Abstract
A persistent design problem for ratiometric microarray studies is selecting the ‘denominator’ RNA cohybridization standard. The ideal standard should be readily available, inexpensive, invariant over time and from laboratory to laboratory, and should represent all genes with a uniform signal. RNA references (both commercial ‘universal’ and experiment- specific types), fall short of these goals. We show here that mouse genomic DNA is a reliable microarray cohybridization standard which can meet these criteria. Genomic DNA was superior in universality of coverage (>98% of genes from a 16 000 feature mouse 70mer microarray) to the Stratagene Universal Mouse Reference RNA standard. Ratios for genes in very low abundance in the Stratagene standard were more unstable with the Stratagene standard than with genomic DNA. Genes with mid-range, and therefore presumably optimal RNA denominator values, showed comparable reproducibility with both standards. Inferred ratios made between two different experimental RNAs using a genomic DNA standard were found to correlate well with companion, directly measured ratios (Spearman correlation coefficient = 0.98). The advantage in array feature coverage of genomic DNA will likely increase as newer generation microarrays include genes which are expressed exclusively in minor tissue or developmental domains that are not represented in mixed tissue RNA standards.
INTRODUCTION
DNA microarrays have quickly become an indispensable tool for transcriptome analysis (1). Mechanical spotting of DNA on glass slides has emerged as a widely used microarray platform because it affords flexibility of array design and relative economy. However, this technology also has some significant shortcomings. Because feature geometry and the amount of DNA per feature vary within a gene chip, and also from one chip to another, measurements must be made as internal ratiometric comparisons of one RNA sample with a reference (or ‘denominator’) RNA (2,3). This is done by simultaneous hybridization of experimental and reference samples, where each RNA population is transcribed into cDNA with a different fluorophore (typically Cy3 for one and Cy5 for the other). While this is very effective for direct comparisons of just two RNA samples, the full power of large-scale expression analysis comes from comparisons of multiple (tens to hundreds or even thousands) of different RNA samples. To do this using spotted microarrays, the ratio observed for each feature on the array is compared across all gene chips in a study, each of which has used the same denominator RNA sample (converted to labeled cDNA or cRNA).
Although this design has proved very successful, the requirement for internal ratiometric measurement presents a thorny set of problems that come from properties of the reference hybridization standard. For example, instability and error is expected for RNAs not represented in the reference or, alternatively, for RNAs so prevalent in the reference that they saturate their corresponding features (detectors). Moreover, the reference RNA sample composition is not standard from one study to another, usually having been selected based on different criteria for each study. Once a standard is selected, the vagaries of biology make it difficult to reproduce precisely from one preparation to another. This means that global comparisons between studies done in the same laboratory over a long time or between different laboratories are compromised. These issues have so far been dealt with using strategies that range from selecting a single tissue standard, such as whole spleen RNA for a study of B cells done by the Alliance for Cell Signaling (AFCS) (http://www.signaling-gateway.org) to making a denominator mixture of RNAs by pooling aliquots from each sample in a given study (4,5), to attempting to make a ‘general mixture’ of RNA from diverse cell lines (e.g. the Stratagene Universal Reference RNA standards) (4,6).
Genomic DNA should, in principle, be a more general, invariant and inexpensive solution (1,7). Major virtues of the genome as a cohybridization ‘standard’ include complete sequence representation, sequence stability over time and from one preparation to another, uniform prevalence for most genes and very low cost. These features mean that it is also applicable to any array, independent of which subset of genes is arrayed or which strand, in the case of oligonucleotides, is represented.
It is also clear that genomic DNA presents problems and challenges of its own. In the large vertebrate genomes that are our principle interest, mRNA coding sequences are highly diluted by non-coding DNA. This is expected to adversely affect absolute signal level, elevate noise, and perhaps generate additional variability from the effects of interspersed repeat sequences. However, recent successes with microarray-based comparative genome hybridization (CGH), in which two human genomic DNA samples are cohybridized to cDNA microarrays (8) suggested that genomic DNA signals could be made reliable enough to act as a universal denominator for expression measurements. Recent reports of genomic DNA as an expression array denominator on various kinds of arrays have been mixed. In small bacterial genomes where most of the complications listed above do not apply, it is very effective (9). For much larger genomes including Arabidopsis and human, the results have generally been less convincing, when compared with RNA standards (see Discussion).
In this study we tested total sheared mouse genomic DNA as a reference in the context of a complex 70mer oligonucleotide microarray (∼16 000 features). Genomic DNA provided superior array feature coverage compared with the Stratagene Mouse Universal Reference RNA standard, and comparable signal stability for all array features in multiple replicate experiments. It has the specific salutary effect of adjusting and stabilizing ratios for genes whose transcripts are present at very low levels in the Stratagene Universal Reference RNA standard used for comparison. As the sequence representation of microarray gene sets becomes more comprehensive, the genomic DNA standard has the potential to provide reliable denominator signals for every sequence represented on an array. Coverage by a mixed RNA reference standard is necessarily limited to those genes expressed in its constituent cell lines.
MATERIALS AND METHODS
Oligonucleotide arrays
70mer oligonucleotides representing 13 443 expressed sequences from the mouse genome (Operon Array Ready Oligo Set version 1.0) were printed on SurModics 3-D Link glass slides using a robotic printing apparatus assembled according to instructions from the Pat Brown Laboratory website (http://cmgm.stanford.edu/pbrown/mguide/index.html). The Operon 70mers were resuspended in SurModics print buffer at a concentration of 20 pmol/µl. Samples of xenotypic DNA (408 features) and sequences informatically determined to be absent from the mouse genome (320 features) served as negative controls. An additional 1436 print buffer features served as blanks for carryover control, and a select group of positive control genes was included for quantitative comparisons and statistical analysis, bringing the final array size to 16 192 features (herein referred to as the 16K array). A 32 pin print head outfitted with MicroQuill 2000 print pins (Majer Precision Engineering) was used to array the features in 32 sectors, each 23 × 22 features in dimension. Slides were post-processed according to the manufacturer’s protocol. Hybridizations were carried out in 5× SSC, 50% formamide and 100 ng/µl yeast tRNA, at 46°C for 72 h. Coverslips were removed in 4× SSC, 0.1% SDS; the slides were then washed twice in 1× SSC, 0.1% SDS at 67°C for 5 min, then in 0.2× SSC at room temperature for 1 min, and again in 0.1× SSC for 1 min at room temperature, before spin drying at 900 r.p.m. for 3 min in an IEC Centra GP8 centrifuge using a 216 rotor.
