Abstract
Variations in the sugar component (ribose or deoxyribose) and the nature of the phosphodiester linkage (3′-5′ or 2′-5′ orientation) have been a challenge for genetic information transfer from the very beginning of evolution. RNA polymerase II (pol II) governs the transcription of DNA into precursor mRNA in all eukaryotic cells. How pol II recognizes DNA template backbone (phosphodiester linkage and sugar) and whether it tolerates the backbone heterogeneity remain elusive. Such knowledge is not only important for elucidating the chemical basis of transcriptional fidelity but also provides new insights into molecular evolution. In this study, we systematically and quantitatively investigated pol II transcriptional behaviors through different template backbone variants. We revealed that pol II can well tolerate and bypass sugar heterogeneity sites at the template but stalls at phosphodiester linkage heterogeneity sites. The distinct impacts of these two backbone components on pol II transcription reveal the molecular basis of template recognition during pol II transcription and provide the evolutionary insight from the RNA world to the contemporary ‘imperfect’ DNA world. In addition, our results also reveal the transcriptional consequences from ribose-containing genomic DNA.
INTRODUCTION
While the Watson–Crick base pairing serves as an important intrinsic feature of nucleic acids underlying faithful template-dependent genetic information transfer (encoded in nucleobase) from one nucleic acid polymer to another, the copying of correct nucleic acids backbone is more permissive, which results in backbone heterogeneity of nucleic acids (heterogeneity of sugar and/or phosphodiester linkage).
Backbone heterogeneity of nucleic acids is likely a persistent issue during the evolution of life from prebiotic RNA synthesis, to primitive RNA world, and to modern DNA-RNA–Protein world. Indeed, non-enzymatic replication of RNA synthesis generates a mixture of 2′-5′ and 3′-5′ RNA phosphodiester linkage (1–8). Additional/external factors such as metal ions and surrounding environment of enzyme active site help to reduce the extent of backbone heterogeneity (9,10). In the contemporary world, the majority of phosphodiester linkages of nucleic acids are 3′-5′ with a few exceptions: a lariat RNA intron containing both 3′-5′ and 2′-5′ linkage (11–13), and 2′-5′ oligomers of adenosine formed by 2′-5′-oligoadenylate synthase during the interferon antiviral response (14,15). It has also been reported that in vitro ligation of an RNA–DNA junction can generate a 2′-5′ linkage in the DNA backbone (16). In sharp contrast, a significant amount of ribonucleotides is embedded in the genomic DNA template. For example, it is estimated that over 10 000 ribonucleotides might be incorporated in a single round of nuclear DNA replication in yeast (17). In RNase H2 deficient mammalian cells, it is estimated that the rNMPs can be over 1 000 000 after one cycle of DNA replication (18–21). These results reveal that backbone heterogeneity has persisted despite of billion years of evolution.
Understanding how nucleic acid enzymes cope with backbone heterogeneity is not only important for elucidating the chemical basis of molecular recognition of backbone during genetic information transfer, but also provides new insights into the impact of backbone heterogeneity during evolution. As a template-dependent contemporary enzyme, RNA polymerase II (pol II) governs the transcription of DNA into precursor mRNA in all eukaryotic cells (22,23) and also has RNA-dependent polymerization activity (24). The observation of abundant ribonucleotides embedded in genomic DNA raises several essential questions: what are the impacts of backbone heterogeneity on pol II transcription? How does pol II recognize DNA template backbone and achieve high transcriptional fidelity for gene expression? Previous studies have revealed the molecular basis of how RNA pol II recognizes functional groups and structural features of nucleic acids during transcription (25–29). Particularly, our very recent work used a synthetic 2′-5′ DNA template as a model to dissect the phosphodiester linkage impact on pol II transcription (30). However, it is unclear how pol II processes the template DNA containing ‘natural’ backbone heterogeneity (3′-5′DNA, 3′-5′ RNA and 2′-5′ RNA), which could exist during evolution and whether pol II recognizes these three types of backbone differently.
In this work, we systematically investigated and compared the impacts of sugar backbone and phosphodiester linkage heterogeneity on pol II transcription. To this end, we generated DNA template containing all possible backbone variants (Figure 1a, 3′-5′ DNA, 3′-5′ RNA and 2′-5′ RNA) during molecular evolution. Among these backbones, we termed the DNA template containing all 3′-5′ linked deoxyribonucleotide as the ‘wild-type’ DNA template (or 3′-5′ dT template), whereas the DNA template containing a 3′-5′ linked ribonucleotide is termed as ‘sugar backbone mutant’ (or 3′-5′ rU template) to mimic the ribonucleotide incorporation in genomic DNA. To investigate the effect of the phosphodiester linkage heterogeneity, we generated the DNA template containing a site-specific phosphodiester linkage backbone mutant (2′-5′ rU template with a 2′-5′ linkage between 2′-OH of embedded ribonucleotide and 5′-end of downstream DNA template) to mimic a natural 2′-5′ phosphodiester linkage at template strand. Additionally, we generated a synthetic model template (2′-5′ dT with a 2′-5′ linkage between 2′-OH of 3′-deoxyribonucleotide and 5′-end of downstream DNA template) to help us distinguish the contribution of phosphodiester linkage change from sugar change. Comparative enzymatic kinetic studies on the effects of these backbone heterogeneities on pol II transcription along with molecular modeling revealed the chemical basis of pol II recognition of template backbone, as well as new insight into genetic information storage and transfer during molecular evolution.
Figure 1.
