Abstract
We previously characterized the link between WNT7A and the progression of ovarian cancer. Other groups have identified FGF1 as a relevant risk factor in ovarian cancer. Here, we show a linkage between these two signaling pathways that may be exploited to improve treatment and prognosis of patients with ovarian cancer. High expression of WNT7A and FGF1 are correlated in ovarian carcinomas and poor overall patient survival. A chromatin immunoprecipitation assay demonstrated that WNT7A/β-catenin signaling directly regulates FGF1 expression via TCF binding elements in the FGF1-1C promoter locus. In vitro gene manipulation studies revealed that FGF1 is sufficient to drive the tumor promoting effects of WNT7A. In vivo xenograft studies confirmed that the stable overexpression of WNT7A or FGF1 induced a significant increase in tumor incidence, while FGF1 knockdown in WNT7A overexpressing cells caused a significant reduction in tumor size. Niclosamide most efficiently abrogated WNT7A/β-catenin signaling in our model, inhibited β-catenin transcriptional activity and cell viability, and increased cell death. Furthermore, niclosamide decreased cell migration following an increase in E-cadherin subsequent to decreased levels of SLUG. The effects of niclosamide on cell functions were more potent in WNT7A overexpressing cells. Oral niclosamide inhibited tumor growth and progression in an intraperitoneal xenograft mouse model representative of human ovarian cancer. Collectively, these results indicate that FGF1 is a direct downstream target of WNT7A/β-catenin signaling and this pathway has potential as a therapeutic target in ovarian cancer. Moreover, niclosamide is a promising inhibitor of this pathway and may have clinical relevance.
Keywords: FGF1, WNT7A, WNT/β-catenin pathway, ovarian cancer, therapeutic target
INTRODUCTION
Ovarian cancer (OvCa) remains the most common cause of death from gynecological malignancies and is the fifth leading overall cause of death from cancer in women. In 2014, approximately 22,000 new OvCa cases and 14,500 deaths are estimated from OvCa in the United States.1 The dearth of specific signs, symptoms, or efficient early detection markers for this disease contributes to its diagnosis at advanced stages, resulting in low overall survival. Indeed, more than 75% of OvCa cases are diagnosed when there is widely metastatic disease in the peritoneal cavity.2 Therefore, it is important to identify therapeutic targets and efficient drugs that can improve current OvCa treatment by preventing its dissemination.
WNT genes encode secreted glycoproteins, acting through frizzled receptor (FZD), that control cell fate, mortality, proliferation, differentiation and tissue growth.3, 4 Gene mutations and changes in the expression of extracellular inhibitors and intranuclear transcription cofactors within the WNT pathway promote tumor progression and metastasis.5, 6 The canonical pathway of WNT signaling results in the nuclear accumulation of β-catenin and transcriptional activation of target genes. WNT/β-catenin signaling plays a role in ovarian tumorigenesis,7 as well as chemoresistance in cancer stem cells of all OvCa subtypes.8 Our recent findings also suggest that the expression of WNT7A during the malignant transformation of OvCa plays a critical role in tumor progression mediated by the WNT/β-catenin signaling pathway.9
FGF1 is one of 23 members of the highly conserved polypeptide fibroblast growth factor family. FGF1 has strong mitogenic effects on a variety of different cell types in various stages of development, morphogenesis and angiogenesis in neoplastic or non-neoplastic tissues.10, 11 FGF1 has been identified as a potential prognostic marker for OvCa.12 Genetic variation of FGF1 has the most significant association with increased OvCa risk within the FGF family.13 Furthermore, FGF1 expression is also a significant determinant of survival and response to platinum-based chemotherapy.14 Thus, modulation of FGF1 by distinct mechanisms in OvCa may be important in ovarian tumor progression.
Niclosamide is an efficacious and minimally toxic, FDA-approved drug for the treatment of helminth parasites, specifically tapeworms, in humans. Several groups have reported that niclosamide is active against cancer cells and targets WNT signaling.15–19 Niclosamide inhibits solid tumor growth in a colon cancer model by promoting FZD endocytosis, leading to the downregulation of DVL, β-catenin stabilization and TCF/LEF activity.17, 20 Niclosamide inhibits tumor growth by targeting S100A4, which is a transcriptional target of WNT signaling,18 and by suppressing LRP6 in prostate and breast cancer cells.15 Given its proven safety record and mechanisms that target WNT signaling raises the possibility of repurposing niclosamide for OvCa treatment.
In the present study, we show that WNT7A and FGF1 expression are highly correlated in ovarian carcinomas, and FGF1 is a direct transcriptional target of WNT7A/β-catenin signaling. Niclosamide was an effective inhibitor of WNT7A/β-catenin signaling, including FGF1 expression, and should be further explored as treatment for OvCa.
RESULTS
Analysis of FGF1 in OvCa
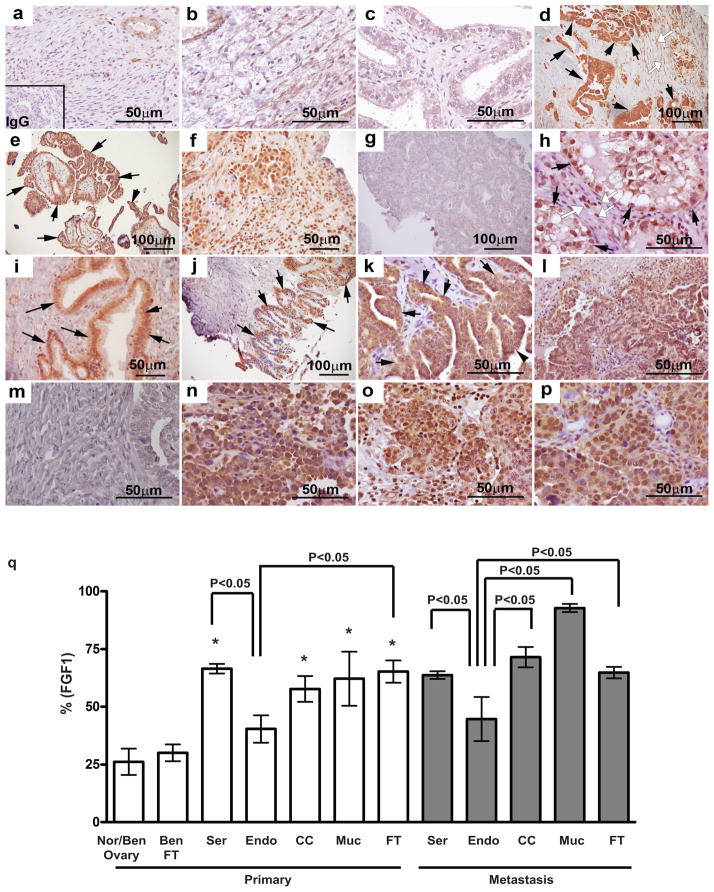
While analysis of FGF1 gene expression or variation has indicated an association with OvCa risk,12–14 the expression pattern of FGF1 in ovarian cancer has not been characterized. Therefore, immunoreactive FGF1 was examined using human OvCa tissues. FGF1 was low in normal (Figure 1a) and benign (Figure 1b) ovary, as well as benign fallopian tube epithelium (Figure 1c). FGF1 was highly detected in high grade invasive serous epithelial carcinomas (Figure 1d, black arrows), and surface epithelial cells of low grade serous carcinomas (Figure 1e, black arrows). Heterogeneous FGF1 was seen in the tumor microenvironment, with lymphocytes positive in serous (Figure 1f), but not in endometrioid carcinomas (Figure 1g). FGF1 was observed in fibroblasts of several histotypes (Figure 1d, h, open white arrows). FGF1 was positive in clear cell carcinomas (Figure 1h, black arrows). FGF1 was specifically detected in non-invasive cells lining the basal membrane (Figure 1i, black arrows) and in the surface epithelial cells of mucinous carcinomas (Figure 1j). Abundant FGF1 was also detected in epithelial fallopian tube carcinomas (Figure 1k, black arrows). FGF1 was significantly higher in serous, clear cell, mucinous and fallopian tube primary carcinomas compared to normal/benign ovarian and fallopian tube tissues. Specifically, serous and fallopian tube carcinomas showed higher levels of FGF1 than endometrioid carcinomas (Figure 1q). FGF1 was observed in metastases from serous (Figure 1l, q), clear cell (Figure 1n, q), mucinous (Figure 1o, q) and fallopian tube (Figure 1p, q) carcinomas, but not from endometrioid carcinomas (Figure 1m, q).
