Abstract
Methanol is already an important carbon feedstock in the chemical industry, but it has found only limited application in biotechnological production processes. This can be mostly attributed to the inability of most microbial platform organisms to utilize methanol as a carbon and energy source. With the aim to turn methanol into a suitable feedstock for microbial production processes, we engineered the industrially important but nonmethylotrophic bacterium Corynebacterium glutamicum toward the utilization of methanol as an auxiliary carbon source in a sugar-based medium. Initial oxidation of methanol to formaldehyde was achieved by heterologous expression of a methanol dehydrogenase from Bacillus methanolicus, whereas assimilation of formaldehyde was realized by implementing the two key enzymes of the ribulose monophosphate pathway of Bacillus subtilis: 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase. The recombinant C. glutamicum strain showed an average methanol consumption rate of 1.7 ± 0.3 mM/h (mean ± standard deviation) in a glucose-methanol medium, and the culture grew to a higher cell density than in medium without methanol. In addition, [13C]methanol-labeling experiments revealed labeling fractions of 3 to 10% in the m + 1 mass isotopomers of various intracellular metabolites. In the background of a C. glutamicum Δald ΔadhE mutant being strongly impaired in its ability to oxidize formaldehyde to CO2, the m + 1 labeling of these intermediates was increased (8 to 25%), pointing toward higher formaldehyde assimilation capabilities of this strain. The engineered C. glutamicum strains represent a promising starting point for the development of sugar-based biotechnological production processes using methanol as an auxiliary substrate.
INTRODUCTION
Highly productive strains of Corynebacterium glutamicum have been developed to produce several million tons of amino acids annually, in particular the feed additive l-lysine and the flavor enhancer l-glutamate. In addition, extensive research has focused on engineering C. glutamicum for the microbial production of a variety of other commercially interesting compounds (1), such as organic acids (2–4), diamines (5–7), and alcohols (8–10). The natural substrate spectrum of C. glutamicum includes sugars, organic acids, and alcohols, but for industrial production processes mainly glucose (from starch) or sucrose and fructose (from molasses) are used as carbon sources (11–13). The availability and price of sugars are influenced by seasonal variations and weather conditions and are also subject to price regulations and import limitations imposed on agricultural products. Furthermore, due to the increasing world population and loss of arable land, the sugar price is expected to rise in the coming decades. Thus, there is demand for an alternative carbon source for the microbial production of valuable compounds (14). In recent years, C. glutamicum has been genetically engineered toward the ability to efficiently utilize several cheap carbon sources, such as the tricarboxylic acid cycle intermediates malate, fumarate, and succinate and the lignocellulosic compounds arabinose and xylose, as well as starch, cellobiose, glycerol, lactose, galactose, and glucosamine (references 11 and 15 and references therein). Cheap raw methanol that may contain impurities, such as ethanol, higher alcohols, or water, is already an important carbon feedstock in the chemical industry and also represents an interesting alternative substrate. At present, methanol is mainly produced from synthesis gas (a mixture of CO and H2), which is obtained by catalytic reforming of coal or natural gas. In addition, procedures are available for producing methanol from renewable carbon sources (16–19). The high availability and low market price of methanol raise the question of whether this C1 compound could also serve as alternative carbon source for microbial production processes (20, 21). Although C. glutamicum harbors an endogenous pathway for oxidation of methanol to CO2 (22, 23), it is a nonmethylotrophic organism and therefore is not able to utilize C1 compounds as the sole carbon and energy source.
A key step in methylotrophic metabolism is the oxidation of methanol to formaldehyde. Whereas Gram-negative methylotrophic bacteria such as Methylobacterium extorquens employ pyrroloquinoline quinone (PQQ)-dependent and periplasmic methanol dehydrogenases (MDHs) to oxidize methanol (24), Gram-positive thermotolerant Bacillus strains usually use NAD+-dependent cytoplasmic methanol dehydrogenases (25). The cytotoxic formaldehyde is either assimilated into cell material or further oxidized to carbon dioxide (26, 27). The assimilation of C1 compounds in methylotrophic bacteria occurs either via the ribulose monophosphate (RuMP) pathway, the serine cycle, or the Calvin-Benson-Bassham cycle. Whereas CO2 is reduced and converted to biomass in the Calvin-Benson-Bassham cycle, assimilation of carbon in the serine cycle occurs at the level of methylene-H4F and CO2 (28). In the RuMP pathway, formaldehyde and ribulose-5-phosphate are condensed to form d-arabino-3-hexulose-6-phosphate, which can be isomerized to fructose-6-phosphate. These reactions are catalyzed by 3-hexulose-6-phosphate synthase (HPS) and 6-phosphate-3-hexuloisomerase (PHI), respectively. Fructose-6-phosphate can be converted to pyruvate via glycolysis or the Entner-Doudoroff pathway, or it can be used to regenerate the formaldehyde acceptor ribulose-5-phosphate by utilizing several reactions of the pentose phosphate pathway (29).
In this work, we describe the functional implementation of methanol oxidation and formaldehyde assimilation via the RuMP pathway in C. glutamicum and present promising approaches to use methanol as an auxiliary substrate during growth in sugar-based defined medium.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used or constructed in the course of this work are listed in Table 1. C. glutamicum was routinely cultivated aerobically in either 500-ml baffled shake flasks with 50 ml medium on a rotary shaker (120 rpm) at 30°C or in 48-well FlowerPlates (m2p-labs, Aachen, Germany) filled with 750 μl medium in a BioLectorBasic apparatus (m2p-labs, Aachen, Germany) at 900 rpm, 30°C, and 80% humidity. Growth in shake flasks was monitored by measuring the optical density at 600 nm (OD600), and growth in the BioLectorBasic was monitored online by measuring the backscatter (BS) at 620 nm (gain, 12). The gravimetric determination of cell dry weight (CDW) was performed in triplicate by centrifugation of 2 ml culture broth in predried and preweighed 2-ml Eppendorf tubes. The pelleted cells were dried for 48 h at 80°C and weighed afterwards. LB medium (30) was used for 5-ml precultures, and main cultures were grown in modified CGXII defined medium (31) containing 55 mM glucose. Methanol was added to the culture medium to a final concentration of 120 mM. For strain construction and maintenance, either LB or BHIS agar plates (brain heart infusion [BHI] agar [Difco, Detroit, MI, USA] supplemented with 0.5 M sorbitol) were used. Escherichia coli DH5α was used for cloning purposes and was grown aerobically on a rotary shaker (170 rpm) at 37°C in 5 ml LB medium or on LB agar plates (LB medium with 1.8% [wt/vol] agar). If appropriate, kanamycin and/or tetracycline was added to final concentrations of 25 μg ml−1 or 5 μg ml−1, respectively (C. glutamicum) or 50 μg ml−1 or 12.5 μg ml−1 (E. coli). Correlations between BS values and the CDW were determined by using the following calibration model formula (32, 33): CDW (g liter−1) = 0.048 g liter−1 × BS − 0.78 g liter−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli DH5α | F− ϕ80dlacΔ(lacZ)M15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 phoA supE44 λ− gyrA96 relA1 | Invitrogen (Karlsruhe, Germany) |
| B. subtilis 168 | Wild type, trpC2, auxotrophic for tryptophan | BGSCa |
| C. glutamicum strains | ||
| ATCC 13032 | Biotin auxotroph, wild-type strain | 36 |
| Δald ΔadhE | Derivative of ATCC 13032 with in-frame deletion of ald (cg3096) and adhE (cg0387) | 22 |
| Plasmids | ||
| pEKEx2 | Kanr; C. glutamicum-E. coli shuttle vector for regulated gene expression; Ptac, lacIq pBL1 oriVCg pUC18 oriVEc | 37 |
| pVWEx2 | Tetr; C. glutamicum-E. coli shuttle vector for regulated gene expression; Ptac, lacIq pCG1 oriVCg pUC18 oriVEc | 38 |
| pVWEx2-Bm(mdh-act) | Tetr; pVWEx2 derivative containing Bm(mdh-act) under control of Ptac | This work |
| pVWEx2-Ptuf-Bm(mdh-act) | Tetr; pVWEx2 derivative containing Bm(mdh-act) under control of Ptuf | This work |
| pVWEx2-Bm(mdh3-act) | Tetr; pVWEx2 derivative containing Bm(mdh3-act) under control of Ptac | This work |
| pVWEx2-Ptuf-Bm(mdh3-act) | Tetr; pVWEx2 derivative containing Bm(mdh3-act) under control of Ptuf | This work |
| pVWEx2-CgadhA | Tetr; pVWEx2 derivative containing CgadhA under control of Ptac | This work |
| pEKEx2-Bs(hps-phi) | Kanr; pEKEx2 derivative containing Bs(hps-phi) under control of Ptac | This work |
| pEKEx2- Ptuf -Bs(hps-phi) | Kanr; pEKEx2 derivative containing Bs(hps-phi) under control of Ptuf | This work |
| pEKEx2-Mg(hps-phi) | Kanr; pEKEx2 derivative containing Mg(hps-phi) under control of Ptac | This work |
BGSC, Bacillus Genetic Stock Center.
