Abstract
Despite their clear importance as a class of regulatory molecules, pinpointing the relevance of individual miRNAs has been challenging. Studies querying miRNA functions by overexpressing or silencing specific miRNAs have yielded data that are often at odds with those collected from loss-of-functions models. In addition, knockout studies suggest that many conserved miRNAs are dispensable for animal development or viability. In this review we discuss these observations in the context of our current knowledge of miRNA biology and review the evidence implicating miRNA-mediated gene regulation in the mechanisms that ensure biological robustness.
Keywords: microRNAs, animal models, paralogs, network motifs, buffering
An elusive role for microRNAs
MicroRNAs (miRNAs) are small non-coding RNAs that regulate protein output post-transcriptionally [1]. Overwhelming evidence accumulated since their discovery [2,3] leaves little doubt regarding their importance. They comprise 1-2% of all genes in worms, flies, and mammals [1], and because each miRNA is predicted to regulate hundreds of targets, the majority of protein coding genes is thought to be under their control [4]. In practice, this means that virtually every biological process is subject to miRNA-dependent regulation. As additional evidence of their functional relevance, miRNAs and their targets often display striking evolutionary conservation [5-7]. Lastly, animals carrying mutations that impair miRNA processing [8-12] are not viable, indicating that complete loss of miRNA activity is incompatible with life.
Despite their clear importance as a class of regulatory molecules, determining the biological relevance of individual miRNAs has proven challenging. For the most part, the physiological functions of specific miRNAs have been inferred from overexpression studies in animals and cultured cells or from studies that used antisense molecules as a means of disrupting their pairing to targets. These experiments have attributed critical roles to miRNAs in processes such as cell proliferation, differentiation, and survival, and have implicated them as crucial players during normal development, homeostasis, and disease [13-17]. Surprisingly, the expectations raised by these early studies have been met by a growing number of knockout animals with very modest or no apparent phenotypes. Furthermore, so far only two miRNA genes (miR-17∼92 and miR-96) have been shown to cause developmental defects in humans when mutated [18,19]. The absence of phenotypic consequences upon ablation of individual miRNAs seems to be the rule rather than the exception. In Caenorhabditis elegans, for example, systematic deletion of miRNAs indicates that less than 10% of them are individually required for normal animal development or viability [20], and this trend seems to be true in mice as well [21] (Table 1).
Table 1. miRNA-knockout phenotypes in mice.
| miRNA gene | Knockout phenotype | Phenotype penetrance | Evidence of functional redundancy with family members | Phenotypes in response to external or internal perturbations | References |
|---|---|---|---|---|---|
| miR-155 | immunodeficiency and increased lung remodeling | increased lung remodeling in ∼56% of miR-155-/- animals | NA | NA | [119] |
| miR-17∼92 | perinatal lethality with heart, lung, B cell and skeletal defects | 100% | co-deletion of miR-17∼92 with miR-106∼25 aggravates developmental phenotypes of miR-17∼92 single deletion | NA | [67], [19] |
| miR-106a∼363 | no obvious phenotype | NA | co-deletion of miR-17∼92 with miR-106∼25 and miR-106a∼363 aggravates developmental phenotypes of miR-17∼92 single deletion | NA | [67] |
| miR-106b∼25 | no obvious phenotype | NA | co-deletion of miR-17∼92 with miR-106∼25 aggravates developmental phenotypes of miR-17∼92 single deletion | NA | [67] |
| miR-15a/16-1 | B cell lymphoproliferative disorders | ∼24% | NA | NA | [120] |
| miR-144/451 | impaired late erythroblast maturation | not specified. assumed to be 100% | NA | NA | [121] |
| miR-150 | B1 cell expansion and increased humoral immune response | not specified. assumed to be 100% | NA | NA | [122] |
| miR-223 | expanded granulocyte compartment | not specified. assumed to be 100% | NA | NA | [123] |
| miR-1-2 | lethality at weaning with heart defects | ∼50% | NA | NA | [124] |
| miR-34a | no obvious phenotype | NA | co-deletion of miR-34 and miR-449 leads to defects in cilliogenesis. Lethal for 60% of animlals. Surviving adults are infertile | loss of miR-34 family members aggravates prostate neoplastic lesions caused by inactivation of trp53; loss of miR-34a improves cardiac function during aging and in response to stress. | [42],[65], [86] [125] |
| miR-34b/c | no obvious phenotype | NA | co-deletion of miR-34 and miR-449 leads to defects in cilliogenesis. Lethal for 60% of animlals. Surviving adults are infertile | loss of miR-34 family members aggravates prostate neoplastic lesions caused by inactivation of trp53 | [42],[65], [86] |