Figure 1.
Analysis of FGF1 in ovarian cancer. (a–p) Representative photomicrographs of immunoreactive FGF1 in normal/benign, primary and metastasized ovarian and fallopian tube tumors (A total of 537 specimens including 10 normal/benign (8 ovaries and 2 fallopian tubes), 147 serous, 20 endometrioid, 22 clear cell, 7 mucinous and 32 fallopian tube primary tumors, and 299 metastasized tumors from 199 serous, 8 endometrioid, 15 clear cell, 4 mucinous and 73 fallopian tubes). (a) Normal ovary. (b) Benign ovary. (c) Benign fallopian tube. (d) Black arrows point to high grade invasive serous epithelial cells. Open white arrows point to fibroblasts. (e) Black arrows point to epithelial cells of low grade serous carcinoma. (f) Serous carcinoma lymphocytes. (g) Endometrioid carcinoma. (h) Black arrows point to epithelial cells of clear cell carcinoma. Open white arrows point to fibroblasts. (i) Black arrows point to non-invasive cells lining the basal membrane of mucinous carcinoma. (j) Black arrows point to surface epithelium of mucinous carcinoma. (k) Black arrows point to epithelial fallopian tube carcinoma. (l) Serous carcinoma metastasis in peritoneum. (m) Endometrioid carcinoma metastasis in omentum. (n) Clear cell carcinoma metastasis in peritoneum. (o) Mucinous carcinoma metastasis in omentum. (p) Fallopian tube carcinoma metastasis in ovary. (q) Immunoreactive FGF1 levels were quantitatively scored by Image J analysis. Nor/Ben, normal/benign; Ser, serous; Endo, endometrioid; CC, clear cell; Muc, mucinous; FT, fallopian tube. *P<0.05 vs. normal/benign ovaries and fallopian tubes.
Correlation between WNT7A and FGF1 levels and overall survival
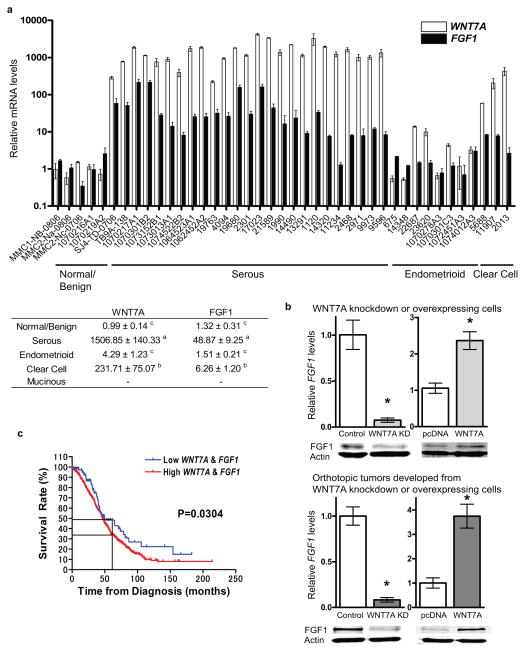
Since FGF family members have previously been identified as downstream targets of WNT signaling,21 and a role for the specific ligand WNT7A was elucidated in tumor growth and progression,9 we next examined whether WNT7A is associated with FGF1 in OvCa. A total of 41 fresh-frozen ovarian samples including 5 normal/benign, 25 serous, 8 endometrioid, 3 clear cell and 0 mucinous were examined for WNT7A and FGF1 mRNA levels (Figure 2a). The expression of WNT7A in serous carcinomas was correlated with high levels of FGF1 expression. Elevated WNT7A was correlated with moderately increased FGF1 levels in clear cell carcinomas. Significantly higher WNT7A was observed in serous (1506-fold) and clear cell (231-fold), but not in endometrioid carcinomas compared to normal/benign ovaries. FGF1 transcripts were significantly higher (48-fold) in serous carcinomas compared to normal/benign ovaries and slightly increased in clear cell carcinomas (6.26-fold), but unchanged in endometrioid carcinomas.
Figure 2.
Expression of FGF1 correlates with WNT7A in ovarian cancer. (a) Relative WNT7A and FGF1 expression. A total of 41 fresh-frozen ovarian samples including 5 normal/benign, 25 serous, 8 endometrioid, 3 clear cell and 0 mucinous were examined for WNT7A and FGF1 levels. Duplicates of each sample were analyzed by qPCR. Values were normalized against RPL19 and are expressed as fold above normal/benign (± SEM), which was arbitrarily given a value of 1. Mean vales from each subtype are shown in table. Different letters denote transcripts that have statistically significant (P<0.01) differences in mean expression levels. (b) Relative FGF1 mRNA and protein expression in WNT7A knockdown SKOV3.ip1 cells and WNT7A overexpressing SKOV3 cells. *P<0.05 vs. control or pcDNA. Relative FGF1 mRNA and protein expression in orthotopic tumors developed from WNT7A knockdown SKOV3.ip1 cells or WNT7A overexpressing SKOV3 cells. *P<0.05 vs. control or pcDNA. (c) WNT7A and FGF1 expression correlates with survival. Overall survival rate was calculated in 151, 68, 185, 557 patients from GSE9891, GSE14764, GSE26712 and TCGA-OV, respectively, in relation to the expression of WNT7A and FGF1 by Kaplan-Meier method using Prism 4.0. The P-value was determined by the log rank test.
Using previously generated cell lines,9 a correlation between FGF1 and WNT7A was observed in cells with knockdown or overexpression of WNT7A, and in orthotopic tumors that developed from these cells (Figure 2b). However, no other FGF family members, nor FGF receptors, appeared to consistently follow WNT7A expression (Supplementary Figures 1 and 2). To further explore the correlation between WNT7A and FGF1 expression in OvCa, we evaluated the prognostic and predictive impact of WNT7A and FGF1 using Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) datasets. A total of 961 primary ovarian tumors with completed data sets were selected for survival analysis. High expression of both WNT7A and FGF1 was significantly correlated with poor survival as determined by log-rank test (P=0.0304, Figure 2c). Furthermore, the 5-year (60 months) survival rate was 34.2% in women with high WNT7A and FGF1 expressing tumors as compared to 49.4% in women with low WNT7A and FGF1 expressing tumors. These findings were similar to previously reported survival rates of advanced OvCa patients 5 years after initial diagnosis.22, 23
FGF1 is a direct transcriptional target of WNT7A/β-catenin signaling
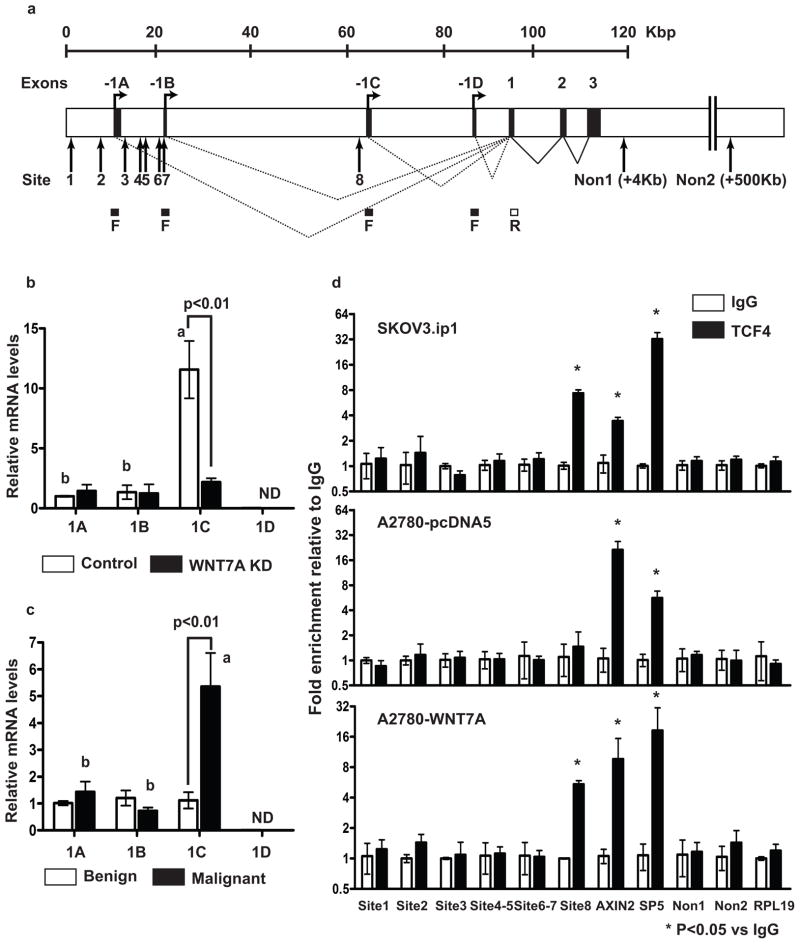
The FGF1 gene is composed of a single protein isoform and has four alternative tissue-specific promoters subject to alternative splicing (Figure 3a).24, 25 Each promoter is coupled with its 5′ untranslated exon, giving rise to four mRNAs with distinct UTRs, containing exons 1A, 1B, 1C and 1D, respectively. Therefore, we examined specific FGF1 transcript(s) in OvCa using 4 different forward primers designed in each untranslated exon and one reverse primer designed in exon 1. SKOV3.ip1 cells, which express high endogenous WNT7A and FGF1 (Supplementary Figure 3), show low or no expression of transcripts 1A, 1B or 1D, and high expression of the 1C transcript (12-fold higher than that of 1A and 1B (Figure 3b)). The 1C transcript was reduced in WNT7A knockdown SKOV3.ip1 cells, suggesting that 1C is specific to OvCa and possibly regulated by WNT7A. Similarly, FGF1-1C was the most abundant transcript in human malignant ovarian tissues (Figure 3c).
