Abstract
Circulating bilirubin, a natural antioxidant, is associated with decreased risk of type 2 diabetes (T2D), but the nature of the relationship remains unknown. We performed Mendelian randomization in a prospective cohort of 3,381 participants free of diabetes at baseline (aged 28-75 years; women, 52.6%). We used rs6742078 located in UDP-glucuronosyltransferase (UGT1A1) locus as instrumental variable (IV) to study a potential causal effect of serum total bilirubin on T2D risk. T2D developed in a total of 210 (6.2%) participants during a median follow-up of 7.8 years. In adjusted analyses, rs6742078, which explained 19.5% of bilirubin variation, was strongly associated with total bilirubin (a 0.68-SD increase in bilirubin levels per T allele; P<1×10−122) and was also associated with T2D risk (OR 0.69 [95%CI, 0.54-0.90]; P=0.006). Per 1-SD increase in log-transformed bilirubin levels, we observed a 25% (OR 0.75 [95%CI, 0.62-0.92]; P=0.004) lower risk of T2D. In Mendelian randomization analysis, the causal risk reduction for T2D was estimated to be 42% (causal ORIVestimation per 1-SD increase in log-transformed bilirubin 0.58 [95%CI, 0.39-0.84]; P=0.005), which was comparable to the observational estimate (Durbin-Wu-Hausman chi-square test Pfor difference =0.19). These novel results provide evidence that elevated bilirubin is causally associated with risk of T2D and support its role as a protective determinant.
Keywords: Bilirubin, liver, epidemiology, type 2 diabetes, Mendelian randomization
INTRODUCTION
Experimental and human studies show that elevated bilirubin levels are associated with decreased risk of type 2 diabetes (T2D) and diabetes related outcomes (1-6). Bilirubin, might antagonize oxidative stress by acting as an antioxidant and cytoprotectant, which may have beneficial effects in diseases related to oxidative stress (7-10). For example, we have previously shown that elevated bilirubin levels are associated with a reduction in mortality in the carriers of iron overload genotypes, which are phenotypically associated with increased oxidative stress (11). It is, however, unclear whether the association between bilirubin and diabetes is free of unobserved confounders. Apart from confounding, bilirubin concentrations could be changed as a consequence of T2D pathology (i.e., reverse causality). Therefore, causal inference of the effect of bilirubin on T2D (i.e., bilirubin being an intermediate phenotype in the etiology of T2D) is uncertain in conventional observational studies (12).
One approach to overcome these aforementioned limitations, is to implement an instrumental variable (IV), which represents bilirubin levels and allows for assessment of the relation to T2D robust to confounding and reverse causality biases (13; 14). Recent genome wide association studies (GWAS) have identified that rs6742078, the single nucleotide polymorphism (SNP) which is mapped to hepatic uridine diphosphate-glucuronyltransferase (UGT1A1), explains 18% of the variation in serum total biliruin levels (15-17). The UGT1A1 gene encodes hepatic uridine diphosphate-glucuronyltransferase, which is a major enzyme in bilirubin conjugation and in regulation of bilirubin levels (5; 15-17). When UGT1A1 is down-regulated, bilirubin levels rise subsequently. Given the fact that inheritance of either a T or a G allele of rs6742078 is randomly defined prior to disease onset (i.e., at time of conception, based on Mendel’s laws), the variation of rs6742078 is robust to unobserved confounding and reverse causality. Therefore, rs6742078 is a suitable candidate to be used as an IV representing bilirubin variation between individuals. The approach of using genetic instruments (i.e., rs6742078) to test causal association of a given intermediate exposure (i.e., bilirubin) with a disease outcome (i.e., T2D) is called Mendelian Randomization (14). We hypothesized that implementing rs6742078 as an IV could provide evidence that the previously reported association between serum bilirubin and T2D is causal or not.
We aimed to investigate whether elevated total bilirubin levels are causally associated with a lower risk of new-onset T2D. By implementation of Mendelian randomization, we examined whether rs6742078(T) is associated with serum total bilirubin levels and T2D risk in a population-based cohort study.
RESEARCH DESIGN AND METHODS
Study population and design
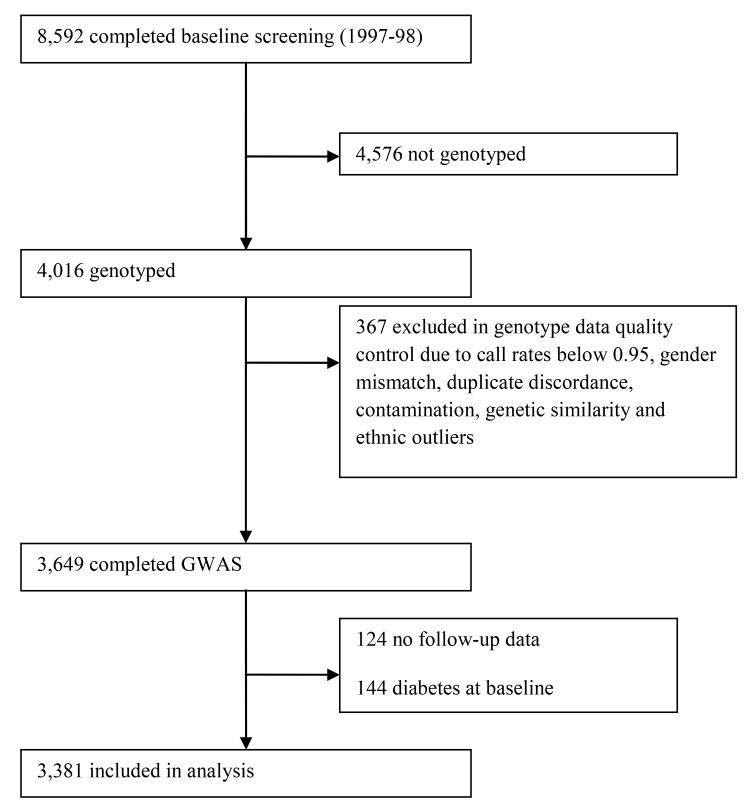
The Prevention of Renal and Vascular End-stage Disease (PREVEND) study is a Dutch population-based cohort for which 8,592 participants (age range, 28-75 years) from the city of Groningen, the Netherlands were recruited between 1997 and 1998. We examined a random sample of 4,016 individuals of the total cohort in which genotyping was performed. Details of the PREVEND study have been published previously (18). Among 4,016 individuals, 367 participants were excluded because of not passing quality control of DNA analyses, 124 participants because of missing follow-up data or information about medication and 144 individuals because of prevalent diabetes at baseline, leaving 3,381 participants eligible for analyses (Figure 1).
Figure 1. Study population sample from the PREVEND study cohort.
For quality control (QC), samples were excluded (n=367) due to call rates below 0.95, sex mismatch, duplicate discordance, contamination and genetic similarity (identity by state > 0.2). Population stratification was assessed by principal component analysis over the sample correlation matrix, based on 16,842 independent (LD-pruned) SNPs. We excluded samples when they diverged from the mean with at least 3 standard deviations (Z-score > 3) for the first five principal components.
The PREVEND study has been approved by the medical ethics committee, University Medical Center Groningen, and conformed to the principles outlined in the Declaration of Helsinki. All participants gave written informed consent.
Baseline study measurements
At baseline and three rounds of screening from 1997-1998 (i.e., baseline examination) until January 1st 2007 (i.e., third examination), study participants underwent two outpatient visits to obtain clinical characteristics and to collect two 24-hour urine samples on two consecutive days (18). Furthermore, information on medication use was substantiated using pharmacy-based data from all community pharmacies in the city of Groningen (19). Blood samples were obtained after overnight fasting stored at −80°C until analyses without previous thawing and refreezing (Supplementary Table 1). Total bilirubin was measured by a colorimetric assay (2,4-dichloroaniline reaction; Merck MEGA, Darmstadt, Germany).
