Abstract
Buruli ulcer (BU) caused by Mycobacterium ulcerans is a devastating skin disease, occurring mainly in remote West African communities with poor access to health care. Early case detection and subsequent antibiotic treatment are essential to counteract the progression of the characteristic chronic ulcerative lesions. Since the accuracy of clinical BU diagnosis is limited, laboratory reconfirmation is crucial. However, currently available diagnostic techniques with sufficient sensitivity and specificity require infrastructure and resources only accessible at a few reference centres in the African endemic countries. Hence, the development of a simple, rapid, sensitive and specific point-of-care diagnostic tool is one of the major research priorities for BU. In this study, we have identified a previously unknown M. ulcerans protein, MUL_3720, as a promising target for antigen capture-based detection assays. We show that MUL_3720 is highly expressed by M. ulcerans and has no orthologs in other prevalent pathogenic mycobacteria. We generated a panel of anti-MUL_3720 antibodies and used them to confirm a cell wall location for MUL_3720. These antibodies could also specifically detect M. ulcerans in infected human tissue samples as well as in lysates of infected mouse footpads. A bacterial 2-hybrid screen suggested a potential role for MUL_3720 in cell wall biosynthesis pathways. Finally, we demonstrate that a combination of MUL_3720 specific antibody reagents in a sandwich-ELISA format has sufficient sensitivity to make them suitable for the development of antigen capture-based diagnostic tests for BU.
Author Summary
According to the recommendations of the World Health Organization, the clinical diagnosis of BU should be reconfirmed by at least two laboratory techniques. However, out of the four currently available tests, three (PCR, histopathology and cultivation of M. ulcerans) can only be performed at centralized reference laboratories; the fourth (microscopic detection of acid fast bacilli) lacks the required sensitivity and specificity. Therefore, a simple tool for early diagnosis of the disease, which can be implemented in rural health care facilities of the endemic countries, is of urgent need. In this study we aimed at the identification of M. ulcerans proteins as potential targets for the development of a simple and rapid diagnostic antigen detection assay. Among 36 proteins, MUL_3720 best met the predefined criteria of being highly expressed by M. ulcerans and not having orthologs in other pathogenic mycobacterial species prevalent in the endemic regions. Here we generated monoclonal and polyclonal antibodies against this protein and carried out pilot studies for the development of an antigen capture-based diagnostic test.
Introduction
Buruli ulcer (BU) is a neglected mycobacterial skin disease, reported from tropical and subtropical countries world-wide with highest incidence rates in Western Africa [1]. Populations in rural areas with limited access to health facilities are most affected and often seek medical advice at late disease stages [2]. Advances in the clinical management of BU have shifted options for treatment from surgical resection to combination antibiotic therapy [1]. While PCR analysis targeting the insertion sequence IS2404 has evolved into the gold standard for laboratory diagnosis of BU, this test is only available at a few reference centres. Therefore, the diagnosis of BU is currently often based on clinical findings and antibiotic therapy is started before laboratory diagnostic results can be obtained. BU has a wide range of clinical manifestations including non-ulcerative forms such as subcutaneous nodules or papules, plaques and oedema, which may progress to chronic ulcerative lesions. Due to this diversity of disease presentations the accuracy of clinical diagnosis is limited [1, 3–5] and thus a significant proportion of patients reporting with skin lesions may not receive adequate treatment. This includes cases of cutaneous tuberculosis which may be misdiagnosed as BU and thus receive the recommended eight week course of Streptomycin/Rifampicin combination chemotherapy for BU [5], which is much too short for the treatment of tuberculosis. As for IS2404 PCR, two of the other three currently applied methods for laboratory reconfirmation of BU—histopathology and cultivation of the extremely slow-growing mycobacteria—equally require expensive equipment and expertise [4, 6–8] not accessible at peripheral health facilities. The only available point-of-care diagnostic test, direct-smear examination by microscopy for the detection of acid fast bacilli (AFB), has limited sensitivity and specificity [6]. Hence, one of the major research priorities for BU is the development of a fast, low-tech, sensitive and specific point-of-care diagnostic test, which can be directly implemented at peripheral health centres.
The development of a specific point-of-care diagnostic test for the detection of M. ulcerans is complicated by the broad antigenic cross-reactivity among the various mycobacterial species. Serological approaches targeting the few M. ulcerans-specific antigens identified, turned out to be not suitable for differentiation between BU patients and exposed control individuals, as both groups may or may not exhibit serum IgG titers against these antigens [9–11].
In recent years, point-of-care tests in the form of antigen capture assays have successfully been developed for tropical infectious diseases [12]. Extensive studies focussing on rapid diagnostic tests for malaria [13–17] have paved the way for the development of antigen capture based assays for other diseases such as dengue fever [18, 19], hepatitis C [20, 21] or visceral leishmaniasis [22] to name but a few.
In the present work we aimed at the identification of targets for the development of an antigen capture test for the diagnosis of M. ulcerans infection by using a proteomics approach.
Materials and Methods
Ethics statement
Ethical clearance for the analysis of clinical specimens was obtained from the Cameroon National Ethics Committee (N°172/CNE/SE/201) and the Ethics Committee of Basel (EKBB, reference no. 53/11). Immunization of mice for the generation of monoclonal antibodies was performed in strict accordance with the rules and regulations for the protection of animal rights (“Tierschutzverordnung”) of the Swiss “Bundesamt für Veterinärwesen”.
All animal infection experiments performed were approved by the animal welfare committee of the Canton of Vaud (authorization number 2261) and were conducted in compliance with the Swiss animal protection law under BSL-3 conditions.
Mycobacterial strains
In this study we analyzed M. ulcerans isolates from Ghana (NM20/02), Côte d’Ivoire (ITM 940511), Togo (ITM 970680), China (ITM 98912), Japan (ITM 8756) and Australia (JS5147) as well as additional mycobacterial strains including M. abscessus (ATCC 19977), M. avium (MAC101), M. chelonae (DSM 43804), M. fortuitum (ATCC 49403), M. gordonae (Pasteur 14021.001), M. haemophilum (ATCC 29548), M. intracellulare (clinical isolate), M. kansasii (NCTC 10268), M. lentiflavum (clinical isolate), M. malmoense (NCTC 11298), M. marinum (ATCC 927), M. scrofulaceum (Pasteur 14022.0031), M. simiae (clinical isolate) M. smegmatis (Pasteur 14133.0001), M. terrae (clinical isolate), M. xenopi, M. bovis (ATCC 35734) and M. tuberculosis (Pasteur 14001.0001). M. ulcerans strains were grown in BacT/Alert culture bottles supplemented with enrichment medium according to the manufacturer’s protocol (bioMérieux).
Preparation of protein lysates
For the preparation of M. ulcerans protein lysates, bacteria (5 ml of culture, OD600~1) were washed in PBS, heat-inactivated at 95°C for 35 min, centrifuged at 10′000 × g for 10 min and resuspended in 400 μl lysis buffer (PBS containing 5% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF) and a protease inhibitor cocktail (complete mini, Roche)). The mix was transferred to lysing tubes (Precellys) and homogenized using a mechanical bead beater device (Precellys 24, Bertin Technologies) twice at 6′800 rpm for 30 s. Beads and non-lysed cells were removed by centrifugation at 10′000 × g for 10 min. The preparation of lysates of other mycobacterial species was described previously [9].