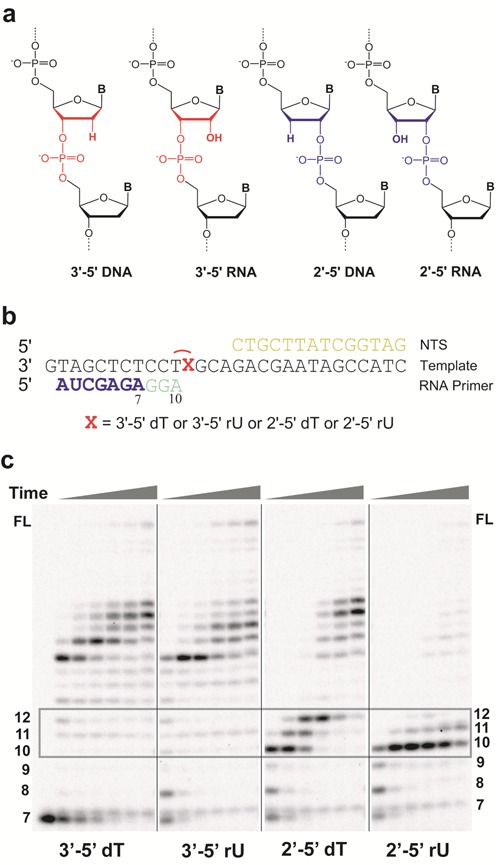
The template backbone variants have distinct impacts on RNA pol II transcription. (a) Four potential DNA template backbone variants derived from phosphodiester linkage alteration and ribose mis-incorporation. (b) Scaffolds used in run-off transcription assays. X refers to different backbone variants; 7mer RNA primer (shown in blue) was used in these scaffolds and the stalled transcript is colored in light green. The template DNA strand is shown in black. NTS stands for non-template strand (shown in yellow). (c) RNA pol II transcription products through different template backbone variants in the presence of 25 μM NTP. Time points are 30 s, 2 min, 5 min, 20 min, 1 h and 2 h from left to right. The transcript length is marked beside the gel. FL means the full length RNA transcript.
MATERIALS AND METHODS
Materials
RNA pol II was purified from Saccharomyces cerevisiae as previously described (31,32). The DNA template and non-template oligonucleotides were purchased from Integrated DNA Technologies (IDT). RNA primers were purchased from TriLink Biotechnologies and radiolabeled using [γ-32P] adenosine triphosphate (ATP) and T4 Polynucleotide Kinase (NEB). The gel-purified 2′-5′-linked DNA oligos (2′-5′-dT and 2′-5′-rU) and 3′-5′-linked DNA oligo containing a ribose (3′-5′-rU) were purchased from Gene Link Inc. The pol II elongation complexes for transcription assays were assembled using established methods (33,34).
In vitro transcription assays
The pol II elongation complexes for transcription assays were assembled using established methods (33,34). Briefly, an aliquot of 5′-32P-labeled RNA was annealed with a 1.5-fold amount of template DNA and two-fold amount of non-template DNA to form RNA/DNA scaffold in elongation buffer (20 mM Tris–HCl (pH 7.5), 40 mM KCl, 5 mM MgCl2). An aliquot of annealed scaffold of RNA/DNA was then incubated with a four-fold excess amount of pol II at room temperature for 10 min to ensure the formation of a pol II elongation complex. The pol II elongation complex is ready for in vitro transcription upon mixing with equal volumes of nucleoside triphosphates (NTPs) solution of various concentrations. The quenched products were analyzed by denaturing polyacrylamide gelelectrophoresis (PAGE) and visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad).
Single turnover nucleotide incorporation assays
The assay was carried out as previously described (33,34). Briefly, nucleotide incorporation assays were conducted by pre-incubating 50 nM scaffold with 200 nM pol II for 10 min in elongation buffer at 22°C. The pre-incubated enzyme:scaffold complex was then mixed with an equal volume of solution containing 40 mM KCl, 20 mM Tris–HCl (pH 7.5), 10 mM dithiothreitol (DTT), 10 mM MgCl2 and two-fold concentrations of various nucleotides. Final reaction concentrations after mixing were 25 nM scaffold, 100 nM pol II, 5 mM MgCl2 and various nucleotide concentrations in elongation buffer. Reactions were quenched at various times by addition of one volume of 0.5 M ethylenediaminetetraacetic acid (EDTA) (pH 8.0).
TFIIS cleavage assays
Recombinant transcription factor IIS (TFIIS) was purified as described (33,34). Cleavage reactions were performed by pre-incubating pol II with various scaffolds as previously described with slight modification. The solution was then mixed with an equal volume of solution containing TFIIS and MgCl2 in elongation buffer. Final reaction conditions were 100 nM pol II, 25 nM scaffold, 1.5 μM TFIIS and 5 mM MgCl2. Reactions were quenched at various time points by addition of one volume of 0.5 M EDTA (pH 8.0). Products were separated by denaturing PAGE as described above.
Kinetic data analysis
Nonlinear-regression data fitting was performed using Prism 6. The time dependence of product formation was fit to a one-phase association equation (1) to determine the observed rate (kobs). The substrate concentration dependence was fit to a hyperbolic equation (2) to obtain values for the maximum rate of NTP incorporation (kpol) and apparent Kd (Kd,app) governing NTP binding essentially as described (Supplementary Figure S3).
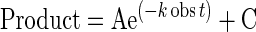 |
(1) |
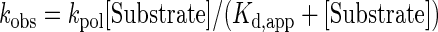 |
(2) |
The specificity constant was determined by kpol/Kd,app. Discrimination was calculated as the ratio of specificity constants governing two different nucleotide incorporation events as described (33,34).
Molecular modeling of the pol II elongation complex
The model was built based on the crystal structure of pol II complex (PDB ID: 2E2H (35)) with bound NTP and closed trigger loop. The 2′-5′ DNA and RNA were introduced based on previous structural studies (36,37). Missing residues were added and built following the same procedure used previously (38). The missing O3′ atom of the primer terminal RNA was added and the coordination modes of the Mg2+ A were fixed (38). To be consistent with the experimental scaffold named ‘10A’, the GTP in the crystal structure was mutated to ATP by atom replacement. Other parts of the crystal structure, including the downstream DNA, DNA–RNA hybrid, Mg2+ B and eight Zn2+ ions, were retained. The parameters for protein, nucleic acids and ions were taken from Amber99sb force field (39). The partial charges of ATP and nucleotide mutant with O2′ were generated by adopting the similar procedure as used in Amber99sb: structural optimization (HF/6-31G**), followed by RESP fitting to the quantum calculation at HF/6-31G* level. The quantum calculations were performed using Gaussian03 (40). The parameters of the bonded terms for ATP and nucleotide mutants were chosen from the existing and similar parameters in Amber99sb force field (39). Each system was solvated in a triclinic box of TIP3P water molecules (41) with the minimum solute-wall box distance of 10Å. Na+ ions were added to neutralize the whole system. A 10 000-steps energy minimization with the steepest descent algorithm was performed for the system using Gromacs 4.5 (42). Similar procedures were applied to build and simulate the pol II complex (with a 2′-5′ phosphodiester linkage connecting i + 1 and i + 2 nucleobases) based on crystal structure (PDB ID: 3HOZ).
