Abstract
Current methods for the isolation of fibroblasts require extended ex vivo manipulation in cell culture. As a consequence, prior studies investigating fibroblast biology may fail to adequately represent cellular phenotypes in vivo. To overcome this problem, we describe a detailed protocol for the isolation of fibroblasts from the dorsal dermis of adult mice that bypasses the need for cell culture, thereby preserving the physiological, transcriptional, and proteomic profiles of each cell. Using the described protocol we characterized the transcriptional programs and the surface expression of 176 CD markers in cultured versus uncultured fibroblasts. The differential expression patterns we observed highlight the importance of a live harvest for investigations of fibroblast biology.
Introduction
Fibroblasts are a principal cell type implicated in organ development and maturation.1,2 Fibroblasts are also the primary cell types that accumulate in diverse medical conditions, including chronic wound healing, tissue and organ fibrosis, atherosclerosis, and formation of atheromatous plaque after blood vessel injury. Fibroblasts also contribute to the progression of cancer by serving as key cellular components in the tumor stroma, a finding that could implicate the tumor-associated fibroblast as an important target for anticancer therapies.3
Current knowledge of fibroblast biology is primarily derived from studying their growth and behavior in vitro on plastic substrates as monolayer cultures. The process of adhesion to a plastic substrate results in an activated state characterized by metabolic changes that promote proliferation and secretion of extracellular matrix (ECM) molecules.4 These activities most resemble the behavior of fibroblasts in an early wound response program. Such behavior is absent or significantly reduced both in vivo and in three-dimensional organotypic models.5,6
Fibroblasts represent a heterogeneous population of cells with diverse functional and phenotypic features.7 Studies have demonstrated fibroblast heterogeneity in surface marker expression, proliferation, immune regulation, and ECM production both within specific tissues and between different anatomic sites.8 However, such diversity remains largely unexplored due to a reliance on cultured populations and a lack of unique surface markers for distinct phenotypic subclasses of fibroblasts.
Fibroblast subpopulations are responsible for distinct contributions to pathologic processes and wound healing. For example, Thy-1-negative fibroblasts in pulmonary fibrosis produce IL-1alpha, express MHC II, and present antigen in response to IFN-gamma whereas Thy-1-positive fibroblasts do not.7 Similarly, Penney et al. showed that lipid-containing fibroblasts proliferate in response to radiation-induced damage of lung tissue whereas nonlipid-containing fibroblasts remain quiescent.9 Gaining a deeper understanding of these fibroblast subtypes will allow for the modulation of their behavior in the context of injury and disease.
Our hypothesis holds that a significant difference exists between in vitro and in vivo environments, particularly in the case of mesenchymal cells (e.g., fibroblasts), where adherence and serum exposure induce significant changes in the cellular phenotype. Current methods for the isolation of dermal fibroblasts from adult mouse skin require at least multiple days in culture.10 A method for harvesting fibroblasts from newborn mouse dermis requires only a short-term overnight culture step; however, this protocol pertains only to newborn mouse skin.11 In comparison, we developed a protocol with a more rigorous digestion step for a same-day isolation of adult dermal fibroblasts without the requirement of cell culture.
Materials and Methods
Live fibroblast harvest reagents
• Collagenase IV (4 mg/mL) in Dulbecco's modified Eagle's medium (DMEM) (0% fetal bovine serum [FBS])
• Elastase (0.12 mg/mL) in DMEM (0% FBS)
• DMEM
• 10% FBS DMEM
• Ammonium Chloride–Potassium (ACK) Lysing Buffer
• 40 μm filters
• 70 μm filters
• 100 μm filters
• CD31 fluorescence-activated cell sorting (FACS) antibody
• CD45 FACS antibody
• Tie2 FACS antibody
• Ter-119 FACS antibody
• EpCAM (CD326) FACS antibody
Live fibroblast harvest procedure
1. Sacrifice mice aged 4–6 weeks. Shave and Nair™ dorsal dermis. Area sprayed with 70% ethanol and wiped clean.
2. Immediately harvest dorsal dermis using dissecting scissors to preserve cell viability.12 Make a transverse cut at the base of the tail and incise upward bilaterally to a cervical transverse cut. Separate along the fascial plane with dissecting scissors.
Actively remove subcutaneous fat from the harvested skin as this can interfere with enzymatic digestion.
3. Rinse dorsal dermis in betadine followed by 5× PBS washes on ice.
4. Cut dorsal dermis from each mouse into two equal-sized pieces and incubate in 0.12 mg/mL elastase (Abcam) in DMEM at 37°C for 25 min. Separate epidermis from dermis under dissecting scope using forceps while pinning epidermal side of skin down to dish surface with a second pair of forceps.13 Discard epidermis.