| miR-449 | no obvious phenotype | NA | co-deletion of miR-34 and miR-449 leads to defects in cilliogenesis. Lethal for 60% of animlals. Surviving adults are infertile | NA | [66], [65] |
| miR-290∼295 | developmental delays; embryonic development outside yolk sac; embryonic lethality; germ cell deficiency in surviving adults | lethality in 75% of animals | NA | NA | [126] |
| miR-10a | no obvious phenotype | NA | NA | NA | [127] |
| miR-208a | no obvious phenotype, but minor cardiac conduction defects reported | 80% | NA | reduced cardiac hypertrophy in response to stress and hypothyroidism | [84], [128] |
| miR-208b | no obvious phenotype | NA | decrease in type I myofibers in soleus muscle when miR-499 is co-deleted | NA | [128] |
| miR-499 | no obvious phenotype | NA | decrease in type I myofibers in soleus muscle when miR-208b is co-deleted | NA | [128] |
| miR-133a-1 | no obvious phenotype | NA | 50% perinatal lethality with ventricular septal defects when miR-133a-2 is co-deleted. Cardiomyopathy and heart failure in adult compound animals. | NA | [129] |
| miR-133a-2 | no obvious phenotype | NA | 50% perinatal lethality with ventricular septal defects when miR-133a-1 is odeleted. Cardiomyopathy and heart failure in adult compound animals. | NA | [129] |
| miR-126 | embryonic/perinatal lethality with angiogenesis defects | 40-50% | NA | adults are prone to myocardial rupture following myocardial infraction | [130], [131] |
| miR-143/145 | no obvious phenotype; but smooth muscle cells display abnormal morphology | NA | NA | miR-143/145-/- have impaired responses to vascular and intestinal injuries. | [132], [83], [80] |
| miR-375 | animals develop hyperglycemia; increased total pancreatic alpha-cells, decreased pancreatic beta-cell mass | not specified. assumed to be 100% | NA | NA | [133] |
| miR-140 | mild skeletal phenotype: short stature, decreased body weight and craniofacial abnormalities | not specified. assumed to be 100% | NA | NA | [134] |
| miR-182 | no obvious phenotype | NA | NA | NA | [135] |
| miR-132/212 | minor synaptic transmission and plasticity defects in the neocortex | not specified. assumed to be 100% | NA | NA | [136] |
| miR-22 | no obvious phenotype | NA | NA | minor defects in cardiac function during acute and chronic hemodynamic stress | [137] |
| miR-21 | no obvious phenotype | NA | NA | NA | [138] |
| miR-29ab1 | reduced lifespan; thymic involution around 9-12 weeks of age | Not specified. assumed to be 100% | NA | NA | [139], [140] |
| miR-29b2c | no obvious phenotype | NA | NA | NA | [139] |
| miR-128 | increased motor activity and fatal epilepsy | Not specified. assumed to be 100% | NA | NA | [52] |
| miR-205 | neonatal lethality with compromised epidermal and hair follicle growth | Not specified. assumed to be 100% | NA | NA | [53] |
| miR-200b | NA | NA | Co-deletion of mir-200b and miR-429 results in female infertility | NA | [141] |
| miR-429 | NA | NA | Co-deletion of mir-200b and miR-429 results in female infertility | NA | [141] |
NA, not available or—to our knowledge—not addressed; grey rows highlight miRNA knockout models with no obvious phenotypes.
In this review we discuss these observations in the context of our current understanding of miRNA-mediated gene regulation and examine the evidence implicating miRNA activity in the processes that ensure robust animal development and homeostasis.
Endogenous miRNAs exert mild repression on many targets
miRNA processing has been extensively reviewed [22] and will be only briefly discussed here. MiRNAs are transcribed as long primary transcripts (pri-miRNAs) and cleaved in the nucleus by the Drosha/DGCR8 microprocessor complex. The resulting ∼70-nucleotide-long hairpin-shaped molecule—the pre-miRNA—is exported into the cytoplasm, where it is further processed by Dicer, bound by an Argonaute protein, and incorporated into an RNA-induced silencing complex (RISC). Metazoan miRNAs typically direct the RISC to target mRNAs through imperfect base pairing to their 3′ untranslated regions (3′UTR), leading to post-transcriptional repression mainly through mRNA destabilization, though a minor component of translation inhibition has also been detected [23] (see Box 1).
Box 1. Mechanisms of miRNA-mediated gene repression.
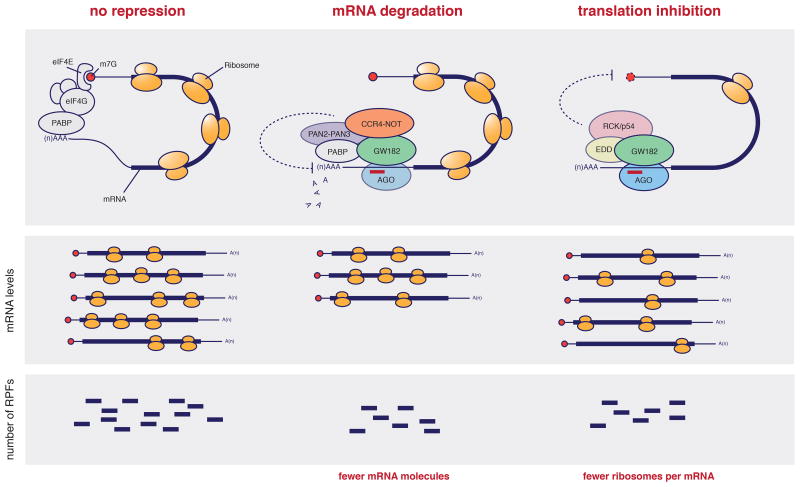
Although it is well understood that miRNAs repress gene expression by recruiting the RISC silencing complex to target mRNAs, the molecular details underlying this repression are not entirely clear [107]. RISC binding to targets has been proposed to elicit both repression of translation and mRNA deadenylation and decay (Figure I). However, the mechanistic basis behind these modes of repression, their relative contribution to the overall action of RISC and the order by which they operate remain a matter of debate.