Hybridized arrays were scanned on an Axon 4000 dual-laser scanning instrument (Axon Instruments) with PMT voltages matched at 600 V. The scanned images were quantified using Axon’s GenePix 3.0 software, and imported to Microsoft Excel for further filtering and analysis after global scaling using the median of ratios values. Comparison of the intensity distributions for all mouse targets and for the collection of negative control features on the chip was performed with an Excel macro. Threshold, percent coverage and receiver operating characteristic (ROC) curve analyses were also performed with Excel macros.
cDNA labeling
Total RNA was isolated from the C2C12 mouse skeletal muscle cell line after 72 h in differentiation medium, using RNeasy columns (Qiagen). Messenger RNA was then extracted from total RNA using oligo dT-coated beads (Oligotex, Qiagen). The same procedures were used to isolate RNA from adult mouse liver. Messenger RNA samples were primed with random hexamers, and then reverse transcribed with SuperScript II (Invitrogen) in the presence of either Cy5 dUTP or Cy3 dUTP (Amersham), for 4 h. Stratagene Universal Mouse Reference RNA samples were primed with anchored oligo dT, and then reverse transcribed with SuperScript II in the presence of Cy3 dUTP. Details of the reverse transcription protocols are as described at http://cmgm.stanford.edu/pbrown/protocols/4_human_RNA.html/.
Genomic DNA shearing
Mus musculus genomic DNA was isolated from adult male and female B6D2F1 mouse kidneys using the MasterPure complete DNA purification technique (Epicentre). We process 8 or more 5 mg preps in a single session. After sequential treatment with RiboShredder (Epicentre), RNAse I and RNAse H, this preparation was sonicated for 45 s in a volume of 233 µl in a 2 ml microcentrifuge tube at setting 18 on a Microson XL 2007 sonicator. The microcentrifuge tube was half submerged in a solution of –20°C ethanol during sonication. Sonicated DNA was then diluted with DNA binding buffer (Zymo), and column purified according to the manufacturer’s protocol (Zymo catalog no. D4005). UV absorbance spectrophotometry was used to estimate the yield of sheared DNA, and gel electrophoresis was performed to verify that sonicated DNA averaged 2–3 kb. Sonicated genomic DNA was frozen in 2 µg single-use aliquots.
Genomic DNA labeling
Two microgram aliquots of randomly sheared mouse genomic DNA were labeled using Klenow fragment (BioPrime; Invitrogen) in the presence of Cy3-labeled dCTP (Amersham), in a 50 µl reaction volume (http://cmgm.stanford.edu/pbrown/protocols/4_genomic.html). The reactions were incubated for 2.5 h in a 37°C oven, respiked with an additional 1 µl of Klenow fragment, and incubated again for 2.5 h. Unlabeled nucleotides from Roche (catalog no. 1 277 049) were frozen in single use 10× aliquots and used at a final reaction concentration of 200 µM. The proportion of unlabeled dCTP to Cy3-labeled dCTP in the final reaction was 100:60 µM. The reaction was terminated with stop buffer (0.5 M Na2EDTA, pH 8.0), and then cleaned up on QiaQuick PCR purification columns using two wash steps (Qiagen). An aliquot representing 5% of the labeled product was reserved for measuring label incorporation via fluorometry (BioRad). The amount of labeled nucleotide incorporated was estimated against a standard dilution curve made with unincorporated Cy-labeled nucleotides. After incorporation was measured, the fluorescent, double strand binding dye Pico Green (Molecular Probes) was added to the sample, and the yield of synthesized DNA estimated against a standard curve of lambda phage DNA. This ‘serial’ method allowed us to measure genomic DNA probe yield and incorporation using only 5% of our reaction product, and eliminates spurious contributions from the Pico Green excitation or emission spectra that overlap with the Cy3 spectra. For each hybridization experiment, three labeling reactions using 2 µg of mouse genomic DNA as template were combined for hybridization to a single array.
Arabidopsis thaliana genomic DNA preparation
Inflorescences, siliques and stem leaves were collected from mature A.thaliana, frozen in liquid nitrogen and ground in a mortar and pestle under liquid nitrogen. Genomic DNA was isolated from the ground tissue using the GenElute Plant Genomic DNA isolation kit (Sigma). After UV absorbance spectrophotometry to estimate yield, the DNA was sonicated as above and column cleaned prior to labeling as described above. Genomic DNA hybridizations using Arabidopsis genomic DNA were carried out using labeled probe from a single reaction using 2 µg of genomic DNA as template.
Data processing and analysis
Before using the genomic DNA based ratio measurements to calculate relative expression ratios, the median of pixel-by-pixel ratios (10) for any given feature is first scaled, using the global scaling algorithm provided with the GenePix software. Briefly, this algorithm assumes that the overall expected mean of expression ratios is equal to 1 for all features on the corresponding array with ratios between 10 and 0.1. The global scaling factor of (1/observed mean) is then applied to the observed median of ratios value from each feature on the corresponding array. After scaling, the inferred ratio of relative gene expression for any given feature on the array (C2C12 cDNA over adult liver cDNA) is calculated as:
![]()
Ratiometric data sets were inspected for normality using an implementation of the Kolmogornov–Smirnoff normality test available in Minitab statistical software (Minitab). Pearson correlation coefficients between replicate data sets were computed with the implementation available in Excel (Microsoft). When raw data sets or log-transformed data sets deviate significantly from normality, the Pearson correlation is less appropriate, since it assumes normality in the distributions. The Spearman correlation coefficients are more appropriate in these cases, and were computed with the implementation available in the Excel plug-in package, Analyse-It (Analyse-It, Leeds, UK).
