Abstract
The reparative mechanism that operates following post-natal cutaneous injury is a fundamental survival function that requires a well-orchestrated series of molecular and cellular events. At the end, the body will have closed the hole using processes like cellular proliferation, migration, differentiation and fusion. These processes are similar to those occurring during embryogenesis and tissue morphogenesis. Palatogenesis, the formation of the palate from two independent palatal shelves growing towards each other and fusing, intuitively, shares many similarities with the closure of a cutaneous wound from the two migrating epithelial fronts. In this review, we summarize the current information on cutaneous development, wound healing, palatogenesis and orofacial clefting and propose that orofacial clefting and wound healing are conserved processes that share common pathways and gene regulatory networks.
Overview
As Paul Martin and colleagues have written: “Embryos are in the business of building tissue, and wound healing is essentially a process of rebuilding what was damaged. To close a wound, all they do is reactivate the machinery they’re using somewhere else in the body anyway” (Wadman, 2005). In wound repair, and particularly cutaneous wound repair, regeneration of the lost tissue requires migration of mesenchymal cells (fibroblasts, endothelial cells, inflammatory cells) that proliferate, differentiate, and then synthesize, deposit and organize a new extracellular matrix. Epithelialization is initiated during the response to injury and leads to keratinocyte proliferation and migration across the regenerating extracellular matrix. The leading epidermal edges appose, and stratification and differentiation take place to restore the barrier function.
For proper palatal formation to occur, the two palatal shelves elongate as neural crest-derived mesenchymal cells migrate and proliferate, and then dynamically elevate above the tongue. As the two shelves appose, the medial edge epithelium (MEE) of each shelf adhere to each other, in part because of the presence of filopodia on the apical surfaces of these epithelial cells. These edges join to form a single epithelial layer, the midline seam. Finally, this seam remodels to allow confluence of the mesenchyme through a combination of processes including cell migration, apoptosis and epithelium-to-mesenchyme transformation (EMT). With the MEE gone, a confluent connective tissue remains and become the secondary palate.
As palatogenesis and wound repair share key cellular behaviors, it is not surprising that conserved embryological and reparative processes involving similar common genes and pathways regulate cutaneous development, wound healing, palatogenesis, and orofacial clefting.
In this review we summarize the current information about the molecular mechanisms involved in normal orofacial and cutaneous development. We propose that wound healing requires many of the same conserved processes necessary for palatal development. We then report on the genetic contribution to orofacial clefting and potential parallels with wound healing.
NORMAL EMBRYOLOGICAL DEVELOPMENT
Orofacial embryogenesis
The face, including the mouth and the nose, form from the oro-pharyngeal region of the early embryo as a result of the complex signaling that occurs between the three primordial cell layers during early embryonic development (Sperber, 2002a). This extremely intricate set of morphogenetic interactions gives rise to the external and internal entrances to the alimentary and respiratory tracts, while functionally separating them to allow normal respiration and deglutition. In human and mice, the growth of placodes yields to pits and processes that fuse to form the nose, the mouth, the lip, the cheeks and the palate.
Early in embryogenesis (before embryonic day (E)10.5 in the mouse and the 6th week in human), the primitive oral cavity consists of a space bound by the forebrain (superiorly), the pericardial cavity (inferiorly) and the first pharyngeal (also called branchial) arch laterally (Sperber, 2002a). The pharyngeal arches are horse-shoe shaped structures that consist of a mesenchymal core covered by an ectodermal epithelium. These arches grow and form embryonic processes that fuse appropriately to turn into the facial and oral structures. The face is derived from seven embryonic processes: i) the medial nasal process, derived from the frontal eminence, that gives rise to the middle part of the nose, including the nasal septum, the middle part of the upper lip, and the primary palate ii, iii) two lateral nasal processes, derived from the frontal eminence, that gives rise to the lateral walls of the nasal cavities iv, v) two maxillary processes, derived from the first pharyngeal arch, that give rise to the upper parts of the cheeks, the upper lip, the maxilla and the secondary palate and vi, vii) two mandibular processes, derived from the first pharyngeal arch, that give rise to the lower part of the face, the lower lip and the mandible (Sperber, 2002b). Chronologically, the external components of the mouth and nose start to form at E9.5 in the mouse, when the nasal placodes develop into pits. Between E10 and E11, the medial and lateral nasal processes extend and meet to form the region around the nostril. A day later, the mandibular processes extend and meet to form the lower jaw and Meckel’s cartilage.
Palatogenesis
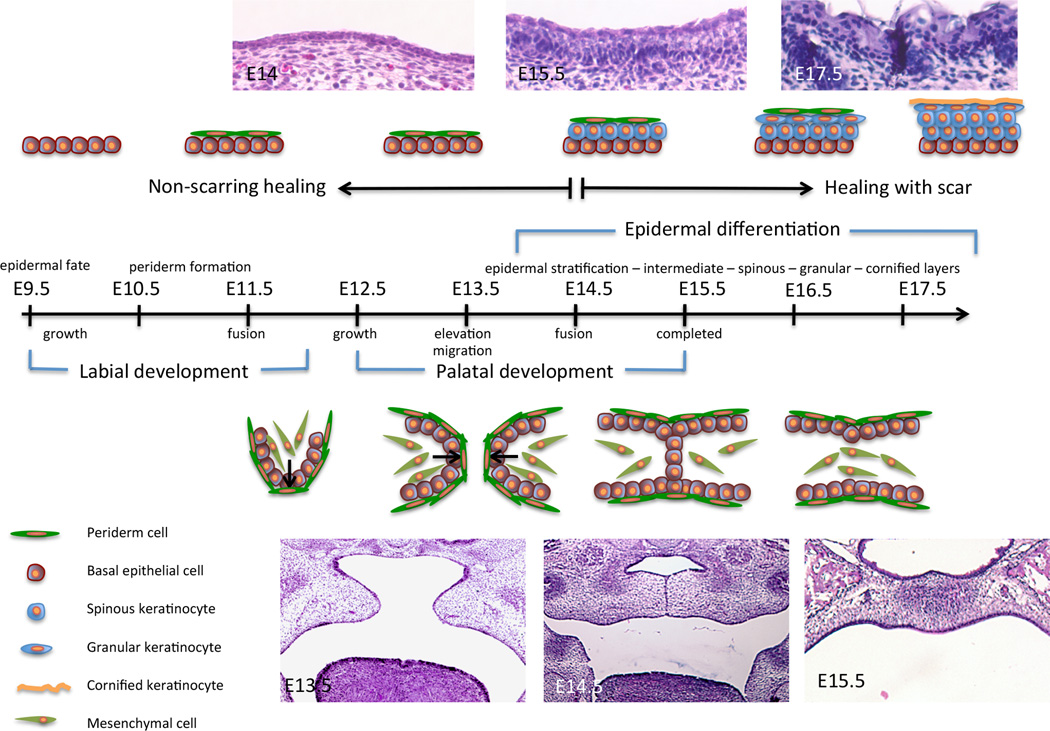
The palate functions to separate the oral and nasal spaces to allow respiration, deglutition, and phonation to occur. It is derived from two embryonic parts: the primary palate is the posterior extension of the medial nasal process and forms the four incisors and the most anterior part of the hard palate (Sperber, 2002a). The secondary palate is derived from the maxillary processes and forms the major part of the hard palate and soft palate (Sperber, 2002b). The first sign of development of the murine secondary palate occurs at E11, followed by the vertical growth of the two palatal shelves down the sides of the developing tongue (E12–13). Each shelf consists of a central core of neural-crest derived mesenchymal cells surrounded by an epithelium composed of cuboidal ectodermal cells (resembling keratinocytes of the epidermis) and a surface layer of periderm cells similar to the periderm in the oral cavity and the epidermis (Fig. 1). Recent data shows the critical presence of these periderm cells for proper craniofacial development, as animal models lacking the periderm exhibit oral adhesions and cleft palate (Casey et al., 2006; Richardson et al., 2009; Peyrard-Janvid et al., 2014; Richardson et al., 2014). About a day later, the tongue descends inferiorly as the mandibular arch lengthens to allow the paired palatal shelves to elevate and reorient from a vertical to a horizontal position and start to extend medially. The palatal shelves finally meet at E14 in the midline where they fuse to one another and to the down-growing nasal septum (Murray, 2002). This fusion is initiated in the middle of the shelves and progresses anteriorly and posteriorly to complete palatogenesis. It results in the formation of the oral and nasal cavities, in which each epithelium will have a different fate; the oral epithelium differentiates into keratinizing epithelium whereas the nasal epithelium differentiates into ciliated epithelium of the respiratory tract (Sperber, 2002b).
Figure 1.
Embryonic development landmarks of the epidermis and the palate in the mouse. Embryonic time scale from E9.5 to E18.5 is represented. Events related to epidermal development are depicted above the time scale. Events related to palatogenesis are depicted below the time scale.
Palatal development involves the complex choreography of cell migration, proliferation, and differentiation that is closely regulated in a spatiotemporal manner. Any interruption of cranial neural crest (CNC) migration into the palatal shelves, failed palatal shelf elevation (due to intrinsic forces within the palatal shelf or extrinsic forces from the tongue blocking elevation), palatal shelf apposition (due to deficient growth), or failure of the MEE to breakdown, results in a cleft of the palate. Not only are these cellular processes required for the formation of the palate, they are also key in the healing of the palate after repair. This is illustrated by the fact that up to 10% of patients develop wound complications following cleft palate surgical repair.
There are multiple genes required for palatal development and like the embryogenesis of this tissue itself, these genes function in temporal and spatial manners (Bush and Jiang, 2012). During the initial migration of the CNC into the first branchial arch, the mesenchymal cells move in response to soluble signals from the adjacent endoderm. Conversely, the mesenchyme secretes soluble factors, which informs epithelial development. Globally, these genes are part of complex regulatory networks that are conserved among the development of other tissues. However, genes that control the development of the anterior and the posterior palate are not identical. This heterogeneity may in part explain the varied clinical phenotypes of patients with cleft palates.
Multiple transcription and growth factors contribute to the development of the anterior palate (for review, see (Bush and Jiang, 2012)). One of the most critical is the Msx1 (Muscle Segment of Homeobox Gene of Drosophila) gene. It is essential for palatal development and complete abrogation of its transcriptional activity in the mouse causes cleft palate; the shelves are able to elevate, but are severely shortened and can not appose (Vastardis et al., 1996). Further investigations reveal that the Msx1 gene is required for the expression of other factors, notably the growth factors Bone Morphogenic Protein (Bmp) 2 and Bmp4 in the adjacent palatal mesenchyme and the Sonic Hedgehog (Shh) gene in the overlying MEE, which then work in a feedback loop to regulate the expression of the Msx1 gene (Zhang et al., 2002). The interruption of the Msx1-Bmp2-Bmp4-Shh signaling loop leads to decreased cellular proliferation and cleft palate in the mouse. Msx1 itself is regulated, in part, by the Hoxa2 (Smith et al., 2009) and the Foxe1 genes (Venza et al., 2011). Themselves, both of these genes contribute to proper palatogenesis: Hoxa2 is required for shelf fusion ex vivo and mutations in the FOXE1 human gene cause Bamforth-Lazarus syndrome (OMIM #241850) and contribute to cleft lip and palate. Signaling of the growth factor Fgf10 within the palatal mesenchyme intersects with the Msx1-Bmp2-Bmp4 pathway to ultimately regulate the expression of Fgfr2a in the adjacent palatal epithelium (Alappat et al., 2005). These two complementary pathways are critical during the elongation, elevation and fusion of the palatal shelves. During mesenchymal differentiation, cells from the shelves undergo osteoblastic commitment to form the hard palate, requiring activation of the Bmp signaling pathway. Specifically, the loss of Bmpr1a signaling in the CNC causes cleft palate and reduces palatine bone formation (Li et al., 2011).
Posterior palatal development remains less well understood, but it appears that genes necessary for its development predominantly regulate cellular proliferation. Amongst them, the Meox2 gene is a transcription factor only detected in the posterior palate. About a third of the mice deficient for Meox2 display a cleft palate, and unlike previously reported clefts, elevated shelves lack epithelial cells in the medial edge area. Although the molecular mechanism underlying this observation is not known, it apparently results from the breakdown of newly fused shelves (Jin and Ding, 2006). The Tbx22 gene, a member of the T-box transcription factor family, is another transcription factor specific to the posterior palatal mucosa. Its expression is regulated by Fgf and Bmp signaling and is an integral component of the molecular network that governs palatogenesis (Fuchs et al., 2010). In mice, deletion of the Tbx22 gene causes submucousal cleft palate that is associated with reduced palatine bone formation and choanal atresia (Pauws et al., 2009). In human, genetic mutations in TBX22 cause X-linked craniofacial syndrome, X-linked cleft palate and ankyloglossia (CPX; OMIM #303400).
The physical process of the adhesion, fusion and dissolution of the MEE after apposition of the palatal shelves, has undergone a great deal of investigation using DiL experiments as well as in vitro and in vivo genetic lineage tracing of the epithelium. Of particular interest are the roles of Interferon Regulatory Factor 6 and Tgfβ3 during this process. The expression of the Irf6 gene intensifies around E13 in the epithelial cells at the tip of the palatal shelves, and even more when the two appose, supporting the notion that the Irf6 gene is necessary for this process (Knight et al., 2006). However, using a reporter murine model, Fakhouri et al showed that Irf6 is not required for MEE fusion (Fakhouri et al., 2012). Absence of Tgfβ3, a growth factor upstream of the Irf6 gene, causes cleft palate in the mouse due to the inability of the periderm of one shelf to firmly attach to the opposing shelf (Kaartinen et al., 1995). Reception of Tgfβ signaling during the development of the MEE was necessary for its dissolution as well, suggesting an autocrine Tgfβ signaling pathway during MEE dissolution (Dudas et al., 2004; Xu et al., 2006). Overall, the scientific community has come to agree that three mechanisms are likely acting in concert during MEE dissolution, including apoptosis, EMT, and cellular migration (Bush and Jiang, 2012), and that these mechanisms are likely working in concert with one another rather than each acting independently. Further live imaging studies are warranted to define the fate of MEE cells during this process.
Epidermal development
The ultimate function of the epidermis is to act as a barrier organ system to protect species from external environmental factors and maintain internal homeostasis. It is also a reservoir of stem cells allowing the constant renewal of this tissue for the life of the organism. The developmental processes and pathways involved in epidermal morphogenesis share strong commonality with processes and genes involved in cutaneous repair. As such, we will describe the gene regulatory networks involved in cutaneous embryogenesis to facilitate our understanding of tissue repair, yet focus on molecules with critical roles in palatogenesis.
Though the three germinative layers are established during gastrulation, it is not until after neurulation that the entire outer surface of the developing embryo adopts an ectodermal fate. This single-layered epithelium consists of loosely integrated ectodermal cells containing keratin intermediate filaments (K8, K18, and K19) characteristic of simple epithelia (Dale et al., 1985). Because of their high keratin content, these cells are presumably keratinocytes.
The surface ectoderm takes on an epidermal fate around E8.5 in the mouse and E20–30 in humans, not by default as it was thought for a long time, but under the influence of mesenchymal signals. Although the nature of the inductive signal is largely unknown, data from animal cap transplantation in amphibians and cultured embryonic stem cell differentiation identify the growth factor Bmp4 as the endogenous neural inhibitor/epidermal inducer (Hemmati-Brivanlou and Melton, 1997; Aberdam, 2004). The acquisition of the epidermal fate is associated with the induction of the p63 gene (Lu et al., 2001; Koster et al., 2004; Koster and Roop, 2007). In fact, the p63 gene is the first transcription factor that is specific for the epidermal lineage (Green et al., 2003). The importance of p63 in epidermal induction is underscored by the phenotype of the p63 knockout mice. Though these mice die shortly after birth, they are born with a single-layered epithelium resembling the surface ectoderm that covers the early embryo, indicating the absence of epidermal specification (Mills et al., 1999; Yang et al., 1999). The p63 gene directly regulates the Tfap2c gene (Koster et al., 2006), a transcription factor previously implicated in the expression of epidermal specific keratin genes, Krt5 and Krt14 (Byrne et al., 1994). By E9.5 and up to E10.5 in the mouse and already at E42 in human, Krt5 and Krt14 are expressed in the yet still single-layered epithelium (Dale et al., 1985; Byrne et al., 1994). The onset of these two genes reflects a commitment towards the formation of a complex epithelium.