RESULTS
Impacts of template backbone heterogeneity on pol II transcription efficiency
To test how site-specific backbone variants (Figure 1b) affect RNA pol II elongation, we first performed a runoff transcription assay in the presence of all four NTPs. Intriguingly, we observed distinct pol II behaviors when pol II encounters and bypasses these backbone variant sites. As shown in Figure 1c, pol II can elongate efficiently along the wild-type 3′-5′ dT template (no obvious pausing between 10–12 nt positions). Interestingly, replacement of a 3′-5′ dT with a 3′-5′ rU has minor effect on pol II transcription elongation: no obvious pausing bands at 10–12 nt positions at 25 μM NTP (Figure 1c) and minor slowdown of pol II transcription at lower NTP concentration (Supplementary Figure S1). Strikingly, as we changed the phosphodiester linkage to 2′-5′ rU, we observed that pol II stalled near the 2′-5′ rU site (10 nt (major stall site, after nucleotide incorporation opposite dT) and 11 nt positions (minor stall sites, after nucleotide incorporation opposite rU)). We also observed that bypass of the 2′-5′ rU site is extremely difficult as little elongation RNA transcripts (longer than 12 nt) were observed even after 2 h incubation, indicating that a single 2′-5′ rU site is a strong block for transcription elongation. It is also important to note that the pol II behavior in processing a 2′-5′ rU site is also in sharp contrast with pol II processing a 2′-5′ dT site. We observed three strong pol II pausing sites near the 2′-5′ dT site (pausing at 10–12 nt positions). However, pol II can eventually bypass the 2′-5′ dT linkage site to generate longer transcripts upon extended incubation as previously observed (30). Taken together, we revealed that pol II tolerates 3′-5′ rU substitution (sugar backbone heterogeneity), whereas a single substitution with 2′-5′ rU causes strong block for pol II elongation.
In order to quantitatively investigate the effects of backbone heterogeneity on RNA pol II transcription, we performed single turnover assays (see four scaffolds in Supplementary Figure S2) to measure the efficiency of nucleotide incorporation opposite to these backbone alteration sites, respectively. We determined the kinetic parameters kpol (catalytic rate constant for substrate incorporation) and Kd,app (apparent substrate dissociation constant), and used kpol/Kd,app as a measurement of enzymatic efficiency. As shown in Figure 2a–c and Supplementary Table S1, a single ribose substitution (3′-5′ rU) only causes ∼25-fold decrease in kpol but ∼4-fold increase in substrate binding, resulting in a ∼5.5-fold decrease in incorporation efficiency (kpol/Kd,app). In sharp contrast, substitution with 2′-5′ rU leads to a sharp decline with ∼3000-fold decrease in kpol and 10-fold reduction in substrate binding affinity (Figure 2 and Supplementary Table S1). As a result, substitution with 2′-5′ rU leads to a 3 × 104-fold reduction of incorporation efficiency. Interestingly, substitution with 2′-5′ dT has a much milder impact in comparison with 2′-5′ rU substitution. It causes ∼45-fold decrease in kpol, ∼6-fold decrease in substrate binding affinity and ∼260-fold reduction of incorporation efficiency (Figure 2c and Supplementary Table S1).
Figure 2.
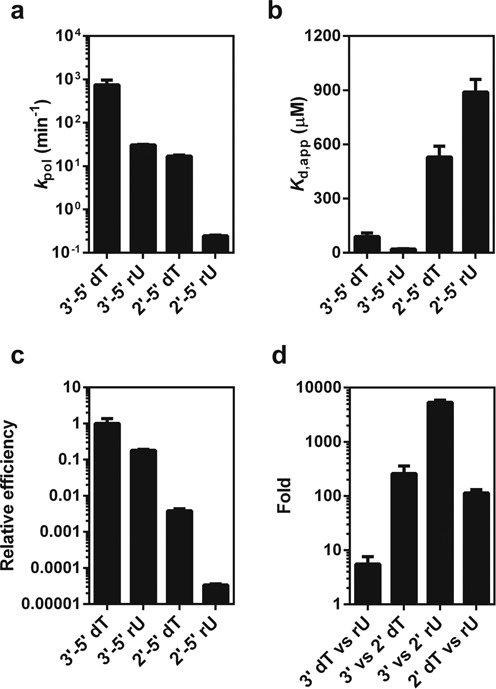
Phosphodiester linkage and sugar heterogeneity has distinct impacts on pol II transcription efficiency. Pol II catalytic rate constants (kpol) (a), substrate dissociation constants (b), relative efficiency of pol II nucleotide incorporation (c) were shown for pol II complex with four template backbone variants. The efficiency was normalized according to the specificity constants (kpol/Kd,app) of pol II transcription through these four templates. (d) Comparisons of phosphodiester linkage and sugar heterogeneity effects on pol II incorporation. The values are calculated based on the specificity constants (kpol/Kd,app).
We then systematically compared the effects of these backbone variants on ATP incorporation efficiency by pol II. Intriguingly, we found the impact of sugar substitution relies on the context of phosphodiester linkage, and vice versa. As shown in Figure 2d, we clearly revealed that the sugar substitution causes a much minor effect (∼5-fold reduction) on pol II transcription (3′-5′ dT versus 3′-5′ rU) with the 3′-5′ linked DNA template, whereas the same sugar substitution has a much more significant effect (∼114-fold reduction) on pol II transcription (2′-5′ dT versus 2′-5′ rU) with the 2′-5′ linked DNA template. On the other hand, the phosphodiester linkage alteration also relies on sugar identity. The 2′-5′ phosphodiester linkage substitution in the context of ribose sugar has the strongest negative impact (∼5400-fold) on pol II transcription (comparison of 2′-5′ versus 3′-5′ rU), whereas the same 2′-5′ phosphodiester linkage substitution in the context of deoxyribose sugar only causes ∼260-fold reduction in pol II transcription (comparison of 2′-5′ versus 3′-5′ dT).
