Plastids import a significant number of proteins in the absence of the chloroplast inner envelope membrane translocon subunit.
Abstract
We report on the characterization of Tic56, a unique component of the recently identified 1-MD translocon at the inner envelope membrane of chloroplasts (TIC) in Arabidopsis (Arabidopsis thaliana) comprising Tic20, Tic100, and Tic214. We isolated Tic56 by copurification with Tandem Affinity Purification-tagged Toc159 in the absence of precursor protein, indicating spontaneous and translocation-independent formation of the translocon at the outer envelope membrane of chloroplasts (TOC) and TIC supercomplexes. Tic56 mutant plants have an albino phenotype and are unable to grow without an external carbon source. Using specific enrichment of protein amino termini, we analyzed the tic56-1 and plastid protein import2 (toc159) mutants to assess the in vivo import capacity of plastids in mutants of an outer and inner envelope component of the anticipated TOC-TIC supercomplex. In both mutants, we observed processing of several import substrates belonging to various pathways. Our results suggest that despite the severe developmental defects, protein import into Tic56-deficient plastids is functional to a considerable degree, indicating the existence of alternative translocases at the inner envelope membrane.
Chloroplast functions depend on the import of several thousand nucleus-encoded proteins that enter the chloroplast through different import pathways (for review, see Jarvis, 2008; Shi and Theg, 2013). The prevalent pathway for most of the proteins with photosynthetic and housekeeping functions operates via complexes in the outer and inner envelope membranes of chloroplasts, designated as TOC and TIC translocon complexes (Schnell et al., 1997). The majority of nucleus-encoded plastid proteins that enter chloroplasts through the TOC/TIC system possess a cleavable N-terminal transit peptide that mediates their specific import into different plastid types. Previous reports suggested that plastid transit peptides contain functional motifs that determine their preference for different plastid types (von Heijne et al., 1989; Pilon et al., 1995; Ivey et al., 2000; Chotewutmontri et al., 2012; Teng et al., 2012; Li and Teng, 2013). These analyses clearly support the view of import specificity that depends on the developmental status of the chloroplast. It is conceivable that this selectivity is mediated through the initial recognition of precursor proteins at the chloroplast surface (i.e. that the selectivity is mediated by different receptor protein complexes at the outer envelope membrane).
Indeed, plastids possess structurally and functionally distinct TOC complexes that differentiate between different client proteins and establish precursor protein selectivity (Jarvis et al., 1998; Bauer et al., 2000; Kubis et al., 2003, 2004; Ivanova et al., 2004). In green Arabidopsis (Arabidopsis thaliana) tissues, the predominant TOC core complex is built from the receptor GTPases Toc159 and Toc33 and the translocation channel Toc75. In alternative TOC complexes, the two GTPases are replaced by their homologs, whereby their substrate selectivity is largely determined by the different receptors of the Toc159 family (Smith et al., 2004; Inoue et al., 2010). The current model suggests that TOC complexes built around Toc159 and Toc90 are mainly involved in the import of photosynthetic proteins (Bauer et al., 2000; Infanger et al., 2011), while the Toc132- and Toc120-containing complexes mediate the import of housekeeping proteins (Ivanova et al., 2004; Kubis et al., 2004). This model explains the ability of chloroplasts to import low-abundance proteins under conditions of high photosynthetic protein import. Proteomic analyses challenged this model because many photosynthetic proteins were found imported in plastid protein import2 (ppi2) plastids lacking Toc159, and the decreased accumulation of photosynthetic complexes is largely explained by their down-regulation at the transcriptional level (Bischof et al., 2011). A recent large-scale split-ubiquitin study suggested overlapping precursor-binding specificities of Toc159 and Toc132, which could explain the occurrence of a subset of photosynthetic proteins in the ppi2 chloroplast proteome (Dutta et al., 2014). Thus, the mechanisms mediating the selective import of preproteins into plastids remain elusive.
Different members of the Toc159 family of receptor proteins differ in the length of a conserved acidic domain, the so-called A domain (Hiltbrunner et al., 2001a). Two studies suggest that the A domains confer specificity to the different members of the Toc159 family (Inoue et al., 2010; Dutta et al., 2014). This could explain why full-length Toc120 or Toc132 fails to complement the ppi2 mutant (Kubis et al., 2004), while a construct of Toc132 lacking the A domain was partially able to do so (Inoue et al., 2010). These data suggest that fully assembled TOC complexes with different members of the Toc159 family must be present in the outer envelope membrane to support specific protein import during chloroplast development, which is in line with a reorganization of the import machinery in phases of changing protein import demand. Indeed, Ling et al. (2012) identified a REALLY INTERESTING NEW GENE ubiquitin ligase as a suppressor of the pale-green phenotype of a Toc33 mutant (ppi1) and showed that the ubiquitin proteasome system controls the assembly of different TOC complexes by regulated proteolysis. As anticipated, this reorganization is especially relevant during etioplast-to-chloroplast and chloroplast-to-gerontoplast differentiation. In the case of etioplast-to-chloroplast conversion, the increasing demand for the import of photosynthetic proteins is apparently accompanied by the disassembly of TOC complexes that have different specificities. However, evidence for the preferential degradation of Toc132 and/or Toc120 during plastid differentiation by the ubiquitin proteasome system is currently missing.
It is currently unclear how far the association of TOC complexes with different TIC components contributes to precursor selectivity. Thus, a full understanding of client protein specificity requires the analysis and identification of translocation-competent TOC-TIC supercomplexes. In contrast to the TOC translocon, the organization and subunit composition of the TIC complexes are less clear and under controversial debate. For Tic20 and Tic110, translocon channel activity has been demonstrated in vitro (Heins et al., 2002; Balsera et al., 2009; Kovács-Bogdan et al., 2011; Kikuchi et al., 2013). However, it is still in question if these proteins are part of two functionally distinct import routes. Recently, a new 1-MD TIC complex was identified that was termed the general import translocon. This complex is built around Arabidopsis Tic20-I and comprises Tic56, Tic100, and the chloroplast-encoded hypothetical chloroplast open reading frame 1.2 gene product termed Tic214 (Kikuchi et al., 2013). Tic110 and Tic40 were absent from this complex, arguing for a separate function of the Tic110 complex. Lack of Tic56 and Tic100 resulted in strongly reduced levels of the 1-MD TIC translocon and in albino phenotypes resembling the phenotype of the tic20-I mutant, further supporting a common function in the 1-MD complex. However, the import capacity of the tic56 and tic100 albino mutants has not been directly tested yet.
Characterization of a system as complex as plastid protein import requires specialized tools that allow an unbiased identification of client proteins for the different receptors. Functional proteomics allows an assessment of protein accumulation in the absence of specific receptors or import components. While a standard proteomics experiment is useful and provides information on plastid protein accumulation, protein N termini are of specific interest because they allow distinguishing imported and correctly processed proteins from accumulated precursors (Bischof et al., 2011). One out of several methods for the analysis of protein N termini is called terminal amine isotopic labeling of substrates (TAILS; Kleifeld et al., 2010; Lange and Overall, 2013). In TAILS, protein N termini are blocked by dimethyl labeling, and newly generated N termini from the subsequent tryptic digestion are removed by their coupling to a polymer. This subtractive approach has been employed successfully for the characterization of protein processing and transport into plastids of the diatom Thalassiosira pseudonana (Huesgen et al., 2013). Here, we used the TAILS method in combination with protoplast import assays and in vitro import kinetics to characterize the contribution of Tic56 and the 1-MD TIC complex to plastid protein import and compare these data with those from the ppi2 mutant deficient in Toc159 (Bauer et al., 2000). Our data reveal a considerable degree of protein import into both mutants and allow pinpointing Toc159- and Tic56-independent client proteins.
