Transcripts encoding polarity factors such as Bni1 and Spa2 are nonrandomly clustered in the cytosol to initiate and maintain sites of polarized growth in the fungus Ashbya gossypii.
Abstract
Dendritic growth in fungi and neurons requires that multiple axes of polarity are established and maintained within the same cytoplasm. We have discovered that transcripts encoding key polarity factors including a formin, Bni1, and a polarisome scaffold, Spa2, are nonrandomly clustered in the cytosol to initiate and maintain sites of polarized growth in the fungus Ashbya gossypii. This asymmetric distribution requires the mRNAs to interact with a polyQ-containing protein, Whi3, and a Pumilio protein with a low-complexity sequence, Puf2. Cells lacking Whi3 or Puf2 had severe defects in establishing new sites of polarity and failed to localize Bni1 protein. Interaction of mRNAs with Whi3 and Puf2 promotes enrichment of transcripts at established sites of polarized growth and clustering of polarity transcripts throughout the cell body. Thus, aggregation-prone proteins make functional assemblies to position polarity transcripts, and nonrandom positioning of transcripts is required for symmetry-breaking events. This reveals a physiological function for polyQ-driven assemblies in regulating cell polarity.
Introduction
Symmetry breaking and the resulting emergence of cell polarity are fundamental to development across the kingdoms of life. The molecules involved are highly conserved and include GTPases, scaffolds, formins, and the cytoskeleton. Work in many model systems has focused on how both positive and negative feedback in the GTPase circuits lead to robust and persistent cell asymmetry (Johnson et al., 2011; Freisinger et al., 2013; Klünder et al., 2013). Additional polarity promotion mechanisms such as asymmetric placement of mRNAs in embryos or the transport of mRNAs to sites of polarized growth indicate that there are likely many mechanisms for generating asymmetries in cells (Lehmann and Nüsslein-Volhard, 1986; Doyle and Kiebler, 2011; Jansen and Niessing, 2012; Baumann et al., 2014). In this study, we uncover a role for protein–RNA assemblies in symmetry breaking within the complex branched cells of a fungal mycelium.
Several recent studies have led to the hypothesis that cytoplasm and nucleoplasm are spatially patterned by phase separation of complexes from bulk solution. Examples include P-granules, stress granules, and the nucleolus (Brangwynne et al., 2009; Weber and Brangwynne, 2012; Lee et al., 2013b). Multivalent associations between components such as low-complexity sequence proteins and mRNAs can drive phase separations in vitro (Han et al., 2012; Kato et al., 2012; Li et al., 2012). We recently showed that the positioning of cyclin mRNAs in syncytial cells relies on polyQ-mediated assembly of RNA–protein complexes that have properties of aggregates or possibly liquids (Lee et al., 2013a). It is likely that the assembly of phase-separated droplets using aggregation-prone proteins and RNAs is a general mechanism for organizing the cytosol for many functions.
Cytoplasm organization using phase separation or physiological aggregation may be especially useful in large cells such as neurons and filamentous fungi. Filamentous fungi must establish new sites of growth in the midst of substantial cytosolic currents and organelle flow (Lew, 2011). The cytoskeleton can provide stable anchors in a turbulent cytosol; however, there must be initiating events that direct the location of cytoskeletal organization. The mechanisms of symmetry breaking for the branched mycelial network are not well understood. We hypothesized that polarity regulators may be localized away from preexisting polarity sites by association with phase-separated assemblies that have restricted diffusivity. If so, polarity factors may be locally produced and concentrated in regions of the mycelium where new polarity sites need to be established.
We recently discovered that protein–RNA complexes, generated by the polyQ-containing protein Whi3, control the nonrandom clustering of G1 cyclin mRNAs in the multinucleate fungus Ashbya gossypii (Lee et al., 2013a). Transcript clustering functionally compartmentalizes the cytosol, enabling asynchronous cycling of nuclei. Here we report that Whi3, along with the pumilio protein Puf2, positions transcripts encoding polarity factors and leads to their localization. The functional aggregation and coassembly of the RNAs and low-complexity proteins are essential for polarity establishment and present a new mechanism for symmetry breaking.
Results and discussion
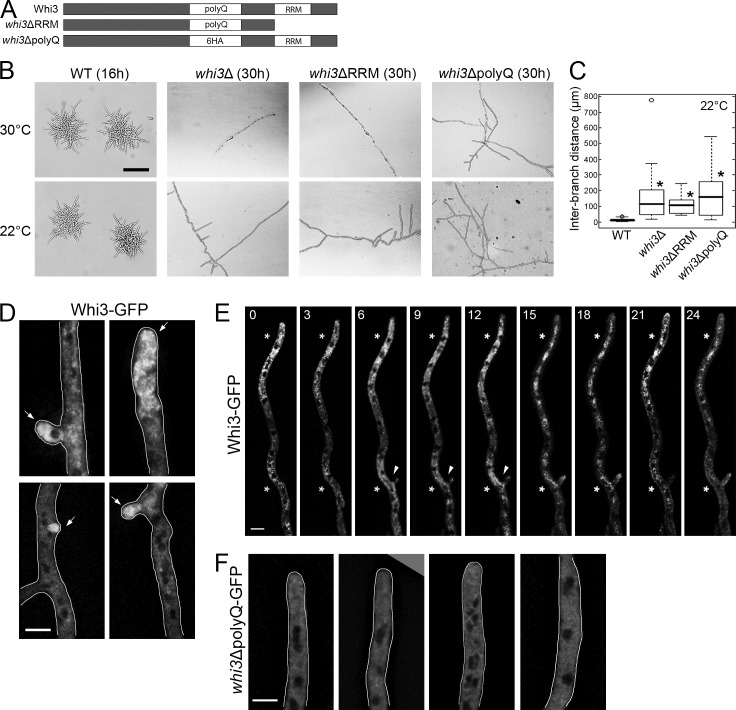
Whi3 is required for cell branching
A. gossypii grows at hyphal tips and generates lateral branches to form mycelia. Lateral branches are new sites of polarity that emerge along an otherwise isotropic sidewall of hyphae and thus are symmetry-breaking events. We observed that cells lacking the entire WHI3 ORF, its RNA recognition motif (RRM), or its polyQ-tract all have severe polarity defects (Fig. 1, A and B). whi3 mutants can establish one primary hypha but either fail or have limited capacity to maintain growth of the main hypha and generate lateral branches, resulting in a significant increase in the distance between lateral branches (Fig. 1, B and C). Consistent with a function in polarity, Whi3 protein is concentrated at nascent lateral branches and hyphal tips (Fig. 1 D). Whi3 remains concentrated on hyphal tips over time and Whi3 assemblies can be seen at incipient branch sites (Fig. 1 E and Video 1). The localization of Whi3 at both locations depends on an ∼120-aa-long polyQ tract, as deletion of it depolarizes and homogenizes the protein localization without changing its levels (Fig. 1 F; Lee et al., 2013a). Thus, the polyQ aggregation domain is required for Whi3′s role in polarity.
Figure 1.
The polyQ tract and RRM domain of Whi3 are required for symmetry breaking. (A) Diagrams of wild-type and mutant Whi3 constructs. Six HA repeats replace the polyQ tract (the same size) for whi3ΔpolyQ. (B) Wild type and whi3 mutants grown at 30 or 22°C. Bar, 200 µm. (C) Distances between neighboring lateral branches on the hypha were measured for each strain (n > 50). *, P < 0.01 compared with wild type using a KS test. For all box-and-whiskers plots in this paper, the top and bottom of box indicate the 75th and 25th quartile, respectively. The thick line in the box shows the median, whiskers indicate maximum and minimum data points, and open circles indicate outliers. (D–F) Whi3 or Whi3ΔpolyQ proteins are localized in the hypha. Arrow, polarization site. Cells are outlined in gray. Bars, 5 µm. (D and F) Images are z-projected (>8 planes). (E) Montage of time-lapse movie. Time points (min) are indicated at the top of the images. Asterisks indicate concentrated Whi3. Arrowheads indicate emergence of a new branch. A single focal plane is shown.
