Abstract
DNA barcodes have been increasingly used in authentication of medicinal plants, while their wide application in materia medica is limited in their accuracy due to incomplete sampling of species and absence of identification for materia medica. In this study, 95 leaf accessions of 23 species (including one variety) and materia medica of three Pharmacopoeia-recorded species of Angelica in China were collected to evaluate the effectiveness of four DNA barcodes (rbcL, matK, trnH-psbA and ITS). Our results showed that ITS provided the best discriminatory power by resolving 17 species as monophyletic lineages without shared alleles and exhibited the largest barcoding gap among the four single barcodes. The phylogenetic analysis of ITS showed that Levisticum officinale and Angelica sinensis were sister taxa, which indicates that L. officinale should be considered as a species of Angelica. The combination of ITS + rbcL + matK + trnH-psbA performed slight better discriminatory power than ITS, recovering 23 species without shared alleles and 19 species as monophyletic clades in ML tree. Authentication of materia medica using ITS revealed that the decoction pieces of A. sinensis and A. biserrata were partially adulterated with those of L. officinale, and the temperature around 80 °C processing A. dahurica decoction pieces obviously reduced the efficiency of PCR and sequencing. The examination of two cultivated varieties of A. dahurica from different localities indicated that the four DNA barcodes are inefficient for discriminating geographical authenticity of conspecific materia medica. This study provides an empirical paradigm in identification of medicinal plants and their materia medica using DNA barcodes.
Keywords: Angelica, authentication, decoction pieces, DNA barcodes, materia medica
Introduction
DNA barcoding uses standardized short DNA regions as tags for rapid and accurate identification of species. This technique has been widely applied in molecular systematics (Hebert et al. 2003), elucidation of cryptic species (Ragupathy et al. 2009), characterization of genetic diversity and conservation of endangered species (Sucher & Carles 2008). Various genes and noncoding regions in the plastid genome had been proposed as the DNA barcodes for plants (Kress & Erickson 2007; Hollingsworth et al. 2011). The CBOL Plant Working Group (2009) initially suggested portions of the plastid coding genes rbcL and matK as the core barcodes to establish a barcoding database for plant species. However, these two core barcodes are not suitable for closely related plant species (Spooner 2009; Ren et al. 2010). The trnH-psbA intergenic spacer is the most variable plastid region in angiosperms and is easily amplified across a broad range of land plants (Kress et al. 2005). The nuclear internal transcribed spacer (ITS) that possesses high rate of nucleotide substitution and relatively high discrimination power have been extensively utilized in plant molecular systematics (Baldwin et al. 1995; Sass et al. 2007; Ren et al. 2010; China Plant BOL Group 2011). Therefore, Kress et al. (2005) recommended ITS and trnH-psbA as potentially usable DNA barcodes for flowering plants. ITS2 (160–320 bp), a subset of ITS, exhibits outstanding advantages in identification of medicinal plants and materia medica due to easier PCR amplification and sequencing (Chiou et al. 2007; Chen et al. 2010) and has been selected as a valuable sequence tag for barcoding termed ‘minibarcode’ (Chen et al. 2010; Yao et al. 2010; Han et al. 2013).
Plant species with medicinal values are abundant in China, of which more than 5000 species are used as materia medica for therapy in traditional Chinese medicine (Heubl 2010). These materia medica have great contribution to the health of Chinese people. However, substantial adulteration with closely related or unrelated materials may occur accidentally or intentionally in materia medica (China Plant BOL Group 2011). Authentication of materia medica is time-consuming and knowledge intensive because they are always in the forms of crude drug and decoction piece, which are air-dried or processed by various methods (stir-frying with adjuvants, baking, charring, steaming, boiling, calcining and so on) and thus with modified morphological and anatomical features. DNA technology is not affected by developmental stages, environmental factors, cultivation area, harvesting period and storage conditions (Heubl 2010) and provides a powerful tool to assist macroscopic, microscopic, and phytochemical analyses for authentication of materia medica to ensure safety, quality, efficacy, and legality of traditional medicines (Barthelson et al. 2006; Coghlan et al. 2012). DNA regions, rbcL, matK, trnH-psbA and ITS, have been generally applied to clarify confusable herbal materials (Kondo et al. 2007; China Plant BOL Group 2011; Kool et al. 2012; Zhang et al. 2012), and a sequence database of commonly used Chinese herbal medicines was established (Chen et al. 2010). These studies advanced the molecular authentication of traditional medicines, whereas the widespread and practical applications of DNA barcodes have been constrained by several issues. First, the identification accuracy for medicinal plants is influenced by incompletely sampling of accessions because the species without medicinal value in the same genus or family, which are the most potential source of adulteration, are generally excluded. DNA barcoding is likely to fail when only a subset species of a genus is examined just for a specific purpose such as medicinal plants identification (Zuo et al. 2011). Therefore, precise verification of DNA barcodes requires a thorough sample collection comprising medicinal and nonmedicinal species in a genus. Second, previous studies focused on materials of original plants but not materia medica and thus are not practical for identification of materia medica. Materia medica is generally difficult to be identified using DNA-based techniques (Heubl 2010). The postharvest processes of materia medica may cause severe DNA degradation and result in exceeding difficulties in barcoding materia medica. Therefore, the impact of processing procedures on PCR amplification and sequencing need to be further addressed for practical demand. Third, application of DNA barcoding in identification of medicinal plants did not meet the actual demand for clarification of geographical authenticity of materia medica. The quality of conspecific materia medica produced in different localities might be affected by genetic variation and geographical differentiation, which makes quality of materia medica associate with localities. This association is called geographical authenticity of materia medica, which is an important parameter evaluating quality of materia medica, and may be verified by identification of genetic variation associated with localities. However, little information was provided to test whether DNA barcodes are suitable markers for exploring the geographical authenticity of materia medica.
Angelica L., an herbaceous perennial genus of Umbelliferae, is extremely polymorphic in fruit anatomy, leaf morphology and subterranean structures (Vasil'eva & Pimenov 1991). Discrimination among Angelica species is highly difficult because of a complex and controversial taxonomic history. There are 45 Angelica species distributed in China, of which 32 are endemic (She et al. 2005; Feng et al. 2009), while some species are known only from a few specimens and extremely rare in the field. Delimitation of 26 species and five varieties in Flora Republicae Popularis Sinicae (http://frps.eflora.cn/frps/Angelica) is more reasonable in practice according to the opinion of Nian-He Wang, the taxonomist of Angelica. Three species of Angelica, A. sinensis (Oliver) Diels (Chinese medicine name: Danggui), A. biserrata (Shan et Yuan) Yuan et Shan (Duhuo) and A. dahurica (Fischer ex Hoffmann) Bentham & J. D. Hooker ex Franchet & Savatier (Baizhi) are Pharmacopoeia-recorded materia medica, namely official materia medica (Chinese Pharmacopoeia Committee of People's Republic of China's 2010). Around 50 000–61 000 tons of the three official materia medica, estimated 1.4–1.7 billions RMB, are produced per year in China, where medicinal materials of A. sinensis, A. biserrata and A. dahurica are from cultivation, field collection and half-cultivation, respectively (Sun et al. 2009; Wang et al. 2002; http://www.zyctd.com/). In addition, 15 species (including one variety) of Angelica are used as herbal medicinal materials in folk remedies (http://frps.eflora.cn/), which are usually collected from the field.