5. Mechanically dice dermis using razor blades and dissecting scissors in Petri dish until sample is of uniform consistency.
6. Divide sample into 50-mL conical tubes containing 20 mL collagenase IV (Gibco) at a concentration of 4 mg/mL in DMEM on the basis of five mice per tube.
7. Agitate samples vigorously in a water-bath shaker at 37°C for 1 h.
8. Remove samples from the water-bath shaker and pass through a 10-mL syringe (no needle) 5× and then pass through an 18.5-gauge syringe 5× using back loading to load the syringe.
9. Place samples back into water bath at 37°C and shake vigorously for 30 min.
10. Remove samples from shaker and pass through an 18.5-gauge syringe 5× using back loading to load syringe.
11. Add 30 mL of 10% FBS DMEM to each tube to bring the total volume to 50 mL and centrifuge at 1250 RPM for 5 min at 4°C.
12. Remove supernatant and resuspend pellets in 30 mL 10% FBS DMEM.
Take care to first remove fat layer before remaining supernatant. This is a critical step to reduce adipocyte contamination.
13. Pass cell suspension through a 100 μm filter using centrifugation at 1250 RPM 4°C if necessary. Rinse filter with 5 mL 10% FBS DMEM.
14. Centrifuge filtered suspension at 1250 RPM for 5 min at 4°C.
15. Remove supernatant and resuspend pellets in 10% FBS DMEM and pass cell suspension through 40 μm filters.
Again, take care to first remove any remaining fat.
16. Centrifuge filtered suspension at 1250 RPM for 5 min at 4°C.
17. Remove supernatant and resuspend pellets in ACK lysis buffer. Incubate for 10 min at room temperature.
18. Add equal volume FACS buffer, mix, and centrifuge at 1250 RPM for 5 min at 4°C.
19. Remove supernatant and resuspend pellets in FACS buffer containing DNAse (10 μg/mL).
20. Incubate suspension on ice with *fluorophore-conjugated CD31 (1:100), CD45 (1:200), Tie2 (1:50), Ter-119 (1:200), and EpCAM (1:100) for 30 min (BioLegend, eBioscience).
*Conjugated fluorophore should be selected based on available wavelengths. In this study we used PacBlue.
21. Double suspension volume with FACS buffer containing DNAse (10 μg/mL) and mix gently. Centrifuge at 1250 RPM for 5 min at 4°C.
22. Remove supernatant and wash with FACS buffer containing DNAse (10 μg/mL).
23. Repeat centrifugation and wash step.
24. Resuspend pellet in FACS buffer containing DNAse (10 μg/mL) and set aside small aliquot as unstained control.
25. Add viability dye of choice to the remaining sample.
Viability dye can be matched with CD31/CD45/Tie2/Ter119/EpCAM antibody conjugates (i.e., DAPI matched with PacBlue) to preserve range of available wavelengths.
26. FACS sort for Viability Dye−/CD31−/CD45−/Tie2−/Ter-119−/EpCAM− cells.
Mice
Mice were bred and maintained in accordance with Stanford University guidelines at the Stanford University Research Animal Facility. Animals were housed in sterile insulators and received ad libitum access to rodent chow and water. CD1 male mice aged 4–6 weeks were obtained from Charles River and used in all experiments.
Lineage negative (Lin−) antibodies
Antibodies used for lineage-negative gating by FACS were: PacBlue-conjugated anti-mouse CD31 (BioLegend), PacBlue-conjugated anti-mouse CD45 (BioLegend), PacBlue-conjugated anti-mouse Tie2 (BioLegend), PacBlue-conjugated anti-mouse Ter119 (BioLegend), eFluor450-conjugated anti-mouse CD326/EpCAM (eBioscience, cat.no. 48-5791).
Cell surface marker screening
To identify changes in the surface marker expression of fibroblasts due to cell culture, we broadly screened the surface marker profiles of both freshly isolated and cultured fibroblasts. Skin fibroblasts were FACS harvested from the dorsal dermis of CD1 mice (n=25) using the live harvest protocol as previously described and either cultured for 2 weeks (three passages) or processed immediately. Approximately 75% of freshly FACS-isolated fibroblasts were immediately used for lyoplate surface marker analysis whereas the remaining cells were expanded in a culture for 2 weeks before being used for lyoplate surface marker analysis.