RESULTS
Coisolation of Tic56 with Affinity-Tagged Toc159
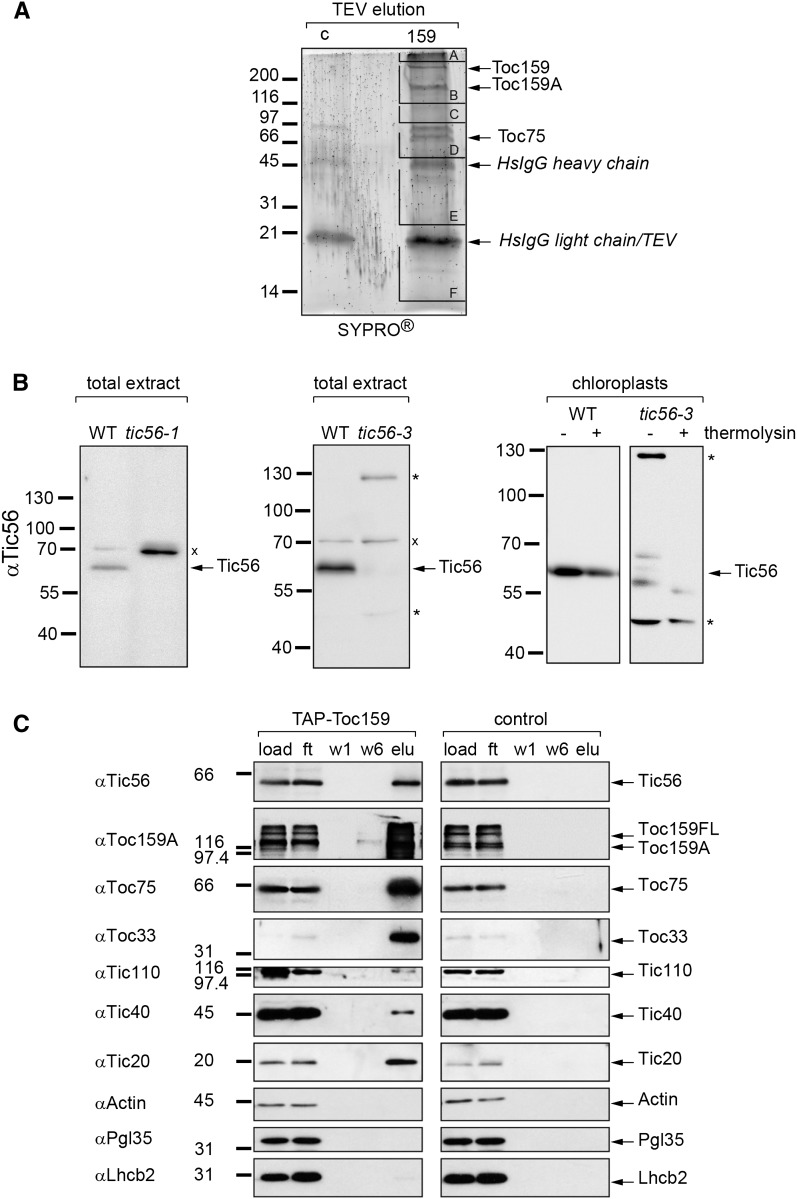
In a precedent study, we used transgenic Arabidopsis plants expressing the Protein A-tagged chloroplast protein import receptor Toc159 to isolate Toc159 from a solubilized whole-cell membrane fraction (Agne et al., 2010). Pursuing this approach to identify new components of the chloroplast protein import machinery, the purification experiment was scaled up and the eluates (Fig. 1A) were subjected to mass spectrometric analysis. Mass spectrometry (data not shown) revealed that Tic56, a component of the recently uncovered 1-MD TIC complex (Kikuchi et al., 2013), specifically copurified with Toc159.
Figure 1.
Association of Toc159 with Tic56, a component of the 1-MD TIC complex. A, SyproRuby stain of an eluate obtained after the purification of TAP-Toc159 from Triton X-100-solubilized membrane proteins of TAP-Toc159:ppi2 plants (159). As a control, the same purification procedure was performed with wild-type plants (c). Subsequent mass spectrometric analysis of gel slices A to F revealed the presence of Tic56 peptides in gel slice D (data not shown). B, Specificity of the anti-Tic56 serum. Fifty micrograms of protein of total protein extracts of the wild type (WT) and of two tic56 T-DNA insertion mutants (tic56-1 on the left and tic56-3 in the center) was analyzed by SDS-PAGE and immunoblotting with anti-Tic56 antiserum. Each x indicates a 70-kD cross reaction of the serum with a protein present in wild-type and mutant samples. To further analyze the additional bands (asterisks) appearing in tic56-3, chloroplasts of 16-d-old wild-type and tic56-3 seedlings were isolated, treated or not with thermolysin (50 µg mL−1), and subjected to the same immunoblotting procedure. C, Confirmation of the copurification of Tic56 with Toc159 by immunoblotting of fractions from a TAP-Toc159 purification. Fifty micrograms of protein of Triton X-100-solubilized membrane proteins loaded to Homo sapiens (Hs)IgG beads (load), 50 µg of the column flow through (ft), and 25% of two different wash fractions (w1 and w6) and the tobacco etch virus protease eluates (elu) were probed with antisera as indicated.
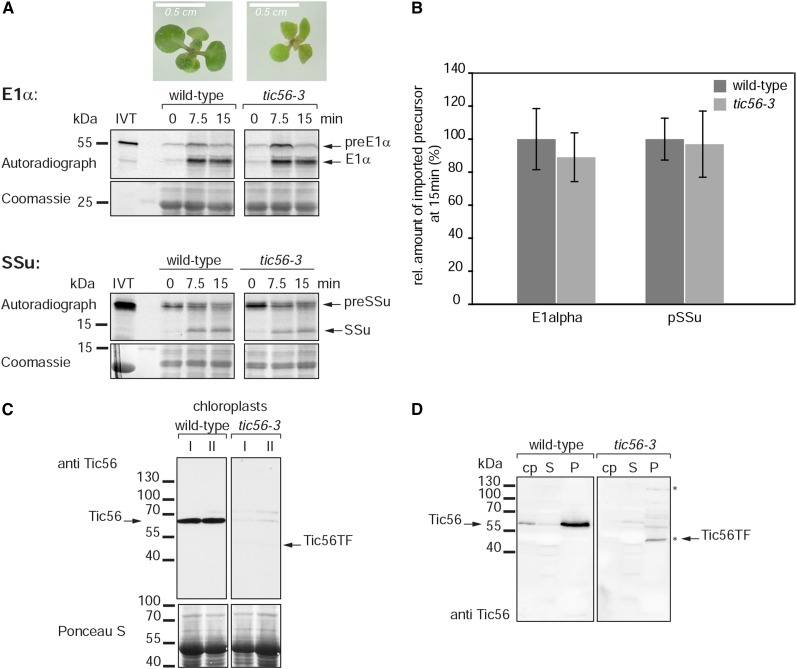
An antiserum was raised against recombinant Tic56. The Tic56-specific antiserum detects a protein at approximately 63 kD by SDS-PAGE that is not detected in protein extracts of the Arabidopsis transfer DNA (T-DNA) insertion mutants tic56-1 and tic56-3 (Fig. 1B), supporting the specificity of this antibody preparation. Tic56 migrates at a higher molecular mass than predicted (56.3 kD), which is probably due to the high content of acidic residues, particularly in its C-terminal part. In tic56-1, which is a null mutant of TIC56 (Kikuchi et al., 2013), only one 70-kD cross-reactive band was detected, whereas in the tic56-3 mutant, additional bands at 130 kD and approximately 48 kD appeared (Fig. 1B, center). These signals are potentially Tic56 specific, as the mutant has the T-DNA insertion in the last exon of the TIC56 gene (Kikuchi et al., 2013; Supplemental Fig. S1). Thus, in tic56-3, elongated or truncated forms of Tic56 can be expected depending on the translation of in-frame coding sequences residing on the T-DNA. Both the 130- and 48-kD proteins can be detected in isolated tic56-3 chloroplasts, but only the 48-kD protein was resistant to thermolysin treatment of chloroplasts and behaved like wild-type Tic56 (Fig. 1B, right). Kikuchi et al. (2013) exclusively detected a truncated form of Tic56 in tic56-3. We assume that the 48-kD protein identified here corresponds to the truncated form described by Kikuchi et al. (2013). It remains undetermined if this truncated form results from a premature stop codon or if it is a stable degradation product of a higher molecular mass Tic56 fusion protein.
