Significance
Carbohydrate uptake in many bacteria is regulated by the phosphotransferase protein IIAGlc, enabling cells to use glucose preferentially over other sugars. Lactose permease (LacY) is one of many sugar permeases regulated by IIAGlc, but the mechanism of inducer exclusion is unclear. We now show by isothermal titration calorimetry that IIAGlc binds to purified LacY with a stoichiometry of one, and that the interaction is driven by favorable solvation entropy. IIAGlc binding inhibits conformational dynamics of LacY and decreases binding affinity for sugar in a manner similar to that observed for melibiose permease (MelB). However, the thermodynamic mechanism by which the inhibitory effect is expressed differs for the two permeases.
Keywords: ITC, PTS, sugar/cation symport, protein–protein interactions, protein conformation
Abstract
In a variety of bacteria, the phosphotransferase protein IIAGlc plays a key regulatory role in catabolite repression in addition to its role in the vectorial phosphorylation of glucose catalyzed by the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS). The lactose permease (LacY) of Escherichia coli catalyzes stoichiometric symport of a galactoside with an H+, using a mechanism in which sugar- and H+-binding sites become alternatively accessible to either side of the membrane. Both the expression (via regulation of cAMP levels) and the activity of LacY are subject to regulation by IIAGlc (inducer exclusion). Here we report the thermodynamic features of the IIAGlc–LacY interaction as measured by isothermal titration calorimetry (ITC). The studies show that IIAGlc binds to LacY with a Kd of about 5 μM and a stoichiometry of unity and that binding is driven by solvation entropy and opposed by enthalpy. Upon IIAGlc binding, the conformational entropy of LacY is restrained, which leads to a significant decrease in sugar affinity. By suppressing conformational dynamics, IIAGlc blocks inducer entry into cells and favors constitutive glucose uptake and utilization. Furthermore, the studies support the notion that sugar binding involves an induced-fit mechanism that is inhibited by IIAGlc binding. The precise mechanism of the inhibition of LacY by IIAGlc elucidated by ITC differs from the inhibition of melibiose permease (MelB), supporting the idea that permeases can differ in their thermodynamic response to binding IIAGlc.
Carbohydrate uptake in bacteria is catalyzed by a collection of sugar permeases that belong to different families of transport proteins. In Escherichia and Salmonella, the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) carries out both catalytic and regulatory functions and plays a key role in catabolite repression resulting in preferential utilization of glucose (a constitutive process) that is transported by vectorial phosphorylation catalyzed by the PTS (1–5). The phosphotransferase protein IIAGlc plays a direct role in this regulation of inducible transport systems. The lac operon (6) with lacZ encoding β-galactosidase and lacY encoding lactose permease (LacY) and the mel operon (7, 8) with melA encoding α-galactosidase and melB encoding melibiose permease (MelB) are subject to IIAGlc regulation (9–13). Both LacY and MelB catalyze electrogenic symport of a galactoside with a cation (14–21). Expression of the structural genes requires the participation of both a global transcriptional activator (the cAMP–CAP complex) and a specific inducer (lactose or melibiose, respectively) (3, 22, 23). IIAGlc regulates both cAMP (24) and inducer levels, and this study focuses on regulation of LacY, which influences inducer entry into the cell.
With maltose permease, an ABC permease also under PTS regulation, two molecules of IIAGlc bind to the cytoplasmic ATPase subunits and consequently prevent the structural rearrangements necessary for ATP hydrolysis (25). Recently, we reported that IIAGlc binds to Escherichia coli MelBEc or Salmonella typhimurium MelBSt with a stoichiometry of one and that enthalpy drives the interaction in the absence or presence of melibiose (26). Furthermore, IIAGlc binding reduces sugar affinity and the conformational entropy of MelBSt, resulting in a block of melibiose entry into the cell, thus preventing induction of the mel operon. With LacY, IIAGlc binding has been extensively studied (9, 10, 27–29). In a direct binding assay, IIAGlc binding to LacY exhibits a dissociation constant (Kd) of 1 μM, and binding requires the presence of sugar substrate; however, a stoichiometry of one molecule of IIAGlc to six molecules of LacY was obtained, which seems highly unusual (10).
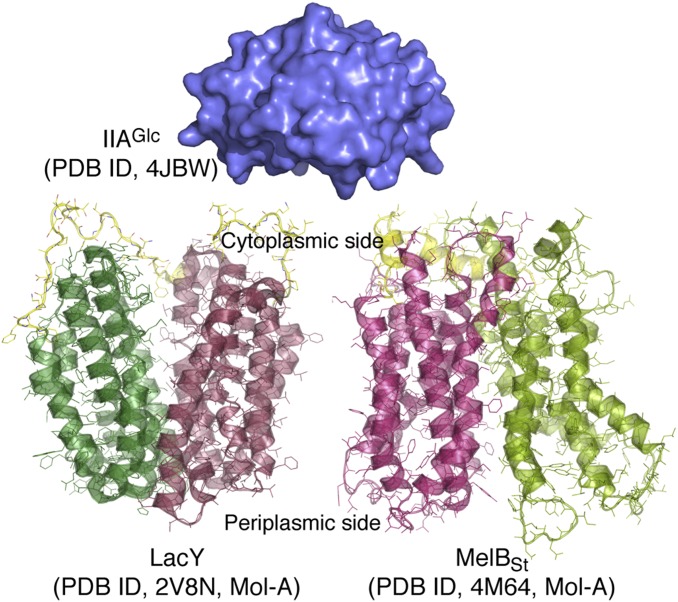
Both X-ray crystallographic and NMR structures of IIAGlc (25, 30, 31) reveal that this soluble protein is structurally rigid (Fig. 1). The crystal structures of LacY (14, 32–36) and MelBSt (37) both exhibit N- and C-terminal domains composed of six largely irregular transmembrane helices positioned pseudosymmetrically and surrounding a deep aqueous cavity (Fig. 1). Both permeases are members of the major facilitator superfamily. WT MelBSt was crystallized in two outward (periplasmic)-open conformations with a sealed cytoplasmic side (37). WT LacY was crystallized in an inward (cytoplasmic)-open conformation (32). Recently, a LacY conformational mutant was captured in an almost occluded, narrowly outward-open conformation with a completely liganded galactoside molecule in the middle of the protein (38). Evidence was also presented indicating that sugar binding involves induced fit, which causes LacY to transition from an open state to an occluded intermediate.