Definition of outcome
During follow-up, participants were classified as new-onset T2D cases if fasting plasma glucose was ≥ 7.0 mmol/L (126 mg/dL), random sample plasma glucose was ≥ 11.1 mmol/L (200 mg/dL), they reported a physician diagnosis of T2D, or received insulin or oral hypoglycemic agents based on a central pharmacy registration. We included cases that were diagnosed with T2D from the first three months after the baseline screening visits (1997-1998) until January 2007 (20; 21).
Genotyping and quality control
Genotyping was performed using Illumina HumanCytoSNP-12 arrays. Genotypes of 3,649 individuals were available pre-imputation (See Figure 1 for quality control of samples). SNPs were called using Illumina Genome Studio software. SNPs were excluded with minor allele frequency <0.01, call rate <0.95, or deviation from Hardy-Weinberg equilibrium (P<1×10−5). Genome-wide genotype imputation was performed using Beagle v. 3.1.0. We used the NCBI build 36 of Phase II HapMap CEU data (release 22) as reference panel (22). Given the expected strong relationship of rs6742078 (imputation accuracy score >0.9) with bilirubin levels (16; 17) and the fact that rs6742078 is in high linkage disequilibrium (LD) (r2=0.88) with the functional UGT1A1*28 polymorphism (17), we decided that rs6742078 is a suitable IV for bilirubin in our study.
Statistical Analysis
We performed analyses in four steps. First, we examined the associations of bilirubin with participants’ characteristics, including age, sex, smoking, family history of diabetes, and associated clinical variables. This was performed using linear regression. Next, we used logistic regression to examine the association of bilirubin with new-onset T2D (model A). This model estimates an observed effect of bilirubin on T2D (βbilirubin-T2D coefficient). The odds ratio (OR) and its corresponding 95% confidence intervals (95% CIs) for the risk of T2D was calculated per 1-SD increase in log-transformed bilirubin levels. Model A was adjusted for age, sex, family history of diabetes, BMI, hypertension, smoking and fasting glucose (21). Second, we tested whether rs6742078 was associated with log-transformed bilirubin levels. We used linear regression under the assumption of an additive genetic model (model B), in agreement with previously published studies (17; 23). Model B was adjusted for the same covariates as those included in model A (14). This analysis provided the observed effect estimate, which was expressed as the change in the number of SDs change in log-transformed bilirubin per each copy increase in number of the T allele (range, 0-2). To estimate the proportion of bilirubin variation explained by the IV of the rs6742078 genotypes, we calculated the difference between r2 values (i.e., explained variance) for model B without the genotype and for model B with the genotype. Third, logistic regression was used to evaluate the association between rs6742078 and T2D (model C). We calculated the OR for T2D per each copy increase in the rs6742078(T) allele. Model C was adjusted for the same covariates as those of model A. Fourth, we used the Wald-type method of IV estimation to test a potential causal relationship between elevated total bilirubin levels and decreased T2D risk. For the Wald type method, the estimated risk is calculated using the following formula:
In this formula, ORgenotupe-T2D is the OR resulting from logistic regression between rs6742078 and T2D, representing risk of T2D per each copy of the rs6742078(T) allele (model C). The ßgenotype-bilirubin coefficient is obtained from the regression of bilirubin on the SNP per each copy of the T allele of rs6742078 (model B). To validate the causal estimate derived from the Wald-type method, we used two alternative methods, called the two-stage least squares (2SLS) method and the multiplicative generalized method of moments (MGMM) (Table 3). The first stage of the 2SLS method is to examine the observational association between rs6742078 and bilirubin by means of linear regression (model 1), saving the predicted values and the residuals. In the second stage, the predicted values of bilirubin from model 1 are used as covariates, with T2D as dependent variable in a logistic regression model, providing the βIV2SLS estimate (14). To obtain the MGMM βIV, the Stata command “ivpois” was used (24). The MGMM does not require distributional assumptions about bilirubin and thus can provide consistent estimation of causal OR (24).
Table 3. Causal estimates for type 2 diabetes and glycemic traits using Mendelian randomization analysis.
| Instrumental variable | Alleles (effect/other) | Mendelian randomization analysis | |||
|---|---|---|---|---|---|
| Method | Causal estimate (95%CI) | P value | Heterogeneity P value | ||
| Incidence of Type 2 diabetes, PREVEND | Per 1-SD log | ||||
| rs6742078 | T/G | Wald-type | 0.58 (0.39 to 0.84)* | 0.005 | - |
| 2SLS | 0.56 (0.38 to 0.81)* | 0.002 | - | ||
| MGMM | 0.50 (0.27 to 0.92)* | 0.027 | - | ||
| rs4149056 | C/T | MGMM | 0.58 (0.05 to 6.08) * | 0.65 | - |
| rs16928809 | A/G | MGMM | 0.17 (0.02 to 2.16) * | 0.17 | - |
| Genetic risk score | - | MGMM | 0.34 (0.15 to 0.77) * | 0.009 | - |
| Type 2 diabetes, DIAGRAM (28) † | Per 1-unit log (umol/L) | ||||
| rs6742078 | T/G | summary statistics | 1.14 (0.96 to 1.35) * | 0.13 | - |
| rs4149056 | C/T | summary statistics | 0.67 (0.28 to 1.62) * | 0.38 | - |
| rs16928809 | A/G | summary statistics | 0.44 (0.15 to 1.35) * | 0.15 | - |
| Genetic risk score | - | summary statistics | 1.09 (0.93 to 1.29) * | 0.28 | 0.14 |
| Glucose, MAGIC (29) | Per 1-unit log (umol/L) | ||||
| rs6742078 | T/G | summary statistics | −0.04 (−0.07 to −0.01) ‡ | 0.02 | - |
| rs4149056 | C/T | summary statistics | 0.01 (−0.19 to 0.20) ‡ | 0.95 | - |
| rs16928809 | A/G | summary statistics | −0.05 (−0.28 to 0.18) ‡ | 0.67 | - |
| Genetic risk score | - | summary statistics | −0.04 (−0.07 to −0.01) ‡ | 0.02 | 0.89 |
| Insulin, MAGIC (29) | Per 1-unit log (umol/L) | ||||
| rs6742078 | T/G | summary statistics | −0.04 (−0.07 to −0.00) ‡ | 0.03 | - |
| rs4149056 | C/T | summary statistics | 0.12 (−0.08 to 0.32) ‡ | 0.25 | - |
| rs16928809 | A/G | summary statistics | −0.04 (−0.27 to 0.20) ‡ | 0.76 | - |
| Genetic risk score | - | summary statistics | −0.03 (−0.07 to −0.00) ‡ | 0.05 | 0.33 |
| HOMA-IR, MAGIC (29) | Per 1-unit log (umol/L) | ||||
| rs6742078 | T/G | summary statistics | −0.04 (−0.08 to −0.00) ‡ | 0.03 | - |
| rs4149056 | C/T | summary statistics | 0.08 (−0.13 to 0.28) ‡ | 0.47 | - |
| rs16928809 | A/G | summary statistics | −0.06 (−0.31 to 0.19) ‡ | 0.63 | - |
| Genetic risk score | − | summary statistics | −0.04 (−0.07 to −0.00) ‡ | 0.04 | 0.55 |
The bilirubin genetic risk score consisted of rs6742078 plus two other bilirubin SNPs in SLCO1B1, rs4149056 and in SLC22A18, rs16928809 (17; 27-29).