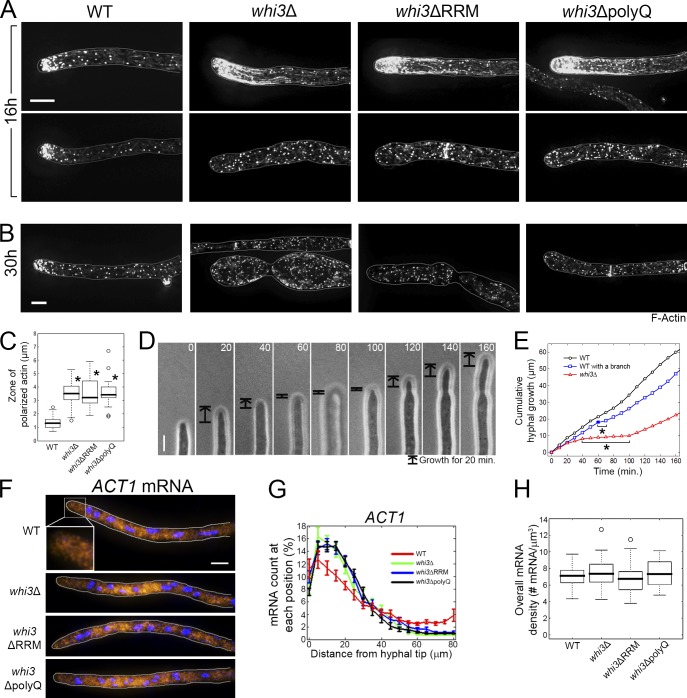
These polarity defects prompted us to examine actin localization in mutants. In wild-type cells, 94% (n = 126) of hyphal tips have concentrated F-actin patches and cables (Fig. 2 A, left). In whi3 mutants, actin at hyphal tips was hyperpolarized or depolarized (Fig. 2, A and C; and Table 1). At later stages (30 h), most mutant cells had exclusively depolarized actin and spherical hyphal tips indicating isotropic growth (Video 1, Fig. 2 B, and Fig. S1 A). Notably, elongating tips in whi3Δ hyphae causes a stutter in their growth, pausing and restarting growth multiple times in the course of a recording, showing that individual tips fluctuate between states of polarized and depolarized growth (Video 2 and Fig. 2, D and E). Despite the transient pauses, the single axis of polarity is “remembered” and cells resume growth in the same location albeit at a slower mean extension rate than in wild-type cells (Figs. 2 E and 5 B).
Figure 2.
F-actin, but not actin transcripts, is mislocalized in whi3 mutants. (A and B) F-actin is visualized in cells grown for 16 h (A) or 30 h (B) at 30°C by Phalloidin staining. Cells are outlined in gray. (A) Top and bottom panels represent hyperpolarized and depolarized actin localization, respectively (n > 105). (C) The length of the zone where actin is polarized (Materials and methods) is measured on the cell growth axis. *, P < 0.01 compared with wild type using a t test. n > 105. (D) Montage of a hyphal growth of whi3Δ. Time points (min) are indicated. (E) Cumulative hyphal growth of wild type with or without branch emergence (closed box), and whi3Δ from D. Asterisks indicate a pause of growth. (D and E) The data shown are from one representative experiment out of 46 (WT) and 6 (whi3Δ). (F) ACT1 mRNAs (yellow) are localized by smFISH, and DNA (blue) is visualized by Hoechst staining. Cells are outlined in gray. The inset shows a 5× magnification. (G) ACT1 mRNAs were counted on every 5-µm distance from the growing tip and summed along each hypha. The mRNA count at each point (5 µm) was converted to the percentage of total number of mRNAs detected in the hypha. Error bars indicate SEM. (G and H) n > 35 for each strain. (H) Overall mRNA density (total number mRNA/total hyphal volume) in each strain. P > 0.39 for all whi3 mutants compared to wild type using a t test. Bars, 5 µm.
Table 1.
F-actin is mislocalized in whi3 mutants
| Strain (total n) | Normal polarizationa (percentage of total n) | Hyperpolarizationa (percentage of total n) | Depolarizationa (percentage of total n) |
| µm | µm | µm | |
| Wild type (126) | 120 (95%) | 0 (0%) | 6 (5%) |
| whi3Δ (160) | 4 (3%) | 70 (44%) | 85 (53%) |
| whi3ΔRRM (106) | 2 (2%) | 48 (45%) | 56 (53%) |
| whi3ΔpolyQ (108) | 2 (2%) | 58 (54%) | 48 (44%) |
These are categorized by the length of the zone of polarized actin (Fig. 2 B). Normal polarization, 0.5–2.5; hyperpolarization, >2.5; and depolarization, no detectable actin polarization.
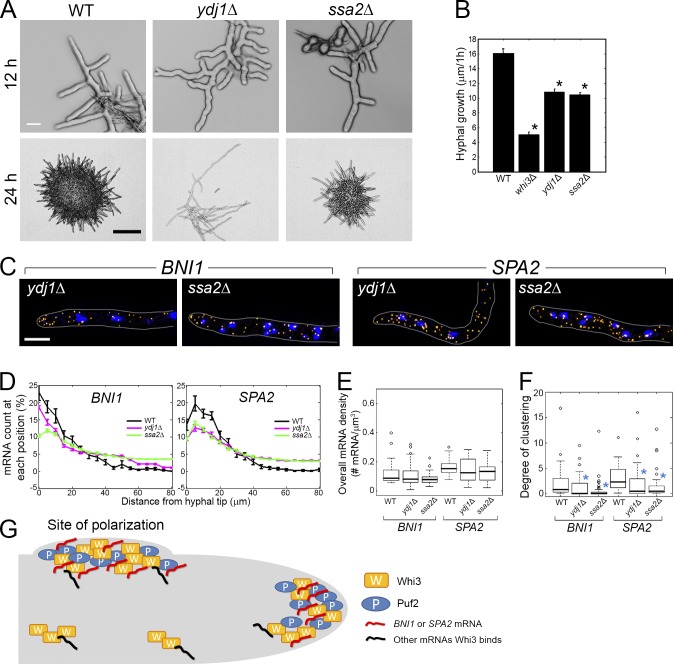
Figure 5.
Ydj1 and Ssa2 affect transcript localization for cell polarity. (A) Cells grown for 12 or 24 h (30°C). Bars: (top) 10 µm; (bottom) 200 µm. (B) The 1-h lateral growth of the hyphal tip was measured with an FITC-ConA pulse. Error bars indicate SEM. *, P < 0.01. (C) BNI1 or SPA2 transcripts (yellow) are localized. Blue, DNA. Cells are outlined in gray. Bar, 5 µm. (D) BNI1 or SPA2 mRNAs were counted from each growing tip. Error bars indicate SEM. (D–F) n > 38 for each strain. (E) Overall mRNA density in mutants is compared with wild type. P > 0.24 for all strains. (F) Degree of clustering in mutants is compared with wild type. *, P < 0.01 by KS test. (G) Working model. Whi3 and Puf2 form complexes at new and preexisting sites of polarization (hyphal tips or nascent lateral branches) in the presence of polarity mRNAs.
Severe defects in the RRM mutants indicate that there is a role for Whi3 mRNA binding for polarity function (Fig. 1, A and B). Because mRNAs encoding actin are polarized in some cell types, we examined the localization of ACT1 mRNA using single-molecule RNA FISH (Shestakova et al., 2001; Buxbaum et al., 2014; Park et al., 2014; Fig. 2 F). ACT1 is asymmetrically localized in both wild-type cells and whi3 mutants with a subtle displacement from the tip in mutants, which is unlikely to account for the dramatic polarity defects (Fig. 2, F–H). G1 cyclins are also unlikely to be responsible because cells that lack CLN3 or express a version of CLN3 that no longer binds Whi3 (cln3-6m) show normal branching, growth, and actin localization (Fig. S1, B and C). Transcripts of the other G1 cyclin, CLN1/2, are enriched at hyphal tips independently of Whi3 or Cln3 (Fig. S1, D and E; Gladfelter et al., 2006). Finally, like CLN3, transcripts encoding the glycolytic housekeeping enzyme Gpm2/3 are not polarized (Fig. S1 F). Thus, it is likely that the polarity defects reflect the loss of function of other mRNAs that are bound to Whi3.