The first step in the process of stratification is a rotation of the plane of cleavage from the vertical position in the single-layer epithelium to a horizontal plane (Smart, 1970). Between E10.5 and E12.5 in the mouse and E35–42 in human, the single-layered epithelium becomes two layers comprised of the basal layer and the periderm (Fig.1). The outermost of the two layers is the periderm, a unique structure that sheds when the epidermis is fully developed underneath. The cells from the periderm are morphologically distinct from the basal layer, with a flattened nucleus and intermediate filaments distributed in small bundles throughout the cytoplasm (Dale et al., 1985). Amongst keratin filaments, K6a, K17, and K19 are expressed in the periderm (Holbrook, 1991; McGowan and Coulombe, 1998; Mazzalupo and Coulombe, 2001). The periderm cells also express Involucrin and Loricrin (Akiyama et al., 1999), which has led to the presentation of the periderm as a stratum corneum substitute during embryonic development. Although its epidermal function is not fully understood, animal models lacking the periderm (i.e. Irf6-, 14-3-3σ-, and Ikkα-deficient) exhibit altered epidermal terminal differentiation, suggesting a potential role for this structure in barrier formation (Richardson et al., 2014).
Beneath the periderm is the basal layer (or stratum basale, stratum germinativum), a germinative layer that provides the other epidermal layers with cells, as epidermal differentiation leads to constant shedding and needs to be replenished. The basal layer is characterized by the expression of K5, K14, and TAp63, yet it still expresses K19 (Dale et al., 1985). Transcription factors AP-1, AP-2 and Ets, in addition to p63 are required for the maintenance of K5 and K14 expression in the basal layer (Byrne et al., 1994; Koster et al., 2004; Romano et al., 2009). The rate of basal keratinocyte proliferation is high (Bickenbach et al., 1986), and these cells will, initially, give rise to the intermediate layer, a unique embryonic-specific layer (E13.5 to E14.5 in the mouse and E70–84 in human).
The addition of a fourth layer is the beginning of the appearance of the spinous layer (E14.5 in mouse, E140 in human). The spinous layer is characterized by keratinocytes withdrawing from the cell cycle, down regulating basal cell markers and expressing markers of differentiation, like K1, Involucrin, Irf6 and 14-3-3σ (Fuchs and Green, 1980; Watt, 1983; Ingraham et al., 2006; Richardson et al., 2006; Bazzi et al., 2007). These changes are orchestrated at the transcriptional, cellular and structural levels. Although many of the molecules and their interactions have not fully been elucidated, a role for miR-203, c/EBP, c-Myc, and Notch in the transition from basal to spinous layers has been identified (for review, see (Blanpain and Fuchs, 2009)). Of particular interest to us are the Irf6, Ikkα, Receptor-interacting protein kinase (Ripk) 4 and 14-3-3σ genes. These four genes, when mutated or deleted from the genome in the mouse, lead to animals with a similar phenotype (Hu et al., 1999; Li et al., 1999; Takeda et al., 1999; Holland et al., 2002; Herron et al., 2005; Li et al., 2005; Ingraham et al., 2006; Richardson et al., 2006). Their skin is taut, and close observation of their epidermis reveals an absence of the granular and cornified layers, demonstrating a deficit in proper terminal differentiation. At the same time, ectopic expression of K14 and p63 proteins in the suprabasal layers suggests an expansion of the basal layer. However, the presence of proliferative cells concomitant with K1 expression is reminiscent of the feature of the intermediate layer rather than the spinous layer. Despite their common phenotypes, no genetic interaction between Ikkα and Irf6 or Ikkα and 14-3-3σ has been identified. Furthermore, Ikkα-deficient and 14-3-3σ-deficient animals were not rescued by mice overexpressing Ripk-4 under the K14 promoter (Rountree et al., 2010). However, the Ikkα gene is a direct target of p63, and genetic interaction between 14-3-3σ and Irf6 and p63 and Irf6 have been demonstrated in vivo (Candi et al., 2006; Richardson et al., 2006; Thomason et al., 2010), suggesting that, in part, these genes belong to the same gene regulatory network.
The first granular cells appear during the 6th month of gestation in human (E160) and at about E15.5-E16.5 in the mouse, thereby initiating the events of normal terminal differentiation (Dale et al., 1985; Hardman et al., 1998). The hallmark of the granular layer is the presence of keratohyalin and lamellar granules, each a precursor of unique structures of the stratum corneum. The keratohyalin granules are composed of keratin filaments compacted with Filaggrin and enzymes like Caspase 14, while lamellar granules contain lipid leaflets (Kuechle et al., 2001). An increase in the extracellular calcium concentration leads to the activation of Protein Kinase C, which in turn downregulates K1 and K10, and upregulates Loricrin, Filaggrin, and Transglutaminase (Denning et al., 1995).
The final process in terminal differentiation occurs when cells migrate outward, lose their nuclei and form the cornified layer (or stratum corneum). It is composed of anucleate corneocytes filled with keratin filaments bound to a peripheral cornified envelope composed of cross-linked proteins. Between the corneocytes are intercellular lipid lamellae, indispensable to the barrier function of the skin (Madison, 2003). Numerous proteins, enzymes, and lipids are used to assemble the cornified layer and have been recently reviewed (Candi et al., 2005). Many of the epidermal-specific differentiation genes belong to the epidermal differentiation complex located on murine Chromosome (Chr) 3 and the syntenic region on human Chr 1q21 (Rothnagel et al., 1994). Amongst them are the Loricrin, Involucrin, Small Proline-Rich Proteins, and Filaggrin genes, along with the S100 protein family of genes. Another cluster termed the stratified epithelium secreted peptides complex on human Chr 19 and murine Chr 7 was identified (Matsui et al., 2004), containing, amongst others, secreted peptides like Dermokine or Suprabasin (Bazzi et al., 2007). In addition, the ontogeny of the stratum corneum is also regulated at the transcriptional level, with a role for Kruppel-like Factor 4 (Klf4), Grainyhead-like 3 (Grhl3), Lim-only protein (LMO4), Ripk-4 and Akt signaling via Pp2a (Segre et al., 1999; Yu et al., 2006; O'Shaughnessy et al., 2009; Rountree et al., 2010).
The completion of the morphogenesis of the epidermis leads to a fully functional epidermis at birth that will allow the transition from the aqueous in utero to the air terrestrial ex utero environment. The embryonic program of differentiation described above is the same as the one taking place throughout adult life when the epidermis continuously renews itself and during cutaneous repair following injury, with the exception that there is no intermediate layer with the basal keratinocytes migrating directly into the spinous layer.
Dermal development
The dermis is the cutaneous tissue below the epidermis and the basement membrane zone, and it consists of connective tissue. The connective tissue layer contains the cutaneous appendages (hair follicles, sebaceous glands, sweat glands), blood vessels and nerves. The dermis is structurally divided into two areas: the superficial dense papillary dermis and the deeper reticular dermis (Odland, 1991). This connective tissue is constituted by an extracellular matrix (collagen, elastin, glycoproteins, proteoglycans, and glycosaminoglycans) in which the largest population of cells is the fibroblast (Odland, 1991). Other cells found within the dermis include monocytes, macrophages, and dermal dendrocytes. The dermis acts to provide structural integrity to the skin, maintain the appendages, and is a critical component during wound healing.
Broadly, the dermis arises from the mesodermal layer of the embryo. Interspecies tissue recombination experiments defined the embryonic origins of the dermis: the dorsal dermis arises from the somatic dermomyotome, the dermis of the lateral and ventral body originates in the lateral plate mesoderm (Mauger, 1972; Christ and Ordahl, 1995; Fliniaux et al., 2004), and the dermis of the head and neck originates from neural crest cells (Sengel, 1990). At the time when the ectoderm acquires its epidermal fate, the mesodermal component of the skin has not been specified and thus is indistinguishable from the subcutaneous mesenchyme. Dermal specification is first noted histologically by the formation of a homogeneous, loose subectodermal mesenchyme between E9.5 and E13 in the mouse and before 3 months in human (Holbrook, 1991; Dhouailly et al., 2004; Driskell et al., 2013). The dermis becomes specified in a wave that starts in the lateral trunk and moves toward the dorsal midline. This acquisition of dermal fate can be visualized by the expression of two mutually exclusive transcription factors: Msx1 and Dermo1. The presumptive dermal cells first express the Msx1 gene in the dorsal mesenchyme around E11, before downregulating it upon expression of the earliest known marker of dermal fate, the Dermo1 gene (also known as Twist2). Dermo1 is first detected around E11 in the lateral trunk and reaches the dorsal midline around E13 (Houzelstein et al., 2000); this pattern of expression correlates with dermal differentiation, hair follicle morphogenesis, and the onset of the first periderm differentiation marker (M'Boneko and Merker, 1988). As cutaneous development continues, the papillary and reticular dermal regions begin to be histologically distinct around E18.5 in the mouse (just before birth) and in the 4th–5th months in human; by P2 (post-natal day 2), the papillary and reticular regions are fully distinguishable from each other. Furthermore, at this time, the lowest part of the dermis, the hypodermis, has formed (Van Exan and Hardy, 1984; Driskell et al., 2013).
Recent molecular analysis of the murine dermis revealed distinct and dynamic expression profiles of the papillary dermis and the reticular dermis (Driskell et al., 2013), with CD26 unique to the papillary dermis and Delta-like homolog 1 (Dlk) specific to the reticular dermis and hypodermis. Flow cytometry sorting of P2 dermal cells and subsequent grafting assays showed that hair follicle formation was dependent on fibroblasts from the papillary dermis, and not from the reticular dermis, demonstrating that these distinct fibroblast lineages have varying developmental potential (Driskell et al., 2013).
Murine models have been used to investigate the molecular pathways of dermal origin and development. Although the Msx1 gene is detected early during dermal specification, is not necessary for dermal development in the mouse. Of interest, Msx1-deficient mice exhibit craniofacial abnormalities, including a complete cleft of the secondary palate (Houzelstein et al., 1997). However, mice deficient for Dermo1 exhibit a decrease in dermal thickness caused by tissue atrophy including a reduction of subcutaneous fat (Sosic et al., 2003). One of the most important signaling pathways for specification of all three dermal regions (lateral and ventral body, the dorsum, and the head) is the Wnt/β-catenin signaling in the subepidermal mesenchyme (Atit et al., 2006; Ohtola et al., 2008; Tran et al., 2010). For example, the Wnt genes are critical for the survival of early ventrum and flank dermal progenitors and for later fate specification (Ohtola et al., 2008). Further, a subset of En1-positive progenitors in the central dermomyotome are responsive to Wnt signaling to become dorsal dermal fibroblasts (Atit et al., 2006). Finally, cranial mesenchymal cells express the Dermo1 protein, rendering them permissive to Wnt activity, which is required for these cells to adopt a dermal fate (Tran et al., 2010).
As the dermis specifies, it communicates with the epidermis to provide molecular cues for cutaneous appendage morphogenesis (for review on appendage development see (Biggs and Mikkola, 2014)). Briefly, the dermis signals to the epidermis, which responds by forming bulges known as placodes. These placodes signal to the dermis, instructing the fibroblasts to condense into a dermal condensate, which is the precursor to the functional dermal unit of the hair follicle known as the dermal papilla. These specified fibroblasts next communicate to the placode and down-growth of the epidermis and subsequent development of the hair follicle occurs. Finally, the dermis contains matrix proteins, the most abundant being Collagens I, III and Fibronectin. These extracellular matrix proteins are secreted by fibroblasts, providing a scaffold for the fibroblasts to populate, and providing structure and integrity to the skin and the body. These proteins eventually become non-randomly distributed around the skin appendages, lending evidence for their role in the development and/or maintenance of the appendages (Sengel, 1990).
WHEN THINGS GO WRONG
Orofacial clefts
Cleft lip and palate (CL/P) is a complex trait with genetic and environmental triggers that affects about 1 in 800 births (Murray, 2002). During facial formation, the CNC migrates into the frontonasal process, grows inferiorly and the paired maxillary arches grow from the side towards the midline. The medial and lateral nasal processes form from the maxillary and frontonasal process, and they all converge to form the nose and upper lip and primary palate. Failure of CNC migration or inability of the epithelium-covered surfaces to fuse, leads to cleft lip and alveolus. The secondary palate develops as an outgrowth of the maxillary prominences as outline above. Interruption of CNC migration, premature epithelial adherence of the palate to the oral cavity, failure of palatal elevation, or inability of the MEE to dissolve leads to cleft palate. CL/P has a wide variability across geographic origin (Mossey, 2002) suggesting underlying genetic differences in etiology. CL/P differs in frequency by sex and side of clefting. There is a 2:1 male to female ratio and a similar 2:1 left-side:right-side ratio for unilateral clefts. Fogh-Andersen (Fogh-Andersen, 1942) in Denmark first proposed the contribution of genetic factors in clefting, which have subsequently been confirmed by segregation analysis (Marazita, 2002) and twin studies (Mitchell et al., 2002). Evidence from genetic studies, animal models, expression data and environmental correlates suggest a variety of candidate genes for CL/P, with a list of about 300 (Jugessur et al., 2009). These genes have been investigated intensively using candidate gene approaches including direct sequencing and association studies. There is strong evidence to show that at least genetic variants in IRF6 (Rahimov et al., 2008), FGFR1, FGFR2 (Riley et al., 2007), BMP4 (Liu et al., 2005; Suzuki et al., 2009), FOXE1 (Vieira et al., 2005), TGFB3 (Lidral et al., 1998), MSX1 (Jezewski et al., 2003), MAFB (Beaty et al., 2010), and at the 8q24 chromosomal locus (Beaty et al., 2010; Mangold et al., 2010) can all result in human clefting (see additional genes in Table 1). Three of these genes, IRF6, TGFB3 and BMP4, have strong evidence to support a role in wound healing as well based on their role in epidermal differentiation (Ingraham et al., 2006; Richardson et al., 2006), scarring (Ferguson and O'Kane, 2004) and spontaneous healing of orofacial clefting (Liu et al., 2005), respectively. Subphenotypes of clefts are increasingly recognized as being important in clinical assessment and gene identification (Marazita, 2007) and include deficiencies of the orbicularis oris muscle, microform clefts such as bifid uvula and submucous clefts and dental anomalies. Deficient wound healing has also been suggested as another manifestation of the underlying contributors to clefting (Suzuki et al., 2009).
Table 1.