Strain construction.
The enzymes for recombinant DNA work were obtained from Fermentas (St. Leon-Rot, Germany) and Merck Millipore (Billerica, MA, USA). E. coli was transformed by using the RbCl method (34), and C. glutamicum ATCC 13032 was transformed via electroporation (35). Routine methods, like PCR, restriction, and ligation, were carried out according to standard protocols (30). The oligonucleotides used for cloning were obtained from Eurofins MWG Operon (Ebersberg, Germany) and are listed in Table S1 in the supplemental material. The genes mdh, mdh3, and act, originating from Bacillus methanolicus MGA3 (ATCC 53907; NCBI gene accession numbers EIJ77596, EIJ80770, and AAM98772, respectively) were optimized for expression in C. glutamicum by adapting the gene sequence to the codon usage of C. glutamicum. These genes, as well as the wild-type coding sequence of the operon consisting of rmpA (here hps) and rmpB (here phi) from M. gastri (NCBI gene accession numbers Q9LBW4 and Q9LBW5, respectively) were ordered from Life Technologies GmbH (Darmstadt, Germany). All genes, synthesized with desired restriction sites for subcloning into C. glutamicum expression vectors, were provided as purified DNA on standard vectors (Life Technologies). The adhA gene (cg3107) was amplified from genomic DNA of C. glutamicum ATCC 13032 by PCR. The genes yckG (here hps) and yckF (here phi) were amplified from genomic DNA of Bacillus subtilis 168 by PCR. For heterologous gene expression under the control of a constitutive promoter, Ptuf (promoter of the elongation factor Tu) was first amplified by PCR from genomic DNA of C. glutamicum (14 bp to 179 bp upstream of the tuf start codon) and subsequently cloned in front of the above-mentioned genes. The correct DNA sequences were verified by DNA sequencing using plasmid-specific primers. Detailed cloning procedures of the constructed vectors are described in the supplemental material.
Determination of methanol and glucose.
The quantitative measurement of methanol and glucose in the cell-free supernatant of C. glutamicum cultures was performed by capillary gas chromatography (GC) using an Agilent 7890A gas chromatograph (Agilent Technologies, Waldbronn, Germany) and by high-performance liquid chromatography (HPLC) analysis using an Agilent 1100 HPLC system (Agilent Technologies, Waldbronn, Germany), respectively, as described previously (22, 39).
Enzyme assays of crude extracts.
Recombinant C. glutamicum strains were cultivated in 100 ml CGXII defined medium with 55 mM glucose and 120 mM methanol at 30°C and 120 rpm in shake flasks. Induction of gene expression was performed with 1.5 mM isopropyl-β-d-thiogalactpyranoside (IPTG) at an OD600 of 1.0. For the determination of enzyme activities in the exponential phase and in the second growth phase, 50 ml of the cell culture was harvested when the OD600 reached 5 and 30 ml was harvested after 12 h of cultivation by centrifugation (4,500 × g, 15 min, 4°C). Cells were washed with 100 mM glycine-KOH buffer, pH 9.4 (for the Mdh/Mdh3 enzyme assay) or with 50 mM potassium phosphate buffer, pH 7.6, containing 1 mM dithiothreitol and 3 mM MgCl2 (for the HPS/PHI enzyme assays). The cell pellets from the exponential phase and the second growth phase were resuspended in 500 μl and 800 μl buffer, respectively. For crude cell extract preparation, mechanical lysis of cells was performed with glass beads (diameter, 0.1 mm; 350 mg in a 1.5-ml screw-cap tube) for three times, 20 s each, using the Precellys24 apparatus (Bertin Technologies, Montigny-le-Bretonneux, France) and centrifuged for 30 min at 4°C and 16.000 × g to remove the cell debris. The supernatant was used for the enzyme assays. The enzyme assays were performed in 96-well plates in 200-μl scale at 30°C by following the increase in absorption at 340 nm with the Infinite M200 Pro instrument (Tecan Group AG, Männedorf, Switzerland). The protein concentration of the cell extracts was determined by the method of Bradford (40) using bovine serum albumin as the standard.
The methanol dehydrogenase assay was performed as described previously (41) with slight modifications. The assay mixtures contained 100 mM glycine-KOH buffer, pH 9.4, 5 mM MgSO4, 1 mM dithiothreitol, 1 mM NAD+, and 40 μl cell extract at different dilutions. The reaction was initiated by the addition of 500 mM methanol and monitored over 3 min. One unit of methanol dehydrogenase activity was defined as the reduction of 1 μmol NAD+ to NADH per minute.
The coupled HPS/PHI assay was performed as described previously (42), but with minor modifications of the protocol. Briefly, determination of HPS/PHI activities through measurement of NADPH formation requires the activity of three additional enzymes in the assay mixture: phosphoriboisomerase (PRI), phosphoglucoisomerase (PGI), and glucose-6-phosphate dehydrogenase (G6PDH). The assay mixtures contained 50 mM potassium phosphate buffer, pH 7.6, 5 mM MgCl2, 5 mM ribose-5-phosphate, 2.5 mM NADP+, 5 U PGI from yeast (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), 5 U G6PDH from yeast (grade II; Roche Diagnostics Deutschland GmbH, Mannheim, Germany), 5 U PRI from spinach (type I, partially purified powder; Sigma-Aldrich, St. Louis, MO, USA), and 40 μl cell extract at different dilutions. The reaction mixture was incubated for 5 min at 30°C to ensure equilibrium between ribose-5-phosphate and ribulose-5-phosphate. Subsequently, formaldehyde (37% formaldehyde solution; Sigma-Aldrich, St. Louis, MO, USA) was added to a final concentration of 5 mM to start the reaction. The reaction was monitored for 15 min. One unit of coupled HPS/PHI activity was defined as the reduction of 1 μmol NADP+ to NADPH per minute.
Determination of intracellular formaldehyde.
Recombinant C. glutamicum strains were cultivated in 100 ml CGXII defined medium with 55 mM glucose and 120 mM methanol at 30°C and 120 rpm in shake flasks. Sampling in the exponential and second growth phases as well as mechanical cell lysis and crude cell extract preparation were performed as described above in “Enzyme assays of crude extracts.” The crude cell extract was analyzed for formaldehyde content via an enzymatic assay described by Nudelman et al. (43).
Determination of 13C-labeled intracellular metabolites and 13CO2.
The assimilation of methanol in recombinant C. glutamicum strains was monitored in [13C]methanol-labeling experiments. Cells were cultivated in 200 ml CGXII defined medium with 55 mM glucose and 120 mM 13C-labeled methanol (99% atom enrichment; Sigma-Aldrich, St. Louis, MO, USA) in a parallel bioreactor system (DASGIP AG, Jülich, Germany). During the cultivation, 13CO2 and 12CO2 off-gas analysis was performed as described previously (22). Analysis of the 13C-labeled intracellular metabolites was performed in the exponential growth phase, in the transition from the exponential growth phase to the second growth phase, and also 12 h and 24 h after reaching the second growth phase. Quenching of the metabolic activity as well as extraction and analysis of intracellular metabolites were performed as previously described (44). Raw mass spectrometry data were corrected for the contribution of all naturally abundant isotopes as well the isotopic impurity of the tracer by using the software IsoCor (45).
RESULTS
Design of a synthetic pathway for the assimilation of methanol-derived carbon in C. glutamicum.
The first step of engineering C. glutamicum toward utilization of methanol was the heterologous expression of a MDHs to oxidize methanol to formaldehyde (Fig. 1). The focus was put on NAD+-dependent MDHs, since PQQ-dependent MDHs are not suitable for expression in C. glutamicum, as this organism does not synthesize PQQ or possess any PQQ-dependent enzymes (46). B. methanolicus MGA3 is a well-known methylotroph that harbors three NAD+-dependent MDHs genes (mdh, mdh2, and mdh3) and the MDH activator protein (Act), which modulates the activity of all three MDHs (47, 48). In the presence of Act and with methanol as the substrate, Mdh and Mdh3 shows the highest activity in vitro (0.5 U/mg and 0.2 U/mg, respectively), whereas Mdh2 shows the lowest catalytic activity (0.14 U/mg) (48). Thus, genes for mdh, mdh3, and act were codon optimized for expression in C. glutamicum and commercially synthesized [Bm(mdh-act) and Bm(mdh3-act)]. In addition, the AdhA of C. glutamicum was previously shown to contribute to endogenous oxidation of methanol to formaldehyde (22). With the aim to simply increase the endogenous methanol oxidation rate, the respective gene (CgadhA) was also chosen as a target for overexpression in C. glutamicum.
