Abstract
The two alleles of a gene can be expressed at different levels, the extreme example being imprinting, a condition in which one allele is totally suppressed. Recently, subtle differences in the expression of the two alleles have been detected in numerous human genes and in a few cases, have been associated with a genetic predisposition to disease. The underlying mechanisms are largely unexplored.
Keywords: allele-specific expression, TGFBR1, APC, DAPK1
Two distinct phenomena seem to dominate the most recent literature on the genetic predisposition to human disease, allele-specific expression (ASE) of coding genes and low-penetrance regulatory loci. The latter are often characterized by anonymous common single-nucleotide polymorphisms (SNPs) in genomic regions ostensibly devoid of genes. In the following, we shall discuss the evidence suggesting that the two phenomena may be related.
Recently, a subtle reduction in the expression of one allele of the transforming growth factor β receptor I gene (TGFBR1) was described in blood cells (germline) in ~ 1% of Caucasian control individuals and in ~10% of patients with colorectal cancer (CRC) from the same population (Valle et al., 2008). This translates into an elevated risk of acquiring CRC for individuals with allele-specific expression compared with individuals without ASE. In other words, ASE is a high-penetrance marker of CRC predisposition. Whether the ASE phenomenon is directly causative of the CRC risk needs further investigation, even though an effect on down-stream SMAD2 and SMAD3 signaling was reported (Valle et al., 2008) resembling the situation in mice haploinsufficient for Tgfbr1 (Zeng et al., 2009). Recent evidence suggests that the extent of ASE for TGFBR1 may be tissue-dependent and the frequency of ASE may be lower than initially observed (Guda et al., 2009).
The first example of ASE in germline cells was described several years ago in individuals with familial adenomatous polyposis in whom no traditional germline mutation of APC had been found (Yan, 2002a). Here, ASE of APC was shown to segregate with the phenotype in families, and the subtle germline reduction of the expression of one allele was accompanied by somatic loss of the other allele in tumors.
A third example comes from the study of a family in which chronic lymphocytic leukemia occurred in seven members of a three-generation family (Lynch et al., 2002). Here, fibroblasts (germline) from the affected individuals displayed relatively pronounced ASE (up to 70% reduction) of the proapoptotic DAPK1 gene. In leukemic cells the expression of both the affected allele and the wild-type allele was knocked out by extensive promoter methylation resulting in total abrogation of DAPK1 activity (Raval et al., 2007).
The three examples of ASE show several similarities. Each of the affected genes belongs to a pathway known to be implicated in the disease. The degree by which the expression of one allele is lowered varies somewhat but can well be considered modest (30–70% reduction). In cells that have not undergone a second hit (for example, somatic mutation, loss of heterozygosity and methylation), the total reduction in the gene’s expression can be as small as 15–25% and is hard to measure, yet apparently has significant downstream effects. Perhaps most intriguingly, in none of the three conditions has any causative mechanism been discovered by sequencing or other means. Yet a genomic change in cis is almost certainly present because in each case, the ASE segregates not only with the phenotype but also with haplotypes covering all or part of the gene and some flanking sequence.
Differences in the level of expression between the two alleles of genes have been recognized for some time (Knight, 2004). The phenomenon is widespread as shown in humans (Lo et al., 2003) and mice (Cowles et al., 2002). Moreover, total abrogation of the expression of one allele (monoallelic expression) was evidenced in 300 out of 4000 human genes studied in clonal cell lines (Gimelbrant et al., 2007). ‘Classic’ examples of total loss of expression of one allele are imprinting of autosomal genes and the selective inactivation of one X chromosome in females. Imprinting is thought to be due to epigenetic phenomena such as DNA methylation and histone acetylation. In the case of non-imprinted autosomal genes (the object of this review), germline epigenetic modifications have not been invoked so far.