The core of the RISC complex is comprised of two components: an Argonaute protein (Ago1-Ago4 in mammals) and a GW182 protein (TNRC6A-C in mammals). While Ago proteins bind to miRNAs and are thus involved in target recognition, GW182 proteins seem to act as a molecular platform to which effector complexes bind to mediate target repression (Figure I, top panel). GW182 can directly bind PABP [108], a poly(A)-tail binding protein involved in the regulation of translation [109], but with a controversial role in miRNA-mediated gene silencing [107]. GW182 also binds (directly or through PABP) to deadenylation complexes [110,111]. Of these, CCR4-NOT has been shown to be required for miRNA-mediated deadenylation [110,112]. Finally, GW182 has been shown to bind to EDD, a protein that associates with the DEAD box helicase RCK/p54, which in turn enhances mRNA decapping and represses cap-dependent translation [113].
But what is the prevalent mechanism of miRNA-mediated repression? Recentgenome wide attempts to clarify this question have examined the influence ofendogenous and exogenous miRNAs [23,114,115] on mRNA levels (measuredby RNA-seq of polyadenylated mRNA; Figure I, middle panel) and translationefficiency (measured by sequencing of ribosome protected fragments or RPFs;Figure I, bottom panel). These studies suggest that the predominant mode ofmiRNA-mediated repression maybe context-dependent. In human cells and latezebrafish embryos the majority of RISC-mediated repression can be attributed toa reduction in the levels of mRNA, with only a minor fraction stemming fromreduced translation efficiency [23,115]. In early stages of zebrafish development (2h and 4h post-fertilization) however, miRNAs seem to predominantly reducetranslation of their targets, while causing no detectable changes in the levels oftheir mRNAs [114,115]. These results are not necessarily in conflict, since arecent study suggests that the prevalence of translational inhibition in earlyzebrafish embryos is caused by a strong coupling of poly(A)-tail length andefficiency of translation in this system [115].
Target recognition is primarily determined by the seed-sequence, a stretch of 6 nucleotides spanning nucleotide 2-7 on the 5′ end of the miRNA [24,25]. Accordingly, targets can be confidently predicted by searching for conserved matches to this sequence in the 3′UTR of messages [26]. Prediction accuracy increases further when this search is restricted to 7-nucleotide-long motifs encompassing the seed [26] and when the sequence context within the 3′UTR is taken into consideration [27]. Non-canonical targeting through sites with mismatches to the seed has also been reported [28-32], but seems to be generally associated with lower levels of repression and its biological relevance remains unclear [4,30]. Because targeting requires the presence of such short conserved sequences, individual miRNAs have the potential to regulate hundreds of targets [4]. These computational predictions have been supported by experimental evidence showing that loss or overexpression of a miRNA in cultured cells results in the deregulation of hundreds of genes [33,34]. In both cases, the deregulated messages are enriched in conserved miRNA binding sites and their expression in vivo tends to be anti-correlated with that of the miRNA [33-36]. These observations suggest that both knockout and ectopic expression studies can give clues into the biological functions of miRNAs.
Nevertheless, in vivo studies tell a cautionary tale against taking these approaches as equivalent. In C. elegans for example, overexpression of miR-61 results in vulval development defects, which seem to stem from its ability to regulate Vav-1 and establish a feedback loop that regulates LIN-12/Notch expression within the vulval precursor cell population [13]. Yet, deletion of miR-61 is compatible with normal vulval development, and even animals carrying co-deletion of miR-61 and the closely related miR-267 are phenotypically normal [20]. In mammals, overexpression of members of the miR-34 family suggests a potent tumor suppressor function downstream of p53 [17,37-41]. However, mice carrying targeted deletions of all miR-34 genes display wild type p53 responses to a variety of cellular insults, including ionizing radiation and oncogenic stress [42,43].
How can we account for these differences? Obviously, ectopic expression studies address the question of whether a miRNA can exert a specific function, while loss-of-function studies test whether it is required for that function. In addition, it is important to keep in mind that the ability of a miRNA to repress its targets crucially depends on its expression levels [44,45].