With the anti-Tic56 antiserum, we confirmed the interaction of Tic56 with Toc159 (Fig. 1C). Tic20, the channel protein and key component of the 1-MD TIC complex, also was found to be enriched in the eluates of the TAP-Toc159 purification experiment. In addition to Tic56 and Tic20, other members of the TOC core complex, Toc33 and Toc75, as well as Tic110 and Tic40, also were detected in the eluates by western blotting (Fig. 1C). Our data indicate that protein import supercomplexes between Toc159 and different TIC complexes can be isolated in the absence of any externally added precursor protein. In the following, we report on the functional characterization of Tic56 in the context of chloroplast biogenesis and the import capacity of TIC56-deficient mutants.
Disturbed Plastid Development in tic56-1 Plants
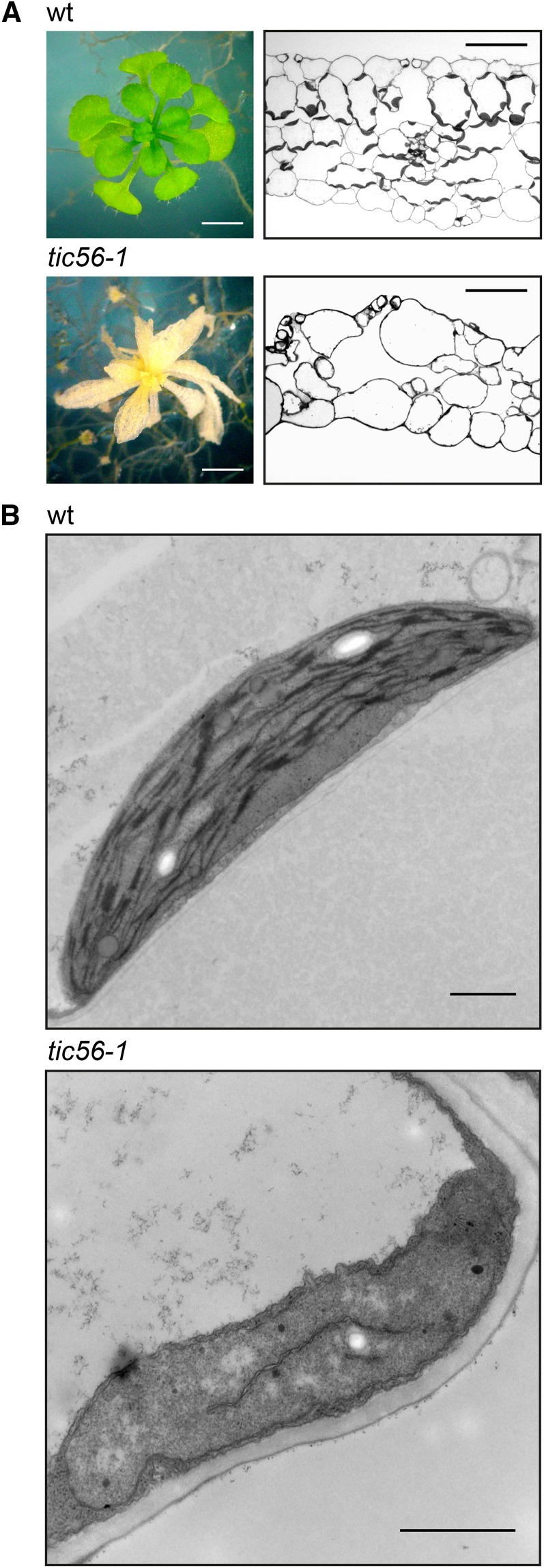
Lack of Toc159 as well as Tic56 results in seedling-lethal albino phenotypes in Arabidopsis (Bauer et al., 2000; Kikuchi et al., 2013; Supplemental Fig. S2). However, the two albino mutants ppi2 (toc159) and tic56-1 are phenotypically not identical. Compared with the Toc159-deficient mutant ppi2, tic56-1 plants have a more severe phenotype with less residual chlorophyll and irregularly shaped, nearly translucent ivory-colored leaves (Fig. 2A; Supplemental Fig. S2). Cross sections of 8-week-old tic56-1 leaves reveal a loss of cellular organization with reduced mesophyll and the absence of mature chloroplasts (Fig. 2A). In transmission electron micrographs, plastids of tic56-1 appear to be two to three times smaller and variable in shape compared with their wild-type counterparts. Furthermore, tic56-1 plastids lack a thylakoid network characteristic of mature chloroplasts (Fig. 2B). Only a few presumably (pro)thylakoid membranes that are associated with plastoglobules can be observed. These results confirm that Tic56 is required for chloroplast development.
Figure 2.
Disturbed leaf morphology and plastid development in tic56-1 plants. A, Leaf cross sections of 8-week-old wild-type (wt) and tic56-1 plants show disordered tissue and lack of chloroplasts of tic56-1 leaves. Bars = 2 mm (plant images) and 50 µm (cross sections). B, Transmission electron micrographs of wild-type and tic56-1 plastids uncover differences in the overall structure, in the shape and size of the organelles as well as in their stroma-localized membrane system. Bars = 1 µm.
Levels of Other TOC and TIC Components in tic56-1
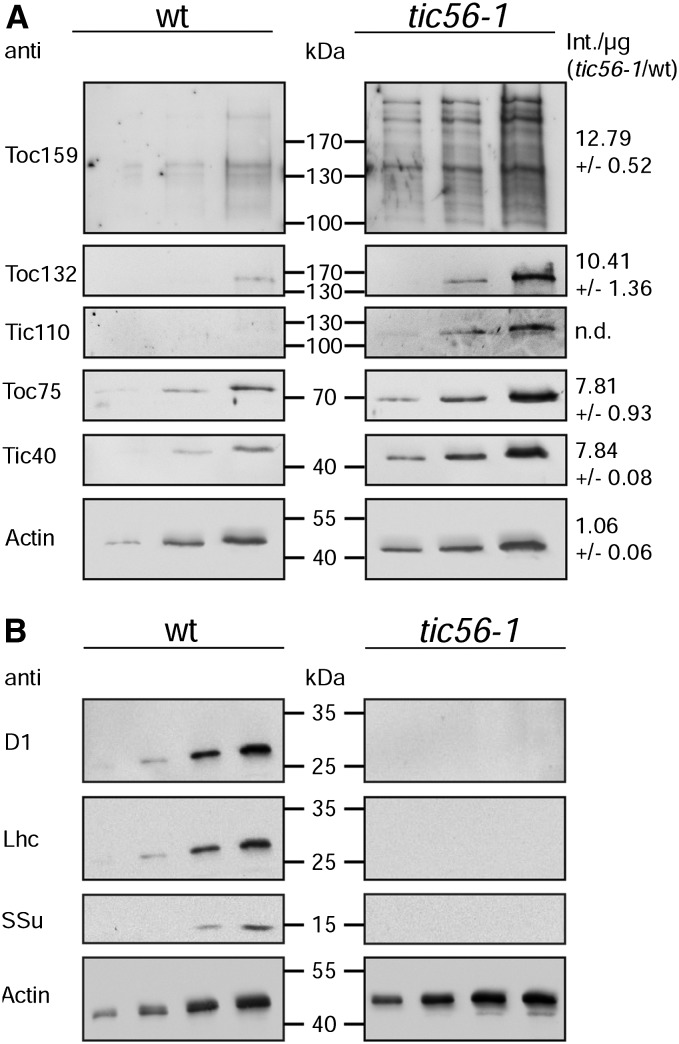
The coisolation of Tic56 with the chloroplast import receptor Toc159 and its presence in the recently identified 1-MD TIC complex suggest that the tic56-1 mutant has a defect in plastid protein import. In tic56-1, Tic56 is lacking and the other components of the 1-MD Tic20 complex (Tic20, Tic100, and Tic214) do not accumulate (Kikuchi et al., 2013). Therefore tic56-1 is ideal for the characterization of plastid protein import in the absence of the 1-MD TIC complex that was designated as the general TIC translocon. Because Tic56 associates with a Toc159-containing complex (Fig. 1), we tested whether the absence of Tic56 perturbs the integrity of the TOC translocon by western blotting (Fig. 3). The TOC components Toc159, Toc132, and Toc75 as well as Tic110 and Tic40 were detected at normal or even higher levels in the mutant plants compared with the wild-type control. Higher TOC/TIC protein levels were detected also in other albino mutants with defects in genes unrelated to chloroplast protein translocation (Motohashi et al., 2012). Thus, our data suggest that a lack of Tic56 perturbs neither the TOC translocon nor the Tic110-Tic40 complex. Toc75 and Tic40 have transit peptides and are processed to their mature size in tic56-1, indicating that, despite the absence of the 1-MD TIC complex, a subset of proteins is still imported. On the contrary, nucleus-encoded light harvesting complex of PSII (Lhcb4) and the small subunit of Rubisco (SSu) as well as plastid-encoded PSII reaction center protein A (PsbA) were not detectable in the tic56-1 total leaf extract, suggesting either substrate specificity in the disturbed import process or a systemic regulation at the level of transcription (Richly et al., 2003).