Impacts of template backbone heterogeneity on pol II nucleotide selection
We then address the question of whether these backbone variants will affect pol II transcriptional fidelity. To this end, we compared correct (ATP) and mismatched (UTP as a representative) incorporation. We measured UMP incorporation specificity constants in the opposite of all of these four templates as shown in Figure 3a. Intriguingly, in sharp contrast to the significant differences in term of specificity constants for the matched AMP incorporation across the four templates, the specificity constants of mismatched UMP incorporations for these four templates are very similar (all in the range of ∼10−5 μM−1 min−1) (Figure 3a and Supplementary Table S2), suggesting that template variation has little effect on the mismatched nucleotide addition. A probable explanation for these behaviors is that mismatched nucleotide incorporation is generally less template-dependent, so the heterogeneity in the DNA template has a much minor effect on mismatched nucleotide incorporation.
Figure 3.
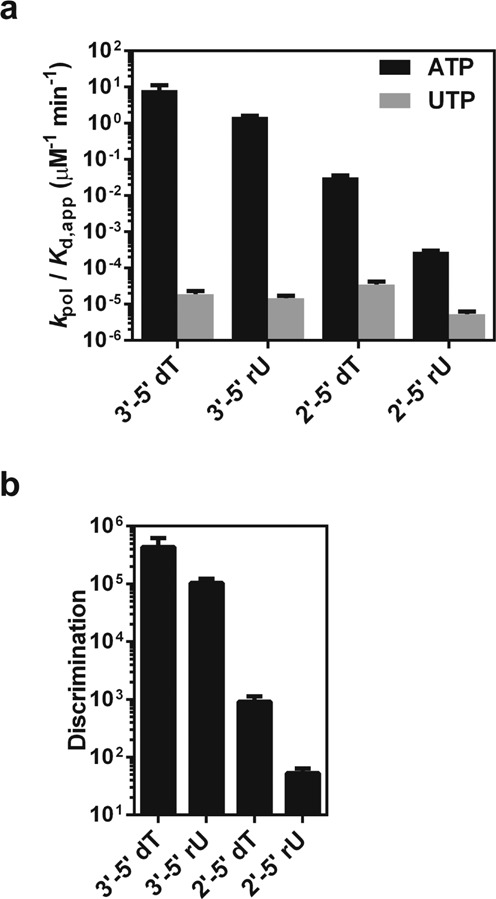
The presence of phosphodiester linkage heterogeneity and sugar heterogeneity in the template has distinct impact on pol II transcriptional fidelity. (a) Impacts of backbone heterogeneity on specificity constants of correct nucleotide incorporation (ATP) and incorrect nucleotide incorporation (UTP) by pol II. (b) Impacts of backbone heterogeneity on pol II discrimination power for the nucleotide selection.
Consequently, the difference in pol II discrimination power (selection of matched nucleotide over mismatched nucleotide for incorporation) among these template variants is mainly determined by correct nucleotide incorporation efficiencies. As shown in Figure 3b, we found that sugar substitution of a 3′-5′ dT with a 3′-5′ rU in the DNA template barely affects pol II's strong selection of ATP over UTP (∼1 × 105 for 3′-5′ rU versus ∼4 × 105 for 3′-5′ dT). In sharp contrast, phosphodiester linkage alteration in both dT and rU templates causes significant decreases in pol II fidelity. Particularly, substitution of a 3′-5′ rU with a 2′-5′ rU leads to a significant reduction of pol II discrimination power of ATP over UTP (decrease by ∼2 × 103-fold). Clearly, while the sugar heterogeneity barely affects pol II nucleotide incorporation fidelity, the phosphodiester linkage alteration significantly abolishes pol II capability of nucleotide selection.
Impacts of template backbone heterogeneity on pol II translocation
As shown in Figure 1, we observed strong pausing or stall when pol II encounters 2′-5′ dT or 2′-5′ rU site, whereas 3′-5′ rU only causes transient slowdown (Supplementary Figure S1). In order to further understand how these different template variants affect pol II translocation states, we further performed TFIIS-mediated cleavage assays. In these assays, we assembled pol II with four scaffolds containing an 11mer RNA primer with a 3′-end AMP opposite to the template backbone variants, respectively (shown in Figure 4a). These pol II elongation complexes mimic the state that ATP has been incorporated opposite to the template backbone variants.
Figure 4.
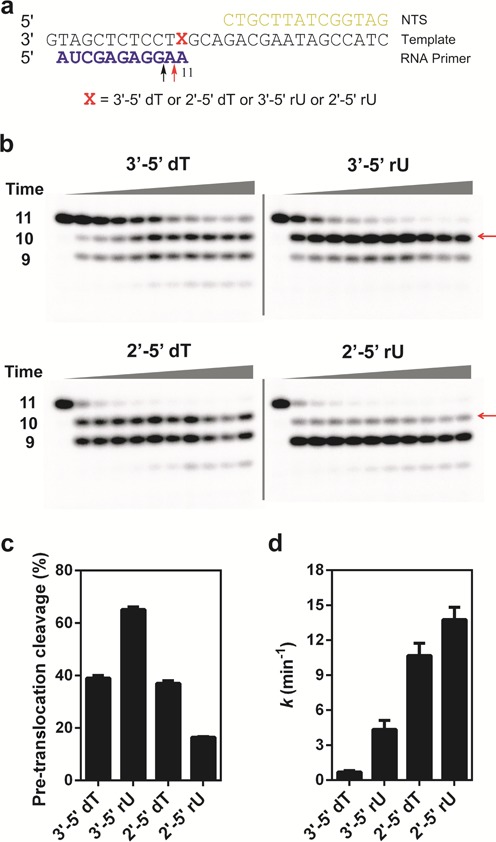
TFIIS-mediated pol II transcript cleavage reveals distinct cleavage patterns and rates. (a) The scaffolds used for TFIIS-mediated pol II transcript cleavage. The 11mer RNA primer is the initial transcript. The red arrow indicates the cleavage product (10mer) from pol II complex at a pre-translocation state. The black arrow indicates the cleavage product (9mer) from pol II complex at a backtracked state. (b) Gels for transcript cleavage for four different templates. The transcript length is marked beside the gel. The red arrow indicates the cleavage product from pol II complex at a pre-translocation state. (c) Percentage of cleavage product (10mer) from pol II complex at the pre-translocation state. (d) The observed overall transcript cleavage rates in these templates are measured by disappearing of 11mer.