Fig. 1.
Crystal structures. All of the depicted structural models are on the same scale. The surface representation of IIAGlc is colored blue [Protein Data Bank (PDB) ID code 4JBW]. LacY (PDB ID code 2V8N; Mol-A) and MelBSt (PDB ID code 4M64; Mol-A) are shown with the cytoplasmic side at the top. The N- and C-terminal domains of both permeases are shown in different colors, and their middle cytoplasmic loops are colored yellow. LacY and MelBSt are in an inward-open and partially outward-open conformation, respectively.
It is generally believed that a crystal structure typically captures a molecule in the lowest free-energy state. With regard to LacY, abundant, independent lines of evidence demonstrate that the inward-open conformation in the absence of bound substrate represents the lowest free-energy state in both the membrane-embedded and detergent-solubilized state (39). LacY functions by exposing its substrate-binding sites (sugar and H+) alternatively to either side of the membrane via an alternating access mechanism (36, 39, 40), and a similar model has also been proposed for MelB (37, 41, 42). With both permeases, it has been well-documented that ligand binding induces conformational changes (39, 41, 43, 44).
The ground-state conformations of protonated LacY and Na+-bound MelB appear to have opposite orientations, LacY inward-open and MelBSt outward-open. To compare IIAGlc binding with the two symporters, we have used isothermal titration calorimetry (ITC). The results indicate that IIAGlc binding inhibits conformational entropy in both transporters, but by different energetic mechanisms.
Results
IIAGlc Binding to WT LacY.
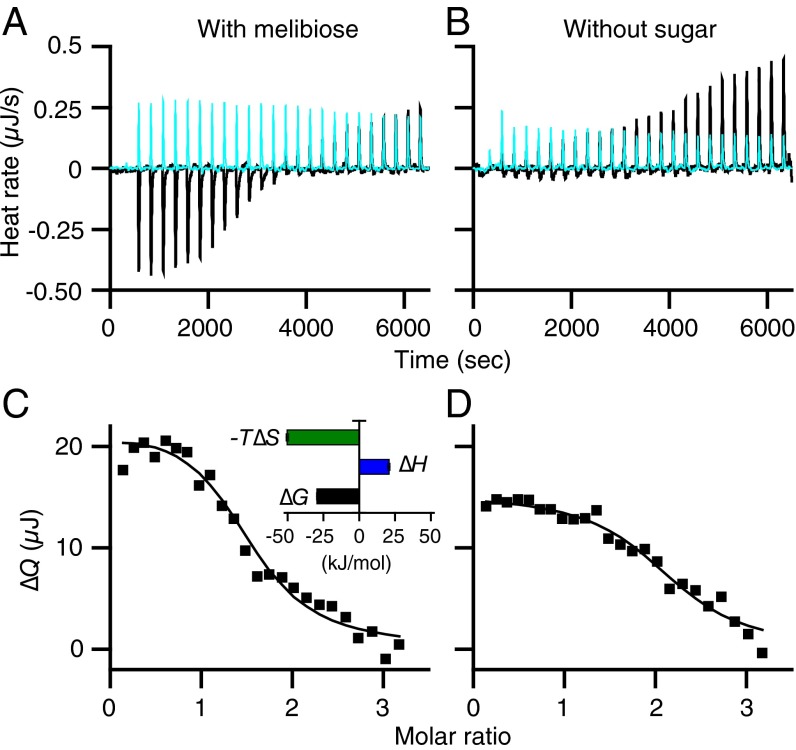
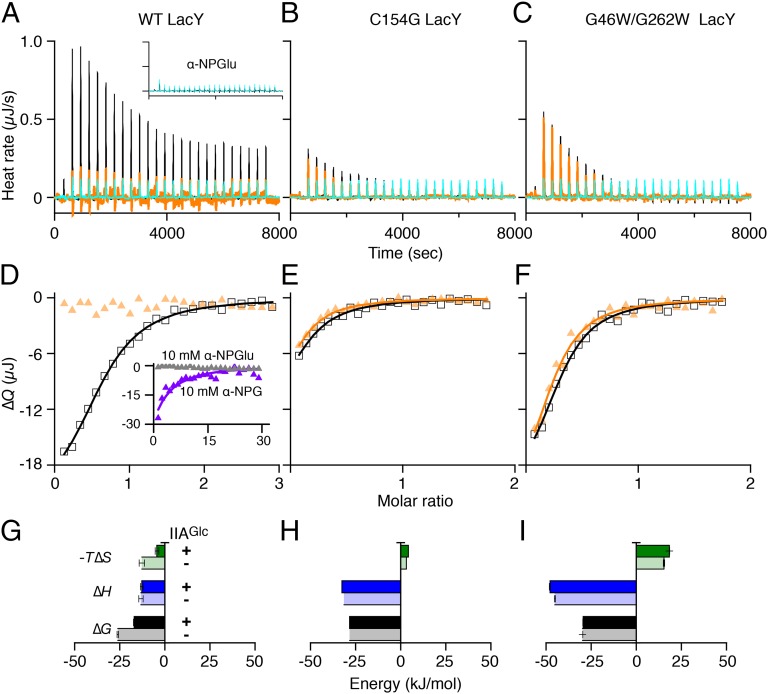
LacY or IIAGlc samples were preequilibrated with melibiose (a substrate for LacY) at a saturating concentration. Titration of LacY with IIAGlc yields an endothermic titration curve (Fig. 2 A, black and C). When injecting IIAGlc into melibiose-containing buffer in the absence of LacY, the thermogram is flat and exothermic (Fig. 2A, cyan). A plot of the accumulated heat change (∆Q) vs. the molar ratio fits a one-site independent binding model (Fig. 2C), which yields a Kd of 5.7 μM (SEM, 1.11; n = 3) and a stoichiometry of unity. The binding affinity is in a range similar to that obtained by direct binding with 125I-labeled IIAGlc (10), as well as the affinity obtained with MelBSt (26). Energetically (Table 1), binding is driven by a favorable entropy change (−T∆S) and opposed by a change in enthalpy (∆H), which is opposite to that found with IIAGlc binding to MelBSt or MelBEc, where binding is driven enthalpically and opposed by an entropic change (26).
