Abstract
Lipid droplets (LDs) are ubiquitous organelles that serve as a neutral lipid reservoir and a hub for lipid metabolism. Manipulating LD formation, evolution, and mobilization in oleaginous species may lead to the production of fatty acid-derived biofuels and chemicals. However, key factors regulating LD dynamics remain poorly characterized. Here we purified the LDs and identified LD-associated proteins from cells of the lipid-producing yeast Rhodosporidium toruloides cultured under nutrient-rich, nitrogen-limited, and phosphorus-limited conditions. The LD proteome consisted of 226 proteins, many of which are involved in lipid metabolism and LD formation and evolution. Further analysis of our previous comparative transcriptome and proteome data sets indicated that the transcription level of 85 genes and protein abundance of 77 proteins changed under nutrient-limited conditions. Such changes were highly relevant to lipid accumulation and partially confirmed by reverse transcription-quantitative PCR. We demonstrated that the major LD structure protein Ldp1 is an LD marker protein being upregulated in lipid-rich cells. When overexpressed in Saccharomyces cerevisiae, Ldp1 localized on the LD surface and facilitated giant LD formation, suggesting that Ldp1 plays an important role in controlling LD dynamics. Our results significantly advance the understanding of the molecular basis of lipid overproduction and storage in oleaginous yeasts and will be valuable for the development of superior lipid producers.
INTRODUCTION
Lipid droplets (LDs), intracellular organelles with deposits of neutral lipids and involved in many cellular activities, are widely present in both eukaryotic and prokaryotic cells (1–4). These organelles consist of a neutral lipid core surrounded by a phospholipid monolayer and associated proteins (3, 5). It has been known that LDs serve as the energy reservoir of cells, which may increase the adaptation by mobilization and degradation of lipids during nutrient deprivation, and also connect with other cellular processes, including lipid transport, membrane biogenesis, lipotoxicity relief, protein storage and degradation, pathogenicity, and autophagy (6–9). Because the biology of LDs is closely linked to some diseases, such as obesity, type 2 diabetes, and atherosclerosis, great progress has been made in elucidating the cellular trafficking, dynamics, and biogenesis of LDs in mammalian cells (2, 7, 10). However, there have been few studies on LDs in other species, especially naturally lipid-producing microorganisms (11–13). Analysis of these microorganisms is motivated by the fact that microbial lipid production holds a great promise to convert waste materials, including lignocellulosic biomass, into fatty acid-derived fuel molecules and chemicals in a scenario of biorefinery and sustainable development (14, 15).
The major components of LDs are neutral lipids, including triacylglycerols (TAGs), sterol esters, and ether lipids (16). Neutral lipids constitute more than 90% of LDs by weight, but the ratio of TAGs to sterol esters can change substantially depending on culture conditions and organisms (13, 17, 18). Polar lipids, mainly phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol, form a micellar structure that stabilizes the hydrophobic center. Besides lipids, purified LDs have also been found to be associated with hundreds of proteins (10). The PAT family proteins are present on the LD surface in evolutionarily distant organisms (19). Other LD proteins include lipid metabolism enzymes, Rab GTPases, coatomer components, ARF-related proteins, and SNARE proteins (20). The presence of functionally diversified proteins in LDs suggests that the biological function of LDs is much more than just an energy reservoir (10, 21).
The basidiomycetous yeast Rhodosporidium toruloides is robust in terms of lipid production under nitrogen-limited (NL) or phosphorus-limited (PL) conditions (15, 22, 23). When R. toruloides cells accumulated substantial lipids, giant LDs occupied almost the entire cells. Our previous analysis of soluble cell lysates of R. toruloides showed that the levels of over 500 proteins were significantly changed during lipid accumulation in response to nitrogen limitation (24). However, the identity and dynamics of LD-associated proteome remained unknown in this and other basidiomycetous yeasts.
In this study, we cultured R. toruloides in nutrient-rich, NL, and PL media, obtained cell samples of different lipid contents, purified LDs, and identified 226 LD-associated proteins. These proteins had diverse biological functions and were involved in lipid metabolism, vesicle traffic, small molecule metabolism, protein synthesis and processing, or mitochondrial function. Further reanalysis of our previous differential transcriptome and proteome data sets (24, 25) indicated that the expression levels of the majority of LD-associated proteins changed in response to culture conditions. Such changes were partially confirmed by reverse transcription-quantitative PCR (RT-qPCR) analysis. Interestingly, we found that RHTO_05627 was an abundant LD-associated protein, and thus it was termed Ldp1 (lipid droplet protein 1). RT-qPCR and quantitative “omic” data indicated that the expression level of LDP1 was higher in cells with more lipids than in cells with fewer lipids. Western blot analysis showed that Ldp1 was present specifically in the LD fraction, and its abundance in LDs increased in lipid-rich cells. We showed that overexpressed Ldp1 localized on the LD surface and facilitated the formation of giant LDs in engineered ascomycetous yeast Saccharomyces cerevisiae strains.
This multidisciplinary study advanced the understanding of the molecular basis of neutral lipid overproduction and storage in oleaginous yeasts. Such information will facilitate LD research in general as well as the development of superior lipid producers for the production of biofuels and chemicals.
MATERIALS AND METHODS
Strain and culture conditions.
R. toruloides CGMCC 2.1389 was obtained from the China General Microbiological Culture Collection (Beijing, China). It was the parent strain of the sequenced haploid NP11 (24). The strain in glycerol stock was revived at 28°C on yeast extract-peptone-dextrose (YPD) agar slants (10 g/liter yeast extract, 10 g/liter peptone, 20 g/liter dextrose, and 15 g/liter agar, pH 6.0). Inocula were grown in YPD liquid medium at 30°C and 200 rpm for 24 h. Cells were separately cultured in nutrient-rich (YPD), NL, or PL medium with 10% (vol/vol) initial inoculum at 30°C and 200 rpm. The NL medium consisted of 70 g/liter glucose, 0.1 g/liter (NH4)2SO4, 0.75 g/liter yeast extract, 1.0 g/liter KH2PO4, 1.5 g/liter MgSO4 · 7H2O, and 1% (vol/vol) trace element solution, pH 6.0. The PL medium consisted of 70 g/liter glucose, 5 g/liter (NH4)2SO4, 2 g/liter peptone, 2.3 g/liter K2SO4, 1.5 g/liter MgSO4 · 7H2O, and 1% (vol/vol) trace element solution, pH 6.0. The trace element solution contained 4.0 g CaCl2 · 2H2O, 0.55 g FeSO4 · 7H2O, 0.52 g citrate · H2O, 0.10 g ZnSO4 · 7H2O, 0.076 g MnSO4 · H2O, and 100 μl of 18 M H2SO4 per liter (25). The initial carbon-to-nitrogen (C/N) molar ratios of YPD, NL, and PL media were 3.9, 309, and 24.8, respectively. The initial carbon-to-phosphorus (C/P) molar ratios of YPD, NL, and PL media were 143, 280, and 10,510, respectively. The culture pH was adjusted to 6.0 with 2 M NaOH once per 24 h. Cell pellets harvested from the culture broth grown in YPD medium at 24 h, in NL medium at 24 h, and in PL medium at 48 h were subsequently used for LD preparation and RT-qPCR analysis.
LD purification.
LDs were isolated at 4°C according to a previously described protocol with minor modifications (26). Briefly, cell pellets harvested from 400 ml of culture broth were washed twice with 30 ml of phosphate-buffered saline (PBS) buffer and then suspended in 30 ml of buffer A (20 mM tricine, 250 mM sucrose, pH 7.8) containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF) and incubated on ice for 20 min. These cell samples were then homogenized by running them four times over a JN3000-type ultrahigh-pressure continuous flow cell disrupter (JNBIO Co., Ltd., Guangzhou, China) at 150 MPa. For cells cultured in YPD, the cell debris of the homogenate was removed by centrifugation at 3,000 × g for 10 min. Eight milliliters of supernatant layered with 2 ml of buffer B (20 mM HEPES, 100 mM KCl, 2 mM MgCl2, pH 7.4) on top was centrifuged in an SW40 rotor at 8,000 × g for 40 min. The floating LDs were collected in a 1.5-ml Eppendorf tube and washed three times with buffer B. For the cells cultured in NL medium or PL medium, the homogenate was centrifuged directly at 4,000 × g for 15 min. The LDs were collected in 1.5-ml Eppendorf tubes and washed three times with buffer A.
Lipid extraction and analysis.
Wet cells were harvested from 30 ml culture broth by centrifugation and washed twice with distilled water. The cell mass, expressed as cell dry weight (CDW), was determined gravimetrically after drying the wet cells at 105°C for 24 h. Dried cells were digested with 4 M HCl at 78°C for 1 h before extraction with chloroform-methanol (1:1, vol/vol). The extracts were washed with 0.1% NaCl solution, dried over anhydrous Na2SO4, and evaporated in vacuo, and finally the residues were dried at 105°C for 24 h to yield the total lipids (27). Lipid content was expressed as gram lipids per gram CDW. To purify lipids from LDs, samples were extracted by chloroform-acetone (1:1, vol/vol) and dried with high-purity nitrogen gas. Neutral lipids and polar phospholipids were separated by silica gel thin-layer chromatography with hexane-diethyl ether-acetic acid (80:20:1, vol/vol/vol) and chloroform-methanol-acetic acid-H2O (75:13:9:3, vol/vol/vol/vol) solvent systems, respectively, and visualized by iodine vapor (11).