Intriguingly, we observed distinct cleavage patterns that are specific to each of the four scaffolds (Figure 4 and Supplementary Table S3). Consistent with previous literature, for pol II complex containing a 11mer RNA and a wild-type 3′-5′ dT template, we observed two major cleavage bands (9mer and 10mer) (Figure 4b). The 10mer cleavage product band represents the specific cleavage product from the pol II complex at pre-translocation state (cleavage of 11mer to 10mer). In contrast, the 9mer cleavage band may reflect either two consecutive rounds of cleavage products from the pol II complex at pre-translocation states (consecutive cleavages of 11mer to 10mer and 10mer to 9mer) or, alternatively, cleavage product from the pol II complex at a backtracked state (direct removal of a dinucleotide from 11mer). To test the first possibility, we also performed TFIIS-mediated cleavage assays for pol II complex with 10mer and found that the generation of 9mer cleavage product from 10mer is significantly slower and weaker (Supplementary Figure S4). In addition, we observed that 10mer cleavage product band (cleavage from 11mer, Figure 4b) does not disappear upon long incubation, suggesting that pol II complex with 10mer is in a stable post-translocation state that is resistant to further cleavage. Therefore, we attributed the 9mer cleavage band (observed from pol II complex with 11mer) is mainly the cleavage product from a backtracked state rather than two consecutive rounds of cleavage products from the pre-translocation states (Figure 4b). Interestingly, we observed the cleavage reaction reaches a plateau in which the percentages of 11mer, 10mer and 9mer are relatively constant at long incubation time points (Supplementary Figure S5). We further revealed distinct cleavage signatures by quantitation of the percentage of 10mer and 9mer cleavage products at the plateau stage, respectively (Figure 4c, Supplementary Figure S6). Intriguingly, for pol II complex with 11mer RNA and the wild-type 3′-5′ dT template, the 10mer (generated from pol II complex at pre-translocation state) and 9mer cleavage products (generate from pol II at backtracked state) are ∼40 and ∼30%, respectively. Interestingly, for pol II complex with 2′-5′ dT template, the 10mer and 9mer cleavage products are ∼40 and ∼50%, respectively. All 11mers disappeared at plateau stage. In sharp contrast, for pol II complex with 3′-5′ rU template, the 10mer band is the major cleavage product (65%), whereas 9mer is the minor cleavage band (∼20%). Conversely, for pol II complex with the 2′-5′ rU template, we observed a major 9mer cleavage band (70%) and a minor 10mer cleavage band (15%), respectively. The difference revealed distinct pol II translocation behaviors in heterogeneous template backbones.
Quantitative analysis of cleavage rates (Figure 4d) revealed that phosphodiester linkage alteration (2′-5′ dT and ribose 2′-5′ rU) greatly accelerated pol II cleavage rates, with ∼15- and ∼20-fold increase, respectively. In contrast, 3′-5′ linked ribose replacement (3′-5′ rU) only modestly increased cleavage activity by ∼6-fold. These cleavage results are highly consistent with nucleotide incorporation efficiency and fidelity results that phosphodiester linkage heterogeneity leads to significant changes in pol II transcription process, whereas the sugar heterogeneity is more tolerated by pol II and has much weaker effect on pol II transcription.
Molecular modeling of pol II complexes encountering template backbone heterogeneity
In order to gain more structural insight, we performed molecular modeling of pol II elongation complexes containing backbone heterogeneity at the active site. The initial models were built based on the crystal structure of pol II complex (PDB ID: 2E2H (35)) with bound correct NTP. The sugar and linkage alteration were introduced based on structural alignment with the structures of 2′-5′ linked nucleic acids (36,37,43). The system was solvated in a triclinic box of TIP3P water molecules (41) with the minimum solute-wall box distance of 10Å. Na+ ions were added to neutralize the whole system. A 10 000-steps energy minimization with the steepest descent algorithm was performed. Details can be found in the experimental section.
As shown in Figure 5, we observed distinct effects of backbone heterogeneity on pol II elongation complex structure. In comparison with the structure of pol II complex with wild-type 3′-5′ dT DNA template, the ribose replacement (3′-5′ rU) has a very minor geometry effect on DNA template (Figure 5b). The incoming ATP can still form canonical Watson–Crick base pair with rU template, which is fully consistent with our kinetic result that ATP incorporation opposite to 3′-5′ rU is almost as efficient as the wild-type 3′-5′ dT template. In addition, further analysis of interaction network revealed that the 2′-OH of rU (i + 1 position) interacts with upstream sugar ring at i − 1 position as well as phosphodiester backbone connecting i − 1 and i + 1 position. These additional interactions further stabilize rU in i + 1 template position and prevent the backtrack of rU, as backtrack of rU to i + 2 position would require the disruption of these additional interactions. This result nicely explains why we observed a significantly higher population of pol II complex with 3′-5′ rU (∼65%) is in pre-translocation state in comparison with pol II complex with 3′-5′ dT (∼40%) (Figure 4c).
Figure 5.
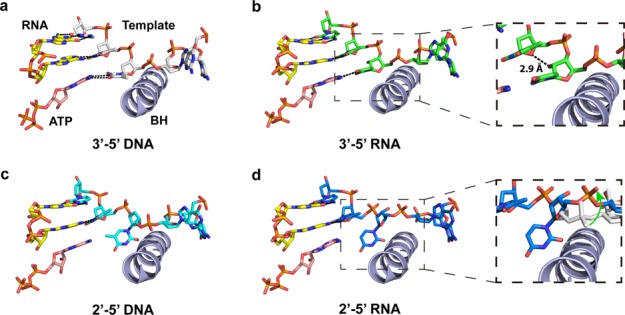
Sugar heterogeneity and phosphodiester linkage heterogeneity have different impacts on the pol II active site. (a) Wild-type RNA pol II elongation complex bound with ATP. RNA primer, DNA template and NTP are marked in the figure. ‘BH’ refers to bridge helix. (b) RNA pol II elongation complex model with ribose incorporated at i + 1 site (3′-5′ rU template). The enlarged area indicates potential hydrogen bonding between 2′-OH of rU and the sugar ring at i − 1 position of template. (c) RNA pol II elongation complex model with 2′-5′ DNA linkage at i + 1 site (2′-5′ dT template). Clearly, the nucleobase orientation at i + 1 site is disrupted due to linkage alteration. (d) RNA pol II elongation complex model with 2′-5′ RNA linkage at i + 1 site (2′-5′ rU template). The enlarged area not only shows the mis-aligned nucloebase but also highlights that the template backbone is shifted away from bridge helix. The light gray shows the 2′-5′ DNA backbone; the green arrow indicates the shift of the 2′-5′ RNA template backbone.