RESULTS
Genomic DNA hybridization signals on 70mer microarrays
Because the vast majority of protein coding sequences are present once in the mouse genome, it is expected that all microarray features representing mouse mRNAs will react with labeled genomic DNA at similar signal levels. However, mammalian genomes are large and only ∼1% is mRNA coding sequence (36 433 genes over 3.1 × 109 bp, average ORF ∼1000 bp) (http://iubio.bio.indiana.edu:8089/). This means that with current technology, genomic DNA is expected to give relatively low absolute hybridization signals, and the vast amount of non-coding sequence is an obvious candidate source of noise. We therefore evaluated signal and background noise distributions for genomic DNA hybridization under our conditions. The microarray used in these studies includes a majority of verified mouse genes represented as ‘long oligos’ (70mers), and includes for comparison a smaller set of typical 0.5–2.5 kb cDNAs as PCR products. Among widely used ratiometric arrays, we expect long oligonucleotides to be the most rigorous test case for genomic DNA reactivity, since longer probe sequences (i.e. cDNA, fosmids, BACs) generally give higher absolute signals than shorter ones (8). However, it is worth noting that this is not likely to be a simple linear relationship with probe length due to other factors such as probe accessibility, molar density and sequence length of labeled DNA, among others.
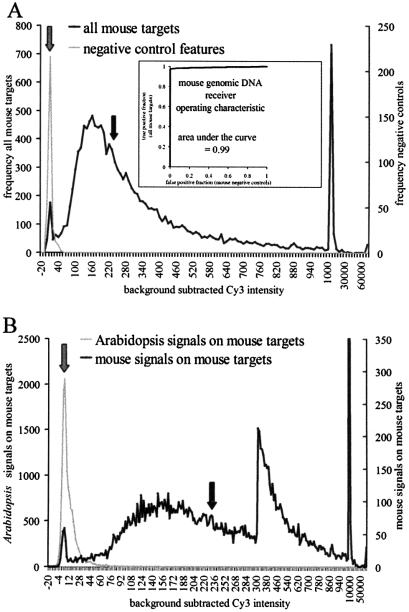
Figure 1A shows signal intensity distributions from an experiment in which labeled genomic DNA (Cy3) was cohybridized to an array with labeled cDNA from the C2C12 mouse skeletal muscle cell line after 72 h of differentiation (Cy5). The distribution of mouse genomic DNA hybridization for 13 915 mouse features (black trace) is compared with a set of 315 70mer negative control sequences (gray trace). The negative control group is comprised of 15 different sequences, each replicated 21 times. These control 70mers were selected because they are absent from the mouse genome and they have melting temperatures similar to gene oligos (http://oligos.qiagen.com/arrays/oligosets_mouse.php). Separation of negative control signals from putative positive signals was evaluated in two ways, both of which lead to the general conclusion that signals from genomic DNA are significantly above background for >98% of features on the array. First, if the distribution of negative control spots is modeled as a Gaussian distribution, the median signal is 3, and two standard deviations above this control median is 20. In contrast the median signal for positive mouse probes on the array is 239 in the experiment shown, and overlap with the negative control distribution is negligible (1.9% of mouse genomic values were below 20). These results were robust over five replicate determinations with the average negative control median 3.3 ± 0.4 and the average genomic DNA median 217 ± 41. The fraction of mouse genes falling within the negative control distribution at 2 SD was, on average, 1.8 ± 0.6%. We observed that the genomic DNA signal distribution was not strictly Gaussian, nor is it expected to be, so we also applied a ROC analysis to quantify separation of the two distributions (11). An average ROC value of 0.99 over the five replicates is very close to 1.0, which defines two entirely non-overlapping distributions (Fig. 1A, inset shows a typical ROC curve).
Figure 1.
Mouse genomic DNA signal intensities compared to negative control signal intensities. (A) The distribution of Cy3 signal intensities for 13 915 putative mouse gene features (black trace), and the distribution of Cy3 intensities for 315 negative control features on the same array (gray trace). Bin widths are adjusted to accommodate the full range of signal intensities on a single plot. Values from –20 to 1000 have a bin width of 5, and values from 1000 to 70 000 have a bin width of 5000. The background-subtracted median positive mouse feature intensity is 239 units (black arrow). Median intensity for the negative control features is 3 units (gray arrow). The inset to (A) shows a ROC curve for the two distributions. Plotted on the vertical axis is the fraction of positive mouse probes at or below a given level of Cy3 intensity; on the horizontal axis is the corresponding fraction of negative control features at or below that same level of Cy3 intensity. The area under the curve indicates separation of the two distributions, with a score of 1.0 indicating perfect separation of the distributions (i.e. no overlap). (B) Complex labeled genomic DNA specificity: Reactivity of labeled plant genomic DNA (gray trace) on the mouse 16K array. The mouse genomic DNA distribution from (A) (black trace) is superimposed for comparison. The horizontal axis is arranged with different bin widths in order to accommodate the full range of signal intensities. Background-subtracted levels from –20 to 300 have a bin width of 2; values from 300 to 1000 have a bin width of 10; and values from 1000 to 70 000 have a bin width of 5000. Median Arabidopsis genomic DNA signal is 6 (gray arrow), and the median mouse genomic DNA signal is 239 (black arrow).
The sequence complexity of the negative control oligo group of Figure 1A is relatively low (15 different 70mers) compared with the full array of mouse gene sequences (∼13 000), and may therefore fail to detect problematic background signals that are peculiar to some (unknown) subset of mouse oligo array features. To address this, we tested the reactivity of labeled genomic DNA from the plant A.thaliana with the entire mouse array. The Arabidopsis genome is complex, with ∼27 000 genes, and since plant and animal genomes have diverged greatly, the distribution of plant signals on the mouse microarray should be a good measure of background from a high complexity DNA source. Figure 1B shows that Arabidopsis genomic DNA reactivity is also very well separated from the distribution of mouse genomic DNA with medians of 6 for Arabidopsis versus 239 for mouse. To validate its use as a negative control, an aliquot of the same labeled Arabidopsis genomic DNA was hybridized with an Arabidopsis cDNA microarray under the same conditions, and it gave strong positive signals (data not shown). Moreover, several features on our mouse array correspond to the Arabidopsis gene apetela2, and they reacted with a median value of 363 with the labeled plant DNA. The ROC score for plant versus mouse DNA on mouse features was 0.98. We conclude that mouse genomic DNA reacts in a sequence-specific manner with 70mer microarrays and that the signals are sufficiently well separated from background to permit the use of genomic DNA as a ratiometric standard.