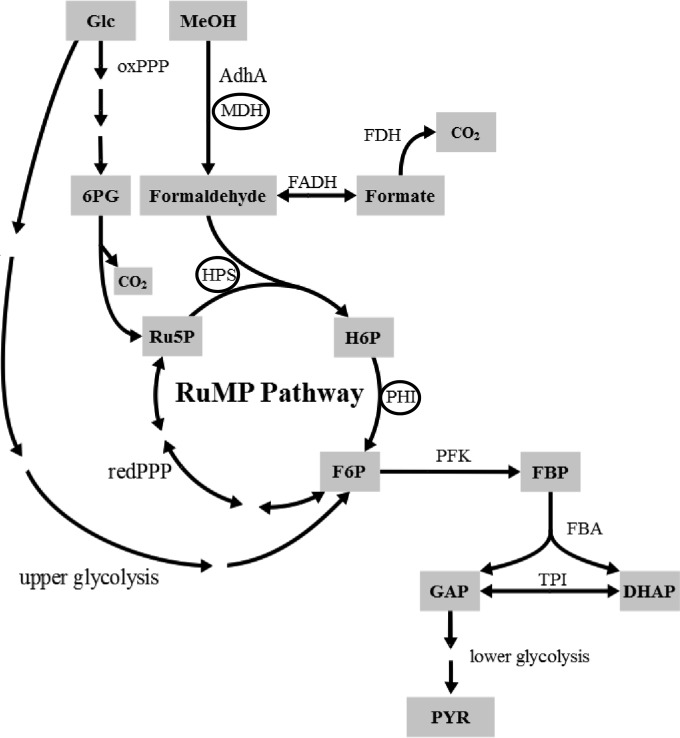
FIG 1.
Simplified overview of the (engineered) oxidation of methanol, the endogenous pathway for formaldehyde oxidation, and the synthetic pathway for the assimilation of formaldehyde via the RuMP pathway in the context of the central carbon metabolism of C. glutamicum. Abbreviations: oxPPP/redPPP, oxidative/reductive pentose phosphate pathway; MDH, methanol dehydrogenase; AdhA, alcohol dehydrogenase; FADH, formaldehyde dehydrogenase; FDH, formate dehydrogenase; HPS, 3-hexulose-6-phosphate synthase; PHI, 6-phosphate-3-hexuloisomerase; PFK, phosphofructokinase; FBA, fructose bisphosphate aldolase; TPI, triosephosphate isomerase; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; H6P, hexulose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; PYR, pyruvate. The three enzymes (MDH, HPS, and PHI) necessary for establishing a synthetic pathway for methanol oxidation and formaldehyde assimilation in C. glutamicum are circled.
We decided to pursue the implementation of the RuMP pathway for methanol assimilation in C. glutamicum in order to establish that this pathway demands only the functional expression of genes for HPS and PHI. All other enzymatic activities required for the conversion of fructose-6-phosphate and regeneration of ribulose-5-phosphate as a C1 acceptor can be recruited from the central metabolism of C. glutamicum (Fig. 1). Furthermore, these two enzymes require no cofactors, and the assimilation of C1 via the RuMP pathway occurs directly on the level of formaldehyde and not on the level of methylene-H4F and/or CO2, as in the serine cycle (28, 29). Hence, we decided to evaluate the functionality of the HPS and PHI from the nonmethylotroph B. subtilis [Bs(hps-phi)] and from the methylotroph Mycobacterium gastri [Mg(hps-phi)] to constitute a functional RuMP pathway in C. glutamicum.
Implementation of a synthetic pathway for methanol oxidation and formaldehyde assimilation in wild-type C. glutamicum.
For growth on methanol, methylotrophic organisms require a pathway for the assimilation of formaldehyde to generate biomass, as well as a pathway for the dissimilation of formaldehyde to generate energy. The dissimilatory pathway also serves as “safety valve” in case of an intracellular accumulation of formaldehyde to toxic amounts. Since the wild-type C. glutamicum already possesses an endogenous pathway for the oxidation of naturally occurring cytotoxic formaldehyde to CO2 (22, 23), we decided to initially express the genes of the synthetic methanol assimilation pathway in this strain. Two plasmids allowing for heterologous gene expression under the control of the IPTG-inducible promoter Ptac were used: one for the expression of the methanol oxidation modules [CgadhA or Bm(mdh-act)] and one for the expression of the formaldehyde assimilation modules [Bs(hps-phi) or Mg(hps-phi)]. Thus, CgadhA or Bm(mdh-act) were individually expressed in combination with Bs(hps-phi) or Mg(hps-phi) in C. glutamicum to constitute a functional pathway for methanol oxidation and subsequent assimilation of the generated formaldehyde into biomass.
First, the activities of the formaldehyde assimilation modules were evaluated in crude extracts of recombinant C. glutamicum strains in in vitro enzyme assays. These assays showed that only the Bs(Hps-Phi) module was functionally active in C. glutamicum (73 ± 25 mU/mg in the exponential phase [mean ± standard deviation]), whereas only negligible activity (1.4 ± 2 mU/mg) could be detected for the Mg(Hps-Phi) module (Table 2). Consequently, strains expressing the Mg(hps-phi) module were not further characterized. The functionality of the methanol oxidation modules was tested by following the methanol oxidation rate of recombinant C. glutamicum strains during shake flask cultivations in defined medium containing 55 mM glucose and 120 mM methanol. The strain expressing CgadhA and Bs(hps-phi) consumed methanol only slightly faster than the control strain C. glutamicum(pVWEx2 pEKEx2) (data not shown). In contrast, the methanol oxidation rate of the recombinant C. glutamicum strain expressing Bm(mdh-act) and Bs(hps-phi) was nearly 3-fold higher (1.3 ± 0.2 mM/h) than in the control strain (0.5 mM/h ± 0.1) (Fig. 2A). Due to these results, we excluded the strain expressing the Cg(adhA) module from further experiments and put our focus on the C. glutamicum strain expressing the Bm(mdh-act) and Bs(hps-phi) modules.
TABLE 2.
Specific activities of Bm(Mdh-Act) and Bm(Mdh3-Act) and coupled specific activities of Bs(Hps-Phi) and Mg(Hps-Phi) in crude cell extracts of C. glutamicum strainsa
| Strain | Coupled HPS-PHI sp act (mU/mg) during: |
Mdh/Mdh3 sp act (mU/mg) during: |
||
|---|---|---|---|---|
| Exponential phase | Second growth phase | Exponential phase | Second growth phase | |
| Recombinant C. glutamicum strains | ||||
| Bm(mdh-act) Bs(hps-phi) | 73.3 ± 24.8 | 22.1 ± 7.9 | 0.4 ± 0.2 | 0.7 ± 0.5 |
| Bm(mdh-act) Mg(hps-phi) | 1.4 ± 1.6 | 1.0 ± 1.4 | ND | ND |
| Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) | 53.1 ± 23.5 | 13.7 ± 1.3 | 3.3 ± 1.2 | 2.0 ± 1.5 |
| Recombinant C. glutamicum Δald ΔadhE strains | ||||
| Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) | 45.3 ± 9.7 | 24.3 ± 15.3 | 5.5 ± 1.1 | 2.2 ± 1.8 |
| Ptuf-Bm(mdh3-act) Ptuf-Bs(hps-phi) | 36.3 ± 4.9 | 22.3 ± 12.1 | 6.0 ± 1.5 | 5.2 ± 1.9 |
| Ptuf-Bm(mdh-act) pEKEx2 | ND | ND | 5.5 ± 1.1 | 3.2 ± 2.3 |
Cells were cultivated in CGXII defined medium with 55 mM glucose and 120 mM methanol at 30°C with shaking at 120 rpm. Genes under the control of Ptac were induced with 1.5 mM IPTG at an OD600 of 1.0. Means and standard deviations were calculated from triplicates, and values were corrected for background activity. The background activity was determined in enzyme assays with the crude cell extracts of C. glutamicum(pVWEx2 pEKEx2) and C. glutamicum Δald ΔadhE(pVWEx2 pEKEx2). ND, not determined.