Polarisome mRNAs require the polyQ domain of Whi3 for localization
We next scanned the A. gossypii genome for loci that contain multiple binding sites for Whi3 (UGCAU) and found several transcripts encoding proteins related to cell polarity and/or cytoskeletal organization that contained more than three predicted sites (Table 2 and Fig. S2 A). We found using an RNA immunoprecipitation (RIP) of Whi3-TAP that the formin BNI1, the polarisome scaffold SPA2, and the majority of predicted mRNA interactors could be amplified from precipitated Whi3 (Fig. 3 A). This is in contrast to both CLN1/2 and ACT1 (Fig. S1 G). Additionally, BNI1 and SPA2 transcripts are enriched at hyphal tips and nascent lateral branches in a Whi3-dependent manner (Fig. 3, B–D). Transcript levels are diminished in the null and RRM mutant but not in the whi3ΔpolyQ cells, which show comparably severe polarity defects to the null or ΔRRM mutant (Fig. S2 B). Thus, polarity-related transcripts are asymmetrically localized and this polarized distribution depends on Whi3 assemblies.
Table 2.
Polarization-related mRNAs that are predicted to be bound to Whi3 in A. gossypii
| mRNA | Number of WBSa | Function | GO categoryb |
| SPA2 | 11 | Scaffold protein (polarisome component) | Actin polarization, Cdc28 substrate |
| SCD5 | 5 | Actin organization, endocytosis | Actin polarization |
| SHE4 | 5 | Myosin function regulator, endocytosis, asymmetric mRNA localization | Actin polarization |
| MYO2/4 | 5 | Type V myosin | Cell polarity, mitochondria |
| BNI1 | 4 | Formin, Actin polymerization | Actin polarization |
| AIM21 | 4 | Mitochondrial migration along actin filament | Actin polarization, mitochondria |
| CDC24 | 4 | Guanine nucleotide exchange factor for Cdc42 | Cell polarity |
Number of Whi3-binding sites (UGCAU)
Gene Ontology in Saccharomyces Genome Database (SGD).
Figure 3.
Whi3 is required for polarized and clustered localization of BNI1 and SPA2 mRNAs. (A and H) mRNAs were immunoprecipitated (RIP) in a strain that expresses Whi3-TAP using α-TAP antibody (A) or in strains labeled using GFP-trap (ChromoTek; H), and identified after RT-PCR. Wild type (untagged Whi3) was used for RIP as a negative control. Total RNA was purified from the lysates (whole cell extract) of indicated strains and amplified after RT-PCR to confirm the presence of target mRNAs as inputs. (B, C, and I) Indicated mRNAs (yellow) are localized at hyphal tips (B and I) or nascent branches (C, arrow). Blue, DNA. Cells are outlined in gray. (D) The number of BNI1 or SPA2 mRNAs were counted at each 5 µm distance from the tip of hyphae and converted to a percentage. Error bars indicate SEM. n > 39. (E) Histograms (percentage of the total number of mRNAs) of NND in wild type, whi3Δ, or BNI1-3m for BNI1 or SPA2 (n > 1,224). NND in wild type are significantly different from the mutants. P < 0.01 by KS test. bni1-3m is expressed on a plasmid and compared with the strain episomally expressing BNI1-mCherry. (F) The degree of mRNA clustering (see Materials and methods) was calculated only with inter-region hyphal images (at least 25 µm away from the tip, n > 32). *, P < 0.01 compared with wild type using a KS test. (G) Whi3 (or ΔpolyQ, green) and Bni1 (or -3m, magenta) were localized in the same cell grown for 16 h (30°C; z-projected). The images shown are representatives (n > 15). (J) Cells were grown at 30°C for 20 h. Bars: (B, C, G, and I) 5 µm; (J) 200 µm.
We next examined the localization pattern of BNI1 and SPA2 mRNAs throughout the hyphae because these regions give rise to lateral branches. Nearest neighbor inter-transcript distances (NNDs) are in a bimodal distribution, which is significantly different from normal distribution, suggesting that groups of mRNAs are located very close together for both BNI1 and SPA2 existence of two or more subpopulations (Fig. 3 E). In contrast, the NND in whi3 null mutants is unimodal. Additionally, using spatial statistics we found that both BNI1 and SPA2 were nonrandomly positioned and were spatially clustered in the cytosol of wild-type cells (Figs. 3 F and S3, A and B). The spatial clustering of BNI1 and SPA2 in the central regions of hyphae away from tips was randomized with elimination of Whi3, its RRM, or polyQ tract (Figs. 3 F and S3, A and B). We speculate that cytosolic clusters can promote efficient local symmetry breaking, possibly through local translation, when environmental cues for branching are received.
We next investigated the consequence of Whi3 on Bni1 protein. Consistent with a functional role for localized transcripts, we find that polarized Bni1 protein localization depends on Whi3 (Fig. 3 G). Bni1 localization is lost or diminished in intensity at hyphal tips and nascent branches in whi3ΔpolyQ cells or when a form of Bni1 is expressed in which silent mutations have been made that eliminate three predicted Whi3 binding sites (bni1-3m) in the transcript (Figs. 3 G and S2 D). The bni1-3m mRNA can no longer detectably associate with Whi3 in an RIP although Whi3′s ability to bind SPA2 was unaffected (Fig. 3 H). bni1-3m also showed the polarity defects in both lateral branch formation, even when expressed as a plasmid, and mRNA localization, very similar to whi3 mutants (Fig. 3, E, F, I, and J; and Fig. S2 C). Thus, Whi3 is required for the spatial patterning of polarity transcripts and proteins.
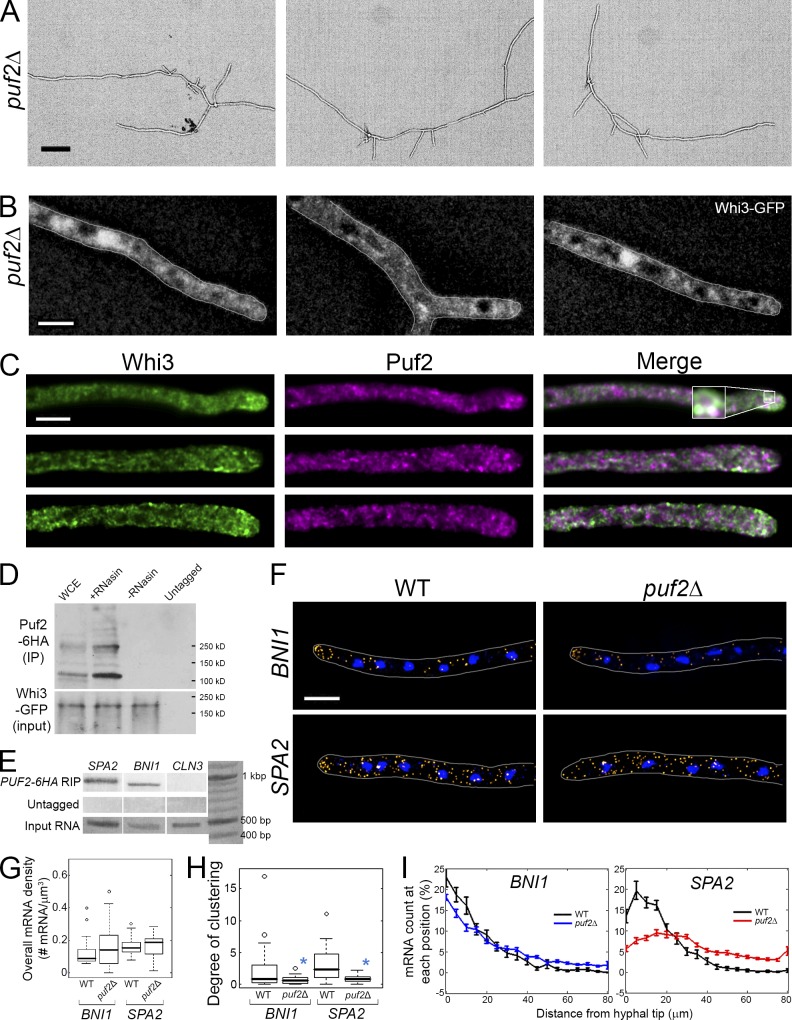
Whi3 associates with Puf2 via mRNAs to regulate polarity
What distinguishes Whi3 complexes that control the distribution of polarity mRNAs from complexes that localize cyclin mRNAs in proximity of nuclei? We hypothesized that the functional specification of different Whi3 complexes originates at the level of the target mRNAs. We found support for this idea in BNI1 and SPA2 mRNAs, which also both contain pumilio-consensus binding sites (Fig. S2 A). Furthermore, A. gossypii Puf2 is predicted to be intrinsically disordered with a substantial low-complexity sequence based on several different structure prediction algorithms (Fig. S2 E; disordered region: IUPred, 48.6%; Tango score, 2492.33; Waltz, 13 regions; see Materials and methods). puf2Δ cells had substantial defects in making lateral branches, and Whi3 is delocalized from nascent polarity sites (Fig. 4, A and B). Additionally, Whi3 and Puf2 frequently colocalize at sites of polarity and Whi3 depends on Puf2 for its polarized localization (Fig. 4, B and C). Puf2 is highly heterogeneous in localization, suggesting that, like Whi3, Puf2 is in some sort of aggregate or higher-order assembly (Fig. 4 C). Whi3 and Puf2 physically associate in coimmunoprecipitation experiments but only when RNAs are preserved (Fig. 4 D). As predicted by bioinformatics, RIP with Puf2-6HA shows the presence of both SPA2 and BNI1 transcripts but not CLN3, a known Whi3 target (Fig. 4 E). This suggests distinct Whi3 complexes that contain cell cycle transcripts and different complexes that contain both Whi3 and Puf2 along with the polarity mRNAs.