Array coverage
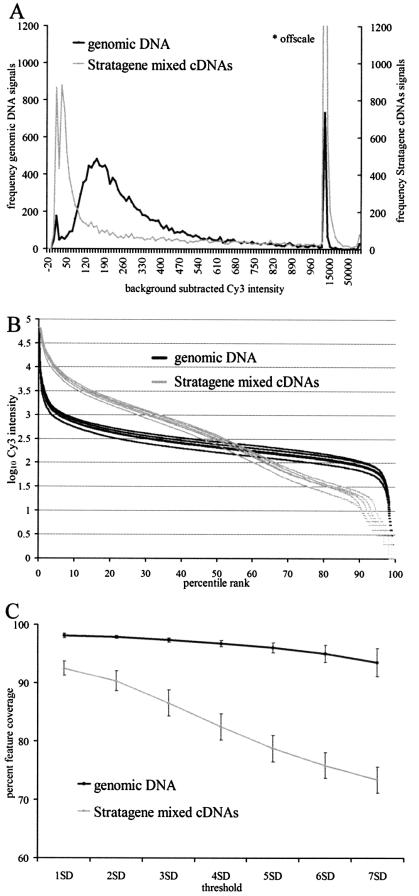
We compared genomic DNA reactivity over the entire array with the Stratagene Mouse Universal RNA standard reactivities to evaluate overall signal distribution and coverage. C2C12 cell line cDNA (Cy5) was co-hybridized with either Cy3-labeled mouse genomic DNA or Cy3-labeled cDNA produced from the Stratagene Mouse Universal RNA mixture. In contrast to the relatively narrow distribution of hybridization signals for DNA, a mixed collection of natural RNA populations is expected to deliver a very broad distribution, reflecting the entire span of prevalence values, from no detectable signal for genes that are not expressed, through intermediate values (2–3 orders of magnitude), to the most prevalent RNAs which can saturate the scanner. Figure 2A shows an example of the Stratagene Universal Mouse Reference distribution (gray trace) compared to the genomic DNA distribution (black trace). As expected, the Stratagene distribution has much larger numbers of genes at both extremes, and a relatively uniform distribution through middle values. In Figure 2B, feature intensities for the entire array are compared for five Stratagene replicates and five genomic DNA replicates. The reproducibly flatter profile of genomic DNA percentile rank plots shows that the vast majority of array features are hybridized within a narrow band of intensities, in contrast to the substantially more variable profile for the mixed RNA standard. This fits the simplest expectation for genomic DNA in which most template sequences are represented at equimolar concentrations. In Figure 2C, the five replicates are evaluated for the fraction of gene features that react above a given background threshold. Although the RNA sources in the Stratagene standard were selected by the manufacturer to explicitly maximize feature coverage, genomic DNA clearly provides more comprehensive coverage of the array features at all threshold values. Furthermore, feature coverage between repeated experiments was more consistent for genomic DNA than for the Stratagene standard.
Figure 2.
Comparison of genomic DNA cohybridization standard (black trace) with the Stratagene Mouse Universal Reference RNA standard (gray trace). (A) Intensity distributions from two separate hybridization experiments are compared. The background-subtracted denominator values are plotted on the horizontal axis, which is arranged with varying bin widths to accommodate the full intensity range. Values between –20 and 1000 have a bin width of 5, and values from 1000 to 70 000 have a bin width of 5000. (B) Comparison of percentile rank plots of Cy3 intensities for five replicate experiments in which genomic DNA (black traces) or Stratagene mixed cDNAs (gray traces) were co-hybridized with Cy5-labeled C2C12 cDNA to the mouse 16K chip. Log10 of the background subtracted intensity values (y-axis) were sorted, and assigned a percentile rank (x-axis). (C) Comparison of array feature coverage for RNA and genomic DNA cohybridization standards. The mean percent mouse feature coverage as a function of increasing threshold value is shown for five replicate experiments. Background thresholds are defined as the median fluorescence intensity for the group of negative control features, plus a multiple of the standard deviation value for the negative controls group (i.e. median negative controls + 1 SD, etc.). The feature coverage percentage is defined as the number of features exceeding a given background threshold value divided by the total number of mouse features on the array, multiplied by 100. Mean and standard deviation for the replicate groups at each threshold are plotted.
Comparison of variation in ratiometric measurements
To compare the reproducibility and stability of measurements using the two different kinds of standards, we made a set of five replicate measurements each for RNA and for genomic DNA standards, all cohybridized with the same ‘numerator’ RNA across the full 16K array. We first analyzed the data using correlation statistics over the entire array. The distribution of ratio values from these experiments roughly resembles a log-normal distribution (12). When transformed by taking the base 2 log, the distribution of ratiometric values appears more normal, but still deviates significantly from normality using the Kolmogorov–Smirnoff test. Under these conditions, the more appropriate metric for correlation between replicate distributions is the Spearman correlation coefficient (13). Averaging over all pair-wise comparisons, the Stratagene RNA experiments gave a mean Spearman correlation coefficient (rs) of 0.94, with a coefficient of variation of 1.04%. In contrast, all pair-wise comparisons of the genomic DNA experiments gave a mean Spearman correlation coefficient of rs = 0.99, with a CV of 0.43%. We conclude that inter-experiment ratiometric measurements are more precise when using the genomic DNA standard.