The materia medica of Angelica are often misused or intentionally substituted in the market for cost concern. Angelica sinensis and A. biserrata are reciprocally substituted, and Levisticum officinale W. D. J. Koch (Oudanggui) is frequently used as their alternatives. Levisticum officinale, native to southwest Asia and Europe, was officially introduced from Europe in 1957 as a substitute to mitigate the large demand of A. sinensis (She et al. 2005) and then was widely cultivated in provinces of Mongolica, Hebei, Shandong, Henan, Shanxi and Shaanxi. Afterwards, pharmacological experiments indicated that the medicinal efficacy of L. officinale is lower than that of A. sinensis, and thus, L. officinale was excluded in Chinese Pharmacopoeia (2005) and prohibited to be used instead of A. sinensis since 1983. However, intentional cultivation of L. officinale as the alternatives for A. sinensis and A. biserrata is prevalent in some provinces, such as Hebei, Shandong and Henan (Sun et al. 2009). In addition, five species of Angelica are used as folk substitutes, A. nitida H. Wolff and A. gigas Nakai for A. sinensis, A. anomala AvÈ-Lallemant and A. porphyrocaulis Nakai & Kitagawa for A. biserrata, and A. polymorpha Maximowicz for A. dahurica, respectively.
The processing procedures for the three official materia medica of Angelica from crude drugs to decoction pieces are different. The decoction pieces of A. sinensis and A. biserrata, smaller and thinner than those of A. dahurica, are usually baked at 60 °C which might have less impact on the performance of DNA barcoding. The decoction pieces of A. dahurica are conducted by baking the slices of crude drugs at 80 °C (Tan et al. 2013) which might cause severe DNA degradation and render PCR amplification and sequencing difficult.
Two cultivated varieties of A. dahurica, A. dahurica (Fischer ex Hoffmann) Bentham & J. D. Hooker ex Franchet & Savatier cv. Qibaizhi and A. dahurica (Fischer ex Hoffmann) Bentham & J. D. Hooker ex Franchet & Savatier cv. Hangbaizhi (http://frps.eflora.cn/), with various levels of quality resulted from different cultivation localities, are the sources of materia medica of A dahurica in the market. However, these materia medica are difficult to be differentiated according to the morphological and micromorphological traits. Evaluation of the efficiency of DNA barcodes in clarifying geographical authenticity of materia medica of A. dahurica will provide deeper and practical insights for further quality control.
This report aims to evaluate the effectiveness of the empirical DNA barcodes rbcL, matK, trnH-psbA and ITS for discrimination of medicinal Angelica species based on phylogeny, elucidates the impacts of processing procedures on barcoding decoction pieces and scrutinizes the feasibility of DNA barcodes for identification of the geographical authenticity of materia medica.
Materials and methods
Leaf accessions
Twenty-three species (including one variety) of Angelica from 46 populations located in 14 provinces, representing around 85% species and covering most of the distribution range of Angelica in China (http://frps.eflora.cn/), were sampled in 2010 (Table S1, Supporting information). The leaf accessions of 1–5 individuals were collected for each population. Leaves of Pleurospermum angelicoides (Wallich ex de Candolle) C. B. Clarke (Umbelliferae) were sampled from Danba county of Sichuan province as an outgroup (Table S1, Supporting information). Fresh leaves were instantly dried with silica gels for DNA extraction. Voucher specimens were deposited in herbaria of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences. The precise geographical location of each sampled population was determined using a Garmin GPS unit.
Decoction pieces of A. sinensis, A. biserrata and A. dahurica
Decoction pieces of A. sinensis, A. biserrata and A. dahurica were purchased from 10 Chinese medicine stores distributed in 10 cities of eight provinces in China: Beijing, Wuhan, Zhoukou, Changsha, Zibo, Binzhou, Dezhou, Qitaihe, Chengdu and Hejing (sample code of 10 medicine stores: A. sinensis: GBJ, GWH, GZK, GCS, GZB, GBZ, GDZ, GQT, GCD, GHJ; A. biserrata: HBJ, HWH, HZK, HCS, HZB, HBZ, HDZ, HQT, HCD, HHJ; A. dahurica: BBJ, BWH, BZK, BCS, BZB, BBZ, BDZ, BQT, BCD, BHJ). Three to eight of decoction pieces for each species from each medicine store were used in the experiments.
Crude drugs of A. dahurica
Crude drugs of two cultivated varieties of A. dahurica were collected from four different traditional producing localities: A. dahurica cv. Qibaizhi was from Anguo of Hebei province and Yuzhou of Henan province (sample code: BAG and BYZ); A. dahurica cv. Hangbaizhi was from Hefei of Anhui province and Suining of Sichuan province (sample code: BHF and BSN). After cleaning, the fresh roots were air-dried prior to use. The roots of three to five individuals from each producing locality were tested in the experiments.
All the decoction pieces and crude drugs were morphologically and microscopically identified by Prof. Shi-Yuan Jin and then deposited in herbaria of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences.
DNA extraction, PCR amplification and sequencing
Total DNA of dried leaves of Angelica species, decoction pieces of official medicinal species A. sinensis, A. biserrata and A. dahurica, and crude drugs of A. dahurica was extracted using 2× CTAB method (Doyle & Doyle 1987). The concentration of DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The representative fragments of different DNA barcodes were amplified using the primers described as follows: 1F (ATGTCACCACAAACAGAAAC) and 724R (TCGCATGTACCTGCAGTAGC) for rbcL (Fay et al. 1997), 3F_KIMf (CGTACAGTACTTTTGTGTTTACGAG) and 1R_KIMr (ACCCAGTCCATCTGGAAATCTTGGTTC) for matK (CBOL Plant Working Group 2009), trnH2 (CGCGCATGGTGGATTCACAATCC) (Tate & Simpson 2003) and psbAF (GTTATGCATGAACGTAATGCTC) (Sang et al. 1997) for trnH-psbA, and ITS5 (GGAAGTAAAAGTCGTAACAAGG) and ITS4 (TCCTCCGCTTATTGATATGC) for ITS (White et al. 1990). The PCR amplification was performed with TC-512 thermocycler (Techne, England). Each 20 μL PCR solution contained 0.5 μL of template DNA (40–80 ng), 2 μL 10× PCR reaction buffer, 1.6 μL 2.5 mm dNTP, 0.5 μL 10 μm forward/reverse primers, 0.2 μL Ex-Taq DNA polymerase (Takara Shuzo Co., Ltd., Otsu, Japan) and 14.7 μL sterile distilled water. The conditions for PCR were 35 cycles of 94 °C for 30 s, 56 °C for 20 s and 72 °C for 20 s. PCR products were examined with 1.5% agarose gel electrophoresis and purified through precipitation with 95% ethanol and 3 m sodium acetate (pH 5.2). All the purified PCR products were subjected to double-stranded sequencing using the DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech) according to the manufacturer's instruction, and analysed with an ABI 377XL automated sequencer.