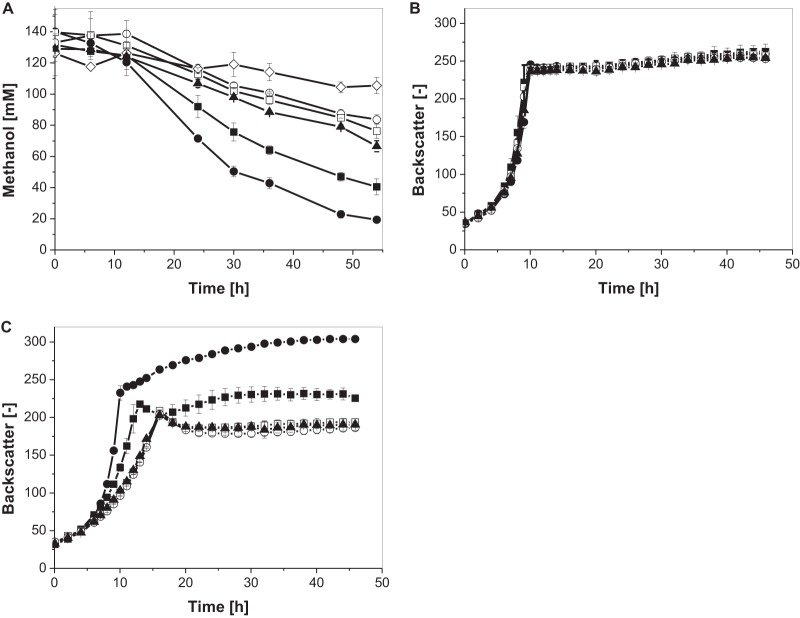
FIG 2.
(A) Methanol consumption in CGXII defined medium supplemented with 55 mM glucose and 120 mM methanol. (B) Growth in CGXII defined medium supplemented with 55 mM glucose. (C) Growth in CGXII defined medium supplemented with 55 mM glucose and 120 mM methanol of recombinant C. glutamicum strains. Induction of gene expression was achieved with 1.5 mM IPTG in the case of Ptac-controlled gene expression. Three independent shake-flask experiments, always accompanied by a methanol evaporation control, were performed for the determination of the methanol concentration (◇). Monitoring of growth was performed in 48-well FlowerPlates by using a BioLectorBasic apparatus (n = 3). Strains: C. glutamicum Bm(mdh-act) Bs(hps-phi) (■), C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) (●), C. glutamicum Ptuf-Bm(mdh-act) pEKEx2 (▲), C. glutamicum pVWEx2 pEKEx2 (+ IPTG) (□), and C. glutamicum pVWEx2 pEKEx2 (- IPTG) (○).
Enzyme assays conducted with crude extracts revealed only negligible MDH activity in the exponential growth phase, whereas slightly higher activity was detected in the second growth phase (0.7 ± 0.5 mU/mg) (Table 2). These results were in line with the observation that methanol oxidation was more pronounced in the second growth phase (Fig. 2A). Since IPTG-inducible expression of Bm(mdh-act) seems not to be suitable due to the low activity of Bm(Mdh-Act) in the exponential growth phase, we also evaluated the application of a strong constitutive promoter to control heterologous gene expression. For this purpose, the expression of both modules, Bm(mdh-act) and Bs(hps-phi), was set under the control of the constitutive promoter Ptuf. Enzyme assays with crude cell extract of the respective strain revealed that the specific MDH activity in the exponential growth phase was significantly increased, from 0.4 ± 0.2 mU/mg to 3.3 ± 1.2 mU/mg (Table 2). During shake flask experiments for comparing the effect of constitutive and inducible heterologous expression of both modules on methanol consumption, no significant difference was observed in the exponential growth phase. This could be explained by the low biomass at the beginning of the cultivations, leading to only a slow consumption of the supplied methanol in the culture supernatant. These differences could not be reliably distinguished from the background of evaporation of this volatile alcohol. However, over the course of the whole cultivation, a higher average methanol consumption rate (1.7 ± 0.3 mM/h) tha in the strain expressing both modules under the control of the inducible promoter Ptac (1.3 ± 0.2 mM/h) was obtained (Fig. 2A). In the second growth phase, these results were even more significant. The specific methanol consumption rates for both strains were 6.49 ± 0.4 μM/(h · BS) and 5.78 ± 0.2 μM/(h · BS), respectively.
Initial experiments showed that all recombinant strains and the control strain C. glutamicum(pVWEx2 pEKEx2) showed comparable growth behaviors in medium containing 55 mM glucose without methanol (Fig. 2B). In the presence of 120 mM methanol, major differences in growth and final backscatter of the cultures were observed (Fig. 2C). The growth of the control strain and of the recombinant strain expressing the methanol oxidation and formaldehyde assimilation modules under the control of Ptac were significantly retarded, revealing an inhibitory effect of methanol. The latter strain could cope slightly better with methanol, as it grew faster during the exponential growth phase. However, this growth advantage cannot be explained by the observed higher methanol oxidation of the recombinant strain (1.3 ± 0.2 mM/h) in comparison to the control strain (0.5 ± 0.1 mM/h) during the course of the cultivation, since both strains hardly consumed methanol in the exponential growth phase (Fig. 2A). At the end of the exponential growth phase, backscatter measurements showed a decrease in cell density in both cultures (Fig. 2C). In contrast to the control culture, the C. glutamicum Bm(mdh-act) Bs(hps-phi) culture showed a second growth phase, leading to a significantly (1.3-fold) higher final backscatter (Fig. 2C) and an increased CDW by 4% (Table 3) in comparison to the control strain. The same was observed for a C. glutamicum strain that exclusively expressed the methanol oxidation module but not the formaldehyde assimilation module (see Fig. S1 in the supplemental material). This led to the assumption that the observed higher final backscatter of the C. glutamicum Bm(mdh-act) Bs(hps-phi) culture compared to the control strain C. glutamicum(pVWEx2 pEKEx2) does not reflect the assimilation of methanol-derived carbon but could be due to a lower methanol concentration in the medium during the second growth phase, thereby diminishing its inhibitory effect.
TABLE 3.
Cell dry weight measures for C. glutamicum strainsa
| Strain | CDW (mg/ml of culture broth) |
|---|---|
| Recombinant C. glutamicum strains | |
| Bm(mdh-act) Bs(hps-phi) | 6.72 ± 0.05 |
| Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) | 8.37 ± 0.03 |
| Control (pVWEx2 pEKEx2) | 6.49 ± 0.13 |
| Recombinant C. glutamicum Δald ΔadhE strains | |
| Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) | 6.75 ± 0.01 |
| Ptuf-Bm(mdh3-act) Ptuf-Bs(hps-phi) | 7.14 ± 0.13 |
| Ptuf-Bm(mdh-act) pEKEx2 | 4.87 ± 0.25 |
| Control (pVWEx2 pEKEx2) | 6.35 ± 0.10 |
Cells were grown in CGXII defined medium containing glucose (55 mM) and methanol (120 mM). The CDW was determined 32 h after inoculation (the second growth phase). Genes under the control of Ptac were induced with 1.5 mM IPTG at an OD600 of 1.0. Means and standard deviations were calculated from triplicates.
In contrast, the C. glutamicum strain expressing the genes for methanol oxidation and formaldehyde assimilation under the control of Ptuf showed similar growth in defined medium with or without methanol. In comparison to the C. glutamicum strain expressing the same genes under the control of Ptac, higher backscatter of the culture at the end of exponential growth was detected. Moreover, an additional increase of the cell density during the second growth phase in methanol-containing medium was observed that led to a significantly higher final backscatter and to a CDW that was increased by 18% (Table 3). Glucose measurements during cultivation of both strains and of the control strain revealed that glucose was completely consumed at the end of the exponential growth phase. Thus, the metabolization of methanol during this phase appears to be a diauxic shift. Remarkably, C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) also grew to a higher final backscatter in the presence of methanol (Fig. 2C) and showed a significantly increased CDW under these conditions (8.4 ± 0.03 versus 8.0 ± 0.05 mg/ml) (Table 3). These results hint toward the ability of this particular C. glutamicum strain to convert methanol-derived carbon into biomass. This was verified by analysis of a C. glutamicum strain expressing only the genes for methanol oxidation under the control of Ptuf [C. glutamicum Ptuf-Bm(mdh-act(pEKEx2)]. This strain grew slower in the presence of methanol than the control strain, which additionally expressed the genes enabling formaldehyde assimilation.
Methanol-derived carbon is assimilated into intracellular metabolites of C. glutamicum.