The causal ORs for T2D were estimated using the Wald type method, the two-stage least squares (2SLS) method, the multiplicative generalized method of moments (MGMM) and the summary statistics (27; 28). The causal OR for T2D was calculated as OR=exp(βIV).
The DIAGRAM GWAS includes both prevalent and incident cases of type 2 diabetes.
The causal β coefficient (95%CI) for glucose (mmol/L), insulin and HOMA-IR was estimated for each SNP separately and the genetic risk score which is based on the combination of these three SNPs.
To examine whether the association between bilirubin and T2D has ‘endogeneity’, we compared the IV estimate with the observed estimate from standard logistic regression analysis (i.e., βbilirubin-T2D coefficient) using the Durbin-Wu-Hausman (DWH) chi-square test, which assesses whether there is a difference between these two estimates (14; 24). We calculated the power of our study to detect a causal effect for bilirubin, given our sample size and bilirubin variation explained by rs6742078 (17; 25).
To account for potential confounding bias, we tested whether rs6742078 is associated with common diabetes risk factors, other variables (Table 2) and 24-hour urine albumin excretion (UAE). The association with UAE was tested because of the enrichment of the PREVEND cohort with microalbuminuric individuals at baseline (18).
Table 2. Associations of baseline variables with serum bilirubin and rs6742078.
| Serum bilirubin | rs6742078 | |||
|---|---|---|---|---|
| Variables | Standardized β coefficients (95%CI) | P value | Standardized β coefficients (95%CI) | P value |
| Age | 0.012 (−0.021 to 0.046) | 0.46 | 0.013 (−0.021 to 0.046) | 0.46 |
| Female sex | −0.215 (−0.245 to −0.181) | <0.001 | 0.001 (−0.033 to 0.034) | 0.97 |
| Smoking | −0.015 (−0.050 to 0.017) | 0.35 | 0.003 (−0.030 to 0.037) | 0.87 |
| Alcohol use | 0.049 (0.015 to 0.082) | 0.01 | 0.02 (−0.013 to 0.054) | 0.24 |
| Family history of diabetes | −0.057 (−0.093 to −0.026) | 0.001 | 0.018 (−0.016 to 0.051) | 0.30 |
| Prior history of CVD | −0.026 (−0.060 to 0.007) | 0.13 | 0.026 (−0.003 to 0.064) | 0.14 |
| BMI | −0.119 (−0.152 to −0.085) | <0.001 | 0.006 (−0.028 to 0.039) | 0.74 |
| Waist circumference | −0.025 (−0.059 to 0.008) | 0.14 | −0.002 (−0.036 to 0.032) | 0.91 |
| Systolic blood pressure | 0.039 (0.00 to 0.072) | 0.02 | −0.007 (−0.040 to 0.027) | 0.70 |
| Diastolic blood pressure | 0.040 (0.003 to 0.070) | 0.03 | −0.003 (−0.036 to 0.031) | 0.88 |
| Hypertension | 0.013 (−0.020 to 0.046) | 0.44 | −0.017 (−0.051 to 0.016) | 0.32 |
| Fasting glucose | −0.021 (−0.055 to 0.013) | 0.22 | 0.025 (−0.009 to 0.058) | 0.15 |
| HDL cholesterol | 0.025 (−0.009 to 0.058) | 0.15 | −0.006 (−0.039 to 0.027) | 0.73 |
| Triglycerides | −0.116 (−0.148 to −0.082) | <0.001 | 0.001 (−0.033 to 0.034) | 0.97 |
| Fasting insulin | −0.106 (−0.140 to −0.072) | <0.001 | −0.002 (−0.036 to 0.032) | 0.87 |
| AST | 0.145 (0.112 to 0.178) | <0.001 | −0.003 (−0.036 to 0.030) | 0.85 |
| ALT | 0.082 (0.049 to 0.115) | <0.001 | 0.021 (−0.013 to 0.054) | 0.22 |
| GGT | 0.030 (−0.007 to 0.060) | 0.12 | 0.010 (−0.023 to 0.043) | 0.55 |
| Albumin | 0.247 (0.212 to 0.282) | <0.001 | −0.014 (−0.052 to 0.023) | 0.44 |
| ALP | −0.022 (−0.054 to 0.013) | 0.24 | 0.022 (−0.015 to 0.060) | 0.23 |
| hs-CRP | −0.115 (−0.147 to −0.080) | <0.001 | −0.010 (−0.044 to 0.023) | 0.55 |
| UAE | −0.031 (−0.066 to 0.002) | 0.07 | 0.006 (−0.027 to 0.040) | 0.72 |
BMI: body mass index, HDL: high-density lipoprotein, GGT: gamma-glutamyltransferase, ALT: alanine aminotransferase, AST: aspartate aminotransferase, ALP: Alkaline phosphatase, hs-CRP: high sensitivity C-reactive protein and UAE: urine albumin excretion.
To test for pleiotropy, we performed logistic regression with bilirubin as independent variable and T2D as dependent variable and saved the residuals. Next, we tested the correlation between the genotype and the residuals that are independent of total bilirubin levels (26). To further dissect potential pleiotropy, we also calculated IV estimates from two additional bilirubin SNPs, namely rs4149056 in SLCO1B1 and rs16928809 in SLC22A18(17), in PREVEND. Additionally, we extracted effect estimates for rs6742078, rs4149056 and rs16928809 from publically-available GWAS data on bilirubin, T2D (DIAbetes Genetics Replication And Meta-analysis [DIAGRAM]) or glycemic traits (Meta-Analyses of Glucose and Insulin-related traits Consortium [MAGIC]) (17)(27-29) (Supplementary Table 2), and we calculated a summary statistics genetic risk score for bilirubin, like described previously (27). We then evaluated the presence of heterogeneity (derived from the likelihood-based method (27)) on causal effects of these three SNPs on T2D, glucose, insulin and HOMA-IR (27-29) (Table 3). The presence of heterogeneity indicates potential pleiotropy (30).
We also performed a series of secondary analyses. First, we repeated regression models for bilirubin levels and rs6742078 by using a weighted method to compensate for the enrichment of the PREVEND cohort with microalbuminuric individuals (18). Second, we added the interaction terms of bilirubin×sex and bilirubin×age to model A or rs6742078×sex and rs6742078×age to model C, to evaluate potential sex or age differences in the association of bilirubin or rs6742078 with T2D, respectively. Third, we examined whether the effect estimates for bilirubin or rs6742078 were consistent across age strata of people <50 or ≥50 years old. Fourth, we accounted for the time between taking the sample for measurement of bilirubin (and determining the genotype) and the development of T2D by using a Cox regression in which we applied the approximate time to the incidence of T2D as the time scale in our analysis. The time to incidence of T2D cannot be ascertained exactly, because we screened for incident T2D periodically. We tested the proportional-hazards assumption indicating that the effect estimates do not vary over time. Fifth, a potential correlation between the drop-out rates and the genotype was tested. Sixth, we examined the association of rs6742078 with prevalent or a combination of both prevalent and incident T2D. A single imputation and predictive mean matching method was applied for missing data (21). For most baseline clinical variables, <1% was missing, whereas this was up to 8% for self-reported variables such as family history of diabetes. All the statistical analyses were carried out using IBM SPSS 19.0, Stata/IC version 11.2 (Stata Corp, College Station, Texas) or R version 2.15.1 (Vienna, Austria) for Windows (http://cran.r-project.org/).