Analysis of data sets
All the sequences excluding primer regions were edited and assembled using contigexpress (vector nti advance version 10.3.1, Invitrogen) and bioedit 7.0.9 assisted with manual correction. Sequence alignments for each region were carried out using clustalx version 2.0 (Larkin et al. 2007) and then modified artificially. Multiple sequence alignments were concatenated into a single file using geneious pro version 4.8.5 (Drummond et al. 2009). To check for shared alleles between species, accessions from each species were merged into one or more private sequences using dambe (Xia & Xie 2001). Pairwise interspecific and intraspecific distances for each locus were analysed based on the Kimura 2-parameter (K2P) model through mega version 4.0 (Tamura et al. 2007). The barcoding gaps were graphed by comparing the distributions of intra- and interspecific divergences of each barcode. Neighbour-joining (NJ) and standard phylogenetic trees, maximum parsimony (MP) and maximum likelihood (ML) trees, were estimated to evaluate whether species were recovered as monophyletic with each barcode and their combinations. NJ tree was analysed based on K2P model with mega version 4.0 (Tamura et al. 2007). MP and ML trees were constructed with paup version 4.0b10 (Swofford 2002). For the ML analysis, the best model of nucleotide substitution was calculated with modeltest 3.7 (Posada & Crandall 1998). Branch support for all analyses was assessed with 10 000 bootstrap replications. ITS or ITS2 sequences of four species of Umbelliferae, Levisticum officinale, Ostericum grosseserratum (Maximowicz) Kitagawa, Peucedanum delavayi Franchet and Peucedanum elegans Komarov retrieved from GenBank and the sequences of four barcodes of Pleurospermum angelicoides (Umbelliferae) obtained from collected leaf accessions were used as the outgroups for NJ and standard phylogenetic analyses (Table S1, Supporting information). The identification efficiency of single and combined barcodes was determined by evaluating the number of species without shared alleles and percentage of monophyletic species in NJ, MP and ML trees.
Results
Character analysis of each barcode
A total of 95 individuals representing 23 species (including one variety) of Angelica were successfully amplified and sequenced using four DNA barcodes. With regard to universality of primers and success of PCR and sequencing, the proportion at each of the four regions was 100% (Table 1). The newly generated sequences in this study were submitted to GenBank (Table S1, Supporting information). ITS sequences ranged from 670 to 676 bp with 166 variable sites, 164 informative sites and 12 indels of 1–4 bp within aligned 687 bp. The trnH-psbA intergenic spacer region ranged from 261 to 307 bp with 31 variable sites, 31 informative sites and 16 indels of 1–24 bp within aligned 373 bp, wherein A/T repeats occurred at 84–102 bp, 115–147 bp and 272–283 bp. The matK and rbcL genes, both without indels, were 834 bp with 38 variable and 35 informative sites, and 703 bp with 12 variable and 11 informative sites, respectively (Table 1).
Table 1.
Evaluation of four DNA barcoding regions, three chloroplast regions (rbcL, matK and trnH-psbA) and the nuclear ITS region
| DNA region | rbcL | matK | trnH-psbA | ITS |
|---|---|---|---|---|
| Universal ability to primer | Yes | Yes | Yes | Yes |
| Percentage PCR success (%) | 100 | 100 | 100 | 100 |
| Percentage sequencing success (%) | 100 | 100 | 100 | 100 |
| Aligned sequence length (bp) | 703 | 834 | 373 | 687 |
| No. indel (length in bp) | 0 | 0 | 16 (1–24) | 12 (1–4) |
| No. informative sites/variable sites | 11/12 | 35/38 | 31/31 | 164/166 |
| No. sampled species (individuals) | 23 (95) | 23 (95) | 23 (95) | 23 (95) |
| Interspecific distance mean (range) (%) | 0.025 (0–0.6) | 0.071 (0–1.7) | 0.158 (0–3.8) | 0.471 (0–11.3) |
| Intraspecific distance mean (range) (%) | 0.004 (0–0.1) | 0.013 (0–0.3) | 0.058 (0–1.4) | 0.008 (0–0.2) |
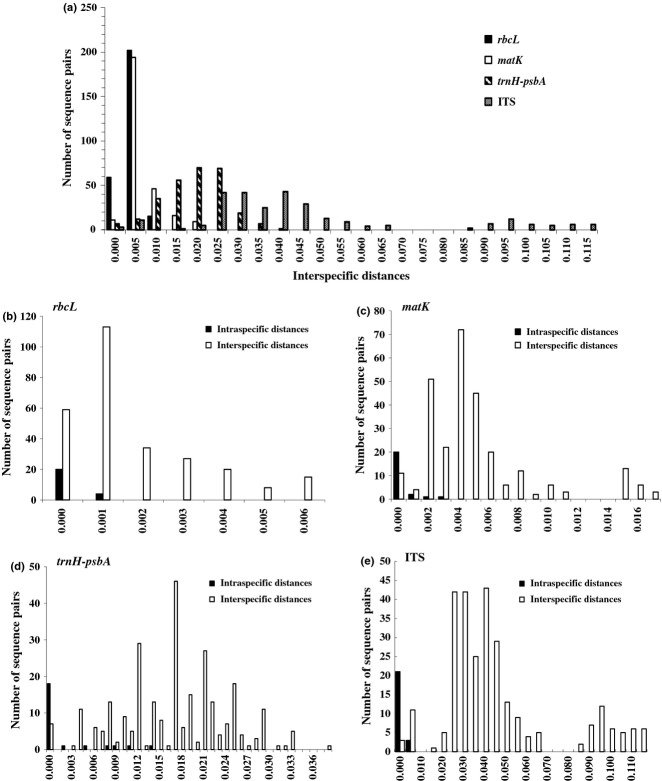
The mean interspecific/intraspecific genetic distances of 23 measurements with K2P were 0.025% (0–0.6%)/0.004% (0–0.1%) for rbcL, 0.071% (0–1.7%)/0.013% (0–0.3%) for matK, 0.158% (0–3.8%)/0.058% (0–1.4%) for trnH-psbA and 0.471% (0–11.3%)/0.008% (0–0.2%) for ITS (Table 1). The distribution of congeneric species distance from the four markers and their individual relative distributions of interspecific and intraspecific distances are shown in Fig. 1. In general, the mean interspecific distances were higher than those of the intraspecific distances for the four markers. ITS showed the largest barcoding gap with more than 50 times higher inter- vs. intraspecific distance, whereas the trnH-psbA region exhibited the highest intraspecific genetic distance in the four DNA regions (Table 1). The rank of mean sequence divergences in Angelica species was ITS > trnH-psbA > matK > rbcL.