LD protein separation and identification.
One milliliter of chloroform-acetone (1:1, vol/vol) was added to the purified LDs to extract lipids and precipitate LD proteins. The pellets were harvested by centrifugation at 20,000 × g for 10 min, dissolved in 2× SDS-PAGE sample buffer, and then denatured at 95°C for 5 min. The total membrane, cytosol, and postnuclear supernatant (PNS) fractions were obtained by following a protocol previously described (26). The protein samples of the LD, membrane, cytosol, and PNS were separated on a 10% SDS-polyacrylamide gel and stained with colloidal blue. For mass spectrometry analysis, the entire lane corresponding to LD proteins from cells cultured in YPD medium was cut into 29 slices. In the NL and PL samples, seven bands were sliced because they were obviously different from those in the YPD sample. These gel slices were destained twice with 200 μl acetonitrile–25 mM ammonium bicarbonate (2:3, vol/vol), dehydrated with acetonitrile, and subjected to reduction and alkylation treatment as described before (11). Gel slices were washed sequentially with 25 mM ammonium bicarbonate, 25 mM ammonium bicarbonate solution–acetonitrile (1:1, vol/vol), and acetonitrile, dried in a SpeedVac vacuum concentrator (ThermoFisher Scientific, Waltham, MA, USA), and incubated in trypsin solution (10 ng/μl in 25 mM ammonium bicarbonate) at 4°C for 30 min. The excess trypsin solution was removed, and 50 μl ammonium bicarbonate (25 mM) was added for overnight digestion at 37°C. The digestion reaction was stopped by adding formic acid to a final concentration of 0.1%.
The peptides extracted from in-gel digestion were analyzed by a nano-liquid chromatography-electrospray ionization-linear trap quadrupole tandem mass spectrometry (LC-ESI-LTQ MS/MS) system as described before (11). The peptide solution loaded onto a C18 trap column by an autosampler was eluted onto a C18 column (100 mm by 100 μm) packed with Sunchrom packing material (SP-120-3-ODS-A, 3 μm) and then subjected to an LTQ linear quadrupole ion trap mass spectrometer (ThermoFisher Scientific) equipped with an electrospray ion source. The LTQ mass spectrometer was operated in a data-dependent mode with the initial MS scan ranging from 400 to 2,000 Da. The five most abundant ions were selected automatically for subsequent collision-activated dissociation. The normalized collision energy was 35, and the activation time was 30 ms. The dynamic exclusion parameters were as follows: repeat count, 1; repeat duration, 30.00; exclusion list size, 500; exclusion duration, 90.00; exclusion mass width by mass, ±1.50 atomic mass units (amu). The peak list files were generated by Extract_msn.exe in a BioWorks package (v3.3.1 SP1) with default parameters. All MS/MS data were searched against the R. toruloides protein database containing 8,171 proteins (http://www.bioconversion.dicp.ac.cn/EWEB/DATA/02/RT.pep.corrected.annotated.nameCH.fa.rar). It should be noted that contaminants such as trypsin and keratins were also included in the database. The search parameters of SEQUEST in the BioWorks suite were set as follows: enzyme, trypsin as specific protease, and two miscleavages allowed; precursor ion mass tolerance, 2.0 Da; and fragment ion mass tolerance, 1.0 Da. The variable modification was set to oxidation of methionine (Met + 15.99 Da). The fixed modification was set to carboxyamidomethylation of cysteine (Cys + 57.02 Da). The search results were filtered with Xcorr versus Charge values as follows: Xcorr (+ 1) > 1.90, Xcorr (+ 2) > 2.50, and Xcorr (+ 3) > 3.75). The peptide assignment probabilities were automatically generated by the BioWorks package using the SRF files. Proteins with at least two distinct peptides were assigned as identified proteins.
Protein function prediction and classification.
The S. cerevisiae orthologs of identified proteins were obtained by protein sequence similarity searching against the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/). An E value below 1E−10 was used as a threshold for homologous candidates. Functional classification of the LD proteins was based on protein annotation and SGD gene description. The orthologs of previously identified LD proteins in humans (21, 28), fly (21, 29), S. cerevisiae (17, 21, 30), Pichia pastoris (18), and Yarrowia lipolytica (13) were assigned by reciprocal BLAST.
Differentially expressed transcripts and proteins.
The transcript and protein levels of the genes coding for LD-associated proteins were retrieved from the database of our previous digital gene expression profiling and semiquantitative proteome study of R. toruloides (24). The sequence tags of digital gene expression profiling (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39023) were cleaned and normalized to tags per million (TPM) and then mapped to annotated transcripts to calculate the transcription level. Optimal criteria implemented with SFOER software (31) were used for peptide assignment filter in the proteome analysis of the soluble cell lysates, which made the false-discovery rate below 1%. Spectral counts (http://www.bioconversion.dicp.ac.cn/EWEB/DATA/03/Peptide_identification.zip) of the identified peptides were collected and normalized as described previously (32). The same statistical algorithms for significance tests as those in previous processing (24) were used. P values of <0.01 and fold changes of >2 were used as the thresholds of the differentially expressed transcripts, while P values of <0.05, G values of >1.35, and fold changes of >2 were used for assignment of the changed LD proteins.
RT-qPCR analysis.
The cell pellets harvested from 1 ml culture broth of R. toruloides were immediately frozen in liquid nitrogen and stored at −70°C. Total RNA was isolated from about 30- to 50-mg cell samples using the FastRNA Pro Red kit and FastPrep instrument (Qbiogen, Inc., Irvine, CA, USA) according to the manufacturer's instructions, and the setting of FastPrep Instrument was 6.0 m/s for 60 s. The RNA concentration and quality were determined by a Nanodrop ND1000 spectrophotometer (ThermoFisher Scientific), while the RNA integrity was assessed by agarose gel electrophoresis. DNA removal and cDNA synthesis were performed with the PrimerScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China). Equal amounts of each RNA (750 ng) were used for reverse transcription. Quantitative PCR (qPCR) experiments were carried out using the SYBR Premix Ex Taq GC kit (TaKaRa) in an Eco real-time PCR system (Illumina, San Diego, CA, USA). Gene-specific primers are listed in Table S1 in the supplemental material. Reactions were performed with appropriate amounts of cDNA templates and 0.2 μM each primer in a total volume of 10 μl. Nonspecific amplicons were not observed by dissociation curves and agarose gel electrophoresis analysis. The amplification efficiencies were calculated from five-point calibration curves based on a 6-fold dilution series using a pool of equal volumes of all cDNA as the template (see Table S1 in the supplemental material). The expression levels were normalized to two reference genes (RHTO_03560, coding for actin, and RHTO_03746, coding for glyceraldehyde 3-phosphate dehydrogenase) by a normalization factor as described before (33). Three biological and two technical replicates were analyzed to obtain the relative expression quantity.
Western blotting.
Anti-Ldp1 polyclonal antibody was raised against two synthesized peptides, NEKQPATDAPLAHEC (corresponding to amino acid residues 5 to 18) and CKKKDEVAETVEEKEGEGEK (corresponding to amino acid residues 246 to 264), selected from the N and C termini of Ldp1. The mixture of these two peptides was used as antigens and injected into two rabbits. After three injections, the antibody activity of the rabbit sera was confirmed by Western blotting. Four cellular fractions (LD, total membrane, cytosol, and PNS) of R. toruloides were prepared according to a previously reported protocol (26). Proteins were separated by 12% SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes, blotted with the anti-Ldp1 polyclonal antibody, and detected with the enhanced chemiluminescence system.
Cloning and expression of LDP1.
The cDNA of Ldp1 (NCBI accession number EMS18697) was amplified from the reverse transcription product with primers PRLP1-L1 (CAAACAACGAGCACAGCGACAC) and PRLP1-R1 (AAACCGAGAAGAAACCCGAAC). The amplification product was first cloned into a pMD-19T vector (TaKaRa) to give pMD-19T:perilipin. The complete open reading frame of Ldp1 was amplified with the primer pair PG-PER-F (GGAATTCAACATGGCCACCGTCAACGAGAAGC), where underlining indicates the EcoRI restriction site, and PG-PER-R (TGCAAGCTTGGCGTAATCATGGTCATCTGCTTCTCCCCCTCGCCCTCCT). The green fluorescent protein (GFP) gene was amplified from pGFPuv (Clontech Laboratories, Inc., Mountain View, CA, USA) with the primer pair PG-GFP-F (GAGAAGGAGGGCGAGGGGGAGAAGCAGATGACCATGATTACGCCAAGCT) and PG-GFP-R (AAAAGGAAAAGCGGCCGCTCATTTGTAGAGCTCATCCATGCCATG), where underlining indicates the NotI restriction site. The amplification products were gel purified and spliced by overlap extension PCR (34). The fusion segment was digested with EcoRI and NotI and inserted into pYES2/CT (Invitrogen Co., Carlsbad, CA, USA) to produce the plasmid pYES2-PG expressing the C-terminal GFP-tagged Ldp1 fusion protein (Ldp1-GFP) under the control of the GAL1 promoter. pYES2-PG was digested with HindIII to remove the LDP1 fragment and self-ligated to generate pYES2-GFP. Both plasmids were transformed by electroporation into S. cerevisiae INVsc1 according to a previous protocol (35). The pYES2-PG plasmid was also transformed to strains expressing Erg6-RFP or Sec13-RFP (36).