Fig. 2.
ITC measurements of IIAGlc binding to LacY. (A and B) Thermograms were recorded at 20 °C. Black curves, titrations of LacY (50 μM) with IIAGlc (455 μM) in the presence or absence of melibiose at 10 mM as indicated. Cyan curves, titrations of buffer in the absence of LacY with IIAGlc under the same conditions. (C and D) Data fitting. Accumulated heat change (∆Q) was plotted against the IIAGlc/LacY molar ratio, and the data were fitted to a one-site independent binding model as described in Materials and Methods. (C, Inset) Histograms showing ∆G, ∆H, and −T∆S as described in Materials and Methods; the values are also presented in Table 1. Error bars denote SEM, n = 3.
Table 1.
IIAGlc binding to LacY
| Temperature, °C (K) | Ka (/mol) | Kd (μM) | ∆G (kJ/mol) | ∆H (kJ/mol) | −T∆S (kJ/mol) | ∆S (J/mol⋅K) |
| 15 (288.15) | 213,900 | 4.68 | −29.41 | 28.06 | −57.46 | 199.40 |
| 20 (293.15) | 187,167 (30,739*) | 5.71 (1.11) | −29.52 (0.45) | 20.63 (1.02) | −50.15 (0.78) | 171.1 (2.65) |
| 25 (298.15) | 173,433 (7,597) | 5.79 (0.24) | −29.90 (0.11) | 11.33 (0.10) | −41.23 (0.04) | 138.27 (0.15) |
| 30 (303.15) | 215,400 | 4.64 | −30.95 | 5.06 | −36.01 | 118.80 |
Under the same experimental conditions without sugar, endothermic binding is also observed (Fig. 2 B and D), but with much smaller heat changes. A best fit of the data suggests a Kd of 6.6 µM (SEM, 0.65; n = 4), similar to that obtained in the presence of melibiose. The finding stands in opposition to previous conclusions that IIAGlc binding requires binding of substrate (10, 27, 45). It is likely that the ITC method is more sensitive than previously used methods.
IIAGlc Binding to LacY Mutants.
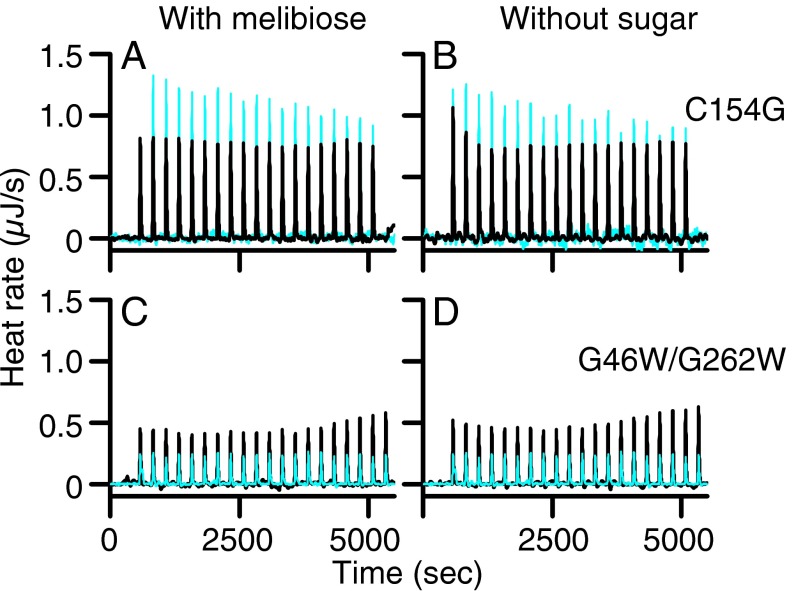
Two structurally resolved LacY mutants, C154G (33, 35) and G46W/G262W (38), bind sugar with an affinity similar to WT LacY but do not catalyze transport (32, 46, 47). In the absence of sugar, mutants C154G and G46W/G262W exhibit inward-open and outward-open conformations, respectively. A previous study reported that C154G LacY does not bind IIAGlc (10). Titration of C154G LacY with IIAGlc (Fig. 3, Upper, black curves) or with buffer (cyan) in the presence (Fig. 3A) or absence (Fig. 3B) of melibiose yields exothermic peaks with flat titration curves. The data are consistent with the conclusion that IIAGlc does not bind to the C154G LacY mutant. Similar results were obtained with mutant G46W/G262W (Fig. 3, Lower, black curves) in the presence (Fig. 3C) or absence (Fig. 3D) of melibiose, indicating that IIAGlc also does not bind to either the outward-open (47) or the almost occluded, outward-open conformation of the conformationally compromised LacY mutant.
Fig. 3.
Titration of LacY mutants with IIAGlc. Thermograms were recorded at 25 °C in the presence or absence of melibiose at 10 mM. (A and B) C154G LacY mutant (50 μM) was titrated with IIAGlc at a concentration of 780 μM. (C and D) G46W/G262W LacY mutant (50 μM) was titrated with IIAGlc at a concentration of 455 μM. Black curves, injections of IIAGlc into LacY mutants. Cyan curves, injections of IIAGlc into buffer.
Heat Capacity Change of IIAGlc Binding to LacY.