Genetic inactivation of a miRNA results in very modest de-repression of its direct targets, typically less than two-fold even for highly abundant miRNAs [33,46]. These differences are well within the range that could be attributed to fluctuations of gene expression between two genetically identical cells or between individuals [47]. For most genes such modest changes in expression can be well tolerated by the organism, which might explain why genetic inactivation of miRNAs often does not have obvious phenotypic consequences. These observations sparked the idea that rather than acting as genetic switches—where strong repression of one or few targets results in a clear phenotypic outcome [48,49]—most miRNA act as rheostats, fine-tuning the expression of hundreds of genes to reinforce cell fate decisions brought about through other mechanisms [47,50,51].
It is important to note however, that even a mild derepression of many targets can have severe phenotypical consequencesly, especially if the targets are functionally linked. For example, in mice, deletion of miR-128 results in fatal epilepsy due de-repression of several components of the MAPK pathway, leading to a significant increase in ERK2 phosphorylation [52]. Similarly, loss of miR-205 results in neonatal lethality in mice with compromised epidermal and hair follicle growth [53], presumably by modulating the expression of multiple components of the PI(3)K signaling pathway. Finally, a recent study has implicated Drosophila's miR-iab-8 in CNV patterning and fly fertility through the regulation of genes whose products act together within a protein complex [54]. Remarkably, there is evidence that even partial relief of miRNA-mediated repression can have phenotypic consequences. For example, hemizygous deletion of miR-17∼92 leads to severe developmental defects in mice and in humans [19].
In contrast to loss-of-function studies, ectopic expression often leads to supra-physiologic levels of the miRNA and stronger repression of its targets [23,33,34]. The magnitude of this repression can bring down to inconsequential levels the expression of genes that would otherwise remain functional even in the presence of the targeting miRNA. Often, these experiments also result in the expression of miRNAs in tissues or cells in which they would normally be absent [55], leading to repression of messages that might not be their biological targets. Thus, despite their widespread use, the propensity to generate a high fraction of false-positive results constitutes a major caveat of miRNA overexpression experiments. This does not mean that such experiments are devoid of value. In fact, much of the knowledge we have accumulated over the years on miRNA biology has depended on them [27,44]. In addition, regardless of the physiologic relevance of these studies, overexpression of miRNA mimics may serve a therapeutic purpose [56-58] (Box 2).
Box 2. miRNA-based therapeutic strategies.
Deregulation of miRNA activity has been frequently implicated in the development and progression of human diseases [116], and this observation has driven much of the research in the field. The discovery that in vivo delivery of oligonucleotides—that mimic or inhibit the activity of specific miRNAs—can have therapeutic effects in animal models has suggested that analogous approaches might be applicable in the clinic as well [117].
Several miRNA-based therapeutic strategies are currently under development, and these can broadly be divided into: (i) miRNA replacement strategies, which attempt to mimic the activity of specific miRNAs by delivering small double-stranded RNA molecules that resemble miRNA duplexes. MRX34 for example, is a mimic of the miR-34 family [56], which is currently in Phase I of clinical trials to test its safety for patients with primary liver cancer or liver metastasis (NCT01829971); (ii) miRNA targeting strategies, which rely on antisense oligonucleotides (ASO) that bind to endogenous miRNAs to prevent their interaction with targets. One such molecule, designed to inhibit miR-122 [118], has also reached clinical trials (Phase II) and is being evaluated for its long-term safety and efficacy in patients with chronicle HCV infection (NCT02031133).
An important aspect to keep in mind when thinking about delivery of oligonucleotides to patients or animal models is that they are generally poorly suited for in vivo applications [117]:
They are substrates of serum nucleases, making them unstable;
They are unable to penetrate the cell membrane;
They have poor tissue distribution when delivered systemically.
These obstacles can be partially overcome by encapsulation in lyposomes or polymer-based nanoparticles and through addition of chemical modifications [117]. Conjugation of cholesterol groups to the 3′ end of the oligonucleotide, for example, improves both entry of the oligo in the cell and its distribution across tissues. Modifications like 2′-O-methyl (2′-OMe), 2′-O-methyoxyethyl (2′-MOE), and locked nucleic acid (LNA), on the other hand, improve the oligonucleotide's resistance to exonucleases. Of note, for miRNA-targeting strategies, LNA modifications have the additional advantage of increasing the affinity of the oligo to its complementary miRNA, leading to more efficient inhibition. Substituting the phosphodiester bond by a phosphorothioate bond can further increase stability of the oligonucleotides, but this modification decreases miRNA-binding affinity.
Functional redundancy among family members
One remarkable aspect of miRNA genes is that a large number of them have obvious paralogs in the genome. Paralog miRNAs arise from both tandem and non-local gene duplication events, which give rise to either duplication of sequences in the same transcript—thus originating miRNA clusters—or on distant loci, typically on different chromosomes [59]. These miRNA ‘copies’ not only retain a high degree of sequence homology but also share the same seed-sequence and are thus by convention grouped into ‘miRNA seed families’ [60]. In C. elegans, about 60% of all miRNAs can be assigned to one of 23 families [61], a percentage significantly higher than that of protein-coding genes with known paralogs, which approximates 25% [62]. Similarly, about one-third of human miRNA genes and almost 40% of those in the mouse can be grouped into families based on sequence similarity [63,64].