2D gel electrophoresis of M. ulcerans lysates
90 μg of trichloroacetic acid (TCA) precipitated M. ulcerans (NM20/02) protein lysate was resuspended in rehydration buffer (8 M urea, 2% 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.5% (v/v) ZOOM Carrier Ampholytes (Invitrogen), 0.002% bromophenol blue and 0.4% dithioerythritol (DTE)). The mix was incubated with a 3–10 pH gradient strip (ZOOM Strip; Invitrogen) over night (ON) at room temperature (RT). First-dimension isoelectric focusing (IEF) was performed on a ZOOM IPG runner (Invitrogen) using a step voltage protocol (175 V for 15 min, 175–2000 V for 45 min, 2000 V for 2 h). After IEF, the strips were incubated for 15 min with equilibration buffer (6 M urea, 50 mM Tris pH 8.8, 30% glycerol, 2% SDS, 30 mM DTE) followed by a 15 min incubation period with alkylating solution (6 M urea, 50 mM Tris (pH 8.8), 30% glycerol, 2% SDS, 0.23 M iodacetamide). Second-dimension gel electrophoresis was performed at 200 V for 35 min using a 10% NuPAGE Novex Bis-Tris ZOOM Gel (Invitrogen). The gel was stained with Coomassie blue (Invitrogen).
In-gel tryptic digestion of proteins
All Coomassie stained protein spots were selected for mass spectrometry analysis. Spots were excised from the 2D gel, placed in a low-binding microcentrifuge tube and destained in 0.1 M ammonium bicarbonate / 30% acetonitrile at 30°C. Gel spots were dried in a SpeedVac concentrator and digested with 4 μl of 10 μg/ml trypsin (trypsin porcine, Roche Applied Science) ON at 37°C. Peptides were extracted from gel pieces with 4 μl of 0.3% trifluoroacetic adic (TFA) / 50% acetonitrile. The samples were desalted and concentrated using ZipTipC18 tips (Millipore). Eluted peptides were loaded onto a MALDI target.
Mass spectrometry (MS) and data analysis
MS analysis was performed using a MALDI-TOF mass spectrometer (Bruker ultraflex III TOF/TOF, Bruker Daltonics Inc.) in the reflector mode. 1 μl of tryptic digest and 1 μl of matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid, 50% acetonitrile, 0.1% TFA) were spotted onto a MALDI target (MTP AnchorChip 600/384, Bruker Daltonics) and allowed to co-crystallize at room temperature. Data were processed using FlexAnalysis software (Bruker Daltonics flexAnalysis 2.4). Spectra were smoothed (Sawitzgy Golay algorithm, 0.2 m/z width, 1 cycle), baseline subtracted (median algorithm, 0.8 flatness) and calibrated using trypsin autocleavage or internal standard peptide mass peaks. A monoisotopic peak list was generated from the spectrum using SNAP algorithm and analyzed with BioTools (Bruker Daltonics BioTools 3.0). Peptide mass fingerprinting searches were performed using the Aldente search engine on the Expasy server.
Recombinant expression of full length MUL_3720 and truncated MUL_3720
The full length MUL_3720 (aa 1–207) and a truncated version of this protein lacking the lectin domain (aa 115–207) were recombinantly expressed in Escherichia coli BL21 Star (DE3, Invitrogen) as N-terminal hexahistidin-tagged fusion proteins. Briefly, PCR was performed on a pUC57 vector containing the DNA sequence of MUL_3720 generated by gene synthesis (Genscript), including NdeI and NotI restriction sites. The amplified sequences were inserted into a TOPO-TA cloning vector using the TOPO Cloning Kit and introduced into E. coli (Top 10, Invitrogen). The vector was digested with NdeI and NotI (New England Biolabs) and the sequence was ligated into a pET28a expression vector using the Rapid DNA Ligation Kit (Roche). E. coli BL21 Star (DE3, Invitrogen) were grown in Luria-Bertani (LB) medium until an OD600 of ~0.5. Protein expression was induced by addition of isopropyl thiogalactoside (IPTG) to a final concentration of 1 mM and subsequent incubation for 3 h at 37°C. Bacteria were lysed by sonication and His-tagged proteins were purified by nickel-nitrilotriacetic acid (Ni-NTA) chromatography.
Construction of a MUL_3720 overexpressing M. ulcerans strain
MUL_3720 was amplified from genomic M. ulcerans DNA and cloned into TOPO vector using NdeI and ScaI restriction sites. Electrocompetent E. coli TOP10 cells (Invitrogen) were transformed with TOPO::MUL_3720 vectors and spread onto LB-Ampicillin (50 μg/ml) agar. Plasmid DNA of the mutant colonies was prepared and inserts with correct size and sequence were excised from TOPO by NdeI/ScaI and ligated into the mycobacterial vector pSD5. Chemically competent E. coli TOP10 were transformed with pSD5::MUL_3720 and grown on LB-Kanamycin (50 μg/ml) plates. Plasmid DNA was prepared and the presence of the insert was confirmed.
M. ulcerans strain NM20/02 was grown in BacT bottles (bioMérieux) containing enrichment medium (bioMérieux). Bacteria were harvested and washed twice with 10% glycerol or distilled water. Competent M. ulcerans bacteria were electroporated (2.5 kV, 1000 Ohm, 25 μF) with varying amounts (50–1000 ng) of DNA, transferred to MGIT-OADC medium (BD) and grown under non-selective conditions for 36 hours at 30°C. After the recovery phase, bacteria were spread on 7H10-Kanamycin (25 μg/ml) agar and incubated for several months at 30°C. Colonies were picked and regrown on selective agar and in BacT bottles (bioMérieux) in order to prepare lysates and stocks.
Generation of antibodies against MUL_3720
For the preparation of mAbs, mice were immunized two times intraperitoneally with 40 μg of recombinant full length MUL_3720 (aa 1–207) emulsified in Immune Easy adjuvant (Qiagen). Two weeks after the second immunization serum antibody titres against MUL_3720 (aa 1–207) as well as against the truncated MUL_3720 (aa 115–207) were determined by ELISA. Based on these results one BALB/c mouse was selected to receive a final intraperitoneal injection of 40 μg of recombinant MUL_3720 (aa 1–207) without adjuvant. Three days after this last booster dose, hybridoma cell lines were generated as described previously [23]. Briefly, the spleen of the selected mouse was removed and the spleen cells were fused with mouse myeloma cells (PAI cells). After a few days, cell culture supernatants were tested for the presence of anti-MUL_3720 (aa 1–207) as well as anti-MUL_3720 (aa 115–207) antibodies. Positive cell lines were cloned by limiting dilution and expanded. MAbs were purified using HiTrap rProtein A column (Amersham Biosciences). Two individual fusion experiments resulted in 24 and 17 MUL_3720-ELISA positive B-cell hybridoma cell lines, respectively. Of these, a total of 5 B-cell hybridoma cell lines (JD3.1, JD3.2, JD3.3, JD3.4, JD3.6, JD3.7) were successfully cloned and expanded for antibody production.
Rabbit polyclonal antibodies were generated and affinity purified by Eurogentech. New Zealand white Rabbits were injected intramuscularly with 20 μg recombinant MUL_3720 (aa 1–27) with Sigma Adjuvant System (SZ3398) or Imject Alum (SZ3403) on day 0, 14, 28 and 56. Total IgG was purified from antiserum collected on day 66 by protein A affinity chromatography.
ELISA
96-well Immulon microtiter plates (Thermo Scientific) were coated with 1 μg recombinant MUL_3720 (aa 1–207) or MUL_3720 (aa 115–207) per well in 100 μl PBS and incubated ON at 4°C. Plates were washed three times with washing buffer (2.5% Tween 20 in dH2O) and blocked with 5% non-fat dry milk in PBS containing 0.1% Tween for 1 h at 37°C. After washing as described above, 100 μl of the primary antibody (mAbs or hybridoma supernatant) was added and incubated for 2 h at 37°C. Following an additional washing step, 100 μl of 1:30′000 diluted goat anti-mouse IgG (γ-chain specific) antibodies coupled to alkaline phosphatase (SouthernBiotech) was added to each well and incubated for 1 h at 37°C. Plates were washed and 100 μl/well of phosphatase substrate solution (1 mg/ml p-Nitrophenyl phosphate in substrate buffer) was added and incubated for 1 h at 37°C. Absorbance at 405 nm was measured with a microplate reader (Tecan Sunrise).