Genes associated with cleftinga and their known role in cutaneous wound healing
| Gene name | Expression in face | Role in CL/P | Expression in skin | Role in Wound Healing |
| RhoGTPase activating protein 29 (ARHGAP29) | In the oral epithelium (Leslie et al., 2012) | Associated with CL/P (Leslie et al., 2012) | In both dermis and epidermis (Biggs et al., 2014) | Genetic variant associated with poor scarring (Smith et al., 2014) |
| Bone Morphogenetic Protein 4 (BMP4) | In the epithelium, then the mesenchyme of the facial primordial (Francis-West et al., 1994) | Mutations in patients with cleft lip (Suzuki et al., 2009). Conditional knockout have cleft lip that spontaneously heal (Liu et al., 2005) | In the hair follicle, fibroblasts, melanocytes and keratinocytes (Yaar et al., 2006; Plikus et al., 2008) | Role in bone repair, no direct studies on cutaneous wound healing |
| Cystein-Rich Secretory Protein LCCL Domain Containing 2 (CRISPLD2) | In developing oropharynx and nasopharynx at e13.5, mandible at e14.5, and cartilage primordial of the nasal bones, palate, and tooth germs at e17.5 (Chiquet et al., 2007) | Associated with CL/P (Chiquet et al., 2007) | Not reported. | Not reported. |
| Fibroblast Groth Factor 8 (FGF8) | In medial and lateral nasal processes of e10.5 and e11.5 mice (Thomason et al., 2008) | Associated with CL/P (Riley et al., 2007) | Not reported. | In amphibian wound blastema (Endo et al., 2000) |
| Fibroblast Growth Factor Receptor 1 (FGFR1) | In the palatal epithelium (Britto et al., 2002) | Associated with CL/P (Riley et al., 2007) | In dermis and epidermis (Takenaka et al., 2002) | Upregulated in wound healing (Komi-Kuramochi et al., 2005) |
| Fibroblast Growth Factor Receptor 2 (FGFR2) | In the palatal epithelium (Britto et al., 2002) | Associated with CL/P (Riley et al., 2007) | In the basal layer of the epidermis (Takenaka et al., 2002) Knockout mice have thinner skin (Revest et al., 2001) | No change in expression during murine cutaneous wound (Komi-Kuramochi et al., 2005) |
| Forkhead/wingled-helix domain transcription factor (FOXE1) | In oral epithelia (palatal, nasal) (Dathan et al., 2002) | Associated with CL/P (Vieira et al., 2005) Knockout mice have cleft palate | In the epidermis and hair follicles (Eichberger et al., 2004) | Not evaluated |
| GLI-Kruppel family member GLI2 (GLI2) | In the palatal shelves (Mo et al., 1997) | Associated with CL/P (Vieira et al., 2005) | In the epidermis and hair follicles (Eichberger et al., 2004) | Not evaluated |
| Grainyheald like-3 (GRHL3) | In the oral cavity (Auden et al., 2006). | Mutations cause VWS (Peyrard-Janvid et al., 2014) Mice have abnormal periderm with oral adhesions and low penetrance cleft (Peyrard-Janvid et al., 2014) | In the periderm, ectoderm, and throughout the epidermis (Auden et al., 2006; de la Garza et al., 2013) | Embryonic wounds open after 24 hr assay (Caddy et al., 2010) |
| Interferon Regulatory Factor 6 (IRF6) | In the medial edge epithelium, palatal epithelium (Knight et al., 2006) | Mutations cause VWS and PPS (Kondo et al., 2002). Knockout mice have orofacial anomalies (Ingraham et al., 2006) | In the spinous layer of the epidermis (Ingraham et al., 2006; Richardson et al., 2006) | Patients with VWS have increased likelihood of worse surgical outcome (Jones et al., 2010) |
| Isthmin 1 (ISM1) | In oral mucosa (Valle-Rios et al., 2014) | Not reported | In human skin (Valle-Rios et al., 2014) | Not reported |
| Jagged 2 (JAG2) | In the oral epithelium (Casey et al., 2006) | Associated with CL/P (Vieira et al., 2005) Knockout mice have craniofacial defects (Jiang et al., 1998) | In fibroblasts | Not reported. |
| LIM homeobox 8 (LHX8) | In mesenchyme of maxillary and mandibular processes during palatogenesis (Zhao et al., 1999) | Associated with CL/P (Vieira et al., 2005) Knockout mice have cleft palate (Zhao et al., 1999) | Not reported. | Not reported. |
| v-Maf musculo aponeurotic fibrosarcoma (MAFB) | In the oral epithelium (Beaty et al., 2010) | Associated with CL/P (Beaty et al., 2010) | In the suprabasal layer of the epidermis (Ogata et al., 2004) and hair follicles (Miyai et al., 2010) | Not reported |
| Muscle Segment Homeobox Gene of Drosophila (MSX1) | Restricted to the anterior of the first upper molar site in the palatal mesenchyme (Zhang et al., 2002) | Associated with CL/P (Jezewski et al., 2003) Knockout mice have cleft palate (Satokata and Maas, 1994) | In embryonic and adult skin, epithelial derived structures (Stelnicki, 1997) (Vieira et al., 2005) In the hair follicle (Ma et al., 2003) | Not evaluated |
| Muscle Segment Homeobox Gene of Drosophila (MSX2) | In the developing palate (Winograd et al., 1997) | Point mutations associated with CL/P (Vieira et al., 2005) Knockout mice have a cleft palate (Winograd et al., 1997) | In the hair follicle (Ma et al., 2003) | Not evaluated |
| Paired box 7 (PAX7) | Not reported | Maternal SNPs associated with CL/P (Beaty et al., 2013; Butali et al., 2013) | Not reported | Lineage tracing show 25% of dermal cells in the healing wound Pax7 expressing progeny (Amini-Nik et al., 2011) |
| Paired box 9 (PAX9) | In the lingual epithelium, medial edge epithelium, and palatal mesenchyme (Peters et al., 1998) | Associated with CL/P Knockout mice exhibit a cleft palate (Peters et al., 1998) | Not expressed in the epidermis (Peters et al., 1998) | Not reported. |
| Platelet-derived Growth Factor C (PDGF C) | In the epithelium of the palatal shelves (Ding et al., 2004) | Associated with CL/P (Choi et al., 2008) Knockout mice have cleft palate (Ding et al., 2004) | In basal layer of the epidermis and hair follicle (Ding et al., 2000) | Promotes wound healing (Gilbertson et al., 2001) |
| Patched Homolog 1 (PTCH1) | In neural crest-derived mesenchyme of facial prominences (Metzis et al., 2013) | Associated with CL/P (Mansilla et al., 2006) Neural crest specific knockout exhibit cleft lip (Metzis et al., 2013) | In dermal papillae and a few follicular and interfollicular keratinocytes (Morgan et al., 1998) | Absence of Ptch1 in basal keratinocytes results in formation of basal cell carcinomas in healing wounds (Kasper et al., 2011) |
| Poliovirus Receptor-Related 1 (PVRL1, nectin-1) | In the medial edge epithelium (Suzuki et al., 2000) | Mutations in human cause autosomal recessive ectodermal dysplasia associated with clefting (CLPED-1) (Suzuki et al., 2000) | In all the layers of the epidermis, stronger in the spinous layer; regulates loricrin expression via ERK pathway (Wakamatsu et al., 2007) | Not reported. |
| Poliovirus Receptor-Related 2 (PVRL2) | Not reported. | Associated with CL/P (Nikopensius et al., 2011) | Not reported. | Not reported. |
| Receptor-Interacting Serine-threonine Kinase 4 (RIPK-4) | Not reported | Mutations cause Bartsocas-Papas Syndrome (Mitchell et al., 2012) and recessive form of popliteal pterygium (Kalay et al., 2012) | In epidermis and necessary for proper epidermal differentiation (Holland et al., 2002; Adams et al., 2007; Rountree et al., 2010). | Downregulated within 1 hour of wounding and remains low throughout healing process (Adams et al., 2007), regulator of inflammation (Rountree et al., 2010) |
| Receptor-like Tyrosine Kinase (RYK) | In lingual and palatal mesenchyme (Halford et al., 2000) | Associated with CL/P (Watanabe et al., 2006) Knockout mice have cleft palate (Halford et al., 2000) | In the basal layer of the epidermis and hair follicles (Serfas and Tyner, 1998) | Not reported. |
| SATB homeobox 2 (SATB2) | In the developing palate (Fitzpatrick et al., 2003) | Association with CL/P (Fitzpatrick et al., 2003) | Not known | Not evaluated |
| SKI-protooncogene (SKI) | At low levels in all tissues, but increased expression in cranial neural cells (Berk et al., 1997) | Associated with CL/P (Vieira et al., 2005) Knockout mice exhibit exencephaly and cleft lip and palate (Berk et al., 1997) | Not expressed in normal skin (Liu et al., 2006) | Upregulated after wounding during epithelialization and maturation phase in fibroblasts and less so in epithelial cells (Liu et al., 2006) |
| Sonic Hedgehog (SHH) | In the ectoderm of the facial primordial (Hu and Helms, 1999) | Mutations cause CL/P (Orioli et al., 2002; Porter, 2006) | In the hair follicle (Chuong et al., 2000) | Promotes wound healing (Le et al., 2008) |
| Sprouty Homolog 2 (SPRY2) | In the epithelium and mesenchyme of the developing palate (Matsumura et al., 2011) | Point mutations may cause CL/P (Vigano et al., 2006) Knockout mice have a cleft at low penetrance (Matsumura et al., 2011) | Not reported | Downregulates angiogenesis during resolution of wound healing (Wietecha et al., 2011) |
| T-Box 10 (TBX10) | In the hindbrain but not in the face (Bush et al., 2003) | Associated with CL/P (Vieira et al., 2005) Spontaneous mutant ‘Dancer’ (Dc), which is the result of ectopic Tbx10 has a cleft palate (Trasler et al., 1984) | Not reported | Not reported |
| Transcription Factor AP2 (TFAP2a) | In the facial mesenchyme (Byrne et al., 1994; Moser et al., 1997) | Mutations cause branchio-oculo-facial syndrome (Milunsky et al., 2008) Knockout mice have severe craniofacial anomalies (Schorle et al., 1996) | Alpha and gamma in the epidermis and hair follicle (Byrne et al., 1994) | Not evaluated |
| Transforming Growth Factor Beta 3 (TGFB3) | In the medial edge epithelium (Fitzpatrick et al., 1990) | Mutations and association with CL/P (Lidral et al., 1998) Knockout mice have cleft palate (Kaartinen et al., 1995) | In all layers of the epidermis (Levine et al., 1993) | Important for scarless wound healing (Ferguson and O'Kane, 2004). Recombinant form in clinical trial (Ferguson et al., 2009). |
| Tumor Protein 63 (TP63) | In the basal cells of the ectoderm in nasal processes, medial edge epithelium, palatal epithelium (Thomason et al., 2008) | Mutations and association with CL/P (McGrath et al., 2001) Knockout mice have orofacial anomalies (Mills et al., 1999; Yang et al., 1999) | In the basal layer of the epidermis (Koster et al., 2004) | No wound problem in cleft repair of p63 ectodermal dysplasia (Cabiling et al., 2007) Splice variant specific function based on spatiotemporal expression in murine wounds (Bamberger et al., 2005) |
| Ventral anterior homeobox 1 (VAX1) | In the first branchial arch and palate (Zhao et al., 2010) | Associated with CL/P (Mangold et al., 2010; Butali et al., 2013) | Not reported | Not reported |
| Wingless type 5a (WNT5a) | In the mesenchyme of palatal shelves at e13.5 in a gradient from high to low in an anterior-posterior direction (He et al., 2008) | SNPs associated with NSCLP (Chiquet et al., 2008) Knockout mice have cleft of secondary palate (He et al., 2008) | In the dermis and inner and outer root sheaths of mature hairs (Reddy et al., 2001) | Upregulated in the mesenchyme after epithelialization (Fathke et al., 2006) |
| Wingless type 9b (WNT9b) | In the maxillary, medial and lateral nasal ectoderm from e9.5 through e11.5 (Lan et al., 2006) | Associated with CL/P (Chiquet et al., 2008) Knockout mice have cleft palate with incomplete penetrance (Carroll et al., 2005) | Not expressed in skin (Reddy et al., 2001) | Not reported |
These 35 genes are not a comprehensive list of clefting genes, but rather a list of genes that are deemed most important and/or most interesting genes as research in the field of craniofacial research moves forward. Its strength is that it was elaborated based on the contribution of these genes either in craniofacial development, in the function of craniofacial structures or in human genetics.
Surgical repair of orofacial clefts
The standard of practice for children born with cleft lip and palate is surgical repair. Usually within the first six months of life, the cleft lip is repaired while the palate is usually repaired between 6 to 18 months of age (Hurwitz and Kathju, 2003). Secondary corrections are often required to improve the function and appearance of the lip. Secondary lip repairs may occur before 5 years of age or later, and corrections of nasal deformity around 14–18 years of age. Alveolar bone grafting improves the bony support of the cleft-adjacent teeth and allows the closure of oronasal and nasolabial fistulae. This procedure is commonly performed around 6–9 years of age. Among reported post-operative complications are oropharyngeal infection, upper respiratory tract infection, airway obstruction, feeding difficulties, flap dehiscence and palatal fistula (Moore et al., 1988). The latter two are particularly relevant as they directly relate to wound healing. They both consist of the reopening of the closed wound along the surgical suture line. The incidence of these post-operative complications varies from 10–30% reported for palatal fistula (Cohen et al., 1991; Muzaffar et al., 2001) up to 63% (Gosman, 2007). A retrospective chart study performed at the University of Iowa suggests that the IRF6 genotype of the individual may be a determinant factor as patients with Van der Woude syndrome (OMIM #119300, haploinsufficient for IRF6) are more likely to have a poorer surgical outcome compared to patients with isolated CL/P with the same degree of clefting (Jones et al., 2010). Using a novel digital imaging system to quantitatively characterize the cleft scar, we identified genetic variants in RhoA GTPase Activating Protein 29 (ARHGAP29) and TGFβ3 as associated with poor scarring (Smith et al., 2014). Expanding the identification of biomarkers (like IRF6) could inform craniofacial surgeons on the risks for such post-operative complications, and may suggest rational interventions.
There have been very few alternatives or adjuvants to the classical surgical repair reported above. The prospect of fetal surgery has been driven by the potential advantages of scar-less healing (due in part to an increased TGFβ3:TGFβ1 ratio in mesenchymal tissues (Schrementi et al., 2008)), repair of the primary deformity, prevention of secondary deformities, prevention of facial growth disturbance due to scarring and normal facial appearance of the infant when presented to the parents at birth. Another alternative surgical adjuvant to surgery is the use of recombinant protein in replacement of autologous bone graft for alveolar repair. Particularly, recombinant human BMP2 has proven to be an efficient substitute in patients with cleft (Chin et al., 2005). The use of mesenchymal stem cells with platelet-rich plasma has also been used as an adjuvant during palatal repair and alveolar bone grafting (Behnia et al., 2012). Collectively, despite the fact that many patients with CL/P suffer complications and require revision surgery, very few options are currently available to them, yet are critically needed.
Cutaneous wound healing
Injury to the skin initiates a cascade of events that, when executed properly, leads to tissue repair (for review, (Singer and Clark, 1999; Werner and Grose, 2003; Shaw and Martin, 2009)). This repair process is artificially compartmentalized into three phases: inflammation, proliferation, and maturation. None of these phases correspond to a precisely defined period of time, and all phases overlap to a certain degree.
Inflammation
Upon wounding, both the skin and the underlying vasculature are injured. The mechanical damage to cells results in a rapid flash of calcium that travels from the wound margin outward and ebbs back toward the wound margin, resolving within minutes (Razzell et al., 2013), yet is sufficient to activate DUOX, and leads to the production of hydrogen peroxide (Niethammer et al., 2009). Reactive oxygen species as well as adenosine triphosphate serve as attractants to induce extravasation of circulating leukocytes, mostly neutrophils, to the site of injury (Hattori et al., 2010; McDonald et al., 2010). A blood clot forms within minutes to restore homeostasis and to secrete mediators of wound healing that attract and activate neutrophils, macrophages and fibroblasts (Singer and Clark, 1999).
Neutrophils extravasate from damaged blood vessels and are the first cells to arrive at the wound site. They are responsible for protecting and cleaning the area of foreign particles and bacteria. In addition, it has been proposed that neutrophils contribute to the resolution of the fibrin clot and provisional extracellular matrix (ECM), to the promotion of angiogenesis, and to epithelialization. After the initial influx of neutrophils, monocytes are recruited to the injured site by chemoattractants such as ECM protein fragments, TGFβ1, PDGFs, and Monocyte Chemoattractant 1, amongst many others (Singer and Clark, 1999; Werner and Grose, 2003). Monocytes appear in two waves: the first one is immediate, resulting from ‘inflammatory monocytes’ while the second consists of tissue resident-derived monocytes that arrive after homing to inflammation (Novak and Koh, 2013). Once outside the circulation, monocytes become activated and differentiate into macrophages. Macrophages are responsible for clearing matrix and cell debris, including spent neutrophils (Eming et al., 2007). Inflammatory cells release cytokines and growth factors and also generate nitric oxide and reactive oxygen species. Together, they clean the wound bed and prepare it for epithelialization, neovascularization and fibroblast migration. These reactive oxygen species also act as signaling molecules to promote wound healing including release of cytokines, angiogenesis, cell motility, and ECM formation (for review see (Sen, 2003)).