Figure 3.
FGF1 is a direct β-catenin/TCF target. (a) Human FGF1 gene and transcripts. Alternative splicing of untranslated exons (1A, 1B, 1C and 1D) to exon 1 will generate mRNAs 1A, 1B, 1C and 1D. A total of 8 consensus TCF/LEF-binding elements (WWCAAWG, W=A/T) at the FGF1 genomic locus are shown. F; forward primers designed in 1A, 1B, 1C or 1D and R; reverse primer designed in exon 1. Relative FGF1 alternative splicing mRNA levels in (b) control and WNT7A knockdown SKOV3.ip1 cells, and (c) human ovarian normal/benign (n=5) and malignant (n=36) tumors. Different letters denote transcripts that have statistically significant (P<0.01) differences in mean expression levels. ND: non detectable. (d) ChIP assay of DNA isolated from SKOV3.ip1, A2780-pcDNA5 and WNT7A overexpressing cells immunoprecipitated with TCF4 antibody. Immunoprecipitated DNA was analyzed by qPCR and normalized to input. TCF4 binding sites at AXIN2 and SP5 loci were used as positive controls. Three irrelevant non TCF4 binding sites on the same chromosome of FGF1 and different chromosome at the RPL19 locus were used as negative controls.
To determine whether FGF1 is likely to be a direct rather than indirect β-catenin/TCF target gene, we searched the FGF1 genomic locus and flanking DNA sequences for consensus TCF/LEF-binding elements (WWCAAWG, W=A/T). We found several putative binding sites at the 1A, 1B and 1C locus, but not at the 1D locus (Figure 3a). Therefore, we performed chromatin immunoprecipitation (ChIP) assays using a TCF4 antibody to analyze chromatin isolated from SKOV3.ip1, parental A2780 (lacking endogenous WNT7A), and A2780 cells overexpressing WNT7A (Figure 3d). DNA from the anti-TCF4 ChIP of SKOV3.ip1 was approximately 8-fold enriched in site 8, which contains the consensus TCF4 binding site located in the 1C locus, when compared to IgG control. Interestingly, site 8 enrichment (5.5-fold) was observed in WNT7A overexpressing A2780 cells, but not in control A2780 cells. TCF/LEF binding sites in the promoters of AXIN2 and SP5, well-known direct β-catenin/TCF target genes, were used as positive controls. Irrelevant sites ~4 kb and ~500 kb downstream of the FGF1 locus (Non1 and Non2) and the RPL19 locus on a different chromosome served as negative controls. Dominant negative (DN)-TCF4 expression in SKOV3.ip1 cells decreased FGF1, but not WNT7A levels (Supplementary Figure 4a). Specifically, FGF-1C transcript expression was decreased in DN-TCF4 expressing cells (Supplementary Figure 4b), confirming the ChIP results in Figure 3d. Collectively, these results suggest that at least one site within the FGF1 promoter is directly regulated by WNT7A/β-catenin/TCF in OvCa.
WNT7A-FGF1 signaling promotes neoplastic transformation in OvCa
To understand whether FGF1 is necessary and/or sufficient for the effects of WNT7A in OvCa tumor progression, we generated stable WNT7A or FGF1 overexpressing or knockdown human OvCa cell lines (Supplementary Figure 5). Relative WNT7A and FGF1 expression in a panel of OvCa cell lines was determined (Supplementary Figure 3) and A2780 cells were chosen for further analysis, as these cells have been widely used for gene manipulation and xenograft study, and showed lack of or low endogenous WNT7A and FGF1. The expression of FGF1 correlated with the upregulation of WNT7A in a genetically manipulated A2780 line, whereas WNT7A expression was not induced by FGF1 overexpression. Therefore, stable FGF1 knockdown in WNT7A overexpressing cells and their control cells were also generated.
Using these stable cell lines, we examined the in vitro effects of WNT7A and/or FGF1 on cell proliferation and adhesion (Supplementary Figure 6), using these assays as indicators of the roles of WNT7A and/or FGF1 in tumor growth in vivo. Cell doubling time was significantly shorter and ability to adhere to plastic was significantly greater in WNT7A overexpressing cells as compared to vector control cells. FGF1 overexpressing cells also exhibited a significant increase in cell adhesion, but no obvious decrease in cell doubling time was seen. However, FGF1 knockdown in WNT7A overexpressing cells showed that the effect of WNT7A on cell doubling time and adhesion was attenuated in the absence of FGF1.
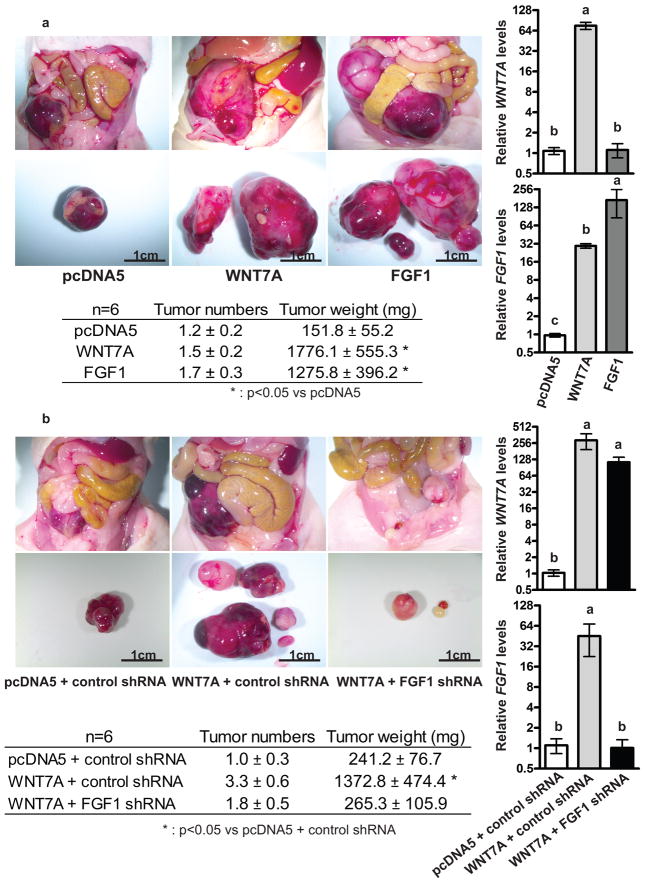
Next, we examined the effects of WNT7A and/or FGF1 on tumor growth in vivo. Nude mice were intraperitoneally inoculated with control, WNT7A overexpressing, or FGF1 overexpressing cells (n=6 each group, Figure 4a). After 5 weeks, a significant increase in tumor burden, but not tumor number, was observed in mice injected with WNT7A or FGF1 overexpressing cells (1776.1 ± 555.3 mg or 1275.8 ± 396.2 mg, respectively compared to pcDNA5, 151.8 ± 55.2 mg), suggesting that both WNT7A and FGF1 have potential roles in tumor growth. The increased tumor growth by WNT7A overexpression was abrogated by inhibiting FGF1 (n=6 each group, Figure 4b), confirming that FGF1 is downstream of WNT7A signaling in OvCa cells. FGF1 knockdown in WNT7A overexpressing cells resulted in reduced tumor burden (265.3 ± 106.5 mg) compared to mice injected with cells overexpressing WNT7A and control shRNA (1372.8 ± 474.4 mg). We confirmed that expression of WNT7A and FGF1 was maintained in orthotopic tumors.