Western blot analysis
2 μg of mycobacterial protein lysates per lane were separated on NuPAGE Novex 4–12% Bis-Tris ZOOM Gels (Invitrogen) using NuPAGE MES SDS Running Buffer (Invitrogen) under reducing conditions. After electrophoresis proteins were transferred onto nitrocellulose membranes using an iBlot gel transfer device (Invitrogen). Membranes were blocked with blocking buffer (5% non-fat dry milk in PBS) ON at 4°C. Membranes were then incubated in blocking buffer containing anti-MUL_3720 IgG (mouse mAbs JD3.2, JD3.4 or rabbit polyclonal IgG SZ3398) or mouse mAb DD3.7 (specific for a conserved mycobacterial protein) serving as loading control for 1 h at RT. After washing, membranes were incubated with secondary goat anti-mouse IgG (γ-chain specific) (HRP, SouthernBiotech) or goat anti-rabbit IgG (Fc fragment specific) (HRP, Milan) for 45 min at RT. After washing, bands were visualized by chemiluminescence using the ECL Western Blotting substrate (Pierce).
Histology
Immunohistochemical analysis was performed on tissue or punch biopsies from different IS2404 qPCR reconfirmed patients. Tissue or punch biopsies of BU patients were removed aseptically and immediately fixed in 10% neutral buffered formalin for 24 hours. Afterwards the tissue was embedded into paraffin, cut into 5 μm thin sections and transferred onto microscopy glass slides. Immunohistochemical staining of the sections was performed after deparaffinisation, rehydration and antigen retrieval with citrate-pretreatment according to standard protocols [24]. Inactivation of endogenous peroxidase as well as prevention of unspecific binding was achieved by incubation in PBS containing 0.3% hydrogen peroxide and 1.5% horse serum for 20 min. Primary anti-MUL_3720 IgG was diluted in PBS containing 0.1% Tween-20 and added to the slides for 1 h at RT or ON at 4°C. After incubation with biotin-conjugated horse anti-mouse IgG, slides were stained using the Vector ABC and NovaRED system. Sections were counterstained with haematoxylin.
JD3.4 and JD3.2 showed a comparable staining in intensity, specificity and sensitivity. JD3.2 gave a slightly lower unspecific background staining of the surrounding tissue and was used for IHC analysis.
Immune fluorescence assay (IFA)
IFA was carried out as described previously [25]. Briefly, a pellet of M. ulcerans bacteria (OD600~0.6) was resuspended in 1.5% low-melting agarose (BioWhittaker Lonza, Basel Switzerland) and transferred to cryomodules (Applied BioSystems). Agarose blocks were embedded into paraffin, cut in 3 μm sections and transferred onto microscopy glass slides (Thermo Scientific). Bacteria were stained with mAb JD3.2 and Alexa fluor488 (Invitrogen) conjugated goat anti-mouse IgG and mounted in ProLong Gold anti-fade reagent containing 4′,6-Diamidino-2-phenylindole (DAPI; Invitrogen).
Mycobacterial protein fragment complementation
The system described for investigating protein interactions by the functional reconstitution of a murine dehydrofolate reductase domain in M. tuberculosis [26] was modified here for use in M. ulcerans. N-terminal and C-terminal fusions of the bait domains to full length MUL_3720 were constructed using pUAB400 or pUAB200. Cloning was facilitated by the MfeI and ClaI restriction sites in the pUAB multiple cloning sites. All cloning was performed using E. coli DH10B and confirmed by Sanger sequencing. Plasmids were extracted from E. coli using mini-prep columns (Qiagen) and plasmid DNA was used to transform M. smegmatis MC2155 by electroporation as previously described [27].
M. ulcerans Agy99 genomic DNA libraries were prepared by partial AciI digestion. Digested DNA between 500 bp and 3 Kbp was purified using a gel purification kit (Qiagen), ligated into ClaI digested pUAB300 and used to transform E. coli DH10B. A number of colonies were randomly selected for PCR using primers F102 (5′-agaaccaccacgaggagctcat-3′) and R102 (5′-tgatgcctggcagtcgatcgta-3′) that flank the multiple cloning site on the vector to check for insertions containing inserts within the desired size range [26]. Approximately 2 × 105 clones were subsequently collected and cultured in LB ON. Plasmid DNA maxi-preps were performed on ON cultures according to the manufacturer’s instructions (Sigma-Aldrich).
Bacteria co-transformed with plasmids containing interacting, complementary mDHFR fragments were selected on 7H11 kanamycin (25 μg/ml) and hygromycin (50 μg/ml) plates. Colonies were patched onto 7H11 kanamycin-hygromycin-trimethoprim plates and colonies resistant to trimethoprim were selected for PCR. Using primers F102 and R102, PCR products were sequenced. The sequences were used to perform BLAST against the M. ulcerans Agy99 genome. Inserts containing open reading frames in the incorrect orientation were discarded. Also removed were inserts that matched non-coding genomic DNA or the dehydrofolate reductase from M. ulcerans.
Bacteria and mouse foot pad infection model
M. ulcerans strain S1013 used for experimental infection of mice was isolated in 2010 from the ulcerative lesion of a Cameroonian BU patient [28]. Bacteria were cultivated in Bac/T medium for 6 weeks, recovered by centrifugation and a stock suspension in sterile PBS of 125 mg/ml wet weight was prepared. 30 μl of a 1:1000 dilution of the stock solution was injected subcutaneously into the left hind foot pad of 14 week old female BALB/c mice. On day 87 after infection, mice were euthanized and foot pads were aseptically removed. Foot pads were dipped into 70% ethanol, dried under the laminar flow, cut into four pieces with a scalpel and transferred to reinforced hard tissue grinding tubes (MK28-R, Precellys) containing 750 μl of Bac/T medium (bioMérieux). Tissue homogenization was performed with a Precellys 24-dual tissue homogenizer (3 × 20 s at 5000 rpm with 30 s break). After transferring the supernatant to a fresh tube, the residual tissue remains were homogenized a second time in 750 μl of Bac/T medium. Tissue lysates were pooled and stored at -80°C until further use. 500 μl of thawed tissue lysate was transferred into tough microorganism lysis tubes (VK05–2ml, Precellys), inactivated for 1 h at 85°C and centrifuged at 17′000 × g for 5 min. The pellet was resuspended in 250 μl PBS containing protease inhibitors (Roche, EDTA—free) and cells were disrupted with Precellys 24-dual tissue homogenizer (2 × 30s at 6800rpm with 1 min break in between). Lysates were cleared by centrifugation and tested by ELISA.
Antigen capture assay
Nunc-Immuno Maxisorp 96-well plates (Thermo Scientific) were coated with 10 μg/ml JD3.4 mAb (50 μl per well) in PBS and incubated ON at 4°C. Plates were washed three times with washing buffer (2.5% Tween 20 in dH2O) prior to incubation with blocking buffer (5% non-fat dry milk in PBS) for 2 h at RT. After washing as described above, 50 μl of different dilutions of the purified recombinant full length MUL_3720, M. ulcerans lysate (NM20/02), or lysates from M. ulcerans infected tissue samples in PBS were added and incubated for 2 h at RT. Following an additional washing step, 50 μl anti-MUL_3720 rabbit IgG (5 μg/ml) in blocking buffer with detergent (0.5% non-fat dry milk in PBS containing 0.05% Tween 20) was added and incubated for 2 h at RT. After washing as described above, 50 μl goat anti-rabbit IgG coupled to horseradish-peroxidase (Milan) diluted 1:10′000 in blocking buffer with detergent was added and incubated for 1 h at RT. Plates were washed and TMB peroxidase substrate solution was added. After 10 min the reaction was stopped with 2 M sulfuric acid and absorbance was measured at 450 nm with a microplate reader (Tecan Sunrise).