Confocal fluorescence microscopy.
Single colonies from the transformation plates were first cultured at 30°C in the maintenance medium (20 g/liter raffinose, 6.7 g/liter yeast nitrogen base without amino acids, and supplementary amino acids as needed, i.e., 100 mg/liter leucine, 20 mg/liter histidine, 20 mg/liter tryptophan, or 30 mg/liter lysine) to an optical density at 600 nm of 3.0. One milliliter of preculture was used to inoculate 10 ml of inducing medium (replacement of raffinose with galactose in the maintenance medium) supplemented with 200 μM oleic acid. After cultivation at 30°C for 30 h, cells were collected and washed twice with PBS and then dropped onto microscope slides pretreated with collagen for 20 min. The samples were stained with a 500-fold-diluted solution of LipidTOX Red (catalog number H34477; Invitrogen) in darkness for 20 min and viewed with an Olympus FV1000 confocal microscope (Olympus Co., Tokyo, Japan).
RESULTS
Lipid production and LD morphology under different culture conditions.
It has been known that oleaginous yeasts accumulate lipids under nutrient-limited conditions and that cellular lipid contents are low (below 20%) under nutrient-rich conditions (22). In the present study, the lipid contents of the inoculum cells and the cells cultured in YPD medium for 24 h were 8.6% and 8.2%, respectively; however, lipid content increased to 51.5% when the cells were grown in NL medium for 72 h (NL-72 h). Similarly, in PL medium, the cells accumulated lipids to 22.3% and 31.7% after 48 h and 72 h, respectively (Fig. 1A), indicating that the cells grew and accumulated lipids slower in PL medium. The lipid accumulation was consistent with the evolution of LD size. Several small LDs were present in low-lipid cells, while one or two large LDs appeared in high-lipid cells (Fig. 1B). To maintain similar lipid contents and thus similar LDs between samples, we used cells cultured for 24 h in NL medium and for 48 h in PL medium for further LD purification.
FIG 1.
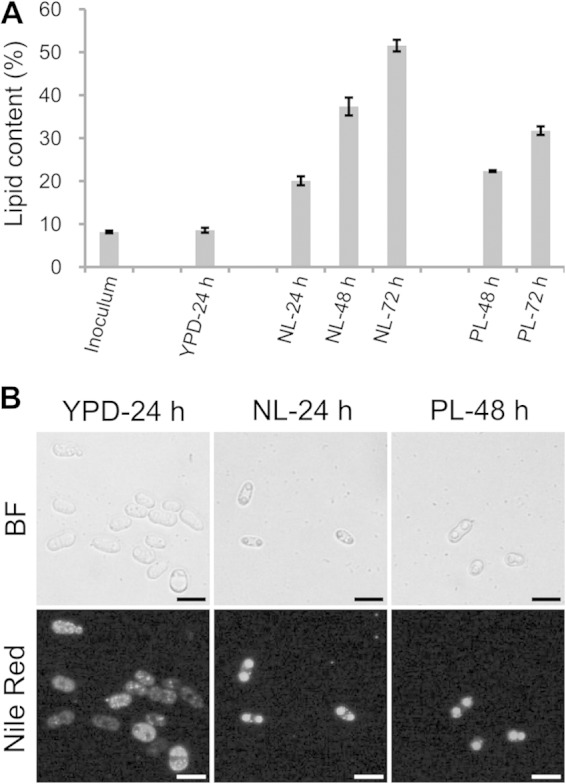
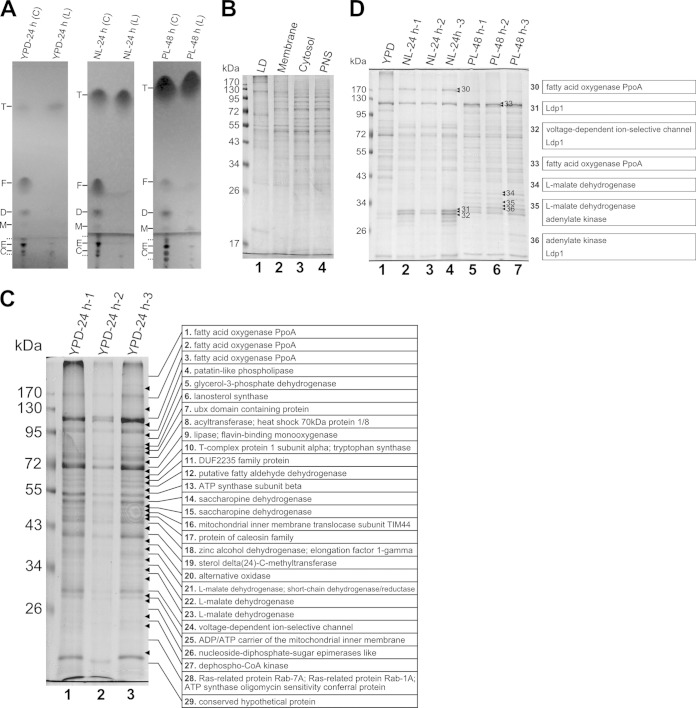
Lipid accumulation by R. toruloides cells. (A) Lipid contents of R. toruloides cells cultured under different conditions. Cells were collected from cultures in YPD, nitrogen-limited (NL), and phosphorus-limited (PL) media. The total lipids were extracted and measured gravimetrically. The error bars represent the standard deviations for three independent samples. (B) Morphology of R. toruloides lipid droplets (LDs). Cells cultured in YPD medium for 24 h, NL medium for 24 h, or PL medium for 48 h were stained with Nile Red and viewed by fluorescence microscope. BF, bright field microscopy; bar, 10 μm.
LD purification and comprehensive proteomic analysis.
To get more insights into lipid accumulation, we purified LDs as well as the lipids within LDs from R. toruloides cells according to a previously described protocol (11, 26). It was found that the lipid samples from the isolated LDs had significantly larger amounts of neutral lipids but smaller amounts of polar lipids than the total lipid samples from the whole cells (Fig. 2A). Such a difference in lipid composition was a persuasive indication of the good purity of the isolated LDs. The protein profile of the LDs was unique compared with other cellular fractions (including total membrane, cytosol, and PNS), which further demonstrated that the isolated LDs were considerably pure (Fig. 2B). It was also clear that the purification procedure was reproducible because the protein patterns of LDs from three independent biological replicates were almost identical (Fig. 2C and D).
FIG 2.
Analysis of the lipids and proteins in purified lipid droplets (LDs) from R. toruloides. (A) Thin-layer chromatography analysis of lipid samples from whole cells (C) and LDs (L) prepared from cultures in YPD, nitrogen-limited (NL), or phosphorus-limited (PL) media. Solid lines and dotted lines on the left of the three panels indicate lipid standards and unknown lipid species, respectively. T, triacylglycerol; F, fatty acid; D, diacylglycerol; M, monoacylglycerol; E, phosphatidylethanolamine; C, phosphatidylcholine. (B) SDS-PAGE analysis of proteins from LD, membrane, cytosol, and postnuclear supernatant (PNS) fractions. (C) SDS-PAGE analysis of LD proteins isolated from three independent samples cultured in YPD medium for 24 h. The whole lane of the YPD-24 h-3 sample was cut into 29 slices and analyzed by LC-MS/MS, and the major proteins of each band are listed. (D) SDS-PAGE analysis of LD proteins isolated from three independent samples cultured in NL medium for 24 h or PL medium for 48 h and comparison of the protein patterns with those from a YPD culture. The seven marked bands were cut for MS analysis, and the major proteins present in each band are listed on the right.