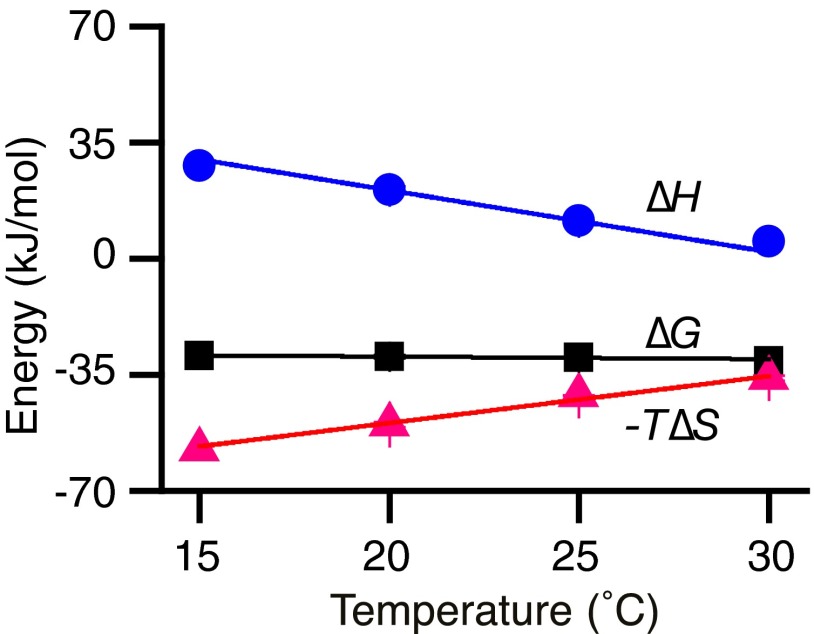
IIAGlc binding to WT LacY in the presence of melibiose was tested further over a range of temperatures from 15 to 30 °C. With increasing temperature, −T∆S becomes less favorable and ∆H becomes less unfavorable (Fig. 4 and Table 1). As a result of the good enthalpy–entropy compensation, there is little change in free energy (∆G) or Kd with temperature. The linear fit of ∆H with temperature reveals a large negative value of heat capacity change (∆Cp) (slope) of about −1,566 J/mol⋅K.
Fig. 4.
Temperature effect and ∆Cp determination. Under the conditions described in the legend to Fig. 2, IIAGlc binding to the WT LacY in the presence of melibiose was tested at 15, 20, 25, and 30 °C. Thermodynamic data are presented in Table 1; ∆G, ∆H, and −T∆S values are plotted against temperature and fitted with a linear function. The slope (∆H/∆T) is ∆Cp, which is negative (−1,566 J/mol⋅K).
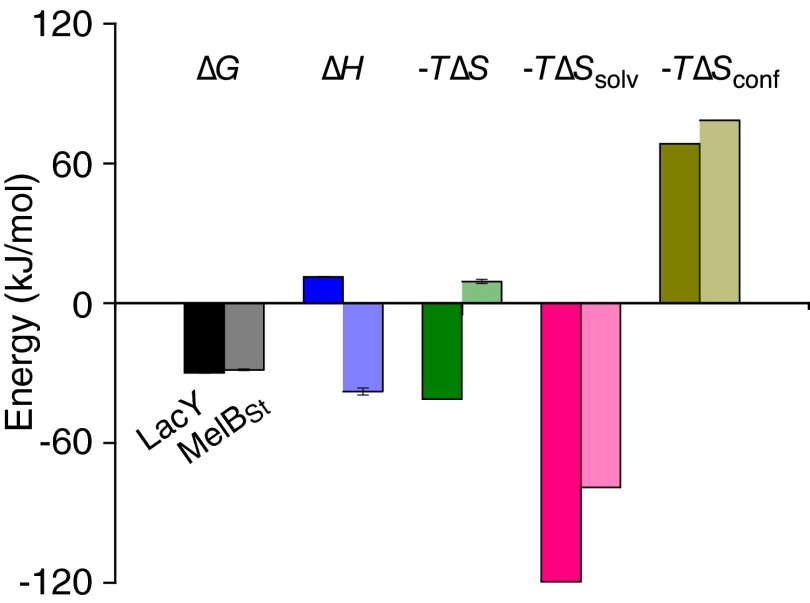
The solvation entropy change (∆Ssolv) is associated with the change in solvation of IIAGlc and LacY upon binding. Because the ∆Cp value has been determined, the ∆Ssolv value at 25 °C can be calculated from the equation ∆Ssolv = ∆Cp ln (298.15/385.15) (48, 49), yielding a value of 400.9 J/mol⋅K. The calculated −T∆Ssolv is −119.53 kJ/mol, suggesting that a very favorable solvation entropy is the major driving force for IIAGlc binding to LacY (Fig. 5 and Table 2). The measured total reaction ∆S is the net value of ∆Smix + ∆Ssolv + ∆Sconf (48, 49). The mixing entropy change ∆Smix can be estimated by ∆Smix = R ln (1/55.5), that is, −33 J/mol⋅K (48, 49). ∆Sconf is the conformational entropy change associated with the change in the side-chain and backbone conformation of IIAGlc and LacY upon binding. Because the ∆S value at 25 °C was also determined (Table 1), ∆Sconf can be calculated from the equation ∆Sconf = ∆S − ∆Ssolv − ∆Smix, which yields a value of −229.63 J/mol⋅K. The calculated −T∆Sconf is 68.46 kJ/mol with a positive sign, suggesting that the conformational entropy for the complex is largely unfavorable (Fig. 5 and Table 2).
Fig. 5.
Comparison of binding energy of IIAGlc to LacY or MelBSt. The thermodynamic data for IIAGlc binding to LacY (first bar in each pair) are from Tables 1 and 2; data for IIAGlc binding to MelBSt (second bar in each pair) are from ref. 26. Error bars denote SEM, n = 2–3.
Table 2.
Parameterization of reaction entropy change ∆S
Inhibition of Galactoside Affinity by IIAGlc.