Figure 4.
FGF1 promotes the tumorigenic functions of WNT7A. (a) WNT7A or FGF1 overexpression increased tumor growth. Nude mice were injected i.p. with vector control cells, WNT7A overexpressing, or FGF1 overexpressing cells. The images provide a direct view of the abdominopelvic cavity and tumors isolated from mice. Total tumor numbers and weight 5 weeks after i.p. injection are shown (n=6 each group). Relative WNT7A or FGF1 expression in orthotopic tumors that developed from vector control, WNT7A overexpressing, or FGF1 overexpressing cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels. (b) FGF1 knockdown inhibits WNT7A-dependent tumor growth. FGF1 knockdown in WNT7A overexpressing cells inhibited tumor growth. Nude mice were injected i.p. with vector control, or control or FGF1 knockdown WNT7A overexpressing cells. The images provide a direct view of the abdominopelvic cavity and tumors isolated from the mice. Total tumor numbers and weight 5 weeks after i.p. injection are shown (n=6 each group). Relative WNT7A or FGF1 expression in orthotopic tumors that developed from vector control, or control or FGF1 knockdown WNT7A overexpressing cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels.
Niclosamide can inhibit WNT7A/β-catenin signaling in OvCa
Several WNT inhibitors have been found to effectively block WNT signaling.26–29 To determine whether WNT7A/β-catenin signaling could be a therapeutic target in the context of OvCa treatment, we searched the LOPAC library containing 1280 pharmacological active compounds (Sigma-Aldrich, St. Louis, MO, USA) to find compounds inhibiting WNT signaling. A total of 14 small molecules were selected to determine WNT7A/β-catenin inhibitory activity in OvCa cell lines (Supplementary Figure 7a). Half of the small molecules significantly inhibited the activity of the TCF/LEF luciferase reporter in A2780 cells stimulated by WNT7A. Niclosamide exhibited the most significant inhibitory effect on WNT7A/β-catenin signaling. Thus, we chose to focus on niclosamide for further study.
Next, we assessed the molecular targets of niclosamide within the WNT/β-catenin signaling pathway in OvCa cells (Supplementary Figure 7bcd). Our results indicated potential regulation of WNT7A production, as niclosamide dose-dependently inhibited WNT7A levels. TCF/LEF reporter activity in cells stimulated by exogenous (A2780) or endogenous (SKOV3.ip1) WNT7A was suppressed by niclosamide. We also confirmed that niclosamide inhibited TCF/LEF activity stimulated by a constitutively active β-catenin (S33Y).
Niclosamide regulates OvCa cell functions and inhibits tumor growth and progression
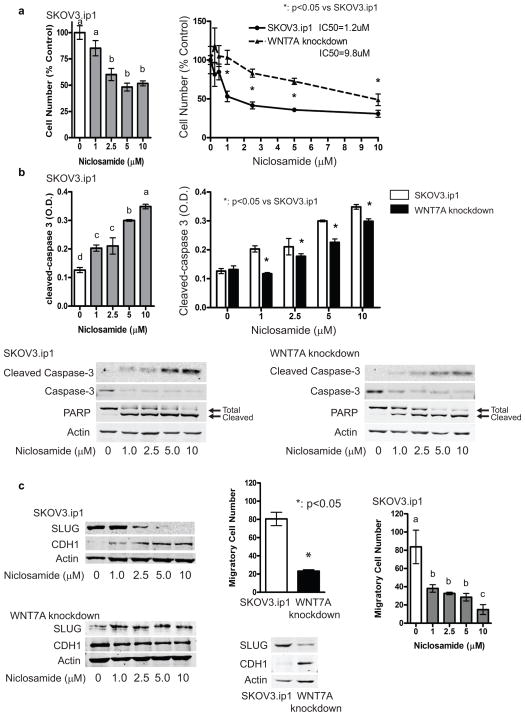
To determine whether niclosamide has therapeutic potential in OvCa, control and WNT7A knockdown SKOV3.ip1 cells 9 were used to assess cell proliferation, apoptosis, and migration (Figure 5). Niclosamide dose-dependently decreased cell number in SKOV3.ip1 cells, whereas cells depleted of WNT7A were less sensitive to the anti-proliferative effects of niclosamide (IC50=1.2 or 9.8 μM in SKOV3.ip1 or WNT7A knockdown cells, respectively, Figure 5a). Similarly, cleaved-caspase 3 was increased up to 2.7-fold by treatment with 10 μM niclosamide in SKOV3.ip1 cells, while WNT7A knockdown cells were less sensitive to this effect (Figure 5b). This result was confirmed by immunoblot analysis, showing that niclosamide increased cleaved caspase-3 and PARP, and decreased total caspase-3 and PARP. Niclosamide inhibited invasive features in SKOV3.ip1 cells (Figure 5c). It is well known that the SKOV3.ip1 cells primarily contain CDH1 (E-cadherin) negative cells. Our results showed that niclosamide dose-dependently increased CDH1 and decreased its transcriptional repressor SLUG in SKOV3.ip1, while niclosamide did not affect CDH1 and SLUG in WNT7A knockdown cells. Consistent with these results, migration was inhibited in WNT7A knockdown cells, and was dose-dependently decreased by niclosamide in parental SKOV3.ip1 cells.
Figure 5.
Niclosamide effectively inhibits ovarian cancer cell functions in WNT7A expressing cells. (a) Effects of niclosamide on cell viability in SKOV3.ip1 control and WNT7A knockdown cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean cell number. (b) Effects of niclosamide on cleavage of the apoptosis effector molecule caspase-3 and its target PARP in SKOV3.ip1 and WNT7A knockdown cells by ELISA and/or western blots. Different letters denote groups that have statistically significant (P<0.05) differences in mean O.D. (c) Effects of niclosamide on CDH1 and SLUG expression levels, and cell migration in SKOV3.ip1 control and WNT7A knockdown cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean cell number.
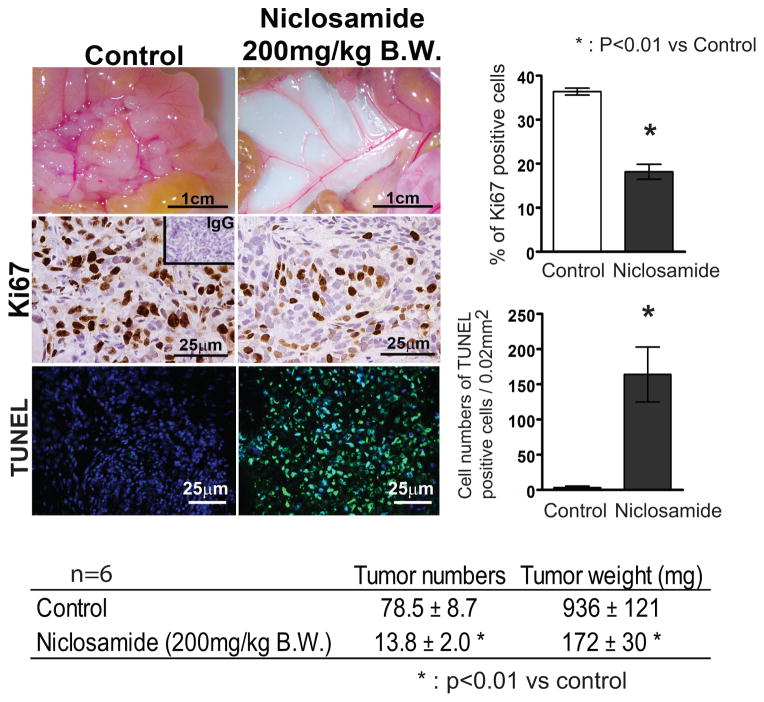
We next determined the effect of niclosamide on ovarian tumor growth and progression (Figure 6). Five weeks after injection with SKOV3.ip1 cells, mice that received vehicle treatment (n=6) developed disseminated abdominal disease mimicking OvCa in patients. Niclosamide treated mice (n=6) had significantly fewer implants on the mesentery and intraperitoneal region (13.8 ± 2.0 vs 78.5 ± 8.7 implants) and their total tumor weight was 172 ± 30 mg vs 936 ± 121 mg. Decreased tumor cell proliferation, as determined by Ki67 immunohistochemistry, was correlated with tumor growth and quantitatively significant. TUNEL analysis revealed a significant number of apoptotic cells in the tumors of mice treated with niclosamide.