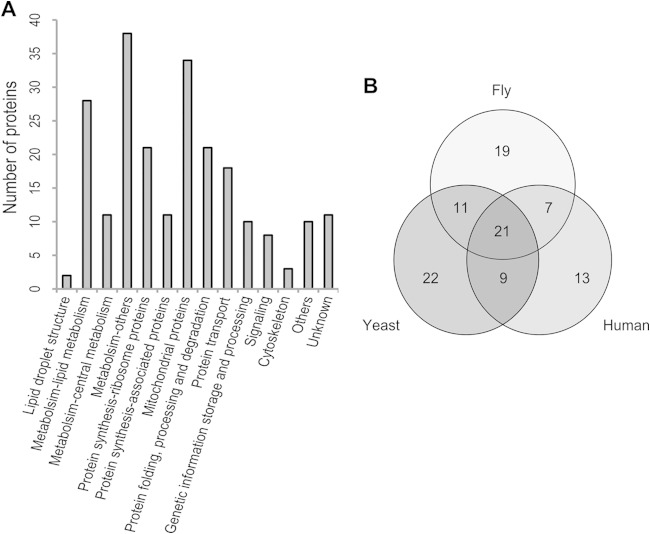
To obtain an accurate LD proteome, a previously published method was applied (20). Briefly, the whole lane of YPD-24 h-3 was cut into 29 slices (Fig. 2C, bands 1 to 29). In order to identify key proteins involved in the maturation of large LDs and to check the comprehensiveness of the LD proteome, 7 visibly different bands that appeared in LDs purified from the NL or PL samples were also collected (Fig. 2D, bands 30 to 36). The gel slices were destained, and the proteins therein were digested according to the in-gel digestion procedure. The peptides were recovered and identified by LC-MS/MS. Thus, 224 proteins were identified from the YPD-24 h-3 sample, and two additional proteins were found in bands 31 and 34. Of these 226 proteins, 172 (76%) had functional orthologs in S. cerevisiae (see Table S2 in the supplemental material). There were 63, 58, and 50 proteins whose orthologs have also been found in the LD proteome of yeasts (S. cerevisiae, P. pastoris, and Y. lipolytica), humans (Homo sapiens), and fly (Drosophila melanogaster), respectively (Fig. 3B), suggesting that at least 102 (45%) were supported as LD proteins by those previous studies (13, 17, 18, 21, 28–30). These observations show that, for the first time, the LD proteome of the lipid-producing yeast R. toruloides was established.
FIG 3.
Function catalogs of the R. toruloides lipid droplet (LD) proteome. (A) Functional classification of the identified proteins. (B) Venn diagram of proteins present in the R. toruloides LD proteome that have homologs in yeasts (S. cerevisiae, P. pastoris, and Y. lipolytica), humans (Homo sapiens), and fly (Drosophila melanogaster). The function of each protein is listed in Table S2 in the supplemental material.
The LD proteome of R. toruloides was classified into 14 groups according to the protein annotation and corresponding orthologs in S. cerevisiae (Fig. 3A; see also Table S2 in the supplemental material). About one-third of the identified proteins were enzymes involved in metabolism. There were 28 proteins associated with lipid metabolism (Table 1), including synthesis and degradation of fatty acid, TAG, and sterol esters. Interestingly, most of them (20 proteins) were also found as LD proteins in other species. The presence of metabolic enzymes indicated that LD is not only a passive reservoir but also an energetic organelle in which metabolic reactions occur. Other large groups included those involved in protein synthesis, folding, processing, degradation, and transportation. There were 32 proteins that were identified as the components of the ribosome or translational machinery and 10 protein-folding-related chaperones, which were also frequently found in the LDs of other species (21). For example, it has been demonstrated that HSP70 of rat adipocytes is localized to LDs upon heat stimulation (37). Rab GTPases, coatomer components, v-SNARE, and ADP ribosylation factor are vesicle-trafficking-related proteins, which play crucial roles in LD formation and size determination. The fact that these proteins were found in LDs in this study was consistent with previous observations (2, 7, 21). We also identified 34 mitochondrial proteins, which might be due to the contamination that resulted from structural or functional interaction between LDs and mitochondria (38). A small proportion of LD proteins, such as two histones, were involved in genetic information storage and processing, which was consistent with the results of a study on Drosophila (39). Moreover, 8 and 3 proteins were related to cellular signaling and cytoskeleton formation, respectively, and 21 proteins were poorly characterized in this LD proteome.
TABLE 1.
Identified LD proteins involved in lipid metabolism
| Function catalog (total no. of proteins) | Identified proteinsa |
|---|---|
| Fatty acid synthesis (4) | RHTO_03915 (ATP citrate synthase), RHTO_02004 (acetyl-CoA carboxylase, Acc1), RHTO_02032 (fatty acid synthase subunit beta, fungus type, Fas1), RHTO_02139 (fatty acid synthase subunit alpha, fungus type, Fas2) |
| TAG synthesis (3) | RHTO_03193 (1-acylglycerone phosphate reductase, Ayr1), RHTO_07665 (glycerol-3-phosphate dehydrogenase, Gut2), RHTO_04314 (glycerol-3-phosphate/dihydroxyacetone phosphate dual substrate-specific sn-1 acyltransferase, Gpt2) |
| Steroid synthesis (4) | RHTO_00856 (delta(24)-sterol C-methyltransferase, Erg6), RHTO_01745 (squalene monooxygenase, Erg1), RHTO_02048 (acetyl-CoA C-acetyltransferase, Erg10), RHTO_04192 (lanosterol synthase, Erg7) |
| Hydrolase (9) | RHTO_01062 (steryl ester hydrolase, Yeh2), RHTO_01389 (lipase, class 3), RHTO_03165 (inositol or phosphatidylinositol phosphatase, Inp51), RHTO_03511 (alpha/beta hydrolase fold protein, Yju3), RHTO_03913 (monoglyceride lipase, homolog of YJU3, Yju3), RHTO_05588 (N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, Fmp30), RHTO_03931 (bifunctional enzyme with triacylglycerol lipase and lysophosphatidic acid acyltransferase activity, Tgl5), RHTO_03669 (esterase/lipase/thioesterase family protein), RHTO_05515 (alpha/beta hydrolase, putative lipase/esterase) |
| Fatty acid oxidation (5) | RHTO_00397 (acyl-CoA dehydrogenase), RHTO_01116 (acyl-CoA thioesterase 8, Tes1), RHTO_03348 (3-oxoacyl-[acyl-carrier protein] reductase), RHTO_07686 (fatty acid oxygenase PpoA), RHTO_05680 (putative fatty aldehyde dehydrogenase, Hfd1) |
| Others (3) | RHTO_07994 (phosphatidylserine decarboxylase, Psd1), RHTO_04135 (lipase/thioesterase family protein, similar to sterol deacetylase Say1), RHTO_03216 (putative acyltransferase with similarity to Eeb1p and Eht1p, YMR210W) |
The names of corresponding orthologs in S. cerevisiae are underlined.
Interestingly, we found two potential LD structural proteins, RHTO_05627 and RHTO_03414. RHTO_05627 was previously annotated as a perilipin-like protein; here, we designated it Ldp1 (lipid droplet protein 1). Fungal perilipin-like protein Mpl1 has been first identified in Metarhizium anisopliae, and its homologs are present only in a small proportion of ascomycetous fungi, such as the Pezizomycotina (40). However, at least one Ldp1 homolog has been found previously in basidiomycetous fungi (24). Moreover, YALI0F24167g, the most abundant protein in the LD proteome of the ascomycetous yeast Y. lipolytica (13), was also the homolog of Ldp1. RHTO_03414 is a caleosin family protein. Caleosins are LD-associated Ca2+ binding proteins that have been found in many plants, several oleaginous fungi, and algae (3, 41). The biological functions of caleosins have been shown to stabilize the oil body (alias of lipid droplet) of cycad megagametophytes (42) and to interact with vacuoles and promote degradation of storage lipids in Arabidopsis seeds (43). These results suggested that LD structural proteins are likely conserved between diverse eukaryotic species.
The proteins identified from the bands with enhanced densitometry in the NL or PL samples are listed in Table 2. Besides the proteins already found in the YPD-24 h-3 sample, RHTO_06504 (conserved hypothetical protein) and RHTO_04226 (actin cortical patch component Lsb4, homolog of S. cerevisiae Lsb3) were newly identified. The protein abundance of Ldp1, which migrated to the bands identical to band 31, was different in these samples. The abundance of Ldp1 was highest in the NL sample while lowest in the YPD sample (Fig. 2D), suggesting that it might be involved in lipid overproduction and giant LD formation. RHTO_07686, a 123-kDa protein similar to Aspergillus nidulans psi factor-producing oxygenase PpoA, was also identified. PpoA is a bifunctional P450 protein that oxidizes linoleic acid to (8R)-8-hydroperoxyoctadecadienoic acid followed by the isomerization to form (5S,8R)-5,8-dihydroxyoctadecadienoic acid (44). The corresponding oxylipin products are hormone-like signaling mediators regulating the balance between asexual and sexual life cycles (44, 45). It has been demonstrated that PpoA is localized to the LDs in A. nidulans (45), but detailed roles of PpoA in LD function and lipid accumulation remained unclear. We also found a higher-molecular-mass PpoA isoform (about 170 kDa) whose abundance was higher in the YPD and NL samples than in the PL sample (Fig. 2D). RHTO_03117, one of the two adenylate kinases and the ortholog of S. cerevisiae cytosolic Adk1, was the major protein of band 36. It was remarkable to find RHTO_03117 in the LD proteome obtained from the PL sample. Under PL conditions, the cellular levels of free phosphate and ATP and the energy charge may be low, and adenylate kinase is responsible for converting two ADP molecules into one ATP and one AMP to maintain ATP homeostasis (46). RHTO_04363, another abundant protein identified in the PL sample, was a mitochondrial l-malate dehydrogenase.
TABLE 2.