Figure 3.
Western-blot analysis of TOC components, Tic110/Tic40 (A) and different plastid proteins involved in photosynthesis (B), in tic56-1 compared with the wild type. Rising protein amounts (top, 10, 20, and 50 µg; bottom, 2, 4, 8, and 12 µg) of 8-week-old wild-type (wt) and tic56-1 leaves were loaded and analyzed with antibodies as indicated. Detection of actin served as a loading control. In A, the ratio between the signal intensities (Int.) per µg of protein of tic56-1 and the wild type is given alongside the blots.
Analysis of Protein Import into tic56-1 Plastids by N-Terminal Peptide Identification
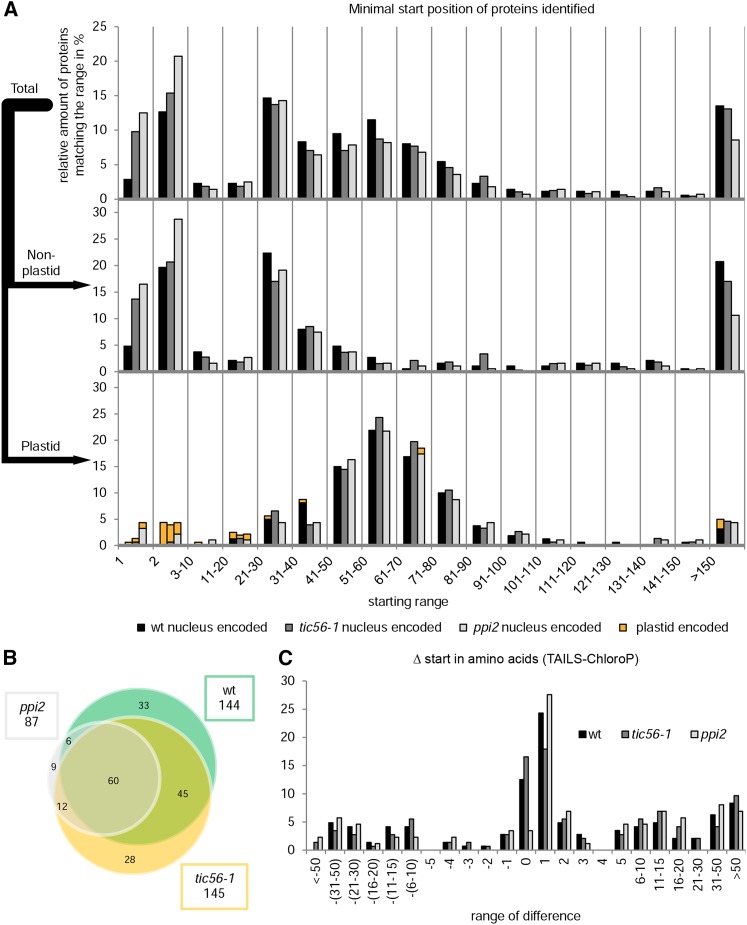
We decided to investigate the import ability and selectivity of tic56-1 mutant plastids by an unbiased experimental approach that provides direct evidence for functional import and is not restricted to a few selected import substrates. The method we have chosen is TAILS, which is based on the specific identification of N-terminal peptides by mass spectrometry. Protein extracts of the wild type, tic56-1, and ppi2 were denatured, and the naturally occurring free N termini of proteins were blocked by dimethylation followed by tryptic digestion. The internal peptides generated by trypsin were subsequently removed by their coupling to a high-molecular-weight dendritic hyperbranched polyglycerol-aldehyde polymer and subsequent filtration. The resulting flow through that is enriched for in vivo N termini was analyzed by liquid chromatography-tandem mass spectrometry on an Orbitrap Velos device (Thermo Scientific). In total, N-terminal peptides of 348 proteins were identified from wild-type, 280 from ppi2, and 481 from tic56-1 plant material (Supplemental Table S1).
Figure 4A (top) shows the relative distribution of the minimal N-terminal peptide of the identified proteins in relation to their corresponding full-length sequences. The proteins were sorted according to the positions of the start amino acids of their most N-terminal peptides. The amount of proteins falling into a starting range is given in percentage of total proteins identified. The pattern of minimal peptide distribution was nearly identical for the three different plant lines, with local maxima at the amino acid position 1 or 2, 21 to 30, and 51 to 60 and at positions greater than 150. Proteins in the 1 or 2 starting bin comprise mostly cytosolic proteins or proteins encoded by organellar genomes. The local maximum at bin 21 to 30 is based on mostly nucleus-encoded processed mitochondrial proteins that generally have shorter N-terminal targeting sequences than plastid proteins (Supplemental Table S2; Huang et al., 2013). Accordingly, these two maxima remain when only nonplastid proteins are plotted in the histogram (Fig. 4A, middle). A closer look at the starting positions of plastid proteins shows that the 21 to 30 bin also comprises several processed plastid proteins. However, most plastid proteins started with amino acids 51 to 60 (Fig. 4A, bottom). The accumulation of proteins with starting positions greater than 150 can be explained by additional proteolytic processing that affects the in vivo accumulation of protein N termini mainly of nonplastid proteins.
Figure 4.
TAILS analyses of tic56-1, ppi2, and the wild type. A, Distribution of the minimal starting positions in the annotated full-length sequences of proteins identified by TAILS. For each identified protein, the most N-terminal peptide identified by the TAILS experiment was determined and grouped into starting ranges as indicated. The amount of proteins falling into a distinct range was set into relation with the total number of proteins of each plant line. In the graph at top, the minimal starting positions of all proteins identified are shown. The middle and bottom graphs show the distribution of starting positions of nonplastid or plastid proteins, respectively. The key at the bottom includes additional information about where the proteins are encoded (nucleus, black, dark gray, and light gray; plastid, orange). The first bar in each group always represents the wild type (wt), the second bar always represents tic56-1, and the third bar always represents ppi2. Chloroplast proteins were classified according to a chloroplast reference proteome (van Wijk and Baginsky, 2011). B, Venn diagram of nucleus-encoded plastid proteins identified for tic56-1, ppi2, and the wild type. C, The difference between the experimental (TAILS) and predicted (ChloroP) starting positions of the proteins was calculated. Positive values signify processing downstream, and negative values signify processing upstream of the theoretical processing site.