Because paralog miRNAs share the same seed-sequence, they are expected to have similar affinities to messages. When expressed in the same cells, these related miRNAs can co-regulate targets, leading to higher levels of repression than those that could be achieved by each miRNA individually. In vertebrate multiciliated cells for example, the six miRNAs that comprise the conserved miR-34/449 family—encoded by the miR-34a, miR-34b/c, and miR-449a/b/c loci—can coordinately repress cp110 expression during ciliogenesis [65]. The existence of miRNAs with redundant functions means that in some instances several members of the family need to be deleted before a phenotypic consequence can be detected (see Table 1). Indeed, animals carrying targeted deletions of single genes of the miR-34/449 family are viable and phenotypically normal [42,65,66], whereas deletion of all three loci leads to high postnatal mortality, with surviving animals displaying an array of phenotypes associated with defective ciliogenesis [65]. Analogously, partial functional overlap has been observed among the miR-17∼92 cluster and its two paralogs: miR-106a∼363 and miR-106b∼25 [67].
This level of redundancy adds considerable complexity to gene knockout studies. For families as numerous as the let-7 family for example—which in mice comprises 12 members distributed across 8 loci [68]—the effort involved in generating compound loss-of-function mutants effectively precludes such studies from being undertaken.
Chemically modified antisense oligonucleotides (Box 2) provide an alternative to genetic miRNA ablation [69,70], and can to some extent circumvent the difficulties posed by the existence of paralog genes. These antisense molecules inactivate miRNAs by binding with high affinity to their mature sequence, thus preventing interaction with targets. Typically, antisense molecules are designed to have full complementarity to the mature miRNA [69,70], but because affinity remains very high even for 8-mers targeting the seed region, this strategy can be adapted to simultaneously inhibit the function of multiple members of the same family [16,71]. This approach has shown that global inhibition of the let-7 family can prevent and treat impaired glucose tolerance in diet-induced obese mice [16]; this result is consistent with data gathered from let-7 transgenic animals [16,72] and from animals carrying gain- or loss-of-function alleles of Lin28a or LIN28B, two proteins that specifically block let-7 maturation [72,73].
Despite their clear utility, designing controls for experiments in which antisense oligos are used is challenging. A common approach is to deliver modified oligonucleotides with sequences that do not match any region in the genome, but these molecules often lead to unwanted effects when delivered systemically to animals. A recent study, for example, reported an increase in liver and spleen size as well as in the number of liver-associated macrophages upon injection of a control oligonucleotide in adult mice. These animals also displayed reduced number of white blood cells and a variety of alterations in blood chemistry [74]. In another example, injection of a control molecule in chick embryos resulted in a wide range of skeletal malformations including vertebral fusions, hemivertebrae, and split vertebrae [75]. Thus, while we can certainly learn a lot from studies with synthetic miRNA antagonists, the intrinsic limitations of this technology need to be carefully considered.
The question of functional redundancy among paralog miRNAs has perhaps been best tackled in C. elegans, where miRNA genes were systematically mutated to generate strains lacking all or most members of 15 of the 23 known miRNA families [61]. As in the cases discussed above, this approach uncovered high levels of redundancy among some paralogs: for example, although deletion of individual components of the miR-35 and miR-51 families did not cause phenotypical abnormalities, mutants for these families died as embryos or early larvae. Similarly, mutants for the miR-58 family showed a variety of phenotypic abnormalities, which were absent in mutants for its individual miRNAs [20]. This is perhaps not unexpected. What is surprising is that mutant animals for the remaining 12 families appeared largely normal even when subjected to a broad panel of phenotypical characterizations [61]. This suggests, that at least in C. elegans, most miRNA families might not be essential for animal development or viability.
miRNAs and biological robustness
Despite the frequent lack of phenotypes in miRNA knockout animals, the strong evolutionary conservation of many miRNAs indicates that they must confer some selective advantage, regardless of our ability to detect it. An important aspect to consider is that miRNAs do not act in isolation. In fact, a common theme among miRNAs is their positioning within gene regulatory networks, in particular within feedback and feedforward loops [76] (Figure 1A). Moreover, though the vast majority of 3′UTRs have a single conserved match for a particular seed, they typically have more than four conserved miRNA binding sites in total, thus allowing for combinatorial and overlapping regulation [1,4]. Redundant components and regulatory loops are two common strategies used to achieve canalization [77], i.e., the stabilization of biological outcomes in spite of genetic, environmental, and stochastic perturbations. This has led to idea that miRNAs play a crucial role in ensuring that small changes arising from such perturbations do not have a detrimental impact on animal development or homeostasis [47,78,79].