Keratinocytes also contribute to this initial phase of tissue repair. They produce and secrete a large number of cytokines, including interleukins, growth factors, colony stimulating factors and chemokines, which can influence local fibroblasts, endothelial cells and regulate the inflammatory response (Werner and Grose, 2003). They also express receptors from the Toll-like family, which are known to mediate the innate and adaptive immune responses. Of note is the prominent role for TLR4 in the production of interleukins and growth factors and ultimately to proper tissue repair (Chen et al., 2013)
The requirement for inflammatory cells in wound repair remains controversial (Martin and Leibovich, 2005). Some studies report faster epithelialization in mice lacking innate immune cells (Dovi et al., 2003; Cooper et al., 2005), others unchanged healing (Martin et al., 2003), while models lacking macrophages demonstrate delayed tissue repair (Goren et al., 2009; Mirza et al., 2009). However, it becomes evident that the disappearance of inflammatory cells is required for healing to proceed, and that the persistence and/or the dysfunction of innate immune cells leads to impaired healing as exemplified by chronic inflammation in diabetic foot ulcers and chronic wounds (Khanna et al., 2010; Pukstad et al., 2010; Miao et al., 2012).
Epithelialization
Epithelialization of the wound is the phase during which the injury is resurfaced by new epithelium derived from both keratinocyte migration at the wound edge and proliferation behind the wound front (for review, see(Coulombe, 2003)). Both of these processes are critical to epithelialization, as chronic wounds exhibit defective keratinocyte migration and ectopic proliferation (Stojadinovic et al., 2008).
Basal keratinocytes normally rest on the basement membrane, a structure composed of Laminin, Entactin, Proteoglycans, and Type IV Collagen (Marinkovich et al., 1993). Wounding destroys this membrane and during epithelialization, keratinocytes migrate on a provisional matrix provided by the clot, such as dermal collagen, unprocessed Laminin 332, and Fibronectin (Nguyen et al., 2000). Keratinocytes themselves secrete unprocessed Laminin 332 at the wound edge, promoting their own migration via Integrin α3β1 binding. Once the α3 chain of Laminin 332 is processed, the α6β4 Integrin, also present on the surface of keratinocytes, binds to processed Laminin 332 and mediates stable adhesion to the basement membrane (Usui et al., 2008). As a consequence, the keratinocytes must change their cell-substrate adhesive properties when migrating over the wound. Integrin α6β4 relocates from the hemidesmosomes to the lamellipodia of the keratinocyte’s leading edge, allowing α3β1 and α6β4 integrins to maintain the directional migratory trajectory by intracellular association with Rac1 (Choma et al., 2004; Pullar et al., 2006).
Exactly which keratinocytes participate in wound epithelialization has been controversial, with studies supporting a sliding model (Radice, 1980), a rolling model (Krawczyk, 1971; Paladini et al., 1996), or a combination of both (Usui et al., 2005). The sliding model posits that the basal keratinocytes release their hemidesmosomal attachments and maintain their desmosomal attachments. They migrate, pulling with them their neighbors and those post-mitotic cells attached above them. The rolling model posits that the suprabasal cells ‘roll over’ the basal keratinocytes, undergoing shape changes and releasing their desmosomes to fill in the hole. Indeed, keratinocytes in their early stages of differentiation are thought to contribute to tissue repair (Mannik et al., 2010). The third model is a combination of the rolling and sliding models, as demonstrated by ectopic expression of K14 and down-regulation of K10 expression in suprabasal cells (Usui et al., 2005). A recent study using cultured human skin equivalents and time-lapse fluorescent labeling found that none of the previously proposed models of epithelialization were true. In fact, they demonstrate that basal cells migrate below the suprabasal cells, which remain in place to act as a shield (Safferling et al., 2013). In this study, proliferating cells in the unwounded tissue, and not the advancing epithelial leading edge, constitute the supply of new cells covering the wound. Although this study provides crucial information as to which cells are participating in wound healing, this in vitro model lacks cutaneous appendages and stem cell niches, two sources of contributing keratinocytes during tissue repair (Ito et al., 2005; Lau et al., 2009). Collectively, it raises the question as to whether this model applies to in vivo situations.
At the single cell level, it is the spatial and temporal reorganization of the filamentous (F-) actin network within the cell and the dynamics of cellular junctions that allows cellular migration (Kardash et al., 2010). The family of small Rho-GTPases, including RhoA, Rac and Cdc42, is the master regulator of the cytoskeleton dynamics and has been extensively studied in vitro and in vivo, with specific roles for each of the enzymes in lamellipodia and filopodia formation as well as overall migration (for review, see (Burridge and Wennerberg, 2004; Hall, 2005). In the context of cutaneous wound healing, RhoA is essential for wound closure by modulating actin cables (Wood et al., 2002), and epidermal-specific deletion of RhoA decreased directed cellular migration (Jackson et al., 2011).
In monolayer epithelial sheets, a finite number of cells are mobilized to close a scratch, and the mobilization of distant cells is independent of the advancement of the leading cells (Matsubayashi et al., 2011). However, in the Drosophila embryo wound model, the cells away from the wound edge stretch toward the wound and show actomyosin-dependent shrinkage of cell-cell junctions with their neighbors to drive cell intercalation events and thus close the wound (Razzell et al., 2014).
Numerous factors regulate the proliferation and migration of keratinocytes over an open wound. Amongst them, a multitude of transcription factors have been described playing a critical role in this process, including AP-1, c-jun/c-fos, PPARβ/δ, c-Myc, E2F-1, Egr-1, Grhl3, HoxA3, HoxD3, Smad2, Smad3, and Stat3, to name just a few (Schafer and Werner, 2007). We recently added Irf6 to the list (Biggs et al. 2014). Rather than enumerate their function in epithelialization, we will elaborate on their role in tissue repair in the context of the potential parallels with palatogenesis at the end of this review.
While the epidermis is undergoing the changes described above to cover the wound, the dermis is also active. It contributes to the closing of the wound by pulling the edges of the wound closer together. First, the fibrin clot is replaced by a new stroma called granulation tissue. Participating cells include migrating fibroblasts from wound margins, circulating fibrocytes, bone marrow progenitor cells, and adipocytes (Hinz, 2007; Schmidt and Horsley, 2013). It is also the time when angiogenesis occurs, leading to the formation of new capillaries. Concomitantly, fibroblasts acquire contractile properties through the inherent formation of actin stress fibers in their cytoplasm. Furthermore, secretion of TGFβ1 and Periostin from the ECM, along with mechanical stress, induce the differentiation of fibroblasts into myofibroblasts (Hinz, 2007; Elliott et al., 2012). Through the action of both wound fibroblasts and myofibroblasts, collagen fibers are synthesized, aligned and bundled, leading to the contraction of the connective tissue (Hinz, 2007). The first fibroblasts to migrate to a wound originate from both the reticular dermis and the hypodermis. Subsequently, the papillary dermis replaces the reticular dermis during epithelialization (Driskell et al., 2013). Migration of the fibroblasts into the wound bed is dependent on the presence of adipocytes (Schmidt and Horsley, 2013). Interestingly, the absence of adipocytes in the wound and subsequent absence of fibroblasts was not detrimental to the contraction of the wound, nor to the epithelialization at first; however, during dermal remodeling, the size of the wound bed increased and the new epithelium reopened (Schmidt and Horsley, 2013), supporting cross-talk between mesenchymal and epithelial cells during tissue repair. Together, these studies provide a rationale for the rapid adipocyte deposition and the lack of hair follicles in newly epithelialized wounds (Ito et al., 2007; Driskell et al., 2013; Schmidt and Horsley, 2013).
Maturation
Upon completion of epithelialization, cellular proliferation and neovascularization cease, scar tissue forms and the wound enters the maturation phase, which lasts several months. Keratinocytes begin again their program of differentiation and granulation tissue turns into scar tissue through the continuous turnover of collagen. Collagen degradation is mediated by matrix metalloproteinases that are secreted by macrophages, epidermal and endothelial cells, and fibroblasts. Wounds slowly regain tensile strength over the ensuing months but never reach the same breaking strength as pre-wound. In fact, wounds are only 70% as strong as normal skin (Levenson et al., 1965). In addition, in human, a scar remains and hair follicles never repopulate the healed tissue (Martin, 1997; Gurtner et al., 2008). However, in the mouse, very large wounds, as well as wounds in which β-catenin was activated in the epidermis before wounding did show de novo hair follicle formation (Ito et al., 2007; Driskell et al., 2013). This suggests different regenerative capacities between the mouse and human.
The scar is the most visible remnant of the cutaneous wound with a wide range of phenotypes, yet healing occurs without a scar before birth. Murine embryos up to E15.5 exhibit scar-less wound healing (Fig. 1), presumably due to immature innate immune cells and the absence of an inflammatory response (Hopkinson-Woolley et al., 1994; Wulff et al., 2012; Chen et al., 2014).
Resolution of inflammation is a critical step for the wound to heal, and its failure results in chronic wounds. The flux of calcium rapidly returns to homeostatic levels through a yet-to-be discovered mechanism while neutrophil –produced myeloperoxidase is required to dampen the production of superoxides (Pase et al., 2012). Further clearing of inflammatory cells occurs through apoptosis of leukocytes and macrophage phagocytosis of neutrophils (Haslett, 1992). Macrophages themselves are deactivated by anti-inflammatory cytokines, glucocorticoids, cell-cell contacts and phagocytosis. In addition, some neutrophils and macrophages return to the vasculature and/or emigrate via lymphatic vessels (Mathias et al., 2006).
Differential wound healing: cutaneous versus oral wound healing
Several studies have demonstrated a marked difference between cutaneous and oral wound healing, most of which concluding that oral wounds heal with much less scarring than cutaneous wounds (Wijdeveld et al., 1991; Larjava et al., 2011). As wound healing can contribute to significant fibrosis and hypertrophic healing, comparing oral with cutaneous wound healing could provide insights into potential therapeutic avenues.
The main environmental difference between cutaneous and oral wound healing is that oral wounds are healing in the presence of saliva. Saliva contains many antibodies, growth factors, and chemokines, including vascular endothelial growth factor (VEGF) (Keswani et al., 2013), epidermal growth factor (EGF) (Noguchi et al., 1991), and basic fibroblastic growth factor (bFGF) (Kagami et al., 2000). Additional factors, including salivary antimicrobial peptide (AMP) and salivary leptin are thought to enhance oral wound closure (Groschl et al., 2005; Oudhoff et al., 2009). The human body makes over a liter of saliva per day, bathing the healing wounds in these, and many more growth factors which are thought to reduce the inflammation, and thus reduce the formation of scar following tissue repair.
Additional intrinsic differences in cutaneous vs oral wound healing include the following observations: i) Greater response to inflammatory stimuli evidenced by high IL-6 and TNF-α (Li et al., 1996), ii) Greater and prolonged expression of Tenasin-C in palatal wounds (Wong et al., 2009), iii) Reduced presence of mast cells, macrophages and neutrophils in oral wounds compared to cutaneous wounds (Kischer et al., 1978; Szpaderska et al., 2003; Mak et al., 2009), iv) Increased levels of Tgfβ3 (antifibrotic) and reduced levels of Tgfβ1 (profibrotic) in oral wounds (Schrementi et al., 2008; Eslami et al., 2009) and v) Increased angiogenesis and gingival-derived mesenchymal stem cells proliferation following oral injury (Zhang et al., 2010).
Collectively, these critical differences between cutaneous and oral wound healing could serve as targets to modulate wound healing in the future, including the use of Tgfβ3 and/or stem cell therapies.
Craniofacial development and cutaneous repair: similar, yet different, conserved mechanisms
It has been proposed and discussed elsewhere that embryogenesis of one tissue or organ and the repair of this same tissue during adulthood share common molecular mechanisms and gene regulatory networks (Ferguson et al., 1998; Wood et al., 2002; Martin and Parkhurst, 2004; Mitsiadis and Rahiotis, 2004; Aller et al., 2012). The parallel between the embryogenesis of one tissue and the repair of a different tissue, however, has not been addressed. Our first observation that patients with Van der Woude syndrome had worse surgical outcome, based on surgical wound healing complications, compared with patients with non-syndromic CL/P lead us to propose that IRF6 plays a role both in craniofacial development and cutaneous wound healing. As we confirmed our hypothesis both clinically and experimentally (Jones et al., 2010; Biggs et al., 2014), we postulated that this concept may be more global and that additional molecules would share commonalities with craniofacial development and wound healing.
The initial thought that one could generate a catalog of shared genes and regulatory network between cleft lip and palate and cutaneous wound healing is somewhat utopic. If the genetic contribution to proper palatogenesis is unquestionable, and the list of genes involved in cleft lip and palate established at about 300 (Jugessur et al., 2009), it has been challenging to generate a workable list of genes involved in wound healing, mainly because of the complexity of the process and the difficulties of obtaining relevant human samples. Nevertheless, a few studies have performed gene expression profiling of cutaneous wound healing (Deonarine et al., 2007; Roupe et al., 2010; Nuutila et al., 2012), or identified a genetic signature unique to non-healing wounds (Charles et al., 2008), yet none provided access to the full list of differentially-regulated genes. Using available information from these studies, we estimated the percentage of reported wound-healing genes that shared a role in CL/P. The results, presented in Table 2, show about 10% commonality in the gene regulatory network between the two processes. To extend this comparison further, we established a list of our top 35 candidate genes for CL/P and asked whether these genes were expressed in cutaneous tissues along with a potential role in wound healing (Table 1). About 50% of them shared a known role in both craniofacial development and wound repair, confirming strong commonality between the two biological processes. The notable difference in shared percentage of genes (Table 1 vs Table 2) may be due to the fact that CL/P occurs during embryonic development involving a broad gene regulatory network, while adult wound healing engages already differentiated tissues and a more narrow and differentiation-oriented gene regulatory network. From these two tables, we chose to discuss a few genes as part of regulatory networks involved in cellular migration and proliferation, two of the most relevant biological mechanisms critical to both the development of the palate and the healing of the skin.
Table 2.
Comparison of overlapping genes between wound healing and CL/P
| Ref | Wound healing genesa | Wound healing and clefting genesb |
|---|---|---|
| Charles et al (Charles et al., 2008) | 58 | 7 (12%) |
| Deonarine et al (Deonarine et al., 2007) | 250 | 18 (7%) |
| Nuutila et al (Nuutila et al., 2012) | 42 | 4 (9%) |
| Eming et al (Eming et al., 2010) | 60 | 4 (7%) |
| Roupé et al (Roupe et al., 2010) | 61 | 3 (5%) |
Number of genes reported up or down regulated in each of the cited wound healing studies
From the genes reported in the previous column, we filtered for genes that have a role in human clefting and/or exhibit a cleft palate in a mouse model deficient for that gene. Data is also expressed in percentage and shown under parenthesis.
The first gene regulatory network involves TGFβ3, IRF6, ARHGAP29, and GRHL3, all of which regulate cellular migration by modulating the actin cytoskeleton. Tgfβ3 induces rapid activation of RhoA GTPase and EMT in vitro (Kaartinen et al., 2002) and mice deficient for Tgfβ3 exhibit a cleft palate (Kaartinen et al., 1995; Proetzel et al., 1995). Tgfβ3 was also required for epithelialization and granulation tissue maturation in a murine model of cutaneous wounds, while dispensable for keratinocyte migration in vitro (Le et al., 2012). In humans, genetic variants in TGFβ3 are associated with CL/P (Lidral et al., 1998), and clinical trials using prophylactic TGFβ3 on cutaneous incisions demonstrated reduction of scarring compared to untreated incisions (Ferguson et al., 2009). Mice deficient for Tgfβ3 also show reduced level of expression of Irf6, both in the medial edge epithelium (Xu et al., 2006) and in the skin (Le et al., 2012), supporting the model that Irf6 is downstream of Tgfβ3.
IRF6 plays an unquestionable role in palatogenesis and human clefting disorders (Kondo et al., 2002; Zucchero et al., 2004; Ingraham et al., 2006; Richardson et al., 2006; Rahimov et al., 2008; de Lima et al., 2009). Recently, we reported a novel function for the Irf6 protein in regulating RhoA GTPase-dependent cellular migration mediated by Arhgap29, a specific inhibitor of RhoA (Biggs et al., 2014). In keratinocytes, lack of Irf6 promotes the formation of actin stress fibers due to increased active RhoA in the cells and decreased level of Arhgap29 protein. Interestingly, the ARHGAP29 human gene was identified as the etiologic gene for the CL/P locus identified by genome-wide association on chromosome 1p22 (Leslie et al., 2012). A genetic variant in ARHGAP29 was also associated with poor scar outcome in patient with CL/P (Smith et al., 2014), demonstrating the conservation of pathways regulating tissue repair and orofacial clefting.
