understanding of its mode of action and its interactions with other proteins.
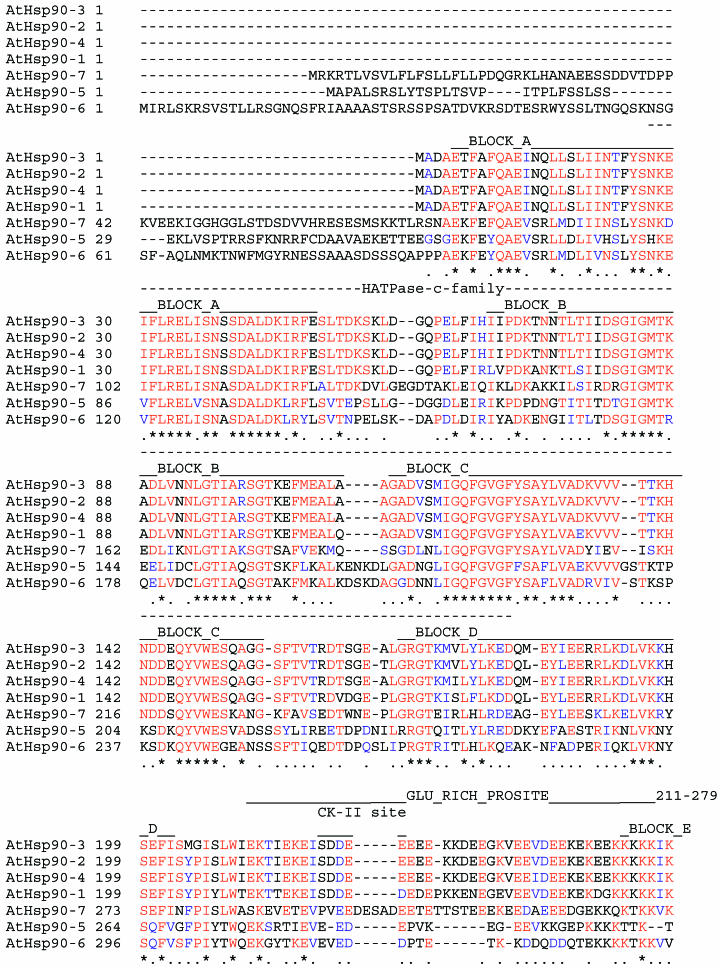
A search for conserved functional domains in A thaliana Hsp90 proteins revealed the presence of an ATP-binding domain in all members (Figure 2). Although ATP binding and hydrolysis by plant Hsp90 remain to be demonstrated, we have previously shown that plant Hsp90 can bind human p23, a component of the animal Hsp90-based cytoplasmic chaperone heterocomplex, in an ATP-dependent manner and that this binding is inhibited by geldanamycin in a temperature-dependent manner (Owens-Grillo et al 1996). The temperature-dependent binding of geldanamycin with plant Hsp90 might be the result of minor differences between the ATP-binding sites of plant and animal Hsp90s.
Fig 2.