Surface markers were characterized using the BD Lyoplate Mouse Cell Surface Marker Screening Panel™ (BD Biosciences, cat.no. 562208) containing 176 purified monoclonal antibodies and corresponding isotype controls. The manufacturer's staining protocol was followed with slight modifications. Cells isolated from the dorsal dermis of CD1 mice were plated into U-bottom 96-well plates at a density of 2.5–5×105 cells per well in FACS buffer. Primary antibody incubation was done in 100 μL volume for 30 min on ice. Incubation with biotinylated secondary antibodies (goat anti-mouse 1:400, goat anti-rat 1:400, goat anti-Syrian hamster 1:400, goat anti-Armenian hamster 1:800) was done in 100 μL volume for 30 min on ice. Incubation with Alexa Fluor 647 Streptavidin (1:4000) was carried out in 100 μL volume for 30 min on ice. Analysis was performed using the flow cytometer BD LSR Fortessa with High Throughput Sampler (HTS).
Microarray analysis
Skin fibroblasts were FACS harvested from the dorsal dermis of CD1 mice (n=5) using the live harvest protocol as previously described and either cultured for 2 weeks (three passages) or processed immediately. This was repeated three times for biological replicates (n=3). Skin fibroblasts from CD1 mice (n=5) were also isolated using a previously published tissue explant technique requiring 2 weeks in culture.10 Cultures were passaged three times. This was repeated for each biologic replicates (n=3).
RNA was precipitated through chloroform–phenol extraction. Samples were processed for cleanup and concentration using the RNeasy MinElute Cleanup Kit (cat.no. 74204, QIAGEN). RNA yield was typically 0.5–1 μg RNA/sorted subpopulations. RNA samples from all sorted populations were converted to cDNA using the SuperScript III first-strand synthesis system for RT-PCR (cat.no. 18080-051, Invitrogen), and hybridized to the Affymetrix Mouse Genome 430 2.0 arrays. Similarity of gene expression between samples was obtained by Pearson product-moment correlation coefficient. Only probesets encoding annotated genes were subjected. If there were multiple probesets for same genes, the probeset that has the widest dynamic range was used. Hierarchical clustering based on the similarity of gene expression was performed by complete linkage method. Differentially expressed genes were obtained by the Significance Analysis of Microarrays (SAM) methodology.14 SAM score higher than FDR 5% threshold and two-fold change were used to identify significantly up- and downregulated genes.
Results
Live fibroblast harvest eliminates cell culture step
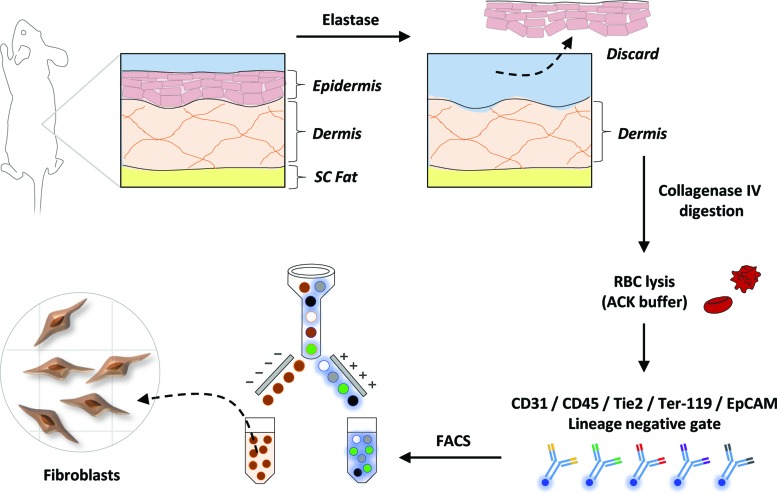
We performed extensive studies testing different combinations of enzymatic and mechanical digestion methods for the harvest of dermal fibroblasts from CD1 mice (Fig. 1). Dermal–epidermal separation was achieved with elastase and epidermis discarded.13 Dermis was digested using either Accutase, Trypsin, Release, Collagenase P, or Collagenase I, II, IV at concentrations of 1, 2, or 4 mg/mL for 1 or 2 h at 37°C. Tissue suspensions were passed through a 20-gauge syringe after 1 h and again at the end (2 h) of the enzymatic digestion. Following digestion, cells were subjected to ACK lysing buffer treatment to facilitate lysis of red blood cells. After ACK lysing buffer treatment, cell viability was assessed using a cell counter and Trypan Blue staining.
FIG. 1.