Results
Identification of MUL_3720
For identification of suitable proteins that could be used as targets in diagnostic test formats, an M. ulcerans whole protein lysate was analysed by 2D gel electrophoresis (S1 Fig.). In total, 384 protein spots were detected, processed and subsequently subjected to MALDI-TOF-MS. Among the 384 spots, 118 peptide fragments were identified and attributed to 36 different genes. In order to select for proteins without orthologs in M. tuberculosis, M. bovis or M. leprae, a BLAST search against the Uniprot database was performed for all 36 proteins, resulting in the identification of three potential targets (MUL_3720, MUL_0343 and MUL_4023) suitable for a selective antigen capture assay. However, MUL_0343 and MUL_4023 presented very weak protein spots in the 2D gel, while MUL_3720 showed a high expression level and was therefore selected for further analysis.
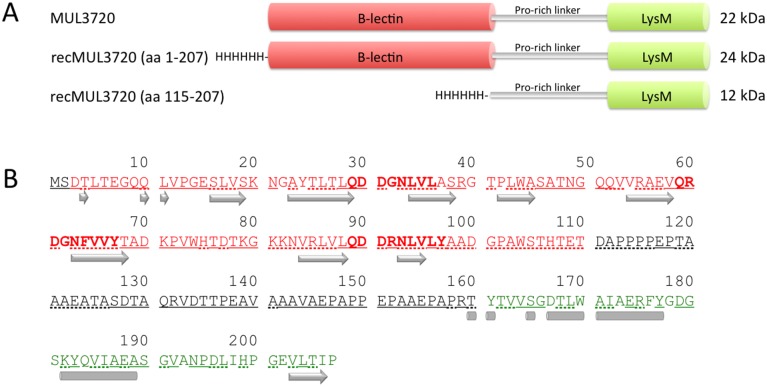
The 624 bp MUL_3720 gene encodes a protein of 207 amino acids, with a molecular mass of 22 kDa. MUL_3720 is predicted to possess an N-terminal bulb-type mannose-specific lectin domain and a C-terminal peptidoglycan-binding Lysin Motif (LysM) linked by a proline-rich sequence (Fig. 1). Database comparisons revealed the presence of orthologs with a similar domain organisation in M. abscessus (MAB_2373), M. avium, M. colombiense, M. fortuitum, M. kansasii (MKAN_05370), M. marinum (MMAR_3773), M. smegmatis (MSMEG_3662) and M. xenopi. The M. marinum ortholog displayed a sequence identity of 99% (S2 Fig.).
Fig 1. Schematic presentation of MUL_3720.
(A) The MUL_3720 protein consists of a N-terminal B-lectin domain and a C-terminal LysM domain linked by a proline rich sequence. MUL_3720 was recombinantly expressed in E. coli either as full length (aa 1–207) or truncated protein (aa 115–207) with a N-terminal hexa-histidine-tag. (B) The predicted amino acid sequence is shown with residues of the B-lectin and the LysM domain marked in red and green, respectively. The consensus sequence motifs QXDXNXVXY involved in alpha-D-mannose recognition are indicated in bold. Strands (arrows), helixes (bars), exposed (underlined) and buried (dotted underlined) regions were predicted with PredictProtein.
Generation and characterization of anti-MUL_3720 antibodies
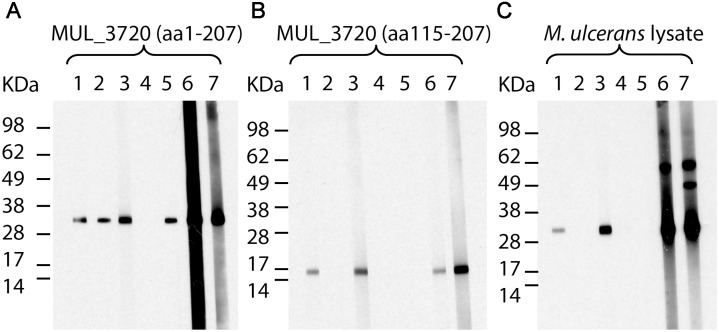
For the generation of antibodies against MUL_3720, required for the detection of the protein in diagnostic assays, we immunized mice and rabbits with the full length protein, recombinantly expressed as a His-tagged fusion protein (predicted molecular mass 24 kDa) in E. coli BL21. Hybridoma cell lines producing antibodies against different epitopes of the protein were identified by analyzing their reactivity against MUL_3720 (aa 1–207) as well as the truncated version of MUL_3720 (aa 115–207), lacking the lectin domain and consisting only of the LysM motif and the proline-rich sequence. Five mAbs (JD3.2, JD3.3, JD3.4, JD3.6 and JD3.7), all of them mouse IgG1(κ) isotype, were generated, purified and further characterized. All the mAbs recognized recombinant full length MUL_3720 (aa 1–207) in ELISA. While all antibodies except for JD3.6 recognized recombinant full length MUL_3720 (aa 1–207) in Western Blot analysis (Fig. 2A), only JD3.2 and JD3.4 also reacted with recombinant truncated MUL_3720 (aa 115–207) (Fig. 2B) and the endogenous protein in M. ulcerans lysates (Fig. 2C) (S1 Table).
Fig 2. Reactivity of MUL_3720-specific antibodies.
Mouse mAbs JD3.2 (1), JD3.3 (2), JD3.4 (3), JD3.6 (4) and JD3.7 (5) and rabbit polyclonal IgG SZ3398 (6) and SZ3403 (7) were tested for their reactivity with MUL_3720 (aa 1–207) (A), truncated MUL_3720 (aa 115–207) (B) and the endogenous MUL_3720 in a total protein lysate of M. ulcerans strain NM20/02 (C) by Western Blot analysis.
Reactivity of the generated antibodies with lysates of different M. ulcerans strains and other mycobacteria
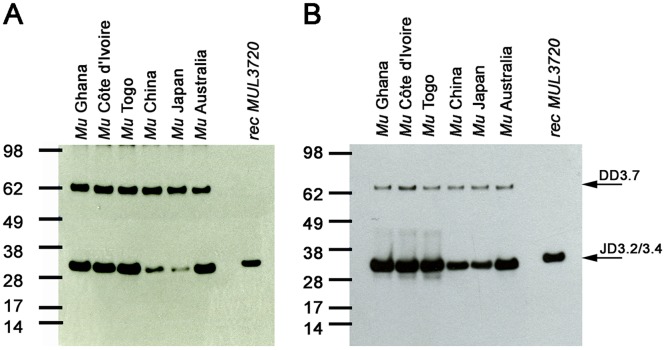
The ability of mouse mAbs JD3.2 and JD3.4 to detect endogenous MUL_3720 in lysates of M. ulcerans strains from different geographical regions was examined by Western Blot analysis. While the protein was recognized by JD3.2 and JD3.4 in all M. ulcerans strains, isolates belonging to the classical M. ulcerans lineage (Ghana, Côte d’Ivoire, Togo and Australia) showed higher expression levels as compared to isolates belonging to the ancestral lineage (China and Japan) (Fig. 3).
Fig 3. MUL_3720 is expressed by M. ulcerans strains of various geographic origin.
Total protein lysates of M. ulcerans strains from Ghana, Côte d’Ivoire, Togo, China, Japan and Australia were analyzed by Western Blot. Detection with mAbs JD3.2 (A) and JD3.4 (B) demonstrates expression of MUL_3720 in strains from different geographical regions, while staining with mAb DD3.7 served as a loading control. MUL_3720 expression levels are lower in M. ulcerans strains belonging to the ancestral (China, Japan) as compared to those of the classical lineage (Africa and Australia).