Identified proteins from bands 30 to 36
| SDS-PAGE band (total no. of proteins in the catalog) | Identified proteinsa |
|---|---|
| Band 30 (1) | RHTO_07686 (fatty acid oxygenase PpoA) |
| Band 31 (3) | LDP1 (perilipin-like), RHTO_06504 (conserved hypothetical protein), RHTO_07648 (voltage-dependent ion-selective channel, Por1) |
| Band 32 (2) | RHTO_07648 (voltage-dependent ion-selective channel, Por1), LDP1 (perilipin-like) |
| Band 33 (1) | RHTO_07686 (fatty acid oxygenase PpoA) |
| Band 34 (3) | RHTO_04363 (l-malate dehydrogenase, mitochondrial, Mdh1), RHTO_00208 (short-chain dehydrogenase/reductase SDR family protein, YOR246C), RHTO_00474 (mitochondrial phosphate carrier protein, Pic2) |
| Band 35 (6) | RHTO_04363 (l-malate dehydrogenase, mitochondrial, Mdh1), RHTO_03117 (adenylate kinase, Adk1), RHTO_06575 (NmrA-like domain containing protein), RHTO_03193 (1-acylglycerone phosphate reductase, Ayr1), RHTO_06960 (protein of glucose/ribitol dehydrogenase family, weak similarity to YKL107W), RHTO_04226 (actin cortical patch component Lsb4, Lsb3), |
| Band 36 (2) | RHTO_03117 (adenylate kinase, Adk1), LDP1 (perilipin-like) |
Proteins in bold are the major proteins for which the most peptides were identified. The names of corresponding orthologs in S. cerevisiae are underlined.
Expression dynamics of the LD proteome.
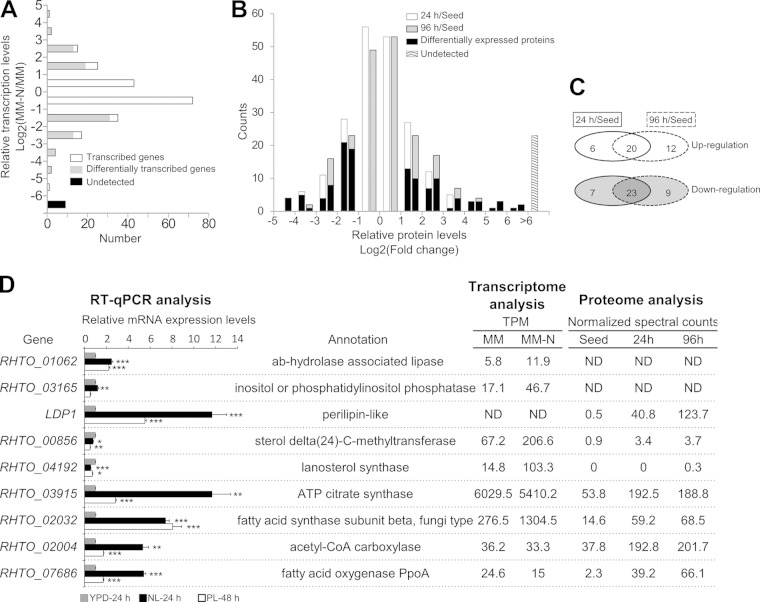
The results showed a dramatic increase in lipid content when the cells were cultured under NL and PL conditions (Fig. 1), indicating that R. toruloides is a good model to study LD dynamics and identify the key proteins involved in the formation of large LDs. The SDS-PAGE results indicated that the LD proteome changed substantially. The most significant change was an enhanced densitometry of bands 31 and 32 in the NL sample and of band 36 in the PL sample compared with those in the YPD sample (Fig. 2D). However, this assay was inadequate to offer more quantitative information. The dynamic changes of the LD proteins might have resulted from the regulation of gene expression and organelle targeting. The information about expression changes of LD-related genes should be included in the global differential transcriptome and proteome data sets. In our previous work, we have sequenced the transcript 3′-end tags and performed a quantitative comparison of the transcriptomes between a sample cultured in minimal medium (MM) containing 47 mM NH4+ and a sample in nitrogen-limited minimal medium (MM-N). The lipid contents of the MM and the MM-N samples were 22.8% and 33.3%, respectively, and the expression levels of over 2,000 genes were significantly different in these two samples (24). Interestingly, in this study we found that more than one-half of the LD-associated genes were expressed within a 2-fold variation between the MM and MM-N samples and only 9 genes were not detected in the 3′-tag digital gene expression profiling analysis (Fig. 4A). Of those with a >2-fold change, 35 and 50 genes were up- and downregulated, respectively, in the MM-N sample (Fig. 4A; see also Tables S3 and S4 in the supplemental material). We also compared the proteomes of cells from the YPD culture (“Seed”), the NL culture for 24 h (“24 h”), and the NL culture for 96 h (“96 h”) (24, 25). Similarly, the lipid contents increased from 9.8% in the “Seed” sample to 63% in the “96 h” sample. The levels of over 500 proteins significantly changed among these samples (24), of which 90% of the LD-associated proteins (203 proteins) were identified in at least one sample (Fig. 4B). By rigorous statistical testing, the proteins that significantly changed in the NL samples in comparison with those in the nutrient-rich sample were found (Fig. 4B). Protein abundances changed similarly in the NL sample over time, because the proportion of overlap between the 24 h/Seed data set and the 96 h/Seed data set was high (Fig. 4C). The changed proteins in the “24 h” and “96 h” samples were merged, and 38 and 39 proteins were up- and downregulated, respectively, under nitrogen-limited conditions (see Tables S3 and S4 in the supplemental material).
FIG 4.
Dynamics of lipid droplet proteome. (A) Relative transcription levels of LD-associated genes revealed from digital gene expression analysis of the cells cultured in minimum medium (MM) and nitrogen-limited minimum medium (MM-N). The MM sample was used as a reference. (B) Relative protein levels of LD proteins revealed from comparative proteome analysis. Protein levels of the cells cultured in nutrient-rich medium (“Seed”) were used as a reference, and relative protein levels and differentially expressed proteins in cells cultured in nitrogen-limited (NL) medium (“24 h” and “96 h”) are shown. (C) Overlap of the differentially expressed LD proteins in the “24 h” and “96 h” samples. (D) Relative mRNA expression levels between cells cultured in nutrient-limited medium (“NL-24 h” and “PL-48 h”) and nutrient-rich medium (“YPD-24 h”) revealed by RT-qPCR. Three biological replicates and two technical replicates were performed. Statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001) was determined by one-tailed Student's t test. The error bars represent the standard errors for three independently cultured samples. The expression levels were normalized to two reference genes (RHTO_03560 and RHTO_03746). The gene annotation and the transcript and protein expression levels, respectively, from comparative transcriptome (panel A) and proteome (panel B) analysis are listed in the right panel. Normalized counts of transcript tags (TPM, tags per million) were used for comparison of transcription levels. The average of the normalized spectral counts from three replicates was used for comparison of the protein levels. The comparative transcriptome and proteome data were retrieved from our previous study (24).
The reanalysis of the previous comparative transcriptome and proteome data clearly revealed that the transcription and expression levels of many LD-related genes changed dynamically during lipid accumulation. In the current work, nitrogen or phosphorus limitation also altered the protein profiles of the LD proteome (Fig. 2D). In order to examine whether the expression of the LD-associated genes was regulated by nutrient limitation, RT-qPCR was performed to analyze the transcriptional levels of 9 selected genes (Fig. 4D). These genes encoded proteins important for LD maintenance, enlargement, and mobilization. It was found that 6 genes were transcriptionally upregulated, which was coincident with the “omic” analysis. The protein abundance of Ldp1 increased more than 200-fold in the “96 h” sample compared with that in the “Seed” sample (Fig. 4D). The RT-qPCR analysis confirmed the upregulation of LDP1 in the MM-N sample (data not shown), although the transcriptome analysis failed to detect the expression level of LDP1, most likely because of the absence of the CATG site to generate the transcript tag. The RT-qPCR results also clearly showed that the transcriptional level of LDP1 was upregulated under both NL and PL conditions. Moreover, the transcription level was higher in the NL sample than in the PL sample (Fig. 4D). Interestingly, protein levels of Acl1 (ATP:citrate lyase) and Acc1 (acetyl-CoA carboxylase), key enzymes for the formation of precursors for lipid biosynthesis (24), were upregulated under the NL condition. The RT-qPCR analysis indicated that the transcription of ACL1 and ACC1 was elevated in both the NL-24 h and the PL-48 h samples and that the transcription levels in the PL sample were lower than that in the NL sample (Fig. 4D). The RT-qPCR results showed that the transcription of FAS1 (fatty acid synthase, beta subunit) was upregulated about 8-fold under NL and PL conditions, which was also in agreement with previous transcriptome and proteome results. Under nutrient-limited conditions, higher protein levels of PpoA, the enzyme for oxylipin production, were also confirmed by RT-qPCR. Although the transcription of 4 genes, RHTO_00856 and RHTO_04192 for steroid synthesis and RHTO_01062 and RHTO_03165 for lipid hydrolysis, were all upregulated in the MM-N sample, RT-qPCR results indicated that only RHTO_01062 was significantly upregulated under NL conditions. The other three were downregulated under NL or PL conditions (Fig. 4D).