As the bulk of chloroplast proteins in all lines started in the range of amino acids 21 to 90 (Fig. 4A, bottom), these data clearly show that many chloroplast proteins are imported into the plastids of both mutants lines and that these are processed to a mature or nearly mature form by transit peptide removal after import. Only a few plastid proteins started with the first or second amino acid, and these are almost exclusively plastid-encoded proteins (Fig. 4A, bottom). Exceptions are two proteins in tic56-1 as well as four proteins in ppi2 that were found as unprocessed precursor proteins (Table I).
Table I. Identified unprocessed precursor proteins in the mutant lines tic56-1 and ppi2.
| Line | Identifier | Description | Start Position | N-Terminal Peptide | Natural Modification |
|---|---|---|---|---|---|
| tic56-1 | AT4G34200.1 | EDA9 (EMBRYO SAC DEVELOPMENT ARREST9) | 2 | SATAAASSSIAVATNSLR | N-Acetyl |
| AT5G56730.1 | Peptidase M16 family | 1 | MDLIAGESSkVLR | N-Acetyl | |
| ppi2 | AT1G60950.1 | FED A (FERREDOXIN2) | 2 | ASTALSSAIVGTSFIR | N-Acetyl |
| AT2G33800.1 | Ribosomal protein S5 family protein | 2 | ATASALSSLSSLSLHTR | N-Acetyl | |
| AT4G17090.1 | CT-BMY (β-AMYLASE3, β-AMYLASE8) | 1 | MELTLNSSSSLIkR | N-Acetyl | |
| AT5G56730.1 | Peptidase M16 family | 1 | MDLIAGESSkVLR | N-Acetyl |
The Venn diagram in Figure 4B gives an overview of the overlap of nucleus-encoded plastid proteins identified in the three different plant lines.
Comparison of Experimental and Predicted N Termini of Imported Plastid Proteins
To validate the correct processing of the imported substrates, their transit peptide length was predicted using ChloroP (Emanuelsson et al., 1999) and the predicted and measured transit peptide lengths were compared (Fig. 4C). As shown in Figure 4C, the measured starting positions for most of the nucleus-encoded plastid proteins in the wild type, ppi2, and tic56-1 matched the prediction by ChloroP perfectly (Δ0) or was shifted by one toward the subsequent amino acid (Δ1). Remarkably, this shift in the starting positions of plastid proteins was much more pronounced in the ppi2 mutant than in the wild type and tic56-1. The observed discrepancy between the ChloroP-predicted and experimental starting amino acids is striking but has been observed in previous studies dealing with the N termini of the plastid proteins (Emanuelsson et al., 1999; Zybailov et al., 2008; Huesgen et al., 2013). A statistical evaluation of the processing site by sequence logo revealed that Cys and Ala are frequently removed compared with the prediction (Supplemental Fig. S3A, bottom, position −1), thus changing the starting amino acids of the affected proteins. Indeed, we observed that among the experimentally determined N termini, Ala occurred much less frequently than predicted (Supplemental Fig. S3B). Furthermore, no protein was found with an N-terminal Cys. In contrast, Ser occurred significantly more often in the experimental data set compared with the prediction (Supplemental Fig. S3B), suggesting a regulatory mechanism to control the N-terminal amino acid in vivo.
Tables II to V present the proteins that are correctly imported and processed in the two albino mutants ppi2 and tic56-1. From these data, we inferred import to the thylakoid lumen. We observed thylakoid proteins with removed bipartite targeting signals, suggesting that both import mutants have functional translocation machineries to transport proteins into the thylakoid lumen. This holds true for substrates of both the secretory (Sec) and the twin arginine translocase (Tat) pathways (Tables II and IV).
Table II. Correctly processed plastid proteins identified with TAILS in ppi2 (thylakoid proteins).
Chloroplast proteins are listed that were identified in their mature (i.e. correctly processed) form on the basis of their N-terminal peptides in ppi2. Most processing events of nucleus-encoded plastid proteins are consistent with the ChloroP prediction or the UniProt entry for thylakoid lumenal proteins, arguing for functional import and functional processing peptidases. Thylakoid proteins were classified according to AT_CHLORO (Ferro et al., 2010). Proteins were classified into pathways using MapMan (Thimm et al., 2004).
| Identifier | Description | Start, TAILS | Start, ChloroP/UniProt | MapMan Classification/Thylakoid Import Pathway |
|---|---|---|---|---|
| AT1G03600.1 | PSB27/PSII family protein | 69 | 68/69 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT1G31330.1 | PSAF/PSI subunit F | 68 | 33/68 | PS.lightreaction.photosystem I.PSI polypeptide subunits/ not assigned |
| AT1G54780.1 | TLP18.3/thylakoid lumen 18.3-kD protein | 85 | 80/85 | not assigned.no ontology/ Sec |
| AT1G71500.1 | Rieske (2Fe-2S) domain-containing protein | 64 | 63/– | misc. other Ferredoxins and Rieske domain/ not assigned |
| AT2G01140.1 | Aldolase superfamily protein | 42 | 41/40 | PS.calvin cycle.aldolase/ not assigned |
| AT2G17630.1 | Pyridoxal phosphate-dependent transferase superfamily protein | 51 | 50/– | amino acid metabolism.synthesis.Ser-Gly-Cys group.Ser.phospho-Ser aminotransferase/ not assigned |
| AT4G02530.1 | Chloroplast thylakoid lumen protein | 74 | 39/74 | not assigned.no ontology/ Tat |
| AT4G04020.1 | FIB/fibrillin | 56 | 56/56 | cell.organization/ not assigned |
| AT4G05180.1 | PSBQ, PSBQ-2, PSII-Q/PSII subunit Q-2 | 83 | 49/85 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT4G09650.1 | ATPD/ATP synthase δ-subunit gene | 48 | 49/– | PS.lightreaction.ATP synthase.delta chain/ not assigned |
| AT4G21280.1 | PSBQ, PSBQA, PSBQ-1/PSII subunit QA | 76 | 45/78 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT5G42650.1 | AOS, CYP74A, DDE2/allene oxide synthase | 34 | 33/22 | hormone metabolism.jasmonate.synthesis-degradation.allene oxidase synthase/ not assigned |
Table V. Correctly processed plastid proteins identified with TAILS in tic56-1 (other chloroplast proteins).
Chloroplast proteins are listed that were identified in their mature (i.e. correctly processed) form on the basis of their N-terminal peptides in tic56-1. Most processing events of nucleus-encoded plastid proteins are consistent with the ChloroP prediction, arguing for functional import and functional processing peptidases. Proteins were classified into pathways using MapMan (Thimm et al., 2004).