Live Fibroblast Harvest Schematic. Schematic outlining the live fibroblast harvest protocol. Color images available online at www.liebertpub.com/tec
Ultimately, a protocol was selected that resulted in the highest yield of live cells. Cells were then stained with PacBlue-conjugated CD31, CD45, Tie2, Ter-119, and EpCAM (CD326). This set of markers was used as a lineage negative (Lin−) gate to exclude cells of endothelial, hematopoietic, erythroid, and epithelial origins. FACS sorting on the basis of these negative selection markers allowed for the isolation of fibroblasts from the dorsal dermis of CD1 mice. The resulting population was assumed to be primarily fibroblastic given that epithelial contamination was limited through elastase-induced dermal–epidermal separation and the Lin- FACS gating strategy allowed for the exclusion of nonmesenchymal lineages within the skin (Fig. 2A).
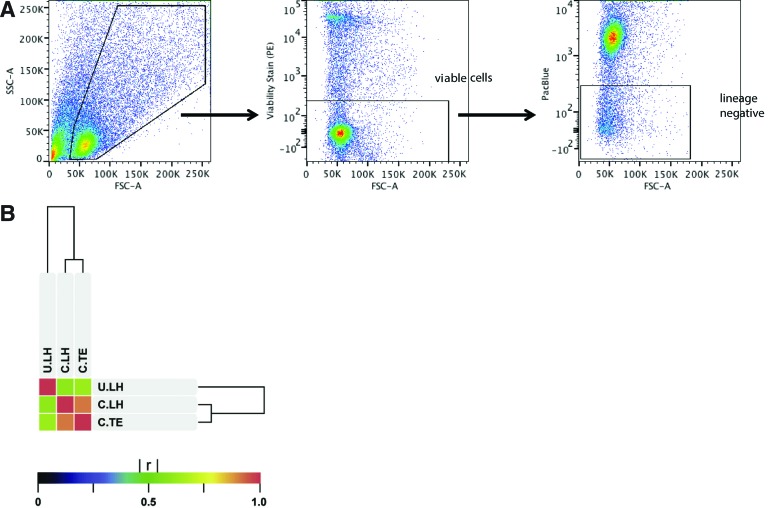
FIG. 2.
Fibroblast Flow Cytometry and Microarray Analysis. (A) Fluorescence-activated cell sorting (FACS) gating strategy showing selection of cells (left plot), selection for viable cells based on PE viability staining (middle plot), and selection of fibroblasts (right plot) on the basis of lineage negativity for CD31, CD45, Tie2, Ter119, and EpCAM. (B) Microarray Analysis of Uncultured Live Harvested (U.LH) versus Cultured Live Harvested (C.LH) versus Cultured Tissue Explant (C.TE) Fibroblasts. Similarity of gene expression between U.LH (n=3), C.LH (n=3), and C.TE (n=3) fibroblast populations as measured by the Pearson product-moment correlation coefficient (r). [C.LH vs. C.TE: r=0.92]; [C.LH vs. U.LH: r=0.61]; [C.TE vs. U.LH: r=0.64] Color images available online at www.liebertpub.com/tec
Differential expression of multiple surface markers in cultured versus uncultured fibroblasts
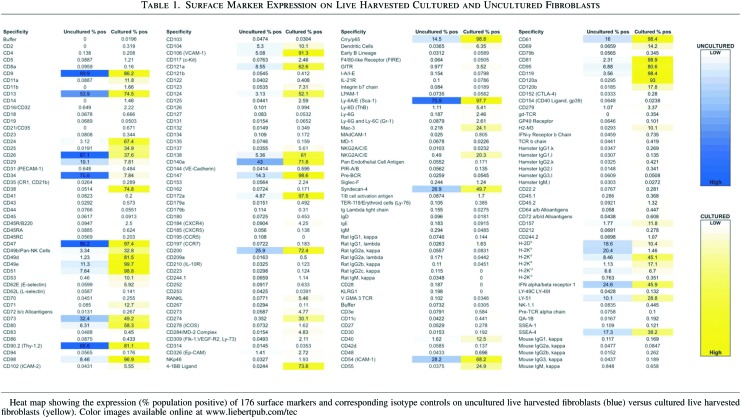
The identification of cellular surface markers is of critical importance to furthering our understanding of cellular biology during development, adult tissue homeostasis, and pathologic states. Using a broad surface marker screening panel, we attempted to document why it is essential that these surface markers be defined for freshly isolated cells that have not been cultured. Fibroblast FACS isolated from the dorsal dermis of CD1 mice using the live harvest protocol (Fig. 1) were either processed immediately according to the BD Lyoplate Mouse Cell Surface Marker Screening Panel (cat.no. 562208) protocol or first expanded in culture. The lyoplates contain purified monoclonal antibodies specific for 176 mouse cell surface markers and their corresponding isotype controls.