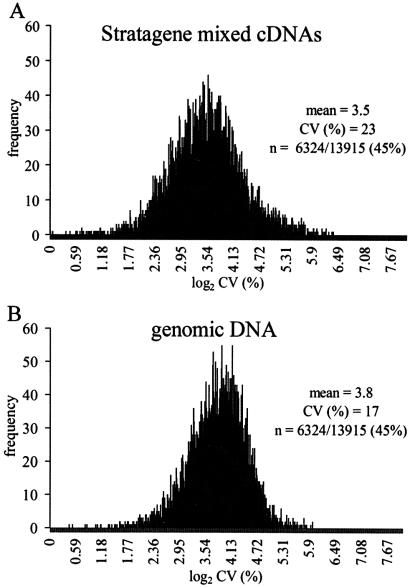
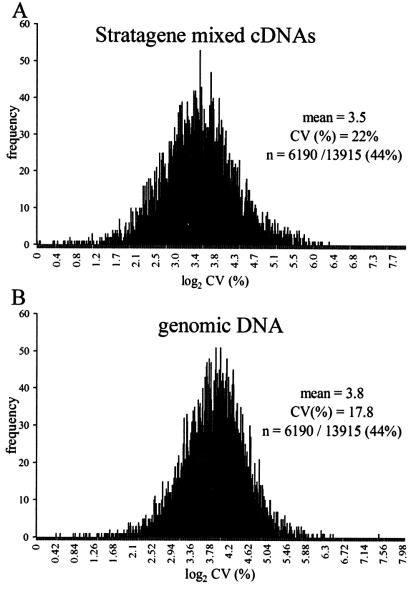
In use, the most critical subset of array data are those genes expressed significantly in at least one sample (numerator) RNA. We therefore focused on all genes with background-subtracted signal values uniformly >250 in the test numerator sample over all five Stratagene replicates (6324/13915 or 45% of the array features) (Fig. 3). The mean ratio, standard deviation and coefficient of variation [CV (%)] were calculated for the Stratagene RNA ratios, and for the corresponding set of genomic DNA ratios. The distribution of CVs for all 6324 features approximated log-normal. The base 2 log transformations are shown in Figure 3A and B. A two-tailed t-test, assuming unequal variances, indicates a significant difference between these two distributions. The mean value for the log2 CV (%) using the Stratagene cohybridization standard is slightly less than that using the genomic DNA method (3.5 for Stratagene versus 3.8 for genomic DNA, corresponding to CVs of 12 vs 14%), indicating that genomic DNA does not introduce a gross variation in the measurement of a ratio at any given feature. However, the smaller spread in the distribution of log2 CV (%) when the genomic DNA method is used (17% for genomic DNA versus 23% for Stratagene), may indicate that inter-experiment variation over the entire array is reduced when genomic DNA is used as the standard.
Figure 3.
Comparison of variation in ratiometric measurements using two different cohybridization standards. Replicate sets of five hybridization experiments were performed, each using mRNA from differentiated C2C12 skeletal muscle cells as the Cy5 numerator signal cohybridized against a denominator standard of either Cy3-labeled cDNA produced from the Stratagene mixed RNAs standard, or Cy3-labeled mouse genomic DNA. C2C12 samples were reverse transcribed separately, then pooled and split equally to minimize variation among the numerator samples applied to the arrays. (A) Plot of the distribution of variation for features with numerator values ≥250 on five separate experiments in which C2C12 72 h cDNA (Cy5) was cohybridized against the Stratagene mixed cDNA standard. Mean, standard deviation and CV (%) were computed for the five measurements for each feature on the array. CV (%) values were then log2 transformed and plotted as a histogram. Overall mean and CV (%) for the log2 CV (%) values are indicated. (B) The distribution of variation in ratios for arrays hybridized with a mouse genomic DNA standard. The same list of features analyzed in (A) was extracted, processed, and plotted for the DNA denominator in (B).
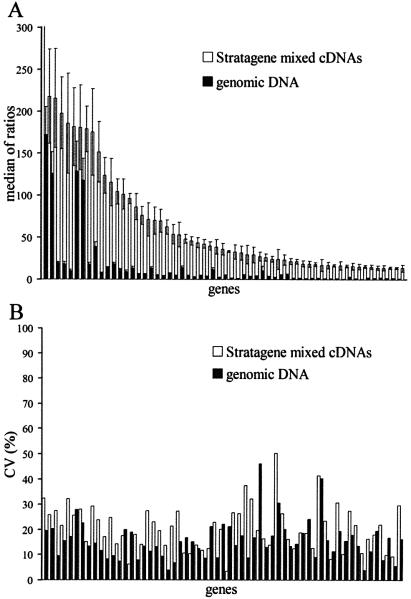
When comparing gene expression levels between two RNA samples, genes that are highly expressed in one sample and not expressed in the other are usually among the most interesting. However, this group is also at greatest risk of delivering highly variable and/or erroneous ratios when using RNA denominators with incomplete array feature coverage. Genes with only background level expression in the denominator standard often generate artificially high relative expression ratios and tend to be more unstable in repeat experiments than those with higher denominator values (see below). This, in turn, has important implications for data quality filtering schemes. For example, setting a reproducibility threshold for each feature could eliminate biologically important differentials, along with truly misleading noise. Denominator signal intensities using the genomic DNA cohybridization standard are, on average, very much lower than the average Stratagene signal intensity, yet they are much higher than the values for several thousand genes missing from the ‘Universal’ RNA mix. To investigate the effects of extremely low denominator values on ratiometric stability, Stratagene ratio measurements from a single chip were re-filtered to collect values with low denominator measurements (<4 SD above the median value for negative control probes) and ratio values >5. In Figure 4A, the ratio values for measurements over five genomic DNA replicates are consistently and appreciably reduced relative to the corresponding values taken over the Stratagene standard. Figure 4B compares coefficients of variation in mean ratio values, and clearly shows that for 44 of the 60 genes shown (73%), the effect of the increased denominator from genomic DNA was to reduce variation in the ratiometric measurement.
Figure 4.
Comparison of mean and variation for a subset of features with very low Stratagene reference values. (A) Shown here are 60 genes with Stratagene denominators <4 SDs above the median negative control value. Mean and standard deviation for the low denominator Stratagene group (open bars) and the corresponding genomic DNA measurements (black bars) are plotted as corresponding pairs. (B) Coefficients of variation for the paired measurements plotted in (A). CV (%) is calculated as (mean/SD) ∗ 100.