RESULTS
Baseline characteristics of the population are shown in Table 1. During a median (Q1-Q3) follow-up for 7.7 (7.4-8.0) years, T2D developed in a total of 210 individuals (6.2%) (from 1997-98 until 2007).
Table 1. Baseline characteristics of the study participants.
| Total (n= 3,381) | Non-converters (n=3,171) | Incident type 2 diabetes cases (n=210) | P value | |
|---|---|---|---|---|
| Variables | ||||
| Age — yr | 49.4±12.4 | 49.0±12.4 | 55.2±9.9 | <0.001 |
| Male sex— no.(%) | 1710 (50.6) | 1573 (49.6) | 137 (65.2) | <0.001 |
| Current smoker— no.(%) | 1198 (35.4) | 1116 (35.2) | 82 (39.0) | 0.13 |
| Ex-smoker— no.(%) | 1240 (36.7) | 1158 (36.5) | 82 (39.0) | |
| Alcohol drinker— no.(%) | 2573 (76.5) | 2416 (76.6) | 157 (75.1) | 0.62 |
| Prior history of CVD — no.(%) | 134 (4.1) | 119 (3.9) | 15 (7.5) | 0.01 |
| Family history of diabetes— no.(%) | 527 (15.6) | 468 (14.8) | 59 (28.1) | <0.001 |
| BMI—kg/m2 | 26.0±4.1 | 25.8±4.0 | 29.3±4.6 | <0.001 |
| Waist circumference—cm | 88.6±13.1 | 87.9±12.8 | 99.5±11.7 | <0.001 |
| Systolic blood pressure— mmHg | 123.9±19.0 | 123.3±18.9 | 133.1±18.6 | <0.001 |
| Diastolic blood pressure–mmHg | 71.8±9.9 | 71.5±9.9 | 76.1±9.6 | <0.001 |
| Hypertension— no.(%) | 936 (27.7) | 833 (26.3) | 103 (49.0) | <0.001 |
| Fasting glucose— mmol/L | 4.7±0.6 | 4.7±0.6 | 5.6±0.8 | <0.001 |
| HDL cholesterol— mmol/L | 1.32±0.40 | 1.34±0.40 | 1.09±0.29 | <0.001 |
| Triglycerides— mmol/L | 1.16 (0.84-1.68) | 1.13 (0.83-1.65) | 1.64 (1.17-2.45) | <0.001 |
| Fasting insulin— mU/L | 7.9 (5.5-11.9) | 7.7 (5.4-11.5) | 13.0 (8.5-21.4) | <0.001 |
| Bilirubin — (μmol/L) | 7.7 ±3.6 | 7.7 ±3.6 | 7.1±3.1 | <0.001 |
| AST— U/L | 25.7±9.5 | 25.6±9.5 | 27.4±8.2 | 0.009 |
| ALT— U/L | 23.8±16.6 | 23.4±16.6 | 29.6±16.4 | <0.001 |
| GGT— U/L | 23.0 (16.0-38.0) | 23.0 (16.0-36.0) | 36.0 (24.7-51.7) | <0.001 |
| Albumin g/L | 45.8±2.8 | 45.8±2.8 | 45.2±3.2 | 0.003 |
| ALP— U/L | 65.3±22.3 | 65.0±22.5 | 70.0±18.3 | 0.002 |
| hs-CRP— mg/L | 1.29 (0.58-2.95) | 1.24 (0.56-2.87) | 2.22 (1.19-4.65) | <0.001 |
| UAE—mg/24h | 10.4 (6.6-20.3) | 10.2 (6.5 -19.7) | 17.2 (8.8-36.5) | <0.001 |
Data were shown as mean± Standard deviation (SD) or median (quartiles 1 and 3 [Q1-Q3]) for continuous variables which were compared by independent t tests or Mann–Whitney U tests as appropriate. Categorical variables are shown as frequencies (percentage) and were compared by χ2 test. BMI: body mass index (the weight in kilogram divided by the square of the height in meters), CVD: cardiovascular disease, HDL: high-density lipoprotein, GGT: gamma-glutamyltransferase, ALT: alanine aminotransferase, AST: aspartate aminotransferase, ALP: Alkaline phosphatase, hs-CRP: high sensitivity C-reactive protein and UAE: urine albumin excretion.
Association between bilirubin and T2D
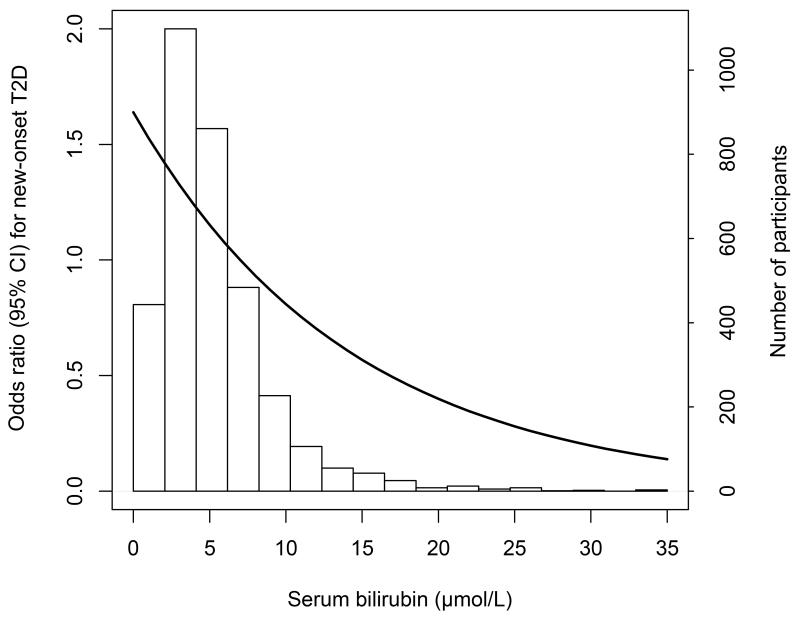
We observed that total bilirubin levels were lower in individuals in whom T2D developed vs. those in whom T2D did not develop (7.1±3.1 and 7.7±3.6 μmol/L, respectively; P<0.001). Figure 2 depicts a graphical representation of the distribution of bilirubin with an incremental decrease in ORs for T2D over the range of bilirubin (2-35 μmol/L). The multivariable adjusted OR for T2D was 0.75 (95%CI, 0.62-0.92), per 1-SD increase in log-transformed bilirubin levels (model A). The associations of bilirubin with baseline variables are shown in Table 2. When adding separately the variables, which were associated with bilirubin, to model A, corresponding ORs for T2D ranged from 0.78 (95%CI, 0.67-0.92) to 0.83 (95%CI, 0.69-0.94).
Figure 2. Histogram of serum total bilirubin levels with odds ratios for new onset (i.e., incident) type 2 diabetes over the range of bilirubin.
The number of participants represents by the white bars which correspond to the right axis. The solid line denotes the odds ratios which were centered on the median value of bilirubin.
Association between rs6742078 and bilirubin
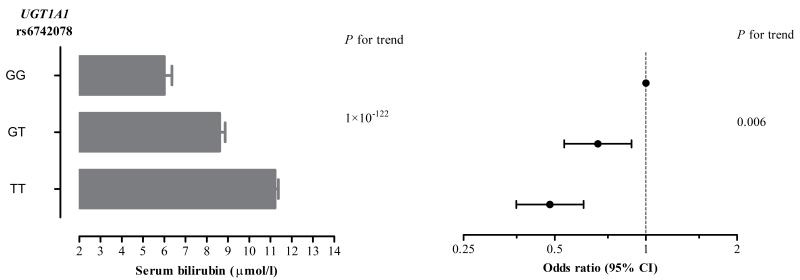
We examined whether rs6742078, which has been previously discovered as the top ranked SNP in a GWAS of total bilirubin (16; 17), was associated with bilirubin levels in our cohort. In model B, the rs6742078*T allele was strongly associated with bilirubin levels (a 0.68-SD increase in log-transformed bilirubin levels per allele; P<1×10−122). The proportion of bilirubin variation explained by rs6742078 was 19.5% (the difference between r2=0.070 for model B without the genotype and r2 =0.265 for model B with the genotype). Using linear regression analysis, the rs6742078*T allele was associated with a 2.607 μmol/L higher total bilirubin level (Figure 3, the left panel).