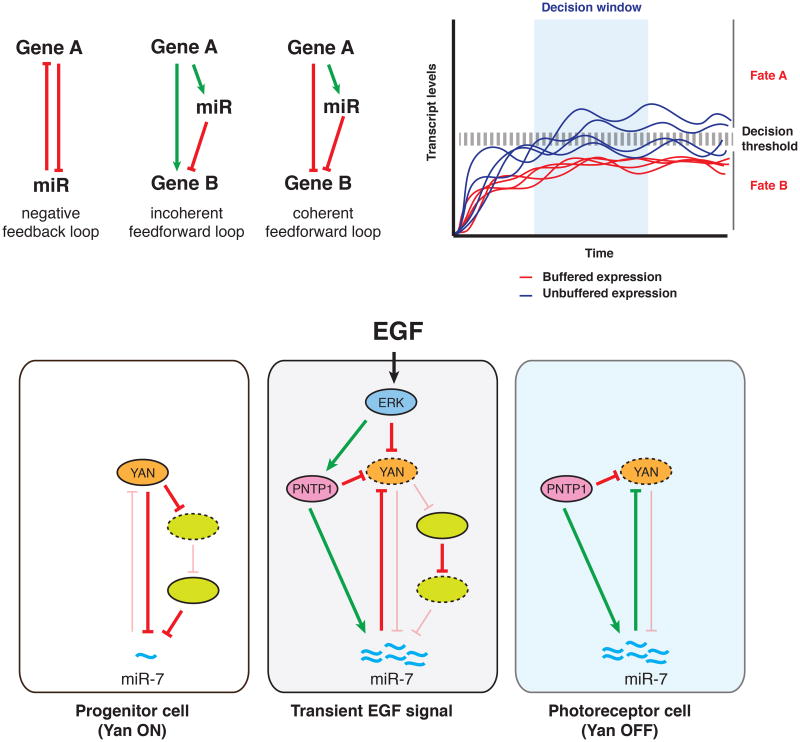
Figure 1. miRNAs participate in mechanisms of biological robustness.
(A)miRNAs are common features of network motifs, some of which are represented here. (B) Regulation of transcripts by miRNAs can be important to ensure robust cell fate decisions by keeping expression levels of a particular gene below a decision threshold (red lines). In the absence of a miRNA, the expression of the gene becomes unbuffered (blue lines), and can lead to ectopic cell fate decisions in a fraction of the population due to stochastic fluctuations in transcript levels. (C) Schematic representation of miR-7's role in the differentiation of photoreceptor cells in Drosophila. Panel (B) and (C) of this figure are inspired by illustrations from [89] and [142], respectively.
In support of this view several miRNA knockout animals display phenotypes only in response to genetic or environmental stresses (Table 1). Under normal conditions, for example, the miR-143/145 cluster can be deleted without apparent consequences to intestinal architecture or epithelial turnover rate. However, miR-143/145-null mice fail to regenerate the intestinal epithelium upon injury, at least in part due to impaired IGF signaling [80], and a similar role for this cluster in the response to blood vessel injury has been reported [81-83]. In another example, mice lacking miR-208 showed no overt cardiac defects under physiologic conditions, but cardiac remodeling was profoundly impaired in response to various stresses [84].
miRNA-dependent phenotypes can also be uncovered by mutations in other genomic loci. In C. elegans, for example, 25 out of 31 miRNA mutations resulted in phenotypic abnormalities only in animals from sensitized backgrounds [85]. In mice, miR-34's role as a tumor suppressor is only uncovered when this miRNA family is co-deleted with trp53 [42,86], an upstream regulator with whom it establishes a coherent feedforward loop that regulates MET expression [86,87]. Finally, it should be noted that a large fraction of miRNA knockout models show phenotypic traits with incomplete penetrance (Table 1), one of the hallmarks of impaired canalization [88-90].
Mechanisms of Canalization by miRNAs
As mentioned in the previous section, a possible mechanism by which miRNA-mediated regulation may confer phenotypic robustness is by buffering against cell-to-cell variability arising from stochastic fluctuations in gene transcription. These fluctuations occur because the biological processes that determine mRNA levels—such as binding of the RNA polymerase to the promoter and mRNA degradation—are inherently noisy, occurring at different times and different rates even among cells from a clonal population [91]. This noise is ultimately amplified by translation, which can give rise to even larger inter-cellular variability [92,93]. Though in many instances organisms can use such variability to control processes like lineage decision [94], noise must generally be buffered to ensure robust animal development. A failure to do so can result in pronounced phenotypic variation among genetically identical individuals [89].