Fig 1.
(a) Relative distribution of interspecific distances between congeneric species from four DNA barcodes; (b–e) Relative distributions of intraspecific and interspecific distances from rbcL, matK, trnH-psbA and ITS, respectively.
Phylogenetic analyses of single or combined locus
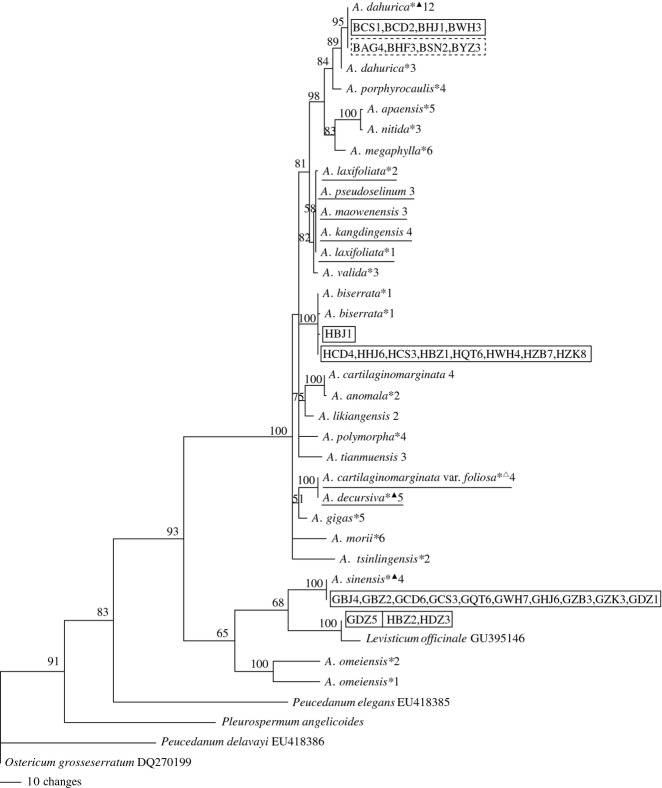
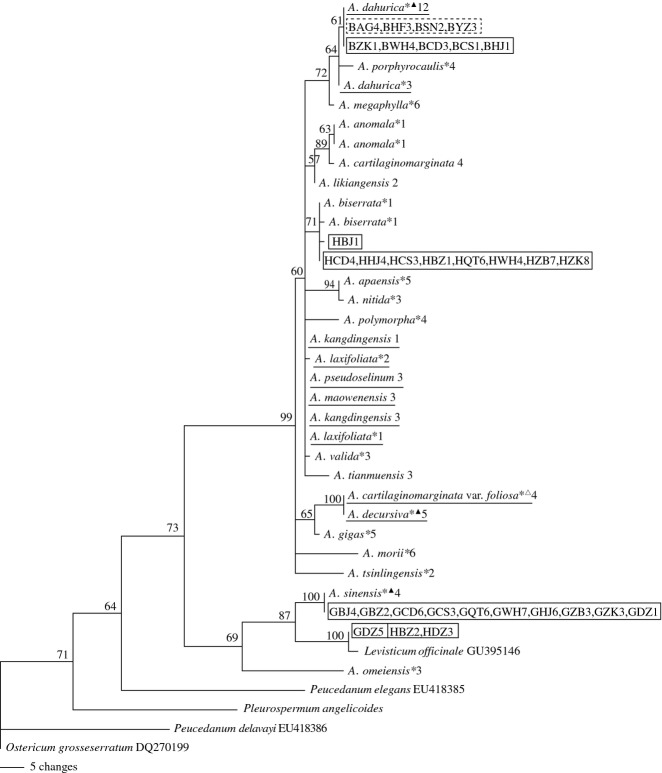
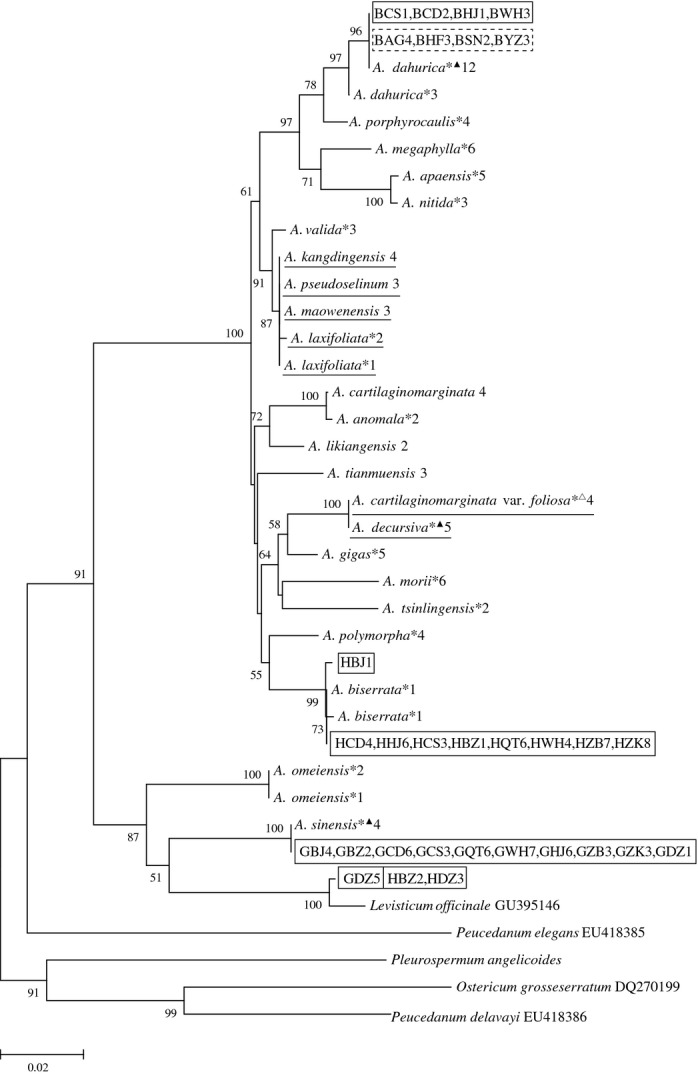
The identification efficiency of single and combined barcodes was determined by evaluating the number (or percentage) of species without shared alleles and monophyletic species in NJ, MP and ML trees. In all cases, NJ analyses discriminated an equal or greater percentage of species resolved as monophyletic clades than did ML analyses, and ML analyses discerned a greater or equal percentage than MP analyses except ITS + matK + trnH-psbA (Table 2). For single barcode, ITS exhibited the highest success rates of identification for Angelica species. Seventeen species were successfully discriminated without shared alleles and grouped together in mutually exclusive monophyletic clades (73.91%) in NJ, MP and ML trees with bootstrap values >80%, among which 11 species are used as herbal medicinal materials in folk remedies and three species (A. sinensis, A. biserrata and A. dahurica) are Pharmacopoeia-recorded materia medica, namely official materia medica (Figs 2 and 3; Fig. S1, Supporting information). Compared to ITS, ITS2 showed lower clade support values in NJ, MP and ML trees (Fig. 4; Figs S2 and S3, Supporting information) and did not assemble A. dahurica as a single monophyletic group in MP tree (Fig. 4), although the number of species without shared alleles and the percentage of monophyletic species in NJ and ML trees were the same as those of ITS (Table 2). Therefore, ITS2 is less suitable as a useful barcode for identification of A. dahurica and its materia medica. In addition, the outgroup Levisticum officinale was nested within Angelica as a sister group of A. sinensis in NJ, MP and ML trees of ITS and ITS2 (Figs 4; Figs S1–S3, Supporting information).