| Identifier | Description | Start, TAILS | Start, ChloroP | MapMan Classification |
|---|---|---|---|---|
| AT1G05385.1 | LPA19, Psb27-H1/PSII 11-kD protein-related | 68 | 65 | PS.lightreaction.photosystem II.PSII polypeptide subunits |
| AT1G08490.1 | ATSUFS, SUFS, ATCPNIFS, ATNFS2, CPNIFS/chloroplastic NIFS-like Cys desulfurase | 38 | 36 | signaling.in sugar and nutrient physiology |
| AT1G08640.1 | CJD1/chloroplast J-like domain1 | 60 | 59 | not assigned.unknown |
| AT1G09795.1 | ATATP-PRT2, HISN1B, ATP-PRT2/ATP phosphoribosyl transferase2 | 57 | 58 | amino acid metabolism.synthesis.His.ATP phosphoribosyl transferase |
| AT1G09830.1 | Glycinamide ribonucleotide (GAR) synthetase | 76 | 75 | nucleotide metabolism.synthesis.purine.GAR Synthetase |
| AT1G19870.1 | iqd32/IQ-domain32 | 56 | 59 | signaling.calcium |
| AT1G24360.1 | NAD(P)-binding Rossmann-fold superfamily protein | 58 | 58 | lipid metabolism.FA synthesis and FA elongation.ACP oxoacyl reductase |
| AT1G50900.1 | GDC1/ankyrin repeat family protein | 66 | 69 | not assigned.unknown |
| AT1G55805.1 | BolA-like family protein | 54 | 51 | not assigned.no ontology |
| AT1G67280.1 | Glyoxalase/bleomycin resistance protein/dioxygenase superfamily protein | 62 | 64 | Biodegradation of Xenobiotics.lactoylglutathione lyase |
| AT1G78630.1 | emb1473/ribosomal protein L13 family protein | 58 | 57 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L13 |
| AT2G14880.1 | SWIB/MDM2 domain superfamily protein | 44 | 44 | not assigned.no ontology |
| AT2G15620.1 | NIR1, NIR, ATHNIR/nitrite reductase1 | 28 | 26 | N-metabolism.nitrate metabolism.nitrite reductase |
| AT2G28000.1 | CPN60A, CH-CPN60A, SLP/chaperonin-60α | 46 | 46 | PS.calvin cycle.Rubisco interacting |
| AT2G35040.1 | AICARFT/IMPCHase bienzyme family protein | 51 | 51 | nucleotide metabolism.synthesis.purine.AICAR transformylase |
| AT2G35450.1 | Catalytics; hydrolases | 47 | 47 | not assigned.no ontology |
| AT2G35500.1 | SKL2/shikimate kinase-like2 | 71 | 72 | amino acid metabolism.synthesis.aromatic aa.chorismate.shikimate kinase |
| AT2G37660.1 | NAD(P)-binding Rossmann-fold superfamily protein | 70 | 70 | not assigned.unknown |
| AT2G40490.1 | HEME2/uroporphyrinogen decarboxylase | 37 | 36 | tetrapyrrole synthesis.uroporphyrinogen decarboxylase |
| AT2G44650.1 | CHL-CPN10, CPN10/chloroplast chaperonin10 | 42 | 40 | protein.folding |
| AT2G45290.1 | Transketolase | 67 | 66 | PS.calvin cycle.transketolase |
| AT3G07480.1 | 2Fe-2S ferredoxin-like superfamily protein | 27 | 26 | not assigned.unknown |
| AT3G08740.1 | Elongation factor P (EF-P) family protein | 52 | 51 | protein.synthesis.elongation |
| AT3G10670.1 | ATNAP7, NAP7/nonintrinsic ABC protein7 | 67 | 67 | protein.assembly and cofactor ligation |
| AT3G14210.1 | ESM1/epithiospecifier modifier1 | 29 | 28 | secondary metabolism.sulfur-containing.glucosinolates.degradation.myrosinase |
| AT3G21200.1 | PGR7/proton gradient regulation7 | 43 | 42 | not assigned.unknown |
| AT3G25920.1 | RPL15/ribosomal protein L15 | 66 | 66 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L15 |
| AT3G32930.1 | Unknown protein | 62 | 62 | not assigned.unknown |
| AT3G48420.1 | Haloacid dehalogenase-like hydrolase (HAD) superfamily protein | 67 | 66 | not assigned.no ontology |
| AT3G51140.1 | Protein of unknown function (DUF3353) | 69 | 67 | not assigned.unknown |
| AT3G54660.1 | GR, EMB2360, ATGR2/glutathione reductase | 75 | 74 | redox.ascorbate and glutathione.glutathione |
| AT3G58010.1 | PGL34/plastoglobulin 34 kD | 56 | 54 | not assigned.unknown |
| AT3G58140.1 | Phenylalanyl-tRNA synthetase class IIc family protein | 54 | 54 | protein.aa activation.Phe-tRNA ligase |
| AT3G58990.1 | IPMI1/isopropylmalate isomerase1 | 57 | 57 | secondary metabolism.sulfur-containing.glucosinolates.synthesis.aliphatic.methylthioalkylmalate isomerase small subunit (MAM-IS) |
| AT3G61440.1 | ATCYSC1, ARATH;BSAS3;1, CYSC1/Cys synthase C1 | 26 | 25 | amino acid metabolism.synthesis.Ser-Gly-Cys group.Cys.OASTL |
| AT4G01310.1 | Ribosomal L5P family protein | 43 | 40 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L5 |
| AT4G03200.1 | Catalytics | 62 | 62 | not assigned.unknown |
| AT4G17560.1 | Ribosomal protein L19 family protein | 72 | 72 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L19 |
| AT4G21445.1 | Unknown protein | 55 | 54 | not assigned.unknown |
| AT4G25370.1 | Double Clp-N motif protein | 65 | 64 | protein.targeting.unknown |
| AT4G25840.1 | GPP1/glycerol-3-phosphatase1 | 56 | 54 | N-metabolism.ammonia metabolism.unspecified |
| AT4G26900.1 | AT-HF, HISN4/HIS HF | 56 | 56 | amino acid metabolism.synthesis.His.N-5-phosphoribosyl-formimino-5-aminoimidazole-4-carboxamide ribonucleotide isomerase |
| AT4G30490.1 | AFG1-like ATPase family protein | 66 | 67 | not assigned.no ontology |
| AT5G01600.1 | ATFER1, FER1/ferretin1 | 49 | 48 | metal handling.binding, chelation and storage |
| AT5G03370.1 | Acylphosphatase family | 65 | 65 | not assigned.no ontology |
| AT5G23040.1 | CDF1/protein of unknown function (DUF3353) | 49 | 48 | cell.cell death.plants |
| AT5G30510.1 | RPS1, ARRPS1/ribosomal protein S1 | 45 | 44 | protein.synthesis.ribosomal protein.prokaryotic.unknown organellar.30S subunit.S1 |
| AT5G47190.1 | Ribosomal protein L19 family protein | 72 | 72 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L19 |
| AT5G48300.1 | ADG1, APS1/ADP Glc pyrophosphorylase1 | 72 | 71 | major CHO metabolism.synthesis.starch.AGPase |
| AT5G51110.1 | Transcriptional coactivator/pterin dehydratase | 51 | 51 | not assigned.unknown |
| AT5G52840.1 | NADH-ubiquinone oxidoreductase-related | 12 | 11 | mitochondrial electron transport / ATP synthesis.NADH-DH.localization not clear |
| AT5G52920.1 | PKP1, PKP-β1, PKP2/plastidic pyruvate kinase β-subunit 1 | 65 | 64 | glycolysis.plastid branch.pyruvate kinase (PK) |
| AT5G54770.1 | THI1, TZ, THI4/thiazole biosynthetic enzyme, chloroplast (ARA6, THI1, THI4) | 46 | 46 | Cofactor and vitamine metabolism.thiamine |
| AT5G54810.1 | TSB1, TRPB, TRP2, ATTSB1/Trp synthase β-subunit 1 | 54 | 53 | amino acid metabolism.synthesis.aromatic aa.Trp.Trp synthase |
| AT5G63980.1 | SAL1, ALX8, ATSAL1, HOS2, FRY1, RON1/inositol monophosphatase family protein | 56 | 54 | nucleotide metabolism.degradation |
| AT5G66120.2 | 3-Dehydroquinate synthase, putative | 59 | 59 | amino acid metabolism.synthesis.aromatic aa.chorismate.3-dehydroquinate synthase |
Table III. Correctly processed plastid proteins identified with TAILS in ppi2 (other chloroplast proteins).