Lyoplate analysis of live harvested–uncultured harvested fibroblasts from the dorsal dermis of CD1 mice revealed positivity (>3%) for CD9, CD13, CD24, CD26, CD29, CD34, CD47, CD49e, CD51, CD54, CD61, CD73, CD90.2, CD95, CD98, CD104, CD106, CD119, CD121a, CD124, CD138, CD147, CD172a, CD200, Syndecan-4, Crry/p65, GITR, Sca-1, Ly-6A/E (Sca-1), H-2Db, H-2Kb, H-2Kd, H-2KQ, IFN α/β R1, SSEA-4, and Ly-51 (Table 1). Expression of many of these markers on dermal fibroblasts has been previously reported.15–18 Among these, known fibroblast markers such as CD90.2 (85.6%) and Sca-1 (75.9%) were expressed on significant fractions of isolated uncultured fibroblasts19–21 (Table 1), validating the fidelity of our FACS sorting definition. Integrins or proteins that complex with integrins were prominent among markers with greater than 5% population positivity (CD9, CD29, CD47, CD54, CD61, CD104) (Table 1). Many of the remaining surface molecules possessed hematopoietic and immune-related functions. As a positive control, additional FACS analyses independently confirmed positivity for several markers identified by the lyoplate analysis, including CD9, CD26, CD47, CD90.2, and CD147.
A standard protocol for the harvest of primary adult fibroblasts requires 2 weeks in culture as fibroblasts migrate out of skin fragments and onto the polystyrene surface of the tissue culture plate.10 We, therefore, harvested fibroblasts after 2 weeks in culture and processed them on the lyoplate panel according to the BD protocol. Lyoplate analysis of cultured fibroblasts from the dorsal dermis of CD1 mice revealed positivity (>3%) for CD9, CD13, CD24, CD25, CD26, CD29, CD34, CD38, CD47, CD49b, CD49d, CD49e, CD51, CD53, CD54, CD61, CD62E, CD71, CD73, CD80, CD81, CD90.2, CD95, CD98, CD104, CD106, CD119, CD120a, CD120b, CD121a, CD123, CD124, CD137, CD138, CD140a, CD147, CD157, CD172a, CD200, CD274, Crry7/p55, 4-1BB ligand, Ly-6A/E, Mao-3, Syndecan 4, IFN-γ Receptorβ chain, IFN α/β R1, Ly-51, SSEA-4, H-2Db, H-2Kd, H-2KK, and H-2KQ (Table 1). Given that the known fibroblast markers such as CD90.219,20 and Sca-119 were broadly expressed (>75%) to similar degrees in both live harvested, uncultured, and cultured fibroblasts (Table 1), we are confident of the purity of uncultured live harvest fibroblasts isolated using our protocol. Individual FACS analyses independently confirmed positivity for several markers identified by the lyoplate analysis, including CD9, CD13, CD47, CD90.2, and CD147. Standard deviations for all values in Table 1 can be found in the Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/tec).
Tissue culture plating of fibroblasts induces transcriptional shift
Although we had observed a shift in surface markers at a protein level in cultured versus uncultured fibroblasts isolated using our live harvest protocol, we hypothesized that a transcriptional shift was likely occurring as well. To test this, fibroblasts FACS-isolated using our live harvest protocol were either harvested immediately for total RNA or plated onto 10-cm polystyrene tissue culture plates. Given that the standard techniques for the harvest of fibroblasts from adult mouse skin require tissue culture plating of tissue explants, we also harvested RNA from cultured fibroblasts isolated using a standard tissue explant technique.10
Whole-transcriptome microarray analysis revealed that cultured fibroblasts isolated by live harvest (C.LH) and by tissue explant (C.TE) techniques have an overall high degree of transcriptome-wide similarity with a Pearson product-moment correlation coefficient (r) of 0.92. In comparison, cultured fibroblasts differed significantly from live harvested uncultured fibroblasts (U.LH) with r=0.61 for C.LH versus U.LH and r=0.64 for C.TE versus U.LH (Fig. 2B).
Whole-transcriptome microarray analysis also revealed that 130 genes were differentially expressed between C.TE and C.LH fibroblasts (Supplementary Table S2). In contrast, 2782 genes were differentially expressed between C.TE and U.LH (Supplementary Table S3), and 3038 genes were differentially expressed between C.LH and U.LH (Supplementary Table S4). A total of 2332 genes were common to both C.LH versus U.LH (3038 genes) and C.TE versus U.LH (2782 genes) lists of differentially expressed genes. Given that these 2332 overlapping genes represented a significant fraction (>76%) of both lists, it is clear that cultured fibroblasts isolated using either the live harvest or tissue explant techniques are significantly different from uncultured fibroblasts in many of the same genes.