The mean and median signals for the Stratagene RNA reference are significantly higher than for genomic DNA. Following the expectation that larger absolute signals are generally more reproducible than low ones, a simple prediction is that genes displaying a denominator signal within an ‘optimal’ signal range for mixed RNA will have more robust and reproducible ratios than corresponding ones derived by using the genomic DNA reference. Thus, a Stratagene ‘sweet spot’ gene set, whose background-subtracted RNA denominator values in all five replicates were >200 and <10 000 (to avoid instability from non-linearity and saturation at very high values) were evaluated for stability. Log2 CV (%) values for 6190 features (44% of the entire array) are shown for the RNA standard (Fig. 5A) and the genomic DNA standard (Fig. 5B). Surprisingly, mean coefficient of variation values for the Stratagene and genomic DNA standards were essentially the same in this ‘RNA sweet spot’ set as for the entire array (mean = 3.5 for Stratagene versus 3.8 for genomic DNA). Even in this selected gene group, there is a wider spread in CV (%) values when the Stratagene Universal Mouse RNA standard is used, compared to the genomic DNA standard (22% for Stratagene versus 18% for genomic DNA). We conclude that, contrary to the naïve expectation, the higher and presumably more ideal universal RNA denominator signals that define this large gene group do not deliver ratios that are considerably more stable or reproducible than those obtained from genomic DNA.
Figure 5.
Signal variation comparisons for features with optimal RNA denominator signals. (A) Cy5-labeled C2C12 cDNA was hybridized against Cy3-labeled Stratagene mixed cDNAs in five replicate experiments. Features with denominator signals >200 and <10 000 in all five Stratagene replicates were extracted [n = 6190/13 916 features (44%)]. Log2 CV (%) for all 6190 features are plotted in the histogram. (B) The list of features analyzed in (A) was used to extract corresponding values from five replicate experiments in which Cy5-labeled C2C12 cDNA was cohybridized against Cy3-labeled genomic DNA. Log2 CV (%) for all 6190 features are plotted.
Use of a common hybridization denominator to infer expression ratios across arrays
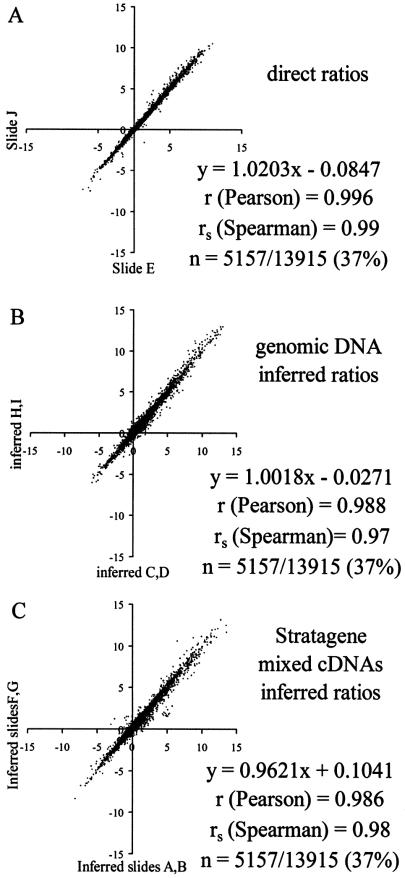
In microarray studies that survey more than two RNA sources, the relationship of any RNA in the experiment with any other (or all other) RNAs is inferred arithmetically through the common denominator. Reproducibility of a direct comparison of muscle and liver RNAs (Fig. 6A) was compared with corresponding indirect determinations, one of which used genomic DNA as the common denominator (Fig. 6B) and the other the Stratagene mixed cDNAs standard (Fig. 6C). To minimize the effects of variation in the numerator signal, the six C2C12 reverse transcription reactions were pooled and split evenly after labeling. Then, only features with numerator values >250 on all six C2C12 muscle cell arrays were included in this analysis (5157 features or 37% of total array features), although the conclusions are the same qualitatively if all data are used. As expected, in two independent replicates, the direct determinations (muscle over liver) were highly reproducible (rs = 0.99). Indirect ratio determinations were found to be only slightly less reproducible than direct ones, with Spearman correlation coefficients of rs = 0.97 for the genomic DNA cohybridization standard, and rs = 0.98 for the Stratagene standard. Given a correlation of 0.99 between two direct ratiometric measurements, 0.98–0.97 values for duplicate inferred ratios are near the theoretical maximum values.
Figure 6.
Correlation plots for replicate experiments of directly measured ratios, and replicate measurements of ratios inferred against either the genomic DNA denominator or the Stratagene mixed cDNAs denominator. (A) Correlation between duplicate arrays for directly measured ratios obtained after C2C12-labeled cDNA (Cy5) was directly cohybridized with adult mouse liver cDNA (Cy3). Only array features with C2C12 Cy5 numerator measurements >250 on all six arrays represented in this figure were included (n = 5157 features). Evaluation of the interslide precision of measurement is given by the Spearman correlation coefficient of rs = 0.99. (B) Correlation between duplicate measurements in which C2C12 over liver ratios were computed using a genomic DNA cohybridization standard. Inter-experiment precision is evaluated by the Spearman correlation coefficient rs = 0.97. (C) Correlation between duplicate C2C12 over liver ratios computed using the Stratagene mixed cDNAs cohybridization standard. Inter-experiment precision is evaluated by the Spearman correlation coefficient rs = 0.98.