Figure 6.
Effects of niclosamide on tumor growth in ovarian cancer. (a) Mice were injected i.p. with SKOV3.ip1 cells and treated with niclosamide (0 and 200 mg/kg/day B.W.) by oral gavage for 5 weeks starting 3 days after i.p. injection. Tumor implant number and total tumor weight 5 weeks after niclosamide treatment are shown (n=6 each group). Cell proliferation and apoptosis were determined by immunoreactive Ki67 staining and TUNEL assay.
While niclosamide reduced FGF1 expression in SKOV3.ip1 cells, FGF1 itself did not influence the efficacy of niclosamide (Supplementary Figure 8). When FGF1 only was overexpressed in A2780 cells, no differences in cell viability or death were observed compared to control following niclosamide exposure, whereas cells overexpressing WNT7A were more sensitive to the effects of niclosamide. Moreover, WNT7A overexpressing cells with and without FGF1 knockdown were equally sensitive to the effects of niclosamide, suggesting that niclosamide targets WNT7A/β-catenin rather than FGF1 directly. However, because FGF1 is a downstream target of aberrant WNT7A/β-catenin signaling and abundant WNT7A and FGF1 are observed in ovarian cancer, indirect downregulation of FGF1 by niclosamide remains clinically relevant.
DISCUSSION
FGF1 has been identified as an adverse prognostic factor in OvCa risk.12–14 In the present study, we show that FGF1 is a downstream target of WNT7A signaling via β-catenin activation. First, FGF1 is abundant in OvCa, correlates with high WNT7A, especially in serous carcinomas. High expression of both WNT7A and FGF1 is associated with poor survival (Figures 1 and 2). Second, the FGF1 gene is directly regulated by WNT7A/β-catenin signaling (Figure 3). Third, FGF1 is sufficient to drive the tumor promoting effects of WNT7A (Figure 4).
Our previous study demonstrated that WNT7A is the sole ligand activating β-catenin/TCF signaling in OvCa.9 We found that the FGF1-1C transcript is specifically regulated in a WNT7A and β-catenin/TCF-dependent manner in OvCa. The ChIP analysis indicates that a single consensus TCF/LEF binding site at the 1C promoter locus is critical for WNT7A-mediated TCF binding, while binding sites within the 1A and 1B loci are not occupied in the context of WNT7A. It has been reported that 1C and 1D transcripts are potential markers for cell proliferation, while 1A and 1B are specific for the maintenance and survival of cells.24 Although FGF1-1C specific functions in OvCa pathogenesis have yet to be determined, FGF1-1C is directly transcribed from activation of β-catenin/TCF signaling stimulated by WNT7A.
FGF1 binds receptors of FGFR1, 2, 3 and 4 with high-affinity.11 Two receptors, FGFR2IIIb and FGFR4 have been implicated in OvCa progression,30, 31 and FGFR4 has been identified as a potential therapeutic target in OvCa.31 Several studies have reported that other members of the FGF family are WNT/β-catenin target genes. FGF9 and FGF20 are indirect and/or direct transcriptional targets of β-catenin/TCF signaling in endometrioid OvCa, in which the WNT pathway is often constitutively activated, usually via missense mutation of CTNNB1.21, 32 However, no members of the FGF family or the FGF receptors, other than FGF1, have been correlated with WNT7A expression in OvCa and none were directly regulated by WNT7A in our model. These results suggest that FGF1 is an important mediator of the tumor promoting events of WNT7A-dependent pathogenesis.
The importance of inappropriate WNT signaling for the development and progression of many cancers, including OvCa, has been well documented.7–9 Therefore, it is plausible to search for therapeutic drugs that target the WNT7A/β-catenin-FGF1 pathway. While several small molecules that target this pathway have been identified33 and, for a few of them, a precise mechanism of inhibition has been determined,27, 33, 34 niclosamide was selected as the most efficient inhibitor of WNT7A-dependent TCF/LEF reporter activity. Niclosamide has recently been identified to target stem-like OvCa-initiating cells by a drug screening method, and gene expression array indicated involvement of WNT hyperactivity.35 In the present study, we found that niclosamide could directly target β-catenin-TCF/LEF transcriptional activity, specifically; the effects of niclosamide on cell functions are more robust in WNT7A overexpressing cells. Our results also revealed that niclosamide reduced tumor growth and progression. We show that WNT7A is highest in serous carcinomas, the most common OvCa subtype,9 and recent studies suggest niclosamide is effective in OvCa stem cells.35 While the importance of WNT7A in OvCa stem cells is not known, WNT7A is one of the critical factors determining cell fate in female reproductive development.36, 37 The regulation of WNT7A in OvCa may lead us to understand the etiology and/or function of stem cells in aggressive serous carcinomas.
Although our results demonstrated that niclosamide efficacy in OvCa depended on WNT7A, but not FGF1 function, other signaling pathways could be additional targets. Niclosamide is reported to target not only WNT signaling but also mTORC1, STAT3 and NFκB pathways in several cancers.38–42 Furthermore, we showed that niclosamide increased E-cadherin and decreased SLUG proteins. E-cadherin establishes cell polarity, mediates inhibition of proliferation and inhibits tumor cell growth.43, 44 E-cadherin is often downregulated during tumor progression, leading to increased tumor invasiveness and metastasis.45, 46 Therefore, it is likely that niclosamide reduces invasive and aggressive features in OvCa. While the potential interaction between these cascades and WNT signaling is an intriguing area for future study, overall niclosamide has broad potential as an effective inhibitor to treat OvCa patients.
In the present study, we determined that WNT7A-FGF1 signaling is capable of inducing tumor growth, indicating a critical role in the aggressive progression of OvCa. Furthermore, we found that niclosamide is an effective therapeutic inhibitor of WNT7A/β-catenin signaling. New drugs are desperately needed to treat OvCa, and niclosamide is FDA-approved with a favorable safety profile. If niclosamide continues to hold promise in further pre-clinical studies, repurposing may ultimately prove to have a tremendous impact on the lives of OvCa patients in the clinical setting.
MATERIALS AND METHODS
Reagents and plasmids
Short hairpin RNAs (shRNA), cDNAs encoding human WNT7A and FGF1, and pcDNA5/FRT/V5-His, pcDNA6/TR and pcDNA5/TO plasmids were purchased from Sigma-Aldrich, Thermo Scientific (Rockford, IL, USA) and Life Technologies (Life Technologies, Grand Island, NY, USA), respectively. Niclosamide, iCRT3, iCRT14, Pyrvinium, Bafilomycin, Quercetin, NSC668036 and LiCl were purchased from Sigma-Aldrich. XAV939 and IWR were purchased from Cayman Chemical (Ann Arbor, MI, USA). IWP and Box5 were purchased from Thermo Scientific. ICG001, CCT031374 and iCRT5 were obtained from R&D Systems (Minneapolis, MN USA).
Tissue samples and cell lines
Tissue microarray paraffin-embedded and fresh-frozen ovarian specimens were obtained from The University of Chicago and Southern Illinois University. Clinical and histopathologic information was collected and verified for each sample by a gynecologic pathologist following International Federation of Gynecology and Obstetrics (FIGO) classifications for stages I–IV.
OVCAR3, OVCAR5, SKOV3, TOV-112D, TOV-21G, OV-90, MDAH 2774 and ES2cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). A2780 and OVCAR4 cells were purchased from Sigma-Aldrich and NCI, respectively. KURAMOCHI, OVKATE and OVSAHO were purchased form JCRB cell bank (Osaka, Japan). HEY, HEYA8, OVCAR8, IGROV-1, OVCA420, OVCA429, OVCA432, OVCA433 and SKOV3.ip1 cells were purchased from the cell bank of The University of Texas MD Anderson Cancer Center. All cells were authenticated by short tandem repeat (STR) analysis and passaged within 6 months of receipt. All cells were tested routinely for cell proliferation and BrdU incorporation as well as mycoplasma contamination, and showed similar growth rates and negative mycoplasma throughout all experiments. TOV-112D, TOV-21G, and OV-90 cells were grown in 1:1 MCDB 105:M199 with 15% FBS and penicillin/streptomycin, other cells were cultured in DMEM with 10% FBS, 200mM glutamine and penicillin/streptomycin. All cell lines were grown at 37 °C in a humidified 5% CO2 incubator.