Discussion
Existing techniques for the harvest of primary adult dermal fibroblasts require tissue culture plating of the skin fragments to allow for the migration of fibroblasts out of the dermis and onto the dish.10 This process requires multiple days in culture. Lichti et al. developed a method for harvesting fibroblasts from newborn mouse dermis requiring only a short-term overnight culture step; however, this protocol pertains only to newborn mouse skin.11 In comparison, our protocol allows for same-day isolation of fibroblasts from both newborn and adult mouse skin with no culture step.
Past studies suggest that serum exposure and the process of migration followed by adherence to a plastic substrate leads to the induction of a specific genetic program in the fibroblast most characteristic of a myofibroblast in an early wound response.22,23 In addition, tissue culture plating imposes a selective pressure that results in preferential expansion of only a subset of fibroblasts that are capable of migrating out of the dermis, adhering to plastic, and proliferating in culture. In this study, we define the surface marker profile of uncultured dermal fibroblasts and show that the immunophenotype of cultured fibroblasts do not recapitulate their surface marker profiles in vivo.
Numerous markers that were minimally expressed in the live harvested uncultured population were ubiquitously expressed within the live harvested cultured population. Prominent among these were the integrins and other surface molecules that mediate cell adhesion (CD24, CD38, CD49d, CD49e, CD51, CD61, CD81, CD98, CD106, CD138, Syndecan-4). This observation is consistent with the importance of cell–cell and cell–matrix contact in the activation of fibroblasts. Fibroblasts are activated by direct cell–cell communication and interactions with the ECM in vivo. The activated state is associated with increased proliferation, expression of alpha-smooth muscle actin, secretion of growth factors, and increased deposition of ECM components. Adhesion to a plastic substrate and exposure to serum in vitro results in a similarly activated state.22,23 In addition to activation, the immunophenotype seen in the cultured population may also be the result of selection for pre-existing clones that are able to thrive in the culture environment.
Activated fibroblasts are defined by increased proliferation, alpha-smooth muscle actin expression, growth factor/ECM secretion, and act as modulators of the immune response following injury by integrating cytokine and chemokine signaling.24,25 It is plausible that fibroblasts activated by tissue injury are primed to respond to cytokines from invading inflammatory cells through the upregulation of cytokine receptors. We observed ubiquitous expression of several cytokine receptors among cultured fibroblasts that were minimally expressed in the uncultured population. Prominent among these were IL-1 receptor (CD121a), IL-4 receptor (CD124), IL-7 receptor (CD172a), IFN-gamma receptor (CD119), and TNF receptor (CD120a). If the cultured phenotype is indeed analogous to the activated phenotype in vivo, such an observation is in keeping with the immunomodulatory role of activated fibroblasts in pathologic/injured states.24,25
CD34 was one of a few markers more highly expressed among uncultured in comparison with cultured fibroblasts. Consistent with these data, CD34 was among the top ten genes transcriptionally upregulated in uncultured compared to cultured fibroblasts. CD34 positivity on fibroblasts is thought to represent an uncommitted cell capable of multidirectional mesenchymal differentiation.26 Prior studies have also suggested that there is an inverse relationship between myofibroblastic differentiation and CD34 positivity.27 The presence of CD34 on 75.6% of live harvested uncultured versus 7.84% of cultured fibroblasts suggests that fibroblasts in a nonactivated resting state within the skin dermis have not undergone myofibroblastic differentiation characteristic of the activated phenotype. A selection event, as a consequence of in vitro culture, may also play a contributory role to the CD34 expression we observed. CD34 positive cells—like most stem cells—do not rapidly proliferate in most environments, whereas proliferation of their daughter cells would be favored in culture. Under this model, there is not necessarily an activation event, but rather a skewing of the surface marker profiles due to selection of specific subpopulations from the live harvest, in this case, the CD34 negative fibroblasts.