Figure 3. Association of the rs6742078 genotypes (TT, TG and GG) with levels of total bilirubin and incidence of type 2 diabetes in the study population.
The mean (s.e.) level of bilirubin for each genotype is represented by the gray bars in the left panel of the figure. The odds ratios (95% confidence intervals) for the association of the genotypes with new onset type 2 diabetes are shown in the right panel of the figure.
Association between rs6742078 and T2D
We observed that one copy increase in number of the rs6742078*T allele was associated with T2D risk with an OR of 0.69 (95%CI, 0.54-0.90; P=0.006), which corresponds to a 31% decreased risk of T2D (Figure 3, the right panel). Adjustment for bilirubin attenuated the association between rs6742078 and T2D risk (OR 0.80 [95%CI, 0.60-1.06]; P=0.13).
Causal estimates for the effect of bilirbuin on T2D risk
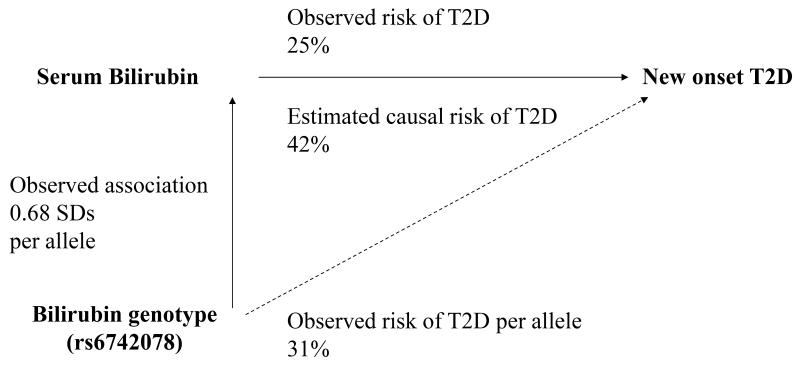
To estimate the causal effect of bilirubin on T2D, we used the Wald-type method, which assesses the ratio of the association of rs6742078 with T2D and that of rs6742078 and bilirubin levels (Figure 4). Based on this, we calculated that the estimated causal effect of total bilirubin (per 1-SD increase in log-transformed bilirubin levels) corresponds to a 42% lower risk of T2D (causal ORIVestimation 0.58 [95%CI, 0.39-0.84]; P=0.005). Given a sample size of 3,381 individuals (including 210 incident cases of T2D) and 19.5% of bilirubin variation explained by rs6742078 in our study, we had 80% power at a significance level of 0.05 to estimate an OR of 0.64 per 1-SD increase in log-transformed bilirubin levels. We observed that direction of the IV and observed estimates were similar with a slightly larger effect size for the IV estimate (Pendogeneity=0.19). The use of two alternative methods for IV estimation provided evidence of causal effect, which was also comparable to the observed OR for T2D (Pendogeneity=0.18 or 0.22; Table 3).
Figure 4. Mendelian randomization analysis for the association of levels of serum total bilirubin and the incidence of type 2 diabetes.
The observed effect of genotype on bilirubin levels (βgenotype-bilirubin) and risk of type 2 diabetes (βgenotype-T2D) was estimated per each copy of the T allele of rs6742078. The observed effect of bilirubin on type 2 diabetes was estimated per 1-SD increase in log-transformed bilirubin levels. The estimated causal risk of type 2 diabetes was calculated by the Wald-type method which is a ratio of βgenotype-T2D to βgenotype-bilirubin, where risk= (1-exp(β))×100.
Confounding and pleiotropy
We found no evidence that rs6742078 is associated with potential confounders like various demographic, lifestyle and clinical variables (Table 2). Thus, we assumed that the association between the IV and T2D was mainly free of confounding. To test for pleiotropy, we assessed the correlation between rs6742078 and the residuals from the logistic regression for T2D (as dependent variable) and bilirubin (as independent variable). There was no evidence for pleiotropy (Pfor correlation=0.11). We consistently observed no heterogeneity for the causal effects of bilirubin on T2D risk or glycemic traits when we used a summary statistics genetic score including rs6742078, rs4149056 and rs16928809 (Table 3). Details of the association of individual SNPs with bilirubin, T2D and glycemic traits are given in Supplementary Table 2. In PREVEND, while rs6742078 explained 19.5% of bilirubin variation, rs4149056 and rs16928809 only explained 0.18% and 0.15% of the variance, respectively. This limits use of these SNPs as a single IV. For rs4149056 and rs16928809, we estimated causal ORs of 0.58 (0.05-6.08) and 0.17 (0.02-2.16) in PREVEND. These estimates lack power for detection of associations as these two SNPs explained only a very small amount of bilirubin variation and therefore are to be considered weak IVs.
Secondary analyses
In subsequent analyses, we repeated the analyses in models A and C, when weighted for individuals with a mean urine albumin >10 mg/L. The results of these complex design analyses were similar to that of the main analyses, with an OR of 0.79 (95%CI, 0.66-0.95; P=0.01) per 1-SD increase in log-transformed bilirubin, and an OR of 0.59 (95%CI, 0.47-0.97; P<0.001) for the rs6742078*T allele. Second, we found no evidence for interactions of sex and age with bilirubin and rs6742078 (Pfor sex-interaction =0.49 and Pfor age-interaction=0.38 with bilirubin; Pfor sex-interaction=0.96 and Pfor age-interaction=0.25 with rs6742078) for the risk of T2D. Third, we observed that both observational and IV estimates were age consistent, with ORs 0.69 (0.51-1.04) and 0.45 (0.21-0.94) in people <50 years, and 0.86 (0.70-1.04) and 0.55 (0.34-0.90) in people ≥50 years, respectively. Fourth, the assumption of proportional-hazards was met for the Cox regression model, indicating there is no evidence of time varying associations of bilirubin and rs6742078 with T2D (P=0.38 and P=0.54, respectively). Fifth, we observed no evidence of an association between the drop-out rates (82.9% and 71.3% of participants who underwent the second and the third round, respectively) and the genotype (Pfor correlation=0.15). Sixth, we found an OR of 0.93 (95%CI, 0.69-1.26) per each T allele of rs6742078 for prevalent T2D; and when we performed a combined analysis of both prevalent and incident T2D, our findings remained materially unchanged (OR 0.77 [95%CI, 0.64-0.93]). In further analysis, we tested for another UGT1A1 SNP, rs887829 (in perfect LD with rs6742978; r2=1.0), which has been shown to be strongly associated with bilirubin levels in GWAS data (31). In our cohort, the rs887829*T-allele was associated with decreased risk of T2D (OR 0.70 [95% CI, 0.54-0.89]), which was comparable to that of the rs6742978*T-allele. Finally, we observed no evidence of systematic differences between the participants who were included in main analysis compared with the rest of cohort participants without diabetes at baseline (data not shown).