Figure 4.
Puf2 associates with Whi3 to control polarity. (A) puf2Δ cells grown for 20 h (30°C). Bar, 200 µm. (B) Whi3 (white) is localized in puf2Δ. Images are z-projected (>9 planes). Cells are outlined in gray. (C) Whi3-GFP and Puf2-6HA are localized in the same cell grown for 16 h (30°C; one focal plane). The inset shows 10× magnification. Pearson’s colocalization coefficient (from the top): 0.84, 0.61, and 0.60 (at hyphal tip, 5 × 5 µm2); 0.71, 0.56, and 0.50 (whole hypha). Costes p-value: 1. (D) Whi3-GFP is immunoprecipitated with GFP-trap in whole cell extract (WCE), WCE with RNasin, or WCE without RNasin (in RNase-free conditions). The untagged strain only expresses Puf2-6HA. Puf2-6HA was blotted with α-HA antibody. Molecular weights: Puf2-6HA, 122.3 kD; Whi3-GFP, 105.7 kD. (E) mRNAs bound to Puf2-6HA are RIPed with α-HA antibody, and cDNA generated from RT-PCR was identified by PCR. (F) BNI1 or SPA2 transcripts (yellow) are localized. Blue, DNA. (G) Overall mRNA density in each strain. P > 0.14 according to a KS test for both strains. (G–I) n > 38 for each strain. (H) Degree of mRNA clustering in each strain. *, P < 0.01 by KS test. (I) SPA2 (top) or BNI1 (bottom) mRNAs were counted and converted to a percentage. Error bars indicate SEM. Bars, 5 µm.
Finally, we examined the role of Puf2 in the localization of polarity transcripts. Both BNI1 and SPA2 were no longer clustered in the cytosol in a puf2Δ mutant but transcript levels were unchanged (Fig. 4, F–H). Interestingly, although SPA2 was completely delocalized from tips in puf2Δ, BNI1 was still enriched at tips, albeit less than in the wild type (Fig. 4 I). BNI1 may have another layer of targeting that promotes tip localization but does not cluster it in the cytosol. This suggests that Puf2 and Whi3 both contribute to the clustered distribution of SPA2 and BNI1 at growing tips and in the cytosol.
Hsp family chaperones contribute to localization of polarity transcripts
Our data lead us to a model in which RNA–protein complexes, in the form of higher-order assemblies, are important for the positioning of transcripts to establish and maintain polarity. Given their dependence on protein-aggregation domains, we predicted that there may be regulated assembly or remodeling of the Whi3/Puf2/RNA complexes using protein chaperones to regulate their distribution. Thus, we generated mutants lacking the chaperones Ydj1 (Hsp40) or Ssa2 (Hsp70). Both ydj1Δ and ssa2Δ cells have substantial defects in polarity and hyphal growth, although not as severe as either whi3Δ or puf2Δ (Fig. 5, A and B). ydj1Δ has significantly fewer lateral branches and wavy hyphae. In ssa2Δ, overall hyphal growth is stunted and the diameter of hyphae is larger and more variable than wild type. Moreover, both ydj1Δ and ssa2Δ show a significant decrease in clustering and some alteration in the polarized distribution of transcripts without any change in the abundance of the transcripts (Fig. 5, C–F). These data suggest that regulated aggregation may play a central role in positioning and ultimate function of BNI1 and SPA2 in polarity establishment and maintenance.
We predict that the biased localization of transcripts promotes local production of proteins that act to stabilize nascent polarity sites, likely established by stochastic activation of Cdc42. Positive and negative feedback loops involving Cdc42 activity are critical to stabilize polarity and maintain singularity of polarity sites in small cells such as Saccharomyces cerevisiae (Irazoqui et al., 2003; Slaughter et al., 2009; Li and Bowerman, 2010; Smith et al., 2013; Kuo et al., 2014). In large mycelial networks, however, symmetry breaking takes place in the context of a larger surface area with substantial cytosolic streaming while the majority of the polarity machinery molecules are already working for rapid tip expansion. There may be substantial competition between stochastically arising Cdc42 activity sites for a limited pool of components that are needed to drive the feedback loops required to stabilize the site. Such limitation and competition would necessitate additional mechanisms such as heterogeneous placement of mRNAs and local synthesis of polarity factors to stabilize a new growth axis in large mycelial networks.
We have shown that transcript localization is required for polarized distribution of Bni1 protein, but what is the mechanistic basis for this relationship? Open questions remain as to how the RNA–protein complexes assemble, localize, and function in translational control of the localized transcripts. The RNA–protein complexes presumably assemble through multivalent interactions, as there are multiple binding sites on both the mRNAs and proteins. Notably, several polarity-related mRNAs that bind to Whi3, such as SHE4, MYO2/4, DYN1, and CDC24, also contain Puf2 binding sites (>4), which suggests there could be coordinated local production of many factors. It is likely that Whi3 has other partners that we have yet to identify that may be involved in positioning/regulating other Whi3-associated mRNAs linked to polarity.
We have unpublished data indicating that the heterogeneous structures of Whi3 behave as demixed liquid droplets. Reconstitution of the Whi3/Puf2 assemblies will be useful for further understanding their biophysical properties as well as localization and function. It is not yet clear how Whi3–Puf2 complexes are positioned at tips and whether the role of the complex in polarity initiation is distinct from the role in polarity maintenance. Distinguishing between these options will require understanding what additional factors cohabitate in the complexes and where and how translation is regulated.
In this study, we show that the nonrandom positioning of mRNAs through the formation of higher-order RNA–protein assemblies promotes local symmetry breaking events. While RNA–protein granules have been shown to be responsive to polarity cues in large cells such as embryos, this work links the positioning of RNA–protein assemblies to polarity establishment outside of development. Given the preponderance of low-complexity sequences associated with RNA-binding domains, we speculate that RNP assemblies built from aggregation-prone sequences are likely to play a role in patterning cytosol to regulate polarity in many cell types.
Materials and methods
Plasmid and strain construction
Methods and protocols for A. gossypii culture and transformation were performed using electroporation as described previously (Wendland et al., 2000). Polymerase chain reactions and DNA manipulation protocols were performed in accordance with Sambrook (2001). All restriction enzymes are from New England BioLabs, Inc., and all plasmids were sequenced at the Dartmouth Molecular Biology Core facility.