In the LD proteome of R. toruloides, several other lipid metabolic enzymes, such as acyl coenzyme A (acyl-CoA) dehydrogenase (RHTO_00397), lipase (RHTO_01389 and RHTO_03913), and fatty aldehyde dehydrogenase (RHTO_05680), were upregulated in the NL samples, while glycerol-3-phosphate dehydrogenase (RHTO_007665), acyl-CoA thioesterase 8 (RHTO_01116), and lipid hydrolase (RHTO_03669) were downregulated. The lipid biosynthesis-associated proteins, such as 3-oxoacyl-ACP reductase (RHTO_03348) and acetyl-CoA C-acetyltransferase (RHTO_02048), were transcriptionally upregulated in the MM-N sample, but the latter was downregulated in the NL sample (see Table S3 in the supplemental material).
Ldp1 localized on LDs and promoted giant LD formation.
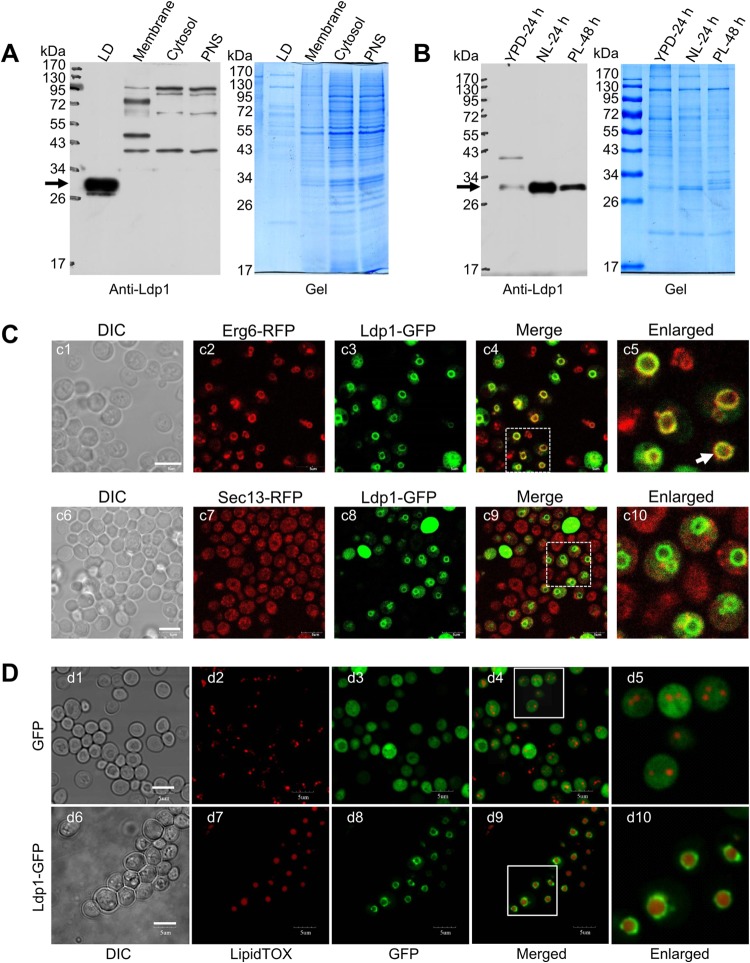
Since Ldp1 has a putative perilipin domain (Pfam number PF03036) and it is a potential LD structural protein, we carried out experiments to verify its LD localization, to examine its cellular distribution, and more importantly, to determine whether it was a key player for giant LD formation. First, we made a polyclonal antibody specifically recognizing Ldp1. To determine the cellular distribution of Ldp1, we fractionated cell lysates into LDs, total membrane, cytosol, and PNS during the LD purification process. Proteins separated by SDS-PAGE were immunoblotted with the anti-Ldp1 antibody, and the results clearly showed that Ldp1 was present only in the LD fraction, even with much lower protein loading than in other cellular fractions (Fig. 5A). These data confirmed the LD localization of Ldp1 and suggested Ldp1 as an LD-specific protein in R. toruloides. We then compared the protein abundance of Ldp1 in the LD fractions prepared from cells cultured under different conditions and found that the protein levels in both the NL-24 h and PL-48 h samples were higher than that in the YPD-24 h sample. Moreover, the NL-24 h sample had a higher level of Ldp1 than the PL-48 h sample (Fig. 5B), which matched well with the densitometry change of the protein bands that migrated identically to band 31 between these samples (Fig. 2D). These results were also in agreement with results from semiquantitative proteome and RT-qPCR analysis (Fig. 4D). Together, these observations clearly demonstrated the dynamic nature of Ldp1 during lipid production. Second, we expressed a C-terminal GFP-tagged Ldp1 (Ldp1-GFP) in S. cerevisiae lacking perilipin-like proteins to morphologically determine the cellular distribution of Ldp1. The Ldp1-GFP colocalized well with the LD marker Erg6-RFP (47) and LipidTOX staining but not with Sec13-RFP, which is a marker protein for endoplasmic reticulum (ER) to Golgi vesicles (Fig. 5C and D), while the expressed GFP distributed in the cytosol space (d1 to d5 in Fig. 5D). We also examined the LD sizes of S. cerevisiae cells overexpressing Ldp1-GFP. Interestingly, most cells that expressed Ldp1-GFP contained one giant LD, whereas the cells that expressed GFP had multiple small LDs (Fig. 5D; see also Fig. S1 in the supplemental material), indicating that the overexpression of Ldp1 was responsible for LD enlargement.
FIG 5.
Localization and function of the lipid droplet (LD) structure protein Ldp1. (A) Distribution of Ldp1 in different cellular fractions. The proteins from LDs (LD), total membrane (Membrane), cytosol (Cytosol), and postnuclear supernatant (PNS) were separated by SDS-PAGE and blotted with anti-Ldp1 antibody. (B) Comparison of Ldp1 protein levels in the LD proteome under different culture conditions. Equal amounts of LD proteins from R. toruloides cells cultured in YPD for 24 h, NL for 24 h, and PL for 48 h were separated by SDS-PAGE and blotted by anti-Ldp1 antibody. The arrows in the left panels indicate the position of Ldp1, and the right panels were colloidal blue stained and used as loading control (A and B). (C) Localization of heterogeneously expressed Ldp1 in S. cerevisiae. The localizations of Ldp1-GFP, Erg6-RFP (LD marker), and Sec13-RFP (the marker for ER to Golgi vesicles) were compared. (D) Overexpressed Ldp1-GFP localized on LDs and facilitated the formation of giant LDs in S. cerevisiae. S. cerevisiae cells expressing GFP or Ldp1-GFP were observed by confocal microscope. LDs were stained with LipidTOX. Bar, 5 μm (C and D).
DISCUSSION
R. toruloides is a basidiomycete fungus of the Pucciniomycotina subphylum. Many species of the Pucciniomycotina clade are plant-parasitic rust fungi that can cause severe forest and agriculture losses (48). It has been known that LDs are connected with pathogen virulence of fungi (40, 49). However, there have been no systematic studies on the LD proteome of basidiomycetes. Moreover, this yeast is well known for the overproduction of lipids and pigments (15, 22, 23, 50) and has been used as a microbial factory for the production of advanced biofuels (15, 51). Manipulating the dynamics of LDs is a potential approach to engineer this yeast for the production of lipids and related metabolites at higher titer, rate, and yield. For example, the ER proteins seipin and fat storage-inducing transmembrane proteins (FIT1 and FIT2) have been found to regulate LD formation and morphology (52), and impairing the expression of seipin can increase TAG synthesis and lead to giant LD formation (53, 54). In addition, higher levels of FIT1 or FIT2 have resulted in more and larger LDs (55). The expression of the LD structure protein caleosin 1 (AtClo1) from Arabidopsis in S. cerevisiae increased lipid accumulation (56). Recently, many efforts have been undertaken to engineer S. cerevisiae or Escherichia coli for the production of lipid-derived metabolites and biofuels, but the efficiency remained fairly low (14, 57). Alternatively, it has been shown that more than 70% of the TAGs in R. toruloides can be converted to fatty acid ethyl esters when incubating lipid-rich cells with 10% ethanol in an aqueous environment (58). It is believed that the LD provides the hydrophobic microenvironment for LD-associated lipases, which traditionally catalyzed the transesterification in nonaqueous media (59). Although the roles of LDs are important in lipid biotechnology, key factors regulating the LD dynamics remain poorly characterized in oleaginous species.
In this study, we established the LD proteome of R. toruloides. By analyzing the protein profiles of the LDs from high-lipid and low-lipid cell samples, we found that the LD proteome changed significantly (Fig. 2D). We speculated that these proteome changes were crucial for lipid accumulation and LD dynamics; thus, we reanalyzed the data sets of our previous comparative transcriptome and proteome study to identify differentially expressed LD-associated genes, as those previous samples were cultured in nitrogen-rich and nitrogen-limited media and accumulated different amounts of lipids (24, 25). The expression levels of about one-third of the LD-associated genes changed (see Table S3 in the supplemental material), while only 25% of the overall genes and 7% of the whole proteome of the cell changed at the transcriptome and proteome levels, respectively. This indicated that the LD proteome changed more significantly than other cellular proteins.