Table 2.
The number (percentage) of species without shared alleles and monophyletic species in 23 Angelica species (including 1 variety) derived from neighbour-joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) trees for four DNA barcodes and their combinations
| Potential barcode | No. species without shared alleles | No. monophyletic species (%) | ||
|---|---|---|---|---|
| NJ | MP | ML | ||
| rbcL | 6 | 6 (26.09) | 6 (26.09) | 6 (26.09) |
| matK | 11 | 10 (43.48) | 10 (43.48) | 10 (43.48) |
| trnH-psbA | 15 | 11 (47.83) | 11 (47.83) | 11 (47.83) |
| ITS | 17 | 17 (73.91) | 17 (73.91) | 17 (73.91) |
| ITS2 | 17 | 17 (73.91) | 16 (69.57) | 17 (73.91) |
| ITS + rbcL | 18 | 18 (78.26) | 16 (69.57) | 16 (69.57) |
| ITS + matK | 19 | 19 (82.61) | 17 (73.91) | 17 (73.91) |
| ITS + trnH-psbA | 23 | 19 (82.61) | 16 (69.57) | 17 (73.91) |
| ITS + rbcL + matK | 21 | 20 (86.96) | 17 (73.91) | 17 (73.91) |
| ITS + rbcL + trnH-psbA | 23 | 19 (82.61) | 16 (69.57) | 18 (78.26) |
| ITS + matK + trnH-psbA | 23 | 19 (82.61) | 18 (78.26) | 16 (69.57) |
| ITS + rbcL + matK + trnH-psbA | 23 | 19 (82.61) | 18 (78.26) | 18 (78.26) |
| matK + rbcL | 13 | 11 (47.83) | 11 (47.83) | 11 (47.83) |
| rbcL + matK + trnH-psbA | 19 | 15 (65.22) | 14 (60.87) | 14 (60.87) |
Fig 2.
Neighbour-joining tree of 23 Angelica species (including one variety) combined with the decoction pieces of three official materia medica (boxed in real line rectangle) and crude drugs of A. dahurica (boxed in dotted line rectangle) based on ITS sequences. The symbols on the top right corner of each species refer to Table S1, and the followed numbers behind each species and materia medica represent the sample size. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig 3.
Maximum parsimony tree of 23 Angelica species (including one variety) based on ITS sequences. The denotation on the right side of each species and the information of materia medica refer to Fig. 2. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig 4.
Maximum parsimony tree of 23 Angelica species (including one variety) based on ITS2 sequences. The denotation on the right side of each species and the information of materia medica refer to Fig. 2. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
The combinations of ITS with plastid regions increased the number of species without shared alleles, while the percentage of monophyletic species was not significantly improved. The combination of ITS + rbcL + matK + trnH-psbA obtained the largest number of species without shared alleles (23 species), and one or two monophyletic species were increased in MP or both NJ and ML trees compared to ITS, while A. dahurica was not assembled as a monophyletic clade in MP tree (Table 2; Figs S4–S6, Supporting information). The combination of ITS + rbcL + matK recovered the highest number of monophyletic species (20 species) in NJ tree, but the number of monophyletic species in MP and ML trees was the same as that of ITS (Table 2). The combination of matK + rbcL exhibited the fewest species without shared alleles (13 species) and the lowest monophyletic species (11 species) in NJ, MP and ML trees in all combinations and ITS alone (Table 2; Figs S7–S9, Supporting information). Therefore, the two-locus combination of matK and rbcL suggested by the consortium for the barcode of life (CBOL Plant Working Group 2009) is comparatively less effective for discrimination of Angelica species.
With regard to universality of primers, success of PCR and sequencing, distribution of intra- and interspecific distances, species without shared alleles and monophyly value, ITS displayed the highest discrimination ability of species and thus is the most promising barcoding region for identification of Angelica species and authentication of their materia medica. The combinations of ITS with plastid regions slightly improved the discrimination ability of species but increased the complexity in practical application which render them unsuitable for widespread use in materia medica authentication.
Authentication of decoction pieces of A. sinensis and A. biserrata
In an attempt to authenticate decoction pieces of A. sinensis and A. biserrata, three to eight pieces of decoction pieces for each species from each one of ten medicine stores were analysed with four DNA regions (Table 3). High success rate of PCR (>88.9% and >89.1%) and sequencing (>85.2% and >84.1%) were obtained from the decoction pieces of A. sinensis and A. biserrata, indicating that the processing procedures for decoction pieces of A. sinensis and A. biserrata have slight impact on their molecular authentication.
Table 3.
Percentage of PCR amplification and sequencing success of the five DNA barcoding regions for the decoction pieces and crude drugs of three official materia medica, Angelica sinensis, A. biserrata and A. dahurica
| Percentage PCR success (%) | Percentage sequencing success (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Decoction pieces | Crude drugs | Decoction pieces | Crude drugs | |||||
| DNA region | Angelica sinensis | Angelica biserrata | Angelica dahurica | Angelica dahurica | Angelica sinensis | Angelica biserrata | Angelica dahurica | Angelica dahurica |
| rbcL | 92.3 | 92.5 | 51.7 | 92.9 | 95.4 | 95.8 | 93.5 | 92.3 |
| matK | 88.9 | 89.1 | 30.0 | 100 | 85.6 | 85.9 | 55.6 | 85.7 |
| trnH-psbA | 96.6 | 95.5 | 40.0 | 85.7 | 91.2 | 91.0 | 62.5 | 91.7 |
| ITS | 92.3 | 91.5 | 46.7 | 100 | 85.2 | 84.1 | 25.0 | 85.7 |
| ITS2 | 51.7 | 32.2 | ||||||
Ninety-one ITS sequences of decoction pieces of A. sinensis and A. biserrata were used to construct NJ and phylogenetic trees with those of leaf accessions of 23 species (including one variety) of Angelica (Fig. 2). All decoction pieces of A. sinensis separately collected from Beijing (GBJ), Hejing (GHJ), Chengdu (GCD), Changsha (GCS), Zibo (GZB), Wuhan (GWH), Binzhou (GBZ), Zhoukou (GZK), Qitaihe (GQT) and one piece from Dezhou (GDZ) fell into a monophyletic clade with leaf accessions of A. sinensis. All decoction pieces of A. biserrata from Beijing (HBJ), Hejing (HHJ), Chengdu (HCD), Changsha (HCS), Zibo (HZB), Wuhan (HWH), Zhoukou (HZK), Qitaihe (HQT) and one piece from Binzhou (HBZ) were grouped into a monophyletic lineage with leaf accessions of A. biserrata. Five decoction pieces of A. sinensis from Dezhou (GDZ) and five decoction pieces of A. biserrata, two pieces from Binzhou (HBZ) and three pieces from Dezhou (HDZ), shared the same allele and formed a monophyletic clade together with Levisticum officinale, indicating that the decoction pieces of A. sinensis and A. biserrata in the market are adulterated with those of L. officinale. The reciprocal replacement of A. sinensis and A. biserrata was not observed.