Using the zebrafish as a model for craniofacial development, de la Garza et al. identified grhl3 as a direct target of irf6 (de la Garza et al., 2013). Grainyhead was discovered in drosophila as a critical regulator of the repair of the cuticle (Mace et al., 2005). Grhl3 is expressed in several epithelia, as well as in the periderm of the skin and of the oral cavity, and its expression is altered in the periderm of Irf6-deficient mice. The Grhl3 gene is required for proper craniofacial development as mice deficient for this transcription factor exhibit cleft palate and oral adhesion (Peyrard-Janvid et al., 2014). Furthermore, patients with Van der Woude syndrome, who were lacking a detectable mutation in IRF6, harbored mutations in GRHL3 (Peyrard-Janvid et al., 2014). Like Irf6, the Grhl3 gene is also required for proper migration of keratinocytes. However, compared to Irf6, the absence of Grhl3 led to decreased level of active RhoA and absence of actin stress fibers (Caddy et al., 2010). In fact, RhoGEF19, an activator of RhoA, was identified as the direct target of Grhl3. It is therefore interesting to think that two genes (Irf6 and Grhl3), one a direct target of the other, both regulate Rho activity in opposite directions to give the same phenotype (migration defect and Van der Woude syndrome). It exemplifies how cellular migration and cytoskeletal dynamics are tightly controlled with positive and negative regulators.
The second gene regulatory network involves WNT, TP63, IRF6, TFAP2, and c-MYC, all of which regulate cellular proliferation. Several studies have demonstrated that mutations in WNT genes are associated with CL/P. The Wnt pathway regulates orofacial development via multiple downstream targets, including Tgfβ3, Lrp6, Irf6, and p63 (Song et al., 2009; He and Chen, 2012; Kurosaka et al., 2014). It is equally essential for cutaneous development, particularly for the maintenance of stem cells (Blanpain and Fuchs, 2009; Lim and Nusse, 2013; Lim et al., 2013). During wound healing, Wnt signaling affects both dermal and epidermal compartments (Amini-Nik et al., 2014). Interestingly, it contributes to the de novo regeneration of hair follicles in large cutaneous wounds (Ito et al., 2007) and enhanced Wnt signaling promotes cutaneous healing in a murine ear model (Whyte et al., 2013), suggesting a potential therapeutic application for this secreted protein.
p63 and Irf6 operate downstream from Wnt signaling and within a regulatory loop to coordinate epithelial proliferation and differentiation during normal epidermal and palatal development (Mills et al., 1999; Yang et al., 1999; Ferretti et al., 2011). Disruption of this pathway because of mutations in TP63 or IRF6 causes CL/P in mice and humans (Brunner et al., 2002; Moretti et al., 2010; Thomason et al., 2010) and mutations in P63 causes a variety of ectodermal dysplasias (Brunner et al., 2002). Although mice deficient for p63 exhibited impaired wound healing (Koster et al., 2007), two patients with ankyloblepharon ectodermal defects-cleft lip and palate syndrome (AEC) appeared to heal properly following surgical repair of their cleft (Cabiling et al., 2007).
Tfap2 is a direct target of p63 (Koster et al., 2006) and binds to an enhancer region of Irf6 containing a genetic variant associated with CL/P (Rahimov et al., 2008). Human mutations in TFAP2 causes branchio-oculo-facial syndrome (OMIM #113620, (Milunsky et al., 2008)), and Tfap2-deficient mice fail to close their cranial neural tube (Zhang et al., 1996). Epidermal-specific removal of Tfap2α and/or γ causes disruption of proper epidermal differentiation (Guttormsen et al., 2008; Wang et al., 2008; McDade et al., 2012), yet a role for this family of transcription factor in cutaneous repair remains to be discovered.
c-Myc is also a downstream target of the p63 gene (Wu et al., 2012). It regulates about 15% of all genes (Gearhart et al., 2007) and may be considered the universal regulator of cellular proliferation. Targeted expression of c-Myc in the epidermis alters keratinocyte proliferation and differentiation, and regulates epidermal homeostasis by depleting the stem cells from the interfollicular compartment (Waikel et al., 1999; Waikel et al., 2001; Frye et al., 2003). It has been found in keratinocytes of the leading edge of chronic wounds, where it inhibits epithelialization (Stojadinovic et al., 2005). c-Myc is activated by Tgfβ during growth of the palatal shelves, and neural crest cell deficiency of c-Myc causes skull frontal bone and middle ear ossicle developmental defects (Wei et al., 2007). Interestingly, c-Myc is physically the closest gene to the Chr. 8q24 cleft susceptibility locus, a 640 kb gene desert chromosomal location which was identified by genome-wide association studies in a population of European ancestry (Birnbaum et al., 2009; Beaty et al., 2010). A recent study in the mouse demonstrates that this interval contains long-range enhancers controlling c-Myc expression in the developing face (Uslu et al., 2014). Deletion of this interval or introduction of genetic variants associated with CL/P at this locus leads to alteration of facial morphology and low penetrance of CL/P in the mouse, supporting the hypothesis that the Chr. 8q24 susceptibility locus regulates c-Myc during craniofacial development.
Differences remain between the closure of a cutaneous wound and the embryogenesis of the palate. Do palatal epithelial cells display an “activated” phenotype identical to keratinocytes migrating during epithelialization? Are the signals sensed by keratinocytes touching each other at epithelial closure similar to signals sensed by epithelial cells from the palatal shelves touching each other and fusing? What drives palatal shelf movement – do cells “rise up” like a bridge, or treadmill or push like in collective cell migration? Answers to these questions will provide further insights into cutaneous wound healing and craniofacial development. Ultimately, as these two biological processes share common pathways and gene regulatory networks, a deeper understanding of one will inform the other to develop new therapeutic strategies.
Acknowledgments
We would like to thank the continuous support of Jeff Murray and Brian Schutte, and to Lisa Harney for establishing the list of clefting genes with members of the Murray Lab. We acknowledge the partial financial support of the National Institute of Health (AR061586 to M.D.). A special thank to the “palate wrecker” for inspirational moments.
Litterature cited
- Aberdam D. Derivation of keratinocyte progenitor cells and skin formation from embryonic stem cells. Int J Dev Biol. 2004;48:203–206. doi: 10.1387/ijdb.15272386. [DOI] [PubMed] [Google Scholar]
- Adams S, Pankow S, Werner S, Munz B. Regulation of NF-kappaB activity and keratinocyte differentiation by the RIP4 protein: implications for cutaneous wound repair. J Invest Dermatol. 2007;127:538–544. doi: 10.1038/sj.jid.5700588. [DOI] [PubMed] [Google Scholar]
- Akiyama M, Smith LT, Yoneda K, Holbrook KA, Hohl D, Shimizu H. Periderm cells form cornified cell envelope in their regression process during human epidermal development. J Invest Dermatol. 1999;112:903–909. doi: 10.1046/j.1523-1747.1999.00592.x. [DOI] [PubMed] [Google Scholar]
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Aller MA, Blanco-Rivero J, Arias JI, Balfagon G, Arias J. The wound-healing response and upregulated embryonic mechanisms: brothers-in-arms forever. Exp Dermatol. 2012;21:497–503. doi: 10.1111/j.1600-0625.2012.01525.x. [DOI] [PubMed] [Google Scholar]
- Amini-Nik S, Cambridge E, Yu W, Guo A, Whetstone H, Nadesan P, Poon R, Hinz B, Alman BA. beta-Catenin-regulated myeloid cell adhesion and migration determine wound healing. J Clin Invest. 2014;124:2599–2610. doi: 10.1172/JCI62059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini-Nik S, Glancy D, Boimer C, Whetstone H, Keller C, Alman BA. Pax7 expressing cells contribute to dermal wound repair, regulating scar size through a beta-catenin mediated process. Stem Cells. 2011;29:1371–1379. doi: 10.1002/stem.688. [DOI] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Auden A, Caddy J, Wilanowski T, Ting SB, Cunningham JM, Jane SM. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Expr Patterns. 2006;6:964–970. doi: 10.1016/j.modgep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Bamberger C, Hafner A, Schamale H, Werner S. Expression of different p63 variants in healing skin wounds suggests a role of p63 in reepithelialization and muscle repair. Wound Rep. Reg. 2005;13:41–50. doi: 10.1111/j.1067-1927.2005.130106.x. [DOI] [PubMed] [Google Scholar]
- Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM. Transcriptional profiling of developing mouse epidermis reveals novel patterns of coordinated gene expression. Dev Dyn. 2007;236:961–970. doi: 10.1002/dvdy.21099. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–531. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, Mangold E, Ludwig KU, Noethen MM, Rubini M, Elcioglu N, Ruczinski I. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet. 2013;132:771–781. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia H, Khojasteh A, Soleimani M, Tehranchi A, Atashi A. Repair of alveolar cleft defect with mesenchymal stem cells and platelet derived growth factors: a preliminary report. J Craniomaxillofac Surg. 2012;40:2–7. doi: 10.1016/j.jcms.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickenbach JR, McCutecheon J, Mackenzie IC. Rate of loss of tritiated thymidine label in basal cells in mouse epithelial tissues. Cell. Tissue Kinet. 1986;19:323–333. doi: 10.1111/j.1365-2184.1986.tb00684.x. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Mikkola ML. Early inductive events in ectodermal appendage morphogenesis. Semin Cell Dev Biol. 2014;25–26:11–21. doi: 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Naridze R, Demali KA, Lusche DF, Kuhl S, Soll DR, Schutte BC, Dunnwald M. Interferon Regulatory Factor 6 regulates keratinocyte migration. J Cell Sci. 2014;127:2840–2848. doi: 10.1242/jcs.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, Mangold E. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto JA, Evans RD, Hayward RD, Jones BM. Toward pathogenesis of Apert cleft palate: FGF, FGFR, and TGF beta genes are differentially expressed in sequential stages of human palatal shelf fusion. Cleft Palate Craniofac J. 2002;39:332–340. doi: 10.1597/1545-1569_2002_039_0332_tpoacp_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Brunner HG, Hamel BC, van Bokhoven H. P63 gene mutations and human developmental syndromes. Am J Med Genet. 2002;112:284–290. doi: 10.1002/ajmg.10778. [DOI] [PubMed] [Google Scholar]
- Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Maltby KM, Cho ES, Jiang R. The T-box gene Tbx10 exhibits a uniquely restricted expression pattern during mouse embryogenesis. Gene Expr Patterns. 2003;3:533–538. doi: 10.1016/s1567-133x(03)00060-7. [DOI] [PubMed] [Google Scholar]
- Butali A, Suzuki S, Cooper ME, Mansilla AM, Cuenco K, Leslie EJ, Suzuki Y, Niimi T, Yamamoto M, Ayanga G, Erkhembaatar T, Furukawa H, Fujiwawa K, Imura H, Petrin AL, Natsume N, Beaty TH, Marazita ML, Murray JC. Replication of genome wide association identified candidate genes confirm the role of common and rare variants in PAX7 and VAX1 in the etiology of nonsyndromic CL(P) Am J Med Genet A. 2013;161A:965–972. doi: 10.1002/ajmg.a.35749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Cabiling DS, Yan AC, McDonald-McGinn DM, Zackai EH, Kirschner RE. Cleft lip and palate repair in Hay-Wells/ankyloblepharon-ectodermal dysplasia-clefting syndrome. Cleft Palate Craniofac J. 2007;44:335–339. doi: 10.1597/06-065. [DOI] [PubMed] [Google Scholar]
- Caddy J, Wilanowski T, Darido C, Dworkin S, Ting SB, Zhao Q, Rank G, Auden A, Srivastava S, Papenfuss TA, Murdoch JN, Humbert PO, Parekh V, Boulos N, Weber T, Zuo J, Cunningham JM, Jane SM. Epidermal wound repair is regulated by the planar cell polarity signaling pathway. Dev Cell. 2010;19:138–147. doi: 10.1016/j.devcel.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, Knight RA, Melino G. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles CA, Tomic-Canic M, Vincek V, Nassiri M, Stojadinovic O, Eaglstein WH, Kirsner RS. A gene signature of nonhealing venous ulcers: Potential diagnostic markers. J Am Acad Dermatol. 2008 doi: 10.1016/j.jaad.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guo S, Ranzer MJ, Dipietro LA. Toll-Like Receptor 4 Has an Essential Role in Early Skin Wound Healing. J Invest Dermatol. 2013;133:258–267. doi: 10.1038/jid.2012.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Schrementi ME, Ranzer MJ, Wilgus TA, DiPietro LA. Blockade of mast cell activation reduces cutaneous scar formation. PLoS One. 2014;9:e85226. doi: 10.1371/journal.pone.0085226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin M, Ng T, Tom WK, Carstens M. Repair of alveolar clefts with recombinant human bone morphogenetic protein (rhBMP-2) in patients with clefts. J Craniofac Surg. 2005;16:778–789. doi: 10.1097/01.scs.0000166802.49021.01. [DOI] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, Ma D, Stal S, Mulliken JB, Hecht JT. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet. 2008;17:2212–2218. doi: 10.1093/hmg/ddn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Lidral AC, Stal S, Mulliken JB, Moreno LM, Arcos-Burgos M, Valencia-Ramirez C, Blanton SH, Hecht JT. CRISPLD2: a novel NSCLP candidate gene. Hum Mol Genet. 2007;16:2241–2248. doi: 10.1093/hmg/ddm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SJ, Marazita ML, Hart PS, Sulima PP, Field LL, McHenry TG, Govil M, Cooper ME, Letra A, Menezes R, Narayanan S, Mansilla MA, Granjeiro JM, Vieira AR, Lidral AC, Murray JC, Hart TC. The PDGF-C regulatory region SNP rs28999109 decreases promoter transcriptional activity and is associated with CL/P. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choma DP, Pumiglia K, DiPersio CM. Integrin alpha3beta1 directs the stabilization of a polarized lamellipodium in epithelial cells through activation of Rac1. J Cell Sci. 2004;117:3947–3959. doi: 10.1242/jcs.01251. [DOI] [PubMed] [Google Scholar]
- Christ B, Ordahl CP. Early stages of chick somite development. Anat Embryol (Berl) 1995;191:381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- Chuong CM, Patel N, Lin J, Jung HS, Widelitz RB. Sonic hedgehog signaling pathway in vertebrate epithelial appendage morphogenesis: perspectives in development and evolution. Cell Mol Life Sci. 2000;57:1672–1681. doi: 10.1007/PL00000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SR, Kalinowski J, LaRossa D, Randall P. Cleft palate fistulas: a multivariate statistical analysis of prevalence, etiology, and surgical management. Plast Reconstr Surg. 1991;87:1041–1047. [PubMed] [Google Scholar]
- Cooper L, Johnson C, Burslem F, Martin P. Wound healing and inflammation genes revealed by array analysis of 'macrophageless' PU.1 null mice. Genome Biol. 2005;6:R5. doi: 10.1186/gb-2004-6-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe PA. Wound epithelialization: accelerating the pace of discovery. Progress in Dermatology. 2003;37:1–12. doi: 10.1046/j.1523-1747.2003.12387.x. [DOI] [PubMed] [Google Scholar]
- Dale BA, Holbrook KA, Kimball JR, Hoff M, Sun T-T. Expression of epidermal keratins and filaggrin during human fetal skin development. J Cell Biol. 1985;101:1257–1269. doi: 10.1083/jcb.101.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathan N, Parlato R, Rosica A, De Felice M, Di Lauro R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev Dyn. 2002;224:450–456. doi: 10.1002/dvdy.10118. [DOI] [PubMed] [Google Scholar]