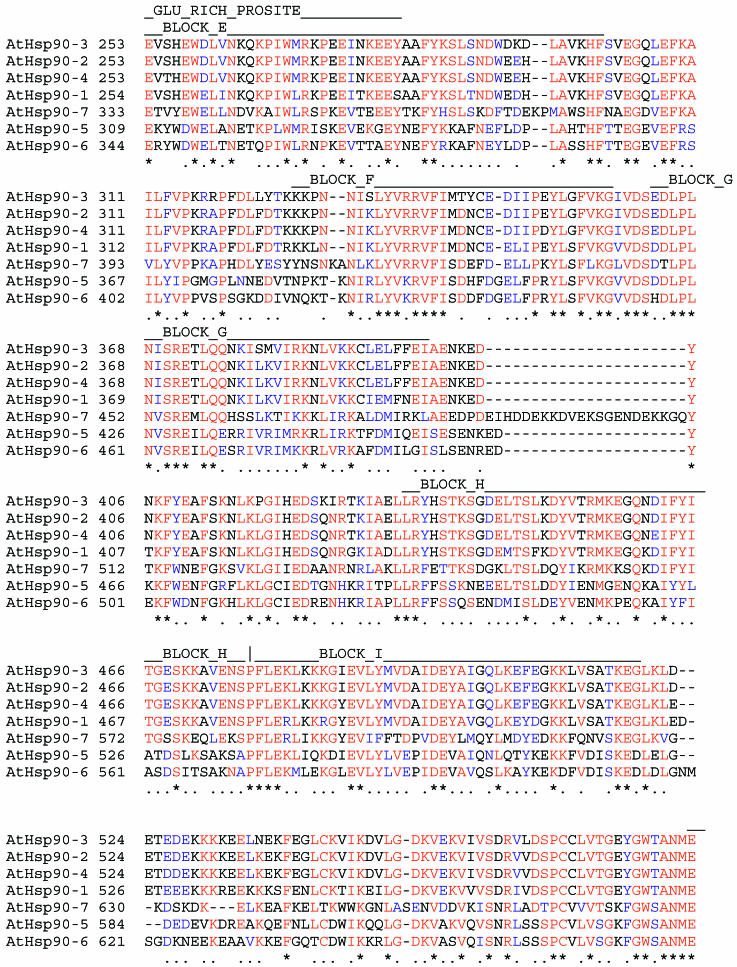
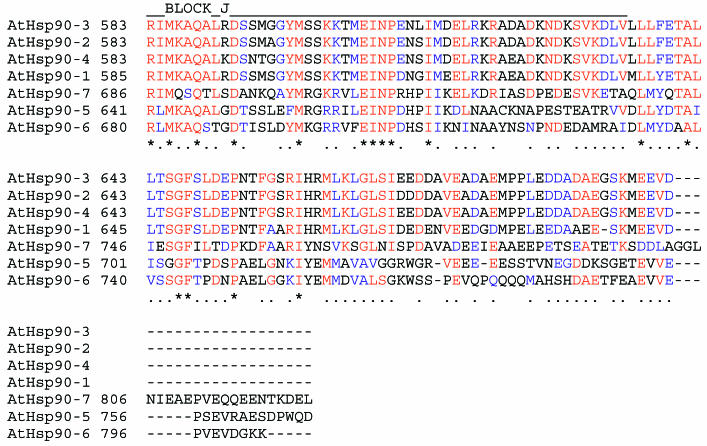
Multiple sequence alignment of members of the A thaliana Hsp90 family. The multiple alignment was generated using CLUSTALX. Positions that share greater than 70% amino acid identities are indicated in red, and positions that share greater than 70% similarity are shown in blue. The positions of conserved functional domains are named above the aligned sequences. Positions of highly related, ungapped sequences that can be considered as being characteristic of the Hsp90 family were determined by searching the BLOCKS database with AtHsp90-2 as a reference sequence. The Blocks, indicated by BLOCK_A to BLOCK_J names, are positioned on the multiple sequence alignment, using AtHsp90-2 sequence as a reference. Only AtHsp90-1 to AtHsp90-4 sequences contain all 10 BLOCKS characteristic of the Hsp90 family of proteins
Proximal to the ATP-binding domain in animal and yeast Hsp90 proteins is the charged linker region, which is variable in both length and composition among isoforms and species. The charged region has been implicated in formation of functional steroid receptor complexes with Hsp90 (Cadepond et al 1994); however, deletion from yeast Hsp90 did not cause any loss of essential functions (Louvion et al 1996). The charged linker region was detected in all Hsp90 members with the exception of AtHsp90-5 and AtHsp90-6, although both these proteins contain the other Hsp90 signature sequences (Fig 2). AtHsp90-5, AtHsp90-6, bacterial HtpG, and animal TRAP1 share a common feature; these proteins either lack or contain a very short charged region. Despite this similarity, it remains unclear if AtHsp90-5 and AtHsp90-6 originated from a common ancestor.
A casein kinase II phosphorylation site is present within the charged region of the AtHsp90 proteins (Fig 2). Although it remains to be seen if plant Hsp90 is phosphorylated similar to animal Hsp90 (Dougherty et al 1987), a recombinant Brassica napus Hsp90 was observed to undergo autophosphorylation and to phosphorylate other proteins, such as histones and casein, in the presence of Mn2+ (Park et al 1998). Hsp90s from different sources also undergo autophosphorylation in vitro, but only a porcine grp94-like protein (Dechert et al 1994), in addition to the B napus protein, has been reported to trans-phosphorylate other proteins in vitro. Further analysis of the phosphotransferase activity of Hsp90 is required to determine its physiological function.