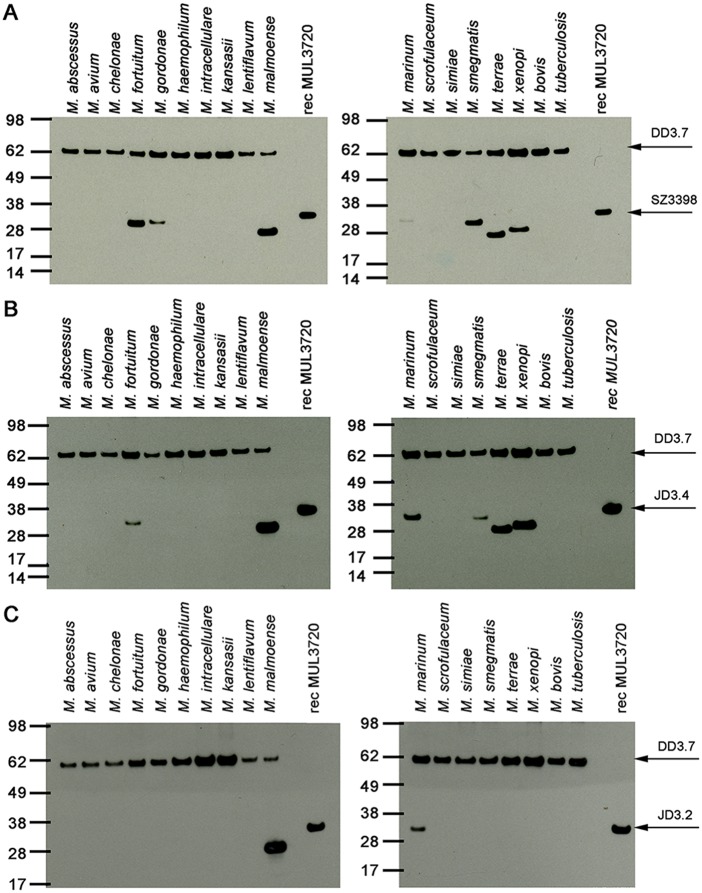
Interspecies cross-reactivity of MUL_3720 was determined by Western Blot analysis with lysates of a range of different mycobacterial species (Fig. 4). In accordance with the BLAST search for MUL_3720 orthologs (S2 Fig.), rabbit polyclonal anti-MUL_3720 IgG reacted with proteins in lysates of M. fortuitum, M. marinum, M. smegmatis and M. xenopi. In agreement with a shorter linker between the lectin and the LysM domains (S2 Fig.), the M. xenopi ortholog was detected at a lower molecular weight. The predicted orthologous proteins in M. abscessus, M. avium and M. kansasii were not recognized by the rabbit polyclonal IgG. Furthermore, a protein band was observed in lysates of M. gordonae, M. malmoense and M. terrae for which no sequence information is available (Fig. 4A). The detected proteins in M. malmoense and M. terrae were slightly smaller than MUL_3720.
Fig 4. Analysis of mycobacterial lysates for the presence of MUL_3720 orthologs.
Total protein lysates of different mycobacterial strains were analyzed by Western Blot. MAb DD3.7 served as a loading control. (A) Detection with pAb SZ3398 demonstrates expression of MUL_3720 orthologs in some of the mycobacteria tested, but not in M. tuberculosis and M. bovis. (B) MUL_3720 orthologs in additional mycobacterial species were detected with JD3.4, indicating the recognition of different protein epitopes. (C) Analysis with mAb JD3.2 led to the detection of MUL_3720 orthologs in M. malmoense and M. marinum.
Analysis with JD3.4 led to a similar staining pattern among the mycobacterial lysates, except for the protein expressed by M. gordonae, which was not recognized (Fig. 4B). JD3.2 only reacted with protein in lysates of M. malmoense and M. marinum, suggesting that the two mAbs JD3.2 and JD3.4 recognize different epitopes of MUL_3720 (Fig. 4C).
Localization of MUL_3720
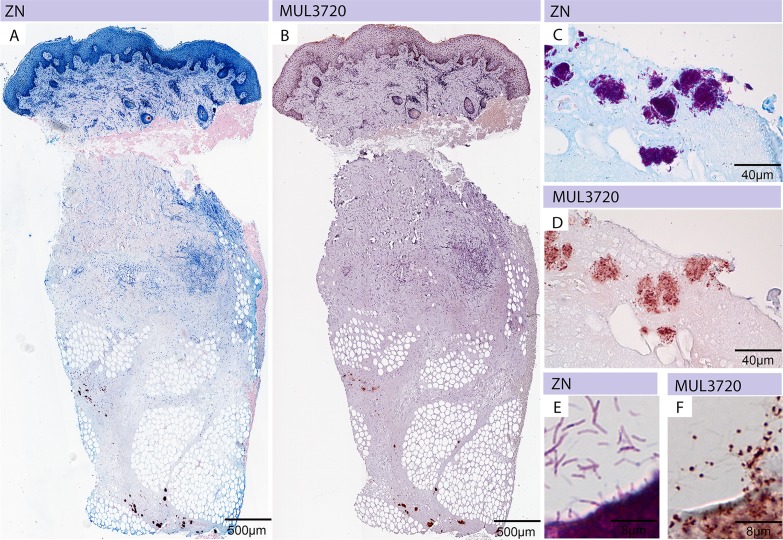
In order to confirm the expression of MUL_3720 by M. ulcerans and the ability of the anti-MUL_3720 mAb JD3.2 to detect the protein in vivo, we performed immunohistochemistry and immunofluorescence stainings. M. ulcerans could be detected in punch biopsies of human BU patients with the mAbs JD3.2 (Fig. 5) and JD3.4. ZN staining (Fig. 5 A, C, E) and JD3.2 staining (Fig. 5 B, D, F) of serial sections showed identical localization at the same tissue region. AFBs detected by ZN staining in tissue sections revealed a homogeneous staining pattern, whereas immuno-staining with mAb JD3.2 exhibited a heterogeneous staining pattern of the bacteria with intensively stained poles (Fig. 5), indicating a higher expression of the protein in these areas in the natural environment of the bacteria. In contrast, IFA of in vitro cultivated bacteria showed a more homogeneous distribution of the protein on the bacterial cell surface. This localization was confirmed in a MUL_3720 overexpressing M. ulcerans strain (Fig. 6).
Fig 5. Immunohistochemical analysis of M. ulcerans using anti-MUL_3720 antibodies.
Histological sections were either stained with carbolfuchsin (counterstain methylenblue) (A, C, E) or with specific monoclonal antibodies (JD3.2) against MUL_3720 (B, D, F). (A) A punch biopsy of a BU patient showing the typical histopathological hallmarks of an active lesion and harbouring large clusters of AFBs inside the subcutaneous tissue (purple clumps) is depicted. (B) Staining of the same tissue specimen with anti-MUL_3720 antibodies. Serial sections were stained by ZN (C) and anti-MUL_3720 antibodies (D). While the whole bacterium is stained by ZN (E) single dots are detected by using anti-MUL_3720 antibodies (F).