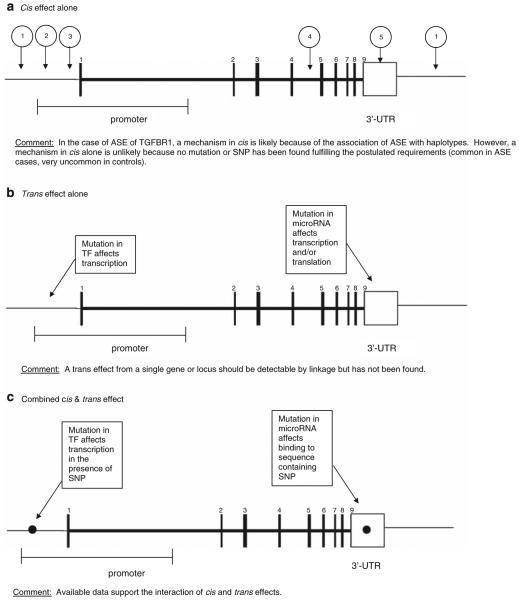
The heritability of gene expression patterns seems to hold the key to our understanding of the phenomenon. As discussed above, in the three examples of genes associated with human disease (TGFBR1, APC and DAPK1) the co-inheritance of ASE with defined haplotypes strongly suggests the existence of a mutation in cis. The same conclusion emerged from the study of ASE of PKD2 and CAPN10 in lymphoblastoid cell lines from families in which the gene expression patterns allowed linkage to be established (Yan, 2002b). However, it is intriguing that the putative mutations in cis have not yet been uncovered. A plausible explanation is that the ASE trait arises only as a consequence of the interaction between a factor in cis and a factor in trans (Figure 1).
Figure 1.
Putative genomic mechanisms leading to allele-specific expression. Schematic representation of the TGFBR1 gene. (a) Cis effect alone: (1) Change in genomic DNA affects enhancer. (2) Change in genomic DNA affects binding of transcription factor. (3) Change in genomic DNA affects antisense transcription. (4) Change in genomic DNA affects mRNA splicing or stability. (5) Change in genomic DNA affects the binding of microRNAs. (b) Trans effect alone. (c) Combined cis and trans effect.
The existence of trans-acting factors (for example, genes) affecting human gene expression has been amply documented in recent years. Treating expression levels as a quantitative trait microarrays allow large numbers of genes to be assessed. The study of large Centre d’Etude du Polymorphisme Humain pedigrees by linkage analyses of 3554 genes detected significant linkage of some 1000 expression phenotypes (Morley et al., 2004). In other words, the causative change (for example, variant) could be localized in the genome. Many phenotypes seemed to be regulated by a single locus, some by multiple loci. Trans-acting loci were more common than cis acting ones. Some ‘hot spots’ of transcription regulation were found that apparently influenced many genes. Subsequently, evidence of heritable expression levels in 85% of 19 648 autosomal genes was found, and linkage analyses favored control mostly in cis (Göring et al., 2007). Yet another study used the allelic association methodology to map the regulatory loci of at least 1348 genes that had association signals in cis and at least 180 in trans (Stranger et al., 2007).
These data reveal a hitherto almost unimaginable extent of heritable, genomic and regulatory functions. It is tempting to say that we are beginning to understand how H. sapiens is able to cope with as few genes as some 30 000. The polymorphic regulation of the expression of protein-encoding genes provides the diversity required by our biological processes in health and disease.