In sharp contrast, the phosphodiester linkage alteration greatly distorts the nucleobase alignment in both the 2′-5′ DNA and 2′-5′ RNA templates. The i + 1 nucleobase is greatly tilted and shifted away from its canonical position because of the 2′-5′ phosphodiester linkage (Figure 5c and d). As a result, this positional shift disrupts the base stacking with i − 1 template and hydrogen bonding base paring with incoming nucleotide. These structures provide structural insights into understanding how the linkage-altered template greatly reduces the nucleotide incorporation efficiency. Interestingly, further analysis revealed that for pol II complex with the 2′-5′ RNA template, the orientation of phosphodiester linkage chain is significantly changed in comparison to that for pol II with the 2′-5′ dT template (Figure 5d). This phosphodiester linkage shift is likely to reduce the steric translocation barrier for the i + 1 nucleobase to crossover the bridge helix to downstream main channel during backtrack. We also simulated pol II complex with a 2′-5′ phosphodiester linkage connecting i + 1 and i + 2 nucleobases (Supplementary Figure S7). Intriguingly, from the energy minimaization, we found that the 3′-OH of sugar in the 2′-5′ rU template may interact with K332 of Rpb1 subunit of pol II, which is located in the switch II region, an important domain for pol II translocation (44,45). This specific interaction provides additional stabilization of the backtracked state for the pol II complex with a 2′-5′ RNA.
Collectively, these molecular modeling results clearly indicate little effect of sugar heterogeneity but strong disruption by linkage heterogeneity in terms of template alignment and conformational change in pol II active site, which is fully consistent with our experimental observations.
DISCUSSION
Distinct impacts of sugar and phosphodiester backbone heterogeneity on pol II transcription
How pol II recognizes nucleic acids is an essential question for elucidating the molecular basis of pol II transcriptional efficiency and fidelity. Structural, genetic, biochemical and computational studies have provided new understanding on the molecular basis of the pol II transcriptional process (25,27,31,46–51). Unlike these conventional approaches, we employed synthetic nucleic acid analogues that ‘chemically modify’ the specific groups or motifs of nucleic acids (‘chemical mutation’) to dissect the individual roles of chemical interactions and the intrinsic structural features of nucleic acids in controlling pol II transcriptional fidelity (29,34,52–54).
In this work, we revealed dramatically distinctive impacts of two different types of backbone heterogeneity (sugar and phosphodiester linkage) on pol II transcription. The ribose replacement (3′-5′ rU) in the template is well tolerated by pol II and only causes a minor effect on transcription, whereas the phosphodiester linkage alteration (2′-5′ rU) leads to a significant decrease in both transcriptional efficiency and fidelity. Structural modeling revealed that the sugar backbone heterogeneity in the template has little effect on the geometry of template strand and polymerase active site. In sharp contrast, the correct 3′-5′ phosphodiester backbone is critical for maintaining the correct template nucleobase alignment and the establishment of effective interaction network, and in turn, ensuring highly efficient transcription.
Potential biological consequences of embedded ribonucleotides on gene transcription
Recent studies revealed high abundance of ribonucleotide incorporation in genomic DNA during replication (17,55,56) with an average of one ribonucleotide per ∼6500 deoxyribonucleotides in yeast (17), and a 1:7600 ratio in the nuclear genome of RNase H2-defective mouse cells (19). The total number of ribonucleotides introduced into DNA genomes exceeds the total of all other canonical DNA lesions. These embedded ribonucleotides can be removed by several pathways including RNAase H2-dependent ribonucleotide excision repair (RER), Top-1/Aprataxin-dependent removal and sometimes by nucleotide excision repair (NER) (56–60). Ribonucleotide incorporation into genomic DNA has obvious negative consequences (61,62), including increased replication stress and genome instability. In human, partial loss of RNase H2 also cause Aicardi-Goutières syndrome (63) and mutations of Aprataxin (important for resolving adenylated RNA–DNA junctions) are linked to Ataxia with Oculomotor Aparxia 1 (AOA1) (64). On the other hand, recent literature also reported that the embedded ribonucleotide in genomic DNA also have positive roles that serve as signals for several cellular DNA transactions (21,56), including mating type switching in Schizosaccharomyces pombe (65), signaling the nascent strand for mismatched repair (66,67) and utilization of ribonucleotide during non-homologous end joining repair (68,69). It appears that genomic DNA with embedded ribonucleotide is likely a feature in our current evolutionary stage.
These embedded ribonucleotides may encounter RNA pol II during transcription if they are not repaired in a timely manner. Little is known about impact of these ribonucleotides on gene transcription. Our results indicate that while pol II can well tolerate ribonucleotides in the DNA template, a single ribonucleotide at DNA template can still cause a ∼25-fold reduction in pol II elongation rate (kpol). Given the fact of the high abundance of ribonucleotides level and likely uneven distributions ribonucleotides in genomic DNA, one would expect stronger effects at those hotspots of ribonucleotides on pol II transcription elongation dynamics. This pausing effect on pol II transcription elongation may serve as a potential signal for fine-tuning co-transcriptional RNA processes and possibly RER in a transcription-coupled manner or transcription-coupled NER (70). Intriguingly, the pattern of how pol II processes ribonucleotide is very similar to how pol II processes 5-formylcytosine and 5-caboxylcytosine (33), indicating additional modulation layer for pol II elongation rates. One interesting direction for future study is to understand how these pol II elongation rate modulators would affect transcription, RNA processing (such as splicing or folding) and repair in vivo.