The whi3Δ (AG570), whi3ΔRRM (AG578), whi3ΔpolyQ (AG636), cln3Δ (AG612), and cln3-6m (AG693) were previously constructed in the laboratory (Lee et al., 2013a). The whi3Δ or cln3Δ have whole ORF (729 aa) deleted by replacing it with a selection marker GEN1 on WHI3 locus by homology recombination. In whi3ΔRRM, aa 591–729 were replaced with GEN1 on the locus. In cln3-6m, all six Whi3 binding sites were silently mutated on CLN3 locus (from TGCAT to CTAGT or TACAC). In the case of protein fusion with partial WHI3, yEGFP and GEN1 were replaced with target sequences for each partial deletion.
To create the puf2Δ;WHI3-GFP (AG728) construct, NAT1 was amplified from pNAT1,AMP (AGB009; backbone: pUC19) with oligos 5′-GTCTTACAGAGCAACCTTCTCATTGGGTAGCCTTTCTGGGTATTAAAAACGACGGCCAGTGAATTCG-3′ and 5′-AAACATTGTAGGCAATAACCGAATGCTACGTTCTGTCTCGTTCAGCATGATTACGCCAAGCTTGC-3′, which contain homology to the promoter and 3′ UTR of PUF2. This DNA fragment was transformed into WHI3-GFP strain (AG569) to delete whole ORF of PUF2 (puf2Δ) via electroporation to yield strain AG728. Genomic DNA was obtained to verify the transformation. Primer sets 5′-GTGGTGAAGGACCCATCCAG-3′, 5′-GCCTCACGCTAGATCTGATG-3′ and 5′-ACATGAGCATGCCCTGCCCC-3′, 5′-AGGGAGTCGCCAAACAAGTG-3′ were used for verification on 5′ and 3′ regions, respectively. The strains were grown in media containing 100 µg/ml Clonat (EMD Millipore) to select the cells that were correctly transformed for microscopy.
To create the PUF2-6HA;WHI3-GFP (AG729) construct, 6HA::NAT1 was amplified from p6HA::NAT1 (AGB030; backbone: pUC19) with oligos 5′-ACTAATACTAATGGATACACAACGCACAACGGGACATTTGGTTACAAAACGACGGCCAGTGAATTCG-3′ and 5′-GGAGGCAGGAGGTCCTCCATATATATACATCTGTTACCACGAACCCATGATTACGCCAAGCTTGC-3′. This DNA fragment was transformed into WHI3-GFP strain (AG569) via electroporation to yield strain AG729. Genomic DNA was obtained to verify the transformation. Primer sets 5′-GTGGTGAAGGACCCATCCAG-3′, 5′-TGCCGTTGCCTCAAATCAGC-3′ and 5′-ACATGAGCATGCCCTGCCCC-3′, 5′-AGGGAGTCGCCAAACAAGTG-3′ were used for verification on 5′ and 3′ regions, respectively. The strains were grown in media containing 100 µg/ml Clonat (EMD Millipore) to select the cells that were correctly transformed for microscopy.
To create the ssa2Δ (AG458) construct, GEN3 was amplified from plasmid pGEN3,AMP (AGB021; backbone: pUC19) with oligos 5′-GGCATTGGGCTCGAAGACTCTTACTATCTGCCATGTTCGTTTGGGGCTAGGGATAACAGGGTAAT-3′ and 5′-CAACAATTCGGGCGTGTGGTCTAGTGGTATGATTCTCGCTTTGGGAGGCATGCAAGCTTAGATCT-3′, which contain homology to the promoter and 3′ UTR of SSA2. This DNA fragment was transformed into wild-type A. gossypii to delete whole ORF of SSA2 (ssa2Δ) via electroporation to yield strain AG458. Genomic DNA was obtained to verify the transformation. Primer sets 5′-GGCCACATACGTTATGGTAG-3′, 5′-GGAGGTAGTTTGCTGATTGG-3′ and 5′-GCTCGATAGCGTCTTTATGG-3′, 5′-CTTGCCATCCTATGGAACTG-3′ were used for verification on 5′ and 3′ regions, respectively. The strains were grown in media containing 200 µg/ml G418 (EMD Millipore) to select the cells that were correctly transformed for microscopy.
To create ydj1Δ (AG457) construct, GEN3 was amplified from plasmid pGEN3,AMP (AGB021) with oligos 5′-TTATCGAATCACGGAGAAGCAACCACCGGGATAGCAAGCTTATAGGCTAGGGATAACAGGGTAAT-3′ and 5′-GCGGCAGTCATTCATCGTGAAGCCAAGCGGTCTTGAGTGAAGGTCAGGCATGCAAGCTTAGATCT-3′, which contain homology to the promoter and 3′ UTR of YDJ1. This DNA fragment was transformed into wild-type A. gossypii to delete whole ORF of YDJ1 (ydj1Δ) via electroporation to yield strain AG457. Genomic DNA was obtained to verify the transformation. Primer sets 5′-CGTACTGGAAGCAGGTTATC-3′, 5′-GGAGGTAGTTTGCTGATTGG-3′ and 5′-TAGAGATCCGGCATACAGGG-3′, 5′-CTTGCCATCCTATGGAACTG-3′ were used for verification on 5′ and 3′ regions, respectively. The strains were grown in media containing 200 µg/ml G418 (EMD Millipore) to select the cells that were correctly transformed for microscopy.
To create WHI3-yEGFP; pBNI1-mCherry (AG751) or whi3ΔpolyQ-GFP; pBNI1-mCherry (AG756), pBNI1-mCherry::NAT1 (backbone: pRS415) plasmid was constructed first and transformed into WHI3-yEGFP or whi3ΔpolyQ-GFP. In the plasmid, BNI1 is driven by an endogenous A. gossypii BNI1 promoter with S. cerevisiae URA3 3′ UTR. To construct the plasmid, mCherry was amplified from pUC19 containing mCherry (AGB048) using primer set 5′-GGTGAACACCCGGAGTCTCGCAAGTCAATGCTCGATGAGCACAAGGGAGCAGGTGCAGGTGCAGGTGCAATGGTGAGCAAGGGCGAGGAG-3′/5′-GGGTTTATCATCTGTATCAAGGTCGTCTGAGGCCGCATCCGGCTGCATGATTACGCCAAGCTTGC-3′. This DNA fragment was fused into the pBNI1::LEU2 plasmid (backbone, pRS415; promoter, endogenous AgBNI1; 3′ UTR, ScURA3) using yeast co-transformation. The plasmid was rescued with Omega EZNA DNA mini preoperative kit and then transformed into NE5a Escherichia coli. The plasmid was verified by sequencing. The strains were grown in media containing 200 µg/ml G418 and 100 µg/ml Clonat (EMD Millipore) to select the cells that were correctly transformed for microscopy.
To create WHI3-yEGFP; pbni1-3m-mCherry (AG753) or whi3ΔpolyQ-GFP; pbni1-3m-mCherry (AG758), pbni1-3m-mCherry::NAT1 plasmid was constructed first and transformed into WHI3-yEGFP or whi3ΔpolyQ-GFP. In the plasmid, bni1-3m is driven by the endogenous AgBNI1 promoter with ScURA3 3′ UTR. To construct the plasmid, a 928-base segment of AgBni1 containing the AgWhi3 binding sites was synthesized by Genscript. This synthesized DNA was amplified using primer set 5′-CCATGGCTGTAGCCTACGAGCCTGCACATC-3′/5′-CCTCATTCTCTAATGCTGGGTACTTG-3′. These DNA fragments and pBNI1-mCherry plasmid (backbone, pRS415; promoter, endogenous AgBNI1; 3′ UTR, ScURA3) were digested by AfeI and NsiI, and fused together by gap-repair yeast co-transformation into NE5a E. coli. The plasmid was verified by sequencing. bni1-3m strains contain three silent mutations on Whi3-binding site (first three sites; total four sites), TGCAT to CGCTT, AGCCT and GGCTT in the order of location from the beginning of ORF. The strains were grown in media containing 200 µg/ml G418 and 100 µg/ml Clonat (EMD Millipore) to select the cells that were correctly transformed for microscopy. All sequencing was performed at the Dartmouth Molecular Biology Core Facility.