Authentication of materia medica of A. dahurica
To investigate the efficiency and feasibility of DNA barcodes in identification of materia medica differently processed, a set of experiments were performed to verify the appropriate barcodes. Three to eight decoction pieces from each one of ten medicine stores and three to five individual crude drugs from each one of four authentic producing localities of A. dahurica were analysed and compared using four DNA regions and ITS2. The PCR amplification/sequencing efficiency of decoction pieces was relatively low using matK (30.0%/55.6%), trnH-psbA (40.0%/62.5%) and ITS (46.7%/25.0%), while rbcL had higher sequencing efficiency (51.7%/93.5%; Table 3). A slight increase in PCR amplification/sequencing efficiency for decoction pieces of A. dahurica (51.7%/32.2%) was detected with the ITS2 region. Higher efficiency (above 85.7%) of PCR amplification and sequencing was obtained for crude drugs using all four DNA barcodes (Table 3). These results indicated that the processing procedures from crude drugs to decoction pieces of A. dahurica dramatically decreased the efficiency and feasibility of DNA barcodes identification, and ITS2 showed slight improvement in PCR amplification/sequencing efficiency for decoction pieces of A. dahurica.
In NJ, MP and ML trees of ITS (Figs 2 and 3; Fig. S1, Supporting information), all decoction pieces and leaf accessions of A. dahurica were grouped into a monophyletic clade with bootstrap values >80%, showing that decoction pieces of A. dahurica analysed were not adulterated.
Authenticating of crude drugs of A. dahurica from different localities
All crude drugs of two cultivated varieties of A. dahurica from four different traditional producing localities, A. dahurica cv. Qibaizhi and A. dahurica cv. Hangbaizhi, shared the same allele in each of the four barcodes and were grouped into a monophyletic clade with leaf accessions of A. dahurica with bootstrap values >80% in NJ, MP and ML trees of ITS (Figs 2 and 3; Fig. S1, Supporting information). Therefore, the four DNA barcodes were not promising to discriminate the materia medica of cultivated varieties of A. dahurica produced in different localities.
Discussion
The most ideal DNA barcode for Angelica species
An ideal DNA barcode should be easily amplified with a pair of universal primers, directly sequenced, quickly aligned without manual editing and highly differentiated between sampled species (CBOL Plant Working Group 2009). Our results demonstrated that ITS region is the most suitable DNA barcode for identification of Angelica species, with 100% PCR and sequencing success, few indels and relatively easy alignment. Moreover, ITS showed the best performance with differentiating 73.91% (17/23) of the species and displayed the largest barcoding gap with more than 50 times higher inter- vs. intraspecific distance (Table 1). These results are consistent with previous studies that ITS can differentiate closely related congeneric species because of its high mutation rate (Kress et al. 2005; Nieto Feliner & Rosselló 2007; Sass et al. 2007). A caveat for ITS as a barcode is that incomplete concerted evolution may complicate the relationship among species (Alvarez & Wendel 2003); however, this study did not find it in Angelica species, echoing that ITS region is a potential DNA barcode for flowering plants (Kress et al. 2005; Sass et al. 2007). The proposed core barcode rbcL + matK discriminated only 47.83% (11/23) of the sampled Angelica species, which is much lower than the previously reported 72% by CBOL Plant Working Group (2009). This result confirms that matK + rbcL does not perform well in Umbelliferae (China Plant BOL Group 2011).
Phylogenetic position of Levisticum officinale
Levisticum officinale and A. sinensis have been classified in different genus; however, the two species could have close relationship because of their affinity in morphological characteristics, chemical constituents and pharmacological efficacy (Zschocke et al. 1998). In a comprehensive molecular systematic studies of Asian Umbelliferae, L. officinale and A. sinensis are sister taxa, which belongs to the Sinodielsia clade (S. R. Downie, D. S. Katz-Downie & M. F. Watson, unpublished data). In our study, the phylogenetic analysis of 23 Angelica species combined with L. officinale using ITS demonstrated that L. officinale was nested within Angelica, sistered to A. sinensis. Therefore, L. officinale should be considered as a species of Angelica.
Necessity of thoroughly sampled phylogeny for identification of medicinal plant using DNA barcodes
The critical issue in DNA barcoding is accuracy. Accuracy depends especially on the extent of separation between intraspecific variation and interspecific divergence in the selected marker, namely barcoding ‘gap’, which cannot be strictly delineated unless a thoroughly sampled phylogeny is available. A larger barcoding ‘gap’ between intra- and interspecific variation was found in initial barcoding evaluation due to greatly undersampled both in evaluation of intraspecific variation (mostly 1–2 individuals per species sampled) and interspecific divergence (because of incomplete or geographically restricted sampling) (May 1997; Stoeckle 2003; Hebert et al. 2004). The incomplete sampling is more noticeable for barcode evaluation in identification of medicinal plants due to limited medicinal species (Chen et al. 2010; Han et al. 2012, 2013) which may artificially lead to overestimate the identification efficiency of barcodes. When intraspecific variation overlaps with interspecific divergence within a phylogenetic tree consisting of reciprocally monophyletic species, the overlap will not affect the identification of unknowns in a thoroughly sampled tree where they should fall within the coalescent of already characterized species (Meyer & Paulay 2005). On the other hand, the overlap will affect the identification of unknowns in an incomplete sampled tree where the coalescent of monophyletic species may fail. Therefore, a successful barcoding project requires comprehensive species and populations sampling to obtain high rates of species resolution. Our data sets, including 23 species (including one variety) of Angelica (covering about 85% of Angelica species) in China, represent a successful barcoding approach. Such barcoding study on the basis of a thoroughly sampled phylogeny is accurate to track species boundaries and to authenticate medicinal species in Angelica.