13C-methanol-labeling experiments were performed to validate that the expression of Ptuf-Bm(mdh-act) and Ptuf-Bs(hps-phi) results in the incorporation of methanol-derived carbon into intracellular metabolites. Two independent batch fermentations were run in CGXII medium supplemented with 55 mM glucose and 120 mM [13C]methanol. Quenching of the metabolic activity as well as analysis of the intracellular metabolites were performed in the exponential growth phase, at the point of transition to the second growth phase, as well as 12 h and 24 h after reaching the second growth phase. In the exponential growth phase, incorporation of [13C]methanol-derived carbon into intracellular metabolites could already be detected, although amounts were quite low (up to 5% labeling fractions in the m + 1 mass isotopomers) (see Fig. S2 in the supplemental material). This showed that small amounts of methanol below the GC detection limit were already oxidized to formaldehyde and subsequently assimilated into biomass in the exponential growth phase. With the transition to the second growth phase, labeling fractions of 3 to 10% in the m + 1 mass isotopomers of various metabolites, such as organic acids (succinate and α-ketoglutarate) and sugar phosphates (glucose-6-phosphate and fructose-1,6-bisphosphate), as well as amino acids (l-serine and l-lysine), were detected (Table 4). For some metabolites, the labeling fractions in the M + 1 mass isotopomers were increased even after a further 12 h of cultivation (see Fig. S4 in the supplemental material). No 13C-labeling that was higher than the natural 13C abundance was detected in any metabolites of the negative-control, C. glutamicum(pVWEx2 pEKEx2) (data not shown). In comparison to this control, a high 13CO2/total CO2 ratio for the strain expressing the methanol oxidation and formaldehyde assimilation modules under the control of Ptuf was observed (Fig. 3). This led us to the assumption that most of the [13C]methanol-derived formaldehyde is oxidized to 13CO2 via the endogenous pathway for formaldehyde dissimilation.
TABLE 4.
Incorporation of [13C]methanol-derived carbon into intracellular metabolites of C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi)a
| Metabolite | m + 1 labeled (%) |
|
|---|---|---|
| BR1 | BR2 | |
| Glucose-6-phosphate | 2.7 | 3.6 |
| 6-Phosphogluconate | 2.0 | ND |
| Sedoheptulose-7-phosphate | 6.0 | 6.2 |
| Fructose-1,6-bisphosphate | 8.2 | 6.2 |
| Dihydroxyacetone phosphate | 3.2 | 3.0 |
| 2-/3-Phosphoglycerate | 4.0 | 5.0 |
| Phosphoenolpyruvate | 1.9 | 4.2 |
| Pyruvate | 5.6 | 5.1 |
| α-Ketoglutarate | 8.0 | 8.3 |
| Succinate | 8.5 | 8.7 |
| Histidine | ND | 6.9 |
| Serine | 6.1 | 8.5 |
| Homoserine | 4.8 | 4.5 |
| Tryptophan | 10.7 | 9.9 |
| Tyrosine | 10.1 | 9.2 |
| Phenylalanine | 7.8 | 7.8 |
| Valine | 6.8 | 5.7 |
| Alanine | 7.0 | 7.1 |
| Proline | 5.6 | 5.2 |
| Arginine | 4.5 | 3.6 |
| Glutamine | 6.0 | 6.2 |
| Lysine | 8.3 | 7.7 |
| Aspartate | 9.3 | 9.1 |
| Threonine | 5.1 | 6.4 |
Two independent batch reactors (BR1 and BR2) were run with CGXII defined medium supplemented with 55 mM glucose and 120 mM 13C-labeled methanol. Quenching of the metabolic activity was performed during the transition to the second growth phase. Mass isotopomer measurements were performed for selected metabolites, and the corresponding M + 1 mass traces are listed. Raw mass spectrometry data were corrected for the contribution of all naturally abundant isotopes. ND, not determined.
FIG 3.
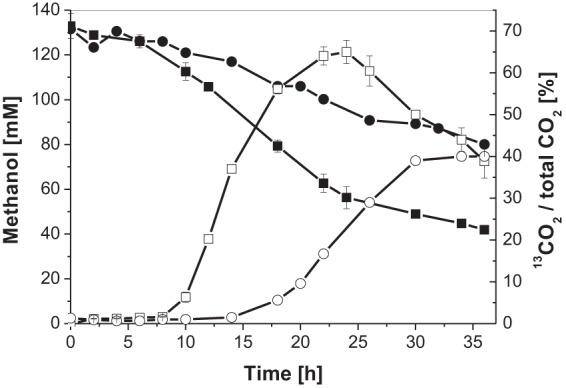
Methanol consumption (filled symbols) and 13CO2 formation, reported as the 13CO2/total CO2 ratio (open symbols) of C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) (■) and C. glutamicum pVWEx2 pEKEx2 (●) during cultivation in CGXII defined medium supplemented with 55 mM glucose and 120 mM 13C-labeled methanol in bioreactors. Two independent experiments were performed, and mean values are shown.
Implementation of methanol oxidation and formaldehyde assimilation in C. glutamicum Δald ΔadhE.
13C-methanol-labeling experiments revealed that only a small amount of [13C]methanol-derived carbon was assimilated into biomass precursors of C. glutamicum genes for methanol oxidation and formaldehyde assimilation (Table 4). Consequently, Ptuf-Bm(mdh-act) as well as Ptuf-Bm(mdh-act) in combination with Ptuf-Bs(hps-phi) were expressed in C. glutamicum Δald ΔadhE, a strain which is strongly impaired in its ability to oxidize formaldehyde to CO2 (22, 23). This approach should prevent loss of methanol-derived formaldehyde as CO2 and boost formaldehyde assimilation. In addition, C. glutamicum Δald ΔadhE(pVWEx2 pEKEx2), only harboring the empty plasmids, was constructed as a control. All strains were analyzed regarding growth and methanol consumption in the presence of methanol to check if the absence of the “safety valve” for formaldehyde oxidation affected growth.
Determination of the in vitro enzyme activities revealed that all enzymes were functionally active in recombinant C. glutamicum Δald ΔadhE strains (Table 2). In glucose-containing medium, growth of all strains was similar (Fig. 4B) and comparable to that of the strains constructed on the basis of wild-type C. glutamicum (Fig. 2B). In comparison, C. glutamicum Δald ΔadhE(pVWEx2 pEKEx2) displayed retarded growth and a 33% reduced final backscatter in the presence of methanol. The C. glutamicum Δald ΔadhE strain expressing only Ptuf-Bm(mdh-act) showed even slower growth in the presence of methanol, and the final backscatter of the culture was reduced by an additional 35% (Fig. 4C). The methanol consumption rate was as low as that observed for the control strain (0.52 ± 0.1 mM/h) (Fig. 4A). This strain possesses the heterologous methanol dehydrogenase for oxidation of methanol to formaldehyde, but due to the absence of Bs(Hps-Phi) and the lack of Ald and AdhE, the toxic accumulation of formaldehyde cannot be avoided, neither via assimilation by the RuMP pathway nor via dissimilation to CO2. However, quantification of intracellular formaldehyde was not successful.
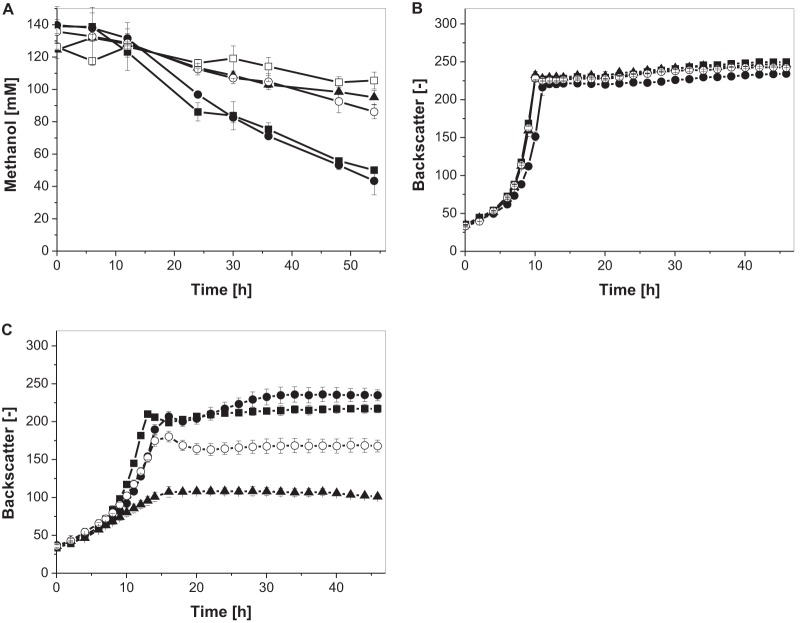
FIG 4.