- de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, Houston DW, Manak JR, Schutte BC, Wagner DS, Cornell RA. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J Invest Dermatol. 2013;133:68–77. doi: 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima RL, Hoper SA, Ghassibe M, Cooper ME, Rorick NK, Kondo S, Katz L, Marazita ML, Compton J, Bale S, Hehr U, Dixon MJ, Daack-Hirsch S, Boute O, Bayet B, Revencu N, Verellen-Dumoulin C, Vikkula M, Richieri-Costa A, Moretti-Ferreira D, Murray JC, Schutte BC. Prevalence and nonrandom distribution of exonic mutations in interferon regulatory factor 6 in 307 families with Van der Woude syndrome and 37 families with popliteal pterygium syndrome. Genet Med. 2009;11:241–247. doi: 10.1097/GIM.0b013e318197a49a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning MF, Dlugosz AA, Williams EK, Szallasi Z, Blumberg PM, Yuspa SH. Specific protein kinase C isoenzymes mediate the induction of keratinocyte differentiation markers by calcium. Cell Growth & Differentiation. 1995;6:149–157. [PubMed] [Google Scholar]
- Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, Norwood C, Wang E, Marincola FM, Stroncek DF. Gene expression profiling of cutaneous wound healing. J Transl Med. 2007;5:11. doi: 10.1186/1479-5876-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D, Olivera-Martinez I, Fliniaux I, Missier S, Viallet JP, Thelu J. Skin field formation: morphogenetic events. Int J Dev Biol. 2004;48:85–91. [PubMed] [Google Scholar]
- Ding H, Wu X, Bostrom H, Kim I, Wong N, Tsoi B, O'Rourke M, Koh GY, Soriano P, Betsholtz C, Hart TC, Marazita ML, Field LL, Tam PP, Nagy A. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat Genet. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Kim I, Tam PP, Koh GY, Nagy A. The mouse Pdgfc gene: dynamic expression in embryonic tissues during organogenesis. Mech Dev. 2000;96:209–213. doi: 10.1016/s0925-4773(00)00425-1. [DOI] [PubMed] [Google Scholar]
- Dovi JV, He LK, DiPietro LA. Accelerated wound closure in neutrophil-depleted mice. J Leukoc Biol. 2003;73:448–455. doi: 10.1189/jlb.0802406. [DOI] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, Watt FM. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas M, Nagy A, Laping NJ, Moustakas A, Kaartinen V. Tgf-beta3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev Biol. 2004;266:96–108. doi: 10.1016/j.ydbio.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Eichberger T, Regl G, Ikram MS, Neill GW, Philpott MP, Aberger F, Frischauf AM. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J Invest Dermatol. 2004;122:1180–1187. doi: 10.1111/j.0022-202X.2004.22505.x. [DOI] [PubMed] [Google Scholar]
- Elliott CG, Wang J, Guo X, Xu SW, Eastwood M, Guan J, Leask A, Conway SJ, Hamilton DW. Periostin modulates myofibroblast differentiation during full-thickness cutaneous wound repair. J Cell Sci. 2012;125:121–132. doi: 10.1242/jcs.087841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Koch M, Krieger A, Brachvogel B, Kreft S, Bruckner-Tuderman L, Krieg T, Shannon JD, Fox JW. Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from normal healing and chronic wounds. J Proteome Res. 2010;9:4758–4766. doi: 10.1021/pr100456d. [DOI] [PubMed] [Google Scholar]
- Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. 2007;127:514–525. doi: 10.1038/sj.jid.5700701. [DOI] [PubMed] [Google Scholar]
- Endo T, Tamura K, Ide H. Analysis of gene expressions during Xenopus forelimb regeneration. Dev Biol. 2000;220:296–306. doi: 10.1006/dbio.2000.9641. [DOI] [PubMed] [Google Scholar]
- Eslami A, Gallant-Behm CL, Hart DA, Wiebe C, Honardoust D, Gardner H, Hakkinen L, Larjava HS. Expression of integrin alphavbeta6 and TGF-beta in scarless vs scar-forming wound healing. J Histochem Cytochem. 2009;57:543–557. doi: 10.1369/jhc.2009.952572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri WD, Rhea L, Du T, Sweezer E, Morrison H, Fitzpatrick D, Yang B, Dunnwald M, Schutte BC. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev Dyn. 2012;241:340–349. doi: 10.1002/dvdy.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathke C, Wilson L, Shah K, Kim B, Hocking A, Moon R, Isik F. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol. 2006;7:4. doi: 10.1186/1471-2121-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C, Miclau T, Hu D, Alpern E, Helms JA. Common molecular pathways in skeletal morphogenesis and repair. Ann NY Acad Sci. 1998;857:33–42. doi: 10.1111/j.1749-6632.1998.tb10105.x. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, Duncan J, Bond J, Bush J, Durani P, So K, Taylor L, Chantrey J, Mason T, James G, Laverty H, Occleston NL, Sattar A, Ludlow A, O'Kane S. Prophylactic administration of avotermin for improvement of skin scarring: three double-blind, placebo-controlled, phase I/II studies. Lancet. 2009;373:1264–1274. doi: 10.1016/S0140-6736(09)60322-6. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, O'Kane S. Scar-free healing: from embryonic mechanisms to adult therapeutic intervention. Philos Trans R Soc London B Biol Sci. 2004;359:839–850. doi: 10.1098/rstb.2004.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, Anderson MJ, Williams T, Dixon J, Dixon MJ, Depew MJ, Selleri L. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21:627–641. doi: 10.1016/j.devcel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, Markham AF, Fantes JA, Bonthron DT. Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum Mol Genet. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick DR, Denhez F, Kondaiah P, Akhurst RJ. Differential expression of TGF beta isoforms in murine palatogenesis. Development. 1990;109:585–595. doi: 10.1242/dev.109.3.585. [DOI] [PubMed] [Google Scholar]
- Fliniaux I, Viallet JP, Dhouailly D. Ventral vs. dorsal chick dermal progenitor specification. Int J Dev Biol. 2004;48:103–106. [PubMed] [Google Scholar]
- Fogh-Andersen P. Inheritance of Harelip and Cleft Palate. Copenhagen: Munksgaard; 1942. [Google Scholar]
- Francis-West PH, Tatla T, Brickell PM. Expression patterns of the bone morphogenetic protein genes Bmp-4 and Bmp-2 in the developing chick face suggest a role in outgrowth of the primordia. Dev Dyn. 1994;201:168–178. doi: 10.1002/aja.1002010207. [DOI] [PubMed] [Google Scholar]
- Frye M, Gardner C, Li ER, Arnold I, Watt FM. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Inthal A, Herrmann D, Cheng S, Nakatomi M, Peters H, Neubuser A. Regulation of Tbx22 during facial and palatal development. Dev Dyn. 2010;239:2860–2874. doi: 10.1002/dvdy.22421. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19:1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- Gearhart J, Pashos EE, Prasad MK. Pluripotency redux--advances in stem-cell research. N Engl J Med. 2007;357:1469–1472. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
- Gilbertson DG, Duff ME, West JW, Kelly JD, Sheppard PO, Hofstrand PD, Gao Z, Shoemaker K, Bukowski TR, Moore M, Feldhaus AL, Humes JM, Palmer TE, Hart CE. Platelet-derived growth factor C (PDGF-C), a novel growth factor that binds to PDGF alpha and beta receptor. J Biol Chem. 2001;276:27406–27414. doi: 10.1074/jbc.M101056200. [DOI] [PubMed] [Google Scholar]
- Goren I, Allmann N, Yogev N, Schurmann C, Linke A, Holdener M, Waisman A, Pfeilschifter J, Frank S. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am J Pathol. 2009;175:132–147. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosman AA. Cleft lip and palate II: Surgical management. Selected Reading Plastic Surgery. 2007;10:1–93. [Google Scholar]
- Green H, Easley K, Iuchi S. Marker succession during the development of keratinocytes from cultured human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2003;100:15625–15630. doi: 10.1073/pnas.0307226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschl M, Topf HG, Kratzsch J, Dotsch J, Rascher W, Rauh M. Salivary leptin induces increased expression of growth factors in oral keratinocytes. J Mol Endocrinol. 2005;34:353–366. doi: 10.1677/jme.1.01658. [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Guttormsen J, Koster MI, Stevens JR, Roop DR, Williams T, Winger QA. Disruption of epidermal specific gene expression and delayed skin development in AP-2 gamma mutant mice. Dev Biol. 2008;317:187–195. doi: 10.1016/j.ydbio.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford MM, Armes J, Buchert M, Meskenaite V, Grail D, Hibbs ML, Wilks AF, Farlie PG, Newgreen DF, Hovens CM, Stacker SA. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat Genet. 2000;25:414–418. doi: 10.1038/78099. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sis P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Haslett C. Resolution of acute inflammation and the role of apoptosis in the tissue fate of granulocytes. Clin Sci (Lond) 1992;83:639–648. doi: 10.1042/cs0830639. [DOI] [PubMed] [Google Scholar]
- Hattori H, Subramanian KK, Sakai J, Jia Y, Li Y, Porter TF, Loison F, Sarraj B, Kasorn A, Jo H, Blanchard C, Zirkle D, McDonald D, Pai SY, Serhan CN, Luo HR. Small-molecule screen identifies reactive oxygen species as key regulators of neutrophil chemotaxis. Proc Natl Acad Sci U S A. 2010;107:3546–3551. doi: 10.1073/pnas.0914351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Chen Y. Wnt signaling in lip and palate development. Front Oral Biol. 2012;16:81–90. doi: 10.1159/000337619. [DOI] [PubMed] [Google Scholar]
- He F, Xiong W, Yu X, Espinoza-Lewis R, Liu C, Gu S, Nishita M, Suzuki K, Yamada G, Minami Y, Chen Y. Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development. 2008;135:3871–3879. doi: 10.1242/dev.025767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton D. Vertebrate embryonic cells will become nerve cells unless told otherwise. Cell. 1997;88:13–17. doi: 10.1016/s0092-8674(00)81853-x. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Liddell RA, Parker A, Grant S, Kinne J, Fisher JK, Siracusa LD. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat Genet. 2005;37:1210–1212. doi: 10.1038/ng1652. [DOI] [PubMed] [Google Scholar]
- Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127:526–537. doi: 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- Holbrook KA. Structure and function of the developing human skin. In: LA Goldsmith., editor. Physiology, biochemistry, and molecular biology of the skin. New-York: Oxford University Press; 1991. pp. 63–110. [Google Scholar]
- Holland P, Willis C, Kanaly S, Glaccum M, Warren A, Charrier K, Murison J, Derry J, Virca G, Bird T, Peschon J. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr Biol. 2002;12:1424–1428. doi: 10.1016/s0960-9822(02)01075-8. [DOI] [PubMed] [Google Scholar]
- Hopkinson-Woolley J, Hughes D, Gordon S, Martin P. Macrophage recruitment during limb development and wound healing in the embryonic and foetal mouse. J Cell Sci. 1994;107(Pt 5):1159–1167. doi: 10.1242/jcs.107.5.1159. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Cheraud Y, Auda-Boucher G, Fontaine-Perus J, Robert B. The expression of the homeobox gene Msx1 reveals two populations of dermal progenitor cells originating from the somites. Development. 2000;127:2155–2164. doi: 10.1242/dev.127.10.2155. [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Cohen A, Buckingham ME, Robert B. Insertional mutation of the mouse Msx1 homeobox gene by an nlacZ reporter gene. Mech Dev. 1997;65:123–133. doi: 10.1016/s0925-4773(97)00065-8. [DOI] [PubMed] [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- Hurwitz DJ, Kathju S. Pediatric plastic surgery of the head and neck. In: Bluestone CD, Stool SE, Alper CM, Arjmand EM, Casselbrant ML, Dohar JE, Yellon RF, editors. Pediatric Otolaryngology. Elsevier Science; 2003. pp. 950–977. [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jackson B, Peyrollier K, Pedersen E, Basse A, Karlsson R, Wang Z, Lefever T, Ochsenbein AM, Schmidt G, Aktories K, Stanley A, Quondamatteo F, Ladwein M, Rottner K, van Hengel J, Brakebusch C. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. Mol Biol Cell. 2011;22:593–605. doi: 10.1091/mbc.E09-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, Romitti PA, Christensen K, Orioli IM, Castilla EE, Machida J, Natsume N, Murray JC. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of Meox-2 mutant mice reveals a novel postfusion-based cleft palate. Dev Dyn. 2006;235:539–546. doi: 10.1002/dvdy.20641. [DOI] [PubMed] [Google Scholar]
- Jones JL, Canady JW, Brookes JT, Wehby GL, L'Heureux J, Schutte BC, Murray JC, Dunnwald M. Wound complications after cleft repair in children with Van der Woude syndrome. J Craniofac Surg. 2010;21:1350–1353. doi: 10.1097/SCS.0b013e3181ec6aad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, Christensen K, Boyles AL, Daack-Hirsch S, Trung TN, Bille C, Lidral AC, Murray JC. Genetic determinants of facial clefting: analysis of 357 candidate genes using two national cleft studies from Scandinavia. PLoS ONE. 2009;4:e5385. doi: 10.1371/journal.pone.0005385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Haataja L, Nagy A, Heisterkamp N, Groffen J. TGFbeta3-induced activation of RhoA/Rho-kinase pathway is necessary but not sufficient for epithelio-mesenchymal transdifferentiation: implications for palatogenesis. Int J Mol Med. 2002;9:563–570. [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-b3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Kagami H, Hiramatsu Y, Hishida S, Okazaki Y, Horie K, Oda Y, Ueda M. Salivary growth factors in health and disease. Adv Dent Res. 2000;14:99–102. doi: 10.1177/08959374000140011601. [DOI] [PubMed] [Google Scholar]
- Kalay E, Sezgin O, Chellappa V, Mutlu M, Morsy H, Kayserili H, Kreiger E, Cansu A, Toraman B, Abdalla EM, Aslan Y, Pillai S, Akarsu NA. Mutations in RIPK4 cause the autosomal-recessive form of popliteal pterygium syndrome. Am J Hum Genet. 2012;90:76–85. doi: 10.1016/j.ajhg.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardash E, Reichman-Fried M, Maitre JL, Boldajipour B, Papusheva E, Messerschmidt EM, Heisenberg CP, Raz E. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat Cell Biol. 2010;12(suppp 41-11):47–53. doi: 10.1038/ncb2003. [DOI] [PubMed] [Google Scholar]
- Kasper M, Jaks V, Are A, Bergstrom A, Schwager A, Barker N, Toftgard R. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc Natl Acad Sci U S A. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswani SG, Balaji S, Le LD, Leung A, Parvadia JK, Frischer J, Yamano S, Taichman N, Crombleholme TM. Role of salivary vascular endothelial growth factor (VEGF) in palatal mucosal wound healing. Wound Repair Regen. 2013;21:554–562. doi: 10.1111/wrr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, Bhasker V, Gordillo GM, Sen CK, Roy S. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLoS One. 2010;5:e9539. doi: 10.1371/journal.pone.0009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischer CW, Bunce H, 3rd, Shetlah MR. Mast cell analyses in hypertrophic scars, hypertrophic scars treated with pressure and mature scars. J Invest Dermatol. 1978;70:355–357. doi: 10.1111/1523-1747.ep12543553. [DOI] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jian R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Komi-Kuramochi A, Kawano M, Oda Y, Asada M, Suzuki M, Oki J, Imamura T. Expression of fibroblast growth factors and their receptors during full-thickness skin wound healing in young and aged mice. J Endocrinol. 2005;186:273–289. doi: 10.1677/joe.1.06055. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, Ferreira de Lima RLL, Daack-Hirsch S, Sander A, McDonald-McGinn D, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, Roop DR. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Huang J, Williams T, Roop DR. TAp63alpha induces AP-2gamma as an early event in epidermal morphogenesis. Dev Biol. 2006;289:253–261. doi: 10.1016/j.ydbio.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes & Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Krawczyk WS. A pattern of epidermal cell migration during wound healing. J Cell Biol. 1971;49:247–263. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechle MK, Predd HM, Fleckman P, Dale BA, Presland RB. Caspase-14, a keratinocyte specific caspase: mRNA splice variants and expression pattern in embryonic and adult mouse. Cell Death Differ. 2001;8:868–870. doi: 10.1038/sj.cdd.4400897. [DOI] [PubMed] [Google Scholar]