Impact of processing procedures on identification of materia medica using DNA barcodes
The bottleneck in materia medica identification using DNA barcodes is that the processing procedure might result in DNA degradation. The aims of processing are to enhance the efficacy and/or reduce the toxicity of crude drugs (Zhao et al. 2010). Traditional baking is an important approach in the processing procedures of decoction pieces for A. sinensis, A. biserrata and A. dahurica. The baking temperature for thicker and bigger decoction pieces of A. dahurica, around 80 °C, is higher than that for A. sinensis and A. biserrata (about 60 °C). Baking temperature higher than 65 °C might cause severe DNA degradation and result in lower success rates in PCR amplification and sequencing. Our results demonstrated that the processing procedure for decoction pieces of A. dahurica has an immediate effect on the efficiency of DNA barcoding. ITS2, with higher success of PCR amplification and sequencing of decoction pieces and crude drugs of A. dahurica (Table 3), could be considered as a supplementary barcode for identification of Angelica materia medica. Recently, a large-scale data set of ITS2 for medicinal plants has been established (Chiou et al. 2007; Chen et al. 2010; Han et al. 2013) to improve the efficiency of authentication for medicinal plants and processed materia medica. However, the accuracy of discrimination using ITS2 should be precisely verified based on a thoroughly sampled phylogeny, because A. dahurica was not grouped into a single monophyletic clade in MP tree of ITS2 (Fig. 4).
Feasibility of DNA barcodes for identification of geographical authenticity of materia medica
Molecular identification of geographical authenticity of conspecific materia medica is practical and meaningful for quality assurance of materia medica. The prerequisite for identification of geographical authenticity is that a DNA region must reflect the genetic differentiation among populations. Although DNA barcodes are mainly used for identification at the species level, they have also intraspecies variation. For example, trnH-psbA harbours the richest intraspecific variation in chloroplast DNA which makes it popular in population genetics or phylogeography (Avise 2000; Hewitt 2001; Petit et al. 2005). However, in our study, no difference was found in all crude drugs of A. dahurica from different producing localities with all four DNA barcodes. Thus, DNA barcodes are not promising for identification of geographical authenticity of A. dahurica. More rapidly evolving markers, such as microsatellites (Sheng et al. 2002), might be hopeful for the identification of geographical authenticity of conspecific materia medica.
Here, we propose ITS as the most ideal barcode for identification of Angelica species and revise L. officinale as a species of Angelica, Angelica officinale. To faciliate the use of DNA barcodes in materia medica identification, we elucidate the necessity of thoroughly sampled phylogeny for barcoding medicinal plant and investigate the impact of processing procedures on materia medica identification using DNA barcodes and the feasibility of DNA barcodes for identification of geographical authenticity of materia medica. This study not only provides a database for discrimination of Angelica species and their materia medica, but also represents an empirical paradigm for accurately identifying medicinal plants and addressing bottleneck issues of barcoding materia medica, which will shed new lights on the future studies in authentication of traditional Chinese medicine using DNA barcodes.
Acknowledgments
The authors thank Qing-mao Fang and Hua-sheng Peng for their field survey and sample collection. We are grateful to Prof. Shi-liang Zhou and Dr Jing Gu for help in data analysis and Prof. Shi-Yuan Jin for introduction in morphological identification of materia medica. This study was funded by the Projects of Large-Scale Scientific Facilities Research Project (2009-LSFGBOWS-01), National Natural Science Foundation of China (NSFC: 81274027, 81130070) and Beijing Joint Project Specific Funds.
Data accessibility
Sampling information and GenBank accessions of ITS (JX022893–022940), matK (KC812767–KC812780 & KC81 2782–KC812786), rbcL (KC812787–KC812798), trnH-psbA (KC812799–KC812804 & KC812945–KC812959) of the test samples, and the sequences retrieved from GenBank are included in Table S1.The input files of phylogenetic analyses are provided (Supporting Information).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1 Maximum likelihood tree of 23 Angelica species (including one variety) combined with the decoction pieces of three official materia medica (boxed in real line rectangle) and crude drugs of A. dahurica (boxed in dotted line rectangle) based on ITS sequences. The symbols on the top right corner of each species refer to Table S1, and the followed numbers behind each species and materia medica represent sample size. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S2–S3 Neighbour-joining (S2) and maximum likelihood (S3) trees of 23 Angelica species (including one variety) based on the combination of ITS2 sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Fig. S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S4–S6 Neighbour-joining (S4), maximum parsimony (S5) and maximum likelihood (S6) trees of 23 Angelica species (including one variety) based on the combination of ITS + rbcL + matK + trnH-psbA sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Table S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S7–S9 Neighbour-joining (S7), maximum parsimony (S8) and maximum likelihood (S9) trees of 23 Angelica species (including one variety) based on the combination of matK + rbcL sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Table S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Table S1. Sampling information of populations and species, and GenBank accession numbers for the sequences of the different molecular markers of Angelica and outgroups.
Data S1 The input files for NJ, MP and ML analyses.
References
- Alvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Avise JC. Phylogeography: The History and Formation of Species. Cambridge, Massachusetts and London, UK: Harvard University Press; 2000. [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Barthelson RA, Sundareshan P, Galbraith DW, Woosley RL. Development of a comprehensive detection method for medicinal and toxic plant species. American Journal of Botany. 2006;93:566–574. doi: 10.3732/ajb.93.4.566. [DOI] [PubMed] [Google Scholar]
- CBOL Plant Working Group. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Yao H, Han JP, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China Plant BOL Group. Comparative analysis of a large dataset indicates that ITS should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Committee of People's Republic of China. Pharmacopoeia of the People's Republic of China. Beijing: Chemical Industry Press; 2005. p. 89. 1. [Google Scholar]
- Chinese Pharmacopoeia Committee of People's Republic of China. Pharmacopoeia of the People's Republic of China. Beijing: Chemical Industry Press; 2010. p. 97. 1, 124, 246. [Google Scholar]
- Chiou SJ, Yen JH, Fang CL, Chen HL, Lin TY. Authentication of medicinal herbs using PCR-amplified ITS2 with specific primers. Planta Medica. 2007;73:1421–1426. doi: 10.1055/s-2007-990227. [DOI] [PubMed] [Google Scholar]
- Coghlan ML, Haile J, Houston J, et al. Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PLoS Genetics. 2012;8:e1002657. doi: 10.1371/journal.pgen.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Ashton B, Cheung M, et al. 2009. Geneious v4.7, Available from http://www.geneious.com.