The LDs are hubs of cellular lipid metabolism and can serve as the reservoir of energy or building blocks of cellular membrane lipids. Acetyl-CoA, a precursor for fatty acid synthesis, is generated from the cleavage of citrate, which permeates out from the mitochondrion in oleaginous yeasts (22). The enzyme catalyzing citrate to acetyl-CoA is Acl1. Acetyl-CoA is activated to malonyl-CoA by acetyl-CoA carboxylase (Acc), and then malonyl-CoA is utilized for fatty acid chain elongation, which is catalyzed by fatty acid synthase (Fas), a multifunctional enzyme complex. These three enzymes were traditionally known as cytoplasmic proteins, but they were found in the LD proteome in this study as well as others (60). It was unknown whether these proteins were translocated to LDs by vesicle transport or were recruited from the cytosol. Other lipid biosynthesis-related proteins were found in the LD proteome, including RHTO_07665, RHTO_03193, and RHTO_04314. All these enzymes constituted a complete pathway from citrate and dihydroxyacetone phosphate to lysophosphatidic acid, a common precursor for TAGs and phospholipids. Although the enzymes responsible for the downstream biosynthetic steps of TAGs and phospholipids, such as Slc1 and Dga1, were found in the LD proteomes of mammalian and yeasts (13, 17, 21, 38, 61), they were not identified in the LD proteome of R. toruloides. It should be noted that a cytosol TAG synthesis machinery has been known in the oleaginous red yeasts (62), but its roles in lipid accumulation and TAG transport remained unclear. Of these LD-associated lipid biosynthesis genes, ACL1, ACC1, FAS1, and FAS2 were all upregulated in lipid-rich cells at the protein level (see Table S3 in the supplemental material) as well as the transcriptional level (Fig. 4D).
The LDs are the major organelles for neutral lipid mobilization in R. toruloides. We found 11 lipolytic enzymes in the LD proteome (Table 1). RHTO_03931 is the ortholog of the yeast bifunctional enzyme Tgl5, which has TAG lipase and lysophosphatidic acid acyltransferase activity. This enzyme could transfer the acyl group from acyl-CoA to lysophosphatidic acid to produce phosphatidic acid, which could promote LD fusion to generate larger LDs (52, 53). This enzyme could also hydrolyze TAG to fatty acid and diacylglycerol, which could then be hydrolyzed to monoacylglycerol by RHTO_02359 (the ortholog of mammalian hormone-sensitive lipase). The monoacylglycerol was hydrolyzed to fatty acid and glycerol by lipases RHTO_03913 and probably RHTO_03511. However, RHTO_02359 was absent in the LD proteome, suggesting that it was not recruited yet at the lipid accumulation stage, because the translocation of the activated and phosphorylated hormone-sensitive lipase onto LDs depends on phosphorylation of the perilipin (Ldp1 might play similar roles in this yeast) by protein kinase A in response to the cellular energy status (19). Released fatty acids may be metabolized, such as the transformation of linoleic acid to oxylipin by PpoA (RHTO_07686). Alternatively, fatty acids may be oxidized via the β-oxidation process. The location of β-oxidization is believed to be in the mitochondrion and peroxisome, which are both in close proximity to LDs. Indeed, mitochondrial and peroxisomal proteins (RHTO_01116, RHTO_03348, and RHTO_00397) were identified in the LD proteome. PpoA was upregulated in lipid-rich cells (Fig. 4D). Interestingly, the expression levels of three genes for lipid degradation (RHTO_03913, RHTO_01116, and RHTO_03348) were upregulated in the MM-N sample (see Table S3 in the supplemental material), in which the β-oxidation was elevated (24).
Membrane trafficking is a delivery process mediated by multiorganelles. The expression patterns of proteins involved in membrane trafficking can influence LD dynamics. RHTO_07779, an ADP-ribosylation factor (ARF1) and a homolog of Drosophila ARF79F, was found in the LD proteome. ARF1 is a small GTPase, which recruits the COPI coatomer subunits and mediates Golgi apparatus-to-ER transport. Knockdown of ARF79F and COPI components has been reported to promote lipid accumulation, partially due to the decrease of the LD targeting of LD lipase ATGL (63, 64). RHTO_07779 was downregulated in the “96 h” sample, which had a lipid content of 63%. Three COPI components, subunits α, β, and γ corresponding to RHTO_00667, RHTO_00612, and RHTO_03548, respectively, were also found in the LD proteome, but the expression of the β subunit was upregulated in the lipid-rich samples (see Table S3 in the supplemental material). It was also known that the COPII controlled by small GTPase SAR1 was involved in protein anterograde transport from the ER to the Golgi apparatus and played similar roles in ATGL translocation to LDs (65). Impaired COPII activity increased lipid storage but in a milder manner. We identified Sar1 GTPase (RHTO_02448) in the LD proteome. This protein was downregulated in the “24 h” and “96 h” samples, but its transcription level increased 2.6-fold in the MM-N sample. Two vesicle fusion-related proteins, RHTO_01157 (Ykt6, v-SNARE) and RHTO_06548 (Sec18, an ortholog to human N-ethylmaleimide-sensitive factor [NSF]) (66), were also present in the LD proteome.
Small Rab GTPases are vesicle-trafficking-regulating G proteins with posttranslationally modified prenyl moieties for membrane anchorage. Seven Rabs were found in the LD proteome, probably due to the presence of ER-derived phospholipid hemimembrane of LDs. Rabs may mediate the interactions between LDs and other organelles, such as the Golgi apparatus, vacuoles, and endosomes (2, 67). The expression of small Rabs also changed during lipid overproduction, such as upregulation of Rab1 (RHTO_06528), Rab8 (RHTO_08047), and Rab11 (RHTO_01102) in the NL sample (see Table S3 in the supplemental material). RHTO_04520, a homolog to Rab18, was upregulated in the MM-N sample. Interestingly, a higher upregulated expression level of Rab18 was found in fatty adipocytes, and recruitment of Rab18 to LDs in association with lipolysis was demonstrated (68). Thus, RHTO_04520 may play a role in LD metabolism. Other Rabs (Rab4, -5, -7, and -11) may be associated with endosomes or vacuoles. Vacuoles are organelles playing a major role in autophagy, the process of “eating” cellular components. It was also demonstrated that autophagy played crucial roles in the formation of giant LDs in adipocytes (69). In our previous study, an elevated autophagy process, which might remodel the cells to make more space for LDs, was revealed along with lipid accumulation (24).
Taken together, this and many recent studies clearly indicate that LD-associated proteins are not simply sticky but structurally and functionally associated with LDs. The presence of metabolic enzymes (involved mainly in lipid metabolism), vesical traffic proteins, Rab GTPases, and others in the LD proteome of different species suggests more-complicated and cryptic roles of these proteins in LD biology. The presence of functionally diverse proteins also suggests that the cytosolic LDs are not simple energy storage particles but more energetic and functional organelles. More importantly, our results showed that the levels of the majority of the LD-associated proteins changed dynamically in response to nutrient availability. Such changes had good correlation with the development of cellular LDs.
In summary, we identified 226 LD-associated proteins, which established a comprehensive LD proteome of the oleaginous yeast R. toruloides. The major LD structure protein Ldp1 was characterized as an LD marker protein, which was important for giant LD formation. Analysis of proteomic data and biochemical results demonstrated the dynamic nature of these proteins during lipid accumulation under different conditions. Our results significantly advance our understanding of the molecular basis of lipid overproduction in oleaginous yeasts. Such information will be valuable for the investigation on LD biology in general and for the development of superior lipid producers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hongyuan Yang for his good suggestions on LD morphology, Erin O'Shea for sharing the strains expressing Erg6-RFP and Sec13-RFP, and Nicolaas A. A. Buijs for critically reading the manuscript.
This work was funded by the Natural Sciences Foundation of China (31370128, 31170060, and 21325627) and the National Basic Research and Development Program of China (2011CB707405 and 2011CBA00906).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00141-14.