Now we shall examine the nature of the regulatory elements (for example, genes). It would seem that the traditional view of gene regulation (promoters, transcription factors, repressors and enhancers,) is not sufficient to account for the described phenotypic diversity of expression (allelic or total). Instead, thanks in great part to the technological developments in genome-wide association studies; it is beginning to seem that regulatory loci often do not comprise genes of traditional (protein-encoding or RNA) type. As an example, genome-wide association studies in CRC performed in the past 2 years have identified at least 10 loci showing significant association with the phenotype (Houlston et al., 2008). Of note, only one of the pinpointed SNPs is located within a known gene (SMAD7). Most are either in so-called gene desert regions or within some 50–300 kb of coding genes. An example is SNP rs6983267 in 8q24. This SNP seems to associate not only with CRC, but also with prostate and possibly other cancers. It is not located in a gene but not far from a pseudogene. Intriguingly, it is only some 350 kb from the oncogene MYC, but as far as is known, no association between the level of MYC transcript and the SNP has been detected. Therefore, what secrets will those disease-associated SNPs eventually reveal? In the absence of facts, speculation is allowable. MicroRNAs are small regulatory molecules that bind to partially or fully complementary sequences in the mRNA of target genes, leading to lowered transcription or inhibition of translation of the genes. Although fully characterized examples of such miR-target gene combinations affecting human phenotypes are not available, it is likely that this concept will soon be better understood. Already close to 1000 microRNAs are known, most of which have dozens or hundreds of target genes by in silico prediction. These facts are compatible with microRNAs having a major role in the regulation of gene expression (either on the RNA or protein level). However, as far as is known, the numerous SNPs that by genome-wide association studies have been found to be associated with disease phenotypes are not usually located in or close to recognized miRs. Instead, one may speculate about the existence of larger, perhaps kilobase-sized or larger regulatory RNA molecules (Carninci, 2009). These non-coding (ncRNA) molecules are largely unexplored but will no doubt figure prominently in the literature of coming years. Finally, antisense RNA tanscribed from the non-coding strand of DNA may turn out to have a major role in gene expression patterns (He et al., 2008).
How are the putative culpable regulatory RNA (and other) genes going to be identified? Two likely ways present themselves. First, concentrating on a single gene showing ASE (such as TGFBR1) associated with a defined haplotype in cis, genetic and molecular biology techniques can be applied to search for sequence variants (mainly SNPs) that occur more often in cases than in controls, followed by functional assays (measured, for example, by luciferase expression) to evaluate such candidate variants. In search of transacting collaborators, the changes in binding affinity of, for example, microRNAs caused by the SNP can be interrogated. Second, in search of trans variants, large families segregating ASE can be subjected to linkage studies in search of candidate loci. Such linkage studies need to account for interactive loci; the one in cis, which is known or at least localized, and the putative one in trans, which is sought. The answers may only be obtained after painstaking research.
In summary, quantitative differences in gene expression levels are widespread, often tissue-specific, allelespecific, subtle and heritable, usually controlled by yet uncharacterized genes or variants in cis or trans or both. This variation in expression adds another level of complexity to the regulation of human development, biological functions and disease. Already a few genes have been shown to exhibit ASE patterns apparently predisposing to cancer. The mechanism causing ASE is unknown but available data suggest a collaboration between genomic variants in cis and trans. Prime candidates for the trans-acting elements are non-coding RNA sequences (genes), some of which may contain the SNPs that have been associated with disease genotypes by genome-wide association studies.
Acknowledgements
Work in the author’s laboratory was supported by NIH grants CA67941, CA16058 and P01 124570.
Footnotes
Conflict of interest
The author declares no conflict of interest.
References
- Carninci P. The long and short of RNAs. Nature. 2009;457:974–975. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nature Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Goöring HHH, Curran JE, Johnson MP, Dyer TD, Charlesworth K, Cole SA, et al. Discovery of expression QTLs using largeskale transcriptional profiling in human lymphocytes. Nature Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Guda K, Natale L, Lutterbaugh J, Wiesner GL, Lewis S, Tanner SM, et al. Infrequent detection of germline allele-specific expression of TGFBR1 in lymphoblasts and tissues of colon cancer patients. Cancer Res. 2009;69:4959–4961. doi: 10.1158/0008-5472.CAN-09-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nature Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, et al. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, Weisenburger DD, Quinn-Laquer B, Watson P, Lynch JF, Sanger WG. Hereditary chronic lymphocytic leukemia: an extended family study and literature review. Am J Med Genet. 2002;115:113–117. doi: 10.1002/ajmg.10686. [DOI] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A, Tanner S, Byrd J, Angerman E, Perko J, Chen S-S, et al. Down-regulation of death associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beasley C, et al. Population genomics of human gene expression. Nature Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle L, Serena-Acedo T, Liyanarachchi S, Hampel H, Comeras I, Li Z, et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. Small changes in expression affect predisposition to tumorigenesis. Nat Genet. 2002a;30:25–26. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- Yan H. Allelic variation in human gene expression. Science. 2002b;297:143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Phukan S, Xu Y, Sadim M, Rosman DS, Pennison M, et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69:678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]