Agreement between inferred ratios methods and directly measured ratios
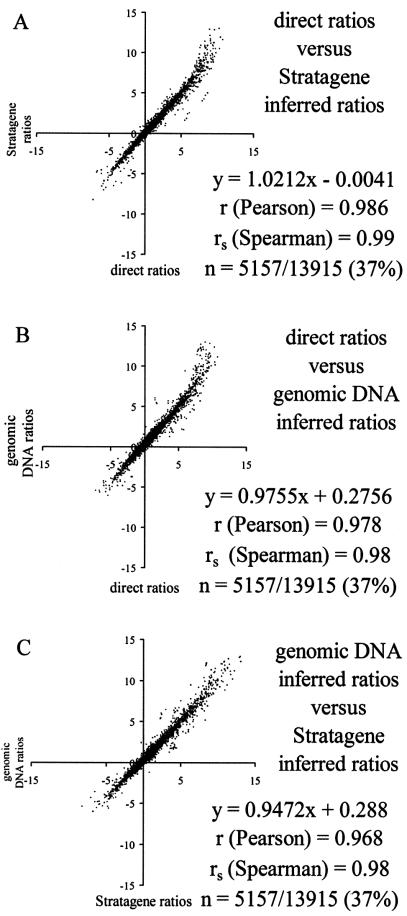
We next asked how similar are arithmetically inferred ratios made using either RNA or genomic DNA standards relative to the corresponding direct cohybridization ratios. We used the Spearman correlation metric, since the distribution of log2 ratio values differed significantly from normality. Experi ments were performed in duplicate, and ratio measurements were averaged prior to correlation comparisons. Spearman correlation values were rs = 0.99 for direct ratios against Stratagene ratios (Fig. 7A), or rs = 0.98 for direct ratios against genomic DNA and for Stratagene against genomic DNA (Fig. 7B and C). We conclude that the inferred ratios from both standards are globally very similar to each other and are also remarkably similar to the directly hybridized C2C12/liver determinations.
Figure 7.
Inter-method comparison of ratiometric values. Duplicate ratio measurements of C2C12 over liver cDNA (either direct or inferred) were averaged and then plotted against one another for correlation. (A) Correlation between direct ratios and ratios inferred against the Stratagene mixed cDNAs cohybridization standard are plotted. Spearman correlation is rs = 0.99. (B) Correlation between directly measured ratios and ratios inferred against the genomic DNA cohybridization standard are plotted. Spearman correlation is rs = 0.98. (C) Correlation between ratios inferred against the Stratagene standard and those inferred against genomic DNA. Spearman correlation is rs = 0.98.
DISCUSSION
This work was motivated by recurring design difficulties surrounding selection and use of the ‘most appropriate’ RNA cohybridization standard in gene expression microarray studies. The practical appeal of a genomic DNA cohybridization standard is considerable because of its universal availability and stability of sequence composition. Our central conclusion is that genomic DNA is a highly viable, and arguably superior, choice as a universal cohybridization standard for mouse gene expression experiments in the context of 70mer gene chips. In principle this should extend readily to microarrays with longer probe features, such as cDNA arrays, and to other genomes of similar or lesser size and sequence complexity (see below). Statistical analysis of replicate arrays confirmed the simple expectation that ratio values for features sequences absent from a reference RNA standard are unstable. In contrast, the presence of those same feature sequences in genomic DNA standards successfully stabilized their ratio values. Moreover, for the especially pertinent set of genes that are expressed above background levels in a typical ‘numerator’ or experimental RNA sample, there was negligible increase in ratio error associated with genomic DNA versus the current Stratagene mixed RNA standard. The error observed with the genomic DNA standard was also more narrowly distributed, which will be helpful for building useful error models. In a practical test of inferred ratios quality for muscle versus liver RNA using the genomic DNA standard, the Spearman correlation value was 0.98 to the directly measured muscle versus liver ratios.
The genomic DNA standard substantially reduced ratio instability for the subset of genes that have extremely low denominator values with the mixed RNA standard. These genes comprise a relatively small proportion of the total array, and therefore have only modest impact on overall statistics, yet they represent an especially important subset of the data that is of high biological interest. Moreover, genes with profiles of this type are likely to constitute a larger fraction of data from fully comprehensive arrays that are now coming into wide use. The oligonucleotide collection used here was an early version that over-represents well known genes, many of which had been found and characterized by classical methods that depended on expression in major adult tissues or cell lines. As a result, these genes should also be well represented in the current mixed RNA standard. In contrast, newer generation arrays that contain essentially all candidate mouse genes, based on the whole genome sequence, have an additional 10 000–15 000 genes that will presumably be biased in favor of RNAs expressed only during development, in specialized adult cell populations or at very low overall levels. We expect that genomic DNA will not have a bias against any of these added genes or against non-coding RNAs. In contrast, a mixed-source RNA standard will increasingly fall short of comprehensive coverage because it will be progressively more difficult to include more and more minor site RNA sources.
Protocol simplicity was emphasized to retain universality and increase robustness. We therefore elected not to fractionate single copy sequences using C0t based techniques, as this could only modestly increase signal, while introducing variability from one DNA preparation to another. Similarly, repeat suppression, before or during the array hybridization, did not significantly improve performance and so was not used.
Our conclusions regarding the efficacy of genomic DNA cohybridization standards are more optimistic than those from Weil et al. (7). In their study, human genomic DNA feature coverage was statistically equivalent (∼80% of spots above threshold) to a mixture of RNA derived from 10 human cell lines. They concluded that market preference would therefore be the driving force for selection of a mixed cell line RNA standard when available. More in-depth comparisons to pinpoint the most meaningful differences between these two studies is difficult because they include many technical differences in labeling and hybridization protocols, microarray types, genomes (mouse versus human), as well as differences in data processing and analysis.
Extension to other genomes
The central problem in developing mammalian genomic DNA normalization was the modest absolute hybridization signal achievable for typical single copy genes. This comes from large genome size and low mRNA sequence density, coupled with the technical limits of the current labeling protocol. A straightforward implication is that—without major technical changes or improvements—genomic DNA normalization for arrays of smaller, more gene dense genomes such as Caenorhabditis elegans, Drosophila melanogaster and Saccharomyces cerevisiae should be even better and more robust. Preliminary work with Arabidopsis, which is ∼20-fold greater in gene density, suggests this is the case (B.A.Williams and B.J.Wold, unpublished results). We observed a difference between mouse and Arabidopsis median probe signal that is close to the ratio predicted by relative genome sizes, and overall uniform feature coverage on Arabidopsis microarrays is consistent with this. However, predicting that genomic DNA normalization will work as well—or better—in Arabidopsis as in mouse should be tempered by a recent study of Arabidopsis genomic DNA normalization by Quackenbush and colleagues (14). In their hands, genomic DNA generally compared unfavorably with RNA. A clearer endorsement is in the literature for small microbial genomes, for which genomic DNA can easily be the most desirable cohybridization standard, as shown for Mycobacterium tuberculosis on cDNA arrays (9), and for Shewanella oneidensis in the 70mer format (B.A.Williams and T.K.Teal, unpublished results).