DN-TCF4 expressing SKOV3.ip1 cells were generated using the T-REx system with pcDNA6/TR and pcDNA5/TO vectors (Life Technologies). A2780 cells were transfected with empty vector (pcDNA5/FRT), or plasmids encoding WNT7A tagged with V5 or FGF1 using the Flp-In system (Life Technologies). Hygromycin resistant stable clones were used for further analysis based on WNT7A and FGF1 expression detected by qPCR and western blot. Furthermore, stable FGF1 knockdown in WNT7A overexpressing cells and their control cells were generated using shRNA gene knockdown methods as described previously.9 Stable clones doubly resistant for hygromycin and puromycin were selected and WNT7A and FGF1 expression were confirmed by qPCR and western blot.
QPCR, western blot, TUNEL and cleaved-caspase assays
Total RNA was isolated from tissues and cells, and cDNA was synthesized from total RNA. Relative gene expression was determined by SYBR green incorporation using a Bio-Rad myCycler as described previously.47 A table of oligonucleotides used for each gene is presented in Supplementary Table 1. Ten micrograms of total protein from whole cell lysates were separated on SDS-PAGE gels and transferred to nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). Membranes were blocked and incubated overnight at 4°C with primary antibodies. Bound antibody was visualized with IRDye 700 or 800 conjugated affinity-purified secondary antibodies (Rockland Immunochemicals, Gilbertsville, PA, USA) using the Odyssey infrared imaging system (LI-COR, Lincoln, NE, USA). A list of antibodies is shown in Supplementary Table 2. The TUNEL assay was performed according to manufacturer’s instructions using ApopTag Fluorescein In Situ Apoptosis Detection Kit (Thermo Scientific). To assess apoptosis, cleaved-caspase 3 was quantitated using a PathScan Cleaved Caspase 3 Sandwich ELISA Kit according to manufacturer’s instructions (Cell Signaling).
Immunohistochemistry
Immunolocalization of FGF1 in a total of 537 human specimens including 10 normal/benign (8 ovaries and 2 fallopian tubes), 147 serous, 20 endometrioid, 22 clear cell, 7 mucinous and 32 fallopian tube primary tumors, and 299 metastases from 199 serous, 8 endometrioid, 15 clear cell, 4 mucinous and 73 fallopian tube tumors; and Ki67 in xenografts from nude mice were examined in cross-sections (5 μm) of paraffin-embedded tissue sections using specific primary antibodies and a Vectastain Elite ABC Kit (Vector laboratories, Burlingame, CA, USA). Immunostaining was semiquantitatively scored by Image J, which is a public domain Java image processing program developed by NIH.
Cell proliferation, adhesion and migration
Cell proliferation, adhesion and migration assays were performed following our previously described methods.9, 48 To assess cell proliferation, cells (2×104/well) were seeded in 24-well plates. Doubling time of cells was calculated from the growth rate during the exponential growth phase (0–72 h) using the formula, Td = 0.693t/ln(Nt/N0), where t is time in days, Nt is cell number at time t, and N0 is cell number at the initial time.49, 50 To determine the effect of niclosamide on cell viability, cells were treated with vehicle (0.1% DMSO) or niclosamide (0–10 μM) for 48 hours before being harvested and counted by trypan blue exclusion. To assess cell adhesion, cells (1×105/well) were seeded in 24-well plates and harvested after 1 h incubation. Cell migration assays were performed using a modified Boyden Chamber method with 8-μm pore size polycarbonate membrane 24-well transwells. Cells were treated with vehicle or niclosamide for 24 hours before being harvested, re-plated (75×103 cells per well in 100 μl of serum-free medium) into upper wells, and placed in lower wells containing 500 μl serum-free medium in order to determine unstimulated migratory ability.
Chromatin immunoprecipitation assay
ChIP assays were performed using the Magna ChIP G-chromatin immunoprecipitation Kit (Thermo Scientific) as described previously.51 Sheared chromatin from 1×106 cells was immunoprecipitated with mouse monoclonal TCF4 antibody or control mouse IgG (Thermo Scientific). Primers for the qPCR are listed in Supplementary Table 3.
Animal experiments
For in vivo animal studies, 7–8 week-old female nude mice (nu/nu BALB/c, Jackson Laboratories) were injected with 1×106 cells i.p. in a total volume of 400 μl. Body weights were monitored every 7 days thereafter. Mice were euthanized and necropsied 5 weeks after injection. Niclosamide (200 mg/kg B.W.) or vehicle control (PEG400) was given daily by oral gavage for 5 weeks. Harvested tumors were either frozen or fixed with 4% paraformaldehyde in PBS for further analysis.
Statistical analyses
Quantitative data were subjected to least-squares ANOVA and differences between individual means were tested by a Tukey multiple-range test using Prism 4.0 (Graphpad, San Diego, CA, USA). QPCR data were corrected for differences in sample loading using the RPL19 data as a covariate. Tests of significance were performed using the appropriate error terms according to the expectation of the mean squares for error. A p-value of 0.05 or less was considered significant. Data are presented as least-square means (LSM) with standard error of the means (SEM). The Kaplan-Meier method was used to calculate the survival rates and was evaluated by the log-rank test using GEO and TCGA datasets: GSE9891,52 GSE14764,53 GSE2671254 and TCGA-OV, that contained 285, 80, 195 and 570 samples of normal and cancerous ovary, peripheral tissues and fallopian tubes, respectively. A total of 961 primary ovarian tumors with completed data sets (151, 68, 185 and 557 from GSE9891, GSE14764, GSE26712 and TCGA-OV, respectively) were selected for survival analysis.
Supplementary Material
FGF gene expression. Relative FGF gene expression in WNT7A knockdown cells, orthotopic tumors that developed from WNT7A knockdown cells, WNT7A overexpressing cells and orthotopic tumors that developed from WNT7A overexpressing cells. N.D. means non-detectable levels by qPCR.
FGF receptor gene expression. Relative FGFR gene expression in WNT7A knockdown cells, orthotopic tumors that developed from WNT7A knockdown cells, WNT7A overexpressing cells and orthotopic tumors that developed from WNT7A overexpressing cells.
Expression of WNT7A and FGF1 in ovarian cancer cell lines. Relative expression of WNT7A and FGF1 was assessed by qPCR in human ovarian cancer cell lines. Data were set to background level of 1 in A2780 cells.
FGF1 expression in DN-TCF4 expressing SKOV3.ip1 cells. (a) DN-TCF4 expressing SKOV3.ip1 cells were generated using the T-REx system with pcDNA6/TR and pcDNA5/TO vectors. Relative expression of WNT7A, FGF1 and AXIN2 as a positive control was assessed by qPCR in control or DN-TCF4 expressing SKOV3.ip1 cells. (b) Relative expression of FGF1 1A, 1B, 1C and 1D transcripts in control or DN-TCF4 expressing SKOV3.ip1 cells. N.D. means non-detectable levels by qPCR.
Gene manipulation of WNT7A and FGF1 in ovarian cancer cells. (a) V5-tagged WNT7A or FGF1 were sub-cloned into pcDNA5/FRT vector. A2780 cells were transfected with empty vector or plasmids encoding WNT7A-V5 or FGF1 using the Flp-In system to establish WNT7A or FGF1 overexpressing cell lines. Relative expression of WNT7A and FGF1 was determined by qPCR in WNT7A or FGF1 overexpressing A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels. WNT7A or FGF1 protein levels were confirmed by western blot analysis using antibodies of V5 for WNT7A and FGF1. (b) WNT7A overexpressing cells were transfected with control shRNA or FGF1 shRNA by lentiviral particles. Relative expression of WNT7A and FGF1 was determined by qPCR in control or FGF1 knockdown WNT7A overexpressing A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels. WNT7A or FGF1 protein levels were confirmed by western blot analysis using antibodies for V5 for WNT7A and FGF1.