Cell culture and preparation for microscopy
A. gossypii cells were grown in 10 ml of Ashbya full media (AFM) with proper antibiotics, such as G418 (200 µg/ml), or Clonat (100 µg/ml) in a 125-ml baffled glass flask, shaking at 30°C for ∼16 h. whi3Δ and whi3ΔRRM cells were grown at 22°C due to temperature sensitivity and for ∼41 h to obtain similar total biomass to wild-type A. gossypii culture. whi3ΔpolyQ and bni1-3m cells were grown at 30°C for ∼21 h to obtain similar biomass to wild-type A. gossypii culture. whi3Δ, whi3ΔRRM, whi3ΔpolyQ, and bni1-3m cells had similar hyphal shape and diameter and rarely showed split-tip growth, which is indicative of the fast cell growth phase. The A. gossypii culture was then transferred to a 15-ml conical tube (Sarstedt) for centrifugation and prepared for RNA FISH as described previously (Lee et al., 2013a) or live imaging. For epifluorescence microscopy, A. gossypii cells were grown and collected by centrifugation at 350 rpm for 1 min in a 15-ml conical tube. AFM was removed and cells were resuspended in 10 ml of mixed media (25% filter-sterilized AFM and 75% 2× low-fluorescence media) to reduce the auto-fluorescence of the medium, placed on a 25% filter-sterilized AFM/75% 2× low fluorescence media gel pad containing 1% dextrose on the top of a slide, covered with a coverslip, and imaged. For time-lapse imaging, a slide with 2× low-fluorescence media–soaked tissues was loosely sealed with transparent plastic wrap to reduce evaporation. Imaging was performed at 30°C for wild-type A. gossypii and 22°C for whi3 mutants. Microscopic images of cells were acquired as described in “Wide-field fluorescence microscopy setup and image processing.”
Wide-field fluorescence microscopy setup and image processing
An upright light microscope (AxioImager-M1; Carl Zeiss) was used and equipped with the following Carl Zeiss oil immersion objectives: EC Plan-Neofluar 40×/1.3 NA, Plan-Apochromat 63×/1.4 NA, Plan-Neofluar 100×/1.3 NA, and α Plan-Fluar 100×/1.45 NA. Chroma filter set 41025 and Carl Zeiss filter set 38HE were used for visualization of GFP. Chroma filter sets 41002B and 41043 were used for visualizing TAMRA and mCherry, respectively. Hoechst staining was visualized using filter set 49 (Carl Zeiss). An Exfo X-Cite 120 lamp was used as the fluorescence light source. An Orca-ER (C4742-80-12AG; Hamamatsu Photonics) CCD camera driven by Volocity 4 (PerkinElmer) was used for acquisition of images. Z stacks of still and time-lapse images were acquired at different slice sizes (0.3–1.5 µm) and the resulting images were processed by fast or iterative deconvolution (100–120 iterations) using calculated point spread functions in Volocity 4. All still images were linearly contrast enhanced in Volocity 4 and Photoshop CS5 (Adobe). All images were acquired through >10 µm along the z axis to ensure that the nuclei and mRNAs in the cell were imaged completely.
F-actin staining and measuring zone of polarized actin
A. gossypii cells were grown in 10 ml AFM with proper antibiotics in a 125-ml baffled glass flask, shaking at 30°C for ∼16 h. Formaldehyde (final concentration: 3.7% vol/vol) was directly added to the culture and treated for 1 h at 30°C on the shaker. Cells were centrifuged at 3,000 rpm for 3 min and washed twice with 1× PBS. Fixed cells were resuspended in 100 µl PBS and 10 µl Alexa Fluor 488 Phalloidin (a dye; final concentration of 6.6 µM; Life Technologies) was treated for 1 h in the dark. Cells were washed twice with PBS and then resuspended in 15 µl mounting media (Prolong Gold) to load on slides for imaging.
To measure the zone of polarized actin, each hyphal image was cropped and turned in order for the hyphae to lie horizontally and all growing tips to be located on the left side of the image. F-actin signal intensity was added up across the hypha (in the vertical direction on the image) as one value on each pixel along the hypha to only consider relative position of polarized actin from the hyphal tip. The edge of the zone of polarized actin was defined as where the intensity difference between neighboring pixels is >10×. The distance between two edges is measured as the zone of polarized actin.
Hyphal growth assay
For measuring the cumulative hyphal growth (Fig. 2 D), A. gossypii cells grown in AFM (for 15 h) were collected by centrifugation at 400 rpm for 1 min. Cells were placed on the microscopic slide loaded with the gel pad (25% filtered AFM and 75% minimal media; Lee et al., 2013a). The slides containing A. gossypii cells were kept in a humid chamber at 30°C for 30 min and then loaded on the wide-field microscope for time-lapse imaging. Phase images were taken every 10 min (or 3 min for Fig. 1 E). The line measurement tool in Volocity 4 or ImageJ was used to measure the hyphal growth.
For measuring the hyphal growth (µm/h; Fig. 5 B), A. gossypii cells grown in AFM (for 15 h) were collected by centrifugation at 400 rpm for 1 min and washed twice with wash buffer (50 mM Tris, pH 7.5, and 150 mM NaCl). Cells were resuspended in 10 ml of wash buffer with 250 µl of 1 mg/ml FITC-Concanavalin A (ConA) for 10 min at room temperature. Then, cells were washed twice with AFM and grown in a baffled flask for 1 h at 30°C on a shaker. Cells then were fixed with 3.7% formaldehyde for 1 h at 30°C and washed with 1× PBS. Fixed cells were loaded on the microscopic slide for imaging. The lateral lengths of hyphae (from the tip) that were not labeled with FITC-ConA were measured for assaying hyphal growth for 1 h.
Single-molecule RNA fluorescence in situ hybridization
Single-molecule RNA FISH was conducted as described previously (Lee et al., 2013a). Cells grown overnight in AFM were fixed with formaldehyde (final concentration 3.7% vol/vol) at 30°C for 1 h on the shaker and washed twice with ice-cold buffer B (1.2 M sorbitol and 0.1 M potassium phosphate, pH 7.5). The fixed cells were resuspended in 1 ml spheroplasting buffer (10 ml buffer B and 2 mM vanadyl ribonucleoside complex) and transferred to a new RNase-free microcentrifuge tube. 1.5 mg Zymolase (MP Biomedicals) was added to the cells and incubated at 37°C for 35 min for wild-type and 10 min for whi3Δ cells, and cells were washed twice with buffer B. Cells were resuspended in 1 ml RNase-free 70% overnight at 4°C. RNA FISH probes (Biosearch Technologies) were resuspended in 20 µl TE buffer (10 mM TrisCl and 1 mM EDTA, pH 8.0). Then probes were diluted 1:10 from initial probe stock (250 µM in TE buffer). On the next day, cells were washed with wash buffer (20× SSC, 10% vol/vol deionized formamide), resuspended in 100 µl hybridization buffer (1 g dextran sulfate, 10 mg E. coli tRNA, 2 mM vanadyl ribonucleoside complex, 2 mg BSA, 20× SSC, and 10% vol/vol deionized formamide) with 2.5–250 nM mRNA FISH probe, and incubated in the dark, overnight, at 37°C. On the third day, cells were washed with wash buffer, resuspended in wash buffer, incubated at 37°C for 30 min, washed again, and incubated in wash buffer with 5 µg/ml Hoechst (Invitrogen) for nuclear counterstaining at room temperature for 10 min. Cells were then washed with wash buffer, mounted on the RNase-free slide with 20 µl mounting media (ProLong Gold antifade reagent; Invitrogen), covered with an RNase-free coverslip, sealed with nail polish, and imaged.