One strategy to manage noise is to couple high rates of transcription with inefficient translation [92,95]. miRNAs limit translation largely by reducing mRNA stability and may therefore provide a simple mechanism to reduce overall fluctuations in protein synthesis. This might be of particular importance when subtle differences in gene expression can determine distinct cellular fates (Figure 1B). In Drosophila's larval wing imaginal disks, differentiation of sensory organ precursors (SOP) requires tight regulation of senseless (sens). While high levels of this transcription factor promote proneural gene expression in cells destined to become SOPs [96], low levels of Sens in non-SOP cells repress proneural genes [97]. Sens is a direct target of miR-9, and in the absence of this miRNA about 33% of larvae develop extra sensory organs [98], presumably because fluctuations in Sens levels inappropriately trigger ectopic cell fate decisions [99]. Interestingly, recent experiments suggest that miR-9 can also minimize the impact of genomic diversity on sensory organ differentiation, thus providing an example of genetic buffering by a miRNA [100].
Integration of miRNAs in incoherent feedforward loops (IFFL) (Figure 1A) provides another mechanism to minimize unwanted oscillations in gene expression. This type of motif is particularly useful because it can counteract the effects of fluctuations in upstream components of the network. In C. elegans, for example, an IFFL involving both lin-4 and its target lin-14 ensures that variation in lin-14 mRNA levels are stabilized by synchronous oscillations in lin-4 [101]. The presence of miRNAs in IFFLs has also been associated with canalization mechanisms in vertebrates. In Zebrafish, for example, an IFFL involving miR-430 plays a significant role in controlling unwanted fluctuations in chemokine receptor signaling, which seems to be important for robust germ cell migration [102].
miRNAs have also been implicated in the mechanisms that buffer gene expression against environmental perturbations. Arguably the best example comes from work in Drosophila, in which miR-7 is involved in cell fate decisions that result in photoreceptor differentiation [103]. Before differentiation into photoreceptors is triggered, progenitor cells in the larval eye imaginal disk are maintained in an undifferentiated state by Yan [104], a direct target of miR-7 [105]. In these cells, Yan binds to the promoter of miR-7 and inhibits its transcription [105], thus establishing a negative feedback loop that reinforces its own expression (Figure 1A, 1C). Yan also inhibits miR-7 indirectly through two other repressors, creating a coherent feedforward loop (CFFL) that is interlocked with the feedback loop [103]. This CFFL buffers miR-7's expression against fluctuations in Yan, ensuring that miR-7 is only turned on in response to a persistent decrease in yan expression (Figure 1A, 1C). Differentiation of progenitors into photoreceptors is triggered by signaling through the EGF receptor [106], which transiently degrades Yan and establishes a new coherent FFL involving miR-7 and Pnt-P1 [103]. This loop ensures that yan is not inappropriately turned on in cells that have committed to the photoreceptor lineage.
Interestingly, under uniform laboratory conditions, regulation of Yan expression is robust enough to ensure normal eye development even in the absence of miR-7 [103,105]. In a classic example of impaired canalization, however, mutant miR-7 flies fail to withstand fluctuations in environmental temperature and display abnormal patterning of sensory organs due to deregulated Yan expression [103].
Concluding Remarks
Over the past two decades we have witnessed a flourishing of studies aimed at defining the biological functions of miRNAs. Converging evidence from computational, biochemical, and genetic experiments have greatly expanded our understanding of their mechanism of action and biological properties. The picture emerging from these studies suggests that miRNAs occupy a very unique position in the hierarchy of gene regulators. By contrast to conventional transcription factors, in most cases miRNAs do not appear to act as master regulators of gene expression. Rather, their mechanism of action allows them to act as fine tuners of transcriptional programs, as components of complex network motifs, and as “post-transcriptional buffers” to confer robustness to transcriptional programs in the face of environmental and genetic variability.
Although investigating individual miRNA-mRNA interactions can and has been useful in some instances, moving forward it will be essential to resist the temptation to reduce the biological functions of individual miRNAs to repression of one or a few “key targets.” Rather, new computational and systems biology approaches will be needed to fully appreciate the intricacy, beauty, and multifaceted roles of gene regulation by small non-coding RNAs.
Figure I. Overview of protein complexes implicated in miRNA-mediated gene silencing and their effect on mRNA levels and translation efficiency.
Highlights.
MicroRNA activity is essential for animal development and viability.
Deletion of individual miRNAs often leads to subtle or no phenotypic consequences.
miRNAs can have high levels of redundancy and are often placed in regulatory loops.
miRNAs can buffer gene expression against internal and external perturbations.