Extension to other microarray formats
We expect that our conclusions will extend from the 70mer oligonucleotide format to arrays of PCR products, although we have not yet tested this on a large scale. Eight different full-length cDNAs were represented in quadruplicate on each of our arrays. The average variation in the ratiometric values across five microarrays was indistinguishable from the variation for the 70mer oligonucleotides. We also note that our hybridization reaction conditions were intentionally optimized for 70mer probes that comprised the bulk of the array, and that more stringent hybridization conditions might further reduce ratiometric signal variation for PCR length products while maintaining or slightly increasing signals. Our results also invite the possibility of using the genomic DNA standard in a third label ‘color’ in the presence of two other labeled RNAs to increase information delivered per array and to gain the benefits of direct comparison while still including a universal standard.
Other denominator strategies
Alternate approaches to the same set of ratio denominator issues include synthesizing a labeled oligonucleotide of known specific activity complementary to a short, common ‘capture’ sequence on each microarray feature (15), synthesizing an oligonucleotide mix that is fully representative and equimolar for all genes, or making a labeled mixture of all cDNA inserts on an array (16,17). The latter method would become comprehensive for all mouse ORFs when (and if) the Mammalian Genome Collection (MGC) becomes complete (http://mgc.nci.nih.gov/), although full MGC completeness is not imminent. These methods have the virtue of allowing one to freely adjust signal by varying the amount of labeled standard in each hybridization. Moreover, dilution with labeled material that is non-reactive, which is a substantial technical issue for genomic DNA cohybridization standards, is not a problem. However, these methods are currently less general and less flexible than genomic DNA because they are not effective for arrays with features lacking the appropriate ‘capture’ sequence, or for arrays with features that are not represented in a given cohybridization custom mixture. Genomic DNA, in contrast, has the advantage of applying to all ratiometric array types and feature content.
On balance, we are optimistic that results similar to those reported here will be possible for smaller model genomes like yeast, C.elegans and Drosophila, and that extension to other large mammalian and plant genomes will be feasible. Benefits for subsets of the data, including genes with very large differences in expression between two RNA samples, as well as overall generality, argue in favor of genomic DNA as a standard of choice.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Joe DeRisi for advice on microarrayer software, Professors Jim Brody and Steve Quake for arrayer assembly, Professor Eric Mjolsness for advice on ROC analysis, and Professor Emmanuel Candes for consultation on the Spearman correlation coefficient. We gratefully acknowledge the generous gift of Arabidopsis microarrays and mature Arabidopsis plants from Dr Frank Wellmer of the Meyerowitz Laboratory. Funding was provided by the L. K. Whittier Foundation and an NCI Director’s Challenge Grant (U01 CA88199). B.A.W. was supported by an NIH/NRSA postdoctoral fellowship.
REFERENCES
- 1.Eisen M.B. and Brown,P.O. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 2.Shalon D., Smith,S.J. and Brown,P.O. (1996) A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res., 6, 639–645. [DOI] [PubMed] [Google Scholar]
- 3.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet. Suppl., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 4.Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,C.A., Spellman,P., Iyer,V., Jeffrey,S.S., van de Rijn,M., Waltham,M. et al. (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet., 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 5.Bergstrom D.A., Penn,B.H., Strand,A., Perry,R.L.S., Rudnicki,M.A. and Tapscott,S.J. (2002) Promoter-specific regulation of myoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell, 9, 587–600. [DOI] [PubMed] [Google Scholar]
- 6.Perou C.M., Sorlie,T., Eisen,M.B., van de Rijn,M., Jeffrey,S.S., Rees,C.A., Pollack,J.R., Ross,D.T., Johnsen,H., Akslen,L.A. et al. (2000) Molecular portraits of human breast tumours. Nature, 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 7.Weil M.R., Macatee,T. and Garner,H.R. (2002) Toward a universal standard: Comparing two methods for standardizing spotted microarray data. Biotechniques, 32, 1310–1314. [DOI] [PubMed] [Google Scholar]
- 8.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 9.Talaat A.M., Howard,S.T., Hale IV,W., Lyons,R., Garner,H.R. and Johnston,S.A. (2002) Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res., 30, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody J.P., Williams,B.A., Wold,B.J. and Quake,S.R. (2002) Significance and statistical errors in the analysis of DNA microarray data. Proc. Natl Acad. Sci. USA, 99, 12975–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metz C.E. (1978) Basic principles of ROC analysis. Semin. Nucl. Med., VIII, 283–298. [DOI] [PubMed] [Google Scholar]
- 12.Limpert E., Stahel,W.A. and Abbt,M. (2001) Log-normal distributions across the sciences: keys and clues. BioScience, 51, 341–352. [Google Scholar]
- 13.Spearman C. (1904) The proof and measurement of association between two things. Am. J. Psychol., 15, 72–101. [PubMed] [Google Scholar]
- 14.Kim H., Zhao,B., Snesrud,E.C., Haas,B.J., Town,C.D. and Quackenbush,J. (2002) Use of RNA and genomic DNA references for inferred comparisons in DNA microarray analyses. Biotechniques, 33, 924–930. [DOI] [PubMed] [Google Scholar]
- 15.Dudley A.M., Aach,J., Steffen,M.A. and Church,G.M. (2002) Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl Acad. Sci. USA, 99, 7554–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y.H., Dudoit,S., Luu,P., Lin,D.M., Peng,V., Ngai,J. and Speed,T.P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res., 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sterrenburg E., Turk,R., Boer,J.M., van Ommen,G.B. and den Dunnen,J.T. (2002) A common reference for cDNA microarray hybridizations. Nucleic Acids Res., 30, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]