Binding of nitrophenyl-α-d-galactopyranoside (α-NPG) to LacY has been determined by a number of methods including ITC (50–52). Titration of WT LacY with a solution of 1 mM α-NPG reveals exothermic peaks (Fig. 6A, black curve). By contrast, injection of nitrophenyl-α-d-glucoside (α-NPGlu), which does not bind to LacY, yields a flat titration curve with small exothermic peaks (Fig. 6A, Inset), strongly supporting the conclusion that the α-NPG titration curve reflects specific binding to LacY. Curve fitting using the one-site independent binding model (Fig. 6D, black curve) reveals a Kd value of ca 27 μM and, energetically, the interaction is driven by both favorable ∆H and −T∆S (Fig. 6G and Table 3), which is consistent with previous observations (51).
Fig. 6.
Sugar-binding energetics and IIAGlc effect. (A–C) Equilibrated to 25 °C, α-NPG at 1 mM was injected into the cell containing WT LacY (120 μM), or α-NPG at 250 μM was titrated into the C154G mutant or G46W/G262W LacY (50 μM), without (black curves) or preequilibrated with IIAGlc at a concentration threefold higher than the LacY concentration (orange curves). DMSO (0.5%) was present in both titrand and titrant to guarantee α-NPG solubility. Cyan curves, injections of α-NPG at 1 mM (A) or 250 μM (B and C) solution into buffers under the same conditions. (A, Inset) Thermograms were recorded from the injection of 1 mM α-NPGlu solution into 120 μM LacY (black curve) or buffer (cyan curve). (D–F) ∆Q was plotted against the α-NPG/LacY molar ratio and fitted to a one-site independent binding model. Empty squares, titration of LacY with α-NPG at 1 mM solution in the absence of IIAGlc; filled yellow triangles, titration of LacY–IIAGlc complex with α-NPG at 1 mM solution. (D, Inset) Data derived from injecting a solution of 10 mM α-NPG (violet curve) or α-NPGlu (gray triangles) into 120 μM LacY/360 μM IIAGlc complex. (G–I) Sugar-binding energy. ∆G, ∆H, and −T∆S values in the absence (histograms filled with lighter colors) and presence of IIAGlc (histograms filled with darker colors) that are from Table 3. Error bars denote SEM, n = 2.
Table 3.
IIAGlc inhibition of α-NPG binding
| Protein | Kd (μM) | ∆G (kJ/mol) | ∆H (kJ/mol) | −T∆S (kJ/mol) |
| WT LacY | 27.50 (4.40*) | −26.06 (0.40) | −13.25 (1.15) | −12.80 (1.55) |
| WT/IIAGlc | 1,013 (114) | −17.11 (0.27) | −12.74 (0.75) | −4.38 (1.02) |
| C154G mutant | 10.80 | −28.33 | −31.56 | 3.23 |
| C154G/IIAGlc | 10.70 | −28.38 | −32.66 | 4.28 |
| G46W/G262W mutant | 6.16 (0.28) | −29.75 (0.11) | −45.10 (0.33) | 15.35 (0.44) |
| G46W/G262W/IIAGlc | 7.05 (0.69) | −29.42 (0.24) | −47.84 (0.31) | 18.42 (0.07) |
ITC measurements were studied at 25 °C; data are presented in Fig. 6.
SEM, number of tests, 2.
Sequential injection of a 1 mM solution of α-NPG into LacY preequilibrated with IIAGlc at a ratio of 3:1 (IIAGlc:LacY) yields small heat changes and a flat titration, suggesting that IIAGlc may significantly inhibit sugar binding (Fig. 6 A and D, orange curve or triangles). To determine the affinity and free energy for binding, a 10 mM solution of α-NPG was used, which yields a Kd value of about 1 mM, an increase of ∼37-fold (Fig. 6D, Inset, violet curve). As a control, injection of α-NPGlu at the higher concentration is flat (Fig. 6D, Inset, gray triangles). Strikingly, the decrease in −T∆S accounts for the decrease in ∆G, because ∆H does not change significantly (Fig. 6G and Table 3).
α-NPG binding to C154G LacY exhibits thermodynamic features similar to those described in previous studies (Fig. 6 B, E, and H and Table 3) (51). Titration of G46W/G262W LacY with α-NPG yields a Kd value of 6 μM (Fig. 6 C, F, and I and Table 3). In both of the conformationally restrained mutants, the free-energy contribution is different from the WT; ∆H drives sugar binding and is opposed by −T∆S, particularly in the G46W/G262W LacY. As further controls for the observed IIAGlc inhibitory effects on sugar binding, both mutants, which do not bind IIAGlc, also do not exhibit decreased α-NPG affinity in the presence of IIAGlc (Fig. 6 B and C, orange curves). The results strongly support the conclusion that IIAGlc inhibits the affinity of LacY for galactoside.
Discussion
The ITC studies presented here show that IIAGlc regulation of LacY and MelB (26) is due to direct interaction with these permeases. In both cases, IIAGlc binding exhibits similar affinities and a stoichiometry of unity. With LacY, binding is endothermic and driven solely by ∆S with compensation by ∆H. In contrast, with MelB, IIAGlc binding is exothermic and driven solely by enthalpic forces with entropic compensation (Fig. 5). The observation that different driving forces for IIAGlc binding are observed for the two conformationally flexible permeases is notable. In either case, linear enthalpy–entropy compensation is obtained, which plays a crucial role in the thermodynamic mechanisms of the protein–protein interactions.
Thus far, the conformation of the IIAGlc-bound state has not been revealed with either MelB or LacY. MelBSt has a closed cytoplasmic surface, and IIAGlc binding is enthalpy-driven. LacY is open on the cytoplasmic side, and IIAGlc binding is entropy-driven, specifically by solvation entropy. A favorable solvation entropy change (−T∆Ssolv with negative value) usually indicates that water molecules, which are localized on the surfaces of the binding molecules, gain degrees of freedom by being released to the bulk solvent upon binding (53). This suggests that LacY must undergo a structural rearrangement(s) to form a surface that binds IIAGlc, accompanied by a large conformational change that involves release of water molecules. Consistently, the two conformationally constrained LacY mutants (C154G in an inside-open conformation and G46W/G262W in an inside-closed configuration) do not bind IIAGlc. Probably, both proteins are unable to form a IIAGlc-binding surface because of restricted conformational flexibility.