(A) Methanol consumption in CGXII defined medium supplemented with 55 mM glucose and 120 mM methanol. (B) Growth in CGXII defined medium supplemented with 55 mM glucose. (C) Growth in CGXII defined medium supplemented with 55 mM glucose and 120 mM methanol of recombinant C. glutamicum Δald ΔadhE strains. Three independent shake-flask experiments, always accompanied by a methanol evaporation control, were performed for the determination of the methanol concentration (◇). Monitoring of growth was performed in 48-well FlowerPlates by using a BioLectorBasic apparatus (n = 3). Strains: C. glutamicum Δald ΔadhE Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) (■), C. glutamicum Ptuf-Bm(mdh3-act) Ptuf-Bs(hps-phi) (●), C. glutamicum Δald ΔadhE Ptuf-Bm(mdh-act) pEKEx2 (▲), and C. glutamicum Δald ΔadhE pVWEx2 pEKEx2 (○).
The C. glutamicum Δald ΔadhE strain possessing the methanol oxidation module [Ptuf-Bm(mdh-act)] and the formaldehyde assimilation module [Ptuf-Bs(hps-phi)] showed an increased methanol oxidation rate (1.25 ± 0.2 mM/h) in comparison to the control strain. The specific methanol consumption rate in the second growth phase for this strain was 5.95 ± 0.3 μM/(h · BS) and for the control strain only 3.25 ± 0.2 μM/(h · BS).
Apparently, the recombinant C. glutamicum Δald ΔadhE strain had a growth advantage over the control strain in the presence of methanol, since the backscatter was found to be 15% higher at the end of the exponential growth phase. This was also reflected by a higher final CDW (6.75 ± 0.00 mg/ml and 4.87 ± 0.25 mg/ml, respectively). In contrast to C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi), the recombinant C. glutamicum Δald ΔadhE strain did not grow in methanol-containing medium as fast as in medium without methanol, and the final backscatter of the culture was not increased in the presence of methanol.
Finally, we also evaluated Mdh3, the other MDH of B. methanolicus, in the C. glutamicum Δald ΔadhE strain background. Ptuf-controlled expression of mdh3 and act in combination with Ptuf-Bs(hps-phi) and cultivation in glucose- and methanol-containing medium revealed methanol consumption, which was similar to the that of C. glutamicum Δald ΔadhE Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) (Fig. 4A). However, with this strain, a slightly higher final backscatter of the culture and an increased CDW was reached (Fig. 4B; Table 3).
Increased assimilation of methanol-derived carbon by C. glutamicum Δald ΔadhE.
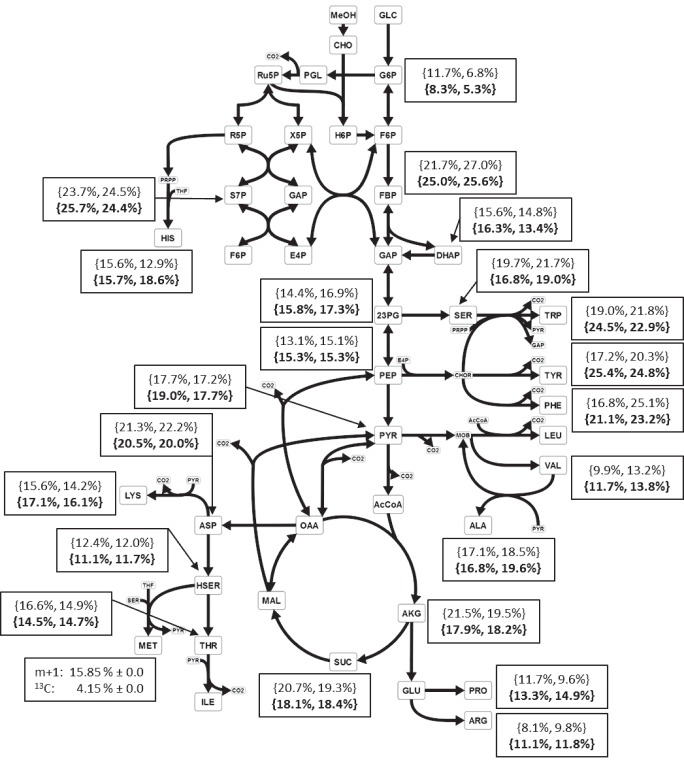
13C-methanol-labeling experiments were performed to answer the question whether the elimination of the endogenous pathway for formaldehyde dissimilation results in increased assimilation of methanol-derived carbon into the biomass, although the determined overall biomass yield was not higher in the presence of methanol. In addition, methanol consumption and generation of 13CO2 was measured (see Fig. S3A and B in the supplemental material). Already in the exponential growth phase, incorporation of [13C]methanol-derived carbon into intracellular metabolites of C. glutamicum Δald ΔadhE Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) and of C. glutamicum Δald ΔadhE Ptuf-Bm(mdh3-act) Ptuf-Bs(hps-phi) was observed. Labeling fractions of up to 11% in the m + 1 mass isotopomers were detected. With the entrance to the second growth phase, the labeling fractions were found to be about 3-fold higher than the wild-type C. glutamicum strain expressing the methanol oxidation and formaldehyde assimilation modules (Table 4; Fig. 5). For the C. glutamicum Δald ΔadhE strain expressing Ptuf-Bm(mdh3-act) and Ptuf-Bs(hps-phi), an average increase of 10% of the labeling fractions in the m + 1 mass isotopomers of intracellular metabolites was observed (see Fig. S4 and S5 in the supplemental material).
FIG 5.
Simplified overview of the central carbon metabolism, including the oxidation of methanol via Bm(Mdh-Act) or Bm(Mdh3-Act) and the assimilation of formaldehyde via the RuMP pathway of recombinant C. glutamicum Δald ΔadhE strains. Additionally, the labeling fractions of the M + 1 mass isotopomers of the measured intracellular metabolites are listed for C. glutamicum Δald ΔadhE Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) (values in the upper row) and C. glutamicum Δald ΔadhE Ptuf-Bm(mdh3-act) Ptuf-Bs(hps-phi) (values in the lower row, bold). For each strain, two independent batch reactors (BR1 and BR2) with CGXII defined medium (55 mM glucose, 120 mM 13C-labeled methanol) were run. Quenching of metabolic activity as well as intracellular metabolic analysis were performed at the transition to the second growth phase, and raw mass spectrometry data were corrected for the contribution of all naturally abundant isotopes. Abbreviations: amino acids are presented according to their 3-letter standard abbreviations; MeOH, methanol; GLC, glucose; CHO, formaldehyde; G6P, glucose-6-phosphate; PGL, phosphogluconolactone; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; X5P, xylulose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrosee-4-phosphate; H6P, hexulose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; 23PG, 2-/3-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl coenzyme A; AKG, α-ketoglutarate; SUC, succinate; MAL, malate; OAA, oxaloacetate; THF, tetrahydrofuran; PRPP, phosphoribosyl pyrophosphate; CHOR, chorismate.
DISCUSSION
Only few methylotrophic organisms, such as B. methanolicus and M. extorquens, have been engineered to produce biotechnologically interesting compounds from methanol (49–51). In this study, we followed the opposite strategy by engineering the nonmethylotrophic platform organism C. glutamicum to utilize methanol instead of turning a methylotrophic organism into a production strain.
The wild-type C. glutamicum is already able to oxidize methanol, predominantly catalyzed by the alcohol dehydrogenase AdhA (22). Nonetheless, we evaluated the implementation of the NAD+-dependent methanol dehydrogenase from B. methanolicus for increasing the methanol oxidation rate. Since wild-type C. glutamicum already possesses an endogenous pathway for the detoxification of naturally occurring cytotoxic formaldehyde to CO2 (22, 23), only a pathway for assimilation of formaldehyde into biomass was required. Heterologous expression of genes for HPS and PHI from methylotrophic organisms, both key enzymes of the RuMP) pathway, has already been performed to confer the capability of formaldehyde assimilation to Burkholderia cepacia and Pseudomonas putida, respectively (20, 52, 53). Surprisingly, in this study heterologous expression of hps and phi from the methylotroph bacterium M. gastri, which is closely related to C. glutamicum, did not result in active enzymes. As an alternative, we tried the expression of hps and phi of B. subtilis, which requires these genes only for formaldehyde detoxification (52, 54), and we were able to complete a functional RuMP pathway in C. glutamicum.