DISCUSSION
In this prospective cohort study, we investigated whether we could provide evidence that bilirubin is causally associated with risk of new-onset T2D. We demonstrated the rs6742078* T in the UGT1A1 locus is strongly associated with elevated levels of endogenous bilirubin and is also associated with new-onset T2D. Several studies have previously demonstrated associations between bilirubin and T2D in animal models and in humans (1-6). However, confounding and reverse causality might have attributed to imply causal relationships in observational studies. To our knowledge, we are the first to report a potential causal association between elevated total bilirubin levels and decreased T2D risk likely free of potential confounders using Mendelian randomization.
Accumulating evidence shows that bilirubin has powerful antioxidant properties (7-10). Bilirubin may compensate the oxidative stress which might be an important factor in the pathophysiology of diabetes (32-36). Oxidative stress is consistently related to increased plasma glucose levels and may contribute to the development of diabetes (37), and micro- and macrovascular complications of diabetes (38; 39). Bilirubin has been shown to prevent against oxidative stress in a number of diseases, including atherosclerosis, cancer and diabetic nephropathy (40; 41). In mice, biliverdin, a precursor of bilirubin, protects against deterioration of glucose tolerance (3), while increased levels of bilirubin reduced streptozotocin-induced pancreatic β-cell damage through attenuating oxidative stress (42), or increasing insulin sensitivity in mice (43). In line with experimental studies, previous observational studies have shown that elevated bilirubin is associated with a 26% to 31% decreased risk of diabetes in humans (2; 44).
In previous studies, variation in the UGT1A1 and SLCO1B1 loci seemed to explain part of inter-individual differences in bilirubin levels (5; 15-17). Johnson et al. (2009) found that rs6742078, very close to the the UGT1A1*28 TATA box polymorphism, accounted for approximately 18% of the variation in total bilirubin levels (P<5.0×10−324) (17). Less powerful, but still present was the association with rs4149056, a functional SNP of the SLCO1B1 locus, which accounted for 0.6% of bilirubin variation (17). In our cohort study, we observed that 19.5% of bilirubin variation was explained by the UGT1A1 rs6742078 SNP. It is likely that the association of rs6742078 with bilirubin can be explained to a large extent by the functional UGT1A1*28 genetic variant, which has been found to be in high LD (r2=0.88) with rs6742078 (17). Therefore, the contribution of genetic variation in UGT1A1 to risk of T2D could be greater than estimated from associations observed for rs6742078. The genetically determined component of protective effects may also vary globally as the TA repeat in the UGT1A1 promoter has other, less common functional variants, e.g., the UGT1A1*6 variant, which is more often observed in Asians with Japanese or Chinese ancestry (45).
From phenotypic perspective, the Framingham heart study has found that the UGT1A1*28 variant was associated with both bilirubin and cardiovascular disease (CVD) (5). However, the relationships of the UGT1A1 SNPs with CVD, glycemic traits and diabetes have been inconsistent, possibly due to differences in design and populations across studies (23). In the present study, we found that the rs6742078 SNP was associated with T2D risk. We tested whether rs6742078 was associated to glycemic traits that were recently evaluated in meta-analysis of 21 GWA studies (29). rs6742078 was associated with fasting glucose (β=−0.009; P=0.02), insulin (β=−0.009; P=0.03) and HOMA-IR (β=−0.009; P=0.03) based on a priori selected single-test association analysis in published GWAS data (29). In a recent GWAS meta-analysis (12,171 cases and 56,862 control subjects), no evidence of an association between rs6742078 and T2D has been reported (P=0.1) (28). The lack of association between rs6742078 and T2D might be due to the heterogeneity in the T2D phenotype (28; 46), as T2D cases have been ascertained using different sources (28). Of note, we identified a similar pattern of absence of an association in the GWAS meta-analysis, while another prospective Mendelian randomization study reported presence of such an association (47). In the latter study, the presence of evidence for the association between two SNPs in the gene encoding sex hormone-binding globulin and T2D could not be confirmed in the case-control GWAS meta-analysis. Case-control studies typically included both prevalent and incident cases which differ with respect to the disease stage, diagnostic criteria and duration of diabetes (46). Also, these discrepancies may partly be explained by differences in data quality control and pre-analysis preparations (28; 47). A prospective cohort design and its accompanying statistical methods will reduce potential biases such as incidence-prevalence bias and non-response bias (46). Although we estimated the risk of future T2D, there still might be a chance of a type I error in our analysis. The current results together with the associations of rs6742078 with glycemic traits in the meta-analysis of GWAS support that elevated bilirubin levels may lead to decreased risk of insulin resistance and T2D. Our finding remains to be confirmed for the potential role of bilirubin in the trajectories of glycemic traits and the development of T2D by prospective cohort studies.
In our study, we used a strong IV, rs6742078, which satisfied requirements for Mendelian randomization (48), to examine potential causal relationship between bilirubin and T2D in a large population-based study. Using this approach, we directly investigated a relationship between genotype and phenotype independent of potential confounders. Other bilirubin SNPs, rs4149056 and rs16928809 (17), may not be selected as a single IV, given the fact that the strength of these instruments is limited (25) because of the small amount of bilirubin variation that is explained by these two SNPs (0.18% and 0.15% in PREVEND, respectively).
Our findings may have implications for prevention or prediction of T2D and its complications. For example, Gilbert syndrome which is characterized by moderate hyperbilirubinemia exists in 5-10% of the general population (5; 16; 49). It has been shown that diabetic patients with Gilbert syndrome have reduced markers of oxidative stress and decreased risk of diabetic nephropathy and CVD (5; 49). One may speculate that modulation of bilirubin levels (e.g., using medical interventions that inhibit UGT1A1) might have protective effects against the risk of T2D (45). A search for the sources of increase in bilirubin levels or its antioxidative capacity (e.g., effective therapeutics with minimal adverse effects) merits further investigations.
One major concern remaining in the context of Mendelian randomization, is that we cannot completely control for the possibility of reverse causality, unobserved confounding and pleiotropy (14; 50). The functional UGT1A1*28 variant (in high LD with rs6742078) which is associated with a 10-35% reduction in UGT1A1 activity is mainly involved in glucuronidation of bilirubin, but also other compounds (e.g., estrogen), and drugs (e.g., irinotecan) (45; 51). Yet, the effects of the genotype on phenotypic traits other than bilirubin and their consequences on disease outcomes (e.g., cancer) remain inconclusive. An extensive knowledge of gene function and the associated biological processes is needed to better understand to what extend such genetic variation in the human genome contributes to lifelong protection against T2D. Our cohort is predominantly comprised of white adults of Dutch descent, and it is therefore unknown whether the findings of our study can be generalized to other populations (18). The PREVEND cohort was enriched for individuals with microalbuminuria at baseline. However, in a secondary analysis, we used a weighted method to compensate this, and this did not affect our results.
In summary, endogenous bilirubin and the most highly associated SNP, rs6742078 in the UGT1A1 locus, are associated with risk of T2D. We observed consistent effect estimates derived from alternative genetic instruments, rs4149056 and rs16928809. However, these two SNPs explain far less of bilirubin variation and therefore the estimates derived from these individual instruments should be interpreted cautiously. Our findings provide evidence that lifelong genetically elevated bilirubin is likely to be causally protective against the development of T2D. Further studies are warranted to validate this finding and elucidate the exact underlying mechanisms of action.
Supplementary Material
Acknowledgements
The authors thank Professor Dr. L.T.W. de Jong van den Berg and Dr. S.T. Visser from the Department of Social Pharmacy, Pharmacoepidemiology, and Pharmacotherapy, Groningen University Institute for Drug Exploration, University of Groningen, University Medical Center Groningen, for providing the data on pharmacy-registered use of insulin or oral hypoglycemic agents.