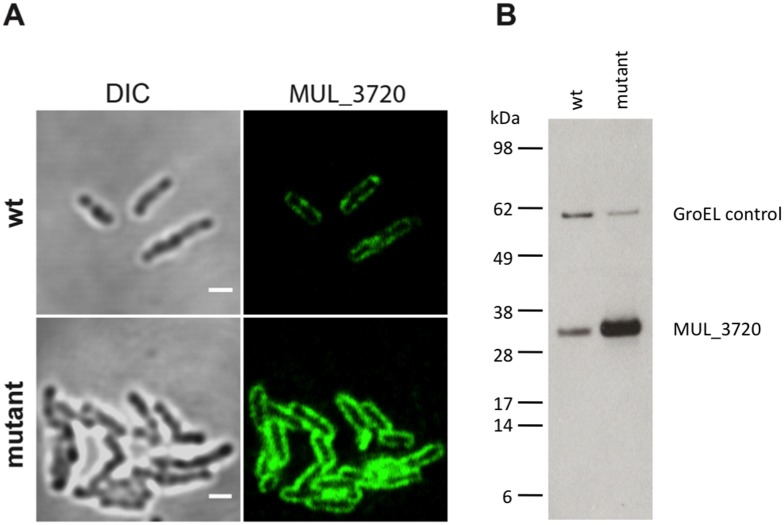
Fig 6. Localization of MUL_3720.
Localization of MUL_3720 in the wildtype or overexpressing M. ulcerans strain. (A) IF analysis (magnification 1000x) shows that MUL_3720 is homogeneously distributed at the cell surface. Primary JD3.2 anti-MUL_3720 antibody was used at a concentration of 0.1 μg/ml. Bars: 2 μm (B) Western Blot analysis of M. ulcerans lysates underlines the high expression of MUL_3720 by the mutant strain. As for the IFA analysis, JD3.2 anti-MUL_3720 antibody was used at a concentration of 0.1 μg/ml. A GroEL-antibody was included as loading control.
Investigation of MUL_3720 function
As a first step to begin to understand the role of MUL_3720 we used a bait and prey approach to identify other M. ulcerans proteins that interacted with this protein. We employed the mycobacterium-specific protein fragment complementation (M-PFC) system. An M-PFC bait clone using a N-terminal fusion of MUL_3720 and co-transformation with a random library of M. ulcerans genomic DNA fragments in pUAB300 (prey) resulted in approximately 150 trimethoprim resistant colonies. Subsequent clones were patched and screened using primers F102 and R102 to determine the identity of the DNA sequence present in pUAB300 (Table 1). Multiple independent clones were identified for sequences encoding DesA1 (MUL_0445) and a PE-PGRS protein (MUL_0572), together with 13 other single-hit CDS, including an interaction with MUL_3720 itself. Many of the putative interacting proteins had a predicted cell wall location or role in cell wall biosynthesis, in line with the localization data for MUL_3720 revealed by mAb staining (Table 1 and Fig. 6). No interacting proteins were identified using the C-terminal MUL_3720 bait fusion, consistent with the predicted cell wall location for this domain of MUL_3720. The M-PFC detects cytoplasmic protein-protein interactions only [26].
Table 1. List of proteins interacting with the N-terminus of MUL_3720 as determined by M-PFC assay.
| Locus Tag | Product description | Predicted Length (aa) | Interacting regions (aa positions) 1 | Predicted location | No. clones 2 identified |
|---|---|---|---|---|---|
| MUL_0445 | Desaturase, DesA1 | 339 | 1–84 | CYT | 3 |
| MUL_0572 | PE-PGRS family protein | 890 | 360–478 | INT | 2 |
| MUL_2853 | PE-PGRS family protein | 403 | 1–236 | INT | 1 |
| MUL_3720 | Conserved protein | 208 | 179–207 | CYT | 1 |
| MUL_4703 | Conserved hypothetical protein | 571 | 1–251 | CYT | 1 |
| MUL_4714 | Conserved integral membrane protein | 464 | 380–463 | INT | 1 |
| MUL_1239 | Transmembrane ATP-binding protein ABC transporter | 691 | 570–690 | INT | 1 |
| MUL_2018 | Phenolpthiocerol synthesis type-I polyketide synthase, PpsB | 1515 | 1411–1514 | INT3 | 1 |
| MUL_2998 | Hydrogen peroxide-inducible genes activator, OxyR | 315 | 1–180 | CYT | 1 |
| MUL_0568 | Alpha-D-glucose-1-phosphate thymidylyl-transferase, RmlA | 289 | 9–288 | CYT | 1 |
| MUL_1214 | Conserved transmembrane protein | 317 | 139–316 | INT3 | 1 |
| MUL_4825 | Conserved protein | 69 | 1–68 | CYT3 | 1 |
| MUL_4327 | Cutinase, Cut5 | 232 | 159–231 | SEC | 1 |
| MUL_0656 | Mannosyltransferase, PimB | 384 | 264–383 | CYT | 1 |
| MUL_4092 | 3-ketoacyl-(acyl-carrier-protein) reductase | 247 | 158–246 | CYT | 1 |
Notes: 1Location is based on BLASTp searches of pUAB300 clones identified as interacting partners and relative to the M. ulcerans Agy99 sequence 2Number identified indicates the number of independent clones generated with this assay. The predicted location is based on at least one cellular location prediction search using psortB, lipoP, signalP and secretomep. 3Detected in the membrane fraction by proteomics (Tafelmeyer ref). CYT: cytoplasm, INT: integral membrane protein, SEC: secreted.
Development of an antigen capture assay using the generated immunological reagents
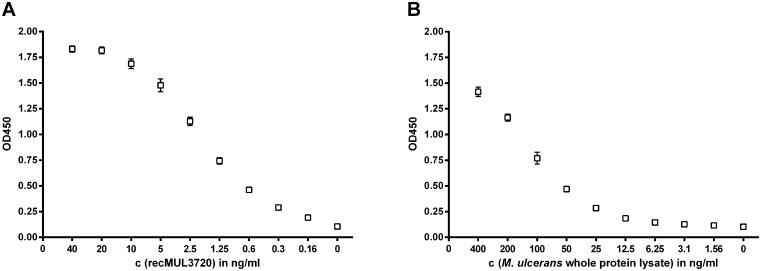
We analyzed different combinations of the generated mAbs and polyclonal IgG as MUL_3720 capturing and detecting antibodies in an antigen capture sandwich ELISA. The application of mAb JD3.4 as capturing and polyclonal rabbit IgG as detecting reagent enabled a highly sensitive detection of recombinant MUL_3720 (Fig. 7A) and the endogenous protein present in lysates of in vitro cultivated M. ulcerans (Fig. 7B).
Fig 7. Sandwich ELISA capturing MUL_3720.
Antigen capture sandwich ELISAs using mAb JD3.4 in combination with polyclonal anti-MUL_3720 rabbit IgG as capturing and detecting antibodies, respectively, were carried out to detect serial dilutions of recombinant MUL_3720 (A) and the endogenous MUL_3720 present in M. ulcerans lysates (B).
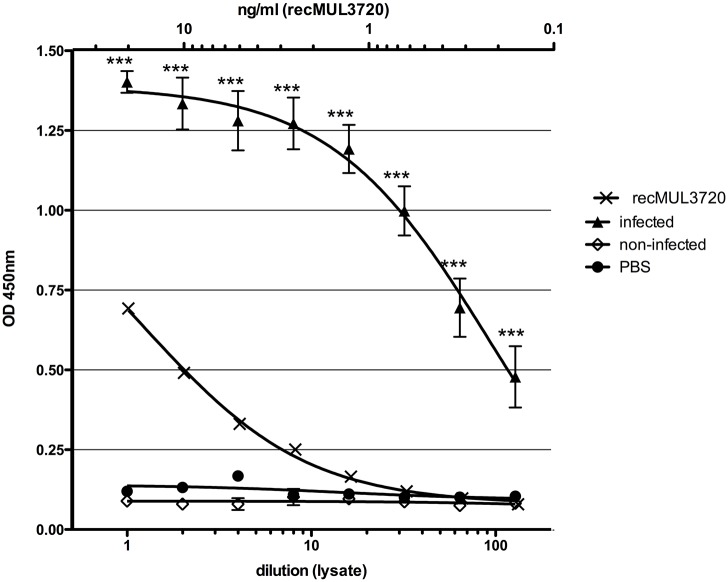
In order to test if the antigen capture ELISA is able to detect MUL_3720 expressed by bacteria in infected tissue samples, we analyzed lysates of M. ulcerans infected mouse foot pads. MUL_3720 could be detected in lysates of all five infected tissue samples analyzed, while only background readouts were obtained for lysates of uninfected foot pads (Fig. 8).