Chloroplast proteins are listed that were identified in their mature (i.e. correctly processed) form on the basis of their N-terminal peptides in ppi2. Most processing events of nucleus-encoded plastid proteins are consistent with the ChloroP prediction, arguing for functional import and functional processing peptidases. Proteins were classified into pathways using MapMan (Thimm et al., 2004).
| Identifier | Description | Start, TAILS | Start, ChloroP | MapMan Classification |
|---|---|---|---|---|
| AT1G09795.1 | ATATP-PRT2, HISN1B, ATP-PRT2/ATP phosphoribosyl transferase2 | 57 | 58 | amino acid metabolism.synthesis.His.ATP phosphoribosyl transferase |
| AT1G24360.1 | NAD(P)-binding Rossmann-fold superfamily protein | 60 | 58 | lipid metabolism.FA synthesis and FA elongation.ACP oxoacyl reductase |
| AT1G73110.1 | P-loop-containing nucleoside triphosphate hydrolase superfamily protein | 41 | 40 | PS.calvin cycle.Rubisco interacting |
| AT2G14880.1 | SWIB/MDM2 domain superfamily protein | 46 | 44 | not assigned.no ontology |
| AT2G28000.1 | CPN60A, CH-CPN60A, SLP/chaperonin-60α | 47 | 46 | PS.calvin cycle.Rubisco interacting |
| AT2G35500.1 | SKL2/shikimate kinase-like2 | 71 | 72 | amino acid metabolism.synthesis.aromatic aa.chorismate.shikimate kinase |
| AT2G37660.1 | NAD(P)-binding Rossmann-fold superfamily protein | 70 | 70 | not assigned.unknown |
| AT3G08740.1 | Elongation factor P (EF-P) family protein | 52 | 51 | protein.synthesis.elongation |
| AT3G21200.1 | PGR7/proton gradient regulation7 | 43 | 42 | not assigned.unknown |
| AT3G25920.1 | RPL15/ribosomal protein L15 | 68 | 66 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L15 |
| AT3G32930.1 | Unknown protein | 63 | 62 | not assigned.unknown |
| AT3G58010.1 | PGL34/plastoglobulin 34 kD | 56 | 54 | not assigned.unknown |
| AT3G61440.1 | ATCYSC1, ARATH;BSAS3;1, CYSC1/Cys synthase C1 | 26 | 25 | amino acid metabolism.synthesis.Ser-Gly-Cys group.Cys.OASTL |
| AT4G01310.1 | Ribosomal L5P family protein | 43 | 40 | protein.synthesis.ribosomal protein.prokaryotic.chloroplast.50S subunit.L5 |
| AT4G14680.1 | APS3/pseudouridine synthase/archaeosine transglycosylase-like family protein | 51 | 50 | S-assimilation.ATPS |
| AT4G21445.1 | Unknown protein | 55 | 54 | not assigned.unknown |
| AT4G23940.1 | FtsH extracellular protease family | 55 | 54 | protein.degradation.metalloprotease |
| AT4G25370.1 | Double Clp-N motif protein | 65 | 64 | protein.targeting.unknown |
| AT4G26900.1 | AT-HF, HISN4/HIS HF | 57 | 56 | amino acid metabolism.synthesis.His.N-5-phosphoribosyl-formimino-5-aminoimidazole-4-carboxamide ribonucleotide isomerase |
| AT4G34350.1 | CLB6, ISPH, HDR/4-hydroxy-3-methylbut-2-enyl diphosphate reductase | 40 | 39 | secondary metabolism.isoprenoids.nonmevalonate pathway.HDR |
| AT5G03370.1 | Acylphosphatase family | 67 | 65 | not assigned.no ontology |
| AT5G23040.1 | CDF1/protein of unknown function (DUF3353) | 49 | 48 | cell.cell death.plants |
| AT5G30510.1 | RPS1, ARRPS1/ribosomal protein S1 | 45 | 44 | protein.synthesis.ribosomal protein.prokaryotic.unknown organellar.30S subunit.S1 |
| AT5G51110.1 | Transcriptional coactivator/pterin dehydratase | 51 | 51 | not assigned.unknown |
| AT5G52840.1 | NADH-ubiquinone oxidoreductase-related | 12 | 11 | mitochondrial electron transport / ATP synthesis.NADH-DH.localization not clear |
| AT5G52920.1 | PKP1, PKP-β1, PKP2/plastidic pyruvate kinase β-subunit 1 | 65 | 64 | glycolysis.plastid branch.pyruvate kinase (PK) |
| AT5G54770.1 | THI1, TZ, THI4/thiazole biosynthetic enzyme, chloroplast (ARA6, THI1, THI4) | 47 | 46 | Cofactor and vitamine metabolism.thiamine |
| AT5G54810.1 | TSB1, TRPB, TRP2, ATTSB1/Trp synthase β-subunit 1 | 54 | 53 | amino acid metabolism.synthesis.aromatic aa.Trp.Trp synthase |
| AT5G63980.1 | SAL1, ALX8, ATSAL1, HOS2, FRY1, RON1/inositol monophosphatase family protein | 56 | 54 | nucleotide metabolism.degradation |
| AT5G66120.2 | 3-Dehydroquinate synthase, putative | 60 | 59 | amino acid metabolism.synthesis.aromatic aa.chorismate.3-dehydroquinate synthase |
Several of the processed substrates were found in both mutants; however, for some of them, the starting amino acid turned out to be different in ppi2 and tic56-1 (Tables II–V). Indeed, whenever the starting amino acid of a protein in ppi2 differed from tic56-1 or the wild type, one or two amino acids were lacking (Tables II–V; Supplemental Table S1). This indicates that the potential regulatory mechanism controlling the N-terminal amino acid in vivo might function differently in the two import mutants.
Correct removal of the transit peptide is strong evidence for the import of a substrate into chloroplasts; however, it is possible that some precursor proteins become processed before they have completely passed across the envelope. To check if plastid proteins in tic56-1 are exposed at the organellar surface, we treated a crude plastid preparation of the wild type and tic56-1 with thermolysin, a protease not penetrating the outer envelope (Cline et al., 1984). Subsequently, the proteome of thermolysin-treated plastids was analyzed by liquid chromatography-tandem mass spectrometry on an Orbitrap Velos device (Thermo Scientific). Here, we found many peptides mapping to the C-terminal region of plastid proteins in the thermolysin-treated tic56-1 sample, indicating that these proteins were fully imported and, therefore, protected against proteolysis (Supplemental Fig. S4A). Some of the chloroplast proteins from the tic56-1 TAILS experiment recurred in this analysis, including 29 proteins that were found correctly processed by TAILS (Tables II–V; Supplemental Table S3). Together, 225 plastid proteins were identified (Supplemental Table S3) in addition to the TAILS data. Therefore, this analysis extends the list of proteins residing in the undeveloped plastids of tic56-1.
Overlap of Substrates Imported by tic56-1 and ppi2
It is still an open question if substrate selectivity exists at the level of the TIC complexes (e.g. if a Tic110-containing complex or different Tic20 complexes select and transport different classes of substrates). Therefore, we analyzed the list of imported proteins from tic56-1. Similar to a previous study with ppi2, the imported proteins are involved in different pathways and no functional category is overrepresented or underrepresented (Bischof et al., 2011; Tables II–V). Furthermore, we could not define any transit peptide properties (physicochemical properties, structural aspects, or sequence conservation) that would mediate a specific import of these proteins into tic56-1. The same is true for ppi2 (Bischof et al., 2011; Tables II–V). Many different substrates were found to be imported, and a remarkable overlap of substrates imported by tic56-1 and ppi2 was observed (Fig. 4B). This is surprising, because the two mutants are blocked at different sites of the chloroplast import machinery, which suggests partial convergence of the import routes dependent on Toc159 and Tic56, supporting their joint function in a supercomplex.
Import Assays with Another Tic56-Deficient Mutant Confirm Functional Import in the Absence of Native Tic56
Taken together, our data demonstrate a remarkable import ability of the tic56-1 mutant. For this reason, we decided to reexamine the import ability of another tic56 mutant, tic56-3 (Kikuchi et al., 2013), by a classical in vitro import assay using the precursor of the small subunit of Rubisco (preSSu) and the α-subunit of pyruvate dehydrogenase E1α (preE1α) as substrates. The tic56-3 mutant has a pale-green phenotype (Kikuchi et al., 2013; Fig. 5A). In this mutant, intact full-length Tic56 is lacking but low levels of truncated (or elongated) Tic56 occur (Kikuchi et al., 2013; Figs. 1B and 5, C and D), which could explain the mild phenotype when compared with tic56-1. We chose this mutant because, in contrast to tic56-1, intact Tic56-deficient chloroplasts can be isolated. Thus, the role of Tic56 in chloroplast import can be assessed directly by in vitro import studies with tic56-3 chloroplasts. We could not observe any drastic import defect of tic56-3 plastids for the two substrates tested (Fig. 5A), although no intact Tic56 and only low levels of modified Tic56 could be detected in the plastids used for the import reactions by western blotting (Fig. 5, C and D). Our data differ from those in the study of Kikuchi et al. (2013), who found that the import rate of preSSu-dihydrofolate reductase into tic56-3 chloroplasts was somewhat reduced. Therefore, we chose a protoplast-based assay as a second test to assess the import capacity of tic56-3 plastids.