Acknowledgments
We apologize to our colleagues whose work could not be cited due to space constraints. We are grateful to all members of the Ventura lab for providing stimulating discussions and to Ciro Bonetti, Carla P. Concepcion, and Yoon-chi Han for critical comments on this manuscript. MicroRNA work in the Ventura lab is funded by grants from NIH/NCI, the STARR Consortium, the Geoffrey Beene Cancer Foundation, and the Gabrielle's Angel Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin R, Smibert P, Yalcin A, Tyler DM, Schafer U, Tuschl T, Lai EC. A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Mol Cell Biol. 2009;29:861–870. doi: 10.1128/MCB.01524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 13.Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 19.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 25.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 29.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 35.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 37.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 40.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 42.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mor E, He L, Torchinsky A, Shomron N. MicroRNA-34a is dispensable for p53 function as teratogenesis inducer. Arch Toxicol. 2014;88:1749–1763. doi: 10.1007/s00204-014-1223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D'Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 50.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 51.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 52.Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O'Carroll D, et al. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342:1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang D, Zhang Z, O'Loughlin E, Wang L, Fan X, Lai EC, Yi R. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol. 2013;15:1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, Sanchez-Herrero E, Lai EC. Homeotic function of Drosophila Bithorax-complex miRNAs mediates fertility by restricting multiple Hox genes and TALE cofactorsin the CNS. Dev Cell. 2014;29:635–648. doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 57.Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, Cai W, Chirino LM, Yang GR, Bronson R, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF. Students of Bioinformatics Computer Labs a: The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 63.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 65.Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, Harland R, Wan Y, Lishko P, Lize M, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, Chen Y, Cao X, Jiang C, Yan W, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 69.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 71.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGlinn E, Yekta S, Mansfield JH, Soutschek J, Bartel DP, Tabin CJ. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci U S A. 2009;106:18610–18615. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 78.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu CI, Shen Y, Tang T. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 2009;19:734–743. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–1116. doi: 10.1016/j.cell.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 85.Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng CY, Hwang CI, Corney DC, Flesken-Nikitin A, Jiang L, Oner GM, Munroe RJ, Schimenti JC, Hermeking H, Nikitin AY. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep. 2014;6:1000–1007. doi: 10.1016/j.celrep.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, Boccaccio C, Thorgeirsson SS, Comoglio PM, Hermeking H, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci U S A. 2011;108:14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutherford S, Hirate Y, Swalla BJ. The Hsp90 capacitor, developmental remodeling, and evolution: the robustness of gene networks and the curious evolvability of metamorphosis. Crit Rev Biochem Mol Biol. 2007;42:355–372. doi: 10.1080/10409230701597782. [DOI] [PubMed] [Google Scholar]
- 89.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 91.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 92.Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- 93.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 94.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 96.Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 97.Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giri R, Carthew RW. microRNAs suppress cellular phenotypic heterogeneity. Cell Cycle. 2014;13:1517–1518. doi: 10.4161/cc.29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cassidy JJ, Jha AR, Posadas DM, Giri R, Venken KJ, Ji J, Jiang H, Bellen HJ, White KP, Carthew RW. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155:1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim D, Grun D, van Oudenaarden A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet. 2013;45:1337–1344. doi: 10.1038/ng.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lai ZC, Fetchko M, Li Y. Repression of Drosophila photoreceptor cell fate through cooperative action of two transcriptional repressors Yan and Tramtrack. Genetics. 1997;147:1131–1137. doi: 10.1093/genetics/147.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 106.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 107.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 108.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 110.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 112.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Su H, Meng S, Lu Y, Trombly MI, Chen J, Lin C, Turk A, Wang X. Mammalian hyperplastic discs homolog EDD regulates miRNA-mediated gene silencing. Mol Cell. 2011;43:97–109. doi: 10.1016/j.molcel.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 118.Hildebrandt-Eriksen ES, Aarup V, Persson R, Hansen HF, Munk ME, Orum H. A locked nucleic acid oligonucleotide targeting microRNA 122 is well-tolerated in cynomolgus monkeys. Nucleic Acid Ther. 2012;22:152–161. doi: 10.1089/nat.2011.0332. [DOI] [PubMed] [Google Scholar]
- 119.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 121.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O'Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 123.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 124.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 125.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 126.Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, White AC, Kirak O, Sharp PA, Page DC, et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci U S A. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stadthagen G, Tehler D, Hoyland-Kroghsbo NM, Wen J, Krogh A, Jensen KT, Santoni-Rugiu E, Engelholm LH, Lund AH. Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet. 2013;9:e1003913. doi: 10.1371/journal.pgen.1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 132.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jin ZB, Hirokawa G, Gui L, Takahashi R, Osakada F, Hiura Y, Takahashi M, Yasuhara O, Iwai N. Targeted deletion of miR-182, an abundant retinal microRNA. Mol Vis. 2009;15:523–533. [PMC free article] [PubMed] [Google Scholar]
- 136.Remenyi J, van den Bosch MW, Palygin O, Mistry RB, McKenzie C, Macdonald A, Hutvagner G, Arthur JS, Frenguelli BG, Pankratov Y. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One. 2013;8:e62509. doi: 10.1371/journal.pone.0062509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, Drumond AL, van Dongen S, Chen Y, Bartonicek N, Enright AJ, Lee B, et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 2012;125:2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith KM, Guerau-de-Arellano M, Costinean S, Williams JL, Bottoni A, Mavrikis Cox G, Satoskar AR, Croce CM, Racke MK, Lovett-Racke AE, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, Zuklys S, Hollander GA, Matthys P, Gray DH, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-alpha receptor. Nat Immunol. 2012;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hasuwa H, Ueda J, Ikawa M, Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–73. doi: 10.1126/science.1237999. [DOI] [PubMed] [Google Scholar]
- 142.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]