In addition to CD34, CD26 stood out as one of the few surface marker molecules that was significantly downregulated in vitro. CD26 was expressed on 87.1% of live harvested uncultured, but only 37.6% of cultured fibroblasts. Consistent with the protein-level downregulation of CD26 in cultured versus uncultured fibroblasts, CD26 was transcriptionally downregulated in cultured versus uncultured fibroblasts. CD26, also known as dipeptidyl peptidase-4 (DPP4), is a cell surface serine exopeptidase. The natural substrates of CD26/DPP4 include a wide range of Ccl/Cxcl chemokines, glucagon-like peptides, and neuropeptide Y.28,29 Interestingly, Cxcl5, a small cytokine belonging to the CXC chemokine family involved in connective tissue remodeling30 and a potential substrate of CD26, was among the top ten genes significantly upregulated in cultured fibroblasts. Cleavage of chemokines by CD26 results in inactivation. This dual CD26/Cxcl5 regulatory scheme suggests a mechanism to tightly control both the production of and response to Cxcl5 by fibroblasts.
GO-term analysis of both cultured live harvest and cultured tissue explant isolated fibroblast microarray data revealed significant enrichment (*p<0.01) for genes involved in the MAPK, Wnt, and TGF-beta pathways, all of which are pathways implicated in fibrosis. Interestingly, plasma membrane proteins were also significantly enriched (*p<0.01) in cultured compared with uncultured fibroblasts. The transcriptional upregulation of membrane-associated proteins is suggestive of two critical points: (1) the culturing of fibroblasts selects for a subpopulation of cells capable of responding to mitogenic growth signals and adhering to plastic and (2) in vitro culture conditions induces a significant shift in plasma membrane proteins. In conjunction with these microarray data, the lyoplate analysis discussed previously confirms that increased expression of surface proteins is occurring at both a transcriptional and protein level.
In conclusion, the ability of flow cytometry to separate cells based on their surface marker expression has become an indispensible tool to furthering our understanding of cell lineage trees. Critical to this effort is the identification of physiologic surface marker profiles. Given the high degree of positivity (>90%) of many markers expressed within the cultured population, it is strikingly apparent that the in vitro culturing of fibroblasts selects for a highly homogenous activated state with a distinct phenotype and surface marker profile. It is, therefore, essential that a live harvest approach be employed as researchers seek to define specific lineages and discrete subpopulations of fibroblasts on the basis of physiological, transcriptional, and surface marker profiles.
Supplementary Material
Acknowledgments
This work was supported, in part, by a grant from the California Institute for Regenerative Medicine (RC1 00354 to I.L.W), the Smith Family Trust (to I.L.W), NIH Grant U01 HL099776 (to M.T.L), the Hagey Laboratory for Pediatric Regenerative Medicine, and The Oak Foundation (to M.T.L). G.G.W was supported by the Stanford School of Medicine, the Stanford Medical Scientist Training Program, and NIGMS Training Grant GM07365. M.S.H. was supported by the California Institute for Regenerative Medicine Clinical Fellow Training Grant TG2-01159.
Disclosure Statement
No competing financial interests exist.
References
- 1.Landsman L., Nijagal A., Whitchurch T.J., Vanderlaan R.L., Zimmer W.E., Mackenzie T.C., and Hebrok M.Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biol 9,e1001143, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sneddon J.B., Borowiak M., and Melton D.A.Self-renewal of embryonic-stem-cell-derived progenitors by organ-matched mesenchyme. Nature 491,765, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Ma Q., Xu Q., Duan W., Lei J., and Wu E.Targeting the cancer-stroma interaction: a potential approach for pancreatic cancer treatment. Curr Pharm Des 18,2404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dike L.E., and Farmer S.R.Cell adhesion induces expression of growth-associated genes in suspension-arrested fibroblasts. Proc Natl Acad Sci U S A 85,6792, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mauch C., Hatamochi A., Scharffetter K., and Krieg T.Regulation of collagen synthesis in fibroblasts within a three-dimensional collagen gel. Exp Cell Res 178,493, 1988 [DOI] [PubMed] [Google Scholar]
- 6.Mio T., Adachi Y., Romberger D.J., Ertl R.F., and Rennard S.I.Regulation of fibroblast proliferation in three-dimensional collagen gel matrix. In vitro Cell Dev Biol Anim 32,427, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Fries K.M., Blieden T., Looney R.J., Sempowski G.D., Silvera M.R., Willis R.A., and Phipps R.P.Evidence of fibroblast heterogeneity and the role of fibroblast subpopulations in fibrosis. Clin Immunol Immunopathol 72,283, 1994 [DOI] [PubMed] [Google Scholar]