- Fay MF, Swensen SM, Chase MW. Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae) Kew Bulletin. 1997;52:111–120. [Google Scholar]
- Feng T, Downie SR, Yu Y, et al. Molecular systematics of Angelica and allied genera (Apiaceae) from the Hengduan Mountains of China based on nrDNA ITS sequences: phylogenetic affinities and biogeographic implications. Journal of Plant Research. 2009;122:403–414. doi: 10.1007/s10265-009-0238-4. [DOI] [PubMed] [Google Scholar]
- Han JP, Chen SL, Chen XC, Lin YL. Comparison of four DNA barcodes in identifying certain medicinal plants of Lamiaceae. Journal of Systematics and Evolution. 2012;50:227–234. [Google Scholar]
- Han JP, Zhu Y, Chen X, et al. The short ITS2 sequence serves as an efficient taxonomic sequence tag in comparison with the full-length ITS. BioMed Research International. 2013:1–7. doi: 10.1155/2013/741476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proceedings. Biological sciences/The Royal Society. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heubl G. New aspects of DNA-based authentication of Chinese medicinal plants by molecular biological techniques. Planta Medica. 2010;76:1963–1974. doi: 10.1055/s-0030-1250519. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Speciation, hybrid zones and phylogeography – or seeing genes in space and time. Molecular Ecology. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Shiba M, Yamaji H, et al. Species identification of licorice using nrDNA and cpDNA genetic markers. Biological & Pharmaceutical Bulletin. 2007;30:1497–1502. doi: 10.1248/bpb.30.1497. [DOI] [PubMed] [Google Scholar]
- Kool A, de Boer HJ, Krüger A, et al. Molecular identification of commercialized medicinal plants in southern Morocco. PLoS One. 2012;7:e39459. doi: 10.1371/journal.pone.0039459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- May RM. The dimensions of life of earth. In: Raven PH, editor. Nature and Human Society: The Quest for a Sustainable World. Washington, DC: National Academy Press; 1997. pp. 30–45. [Google Scholar]
- Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Feliner G, Rosselló JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics Evolution. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V. DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Molecular Ecology Resources. 2009;9(Suppl. 1):164–171. doi: 10.1111/j.1755-0998.2009.02641.x. [DOI] [PubMed] [Google Scholar]
- Ren BQ, Xiang XG, Chen ZD. Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Molecular Ecology Resources. 2010;10:594–605. doi: 10.1111/j.1755-0998.2009.02815.x. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Sass C, Little DP, Stevenson DW, Specht CD. DNA barcoding in the cycadales: testing the potential of proposed barcoding markers for species identification of Cycads. PLoS One. 2007;2:e1154. doi: 10.1371/journal.pone.0001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She ML, Pu FT, Pan ZH, et al. Flora of China Editorial Committee (ED), Flora of China. Vol. 14. St. Louis, Missouri: Missouri Botanical Garden Press; 2005. Apiaceae; pp. 1–205. [Google Scholar]
- Sheng Y, Zheng WH, Pei KQ, Ma KP. Applications of microsatellites in population biology. Acta Phytoecologica Sinica. 2002;26:119–126. [Google Scholar]
- Spooner DM. DNA barcoding will frequently fail in complicated groups: an example in wild potatoes. American Journal of Botany. 2009;96:1177–1189. doi: 10.3732/ajb.0800246. [DOI] [PubMed] [Google Scholar]
- Stoeckle M. Taxonomy, DNA, and the barcode of life. BioScience. 2003;53:796–797. [Google Scholar]
- Sucher NJ, Carles MC. Genome-based approaches to the authentication of medicinal plants. Planta Medica. 2008;74:603–623. doi: 10.1055/s-2008-1074517. [DOI] [PubMed] [Google Scholar]
- Sun H, Zhang B, Qi Y, Zhang Z, Zengxiang G. Survey and analysis on the resource of Angelica sinensis. Chinese Agricultural Science Bulletin. 2009;23:437–441. [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods), Beta Version 4b10. Sinauer Associates, Sunderland.
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan QS, Zhang WW, Li L, et al. Effects of various processings on quality of Angelica dahurica. Chinese Journal of Information on Traditional Medicine. 2013;20:49–51. [Google Scholar]
- Tate JA, Simpson BB. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploidy species. Systematic Botany. 2003;28:723–737. [Google Scholar]
- Vasil'eva MG, Pimenov MG. Karyo taxonomical analysis in the genus Angelica (Umbelliferae) Plant Systematics and Evolution. 1991;177:117–138. [Google Scholar]
- Wang MY, Xiong Y, Jia MR, et al. Investigation on production and marketing of Radix Angelica dahuricae. West China Journal of Pharmaceutcal Sciences. 2002;17:305–306. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yao H, Song JY, Liu C, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CY, Wang FY, Yan HF, et al. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae) Molecular Ecology Resources. 2012;12:98–108. doi: 10.1111/j.1755-0998.2011.03076.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Liang Z, Chan K, et al. A unique issue in the standardization of Chinese materia medica: processing. Planta Medica. 2010;76:1975–1986. doi: 10.1055/s-0030-1250522. [DOI] [PubMed] [Google Scholar]
- Zschocke S, Liu JH, Stuppner H, Bauer R. Comparative study of roots of Angelica sinensis and Related Umbelliferous drugs by thin layer chromatography, high-performance liquid chromatography, and liquid chromatography - mass spectrometry. Phytochemical analysis. 1998;9:283–290. [Google Scholar]
- Zuo YJ, Chen ZJ, Kondo K, et al. DNA barcoding of Panax species. Planta Medica. 2011;77:182–187. doi: 10.1055/s-0030-1250166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Maximum likelihood tree of 23 Angelica species (including one variety) combined with the decoction pieces of three official materia medica (boxed in real line rectangle) and crude drugs of A. dahurica (boxed in dotted line rectangle) based on ITS sequences. The symbols on the top right corner of each species refer to Table S1, and the followed numbers behind each species and materia medica represent sample size. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S2–S3 Neighbour-joining (S2) and maximum likelihood (S3) trees of 23 Angelica species (including one variety) based on the combination of ITS2 sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Fig. S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S4–S6 Neighbour-joining (S4), maximum parsimony (S5) and maximum likelihood (S6) trees of 23 Angelica species (including one variety) based on the combination of ITS + rbcL + matK + trnH-psbA sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Table S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Fig. S7–S9 Neighbour-joining (S7), maximum parsimony (S8) and maximum likelihood (S9) trees of 23 Angelica species (including one variety) based on the combination of matK + rbcL sequences, respectively. The denotation on the right side of each species and the information of materia medica refer to Table S1. The species underlined are not discriminated. Supporting values (>50%) are indicated above or below the relevant branches.
Table S1. Sampling information of populations and species, and GenBank accession numbers for the sequences of the different molecular markers of Angelica and outgroups.
Data S1 The input files for NJ, MP and ML analyses.
Data Availability Statement
Sampling information and GenBank accessions of ITS (JX022893–022940), matK (KC812767–KC812780 & KC81 2782–KC812786), rbcL (KC812787–KC812798), trnH-psbA (KC812799–KC812804 & KC812945–KC812959) of the test samples, and the sequences retrieved from GenBank are included in Table S1.The input files of phylogenetic analyses are provided (Supporting Information).