Effects of WNT7A and/or FGF1 on cell proliferation and adhesion. (a) Overexpression of WNT7A or FGF1 affects cell doubling time and cell adhesion. Different letters denote groups that have statistically significant (P<0.05) differences in mean time or O.D. (b) Loss of FGF1 attenuates WNT7A-mediated cell doubling time and cell adhesion. Different letters denote groups that have statistically significant (P<0.05) differences in mean time or O.D.
Inhibitory mechanisms of niclosamide in ovarian cancer cells. (a) Niclosamide is the most effective WNT7A inhibitor in ovarian cancer. A total of 14 small molecules were examined for TCF/LEF luciferase promoter (TOPFLASH) inhibitory activity in A2780 cells stimulated with WNT7A. Three doses of each compound were used (bafilomycin; 1, 5 and 10 nM, and all others; 1, 5 and 10 μM), based on manufacturer recommendation. Data shown is the result for 5 nM of bafilomycin and 5 μM of the others. Different letters denote inhibitors that have statistically significant (P<0.05) differences in mean TOPFLASH activity. (b) WNT7A expression is inhibited by niclosamide in SKOV3.ip1 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression. (c) Dose dependent effect of niclosamide on TOPFLASH activity in A2780 cells stimulated by WNT7A or in SKOV3.ip1 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean TOPFLASH activity. (d) Effect of niclosamide on TOPFLASH activity stimulated by a constitutively active β-catenin (S33Y, codon 33 substitution of tyrosine for serine) in A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean TOPFLASH activity.
Effects of niclosamide are independent from FGF1 status. (a) FGF1 mRNA and protein expression following niclosamide exposure in SKOV3.ip1. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression. (b) Effects of niclosamide on cell viability and death in WNT7A or FGF1 overexpressing A2780 cells. (c) Effects of niclosamide on cell viability and death in control or FGF1 knockdown WNT7A overexpressing A2780 cells.
Acknowledgments
We thank Gail Isenberg for editing the manuscript. This work was supported by NIH/NCI CA179214 and ACS-IL 139038 (to KH), and NIH/NICHD HD065584 (to JAM).
Footnotes
No potential conflicts of interest were declared.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Goff BA, Mandel LS, Melancon CH, Muntz HG. Frequency of symptoms of ovarian cancer in women presenting to primary care clinics. Jama. 2004;291:2705–2712. doi: 10.1001/jama.291.22.2705. [DOI] [PubMed] [Google Scholar]
- 3.Teo R, Mohrlen F, Plickert G, Muller WA, Frank U. An evolutionary conserved role of Wnt signaling in stem cell fate decision. Developmental biology. 2006;289:91–99. doi: 10.1016/j.ydbio.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Polakis P. Wnt signaling and cancer. Genes & development. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 5.Liu W, Xing F, Iiizumi-Gairani M, Okuda H, Watabe M, Pai SK, et al. N-myc downstream regulated gene 1 modulates Wnt-beta-catenin signalling and pleiotropically suppresses metastasis. EMBO molecular medicine. 2012;4:93–108. doi: 10.1002/emmm.201100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polakis P. The many ways of Wnt in cancer. Current opinion in genetics & development. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Gatcliffe TA, Monk BJ, Planutis K, Holcombe RF. Wnt signaling in ovarian tumorigenesis. Int J Gynecol Cancer. 2008;18:954–962. doi: 10.1111/j.1525-1438.2007.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arend RC, Londono-Joshi AI, Straughn JM, Jr, Buchsbaum DJ. The Wnt/beta-catenin pathway in ovarian cancer: a review. Gynecologic oncology. 2013;131:772–779. doi: 10.1016/j.ygyno.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 9.Yoshioka S, King ML, Ran S, Okuda H, MacLean JA, 2nd, McAsey ME, et al. WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/beta-catenin pathway. Mol Cancer Res. 2012;10:469–482. doi: 10.1158/1541-7786.MCR-11-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friesel RE, Maciag T. Molecular mechanisms of angiogenesis: fibroblast growth factor signal transduction. Faseb J. 1995;9:919–925. doi: 10.1096/fasebj.9.10.7542215. [DOI] [PubMed] [Google Scholar]
- 11.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocrine-related cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 12.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 13.Meng QH, Xu E, Hildebrandt MA, Liang D, Lu K, Ye Y, et al. Genetic variants in the fibroblast growth factor pathway as potential markers of ovarian cancer risk, therapeutic response, and clinical outcome. Clinical chemistry. 2014;60:222–232. doi: 10.1373/clinchem.2013.211490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith G, Ng MT, Shepherd L, Herrington CS, Gourley C, Ferguson MJ, et al. Individuality in FGF1 expression significantly influences platinum resistance and progression-free survival in ovarian cancer. British journal of cancer. 2012 doi: 10.1038/bjc.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu W, Lin C, Roberts MJ, Waud WR, Piazza GA, Li Y. Niclosamide suppresses cancer cell growth by inducing Wnt co-receptor LRP6 degradation and inhibiting the Wnt/beta-catenin pathway. PloS one. 2011;6:e29290. doi: 10.1371/journal.pone.0029290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mook RA, Jr, Chen M, Lu J, Barak LS, Lyerly HK, Chen W. Small molecule modulators of Wnt/beta-catenin signaling. Bioorganic & medicinal chemistry letters. 2013;23:2187–2191. doi: 10.1016/j.bmcl.2013.01.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osada T, Chen M, Yang XY, Spasojevic I, Vandeusen JB, Hsu D, et al. Antihelminth compound niclosamide downregulates Wnt signaling and elicits antitumor responses in tumors with activating APC mutations. Cancer research. 2011;71:4172–4182. doi: 10.1158/0008-5472.CAN-10-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sack U, Walther W, Scudiero D, Selby M, Kobelt D, Lemm M, et al. Novel effect of antihelminthic Niclosamide on S100A4-mediated metastatic progression in colon cancer. Journal of the National Cancer Institute. 2011;103:1018–1036. doi: 10.1093/jnci/djr190. [DOI] [PubMed] [Google Scholar]
- 19.Wieland A, Trageser D, Gogolok S, Reinartz R, Hofer H, Keller M, et al. Anticancer effects of niclosamide in human glioblastoma. Clin Cancer Res. 2013;19:4124–4136. doi: 10.1158/1078-0432.CCR-12-2895. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, Wang J, Lu J, Bond MC, Ren XR, Lyerly HK, et al. The anti-helminthic niclosamide inhibits Wnt/Frizzled1 signaling. Biochemistry. 2009;48:10267–10274. doi: 10.1021/bi9009677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer research. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 22.Aletti GD, Gallenberg MM, Cliby WA, Jatoi A, Hartmann LC. Current management strategies for ovarian cancer. Mayo Clinic proceedings. 2007;82:751–770. doi: 10.4065/82.6.751. [DOI] [PubMed] [Google Scholar]
- 23.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 24.Chotani MA, Chiu IM. Differential regulation of human fibroblast growth factor 1 transcripts provides a distinct mechanism of cell-specific growth factor expression. Cell Growth Differ. 1997;8:999–1013. [PubMed] [Google Scholar]
- 25.Madiai F, Hackshaw KV, Chiu IM. Characterization of the entire transcription unit of the mouse fibroblast growth factor 1 (FGF-1) gene. Tissue-specific expression of the FGF-1. A mRNA The Journal of biological chemistry. 1999;274:11937–11944. doi: 10.1074/jbc.274.17.11937. [DOI] [PubMed] [Google Scholar]
- 26.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nature reviews. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 27.Chen W, Chen M, Barak LS. Development of small molecules targeting the Wnt pathway for the treatment of colon cancer: a high-throughput screening approach. American journal of physiology Gastrointestinal and liver physiology. 2010;299:G293–300. doi: 10.1152/ajpgi.00005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodge ME, Lum L. Drugging the cancer stem cell compartment: lessons learned from the hedgehog and Wnt signal transduction pathways. Annual review of pharmacology and toxicology. 2011;51:289–310. doi: 10.1146/annurev-pharmtox-010510-100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer L, Flajolet M, Greengard P. Pharmacological inhibitors of glycogen synthase kinase 3. Trends in pharmacological sciences. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Steele IA, Edmondson RJ, Bulmer JN, Bolger BS, Leung HY, Davies BR. Induction of FGF receptor 2-IIIb expression and response to its ligands in epithelial ovarian cancer. Oncogene. 2001;20:5878–5887. doi: 10.1038/sj.onc.1204755. [DOI] [PubMed] [Google Scholar]