Distinct tolerance for template backbone heterogeneity implies the potential evolutionary pathway
From the molecular evolution perspective, we found that the significant difference between sugar and phosphodiester linkage impacts on pol II efficiency may reflect adaptability during the molecular evolution pathway. The phosphodiester linkage backbone heterogeneity represents a major issue from the prebiotic to the RNA world. This backbone heterogeneity is largely resolved by evolution from RNA world to contemporary DNA–RNA–Protein world, as DNA is the major form of genetic storage, which is composed of a single form of phosphodiester linkage (3′-5′). The contemporary template-dependent nucleic acid polymerases (DNA and RNA polymerases) are evolved to recognize templates with 3′-5′ phosphodiester linkage and catalyze polymerization of 3′-5′ linked nucleic acids. In sharp contrast, the sugar heterogeneity is a relatively new issue during molecular evolution from the RNA to DNA–RNA–Protein world, which still exists in our contemporary world (Figure 6a).
Figure 6.
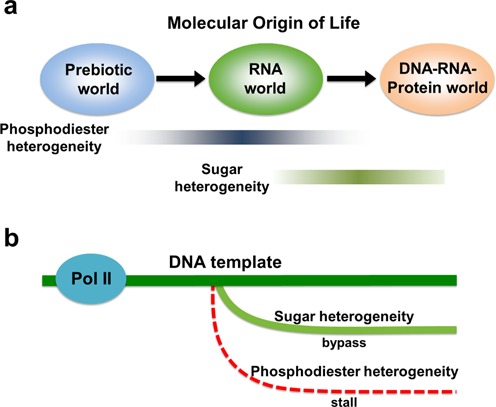
RNA pol II is much more tolerant to sugar heterogeneity than phosphodiester heterogeneity in DNA template. (a) Potential evolution for genetic materials. Phosphodiester heterogeneity may be a major form of backbone heterogeneity in early stages of evolution (prebiotic to RNA world), which has been then largely eliminated for genetic information storage and transfer in the DNA world. On the other hand, sugar heterogeneity represents a major form of backbone heterogeneity that still widely exists in the contemporary DNA world. (b) RNA pol II can smoothly bypass the sugar backbone heterogeneous site with relatively high fidelity; in sharp contrast, pol II stalls at the phosphodiester linkage alteration site and the transcriptional fidelity is significantly decreased.
Consequently, RNA pol II, as one of the most important readers for genomic DNA in all eukaryotes, may also possess this evolutionary imprint. Indeed, as we revealed in this work, pol II has strong discrimination of 3′-5′ phosphodiester linkage over 2′-5′ linkage in the template, whereas pol II is much more tolerant to sugar heterogeneity in the template (ribose versus deoxyribose) (Figure 6b). The recognition of embedded ribonucleotides by pol II may also serve as a signal for DNA transactions in a transcription-coupled manner. This result may reflect the ‘imperfect’ stage of DNA world with redundant embedded ribonucleotides in the genomic DNA as a unique signature (56).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
National Institutes of Health (NIH) [GM102362 to D.W.]; Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (to D.W.); Research Grant Council of Hong Kong [GRF 16302214 to X.H.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ekland E.H., Bartel D.P. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature. 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 2.Ertem G., Ferris J.P. Synthesis of RNA oligomers on heterogeneous templates. Nature. 1996;379:238–240. doi: 10.1038/379238a0. [DOI] [PubMed] [Google Scholar]
- 3.Inoue T., Orgel L.E. Oligomerization of (guanosine 5′-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J. Mol. Biol. 1982;162:201–217. doi: 10.1016/0022-2836(82)90169-3. [DOI] [PubMed] [Google Scholar]
- 4.Usher D.A., McHale A.H. Nonenzymic joining of oligoadenylates on a polyuridylic acid template. Science. 1976;192:53–54. doi: 10.1126/science.1257755. [DOI] [PubMed] [Google Scholar]
- 5.Usher D.A., McHale A.H. Hydrolytic stability of helical RNA: a selective advantage for the natural 3′,5′-bond. Proc. Natl. Acad. Sci. U.S.A. 1976;73:1149–1153. doi: 10.1073/pnas.73.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris J.P., Ertem G. Oligomerization of ribonucleotides on montmorillonite: reaction of the 5′-phosphorimidazolide of adenosine. Science. 1992;257:1387–1389. doi: 10.1126/science.1529338. [DOI] [PubMed] [Google Scholar]
- 7.Engelhart A.E., Powner M.W., Szostak J.W. Functional RNAs exhibit tolerance for non-heritable 2′-5′ versus 3′-5′ backbone heterogeneity. Nat. Chem. 2013;5:390–394. doi: 10.1038/nchem.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowler F.R., Chan C.K., Duffy C.D., Gerland B., Islam S., Powner M.W., Sutherland J.D., Xu J. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 2013;5:383–389. doi: 10.1038/nchem.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adamala K., Szostak J.W. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trevino S.G., Zhang N., Elenko M.P., Luptak A., Szostak J.W. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13492–13497. doi: 10.1073/pnas.1107113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984;39:423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- 13.Carlomagno T., Amata I., Codutti L., Falb M., Fohrer J., Masiewicz P., Simon B. Structural principles of RNA catalysis in a 2′-5′ lariat-forming ribozyme. J. Am. Chem. Soc. 2013;135:4403–4411. doi: 10.1021/ja311868t. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann R., Justesen J., Sarkar S.N., Sen G.C., Yee V.C. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 15.Hovanessian A.G., Justesen J. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie. 2007;89:779–788. doi: 10.1016/j.biochi.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Vengrova S., Dalgaard J.Z. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7:59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Lundstrom E.B., Burgers P.M., Johansson E., Chabes A., Kunkel T.A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clausen A.R., Zhang S., Burgers P.M., Lee M.Y., Kunkel T.A. Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase delta. DNA Repair (Amst) 2013;12:121–127. doi: 10.1016/j.dnarep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F., et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goksenin A.Y., Zahurancik W., LeCompte K.G., Taggart D.J., Suo Z., Pursell Z.F. Human DNA polymerase epsilon is able to efficiently extend from multiple consecutive ribonucleotides. J. Biol. Chem. 2012;287:42675–42684. doi: 10.1074/jbc.M112.422733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potenski C.J., Klein H.L. How the misincorporation of ribonucleotides into genomic DNA can be both harmful and helpful to cells. Nucleic Acids Res. 2014;42:10226–10234. doi: 10.1093/nar/gku773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cramer P. RNA polymerase II structure: from core to functional complexes. Curr. Opin. Genet. Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Cramer P. Structure and function of RNA polymerase II. Adv. Protein Chem. 2004;67:1–42. doi: 10.1016/S0065-3233(04)67001-X. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann E., Brueckner F., Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan C.D., Larsson K.