Eukaryotic Hsp90 exists predominantly in the dimeric form, with its dimerization site being located in the 200 C-terminal residues (Minami et al 1994). A Hsp90 C-terminal truncation mutant that could not dimerize was unable to rescue a yeast strain that had both Hsp90 genes deleted (Minami et al 1994). This finding suggests that dimerization is essential for the biological activity of Hsp90. An examination of the native state of Hsp90 by nondenaturing gel electrophoresis showed that Hsp90 exists as a monomer, dimer, and high-molecular-mass complex in preparations from spinach leaves, B napus seedlings, and wheat germ, with the monomeric form being predominant (Krishna et al 1997). The functional relevance of the different forms of Hsp90 in plants and of the site directing dimerization remains to be determined.
In silico localization of the A thaliana Hsp90 proteins
Each of the AtHsp90-1 through AtHsp90-4 proteins includes the C-terminal pentapeptide MEEVD, which is diagnostic of cytoplasmic Hsp90 proteins from both plants and animals (Fig 2). Results represented in Table 2 and Figure 1 show that AtHsp90-5 and AtHsp90-6 are more closely related to one another than they are to other members of the AtHsp90 family. Unlike the genes encoding cytoplasmic Hsp90 proteins, which contain 2 to 3 introns, the AtHsp90-5 and AtHsp90-6 genes include 18 to 19 introns (Milioni and Hatzopoulos 1997). Comparison of the sequence of the plastid-specific Hsp90 homologue of rye (Schmitz et al 1996) with those of the AtHsp90 family revealed greatest similarity with AtHsp90-5 (76% identity) followed by AtHsp90-6 (64% identity). Both AtHsp90-5 and AtHsp90-6 contain additional amino acids on the N- and C-termini compared to their cytosolic counterparts. Prediction of subcellular localization of these proteins using several algorithms (PSORT, TargetP, Predotar, and MITOPROT) suggests that AtHsp90-5 is chloroplast localized and AtHsp90-6 is mitochondria localized. A 60-residue transit peptide for import into chloroplast and a 48-residue mitochondrial targeting peptide were identified in the N-terminal regions of AtHsp90-5 and AtHsp90-6, respectively. Previously, Milioni and Hatzopoulos (1997) predicted a chloroplast localization for AtHsp90-5. Since the chloroplast localization of the rye plastid protein, the closest relative of AtHsp90-5, has been experimentally confirmed (Schmitz et al 1996), there is little doubt about the location of AtHsp90-5. However, intracellular localization of AtHsp90-6 remains to be confirmed experimentally. The AtHsp90-7 sequence contains a putative 18- to 30-residue N-terminal signal sequence plus a C-terminal KDEL ER-retention motif. Additionally, the AtHsp90-7 sequence is most similar to the 2 previously identified ER-specific plant Hsp90 orthologs (Schroder et al 1993; Walther-Larsen et al 1993).
Expression of Hsp90 in A thaliana
Members of the prokaryotic Hsp90 family, HtpG, are true Hsps: present at very low levels during constitutive growth but induced to high levels by heat shock or other environmental stress conditions (Mason et al 1999). In contrast, the constitutive expression of Hsp90 in mammalian cells is relatively high and is only modestly increased by stress (Buchner 1999). Expression of Hsp90 in plant cells is developmentally regulated (Koning et al 1992; Marrs et al 1993; Krishna et al 1995; Reddy et al 1998a) and additionally responsive to cold stress (Krishna et al 1995) and light and dark transitions (Felsheim and Das 1992). Treatment of plants with the growth regulator 24-epibrassinolide (a brassinosteroid) led to increases in both transcript and protein levels (Wilen et al 1995; Dhaubhadel et al 1999).