- Kurosaka H, Iulianella A, Williams T, Trainor PA. Disrupting hedgehog and WNT signaling interactions promotes cleft lip pathogenesis. J Clin Invest. 2014;124:1660–1671. doi: 10.1172/JCI72688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larjava H, Wiebe C, Gallant-Behm C, Hart DA, Heino J, Hakkinen L. Exploring scarless healing of oral soft tissues. J Can Dent Assoc. 2011;77:b18. [PubMed] [Google Scholar]
- Lau K, Paus R, Tiede S, Day P, Bayat A. Exploring the role of stem cells in cutaneous wound healing. Exp Dermatol. 2009;18:921–933. doi: 10.1111/j.1600-0625.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- Le H, Kleinerman R, Lerman OZ, Brown D, Galiano R, Gurtner GC, Warren SM, Levine JP, Saadeh PB. Hedgehog signaling is essential for normal wound healing. Wound Repair Regen. 2008;16:768–773. doi: 10.1111/j.1524-475X.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- Le M, Naridze R, Morrisson J, Biggs LC, Rhea L, Schutte BC, Kaartinen V, Dunnwald M. Transforming growth factor beta 3 is required for proper excisional wound repair in vivo. PLoS ONE. 2012;7:e48040. doi: 10.1371/journal.pone.0048040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, Dunnwald M, Lidral AC, Marazita ML, Beaty TH, Murray JC. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 2012;94:934–942. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson SM, Geever EF, Crowley LV, Oates JF, 3rd, Berard CW, Rosen H. The Healing of Rat Skin Wounds. Ann Surg. 1965;161:293–308. doi: 10.1097/00000658-196502000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Moses HL, Gold LI, Nanney LB. Spatial and temporal patterns of immunoreactive transforming growth factor beta1, beta2 and beta3 during excisional wound repair. Am J Pathol. 1993;143:368–380. [PMC free article] [PubMed] [Google Scholar]
- Li J, Farthing PM, Ireland GW, Thornhill MH. IL-1 alpha and IL-6 production by oral and skin keratinocytes: similarities and differences in response to cytokine treatment in vitro. J Oral Pathol Med. 1996;25:157–162. doi: 10.1111/j.1600-0714.1996.tb00213.x. [DOI] [PubMed] [Google Scholar]
- Li L, Lin M, Wang Y, Cserjesi P, Chen Z, Chen Y. BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev Biol. 2011;349:451–461. doi: 10.1016/j.ydbio.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Estepa G, Verma IM. Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc Natl Acad Sci U S A. 2005;102:15977–15982. doi: 10.1073/pnas.0508310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee K-F, Izpisua-Belmonte JC, Verma IM. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes & Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, Semina EV, Johnson LR, Machida J, Burds A, Parnell TJ, Rubenstein JL, Murray JC. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X, Nusse R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WL, Chau RM, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun X, Braut A, Mishina Y, Behringer RR, Mina M, Martin JF. Distinct functions for Bmp signaling in lip and palate fusion in mice. Development. 2005;132:1453–1461. doi: 10.1242/dev.01676. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang E, Li P, Liu J, Zhou P, Gu DY, Chen X, Cheng T, Zhou Y. Expression and possible mechanism of c-ski, a novel tissue repair-related gene during normal and radiation-impaired wound healing. Wound Repair Regen. 2006;14:162–171. doi: 10.1111/j.1743-6109.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- Lu P, Barad M, Vize PD. Xenopus p63 expression in early ectoderm and neurectoderm. Mech Dev. 2001;102:275–278. doi: 10.1016/s0925-4773(01)00315-x. [DOI] [PubMed] [Google Scholar]
- M'Boneko V, Merker HJ. Development and morphology of the periderm of mouse embryos (days 9–12 of gestation) Acta Anat (Basel) 1988;133:325–336. doi: 10.1159/000146662. [DOI] [PubMed] [Google Scholar]
- Ma L, Liu J, Wu T, Plikus M, Jiang TX, Bi Q, Liu YH, Muller-Rover S, Peters H, Sundberg JP, Maxson R, Maas RL, Chuong CM. 'Cyclic alopecia' in Msx2 mutants: defects in hair cycling and hair shaft differentiation. Development. 2003;130:379–389. doi: 10.1242/dev.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace KA, Pearson JC, McGinnis W. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science. 2005;308:381–385. doi: 10.1126/science.1107573. [DOI] [PubMed] [Google Scholar]
- Madison KC. Barrier function of the skin: Ȣla raison d'etre" of the epidermis. J. Invest. Dermatol. 2003;121:231–241. doi: 10.1046/j.1523-1747.2003.12359.x. [DOI] [PubMed] [Google Scholar]
- Mak K, Manji A, Gallant-Behm C, Wiebe C, Hart DA, Larjava H, Hakkinen L. Scarless healing of oral mucosa is characterized by faster resolution of inflammation and control of myofibroblast action compared to skin wounds in the red Duroc pig model. J Dermatol Sci. 2009;56:168–180. doi: 10.1016/j.jdermsci.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, Steffens M, Barth S, Kluck N, Paul A, Becker J, Lauster C, Schmidt G, Braumann B, Scheer M, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Moebus S, Krawczak M, Schreiber S, Meitinger T, Wichmann HE, Steegers-Theunissen RP, Kramer FJ, Cichon S, Propping P, Wienker TF, Knapp M, Rubini M, Mossey PA, Hoffmann P, Nothen MM. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Mannik J, Alzayady K, Ghazizadeh S. Regeneration of multilineage skin epithelia by differentiated keratinocytes. J Invest Dermatol. 2010;130:388–397. doi: 10.1038/jid.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla MA, Cooper ME, Goldstein T, Castilla EE, Lopez Camelo JS, Marazita ML, Murray JC. Contributions of PTCH gene variants to isolated cleft lip and palate. Cleft Palate Craniofac J. 2006;43:21–29. doi: 10.1597/04-169R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita M. Segregation analysis. In: Wyszynski DF, editor. Cleft lip and palate: from origin to treatment. Oxford University Press; 2002. pp. 222–233. [Google Scholar]
- Marazita ML. Subclinical features in non-syndromic cleft lip with or without cleft palate (CL/P): review of the evidence that subepithelial orbicularis oris muscle defects are part of an expanded phenotype for CL/P. Orthod Craniofac Res. 2007;10:82–87. doi: 10.1111/j.1601-6343.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- Marinkovich MP, Keene DR, Rimberg CS, Burgeson RE. Cellular origin of the dermal-epidermal basement membrane. Developmental dynamics. 1993;197:255–267. doi: 10.1002/aja.1001970404. [DOI] [PubMed] [Google Scholar]
- Martin P. Wound healing-Aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- Martin P, D'Souza D, Martin J, Grose R, Cooper L, Maki R, McKercher SR. Wound healing in the PU.1 null mouse--tissue repair is not dependent on inflammatory cells. Curr Biol. 2003;13:1122–1128. doi: 10.1016/s0960-9822(03)00396-8. [DOI] [PubMed] [Google Scholar]
- Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. 2005;15:599–607. doi: 10.1016/j.tcb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Martin P, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80:1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Razzell W, Martin P. 'White wave' analysis of epithelial scratch wound healing reveals how cells mobilise back from the leading edge in a myosin-II-dependent fashion. J Cell Sci. 2011;124:1017–1021. doi: 10.1242/jcs.080853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Hayashi-Kisumi F, Kinoshita Y, Katahira S, Morita K, Miyachi Y, Ono Y, Imai T, Tanigawa Y, Komiya T, Tsukita S. Identification of novel keratinocyte-secreted peptides dermokine-alpha/-beta and a new stratified epithelium-secreted protein gene complex on human chromosome 19q13.1. Genomics. 2004;84:384–397. doi: 10.1016/j.ygeno.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Taketomi T, Yoshizaki K, Arai S, Sanui T, Yoshiga D, Yoshimura A, Nakamura S. Sprouty2 controls proliferation of palate mesenchymal cells via fibroblast growth factor signaling. Biochem Biophys Res Commun. 2011;404:1076–1082. doi: 10.1016/j.bbrc.2010.12.116. [DOI] [PubMed] [Google Scholar]
- Mauger A. [The role of somitic mesoderm in the development of dorsal plumage in chick embryos. I. Origin, regulative capacity and determination of the plumage-forming mesoderm] J Embryol Exp Morphol. 1972;28:313–341. [PubMed] [Google Scholar]
- Mazzalupo S, Coulombe PA. A reporter transgene based on a human keratin 6 gene promoter is specifically expressed in the periderm of mouse embryos. Mech Dev. 2001;100:65–69. doi: 10.1016/s0925-4773(00)00489-5. [DOI] [PubMed] [Google Scholar]
- McDade SS, Henry AE, Pivato GP, Kozarewa I, Mitsopoulos C, Fenwick K, Assiotis I, Hakas J, Zvelebil M, Orr N, Lord CJ, Patel D, Ashworth A, McCance DJ. Genome-wide analysis of p63 binding sites identifies AP-2 factors as co-regulators of epidermal differentiation. Nucleic Acids Res. 2012;40:7190–7206. doi: 10.1093/nar/gks389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CC, Beck PL, Muruve DA, Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- McGowan KM, Coulombe PA. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J Cell Biol. 1998;143:469–486. doi: 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, Duijf PH, Doetsch V, Irvine AD, de Waal R, Vanmolkot KR, Wessagowit V, Kelly A, Atherton DJ, Griffiths WA, Orlow SJ, van Haeringen A, Ausems MG, Yang A, McKeon F, Bamshad MA, Brunner HG, Hamel BC, van Bokhoven H. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- Metzis V, Courtney AD, Kerr MC, Ferguson C, Rondon Galeano MC, Parton RG, Wainwright BJ, Wicking C. Patched1 is required in neural crest cells for the prevention of orofacial clefts. Hum Mol Genet. 2013;22:5026–5035. doi: 10.1093/hmg/ddt353. [DOI] [PubMed] [Google Scholar]
- Miao M, Niu Y, Xie T, Yuan B, Qing C, Lu S. Diabetes-impaired wound healing and altered macrophage activation: a possible pathophysiologic correlation. Wound Repair Regen. 2012;20:203–213. doi: 10.1111/j.1524-475X.2012.00772.x. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, Burch MN, Clemens M, Mulliken JB, Smith R, Lin AE. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza R, DiPietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am J Pathol. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell K, O'Sullivan J, Missero C, Blair E, Richardson R, Anderson B, Antonini D, Murray JC, Shanske AL, Schutte BC, Romano RA, Sinha S, Bhaskar SS, Black GC, Dixon J, Dixon MJ. Exome sequence identifies RIPK4 as the Bartsocas-Papas syndrome locus. Am J Hum Genet. 2012;90:69–75. doi: 10.1016/j.ajhg.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE, Beaty TH, Lidral AC, Munger RG, Murray JC, Saal HM, Wyszynski DF. Guidelines for the design and analysis of studies on nonsyndromic cleft lip and cleft palate in humans: summary report from a Workshop of the International Consortium for Oral Clefts Genetics. Cleft Palate Craniofac J. 2002;39:93–100. doi: 10.1597/1545-1569_2002_039_0093_gftdaa_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Rahiotis C. Parallels between tooth development and repair: conserved molecular mechanisms following carious and dental injury. J Dent Res. 2004;83:896–902. doi: 10.1177/154405910408301202. [DOI] [PubMed] [Google Scholar]
- Miyai M, Tanaka YG, Kamitani A, Hamada M, Takahashi S, Kataoka K. c-Maf and MafB transcription factors are differentially expressed in Huxley's and Henle's layers of the inner root sheath of the hair follicle and regulate cuticle formation. J Dermatol Sci. 2010;57:178–182. doi: 10.1016/j.jdermsci.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Mo R, Freer AM, Zinyk DL, Crackower MA, Michaud J, Heng HH, Chik KW, Shi XM, Tsui LC, Cheng SH, Joyner AL, Hui C. Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development. 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- Moore MD, Lawrence WT, Ptak JJ, Trier WC. Complications of primary palatoplasty: a twenty-one-year review. Cleft Palate J. 1988;25:156–162. [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, Ballaro C, Alema S, Chimenti S, Guerrini L, Costanzo A. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Orkin RW, Noramly S, Perez A. Stage-specific effects on sonic hedgehog expression in the epidermis. Dev. Biol. 1998;201:1–12. doi: 10.1006/dbio.1998.8969. [DOI] [PubMed] [Google Scholar]
- Moser M, Ruschoff J, Buettner R. Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Dev Dyn. 1997;208:115–124. doi: 10.1002/(SICI)1097-0177(199701)208:1<115::AID-AJA11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Mossey P. Dental education and CPD: are you being served? Br Dent J. 2002;(Suppl):3–4. [PubMed] [Google Scholar]
- Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- Muzaffar AR, Byrd HS, Rohrich RJ, Johns DF, LeBlanc D, Beran SJ, Anderson C, Papaioannou a AA. Incidence of cleft palate fistula: an institutional experience with two-stage palatal repair. Plast Reconstr Surg. 2001;108:1515–1518. doi: 10.1097/00006534-200111000-00011. [DOI] [PubMed] [Google Scholar]
- Nguyen BP, Gil SG, Carter WG. Deposition of laminin 5 by keratinocytes regulates integrin adhesion and signaling. J Biol Chem. 2000;275:31896–31907. doi: 10.1074/jbc.M006379200. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikopensius T, Kempa I, Ambrozaityte L, Jagomagi T, Saag M, Matuleviciene A, Utkus A, Krjutskov K, Tammekivi V, Piekuse L, Akota I, Barkane B, Krumina A, Klovins J, Lace B, Kucinskas V, Metspalu A. Variation in FGF1, FOXE1, and TIMP2 genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2011;91:218–225. doi: 10.1002/bdra.20791. [DOI] [PubMed] [Google Scholar]
- Noguchi S, Ohba Y, Oka T. Effect of salivary epidermal growth factor on wound healing of tongue in mice. Am J Physiol. 1991;260:E620–625. doi: 10.1152/ajpendo.1991.260.4.E620. [DOI] [PubMed] [Google Scholar]
- Novak ML, Koh TJ. Macrophage phenotypes during tissue repair. J Leukoc Biol. 2013;93:875–881. doi: 10.1189/jlb.1012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuutila K, Siltanen A, Peura M, Bizik J, Kaartinen I, Kuokkanen H, Nieminen T, Harjula A, Aarnio P, Vuola J, Kankuri E. Human skin transcriptome during superficial cutaneous wound healing. Wound Repair Regen. 2012;20:830–839. doi: 10.1111/j.1524-475X.2012.00831.x. [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy RF, Welti JC, Sully K, Byrne C. Akt-dependent Pp2a activity is required for epidermal barrier formation during late embryonic development. Development. 2009;136:3423–3431. doi: 10.1242/dev.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odland GF. Structure of the skin. In: Goldsmitth LA, editor. Physiology, biochemistry, and molecular biology of the skin. sedcond ed. Oxford University Press NY; 1991. [Google Scholar]
- Ogata A, Shimizu T, Abe R, Shimizu H, Sakai M. Expression of c-maf and mafB genes in the skin during rat embryonic development. Acta Histochem. 2004;106:65–67. doi: 10.1016/j.acthis.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Ohtola J, Myers J, Akhtar-Zaidi B, Zuzindlak D, Sandesara P, Yeh K, Mackem S, Atit R. beta-Catenin has sequential roles in the survival and specification of ventral dermis. Development. 2008;135:2321–2329. doi: 10.1242/dev.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli IM, Vieira AR, Castilla EE, Ming JE, Muenke M. Mutational analysis of the Sonic Hedgehog gene in 220 newborns with oral clefts in a South American (ECLAMC) population. Am J Med Genet. 2002;108:12–15. doi: 10.1002/ajmg.10204. [DOI] [PubMed] [Google Scholar]
- Oudhoff MJ, van den Keijbus PA, Kroeze KL, Nazmi K, Gibbs S, Bolscher JG, Veerman EC. Histatins enhance wound closure with oral and non-oral cells. J Dent Res. 2009;88:846–850. doi: 10.1177/0022034509342951. [DOI] [PubMed] [Google Scholar]
- Paladini RD, Takahashi K, Bravo NS, Coulombe PA. Onset of re-organization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J Cell Biol. 1996;132:381–397. doi: 10.1083/jcb.132.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pase L, Layton JE, Wittmann C, Ellett F, Nowell CJ, Reyes-Aldasoro CC, Varma S, Rogers KL, Hall CJ, Keightley MC, Crosier PS, Grabher C, Heath JK, Renshaw SA, Lieschke GJ. Neutrophil-delivered myeloperoxidase dampens the hydrogen peroxide burst after tissue wounding in zebrafish. Curr Biol. 2012;22:1818–1824. doi: 10.1016/j.cub.2012.07.060. [DOI] [PubMed] [Google Scholar]
- Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, Martinez-Barbera JP, Moore GE, Stanier P. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 2009;18:4171–4179. doi: 10.1093/hmg/ddp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, Rautio J, Pegelow M, Karsten A, Basel-Vanagaite L, Gordon W, Andersen B, Svensson T, Murray JC, Cornell RA, Kere J, Schutte BC. Dominant Mutations in GRHL3 Cause Van der Woude Syndrome and Disrupt Oral Periderm Development. Am J Hum Genet. 2014;94:23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong CM. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD. Cholesterol precursors and facial clefting. J Clin Invest. 2006;116:2322–2325. doi: 10.1172/JCI29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukstad BS, Ryan L, Flo TH, Stenvik J, Moseley R, Harding K, Thomas DW, Espevik T. Non-healing is associated with persistent stimulation of the innate immune response in chronic venous leg ulcers. J Dermatol Sci. 2010;59:115–122. doi: 10.1016/j.jdermsci.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Pullar CE, Baier BS, Kariya Y, Russell AJ, Horst BA, Marinkovich MP, Isseroff RR. beta4 integrin and epidermal growth factor coordinately regulate electric field-mediated directional migration via Rac1. Mol Biol Cell. 2006;17:4925–4935. doi: 10.1091/mbc.E06-05-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice GP. The spreading of epithelial cells during wound closure in Xenopus larvae. Dev Biol. 1980;76:26–46. doi: 10.1016/0012-1606(80)90360-7. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell W, Evans IR, Martin P, Wood W. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr Biol. 2013;23:424–429. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell W, Wood W, Martin P. Recapitulation of morphogenetic cell shape changes enables wound re-epithelialisation. Development. 2014;141:1814–1820. doi: 10.1242/dev.107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Characterization of Wnt gene expression in developing and posnatal hair follicles and identification of Wnt 5a as a target of sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/s0925-4773(01)00452-x. [DOI] [PubMed] [Google Scholar]
- Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, Dixon MJ. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman M, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006 doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, Dixon MJ. Periderm prevents pathological epithelial adhesions during embryogenesis. J Clin Invest. 2014;124:3891–3900. doi: 10.1172/JCI71946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M, Marazita ML, Murray JC. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS ONE. 2009;4:e5623. doi: 10.1371/journal.pone.0005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothnagel JA, Longley MA, Bundman DS, Naylor SL, Lalley PA, Jenkins NA, Gilbert DJ, Copeland NG, Roop DR. Characterization of the mouse loricrin gene: linkage with profilaggrin and the flaky tail and soft coat mutant loci on chromosome 3. Genomics. 1994;23:450–456. doi: 10.1006/geno.1994.1522. [DOI] [PubMed] [Google Scholar]
- Rountree RB, Willis CR, Dinh H, Blumberg H, Bailey K, Dean C, Jr, Peschon JJ, Holland PM. RIP4 regulates epidermal differentiation and cutaneous inflammation. J Invest Dermatol. 2010;130:102–112. doi: 10.1038/jid.2009.223. [DOI] [PubMed] [Google Scholar]
- Roupe KM, Alberius P, Schmidtchen A, Sorensen OE. Gene expression demonstrates increased resilience toward harmful inflammatory stimuli in the proliferating epidermis of human skin wounds. Exp Dermatol. 2010;19:e329–332. doi: 10.1111/j.1600-0625.2009.01038.x. [DOI] [PubMed] [Google Scholar]
- Safferling K, Sutterlin T, Westphal K, Ernst C, Breuhahn K, James M, Jager D, Halama N, Grabe N. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. J Cell Biol. 2013;203:691–709. doi: 10.1083/jcb.201212020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Schafer M, Werner S. Transcriptional control of wound repair. Annu Rev Cell Dev Biol. 2007;23:69–92. doi: 10.1146/annurev.cellbio.23.090506.123609. [DOI] [PubMed] [Google Scholar]
- Schmidt BA, Horsley V. Intradermal adipocytes mediate fibroblast recruitment during skin wound healing. Development. 2013;140:1517–1527. doi: 10.1242/dev.087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Schrementi ME, Ferreira AM, Zender C, Dipietro LA. Site-specific production of TGF-beta in oral mucosal and cutaneous wounds. Wound Repair Regen. 2008;16:80–86. doi: 10.1111/j.1524-475X.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sen CK. The general case for redox control of wound repair. Wound Repair Regen. 2003;11:431–438. doi: 10.1046/j.1524-475x.2003.11607.x. [DOI] [PubMed] [Google Scholar]
- Sengel P. Pattern formation in skin development. Int J Dev Biol. 1990;34:33–50. [PubMed] [Google Scholar]
- Serfas MS, Tyner AL. Ryk is expressed in a differentiation-specific manner in epithelial tissues and is strongly induced in decidualizing uterine stroma. Oncogene. 1998;17:3435–3444. doi: 10.1038/sj.onc.1202266. [DOI] [PubMed] [Google Scholar]
- Shaw TJ, Martin P. Wound repair at a glance. J Cell Sci. 2009;122:3209–3213. doi: 10.1242/jcs.031187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AJ, Clark RAF. Cutaneous wound healing. N. Engl. J. Med. 1999;341:738–746. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- Smart IH. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br J Dermatol. 1970;82:276–282. doi: 10.1111/j.1365-2133.1970.tb12437.x. [DOI] [PubMed] [Google Scholar]
- Smith BJ, Nidey N, Miller SF, Moreno Uribe LM, Baum CL, Hamilton GS, 3rd, Wehby GL, Dunnwald M. Digital imaging analysis to assess scar phenotype. Wound Repair Regen. 2014;22:228–238. doi: 10.1111/wrr.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, Wang X, Zhang W, Kulyk W, Nazarali AJ. Hoxa2 plays a direct role in murine palate development. Dev Dyn. 2009;238:2364–2373. doi: 10.1002/dvdy.22040. [DOI] [PubMed] [Google Scholar]
- Song L, Li Y, Wang K, Wang YZ, Molotkov A, Gao L, Zhao T, Yamagami T, Wang Y, Gan Q, Pleasure DE, Zhou CJ. Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development. 2009;136:3161–3171. doi: 10.1242/dev.037440. [DOI] [PubMed] [Google Scholar]
- Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Sperber GH. Formation of the primary palate. In: Wysznyski D, editor. Cleft lip and palate: from origin to treatment. New York: Oxford University Press; 2002a. pp. 5–13. [Google Scholar]
- Sperber GH. Palatogenesis: closure of the secondary palate. In: Wysznyski D, editor. Cleft lip and palate: from origin to treatment. New York: Oxford University Press; 2002b. pp. 14–24. [Google Scholar]
- Stojadinovic O, Brem H, Vouthounis C, Lee B, Fallon J, Stallcup M, Merchant A, Galiano RD, Tomic-Canic M. Molecular pathogenesis of chronic wounds: the role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am J Pathol. 2005;167:59–69. doi: 10.1016/s0002-9440(10)62953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojadinovic O, Pastar I, Vukelic S, Mahoney MG, Brennan D, Krzyzanowska A, Golinko M, Brem H, Tomic-Canic M. Deregulation of keratinocyte differentiation and activation: a hallmark of venous ulcers. J Cell Mol Med. 2008;12:2675–2690. doi: 10.1111/j.1582-4934.2008.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Hu D, Bustos T, Zlotogora J, Richieri-Costa A, Helms JA, Spritz RA. Mutations of PVRL1, encoding a cell-cell adhesion molecule/herpesvirus receptor, in cleft lip/palate ectodermal dysplasia. Nat Genet. 2000;25:427–430. doi: 10.1038/78119. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Marazita ML, Cooper ME, Miwa N, Hing A, Jugessur A, Natsume N, Shimozato K, Ohbayashi N, Suzuki Y, Niimi T, Minami K, Yamamoto M, Altannamar TJ, Erkhembaatar T, Furukawa H, Daack-Hirsch S, L'Heureux J, Brandon CA, Weinberg SM, Neiswanger K, Deleyiannis FW, de Salamanca JE, Vieira AR, Lidral AC, Martin JF, Murray JC. Mutations in BMP4 are associated with subepithelial, microform, and overt cleft lip. Am J Hum Genet. 2009;84:406–411. doi: 10.1016/j.ajhg.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpaderska AM, Zuckerman JD, DiPietro LA. Differential injury responses in oral mucosal and cutaneous wounds. J Dent Res. 2003;82:621–626. doi: 10.1177/154405910308200810. [DOI] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKKa. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- Takenaka H, Yasuno H, Kishimoto S. Immunolocalization of fibroblast growth factor receptors in normal and wounded human skin. Arch Dermatol Res. 2002;294:331–338. doi: 10.1007/s00403-002-0333-z. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Dixon MJ, Dixon J. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev Biol. 2008;321:273–282. doi: 10.1016/j.ydbio.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 2010;120:1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Jarrell A, Zentner GE, Welsh A, Brownell I, Scacheri PC, Atit R. Role of canonical Wnt signaling/ss-catenin via Dermo1 in cranial dermal cell development. Development. 2010;137:3973–3984. doi: 10.1242/dev.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasler DG, Kemp D, Trasler TA. Increased susceptibility to 6-aminonicotinamide-induced cleft lip of heterozygote Dancer mice. Teratology. 1984;29:101–104. doi: 10.1002/tera.1420290112. [DOI] [PubMed] [Google Scholar]
- Uslu VV, Petretich M, Ruf S, Langenfeld K, Fonseca NA, Marioni JC, Spitz F. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat Genet. 2014 doi: 10.1038/ng.2971. [DOI] [PubMed] [Google Scholar]
- Usui ML, Mansbridge JN, Carter WG, Fujita M, Olerud JE. Keratinocyte migration, proliferation, and differentiation in chronic ulcers from patients with diabetes and normal wounds. J Histochem Cytochem. 2008;56:687–696. doi: 10.1369/jhc.2008.951194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui ML, Underwood RA, Mansbridge JN, Muffley LA, Carter WG, Olerud JE. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Rep. Reg. 2005;13:468–479. doi: 10.1111/j.1067-1927.2005.00067.x. [DOI] [PubMed] [Google Scholar]
- Valle-Rios R, Maravillas-Montero JL, Burkhardt AM, Martinez C, Buhren BA, Homey B, Gerber PA, Robinson O, Hevezi P, Zlotnik A. Isthmin 1 Is a Secreted Protein Expressed in Skin, Mucosal Tissues, and NK, NKT, and Th17 Cells. J Interferon Cytokine Res. 2014 doi: 10.1089/jir.2013.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Exan RJ, Hardy MH. The differentiation of the dermis in the laboratory mouse. Am J Anat. 1984;169:149–164. doi: 10.1002/aja.1001690204. [DOI] [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- Venza I, Visalli M, Parrillo L, De Felice M, Teti D, Venza M. MSX1 and TGF-beta3 are novel target genes functionally regulated by FOXE1. Hum Mol Genet. 2011;20:1016–1025. doi: 10.1093/hmg/ddq547. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O'Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigano MA, Lamartine J, Testoni B, Merico D, Alotto D, Castagnoli C, Robert A, Candi E, Melino G, Gidrol X, Mantovani R. New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 2006;25:5105–5116. doi: 10.1038/sj.emboj.7601375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M. The healing touch. Nature. 2005;436:1079–1080. doi: 10.1038/4361079a. [DOI] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang X-J, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- Waikel RL, Wang X-J, Roop DR. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene. 1999;18:4870–4878. doi: 10.1038/sj.onc.1203040. [DOI] [PubMed] [Google Scholar]
- Wakamatsu K, Ogita H, Okabe N, Irie K, Tanaka-Okamoto M, Ishizaki H, Ishida-Yamamoto A, Iizuka H, Miyoshi J, Takai Y. Up-regulation of loricrin expression by cell adhesion molecule nectin-1 through Rap1-ERK signaling in keratinocytes. J Biol Chem. 2007;282:18173–18181. doi: 10.1074/jbc.M611159200. [DOI] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E. AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol. 2008;183:37–48. doi: 10.1083/jcb.200804030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Akita S, Tin NT, Natsume N, Nakano Y, Niikawa N, Uchiyama T, Yoshiura K. A mutation in RYK is a genetic factor for nonsyndromic cleft lip and palate. Cleft Palate Craniofac J. 2006;43:310–316. doi: 10.1597/04-145.1. [DOI] [PubMed] [Google Scholar]
- Watt FM. Involucrin and other markers of keratinocyte terminal differentiation. J Invest Dermatol. 1983;81:100s–103s. doi: 10.1111/1523-1747.ep12540786. [DOI] [PubMed] [Google Scholar]
- Wei K, Chen J, Akrami K, Galbraith GC, Lopez IA, Chen F. Neural crest cell deficiency of c-myc causes skull and hearing defects. Genesis. 2007;45:382–390. doi: 10.1002/dvg.20304. [DOI] [PubMed] [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Whyte JL, Smith AA, Liu B, Manzano WR, Evans ND, Dhamdhere GR, Fang MY, Chang HY, Oro AE, Helms JA. Augmenting endogenous Wnt signaling improves skin wound healing. PLoS One. 2013;8:e76883. doi: 10.1371/journal.pone.0076883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietecha MS, Chen L, Ranzer MJ, Anderson K, Ying C, Patel TB, DiPietro LA. Sprouty2 downregulates angiogenesis during mouse skin wound healing. Am J Physiol Heart Circ Physiol. 2011;300:H459–467. doi: 10.1152/ajpheart.00244.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeveld MG, Maltha JC, Grupping EM, De Jonge J, Kuijpers-Jagtman AM. A histological study of tissue response to simulated cleft palate surgery at different ages in beagle dogs. Arch Oral Biol. 1991;36:837–843. doi: 10.1016/0003-9969(91)90033-q. [DOI] [PubMed] [Google Scholar]
- Winograd J, Reilly MP, Roe R, Lutz J, Laughner E, Xu X, Hu L, Asakura T, vander Kolk C, Strandberg JD, Semenza GL. Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum Mol Genet. 1997;6:369–379. doi: 10.1093/hmg/6.3.369. [DOI] [PubMed] [Google Scholar]
- Wong JW, Gallant-Behm C, Wiebe C, Mak K, Hart DA, Larjava H, Hakkinen L. Wound healing in oral mucosa results in reduced scar formation as compared with skin: evidence from the red Duroc pig model and humans. Wound Repair Regen. 2009;17:717–729. doi: 10.1111/j.1524-475X.2009.00531.x. [DOI] [PubMed] [Google Scholar]
- Wood W, Jacinto A, Grose R, Woolner S, Gale J, Wilson C, Martin P. Wound healing recapitulates morphogenesis in Drosophila embryos. Nat Cell Biol. 2002;4:907–912. doi: 10.1038/ncb875. [DOI] [PubMed] [Google Scholar]
- Wu N, Rollin J, Masse I, Lamartine J, Gidrol X. p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network. J Biol Chem. 2012;287:5627–5638. doi: 10.1074/jbc.M111.328120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff BC, Parent AE, Meleski MA, DiPietro LA, Schrementi ME, Wilgus TA. Mast cells contribute to scar formation during fetal wound healing. J Invest Dermatol. 2012;132:458–465. doi: 10.1038/jid.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Yaar M, Wu C, Park HY, Panova I, Schutz G, Gilchrest BA. Bone morphogenetic protein-4, a novel modulator of melanogenesis. J Biol Chem. 2006;281:25307–25314. doi: 10.1074/jbc.M600580200. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, Lu Z, Gill GN, Roop DR, Wertz P, Andersen B. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- Zhang QZ, Su WR, Shi SH, Wilder-Smith P, Xiang AP, Wong A, Nguyen AL, Kwon CW, Le AD. Human gingiva-derived mesenchymal stem cells elicit polarization of m2 macrophages and enhance cutaneous wound healing. Stem Cells. 2010;28:1856–1868. doi: 10.1002/stem.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- Zhao L, Saitsu H, Sun X, Shiota K, Ishibashi M. Sonic hedgehog is involved in formation of the ventral optic cup by limiting Bmp4 expression to the dorsal domain. Mech Dev. 2010;127:62–72. doi: 10.1016/j.mod.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, Huang S, Westphal H. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci U S A. 1999;96:15002–15006. doi: 10.1073/pnas.96.26.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]