Detection of nuclei/mRNAs and coordinate reconstruction
Deconvolved z stack images were imported into ImageJ (Fiji 1.46a) and cropped so that each image contains a single hypha. Customized C++ code was used for object detection in a fully automated and unbiased manner in ImageJ as described previously (Lee et al., 2013a). Nuclei, mRNAs, and cell boundaries were detected using a customized ImageJ macro. x, y, and z coordinates of nuclei, mRNAs, cell boundary, and center of hypha including image information were imported into MATLAB 2012b (MathWorks) for reconstructing hyphal images in 3D. Hyphae were considered as curvy cylinders with changing radii. Using the customized MATLAB code, metrics such as overall mRNA density and mRNA count at each point were calculated. For statistical analysis, equal variance and normality tests were conducted to determine whether a (parametric) t test or (nonparametric) Kolmogorov-Smirnov test (KS test) should be used.
mRNA distribution line-plot and spatial mRNA pattern analysis (degree of clustering)
The distance between the growing tip of hypha and each mRNA was recorded for each single molecule FISH (smFISH) image. All mRNAs were considered to be on the center of hypha to simply estimate the number of mRNAs along the hypha. A percentage of mRNA count was binned by 5 µm and plotted for the distance from each hyphal tip (0, 5, 10 µm, and so on). Standard error was calculated on each distance with all data in the same category.
To quantitatively assess whether there is any structural pattern to the localization of transcripts within the cell, we used the method of spatial point pattern analysis called Ripley’s K function (Ripley, 1981) as described previously (Lee et al., 2013a). In this method, random distributions are simulated for comparison to the actual data for each image based on the mRNA density in three dimensions. Thus, the statistical evaluation of whether the pattern is spatially random controls for variation in the abundance of given mRNAs between cells or strains. Ripley’s K is defined as the ratio of the mean number of mRNA points within a distance d of each mRNA to the overall mRNA density of the cell. Ripley’s K, as a function of distance, can be estimated from data as:
where n is the number of mRNA spots, V is the total volume of the cytosol in which transcripts can locate, w is an edge correction function, I is a (0,1) indicator function, and D is the Euclidean distance.
To test the null hypothesis of complete spatial randomness (CSR), 100 random (Poisson-distributed) sets of mRNA locations were simulated for each observed cell using overall mRNA density, cell geometry, and nuclear positions that matched those recorded for the cell. The mean and 95% outer limits of these randomly generated K functions were determined for each distance d. If the K(d) function for an actual, observed mRNA pattern extends above the outer bounds of the random distribution at any given distance d, then statistically significant clustering at that distance can be concluded for that cell.
Under the CSR assumption, the expected value of K(d) equals , the volume of a sphere of radius d. Therefore, to enhance graphical representation, we use the following transformation:
where KS(d) is Ripley’s K function for the simulations representing CSR. By normalizing H(d) functions from each cell by the upper bounds of the respective random simulations, plots of all observed cells on a common scale can be shown. We summarize the results across the many observed cells by plotting the median of these rescaled observed H(d) functions, or H(d)* to calculate median clustering index. This line breaching the CSR 95% confidence envelope (bounded by ±1.0 after the rescaling) indicates statistically significant clustering across the many observed cells at that distance. The space between median clustering index line and random boundary was calculated to obtain the degree of clustering or clustering index, which summarizes all the peak H(d) values for each image for a given transcript or genotype. Customized MATLAB code was used for calculations above and plots were generated in MATLAB 2012b.
RIP
The cell cultures (50 ml) were grown up overnight (∼16 h) from the dirty spores, and 1% final concentration of formaldehyde was added directly to the culture and incubated for 15 min at room temperature with slow shaking to cross-link the protein–mRNA complex. Then, 0.2 M glycine was added immediately and the cells were incubated an additional 5 min at room temperature. Cells were harvested by centrifugation for 5 min at 3,000 g at 4°C. For lysis, 500 mg of cells were resuspended in 1 ml ice-cold lysis buffer (50 mM Hepes-KOH, pH 7.6, 140 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% Tween-20, 5% glycerol, 40 U RNasin [Promega] per milliliter, and 25× protease inhibitor cocktail). Cells were lysed by bead-beating with 0.5 mm zirconia/silica beads in a MiniBeadbeater-8 (BioSpec Products) at top speed for five intervals of 90 s with 30 s of rest. Cell lysates were divided into supernatant and pellet by centrifugation for 10 min at 13,200 rpm. Protein concentrations were determined with a Bradford assay.
The lysates were incubated with 30 µl Dynabeads (Invitrogen) coupled to rabbit anti-Tap antibody (ab290; Abcam) for Whi3 RIP or mouse anti-HA antibody (Abcam) for Puf2 RIP according to the manufacturer’s instructions for 2 h at 4°C. Beads were washed three times with wash buffer (50 mM Hepes-KOH, pH 7.5, 200 mM KCl, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, 0.01% Tween-20, and 40 U RNasin [Promega] per milliliter) on a magnet and eluted in 1× sample buffer at 100°C for 5 min. The sample was diluted in 150 µl sample buffer in total, and 200 mM NaCl was added together with 20 µg proteinase K (New England BioLabs, Inc.), then incubated at 42°C for 1 h and 65°C for 1 h sequentially. 30 µl of mRNA solution was eluted using an RNeasy kit (QIAGEN) and cDNA was synthesized using a SuperScript III first-strand kit (Invitrogen) according to the manufacturer’s instructions. cDNA was amplified using oligo sets to identify Whi3 or Puf2 target mRNAs. Below is the target gene name and oligo set used to identify the target that is pulled down.
CLN3, 5′-AGGTTTCGCTGCTACAACTG-3′/5′-GGATAGCTCGCAAAGAATCG-3′; SPA2, 5′-AGGAGCTACGTGGAGGACAG-3′/5′-GGCTATCAGCTGCGGTATCC-3′; SCD5, 5′-ACCGGGTCTGAATGTTAAGG-3′/5′-GAGAATAGACCGCGGCTCAG-3′; BNI1, 5′-GACCTCGGATATTTCGTCTC-3′/5′-ACTCATCAGCTCGCGATGAC-3′; SHE4, 5′-GCGAGGACAAGGTTCAGATG-3′/5′-CCGTGCAAATAGCACCAATG-3′; CDC24, 5′-TTTCGACGGCGACACAACAG-3′/5′-CGAAACAACGGGAAGTTTGC-3′; MYO2, 5′-TGGTGGGCGACAAGTACCAG-3′/5′-CCCACTAGATTGCTCTGTTC-3′; DYN1, 5′-ACGTAGCGAAGCTGTTTCTC-3′/5′-TGATGTCGGGTACCTCGATG-3′; SEC3, 5′-GACAATGGGTCGGGACACTC-3′/5′-TCCAGGTCGCGCTCAATCAG-3′; ACT1, 5′-ATCTTGACGTTGCGTTACCC-3′/5′-AACACTGGGTGTTCCTCTGG-3′; CLN1/2, 5′-TCGCGAGCCCATTTATACCC-3′/5′-TGGATGCCGCTATGTCAACC-3′; mCherry (for BNI1 or bni1-3m), 5′-GCAAGGGCGAGGAGGATAAC-3′/5′-TCGGCGCGTTCGTACTGTTC-3′.
Protein coimmunoprecipitation
The cell cultures (50 ml) were grown overnight (∼16 h) at 30°C from the dirty spores. Cells were harvested by centrifugation for 5 min at 3,000 g at 4°C. For lysis, 500 mg of cells were resuspended in 1 ml of ice-cold lysis buffer (50 mM Hepes-KOH, pH 7.6, 140 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% Tween-20, 5% glycerol, and 25× protease inhibitor cocktail) with or without 40U RNasin (Promega) per milliliter. Cells were lysed by bead-beating with 0.5 mm zirconia/silica beads in a MiniBeadbeater-8 (BioSpec Products) at top speed for five intervals of 90 s with 30 s rest. Cell lysates were centrifuged for 2 min at 13,200 rpm and supernatant was used for further steps. Protein concentration was determined with a Bradford assay.