Transcripts for AtHsp90-1 could be detected only in roots of control A thaliana plants but were abundant in all organs after heat shock or treatment with heavy metals (Yabe et al 1994). AtHsp90-2 and AtHsp90-3 transcripts could be detected in all plant organs but were abundant only in roots and flowers (Yabe et al 1994). The AtHsp90-2 and AtHsp90-3 transcript levels increased modestly after heat shock, but were substantially increased after treatment of plants with the phytohormone indoleacetic acid, 0.1 M sodium chloride, or heavy metals. Analysis of AtHsp90-5 and AtHsp90-6 expression revealed that the former is mildly induced by heat shock and that the latter is barely induced by heat shock (Milioni and Hatzopoulos 1997).
The Hsp90 chaperone complex in plants
In mammalian cells, Hsp90 promotes folding and activation of its client proteins in cooperation with a number of cochaperones (reviewed in Toft 1998; Buchner 1999; Pearl and Prodromou 2000). The Hsp90-based chaperone complex contains Hsp90, p60/Sti1/Hop, Hsp70, Hsp40 (a J-domain protein), the Hsp70 interacting protein p48/Hip, a high Mr immunophilin (immunosuppressant drug-binding protein), and p23. An additional component, p50/Cdc37, has only been detected in Hsp90 complexes with protein kinases. Although the immunophilin possesses peptidylprolyl cis-trans isomerase activity, this appears to not be required for folding and assembly of client proteins. The immunophilin is postulated to have a role in the targeted movement of the complexes. Both p60 and the immunophilin of the chaperone heterocomplex contain several copies of a degenerate 34-residue motif known as a tetratricopeptide repeat (TPR). The binding site on Hsp90 for TPR domains has been localized to the C-terminus, with the MEEVD pentapeptide being critical for binding. The p60/Sti1/Hop protein links Hsp90 and Hsp70 within the complex and inhibits adenosine triphosphatase activity of Hsp90. The p23 protein binds to ATP-ligated Hsp90 and, although it appears to stabilize Hsp90-steroid receptor complexes in vitro, deletion in yeast revealed that it is not essential for viability.
There is a growing catalog of client proteins for animal cell Hsp90, which includes several protein kinases and transcription factors (listed in Csermely et al 1998; Buchner 1999). No client protein for plant Hsp90 has been reported as yet, but an Hsp90-based chaperone complex has been identified in plant cells. Hsp70, a p60/Sti1/Hop ortholog, and high-molecular-weight immunophilins have been detected in the Hsp90 heterocomplexes (Owens-Grillo et al 1996; Stancato et al 1996; Reddy et al 1998b; Krishna et al, in preparation).
The availability of the complete A thaliana genome sequence (The Arabidopsis Genome Initiative 2000) has allowed detection of genes encoding proteins corresponding to all of the components of the mammalian Hsp90 chaperone complex. In addition to Hsp90, these include Hsp70 (Lin et al 2001), the J-domain proteins (Miernyk 2001), and Hip (Webb et al 2001). There have also been genomic analyses of the A thaliana immunophilins (Galat 2000). Furthermore, the genome includes sequences for at least 3 p60/Sti1/Hop orthologs (AtHop-1, AL080318; AtHop-2, AC007190; and AtHop-3, AC025416) and 1 p23-like protein (AL161494). Thus, all of the molecular “equipment” necessary for the Hsp90 chaperone complex is present and accounted for in A thaliana.
The identification of an Hsp90-containing chaperone complex in plant cells similar to that of animal cells suggests a role in plant cell signal transduction. The A thaliana genome contains a plethora of genes encoding protein kinases, many of which are likely to participate in control of growth and development, and these are obvious potential clients for the Hsp90 chaperone complex (Chory and Wu 2001). Plants also produce an array of steroids, including brassinosteroids, which have a demonstrated role in signaling pathways controlling growth and development (Schumacher and Chory 2000). In contrast to nuclear steroid receptors found in animals, a transmembrane receptor kinase has been shown to transduce the brassinosteroid signal in plants (He et al 2000). Nevertheless, the possibility remains that Hsp90 is involved in some aspect of the brassinosteroid signaling pathway. Future studies will focus on which of the 25 498 A thaliana gene products are clients of the Hsp90 chaperone complex.
Fig 2.
Continued
Fig 2.
Continued
Acknowledgments
We thank Professors Mark Perry and Jan Miernyk for helpful comments.