Regardless of the different thermodynamic mechanisms responsible for IIAGlc binding to LacY and MelB, a common feature is observed. The observed large unfavorable ∆Sconf indicates that the IIAGlc-bound LacY or MelB loses degrees of freedom with respect to conformational changes. This is consistent with the functionality of IIAGlc. On the other hand, sugar binding to LacY involves induced fit (38) in which there is a conformational transition from an open conformer to an occluded state that can open to either side of the membrane. We have shown that IIAGlc decreases α-NPG affinity by more than 30-fold with LacY (Table 3) and by 5-fold with MelBSt (26). IIAGlc binding prevents both permeases from interacting optimally with sugars and thereby inhibits the induced-fit process with both regulated permeases. This conclusion is also supported by the energetic change observed with respect to the driving force for sugar binding. With the LacY–IIAGlc complex, the large decrease in sugar-binding affinity is due solely to a decrease in favorable entropy change (Fig. 6). It appears that the initial contacts involving the specificity of sugar binding are enthalpically driven, probably through polar or hydrophilic interactions. Taken together, the thermodynamic insight into the mechanism of IIAGlc regulation of disaccharide uptake by LacY and MelB involves blocking conformational transitions and trapping both permeases in a low-affinity state.
When any PTS sugar is being transported, IIAGlc is predominantly in the dephosphorylated form, which can bind to and inhibit the activity of non-PTS permeases. In the absence of PTS sugars, IIAGlc is mainly in the phosphorylated form, which does not bind to or inhibit permeases (3, 5). Thus, the global mechanism for regulation is clearly dependent on the functionality of the PTS. The regulatory protein IIAGlc blocks inducer entry into the cell, which prevents activation of a specific operon. As a consequence, cells preferentially use glucose by the PTS via a mechanism defined previously as inducer exclusion (54).
A comparison (Fig. 5) of the current study on LacY with the previous (26) study on MelB indicates that the objective of permease inhibition by IIAGlc can be accomplished by a variety of thermodynamic mechanisms.
Materials and Methods
Reagents.
Nitrophenyl-α-galactoside and nitrophenyl-α-glucoside were purchased from Sigma-Aldrich. The detergents undecyl-β-d-maltopyranoside and dodecyl-β-d-maltopyranoside were from Anatrace.
Plasmids and Strains.
The T7-based expression plasmid p7XNH3/IIA-NH10 encoding E. coli IIAGlc with a 10-His tag and a 9-residue linker (MHHHHHHHHHHLEVLFQGPS) at the N terminus was constructed as described (26). Construction of plasmids pT7-5 LacY (32), pT7-5/C154G LacY (33), and pT7-5/G46W/G262W LacY (38), each with a His tag at the C terminus to allow metal-affinity purification, has been described. Overexpression of IIAGlc or LacY was performed in the E. coli T7 express strain (NEB) or XL1 Blue (Stratagene), respectively.
IIAGlc Expression and Purification.
The expression and purification of IIAGlc were carried out as described (26). Briefly, cells were grown in LB containing 5 mM glucose and induced with isopropyl β-d-1-thiogalactopyranoside. IIAGlc was purified by cobalt-affinity chromatography and concentrated to ∼100 mg/mL using a Vivaspin 20 (5,000 MWCO PES; Millipore) in a buffer containing 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, and 10% (vol/vol) glycerol. Protein was measured by the Micro BCA Protein Assay (Pierce Biotechnology). Purified IIAGlc was in the unphosphorylated form as analyzed with both SDS/PAGE and Phos-tag SDS/PAGE (26).
LacY Expression and Purification.
Cell growth, overexpression, and purification of LacY proteins were carried out as described for LacY (32) with a few modifications. The buffer used for the WT LacY contained 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 0.035% undecyl-β-d-maltopyranoside, and 10% glycerol. The buffer used for the LacY mutants was the same except that dodecyl-β-d-maltopyranoside (0.01%) was used instead of undecyl-β-d-maltopyranoside. Proteins were concentrated to 15–20 mg/mL, aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C.
ITC.
ITC measurements were performed on a Nano Isothermal Titration Calorimeter (TA Instruments). Purified LacY was placed in the sample cell with a reaction volume of 163 μL. IIAGlc was prepared in the same buffer by dilution from a highly concentrated sample, and then 2- or 2.5-μL aliquots were injected incrementally into the sample cell at an interval of 250 or 300 s with constant stirring at 250 rpm.
For sugar binding to LacY or the LacY–IIAGlc complex, α-NPG or α-NPGlu was dissolved in dimethyl sulfoxide (DMSO) and diluted with assay buffer to 1 or 10 mM containing 0.5% DMSO. DMSO was added to the protein samples for buffer matching. When testing for the IIAGlc effect, IIAGlc was incubated with LacY at a 3:1 ratio for 3 h before injection of sugar.
The raw thermogram was applied for a baseline correction. A blank correction for the heat of dilution was made by subtracting the integrated peak area from a constant estimated from the convergence value of the final injections. The corrected heat change (∆Q) was plotted against the molar ratio of IIAGlc/LacY or sugar/LacY. Binding stoichiometry, the association constant (Ka), and ∆H values were directly determined by fitting the data using the one-site independent binding model (55) provided by NanoAnalyze version 2.3.6 software (TA Instruments). The dissociation constant is 1/Ka. ∆S values were obtained by calculation using the equation ∆G = −RT ln Ka and T∆S = ∆H − ∆G. R, gas constant, 8.315 J/mol⋅K; T, absolute temperature. Parameterization of ∆S was calculated as described previously (49, 53). Total ∆S = ∆Smix + ∆Ssolv + ∆Sconf. ∆Smix was calculated as reported (48), which reflects the mixing of solute and solvent molecules, and entropy change due to changes in translational/rotational degrees of freedom. Based on the bimolecular binding reaction using a 1 M standard state, ∆Smix = R ln (1/55.5) = −33 J/mol⋅K. The entropy of polar and apolar solvation is close to zero at a temperature of near 385 K, and then solvent entropy change at 25 °C is given as ∆Ssolv = ∆Cp ln (298.15/385.15) (48). ∆Cp of the binding was obtained by fitting ∆H versus temperature. ∆Sconf = ∆S − ∆Smix − ∆Ssolv.