Fig 8. Detection of MUL_3720 in M. ulcerans infected mouse foot pads.
Antigen capture sandwich ELISAs using mAb JD3.4 in combination with polyclonal anti-MUL_3720 rabbit IgG as capturing and detecting antibodies, respectively, were carried out to detect MUL_3720 in lysates of mouse foot pads infected with M. ulcerans. Lysates of non-infected mouse foot pads served as negative control. Purified recombinant MUL_3720 was included as positive control. Asterisks indicate significant differences (P < 0.0001) between mean ODs of infected and non-infected mouse foot pad lysates using a two-tailed Student’s t test.
Discussion
Attempts to develop a diagnostic tool based on serological approaches have been equivocal [9–11], so we decided to focus on direct detection of M. ulcerans antigens in BU patient specimens. In the present study, we identified the MUL_3720 protein as a promising target in antigen capture-based diagnostic tests for M. ulcerans. Based on 2D gel electrophoretic analyses, MUL_3720 is one of the most highly expressed proteins in vitro. The high expression of MUL_3720 is considered an advantage with respect to developing a sensitive antigen detection test for the diagnosis of BU.
While the biological role of MUL_3720 is not known, clues to its function are suggested by its two-domain structure—a conserved bulb-type mannose-binding lectin domain and a Lysin Motif (LysM) domain—predicted to be involved in alpha-D-mannose recognition and in binding to peptidoglycan, respectively. Some bacterial species retain certain proteins attached to peptidoglycan by their LysM domains [29]. We used a mycobacteria-specific two-hybrid system to search for M. ulcerans proteins interacting with MUL_3720 and we had hits to a range of proteins known or predicted to be cell-wall associated or involved in cell wall synthesis (Table 1). Many of the interacting proteins—such as DesA1 (Table 1)—are involved in biosynthesis or modification of cell wall molecules. In other mycobacteria, the resulting double bonds from the DesA1-mediated catalysis of a desaturation reaction of saturated alkyl chains that arise during mycolic acid synthesis are required for subsequent position specific modifications such as epoxidation and cyclopropanation of this key cell wall metabolite [30, 31]. MUL_3720 appears to be arranged in an operon structure with two adjacent putative cell wall-associated protein coding genes (MUL_3721, MUL_3722). Immunofluorescence stainings of M. ulcerans bacilli confirmed the cell wall localization of MUL_3720.
With its cell wall location, the two-domain structure including a mannose-binding N-terminal cytoplasmic component and C-terminal peptidoglycan-binding component, its operon structure and a substantial list of potential interacting proteins, MUL_3720 may be an adaptor protein for multiple cell wall biosynthetic pathways. MUL_3720 might play a role in cell attachment and cell-cell interactions given its presence at the cell surface as revealed by immunofluorescence microscopy and immunohistochemical analyses.
The cell-surface localization of MUL_3720 is an additional advantage with respect to developing a sensitive diagnostic test, since the protein is expected to be easily accessible and detectable in tissue specimens of BU lesions. Potential shedding of the protein from the cell surface may facilitate ready detection in body fluids, which will be examined in future experiments.
Monoclonal and polyclonal antibodies against MUL_3720 were generated for the development of antigen capture assays. These antibodies recognized in vitro grown M. ulcerans bacilli as well as bacteria in biopsies of human BU patients, proving the expression of MUL_3720 in BU lesions.
Since these antibodies did not react with orthologs of MUL_3720 in other pathogenic mycobacterial species prevalent in the BU endemic regions, prospects for the development of a test with the desired specificity, excluding in particular cutaneous tuberculosis [5], are good. The monoclonal antibodies used for the antigen capture test bind to an epitope on the proline/rich linker and/or the LysM domain. Since the LysM domain is a widespread protein module present in more than 4000 proteins of both prokaryotes and eukaryotes [29], the potential cross-reactivity of the anti-MUL_3720 antibodies with those proteins remains to be analyzed.
Importantly, this capture assay specifically detected MUL_3720 protein in tissue lysates of M. ulcerans infected mouse footpads. Furthermore, initial results revealed that MUL_3720 could be detected in swab samples from human BU lesions with a high bacterial burden (manuscript in preparation). Ongoing optimization of the applied reagents as well as the assay format is aiming at the development of a simple test format appropriate for low-resource laboratory settings with suitable test sensitivity.
Antibiotic treatment of BU in its early stages leads in most of the cases to complete healing of the lesions with little or no trauma, whereas treatment at later stages often requires adjunct surgical treatment and is associated with prolonged hospitalization and long-term sequelae. The development of a simple and rapid diagnostic test, whose key elements are provided in the work presented here, will be of immediate benefit to BU patients in rural endemic communities. Clinical findings could directly be reconfirmed by this point-of-care test helping to avoid a false diagnosis and to facilitate a prompt onset of adequate treatment.
Supporting Information
2D gel of M. ulcerans total protein lysate run on a pH3-10 first dimension IPG strip and subsequently run on a 10% SDS-PAGE gel. Coomassie stained protein spots were excised, in-gel digested with trypsin, and subjected to MALDI-TOF MS. Identified proteins are marked as green dots and labelled with the corresponding accession number. Red dots represent proteins that could not be identified. The spots representing MUL_3720 are indicated with an arrow.
(TIF)
Multiple sequence alignment of M. ulcerans MUL_3720 and its orthologs in M. kansasii, M. abscessus, M. colombiense, M. fortuitum, M. smegmatis, M. marinum, M. avium and M. xenopi.