REFERENCES
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant. Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Buchner J. Hsp90 & Co.: a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- Cadepond F, Jibard N, Binart N, Schweizer-Groyer G, Segard-Maurel I, Baulieu E-E. Selective deletions in the 90 kDa heat shock protein (Hsp90) impede heterooligomeric complex formation with the glucocorticosteroid receptor (GR) or hormone binding by GR. J Steroid Biochem Mol Biol. 1994;48:361–367. doi: 10.1016/0960-0760(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Chory J, Wu D. Weaving the complex web of signal transduction. Plant Physiol. 2001;125:77–80. doi: 10.1104/pp.125.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Conner TW, LaFayette PR, Nagao RT, Key JL. Sequence and expression of a HSP83 from. Arabidopsis thaliana. Plant Physiol. 1990;94:1689–1695. doi: 10.1104/pp.94.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function and clinical applications: a comprehensive review. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- Dechert U, Weber P, Konig B, Ortwein C, Nilson I, Linxweiler W, Wollny E, Gassen HG. A protein kinase isolated from porcine brain microvessels is similar to a class of heat shock proteins. Eur J Biochem. 1994;225:805–809. doi: 10.1111/j.1432-1033.1994.0805b.x. [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Chaudhary S, Dobinson KF, Krishna P. Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol Biol. 1999;40:333–342. doi: 10.1023/a:1006283015582. [DOI] [PubMed] [Google Scholar]
- Dougherty JJ, Rabideau DA, Ianotti AM, Sullivan WP, Toft DO. Identification of the 90 kDa substrate of rat liver type II casein kinase with the heat shock protein which binds steroid receptors. Biochim Biophys Acta. 1987;927:74–80. doi: 10.1016/0167-4889(87)90067-x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Felsheim RF, Das A. Structure and expression of a heat-shock protein 83 gene of Pharbitis nil. Plant Physiol. 1992;100:1764–1771. doi: 10.1104/pp.100.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts SJ, Owen BAL, Nguyen PM, Trepel J, Donner DB, Toft DO. The Hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J Biol Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- Galat A. Sequence diversification of the FK506-binding proteins in several different genomes. Eur J Biochem. 2000;267:4945–4959. doi: 10.1046/j.1432-1327.2000.01509.x. [DOI] [PubMed] [Google Scholar]
- He Z, Wang Z-Y, Li J, Ahu Q, Lamb C, Ronald P, Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning AJ, Rose R, Comai L. Developmental expression of tomato heat-shock cognate protein 80. Plant Physiol. 1992;100:801–811. doi: 10.1104/pp.100.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Reddy RK, Sacco M, Frappier JR, Felsheim RF. Analysis of the native forms of the 90 kDa heat shock protein (Hsp90) in plant cytosolic extracts. Plant Mol Biol. 1997;33:457–466. doi: 10.1023/a:1005709308096. [DOI] [PubMed] [Google Scholar]
- Krishna P, Sacco M, Cherutti JF, Hill S. Cold-induced accumulation of Hsp90 transcripts in Brassica napus. Plant Physiol. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B-L, Wang J-S, Liu H-C, Chen R-W, Meyer Y, Barakat A, Delseny M. Genomic analysis of the HSP70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 6:201–208. doi: 10.1379/1466-1268(2001)006<0201:gaoths>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvion JF, Warth R, Picard D. Two eukaryote-specific regions of hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc Natl Acad Sci U S A. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA, Casey ES, Capitant SA, Bouchard RA, Dietrich PS, Mettler IJ, Sinibaldi RM. Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis and pollen development. Dev Genet. 1993;14:27–41. doi: 10.1002/dvg.1020140105. [DOI] [PubMed] [Google Scholar]
- Mason CA, Dunner J, Indra P, Colangelo T. Heat-induced expression and chemically induced expression of the Escherichia coli stress protein HtpG are affected by the growth environment. Appl Environ Microbiol. 1999;65:3433–3440. doi: 10.1128/aem.65.8.3433-3440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones. 2001;6:209–218. doi: 10.1379/1466-1268(2001)006<0209:tjdpoa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milioni D, Hatzopoulos P. Genomic organization of Hsp90 gene family in Arabidopsis. Plant Mol Biol. 1997;35:955–961. doi: 10.1023/a:1005874521528. [DOI] [PubMed] [Google Scholar]