Functional expression of the modules for methanol oxidation [Bm(Mdh-Act)] and formaldehyde assimilation [Bs(Hps-Phi)] in wild-type C. glutamicum, as well as the selection of a suitable promoter for gene expression (Ptuf), resulted in a 3-fold-increased methanol oxidation rate (1.7 ± 0.2 mM/h) in comparison to the C. glutamicum(pVWEx2 pEKEx2) control strain (0.5 ± 0.1 mM/h). Furthermore, the inhibitory effects of methanol on growth and biomass formation of this control strain could be compensated by C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi). This strain showed the same growth in medium with and without methanol. Furthermore, at the end of the exponential growth phase, a higher backscatter than with the control strain was observed. This effect could be explained by the generation of additional NADH due to the methanol dehydrogenase-catalyzed oxidation of methanol in the exponential growth phase and subsequent generation of ATP via oxidative phosphorylation. Thus, less glucose has to be dissimilated for energy generation and can be used for synthesis of biomass precursors. This effect on biomass formation was also observed during coutilization of formate with glucose by Candida utilis and Penicillium chrysogenum (55, 56), as well as during coutilization of formaldehyde with glucose by Saccharomyces cerevisiae expressing a formaldehyde dehydrogenase and a formate dehydrogenase from Hansenula polymorpha (57). Furthermore, the increased biomass formation in recombinant C. glutamicum strains could also be explained by assimilation of methanol-derived formaldehyde via HPS- and PHI-catalyzed reactions. Indeed, [13C]methanol-labeling experiments revealed up to 5% labeling fractions in the m + 1 mass isotopomers of various intracellular metabolites in samples taken from the exponential growth phase.
After exponential growth in glucose-methanol medium, recombinant C. glutamicum cells seem to enter a second growth phase, represented by slowly increasing backscatter and an increased CDW (8.4 ± 0.03 mg/ml versus 8.0 ± 0.05 mg/ml in medium without methanol). Again, [13C]methanol-labeling experiments showed 3 to 10% m + 1 labeling of intracellular metabolites, indicating assimilation of methanol-derived carbon into biomass. However, C. glutamicum Ptuf-Bm(mdh-act) Ptuf-Bs(hps-phi) consumed about 40% more methanol and produced 31% more CO2 than did C. glutamicum(pVWEx2 pEKEx2) when we calculated the amounts until the respective maximal CO2 production was reached. This means that most of the consumed methanol (about 78%) in the recombinant strain is oxidized to CO2 via the endogenous pathway for formaldehyde oxidation and not assimilated via the RuMP pathway. Since implementation of the modules for methanol oxidation and formaldehyde assimilation in wild-type C. glutamicum resulted in a high flux toward CO2 formation, we also engineered C. glutamicum Δald ΔadhE, a strain severely impaired in its ability to detoxify formaldehyde, for assimilation of methanol. As observed for recombinant wild-type C. glutamicum strains, the implementation of the modules for methanol oxidation, [Ptuf-Bm(mdh-act)] or [Ptuf-Bm(mdh3-act)], and formaldehyde assimilation [Ptuf-Bs(hps-phi)] in C. glutamicum Δald ΔadhE resulted in a 15% higher backscatter at the end of the exponential growth phase in methanol-containing medium compared to C. glutamicum Δald ΔadhE(pVWEx2 pEKEx2). As discussed before, this effect might be due to the generation of additional NADH during methanol oxidation. However, in contrast to recombinant wild-type C. glutamicum strains, oxidation of methanol in recombinant C. glutamicum Δald ΔadhE is accompanied by channeling of more methanol-derived formaldehyde into the synthetic RuMP cycle, because C. glutamicum Δald ΔadhE does not have the “safety valve” for formaldehyde detoxification to CO2. Without the assimilation of formaldehyde, toxic amounts of this intermediate would accumulate and inhibit cell growth. Previously, we showed that 4 mM formaldehyde completely inhibited cell growth of C. glutamicum (22). Indeed, [13C]methanol-labeling experiments revealed that the M + 1 labeling of intracellular metabolites in the exponential growth phase was doubled in comparison to the wild-type C. glutamicum strain expressing the same genes; in the transition to the second growth phase, it was even higher (3-fold higher). However, elimination of the “safety valve” also led to a reduced methanol consumption rate, retarded growth, and a lowered final CDW, most probably due to accumulation of formaldehyde. Possibly, synthetic formaldehyde assimilation is currently not efficient enough to keep the intracellular formaldehyde concentrations below the toxicity threshold.
In summary, for the first time C. glutamicum was engineered to utilize methanol as a carbon and energy source during growth in sugar-based defined medium. Furthermore, detection of 13C-labeled amino acids indicates that methanol can also be used as an auxiliary substrate for the production of these value-added compounds. For a more efficient use of methanol during sugar-based fermentation processes, methanol oxidation to formaldehyde has to be increased, whereas accumulation of toxic formaldehyde has to be minimized. Thus, future experiments should aim at balancing the synthetic formaldehyde assimilation and the endogenous dissimilation. This could be achieved by metabolic engineering or by employing process engineering strategies, e.g., by developing a fed-batch process with continuous methanol feeding. This would ensure constant methanol availability during the cultivation, as has been already realized for B. methanolicus MGA3 (21, 58).
Supplementary Material
ACKNOWLEDGMENTS
Sabrina Witthoff is an associated fellow of the CLIB-Graduate Cluster Industrial Biotechnology.
We thank Meike Baumgart (Forschungszentrum Jülich) for carefully reading the manuscript and Petra Geilenkirchen (Forschungszentrum Jülich) for the LC tandem MS measurements.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03110-14.
REFERENCES
- 1.Becker J, Wittmann C. 2012. Bio-based production of chemicals, materials and fuels: Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. 2008. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- 3.Litsanov B, Brocker M, Bott M. 2012. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol 78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wieschalka S, Blombach B, Bott M, Eikmanns BJ. 2013. Bio-based production of organic acids with Corynebacterium glutamicum. Microb Biotechnol 6:87–102. doi: 10.1111/1751-7915.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider J, Wendisch VF. 2011. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol 91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- 6.Mimitsuka T, Sawai H, Hatsu M, Yamada K. 2007. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem 71:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- 7.Kind S, Wittmann C. 2011. Bio-based production of the platform chemical 1,5-diaminopentane. Appl Microbiol Biotechnol 91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- 8.Inui M, Kawaguchi H, Murakami S, Vertes AA, Yukawa H. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol 8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- 9.Smith KM, Cho KM, Liao JC. 2010. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. 2011. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahoor A, Lindner SN, Wendisch VF. 2012. Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput Struct Biotechnol J 3:e201210004. doi: 10.5936/csbj.201210004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelle R, Hermann T, Bathe B. 2005. Lysine production, p 467–490. In Eggeling L, Bott M (ed), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL. [Google Scholar]
- 13.Kimura E. 2005. Glutamate production, p 441–465. In Eggeling L, Bott M (ed), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL. [Google Scholar]
- 14.Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, Marx A, Vorholt JA. 2009. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol 27:107–115. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Uhde A, Youn JW, Maeda T, Clermont L, Matano C, Krämer R, Wendisch VF, Seibold GM, Marin K. 2013. Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl Microbiol Biotechnol 97:1679–1687. doi: 10.1007/s00253-012-4313-8. [DOI] [PubMed] [Google Scholar]
- 16.Olah GA, Prakash GK, Goeppert A, Czaun M, Mathew T. 2013. Self-sufficient and exclusive oxygenation of methane and its source materials with oxygen to methanol via metgas using oxidative bi-reforming. J Am Chem Soc 135:10030–10031. doi: 10.1021/ja405439c. [DOI] [PubMed] [Google Scholar]
- 17.Olah GA, Goeppert A, Czaun M, Prakash GK. 2013. Bi-reforming of methane from any source with steam and carbon dioxide exclusively to metgas (CO2-H2) for methanol and hydrocarbon synthesis. J Am Chem Soc 135:648–650. doi: 10.1021/ja311796n. [DOI] [PubMed] [Google Scholar]