Acknowledgments
This work was supported by the National Science Foundation (Grant MCB-1158085 to L.G.), the National Institutes of Health (Grants R01 DK051131 and R01 DK069463 to H.R.K.; Grant R01 GM095538 to L.G.), and the National Heart, Lung, and Blood Institute (the intramural research program) (A.P.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Kaback HR. The role of the phosphoenolpyruvate-phosphotransferase system in the transport of sugars by isolated membrane preparations of Escherichia coli. J Biol Chem. 1968;243(13):3711–3724. [PubMed] [Google Scholar]
- 2.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengeler JW, Jacobson GR, editors. Postma PW. Am. Soc. Microbiol; Washington, DC: 1996. pp. 1149–1174. [Google Scholar]
- 4.Guan L, Kaback HR. Glucose/sugar transport in bacteria. In: Lennarz WJ, Lane MD, editors. Encyclopedia of Biological Chemistry. 2nd Ed. Elsevier; Oxford: 2013. pp. 387–390. [Google Scholar]
- 5.Deutscher J, et al. The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev. 2014;78(2):231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller-Hill B. The lac Operon: A Short History of a Genetic Paradigm. Walter de Gruyter; Berlin: 1996. [Google Scholar]
- 7.Hanatani M, et al. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J Biol Chem. 1984;259(3):1807–1812. [PubMed] [Google Scholar]
- 8.Mizushima K, et al. Cloning and sequencing of the melB gene encoding the melibiose permease of Salmonella typhimurium LT2. Mol Gen Genet. 1992;234(1):74–80. doi: 10.1007/BF00272347. [DOI] [PubMed] [Google Scholar]
- 9.Osumi T, Saier MH., Jr Regulation of lactose permease activity by the phosphoenolpyruvate:sugar phosphotransferase system: Evidence for direct binding of the glucose-specific enzyme III to the lactose permease. Proc Natl Acad Sci USA. 1982;79(5):1457–1461. doi: 10.1073/pnas.79.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sondej M, Weinglass AB, Peterkofsky A, Kaback HR. Binding of enzyme IIAGlc, a component of the phosphoenolpyruvate:sugar phosphotransferase system, to the Escherichia coli lactose permease. Biochemistry. 2002;41(17):5556–5565. doi: 10.1021/bi011990j. [DOI] [PubMed] [Google Scholar]
- 11.Hogema BM, Arents JC, Bader R, Postma PW. Autoregulation of lactose uptake through the LacY permease by enzyme IIAGlc of the PTS in Escherichia coli K-12. Mol Microbiol. 1999;31(6):1825–1833. doi: 10.1046/j.1365-2958.1999.01319.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda M, Osaki N, Tsuda M, Tsuchiya T. Preferential utilization of glucose over melibiose, and vice versa, in a pts mutant of Salmonella typhimurium. Chem Pharm Bull (Tokyo) 1992;40(6):1637–1640. doi: 10.1248/cpb.40.1637. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda M, Wilson TH, Tsuchiya T. Regulation of galactoside transport by the PTS. J Mol Microbiol Biotechnol. 2001;3(3):381–384. [PubMed] [Google Scholar]
- 14.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaback HR. The passion of the permease. In: Wikström M, editor. Biophysical and Structural Aspects of Bioenergetics. R Soc Chem; Cambridge, UK: 2005. pp. 359–373. [Google Scholar]
- 16.Wilson DM, Wilson TH. Cation specificity for sugar substrates of the melibiose carrier in Escherichia coli. Biochim Biophys Acta. 1987;904(2):191–200. doi: 10.1016/0005-2736(87)90368-3. [DOI] [PubMed] [Google Scholar]
- 17.Tokuda H, Kaback HR. Sodium-dependent methyl 1-thio-β-D-galactopyranoside transport in membrane vesicles isolated from Salmonella typhimurium. Biochemistry. 1977;16(10):2130–2136. doi: 10.1021/bi00629a013. [DOI] [PubMed] [Google Scholar]
- 18.Tsuchiya T, Raven J, Wilson TH. Co-transport of Na+ and methyl-beta-D-thiogalactopyranoside mediated by the melibiose transport system of Escherichia coli. Biochem Biophys Res Commun. 1977;76(1):26–31. doi: 10.1016/0006-291x(77)91663-1. [DOI] [PubMed] [Google Scholar]
- 19.Bassilana M, Pourcher T, Leblanc G. Facilitated diffusion properties of melibiose permease in Escherichia coli membrane vesicles. Release of co-substrates is rate limiting for permease cycling. J Biol Chem. 1987;262(35):16865–16870. [PubMed] [Google Scholar]
- 20.Guan L, Nurva S, Ankeshwarapu SP. Mechanism of melibiose/cation symport of the melibiose permease of Salmonella typhimurium. J Biol Chem. 2011;286(8):6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan L, Jakkula SV, Hodkoff AA, Su Y. Role of Gly117 in the cation/melibiose symport of MelB of Salmonella typhimurium. Biochemistry. 2012;51(13):2950–2957. doi: 10.1021/bi300230h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahramanoglou C, Webster CL, El-Robh MS, Belyaeva TA, Busby SJ. Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon. J Bacteriol. 2006;188(9):3199–3207. doi: 10.1128/JB.188.9.3199-3207.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJ. Binding of the Escherichia coli MelR protein to the melAB promoter: Orientation of MelR subunits and investigation of MelR-DNA contacts. Mol Microbiol. 2003;48(2):335–348. doi: 10.1046/j.1365-2958.2003.t01-1-03434.x. [DOI] [PubMed] [Google Scholar]