Funding: This work was supported by the Netherlands Heart Foundation, Dutch Diabetes Research Foundation, Dutch Kidney Foundation, the Netherlands Organization for Scientific Research project (NWO), and the Medical Research Council UK (grant no. MC_U106179471). A.A. is supported by a Rubicon grant from the NWO (Project no. 825.13.004)
Footnotes
Duality of interest: No potential conflicts of interest relevant to this article were reported. None of the study sponsors had a role in the study design; data collection, analysis, and interpretation; report writing; or the decision to submit the report for publication.
Publisher's Disclaimer: “This is an author-created, uncopyedited electronic version of an article accepted for publication in Diabetes. The definitive publisher-authenticated version will be available in a future issue of Diabetes in print and online at http://diabetes.diabetesjournals.org/ .”
References
- 1.Lin R, Wang Y, Wang Y, Fu W, Zhang D, Zheng H, Yu T, Wang Y, Shen M, Lei R, Wu H, Sun A, Zhang R, Wang X, Xiong M, Huang W, Jin L. Common variants of four bilirubin metabolism genes and their association with serum bilirubin and coronary artery disease in Chinese Han population. Pharmacogenet Genomics. 2009;19:310–318. doi: 10.1097/FPC.0b013e328328f818. [DOI] [PubMed] [Google Scholar]
- 2.Cheriyath P, Gorrepati VS, Peters I, Nookala V, Murphy ME, Srouji N, Fischman D. High Total Bilirubin as a Protective Factor for Diabetes Mellitus: An Analysis of NHANES Data From 1999 - 2006. J Clin Med Res. 2011;2:201–206. doi: 10.4021/jocmr425w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda N, Inoguchi T, Sonoda N, Fujii M, Takei R, Hirata E, Yokomizo H, Zheng J, Maeda Y, Kobayashi K, Takayanagi R. Biliverdin protects against the deterioration of glucose tolerance in db/db mice. Diabetologia. 2011;54:2183–2191. doi: 10.1007/s00125-011-2197-2. [DOI] [PubMed] [Google Scholar]
- 4.Horsfall LJ, Nazareth I, Petersen I. Cardiovascular events as a function of serum bilirubin levels in a large, statin-treated cohort. Circulation. 2012;126:2556–2564. doi: 10.1161/CIRCULATIONAHA.112.114066. [DOI] [PubMed] [Google Scholar]
- 5.Lin JP, O’Donnell CJ, Schwaiger JP, Cupples LA, Lingenhel A, Hunt SC, Yang S, Kronenberg F. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation. 2006;114:1476–1481. doi: 10.1161/CIRCULATIONAHA.106.633206. [DOI] [PubMed] [Google Scholar]
- 6.Jenko-Praznikar Z, Petelin A, Jurdana M, Ziberna L. Serum bilirubin levels are lower in overweight asymptomatic middle-aged adults: An early indicator of metabolic syndrome? Metabolism: clinical and experimental. 2013 doi: 10.1016/j.metabol.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Duann P, Lianos EA. GEC-targeted HO-1 expression reduces proteinuria in glomerular immune injury. Am J Physiol Renal Physiol. 2009;297:F629–638. doi: 10.1152/ajprenal.00213.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham NG, Asija A, Drummond G, Peterson S. Heme oxygenase −1 gene therapy: recent advances and therapeutic applications. Curr Gene Ther. 2007;7:89–108. doi: 10.2174/156652307780363134. [DOI] [PubMed] [Google Scholar]
- 9.Baranano DE, Rao M, Ferris CD, Snyder SH. Biliverdin reductase: a major physiologic cytoprotectant. Proc Natl Acad Sci U S A. 2002;99:16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 11.Alizadeh BZ, Njajou OT, Houwing-Duistermaat JJ, de Jong G, Vergeer JM, Hofman A, Pols HA, van Duijn CM. Does bilirubin protect against hemochromatosis gene (HFE) related mortality? Am J Med Genet A. 2004;129A:39–43. doi: 10.1002/ajmg.a.30163. [DOI] [PubMed] [Google Scholar]
- 12.Iribarren C, Reed DM, Chen R, Yano K, Dwyer JH. Low serum cholesterol and mortality. Which is the cause and which is the effect? Circulation. 1995;92:2396–2403. doi: 10.1161/01.cir.92.9.2396. [DOI] [PubMed] [Google Scholar]
- 13.Thanassoulis G, O’Donnell CJ. Mendelian randomization: nature’s randomized trial in the postgenome era. Jama. 2009;301:2386–2388. doi: 10.1001/jama.2009.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 15.Buch S, Schafmayer C, Volzke H, Seeger M, Miquel JF, Sookoian SC, Egberts JH, Arlt A, Pirola CJ, Lerch MM, John U, Franke A, von Kampen O, Brosch M, Nothnagel M, Kratzer W, Boehm BO, Broring DC, Schreiber S, Krawczak M, Hampe J. Loci from a genome-wide analysis of bilirubin levels are associated with gallstone risk and composition. Gastroenterology. 2010;139:1942–1951 e1942. doi: 10.1053/j.gastro.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Lin JP, Vitek L, Schwertner HA. Serum bilirubin and genes controlling bilirubin concentrations as biomarkers for cardiovascular disease. Clin Chem. 2010;56:1535–1543. doi: 10.1373/clinchem.2010.151043. [DOI] [PubMed] [Google Scholar]
- 17.Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, Lin JP, van Duijn CM, Harris TB, Cupples LA, Uitterlinden AG, Launer L, Hofman A, Rivadeneira F, Stricker B, Yang Q, O’Donnell CJ, Gudnason V, Witteman JC. Genome-wide association meta-analysis for total serum bilirubin levels. Human molecular genetics. 2009;18:2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambers Heerspink HJ, Brantsma AH, de Zeeuw D, Bakker SJ, de Jong PE, Gansevoort RT. Albuminuria assessed from first-morning-void urine samples versus 24-hour urine collections as a predictor of cardiovascular morbidity and mortality. Am J Epidemiol. 2008;168:897–905. doi: 10.1093/aje/kwn209. [DOI] [PubMed] [Google Scholar]
- 19.Monster TB, Janssen WM, de Jong PE, de Jong-van den Berg LT. REnal PSGPo, Vascular ENTSD: Pharmacy data in epidemiological studies: an easy to obtain and reliable tool. Pharmacoepidemiology and drug safety. 2002;11:379–384. doi: 10.1002/pds.722. [DOI] [PubMed] [Google Scholar]
- 20.Abbasi A, Corpeleijn E, Meijer E, Postmus D, Gansevoort RT, Gans RO, Struck J, Hillege HL, Stolk RP, Navis G, Bakker SJ. Sex differences in the association between plasma copeptin and incident type 2 diabetes: the Prevention of Renal and Vascular Endstage Disease (PREVEND) study. Diabetologia. 2012;55:1963–1970. doi: 10.1007/s00125-012-2545-x. [DOI] [PubMed] [Google Scholar]
- 21.Abbasi A, Peelen LM, Corpeleijn E, van der Schouw YT, Stolk RP, Spijkerman AM, van der AD, Moons KG, Navis G, Bakker SJ, Beulens JW. Prediction models for risk of developing type 2 diabetes: systematic literature search and independent external validation study. BMJ. 2012;345:e5900. doi: 10.1136/bmj.e5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verweij N, Mahmud H, Mateo Leach I, de Boer RA, Brouwers FP, Yu H, Asselbergs FW, Struck J, Bakker SJ, Gansevoort RT, Munroe PB, Hillege HL, van Veldhuisen DJ, van Gilst WH, Sillje HH, van der Harst P. Genome-wide association study on plasma levels of midregional-proadrenomedullin and Cterminal-pro-endothelin-1. Hypertension. 2013;61:602–608. doi: 10.1161/HYPERTENSIONAHA.111.203117. [DOI] [PubMed] [Google Scholar]
- 23.Stender S, Frikke-Schmidt R, Nordestgaard BG, Grande P, Tybjaerg-Hansen A. Genetically elevated bilirubin and risk of ischaemic heart disease: three Mendelian randomization studies and a meta-analysis. Journal of internal medicine. 2013;273:59–68. doi: 10.1111/j.1365-2796.2012.02576.x. [DOI] [PubMed] [Google Scholar]