- 8.Ivarsson M., McWhirter A., Borg T.K., and Rubin K.Type I collagen synthesis in cultured human fibroblasts: regulation by cell spreading, platelet-derived growth factor and interactions with collagen fibers. Matrix Biol 16,409, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Penney D.P., Keng P.C., Derdak S., and Phipps R.P.Morphologic and functional characteristics of subpopulations of murine lung fibroblasts grown in vitro. Anat Rec 232,432, 1992 [DOI] [PubMed] [Google Scholar]
- 10.Seluanov A., Vaidya A., and Gorbunova V.Establishing primary adult fibroblast cultures from rodents. J Vis Exp 44, pii: 2033, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichti U., Anders J., and Yuspa S.H.Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat Protoc 3,799, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., and Campisi J.Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5,741, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein M., and Fitzgerald L.R.Enzymatic separation of intact epidermal sheets from mouse skin. J Invest Dermatol 39,111, 1962 [DOI] [PubMed] [Google Scholar]
- 14.Tusher V.G., Tibshirani R., and Chu G.Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98,5116, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan H., Abraham A., Phillips J.R., Pringle J.H., Walker R.A., and Jones J.L.There is more than one kind of myofibroblast: analysis of CD34 expression in benign, in situ, and invasive breast lesions. J Clin Pathol 56,271, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reilkoff R.A., Bucala R., and Herzog E.L.Fibrocytes: emerging effector cells in chronic inflammation. Nat Rev Immunol 11,427, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridger V.C., Wagner B.E., Wallace W.A., and Hellewell P.G.Differential effects of CD18, CD29, and CD49 integrin subunit inhibition on neutrophil migration in pulmonary inflammation. J Immunol 166,3484, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Scanlan M.J., Raj B.K., Calvo B., Garin-Chesa P., Sanz-Moncasi M.P., Healey J.H., Old L.J., and Rettig W.J.Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci U S A 91,5657, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Driskell R.R., Lichtenberger B.M., Hoste E., Kretzschmar K., Simons B.D., Charalambous M., Ferron S.R., Herault Y., Pavlovic G., Ferguson-Smith A.C., and Watt F.M.Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504,277, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihara A., Hou Y., and Jacobson K.The Thy-1 antigen exhibits rapid lateral diffusion in the plasma membrane of rodent lymphoid cells and fibroblasts. Proc Natl Acad Sci U S A 84,1290, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pont S.Thy-1: a lymphoid cell subset marker capable of delivering an activation signal to mouse T lymphocytes. Biochimie 69,315, 1987 [DOI] [PubMed] [Google Scholar]
- 22.Chang H.Y., Sneddon J.B., Alizadeh A.A., Sood R., West R.B., Montgomery K., Chi J.T., van de Rijn M., Botstein D., and Brown P.O.Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2,E7, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer V.R., Eisen M.B., Ross D.T., Schuler G., Moore T., Lee J.C., Trent J.M., Staudt L.M., Hudson J., Jr., Boguski M.S., Lashkari D., Shalon D., Botstein D., and Brown P.O.The transcriptional program in the response of human fibroblasts to serum. Science 283,83, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Rollins B.J., Stier P., Ernst T., and Wong G.G.The human homolog of the JE gene encodes a monocyte secretory protein. Mol Cell Biol 9,4687, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strieter R.M., Wiggins R., Phan S.H., Wharram B.L., Showell H.J., Remick D.G., Chensue S.W., and Kunkel S.L.Monocyte chemotactic protein gene expression by cytokine-treated human fibroblasts and endothelial cells. Biochem Biophys Res Commun 162,694, 1989 [DOI] [PubMed] [Google Scholar]
- 26.Magro G., Michal M., and Bisceglia M.Benign spindle cell tumors of the mammary stroma: diagnostic criteria, classification, and histogenesis. Pathol Res Pract 197,453, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Silverman J.S., and Tamsen A.Mammary fibroadenoma and some phyllodes tumour stroma are composed of CD34+ fibroblasts and factor XIIIa+ dendrophages. Histopathology 29,411, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Abbott C.A., McCaughan G.W., Levy M.T., Church W.B., and Gorrell M.D.Binding to human dipeptidyl peptidase IV by adenosine deaminase and antibodies that inhibit ligand binding involves overlapping, discontinuous sites on a predicted beta propeller domain. Eur J Biochem 266,798, 1999 [DOI] [PubMed] [Google Scholar]
- 29.Ajami K., Abbott C.A., Obradovic M., Gysbers V., Kahne T., McCaughan G.W., and Gorrell M.D.Structural requirements for catalysis, expression, and dimerization in the CD26/DPIV gene family. Biochemistry 42,694, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Persson T., Monsef N., Andersson P., Bjartell A., Malm J., Calafat J., and Egesten A.Expression of the neutrophil-activating CXC chemokine ENA-78/CXCL5 by human eosinophils. Clin Exp Allergy 33,531, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.