- 24.Park YH, Lee BR, Seok YJ, Peterkofsky A. In vitro reconstitution of catabolite repression in Escherichia coli. J Biol Chem. 2006;281(10):6448–6454. doi: 10.1074/jbc.M512672200. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Oldham ML, Davidson AL, Chen J. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature. 2013;499(7458):364–368. doi: 10.1038/nature12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hariharan P, Guan L. Insights into the inhibitory mechanisms of the regulatory protein IIA(Glc) on melibiose permease activity. J Biol Chem. 2014;289(47):33012–33019. doi: 10.1074/jbc.M114.609255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondej M, Vázquez-Ibar JL, Farshidi A, Peterkofsky A, Kaback HR. Characterization of a lactose permease mutant that binds IIAGlc in the absence of ligand. Biochemistry. 2003;42(30):9153–9159. doi: 10.1021/bi034406a. [DOI] [PubMed] [Google Scholar]
- 28.Sondej M, Sun J, Seok YJ, Kaback HR, Peterkofsky A. Deduction of consensus binding sequences on proteins that bind IIAGlc of the phosphoenolpyruvate:sugar phosphotransferase system by cysteine scanning mutagenesis of Escherichia coli lactose permease. Proc Natl Acad Sci USA. 1999;96(7):3525–3530. doi: 10.1073/pnas.96.7.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sondej M, et al. Topography of the surface of the Escherichia coli phosphotransferase system protein enzyme IIAglc that interacts with lactose permease. Biochemistry. 2000;39(11):2931–2939. doi: 10.1021/bi9919596. [DOI] [PubMed] [Google Scholar]
- 30.Hurley JH, et al. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science. 1993;259(5095):673–677. [PubMed] [Google Scholar]
- 31.Wang G, et al. Solution structure of the phosphoryl transfer complex between the signal transducing proteins HPr and IIA(glucose) of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. EMBO J. 2000;19(21):5635–5649. doi: 10.1093/emboj/19.21.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104(39):15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 34.Chaptal V, et al. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc Natl Acad Sci USA. 2011;108(23):9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25(6):1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112(5):1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ethayathulla AS, et al. Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar H, et al. Structure of sugar-bound LacY. Proc Natl Acad Sci USA. 2014;111(5):1784–1788. doi: 10.1073/pnas.1324141111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J Membr Biol. 2011;239(1-2):85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer-Lipp K, et al. The inner interhelix loop 4-5 of the melibiose permease from Escherichia coli takes part in conformational changes after sugar binding. J Biol Chem. 2006;281(36):25882–25892. doi: 10.1074/jbc.M601259200. [DOI] [PubMed] [Google Scholar]
- 42.Yousef MS, Guan L. A 3D structure model of the melibiose permease of Escherichia coli represents a distinctive fold for Na+ symporters. Proc Natl Acad Sci USA. 2009;106(36):15291–15296. doi: 10.1073/pnas.0905516106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.León X, Lórenz-Fonfría VA, Lemonnier R, Leblanc G, Padrós E. Substrate-induced conformational changes of melibiose permease from Escherichia coli studied by infrared difference spectroscopy. Biochemistry. 2005;44(9):3506–3514. doi: 10.1021/bi048301z. [DOI] [PubMed] [Google Scholar]
- 44.Ganea C, et al. G117C MelB, a mutant melibiose permease with a changed conformational equilibrium. Biochim Biophys Acta. 2011;1808(10):2508–2516. doi: 10.1016/j.bbamem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Seok YJ, Sun J, Kaback HR, Peterkofsky A. Topology of allosteric regulation of lactose permease. Proc Natl Acad Sci USA. 1997;94(25):13515–13519. doi: 10.1073/pnas.94.25.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smirnova IN, Kaback HR. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 2003;42(10):3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 47.Smirnova I, Kasho V, Sugihara J, Kaback HR. Trp replacements for tightly interacting Gly-Gly pairs in LacY stabilize an outward-facing conformation. Proc Natl Acad Sci USA. 2013;110(22):8876–8881. doi: 10.1073/pnas.1306849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker BM, Murphy KP. Dissecting the energetics of a protein-protein interaction: The binding of ovomucoid third domain to elastase. J Mol Biol. 1997;268(2):557–569. doi: 10.1006/jmbi.1997.0977. [DOI] [PubMed] [Google Scholar]
- 49.Zakariassen H, Sorlie M. Heat capacity changes in heme protein-ligand interactions. Thermochim Acta. 2007;464(1-2):24–28. [Google Scholar]
- 50.Sahin-Tóth M, Gunawan P, Lawrence MC, Toyokuni T, Kaback HR. Binding of hydrophobic D-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry. 2002;41(43):13039–13045. doi: 10.1021/bi0203076. [DOI] [PubMed] [Google Scholar]
- 51.Nie Y, Smirnova I, Kasho V, Kaback HR. Energetics of ligand-induced conformational flexibility in the lactose permease of Escherichia coli. J Biol Chem. 2006;281(47):35779–35784. doi: 10.1074/jbc.M607232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smirnova IN, Kasho VN, Kaback HR. Direct sugar binding to LacY measured by resonance energy transfer. Biochemistry. 2006;45(51):15279–15287. doi: 10.1021/bi061632m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velázquez Campoy A, Freire E. ITC in the post-genomic era...? Priceless. Biophys Chem. 2005;115(2-3):115–124. doi: 10.1016/j.bpc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Saier MH., Jr Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989;53(1):109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freire E, Mayorga OL, Straume M. Isothermal titration calorimetry. Anal Chem. 1990;62(18):950A–959A. [Google Scholar]