- 31.Zaid TM, Yeung TL, Thompson MS, Leung CS, Harding T, Co NN, et al. Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin Cancer Res. 2013;19:809–820. doi: 10.1158/1078-0432.CCR-12-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. The EMBO journal. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtin JC, Lorenzi MV. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget. 2010;1:563–577. doi: 10.18632/oncotarget.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 35.Yo YT, Lin YW, Wang YC, Balch C, Huang RL, Chan MW, et al. Growth inhibition of ovarian tumor-initiating cells by niclosamide. Molecular cancer therapeutics. 2012;11:1703–1712. doi: 10.1158/1535-7163.MCT-12-0002. [DOI] [PubMed] [Google Scholar]
- 36.Cooke PS, Spencer TE, Bartol FF, Hayashi K. Uterine glands: development, function and experimental model systems. Molecular human reproduction. 2013;19:547–558. doi: 10.1093/molehr/gat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassoon D. Wnt genes and endocrine disruption of the female reproductive tract: a genetic approach. Mol Cell Endocrinol. 1999;158:1–5. doi: 10.1016/s0303-7207(99)00170-7. [DOI] [PubMed] [Google Scholar]
- 38.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PloS one. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fonseca BD, Diering GH, Bidinosti MA, Dalal K, Alain T, Balgi AD, et al. Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. The Journal of biological chemistry. 2012;287:17530–17545. doi: 10.1074/jbc.M112.359638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Y, Lu Z, Ding K, Li J, Du X, Chen C, et al. Antineoplastic mechanisms of niclosamide in acute myelogenous leukemia stem cells: inactivation of the NF-kappaB pathway and generation of reactive oxygen species. Cancer research. 2010;70:2516–2527. doi: 10.1158/0008-5472.CAN-09-3950. [DOI] [PubMed] [Google Scholar]
- 41.Li R, Hu Z, Sun SY, Chen ZG, Owonikoko TK, Sica GL, et al. Niclosamide overcomes acquired resistance to erlotinib through suppression of STAT3 in non-small cell lung cancer. Molecular cancer therapeutics. 2013;12:2200–2212. doi: 10.1158/1535-7163.MCT-13-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.You S, Li R, Park D, Xie M, Sica GL, Cao Y, et al. Disruption of STAT3 by niclosamide reverses radioresistance of human lung cancer. Molecular cancer therapeutics. 2014;13:606–616. doi: 10.1158/1535-7163.MCT-13-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau MT, Klausen C, Leung PC. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via beta-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30:2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 45.Perl AK, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 46.Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Current opinion in cell biology. 1993;5:806–811. doi: 10.1016/0955-0674(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi K, Erikson DW, Tilford SA, Bany BM, Maclean JA, 2nd, Rucker EB, 3rd , et al. Wnt genes in the mouse uterus: potential regulation of implantation. Biology of reproduction. 2009;80:989–1000. doi: 10.1095/biolreprod.108.075416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi K, Burghardt RC, Bazer FW, Spencer TE. WNTs in the ovine uterus: potential regulation of periimplantation ovine conceptus development. Endocrinology. 2007;148:3496–3506. doi: 10.1210/en.2007-0283. [DOI] [PubMed] [Google Scholar]
- 49.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer research. 2009;69:765–774. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 50.Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer research. 2004;64:622–630. doi: 10.1158/0008-5472.can-03-2636. [DOI] [PubMed] [Google Scholar]
- 51.Maclean JA, 2nd, Hu Z, Welborn JP, Song HW, Rao MK, Wayne CM, et al. The RHOX Homeodomain Proteins Regulate the Expression of Insulin and Other Metabolic Regulators in the Testis. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.486340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 53.Denkert C, Budczies J, Darb-Esfahani S, Gyorffy B, Sehouli J, Konsgen D, et al. A prognostic gene expression index in ovarian cancer - validation across different independent data sets. The Journal of pathology. 2009;218:273–280. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- 54.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer research. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FGF gene expression. Relative FGF gene expression in WNT7A knockdown cells, orthotopic tumors that developed from WNT7A knockdown cells, WNT7A overexpressing cells and orthotopic tumors that developed from WNT7A overexpressing cells. N.D. means non-detectable levels by qPCR.
FGF receptor gene expression. Relative FGFR gene expression in WNT7A knockdown cells, orthotopic tumors that developed from WNT7A knockdown cells, WNT7A overexpressing cells and orthotopic tumors that developed from WNT7A overexpressing cells.
Expression of WNT7A and FGF1 in ovarian cancer cell lines. Relative expression of WNT7A and FGF1 was assessed by qPCR in human ovarian cancer cell lines. Data were set to background level of 1 in A2780 cells.
FGF1 expression in DN-TCF4 expressing SKOV3.ip1 cells. (a) DN-TCF4 expressing SKOV3.ip1 cells were generated using the T-REx system with pcDNA6/TR and pcDNA5/TO vectors. Relative expression of WNT7A, FGF1 and AXIN2 as a positive control was assessed by qPCR in control or DN-TCF4 expressing SKOV3.ip1 cells. (b) Relative expression of FGF1 1A, 1B, 1C and 1D transcripts in control or DN-TCF4 expressing SKOV3.ip1 cells. N.D. means non-detectable levels by qPCR.
Gene manipulation of WNT7A and FGF1 in ovarian cancer cells. (a) V5-tagged WNT7A or FGF1 were sub-cloned into pcDNA5/FRT vector. A2780 cells were transfected with empty vector or plasmids encoding WNT7A-V5 or FGF1 using the Flp-In system to establish WNT7A or FGF1 overexpressing cell lines. Relative expression of WNT7A and FGF1 was determined by qPCR in WNT7A or FGF1 overexpressing A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels. WNT7A or FGF1 protein levels were confirmed by western blot analysis using antibodies of V5 for WNT7A and FGF1. (b) WNT7A overexpressing cells were transfected with control shRNA or FGF1 shRNA by lentiviral particles. Relative expression of WNT7A and FGF1 was determined by qPCR in control or FGF1 knockdown WNT7A overexpressing A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression levels. WNT7A or FGF1 protein levels were confirmed by western blot analysis using antibodies for V5 for WNT7A and FGF1.
Effects of WNT7A and/or FGF1 on cell proliferation and adhesion. (a) Overexpression of WNT7A or FGF1 affects cell doubling time and cell adhesion. Different letters denote groups that have statistically significant (P<0.05) differences in mean time or O.D. (b) Loss of FGF1 attenuates WNT7A-mediated cell doubling time and cell adhesion. Different letters denote groups that have statistically significant (P<0.05) differences in mean time or O.D.
Inhibitory mechanisms of niclosamide in ovarian cancer cells. (a) Niclosamide is the most effective WNT7A inhibitor in ovarian cancer. A total of 14 small molecules were examined for TCF/LEF luciferase promoter (TOPFLASH) inhibitory activity in A2780 cells stimulated with WNT7A. Three doses of each compound were used (bafilomycin; 1, 5 and 10 nM, and all others; 1, 5 and 10 μM), based on manufacturer recommendation. Data shown is the result for 5 nM of bafilomycin and 5 μM of the others. Different letters denote inhibitors that have statistically significant (P<0.05) differences in mean TOPFLASH activity. (b) WNT7A expression is inhibited by niclosamide in SKOV3.ip1 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression. (c) Dose dependent effect of niclosamide on TOPFLASH activity in A2780 cells stimulated by WNT7A or in SKOV3.ip1 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean TOPFLASH activity. (d) Effect of niclosamide on TOPFLASH activity stimulated by a constitutively active β-catenin (S33Y, codon 33 substitution of tyrosine for serine) in A2780 cells. Different letters denote groups that have statistically significant (P<0.05) differences in mean TOPFLASH activity.
Effects of niclosamide are independent from FGF1 status. (a) FGF1 mRNA and protein expression following niclosamide exposure in SKOV3.ip1. Different letters denote groups that have statistically significant (P<0.05) differences in mean expression. (b) Effects of niclosamide on cell viability and death in WNT7A or FGF1 overexpressing A2780 cells. (c) Effects of niclosamide on cell viability and death in control or FGF1 knockdown WNT7A overexpressing A2780 cells.