REFERENCES
- 1.Wältermann M, Steinbüchel A. 2005. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J Bacteriol 187:3607–3619. doi: 10.1128/JB.187.11.3607-3619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walther TC, Farese RV Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy DJ. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40:325–438. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 4.Martin S, Parton RG. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. 2008. Lipid droplets: a classic organelle with new outfits. Histochem Cell Biol 130:263–279. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farese RV Jr, Walther TC. 2009. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saka HA, Valdivia R. 2012. Emerging roles for lipid droplets in immunity and host-pathogen interactions. Annu Rev Cell Dev Biol 28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- 8.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. 2009. Autophagy regulates lipid metabolism. Nature 458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. 2009. A role for lipid droplets in inter-membrane lipid traffic. Proteomics 9:914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Ding Y, Chen Y, Zhang S, Huo C, Wang Y, Yu J, Zhang P, Na H, Zhang H, Ma Y, Liu P. 2012. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J Lipid Res 53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, Yang L, Zhang S, Wang Y, Du Y, Pu J, Peng G, Chen Y, Zhang H, Yu J, Hang H, Wu P, Yang F, Yang H, Steinbuchel A, Liu P. 2012. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res 53:399–411. doi: 10.1194/jlr.M021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Athenstaedt K, Jolivet P, Boulard C, Zivy M, Negroni L, Nicaud JM, Chardot T. 2006. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics 6:1450–1459. doi: 10.1002/pmic.200500339. [DOI] [PubMed] [Google Scholar]
- 14.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Zhao Z, Bai F. 2007. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb Technol 41:312–317. doi: 10.1016/j.enzmictec.2007.02.008. [DOI] [Google Scholar]
- 16.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res 48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta 1811:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivashov VA, Grillitsch K, Koefeler H, Leitner E, Baeumlisberger D, Karas M, Daum G. 2013. Lipidome and proteome of lipid droplets from the methylotrophic yeast Pichia pastoris. Biochim Biophys Acta 1831:282–290. doi: 10.1016/j.bbalip.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bickel PE, Tansey JT, Welte MA. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res 6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 21.Hodges BD, Wu CC. 2010. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res 51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratledge C, Wynn JP. 2002. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 51:1–51. doi: 10.1016/S0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Hu C, Jin G, Zhao X, Zhao ZK. 2010. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol 101:6124–6129. doi: 10.1016/j.biortech.2010.02.111. [DOI] [PubMed] [Google Scholar]
- 24.Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H, Zhao ZK. 2012. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun 3:1112. doi: 10.1038/ncomms2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Zhao X, Wang F, Li Y, Jiang X, Ye M, Zhao ZK, Zou H. 2009. Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast 26:553–566. doi: 10.1002/yea.1706. [DOI] [PubMed] [Google Scholar]
- 26.Ding Y, Zhang S, Yang L, Na H, Zhang P, Zhang H, Wang Y, Chen Y, Yu J, Huo C, Xu S, Garaiova M, Cong Y, Liu P. 2013. Isolating lipid droplets from multiple species. Nat Protoc 8:43–51. doi: 10.1038/nprot.2012.142. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Kong X, Hua Y, Feng B, Zhao ZK. 2008. Medium optimization for lipid production through co-fermentation of glucose and xylose by the oleaginous yeast Lipomyces starkeyi. Eur J Lipid Sci Technol 110:405–412. doi: 10.1002/ejlt.200700224. [DOI] [Google Scholar]
- 28.Su W, Wang Y, Jia X, Wu W, Li L, Tian X, Li S, Wang C, Xu H, Cao J, Han Q, Xu S, Chen Y, Zhong Y, Zhang X, Liu P, Gustafsson JA, Guan Y. 2014. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci U S A 111:11437–11442. doi: 10.1073/pnas.1410741111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Walther TC. 2013. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol Cell Proteomics 12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res 55:1465–1477. doi: 10.1194/jlr.M050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Jiang X, Han G, Ye M, Zou H. 2007. Optimization of filtering criterion for SEQUEST database searching to improve proteome coverage in shotgun proteomics. BMC Bioinformatics 8:323. doi: 10.1186/1471-2105-8-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan J, Roth AF, Bailey AO, Davis NG. 2007. Palmitoylated proteins: purification and identification. Nat Protoc 2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 33.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 35.Yang F, Zhang S, Zhou YJ, Zhu Z, Lin X, Zhao ZK. 2012. Characterization of the mitochondrial NAD+-dependent isocitrate dehydrogenase of the oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol 94:1095–1105. doi: 10.1007/s00253-011-3820-3. [DOI] [PubMed] [Google Scholar]
- 36.Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, He J, Pu S, Tang C, Xu G. 2007. Heat shock protein 70 is translocated to lipid droplets in rat adipocytes upon heat stimulation. Biochim Biophys Acta 1771:66–74. doi: 10.1016/j.bbalip.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Wang Y, Li J, Yu J, Pu J, Li L, Zhang H, Zhang S, Peng G, Yang F, Liu P. 2011. Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein A-I. J Proteome Res 10:4757–4768. doi: 10.1021/pr200553c. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Thiel K, Thul PJ, Beller M, Kuhnlein RP, Welte MA. 2012. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol 22:2104–2113. doi: 10.1016/j.cub.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, St Leger RJ. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J Biol Chem 282:21110–21115. doi: 10.1074/jbc.M609592200. [DOI] [PubMed] [Google Scholar]
- 41.Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. 2000. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44:463–476. doi: 10.1023/A:1026564411918. [DOI] [PubMed] [Google Scholar]
- 42.Jiang PL, Chen JC, Chiu ST, Tzen JT. 2009. Stable oil bodies sheltered by a unique caleosin in cycad megagametophytes. Plant Physiol Biochem 47:1009–1016. doi: 10.1016/j.plaphy.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 43.Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC. 2006. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47:917–933. doi: 10.1111/j.1365-313X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- 44.Brodhun F, Gobel C, Hornung E, Feussner I. 2009. Identification of PpoA from Aspergillus nidulans as a fusion protein of a fatty acid heme dioxygenase/peroxidase and a cytochrome P450. J Biol Chem 284:11792–11805. doi: 10.1074/jbc.M809152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsitsigiannis DI, Zarnowski R, Keller NP. 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J Biol Chem 279:11344–11353. doi: 10.1074/jbc.M310840200. [DOI] [PubMed] [Google Scholar]
- 46.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. 2010. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell 21:198–211. doi: 10.1091/mbc.E09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol 181:6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aime MC, Matheny PB, Henk DA, Frieders EM, Nilsson RH, Piepenbring M, McLaughlin DJ, Szabo LJ, Begerow D, Sampaio JP, Bauer R, Weiss M, Oberwinkler F, Hibbett D. 2006. An overview of the higher level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rDNA sequences. Mycologia 98:896–905. doi: 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen LN, Hamari Z, Kadereit B, Trofa D, Agovino M, Martinez LR, Gacser A, Silver DL, Nosanchuk JD. 2011. Candida parapsilosis fat storage-inducing transmembrane (FIT) protein 2 regulates lipid droplet formation and impacts virulence. Microbes Infect 13:663–672. doi: 10.1016/j.micinf.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Buzzini P, Innocenti M, Turchetti B, Libkind D, van Broock M, Mulinacci N. 2007. Carotenoid profiles of yeasts belonging to the genera Rhodotorula, Rhodosporidium, Sporobolomyces, and Sporidiobolus. Can J Microbiol 53:1024–1031. doi: 10.1139/W07-068. [DOI] [PubMed] [Google Scholar]
- 51.Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. 2011. Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol 90:1219–1227. doi: 10.1007/s00253-011-3200-z. [DOI] [PubMed] [Google Scholar]
- 52.Yang H, Galea A, Sytnyk V, Crossley M. 2012. Controlling the size of lipid droplets: lipid and protein factors. Curr Opin Cell Biol 24:509–516. doi: 10.1016/j.ceb.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, Huang X, Parton RG, Wenk MR, Walther TC, Yang H. 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet 7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fei W, Li H, Shui G, Kapterian TS, Bielby C, Du X, Brown AJ, Li P, Wenk MR, Liu P, Yang H. 2011. Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J Lipid Res 52:2136–2147. doi: 10.1194/jlr.M017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL. 2008. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci U S A 105:94–99. doi: 10.1073/pnas.0708579105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Froissard M, D'Andrea S, Boulard C, Chardot T. 2009. Heterologous expression of AtClo1, a plant oil body protein, induces lipid accumulation in yeast. FEMS Yeast Res 9:428–438. doi: 10.1111/j.1567-1364.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- 57.Shi S, Valle-Rodriguez JO, Khoomrung S, Siewers V, Nielsen J. 2012. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels 5:7. doi: 10.1186/PREACCEPT-1932279820621895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin G, Zhang Y, Shen H, Yang X, Xie H, Zhao ZK. 2013. Fatty acid ethyl esters production in aqueous phase by the oleaginous yeast Rhodosporidium toruloides. Bioresour Technol 150:266–270. doi: 10.1016/j.biortech.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 59.Bajaj A, Lohan P, Jha PN, Mehrotra R. 2010. Biodiesel production through lipase catalyzed transesterification: an overview. J Mol Catal B Enzym 62:9–14. doi: 10.1016/j.molcatb.2009.09.018. [DOI] [Google Scholar]
- 60.Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. 2006. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics 5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Kuerschner L, Moessinger C, Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic 9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 62.Gangar A, Karande AA, Rajasekharan R. 2001. Isolation and localization of a cytosolic 10 S triacylglycerol biosynthetic multienzyme complex from oleaginous yeast. J Biol Chem 276:10290–10298. doi: 10.1074/jbc.M009550200. [DOI] [PubMed] [Google Scholar]
- 63.Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol 6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. 2009. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, Fernandez-Rodriguez J, Ericson J, Nilsson T, Boren J, Olofsson SO. 2007. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol 9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- 67.Hutagalung AH, Novick PJ. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem 280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 69.Rodriguez-Navarro JA, Cuervo AM. 2010. Autophagy and lipids: tightening the knot. Semin Immunopathol 32:343–353. doi: 10.1007/s00281-010-0219-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.