- 24.Palmer TM, Sterne JA, Harbord RM, Lawlor DA, Sheehan NA, Meng S, Granell R, Smith GD, Didelez V. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol. 173:1392–1403. doi: 10.1093/aje/kwr026. [DOI] [PubMed] [Google Scholar]
- 25.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. International journal of epidemiology. 2014 doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, van der Sijde MR, Study LC, Bakker SJ, Dullaart RP, van der Harst P, Gansevoort RT, Elbers CC, Wijmenga C, Snieder H, Hofker MH, Fu J. Pleiotropic effects of lipid genes on plasma glucose, HbA1c and HOMA-IR levels. Diabetes. 2014 doi: 10.2337/db13-1800. [DOI] [PubMed] [Google Scholar]
- 27.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genetic epidemiology. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, Mirza G, Muhleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurethsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvanen AC, Eriksson JG, Peltonen L, Nothen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Wellcome Trust Case Control C. Meta-Analyses of G. Insulin-related traits Consortium I. Genetic Investigation of ATC. Asian Genetic Epidemiology Network-Type 2 Diabetes C. South Asian Type 2 Diabetes C. Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njolstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyovalti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jockel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI, Replication DIG. Meta-analysis C: Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, BenShlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Consortium D, Consortium G, Global BC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Anders Hamsten on behalf of Procardis C. investigators M. Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. International journal of epidemiology. 2013;42:1134–1144. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanna S, Busonero F, Maschio A, McArdle PF, Usala G, Dei M, Lai S, Mulas A, Piras MG, Perseu L, Masala M, Marongiu M, Crisponi L, Naitza S, Galanello R, Abecasis GR, Shuldiner AR, Schlessinger D, Cao A, Uda M. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Human molecular genetics. 2009;18:2711–2718. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Campenhout A, Van Campenhout C, Lagrou AR, Abrams P, Moorkens G, Van Gaal L, Manuely-Keenoy B. Impact of diabetes mellitus on the relationships between iron-, inflammatory- and oxidative stress status. Diabetes Metab Res Rev. 2006;22:444–454. doi: 10.1002/dmrr.635. [DOI] [PubMed] [Google Scholar]
- 33.Robertson RP, Harmon JS. Diabetes, glucose toxicity, and oxidative stress: A case of double jeopardy for the pancreatic islet beta cell. Free Radic Biol Med. 2006;41:177–184. doi: 10.1016/j.freeradbiomed.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 34.Seppen J, Bosma P. Bilirubin, the gold within. Circulation. 2012;126:2547–2549. doi: 10.1161/CIRCULATIONAHA.112.147082. [DOI] [PubMed] [Google Scholar]
- 35.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li N, Frigerio F, Maechler P. The sensitivity of pancreatic beta-cells to mitochondrial injuries triggered by lipotoxicity and oxidative stress. Biochem Soc Trans. 2008;36:930–934. doi: 10.1042/BST0360930. [DOI] [PubMed] [Google Scholar]
- 37.Yamagishi S. [Role of advanced glycation end products (AGE) and soluble receptor for AGE (sRAGE) in vascular complications in diabetes] Nihon Rinsho. 2012;70(Suppl 5):243–247. [PubMed] [Google Scholar]
- 38.Baynes JW, Thorpe SR. Role of oxidative stress in diabetic complications: a new perspective on an old paradigm. Diabetes. 1999;48:1–9. doi: 10.2337/diabetes.48.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 40.Vitek L. The role of bilirubin in diabetes, metabolic syndrome, and cardiovascular diseases. Front Pharmacol. 2012;3:55. doi: 10.3389/fphar.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riphagen IJ, Deetman PE, Bakker SJ, Navis G, Cooper ME, Lewis JB, de Zeeuw D, Lambers Heerspink HJ. Bilirubin and Progression of Nephropathy in Type 2 Diabetes: A Post-Hoc Analysis of RENAAL with Independent Replication in IDNT. Diabetes. 2014 doi: 10.2337/db13-1652. [DOI] [PubMed] [Google Scholar]
- 42.Fu YY, Kang KJ, Ahn JM, Kim HR, Na KY, Chae DW, Kim S, Chin HJ. Hyperbilirubinemia reduces the streptozotocin-induced pancreatic damage through attenuating the oxidative stress in the Gunn rat. Tohoku J Exp Med. 2010;222:265–273. doi: 10.1620/tjem.222.265. [DOI] [PubMed] [Google Scholar]
- 43.Dong H, Huang H, Yun X, Kim DS, Yue Y, Wu H, Sutter A, Chavin KD, Otterbein LE, Adams DB, Kim YB, Wang H. Bilirubin increases insulin sensitivity in leptin-receptor deficient and diet-induced obese mice through suppression of ER stress and chronic inflammation. Endocrinology. 2014;155:818–828. doi: 10.1210/en.2013-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung CH, Lee MJ, Kang YM, Hwang JY, Jang JE, Leem J, Park JY, Kim HK, Lee WJ. Higher serum bilirubin level as a protective factor for the development of diabetes in healthy Korean men: A 4year retrospective longitudinal study. Metabolism: clinical and experimental. 2014;63:87–93. doi: 10.1016/j.metabol.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Erlinger S, Arias IM, Dhumeaux D. Inherited Disorders of Bilirubin Transport and Conjugation: New Insights Into Molecular Mechanisms and Consequences. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.03.047. [DOI] [PubMed] [Google Scholar]
- 46.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nature reviews Genetics. 2006;7:812–820. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 47.Ding EL, Song Y, Manson JE, Hunter DJ, Lee CC, Rifai N, Buring JE, Gaziano JM, Liu S. Sex hormonebinding globulin and risk of type 2 diabetes in women and men. The New England journal of medicine. 2009;361:1152–1163. doi: 10.1056/NEJMoa0804381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. International journal of epidemiology. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maruhashi T, Soga J, Fujimura N, Idei N, Mikami S, Iwamoto Y, Kajikawa M, Matsumoto T, Kihara Y, Chayama K, Noma K, Nakashima A, Tomiyama H, Takase B, Yamashina A, Higashi Y. Hyperbilirubinemia, augmentation of endothelial function, and decrease in oxidative stress in Gilbert syndrome. Circulation. 2012;126:598–603. doi: 10.1161/CIRCULATIONAHA.112.105775. [DOI] [PubMed] [Google Scholar]
- 50.Pfister R, Sharp S, Luben R, Welsh P, Barroso I, Salomaa V, Meirhaeghe A, Khaw KT, Sattar N, Langenberg C, Wareham NJ. Mendelian randomization study of B-type natriuretic peptide and type 2 diabetes: evidence of causal association from population studies. PLoS Med. 2011;8:e1001112. doi: 10.1371/journal.pmed.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stender S, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Extreme bilirubin levels as a causal risk factor for symptomatic gallstone disease. JAMA internal medicine. 2013;173:1222–1228. doi: 10.1001/jamainternmed.2013.6465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.