(TIF)
All antibodies recognized recombinant full length MUL_3720 (aa 1–207), while JD3.2 and JD3.4 as well as the polyclonal IgG also reacted with recombinant truncated MUL_3720 (aa 115–207) and the endogenous protein in M. ulcerans lysates.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Stop Buruli Initiative funded by the UBS-Optimus Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Junghanss T, Johnson R, Pluschke G (2014) Mycobacterium ulcerans disease. In: Farrar J, Hotez P, Junghanss T, Kang G, Lalloo D et al. , editors. Manson’s tropical diseases. 23rd ed [Edinburgh: ]: Saunders; pp. 519–531. [Google Scholar]
- 2. Sizaire V, Nackers F, Comte E, Portaels F (2006) Mycobacterium ulcerans infection: control, diagnosis, and treatment. Lancet Infect Dis 6: 288–296. 10.1016/S1473-3099(06)70464-9 [DOI] [PubMed] [Google Scholar]
- 3. Evans MR, Phillips R, Etuaful SN, Amofah G, Adomako J, et al. (2003) An outreach education and treatment project in Ghana for the early stage of Mycobacterium ulcerans disease. Trans R Soc Trop Med Hyg 97: 159–160. 10.1016/S0035-9203(03)90105-2 [DOI] [PubMed] [Google Scholar]
- 4. Guarner J, Bartlett J, Whitney EA, Raghunathan PL, Stienstra Y, et al. (2003) Histopathologic features of Mycobacterium ulcerans infection. Emerg Infect Dis 9: 651–656. 10.3201/eid0906.020485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bratschi MW, Njih Tabah E, Bolz M, Stucki D, Borrell S, et al. (2012) A case of cutaneous tuberculosis in a Buruli ulcer-endemic area. PLoS Negl Trop Dis 6: e1751 10.1371/journal.pntd.0001751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yeboah-Manu D, Asante-Poku A, Asan-Ampah K, Ampadu ED, Pluschke G (2011) Combining PCR with microscopy to reduce costs of laboratory diagnosis of Buruli ulcer. Am J Trop Med Hyg 85: 900–904. 10.4269/ajtmh.2011.11-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bretzel G, Siegmund V, Nitschke J, Herbinger KH, Thompson W, et al. (2007) A stepwise approach to the laboratory diagnosis of Buruli ulcer disease. Trop Med Int Health 12: 89–96. [DOI] [PubMed] [Google Scholar]
- 8. Eddyani M, Debacker M, Martin A, Aguiar J, Johnson CR, et al. (2008) Primary culture of Mycobacterium ulcerans from human tissue specimens after storage in semisolid transport medium. J Clin Microbiol 46: 69–72. 10.1128/JCM.00301-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diaz D, Döbeli H, Yeboah-Manu D, Mensah-Quainoo E, Friedlein A, et al. (2006) Use of the immunodominant 18-kiloDalton small heat shock protein as a serological marker for exposure to Mycobacterium ulcerans. Clin Vaccine Immunol 13: 1314–1321. 10.1128/CVI.00254-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yeboah-Manu D, Röltgen K, Opare W, Asan-Ampah K, Quenin-Fosu K, et al. (2012) Sero-epidemiology as a tool to screen populations for exposure to Mycobacterium ulcerans. PLoS Negl Trop Dis 6: e1460 10.1371/journal.pntd.0001460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pidot SJ, Porter JL, Marsollier L, Chauty A, Migot-Nabias F, et al. (2010) Serological evaluation of Mycobacterium ulcerans antigens identified by comparative genomics. PLoS Negl Trop Dis 4: e872 10.1371/journal.pntd.0000872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitt S (2012) Point-of-care testing for infectious tropical diseases. The biomedical scientist.
- 13. Uguen C, Rabodonirina M, De Pina JJ, Vigier JP, Martet G, et al. (1995) ParaSight-F rapid manual diagnostic test of Plasmodium falciparum infection. Bull World Health Organ 73: 643–649. [PMC free article] [PubMed] [Google Scholar]
- 14. Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH (2007) A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77: 119–127. [PubMed] [Google Scholar]
- 15. Moody A, Hunt-Cooke A, Gabbett E, Chiodini P (2000) Performance of the OptiMAL malaria antigen capture dipstick for malaria diagnosis and treatment monitoring at the Hospital for Tropical Diseases, London. Br J Haematol 109: 891–894. 10.1046/j.1365-2141.2000.01974.x [DOI] [PubMed] [Google Scholar]
- 16. Moody AH, Chiodini PL (2002) Non-microscopic method for malaria diagnosis using OptiMAL IT, a second-generation dipstick for malaria pLDH antigen detection. Br J Biomed Sci 59: 228–231. [DOI] [PubMed] [Google Scholar]
- 17. Drakeley C, Reyburn H (2009) Out with the old, in with the new: the utility of rapid diagnostic tests for malaria diagnosis in Africa. Trans R Soc Trop Med Hyg 103: 333–337. 10.1016/j.trstmh.2008.10.003 [DOI] [PubMed] [Google Scholar]
- 18. Blacksell SD, Newton PN, Bell D, Kelley J, Mammen MP, et al. (2006) The comparative accuracy of 8 commercial rapid immunochromatographic assays for the diagnosis of acute dengue virus infection. Clin Infect Dis 42: 1127–1134. 10.1086/501358 [DOI] [PubMed] [Google Scholar]
- 19. Tricou V, Vu HT, Quynh NV, Nguyen CV, Tran HT, et al. (2010) Comparison of two dengue NS1 rapid tests for sensitivity, specificity and relationship to viraemia and antibody responses. BMC Infect Dis 10: 142 10.1186/1471-2334-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian L, Wang RE, Chang YH (2012) Molecular PINCERs for biomarker analysis and their potential application in hepatitis C diagnosis. Antivir Ther 17: 1437–1442. 10.3851/IMP2469 [DOI] [PubMed] [Google Scholar]
- 21. Shivkumar S, Peeling R, Jafari Y, Joseph L, Pant Pai N (2012) Accuracy of rapid and point-of-care screening tests for hepatitis C: a systematic review and meta-analysis. Ann Intern Med 157: 558–566. 10.7326/0003-4819-157-8-201210160-00006 [DOI] [PubMed] [Google Scholar]
- 22. Maia Z, Lírio M, Mistro S, Mendes CM, Mehta SR, et al. (2012) Comparative study of rK39 Leishmania antigen for serodiagnosis of visceral leishmaniasis: systematic review with meta-analysis. PLoS Negl Trop Dis 6: e1484 10.1371/journal.pntd.0001484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pluschke G, Joss A, Marfurt J, Daubenberger C, Kashala O, et al. (1998) Generation of chimeric monoclonal antibodies from mice that carry human immunoglobulin Cgamma1 heavy of Ckappa light chain gene segments. J Immunol Methods 215: 27–37. 10.1016/S0022-1759(98)00041-6 [DOI] [PubMed] [Google Scholar]
- 24. WHO (2014) Annex 7. In: Portaels F, editor. Laboratory diagnosis of buruli ulcer. WHO/HTM/NTD/IDM/2014. 1 ed. [Google Scholar]
- 25. Vettiger A, Scherr N, Ruf M, Röltgen K, Pluschke G (2014) Localization of Mycobacterial Antigens by Immunofluorescence Staining of Agarose Embedded Cells. Mycobacterial Diseases 4:3 10.4172/2161-1068.1000150 [DOI] [Google Scholar]
- 26. Singh A, Mai D, Kumar A, Steyn AJ (2006) Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc Natl Acad Sci U S A 103: 11346–11351. 10.1073/pnas.0602817103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4: 1911–1919. 10.1111/j.1365-2958.1990.tb02040.x [DOI] [PubMed] [Google Scholar]
- 28. Bratschi MW, Bolz M, Minyem JC, Grize L, Wantong FG, et al. (2013) Geographic distribution, age pattern and sites of lesions in a cohort of Buruli ulcer patients from the Mapé Basin of Cameroon. PLoS Negl Trop Dis 7: e2252 10.1371/journal.pntd.0002252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buist G, Steen A, Kok J, Kuipers OP (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68: 838–847. 10.1111/j.1365-2958.2008.06211.x [DOI] [PubMed] [Google Scholar]
- 30. Yuan Y, Lee RE, Besra GS, Belisle JT, Barry CE (1995) Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 92: 6630–6634. 10.1073/pnas.92.14.6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dubnau E, Chan J, Raynaud C, Mohan VP, Lanéelle MA, et al. (2000) Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol 36: 630–637. 10.1046/j.1365-2958.2000.01882.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2D gel of M. ulcerans total protein lysate run on a pH3-10 first dimension IPG strip and subsequently run on a 10% SDS-PAGE gel. Coomassie stained protein spots were excised, in-gel digested with trypsin, and subjected to MALDI-TOF MS. Identified proteins are marked as green dots and labelled with the corresponding accession number. Red dots represent proteins that could not be identified. The spots representing MUL_3720 are indicated with an arrow.
(TIF)
Multiple sequence alignment of M. ulcerans MUL_3720 and its orthologs in M. kansasii, M. abscessus, M. colombiense, M. fortuitum, M. smegmatis, M. marinum, M. avium and M. xenopi.
(TIF)
All antibodies recognized recombinant full length MUL_3720 (aa 1–207), while JD3.2 and JD3.4 as well as the polyclonal IgG also reacted with recombinant truncated MUL_3720 (aa 115–207) and the endogenous protein in M. ulcerans lysates.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.