The lysates were incubated with 20 µl GFP-Trap_M (ChromoTek) according to the manufacturer’s instructions for 2 h at 4°C. Beads were washed seven times with wash buffer (50 mM Hepes-KOH, pH 7.5, 200 mM KCl, 1 mM MgCl2, 1 mM EGTA, 5% glycerol, and 0.01% Tween-20) with or without 40U RNasin (Promega) per milliliter on a magnet and eluted in 1× sample buffer at 100°C for 5 min. The supernatant was obtained for Western blotting. 20 µg of total protein was diluted in 1× sample buffer, boiled for 5 min, loaded onto a 10% polyacrylamide gel, and separated by SDS-PAGE for 90 min at a constant voltage of 165 V. Proteins were transferred to PVDF membrane (Bio-Rad Laboratories). Membranes were probed with rabbit polyclonal anti-GFP (ab290; Abcam) at a dilution of 1:1,000 or mouse monoclonal anti-HA antibody (ab290, Abcam) at a 1:500 dilution. All primary antibodies were diluted in PBS–Tween-20 (0.1%) with 3% milk. Alkaline phosphatase–conjugated secondary antibodies (Bio-Rad Laboratories) were used at a 1:10,000 dilution in PBS–Tween-20 (0.1%). Blots were developed with the enhanced chemiluminescence (ECL) detection system using Immun-Star AP substrate (Bio-Rad Laboratories) and an X-omat 1000A processor (Kodak).
Prediction of low-complexity sequence protein
IUPred is web-based software (http://iupred.enzim.hu/) that was used to predict if A. gossypii Whi3 or Puf2 are intrinsically unstructured proteins. Parameters used in this study are as follows. Prediction type: long disorder. In the result, >9 consecutive residues that have disorder tendency >0.5 was considered to be an unstructured region. Tango is available at http://tango.crg.es/ and was used to predict aggregating regions in Whi3 or Puf2. Parameters used in this study are as follows. Nterm protected, no; Cterm protected, no; pH, 7; temperature, 298.15 K; and ionic strength, 0.02. Agg scores were calculated and compared with nonaggregating proteins (e.g., Cln3, Gpm2/3). Waltz (waltz.switchlab.org) was used to predict amylogenic regions in Whi3 or Puf2. It outputs the number and size of amylogenic regions.
Protein colocalization test
All colocalization tests in this study have been done with single z-slice images. Microscopic images were imported into ImageJ (Fiji 1.46a) and the plugin Coloc 2 was applied to calculate the Pearson’s colocalization coefficient, Manders’ statistics, and Costes p-value. The background to background (zero to zero) colocalization was excluded from calculations and the p-values after image threshold were reported.
Signal profile plot for assessment of fluorescence intensity heterogeneity
The single z-slice images with multiple channels were imported into MATLAB. Pixels in the field out of the cell or in the nucleus were eliminated by thresholding in order to consider only the fluorescence signal in the cytoplasmic area. Means of signal intensity were calculated in the direction of hyphal width on each pixel along the hypha and plotted on signal profile plots. A moving average over 10 pixels (1 mm) was applied to reduce small noise pikes on the plot. Customized MATLAB code was used for these processes.
Online supplemental material
Fig. S1 contains controls of experiments shown in Figs. 1, 2, and 3. Fig. S2 is related to Figs. 3 and 4, and has additional information about mRNA and proteins focused in this study. Fig. S3 is related to Figs. 3 and 5, and shows results from spatial pattern analysis, Ripley’s H(d), of mRNA used in this study. Video 1 shows Whi3-GFP, which is concentrated on polarization sites, in a wild-type hyphae. Videos 2 and 3 show growth of whi3Δ, which has a growth defect. All MATLAB codes and ImageJ macros are available in a single zip file. 3D_OD_mask_ImagejMacro.ijm is for detecting nuclei, mRNA, and cell boundary in three dimensions and recording their information. main.m is used to sequentially execute smFISH3D_pro1.m and smFISH3D_pro2.m in MATLAB for complete smFISH image analyses. mRNA_lineProfilePlot.m is for generating mRNA profile plots in Figs. 2 G, 3 D, 4 I, 5 D, S1 E, S1 F, and S2 C. FluorSig_heterogeneity_test.m is for plotting the line plots in Fig. 3 G. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407105/DC1.
Supplementary Material
Acknowledgments
We would like to thank the Gladfelter Lab for useful discussions and Jamie Moseley for reading the manuscript. We also thank the Kimble laboratory at the University of Wisconsin-Madison for providing laboratory space and materials for some of the experiments.
This work was supported by National Institutes of Health grant R01-GM081506 to A.S. Gladfelter.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AFM
- Ashbya full media
- CSR
- complete spatial randomness
- KS test
- Kolmogorov-Smirnov test
- NND
- neighbor inter-transcript distance
- RIP
- RNA immunoprecipitation
- RRM
- RNA recognition motif
- smFISH
- single molecule FISH
References
- Baumann S., König J., Koepke J., and Feldbrügge M.. 2014. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 15:94–102 10.1002/embr.201338037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Buxbaum A.R., Wu B., and Singer R.H.. 2014. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 343:419–422 10.1126/science.1242939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M., and Kiebler M.A.. 2011. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30:3540–3552 10.1038/emboj.2011.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisinger T., Klünder B., Johnson J., Müller N., Pichler G., Beck G., Costanzo M., Boone C., Cerione R.A., Frey E., and Wedlich-Söldner R.. 2013. Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat. Commun. 4:1807 10.1038/ncomms2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A.S., Hungerbuehler A.K., and Philippsen P.. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347–362 10.1083/jcb.200507003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S.L.. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Irazoqui J.E., Gladfelter A.S., and Lew D.J.. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062–1070 10.1038/ncb1068 [DOI] [PubMed] [Google Scholar]
- Jansen R.P., and Niessing D.. 2012. Assembly of mRNA-protein complexes for directional mRNA transport in eukaryotes—an overview. Curr. Protein Pept. Sci. 13:284–293 10.2174/138920312801619493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.M., Jin M., and Lew D.J.. 2011. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 21:740–746 10.1016/j.gde.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klünder B., Freisinger T., Wedlich-Söldner R., and Frey E.. 2013. GDI-mediated cell polarization in yeast provides precise spatial and temporal control of Cdc42 signaling. PLOS Comput. Biol. 9:e1003396 10.1371/journal.pcbi.1003396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.C., Savage N.S., Chen H., Wu C.F., Zyla T.R., and Lew D.J.. 2014. Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr. Biol. 24:753–759 10.1016/j.cub.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Zhang H., Baker A.E., Occhipinti P., Borsuk M.E., and Gladfelter A.S.. 2013a. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev. Cell. 25:572–584 10.1016/j.devcel.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.F., Brangwynne C.P., Gharakhani J., Hyman A.A., and Jülicher F.. 2013b. Spatial organization of the cell cytoplasm by position-dependent phase separation. Phys. Rev. Lett. 111:088101 10.1103/PhysRevLett.111.088101 [DOI] [PubMed] [Google Scholar]
- Lehmann R., and Nüsslein-Volhard C.. 1986. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 47:141–152 10.1016/0092-8674(86)90375-2 [DOI] [PubMed] [Google Scholar]
- Lew R.R.2011. How does a hypha grow? The biophysics of pressurized growth in fungi. Nat. Rev. Microbiol. 9:509–518 10.1038/nrmicro2591 [DOI] [PubMed] [Google Scholar]
- Li R., and Bowerman B.. 2010. Symmetry breaking in biology. Cold Spring Harb. Perspect. Biol. 2:a003475 10.1101/cshperspect.a003475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature. 483:336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.Y., Lim H., Yoon Y.J., Follenzi A., Nwokafor C., Lopez-Jones M., Meng X., and Singer R.H.. 2014. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 343:422–424 10.1126/science.1239200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley B.D.1981. Spatial Statistics. Wiley, Hoboken, NJ: 252 pp. [Google Scholar]
- Sambrook J.D.R.2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor: 2,344 pp. [Google Scholar]
- Shestakova E.A., Singer R.H., and Condeelis J.. 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA. 98:7045–7050 10.1073/pnas.121146098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter B.D., Smith S.E., and Li R.. 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb. Perspect. Biol. 1:a003384 10.1101/cshperspect.a003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.E., Rubinstein B., Mendes Pinto I., Slaughter B.D., Unruh J.R., and Li R.. 2013. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J. Cell Biol. 202:1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., and Brangwynne C.P.. 2012. Getting RNA and protein in phase. Cell. 149:1188–1191 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., and Philippsen P.. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 242:381–391 10.1016/S0378-1119(99)00509-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.