- 18.Wesselbaum S, Vom Stein T, Klankermayer J, Leitner W. 2012. Hydrogenation of carbon dioxide to methanol by using a homogeneous ruthenium-phosphine catalyst. Angew Chem Int Ed Engl 51:7499–7502. doi: 10.1002/anie.201202320. [DOI] [PubMed] [Google Scholar]
- 19.Law K, Rosenfeld J, Jackson M. 2013. Methanol as a renewable energy resource. Tiax LLC, Cupertino, CA. [Google Scholar]
- 20.Koopman FW, de Winde JH, Ruijssenaars HJ. 2009. C1 compounds as auxiliary substrate for engineered Pseudomonas putida S12. Appl Microbiol Biotechnol 83:705–713. doi: 10.1007/s00253-009-1922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brautaset T, Jakobsen OM, Josefsen KD, Flickinger MC, Ellingsen TE. 2007. Bacillus methanolicus: a candidate for industrial production of amino acids from methanol at 50°C. Appl Microbiol Biotechnol 74:22–34. doi: 10.1007/s00253-006-0757-z. [DOI] [PubMed] [Google Scholar]
- 22.Witthoff S, Mühlroth A, Marienhagen J, Bott M. 2013. C1 metabolism in Corynebacterium glutamicum: an endogenous pathway for oxidation of methanol to carbon dioxide. Appl Environ Microbiol 79:6974–6983. doi: 10.1128/AEM.02705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lessmeier L, Hoefener M, Wendisch VF. 2013. Formaldehyde degradation in Corynebacterium glutamicum involves acetaldehyde dehydrogenase and mycothiol-dependent formaldehyde dehydrogenase. Microbiology 159:2651–2662. doi: 10.1099/mic.0.072413-0. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, Hayakawa T, Kawai K. 2012. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One 7:e50480. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arfman N, Watling EM, Clement W, van Oosterwijk RJ, de Vries GE, Harder W, Attwood MM, Dijkhuizen L. 1989. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol 152:280–288. doi: 10.1007/BF00409664. [DOI] [PubMed] [Google Scholar]
- 26.Vorholt JA. 2002. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol 178:239–249. doi: 10.1007/s00203-002-0450-2. [DOI] [PubMed] [Google Scholar]
- 27.Yurimoto H, Kato N, Sakai Y. 2005. Assimilation, dissimilation, and detoxification of formaldehyde, a central metabolic intermediate of methylotrophic metabolism. Chem Rec 5:367–375. doi: 10.1002/tcr.20056. [DOI] [PubMed] [Google Scholar]
- 28.Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ Microbiol 13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 29.Kato N, Yurimoto H, Thauer RK. 2006. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci Biotechnol Biochem 70:10–21. doi: 10.1271/bbb.70.10. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol 67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohe P, Venkanna D, Kleine B, Freudl R, Oldiges M. 2012. An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb Cell Fact 11:144. doi: 10.1186/1475-2859-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radek A, Krumbach K, Gätgens J, Wendisch VF, Wiechert W, Bott M, Noack S, Marienhagen J. 2014. Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of d-xylose-containing substrates. J Biotechnol 192:156–160. doi: 10.1016/j.jbiotec.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 35.van der Rest ME, Lange C, Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545. doi: 10.1007/s002530051557. [DOI] [PubMed] [Google Scholar]
- 36.Abe S, Takayama K, Kinoshita S. 1967. Taxonomical studies on glutamic acid producing bacteria. J Gen Appl Microbiol 13:279–301. doi: 10.2323/jgam.13.279. [DOI] [Google Scholar]
- 37.Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. 1991. A family of Corynebacterium glutamicum-Escherichia coli shuttle vectors for cloning, controlled gene-expression, and promoter probing. Gene 102:93–98. doi: 10.1016/0378-1119(91)90545-M. [DOI] [PubMed] [Google Scholar]
- 38.Wendisch VF. 1997. Ph.D. thesis Heinrich-Heine University, Düsseldorf, Germany. [Google Scholar]
- 39.Witthoff S, Eggeling L, Bott M, Polen T. 2012. Corynebacterium glutamicum harbours a molybdenum cofactor-dependent formate dehydrogenase which alleviates growth inhibition in the presence of formate. Microbiology 158:2428–2439. doi: 10.1099/mic.0.059196-0. [DOI] [PubMed] [Google Scholar]
- 40.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 41.Hektor HJ, Kloosterman H, Dijkhuizen L. 2002. Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus. J Biol Chem 277:46966–46973. doi: 10.1074/jbc.M207547200. [DOI] [PubMed] [Google Scholar]
- 42.Arfman N, Bystrykh L, Govorukhina NI, Dijkhuizen L. 1990. 3-Hexulose-6-phosphate synthase from thermotolerant methylotroph Bacillus C1. Methods Enzymol 188:391–397. doi: 10.1016/0076-6879(90)88062-F. [DOI] [PubMed] [Google Scholar]
- 43.Nudelman A, Levovich I, Cutts SM, Phillips DR, Rephaeli A. 2005. The role of intracellularly released formaldehyde and butyric acid in the anticancer activity of acyloxyalkyl esters. J Med Chem 48:1042–1054. doi: 10.1021/jm049428p. [DOI] [PubMed] [Google Scholar]
- 44.van Ooyen J, Noack S, Bott M, Reth A, Eggeling L. 2012. Improved L-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng 109:2070–2081. doi: 10.1002/bit.24486. [DOI] [PubMed] [Google Scholar]
- 45.Millard P, Letisse F, Sokol S, Portais JC. 2012. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28:1294–1296. doi: 10.1093/bioinformatics/bts127. [DOI] [PubMed] [Google Scholar]
- 46.Shen YQ, Bonnot F, Imsand EM, RoseFigura JM, Sjolander K, Klinman JP. 2012. Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51:2265–2275. doi: 10.1021/bi201763d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heggeset TM, Krog A, Balzer S, Wentzel A, Ellingsen TE, Brautaset T. 2012. Genome sequence of thermotolerant Bacillus methanolicus: features and regulation related to methylotrophy and production of L-lysine and L-glutamate from methanol. Appl Environ Microbiol 78:5170–5181. doi: 10.1128/AEM.00703-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krog A, Heggeset TM, Müller JE, Kupper CE, Schneider O, Vorholt JA, Ellingsen TE, Brautaset T. 2013. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS One 8:e59188. doi: 10.1371/journal.pone.0059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brautaset T, Jakobsen OM, Degnes KF, Netzer R, Naerdal I, Krog A, Dillingham R, Flickinger MC, Ellingsen TE. 2010. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for L-lysine production from methanol at 50°C. Appl Microbiol Biotechnol 87:951–964. doi: 10.1007/s00253-010-2559-6. [DOI] [PubMed] [Google Scholar]
- 50.Mokhtari-Hosseini ZB, Vasheghani-Farahani E, Heidarzadeh-Vazifekhoran A, Shojaosadati SA, Karimzadeh R, Khosravi Darani K. 2009. Statistical media optimization for growth and PHB production from methanol by a methylotrophic bacterium. Bioresour Technol 100:2436–2443. doi: 10.1016/j.biortech.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 51.Orita I, Nishikawa K, Nakamura S, Fukui T. 2014. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions. Appl Microbiol Biotechnol 98:3715–3725. doi: 10.1007/s00253-013-5490-9. [DOI] [PubMed] [Google Scholar]
- 52.Mitsui R, Kusano Y, Yurimoto H, Sakai Y, Kato N, Tanaka M. 2003. Formaldehyde fixation contributes to detoxification for growth of a nonmethylotroph, Burkholderia cepacia TM1, on vanillic acid. Appl Environ Microbiol 69:6128–6132. doi: 10.1128/AEM.69.10.6128-6132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yurimoto H, Kato N, Sakai Y. 2009. Genomic organization and biochemistry of the ribulose monophosphate pathway and its application in biotechnology. Appl Microbiol Biotechnol 84:407–416. doi: 10.1007/s00253-009-2120-7. [DOI] [PubMed] [Google Scholar]
- 54.Yasueda H, Kawahara Y, Sugimoto S. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J Bacteriol 181:7154–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruinenberg PM, Jonker R, Vandijken JP, Scheffers WA. 1985. Utilization of formate as an additional energy-source by glucose-limited chemostat cultures of Candida utilis Cbs-621 and Saccharomyces cerevisiae Cbs-8066: evidence for the absence of transhydrogenase activity in yeasts. Arch Microbiol 142:302–306. doi: 10.1007/BF00693408. [DOI] [Google Scholar]
- 56.Harris DM, van der Krogt ZA, van Gulik WM, van Dijken JP, Pronk JT. 2007. Formate as an auxiliary substrate for glucose-limited cultivation of Penicillium chrysogenum: impact on penicillin G production and biomass yield. Appl Environ Microbiol 73:5020–5025. doi: 10.1128/AEM.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baerends RJ, de Hulster E, Geertman JM, Daran JM, van Maris AJ, Veenhuis M, van der Klei IJ, Pronk JT. 2008. Engineering and analysis of a Saccharomyces cerevisiae strain that uses formaldehyde as an auxiliary substrate. Appl Environ Microbiol 74:3182–3188. doi: 10.1128/AEM.02858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pluschkell SB, Flickinger MC. 2002. Dissimilation of [13C]methanol by continuous cultures of Bacillus methanolicus MGA3 at 50°C studied by 13C NMR and isotope-ratio mass spectrometry. Microbiology 148:3223–3233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.