- Minami Y, Kimura Y, Kawasaki H, Suzuki K, Yahara I. The carboxy-terminal region of mammalian HSP90 is required for its dimerization and function in vivo. Mol Cell Biol. 1994;14:1459–1464. doi: 10.1128/mcb.14.2.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Stancato LF, Hoffmann K, Pratt WB, Krishna P. Binding of immunophilins to the 90 kDa heat shock protein (Hsp90) via a tetratricopeptide repeat domain is a conserved protein interaction in plants. Biochemistry. 1996;35:15249–15255. doi: 10.1021/bi9615349. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Kang CY, Krishna P. Brassica napus Hsp90 can autophosphorylate and phosphorylate other protein substrates. Mol Cell Biochem. 1998;185:33–38. doi: 10.1023/a:1006884306169. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The functions of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Effective protein sequence comparison. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- Reddy RK, Chaudhary S, Patil P, Krishna P. The 90 kDa heat shock protein (Hsp90) is expressed throughout Brassica napus seed development and germination. Plant Sci. 1998a;131:131–137. [Google Scholar]
- Reddy RK, Kurek I, Silverstein AM, Chinkers M, Breiman A, Krishna P. High molecular weight FK506-binding proteins are components of heat shock protein 90 heterocomplexes in wheat germ lysate. Plant Physiol. 1998b;118:1395–1401. doi: 10.1104/pp.118.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Schmidt M, Feierabend J. Characterization of a plastid-specific Hsp90 homologue: identification of a cDNA sequence, phylogenetic descendence and analysis of its mRNA and protein expression. Plant Mol Biol. 1996;30:479–492. doi: 10.1007/BF00049326. [DOI] [PubMed] [Google Scholar]
- Schroder G, Beck M, Eichel J, Vetter H-P, Schroder J. Hsp90 homologue from Madagascar periwinkle (Catharanthus roseus): cDNA sequence, regulation of protein expression and location in the endoplasmic reticulum. Plant Mol Biol. 1993;23:583–594. doi: 10.1007/BF00019305. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Chory J. Brassinosteroid signal transduction: still casting the actors. Curr Opin Plant Biol. 2000;3:79–84. doi: 10.1016/s1369-5266(99)00038-2. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Stancato LF, Hutchison KA, Krishna P, Pratt WB. Animal and plant cell lysates share a conserved chaperone system that assembles the glucocorticoid receptor into a functional heterocomplex with Hsp90. Biochemistry. 1996;35:554–561. doi: 10.1021/bi9511649. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Naito S, Komeda Y. Isolation and analysis of the expression of two genes for the 81-kilodalton heat-shock proteins from Arabidopsis. Plant Physiol. 1992;99:383–390. doi: 10.1104/pp.99.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft D. Recent advances in the study of Hsp90 structure and mechanism of action. Trends Endocrinol Metabol. 1998;9:238–243. doi: 10.1016/s1043-2760(98)00060-5. [DOI] [PubMed] [Google Scholar]
- Walther-Larsen H, Brandt J, Collinge DB, Thordal-Christensen H. A pathogen-induced gene of barley encodes a HSP90 homologue showing striking similarity to vertebrate forms resident in the endoplasmic reticulum. Plant Mol Biol. 1993;21:1097–1108. doi: 10.1007/BF00023606. [DOI] [PubMed] [Google Scholar]
- Webb MA, Cavaletto J, Klanrit P, Thompson G. Arabidopsis thaliana orthologs of the Hsp70 interacting protein, Hip. Cell Stress Chaperones. 2001;6:247–255. doi: 10.1379/1466-1268(2001)006<0247:oiatot>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen RW, Sacco M, Gusta LV, Krishna P. Effects of 24-epibrassinolide on freezing and thermotolerance of bromegrass (Bromus inermis) cell cultures. Physiol Plant. 1995;95:195–202. [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yabe N, Takahashi T, Komeda Y. Analysis of tissue-specific expression of Arabidopsis thaliana Hsp90-family gene HSP81. Plant Cell Physiol. 1994;35:1207–1219. doi: 10.1093/oxfordjournals.pcp.a078715. [DOI] [PubMed] [Google Scholar]