-M., Kornberg R.D. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol. Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kireeva M.L., Nedialkov Y.A., Cremona G.H., Purtov Y.A., Lubkowska L., Malagon F., Burton Z.F., Strathern J.N., Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuzenkova Y., Bochkareva A., Tadigotla V.R., Roghanian M., Zorov S., Severinov K., Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erie D.A., Hajiseyedjavadi O., Young M.C., von Hippel P.H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 29.Su Zhang D.W. Understanding the Molecular Basis of RNA Polymerase II Transcription. Isr. J. Chem. 2013;53:442–449. doi: 10.1002/ijch.201300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L., Zhang L., Chong J., Xu J., Huang X., Wang D. Strand-specific (asymmetric) contribution of phosphodiester linkages on RNA polymerase II transcriptional efficiency and fidelity. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3269–E3276. doi: 10.1073/pnas.1406234111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellinger M.W., Song C.X., Chong J., Lu X.Y., He C., Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellinger M.W., Ulrich S., Chong J., Kool E.T., Wang D. Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J. Am. Chem. Soc. 2012;134:8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson H., Jung K.-E., Switzer C., Wang A.H.-J. DNA with 2′-5′ phosphodiester bonds forms a duplex structure in the A-type conformation. J. Am. Chem. Soc. 1995;117:837–838. [Google Scholar]
- 37.Sheng J., Li L., Engelhart A.E., Gan J., Wang J., Szostak J.W. Structural insights into the effects of 2′-5′ linkages on the RNA duplex. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3050–3055. doi: 10.1073/pnas.1317799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Da L.T., Wang D., Huang X.H. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, et al., editors. Gaussion03. Wallingford CT: Gaussian, Inc; 2004. [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Szostak J.W. The free energy landscape of pseudorotation in 3′-5′ and 2′-5′ linked nucleic acids. J. Am. Chem. Soc. 2014;136:2858–2865. doi: 10.1021/ja412079b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kettenberger H., Armache K.-J., Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 45.Majovski R.C., Khaperskyy D.A., Ghazy M.A., Ponticelli A.S. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J. Biol. Chem. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 46.Liu X., Bushnell D.A., Kornberg R.D. RNA polymerase II transcription: structure and mechanism. Biochim. Biophys. Acta. 2013;1829:2–8. doi: 10.1016/j.bbagrm.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Rucobo F.W., Cramer P. Structural basis of transcription elongation. Biochim. Biophys. Acta. 2013;1829:9–19. doi: 10.1016/j.bbagrm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Svejstrup J.Q. RNA polymerase II transcript elongation. Biochim. Biophys. Acta. 2013;1829:1. doi: 10.1016/j.bbagrm.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Svetlov V., Nudler E. Basic mechanism of transcription by RNA polymerase II. Biochim. Biophys. Acta. 2013;1829:20–28. doi: 10.1016/j.bbagrm.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas M.J., Platas A.A., Hawley D.K. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 51.Huang X., Wang D., Weiss D.R., Bushnell D.A., Kornberg R.D., Levitt M. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benner S.A., Sismour A.M. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu L., Plouffe S.W., Chong J., Wengel J., Wang D. A chemical perspective on transcriptional fidelity: dominant contributions of sugar integrity revealed by unlocked nucleic acids. Angew. Chem. Int. Ed. Engl. 2013;52:12341–12345. doi: 10.1002/anie.201307661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu L., Butler K.V., Chong J., Wengel J., Kool E.T., Wang D. Dissecting the chemical interactions and substrate structural signatures governing RNA polymerase II trigger loop closure by synthetic nucleic acid analogues. Nucleic Acids Res. 2014;42:5863–5870. doi: 10.1093/nar/gku238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundstrom E.B., Johansson E., Chabes A., Kunkel T.A. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams J.S., Kunkel T.A. Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst) 2014;19:27–37. doi: 10.1016/j.dnarep.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lazzaro F., Novarina D., Amara F., Watt D.L., Stone J.E., Costanzo V., Burgers P.M., Kunkel T.A., Plevani P., Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 2012;45:99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sparks J.L., Chon H., Cerritelli S.M., Kunkel T.A., Johansson E., Crouch R.J., Burgers P.M. RNase H2-initiated ribonucleotide excision repair. Mol. Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams J.S., Smith D.J., Marjavaara L., Lujan S.A., Chabes A., Kunkel T.A. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol. Cell. 2013;49:1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potenski C.J., Niu H., Sung P., Klein H.L. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caldecott K.W. Molecular biology. Ribose–an internal threat to DNA. Science. 2014;343:260–261. doi: 10.1126/science.1248234. [DOI] [PubMed] [Google Scholar]
- 62.Dalgaard J.Z. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28:592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 64.Tumbale P., Williams J.S., Schellenberg M.J., Kunkel T.A., Williams R.S. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh G., Klar A.J. Mutations in deoxyribonucleotide biosynthesis pathway cause spreading of silencing across heterochromatic barriers at the mating-type region of the fission yeast. Yeast. 2008;25:117–128. doi: 10.1002/yea.1569. [DOI] [PubMed] [Google Scholar]
- 66.Lujan S.A., Williams J.S., Clausen A.R., Clark A.B., Kunkel T.A. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013;50:437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghodgaonkar M.M., Lazzaro F., Olivera-Pimentel M., Artola-Boran M., Cejka P., Reijns M.A., Jackson A.P., Plevani P., Muzi-Falconi M., Jiricny J. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol. Cell. 2013;50:323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin M.J., Garcia-Ortiz M.V., Esteban V., Blanco L. Ribonucleotides and manganese ions improve non-homologous end joining by human Polmu. Nucleic Acids Res. 2013;41:2428–2436. doi: 10.1093/nar/gks1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bartlett E.J., Brissett N.C., Doherty A.J. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1984–E1991. doi: 10.1073/pnas.1302616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