Figure 5.
Import ability of chloroplasts isolated from the pale-green tic56-3 mutant. A, In vitro chloroplast protein import assay with chloroplasts isolated from 27-d-old wild-type and tic56-3 seedlings. For the import reactions, the chloroplast suspensions of the two plant types were adjusted to equal protein levels. The chloroplasts were incubated with two different radiolabeled import substrates, preE1α (top) and preSSu (bottom), and import was allowed to proceed for 0, 7.5, or 15 min. The samples were analyzed by SDS-PAGE and autoradiography. As a loading control, part of the Coomassie Blue-stained SDS-PAGE gel is shown underneath the corresponding autoradiograph. IVT, Product of in vitro translation. The photographs above the gels show the phenotypes of the tic56-3 Arabidopsis mutant in comparison with the wild type (ecotype Wassilewskija) grown on Murashige and Skoog (MS) agar supplemented with 0.8% (w/v) Suc for 14 d under an 8-h photoperiod. B, Quantification of the imported, processed form of the substrates at 15 min of import. The amount of imported substrate in the wild-type sample was set to 100%. Data were derived from three independent experiments. C, Chloroplasts isolated from 27-d-old tic56-3 seedlings have strongly reduced levels of Tic56 or the truncated form of Tic56 as monitored by western blotting. Fifty micrograms of chloroplast protein of tic56-3 and wild-type chloroplasts from two independent preparations was analyzed by western blotting with anti-Tic56 antiserum. D, Chloroplast were lysed hypotonically and separated into soluble (S) and membrane protein (P) fractions by centrifugation. Chloroplasts (cp) and subfractions were subjected to western-blot analysis with anti-Tic56 antiserum. The 130-kD protein and the truncated form of Tic56 (Tic56-TF) became apparent in the membrane protein fraction of tic56-3 chloroplasts (asterisks; compare with Fig. 1B, 16-d-old plants).
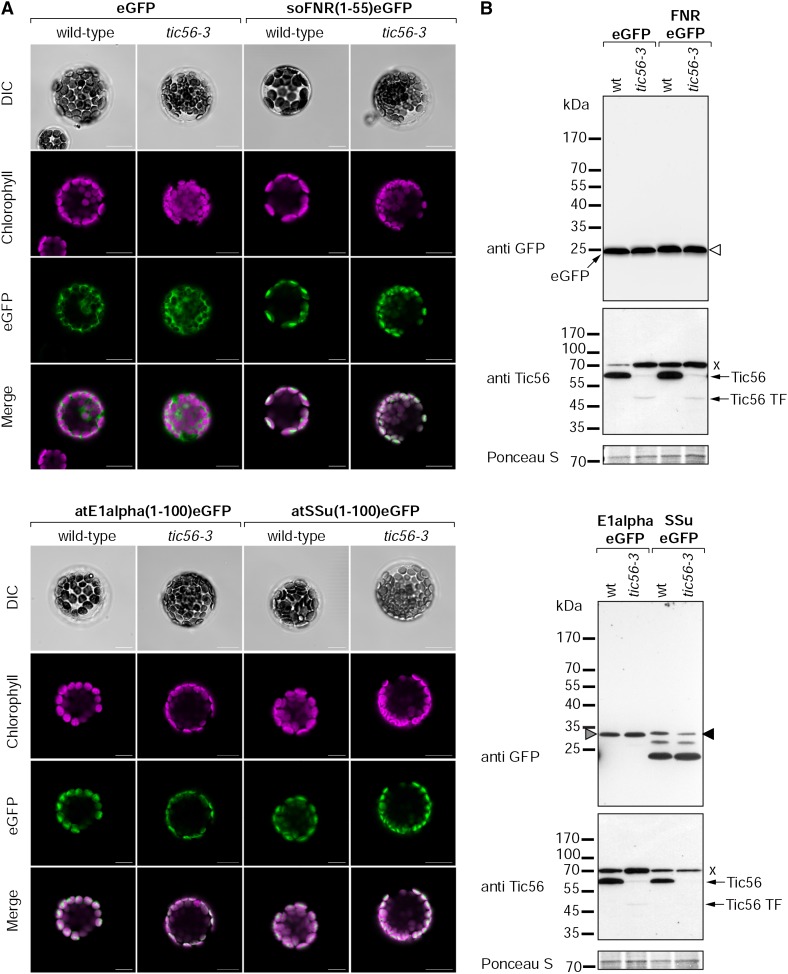
Here, protoplasts of the wild type and tic56-3 were transformed with plasmids coding for the most N-terminal amino acids of three different import substrates fused to enhanced GFP (eGFP) under the control of the strong cauliflower mosaic virus 35S promotor. We employed eGFP fusions of Arabidopsis SSu (amino acids 1–100), the α-subunit of pyruvate dehydrogenase E1α (Arabidopsis E1α; amino acids 1–100), and ferredoxin-NADP+-oxidoreductase (spinach [Spinacia oleracea] FNR; amino acids 1–55). As control, protoplasts were transformed with a plasmid coding for eGFP only. Twenty hours after transformation, the eGFP signals of all three import substrates localized nearly completely within the chloroplasts in wild-type as well as tic56-3 protoplasts (Fig. 6A). Western-blot analyses of protoplast protein extracts with anti-GFP revealed no difference in the migration pattern of the substrates between the wild type and tic56-3 (Fig. 6B). In both samples, the substrates were running at the expected sizes of the processed forms lacking the transit peptide (Fig. 6B, anti GFP, triangles). In protoplasts transformed with SSu(1-100)eGFP, extra bands were detected with anti-GFP, which are most likely the result of successive degradation of the artificial import substrate inside plastids. We monitored the level of Tic56 in the transformed protoplasts by western blotting with anti-Tic56 antiserum and detected only the truncated form of Tic56 at drastically reduced levels when compared with Tic56 detected in the wild-type samples (Fig. 6B, anti Tic56). The faint band detected with anti-Tic56 in the tic56-3 samples running at the same size of Tic56 represents a cross reaction of the antiserum with a stromal protein (Fig. 5D). All in all, our data demonstrate that, despite the high expression level of precursor proteins and the lack of Tic56 in tic56-3 protoplasts, the substrates tested were efficiently imported into chloroplasts and did not accumulate as unprocessed precursor proteins in the cytosol. This is in line with the in vitro chloroplast import assays (Fig. 5) and the tic56-1 TAILS data (Fig. 4; Tables II–V), thus arguing for functional import in the absence of Tic56 and, in case of tic56-1, in the absence of the 1-MD complex. Therefore, our data illustrate the complexity of the plastid import machinery and suggest that alternative TOC/TIC import supercomplexes exist or may form.
Figure 6.
In vivo chloroplast targeting of three different import substrates in wild-type (wt) and tic56-3 protoplasts. A, Arabidopsis protoplasts were transformed with constructs encoding eGFP or N-terminal fragments of spinach FNR (amino acids 1–55), E1α (amino acids 1–100), and SSu (amino acids 1–100) fused to eGFP. The localization of the reporter proteins was examined by confocal laser scanning microscopy 20 h after transformation. Chlorophyll, Chlorophyll autofluorescence; DIC, differential interference contrast; eGFP, eGFP fluorescence; Merge, superposition of chlorophyll and eGFP signals. Bars = 10 µm. B, Western-blot analysis of transformed protoplasts with an anti-GFP antibody and anti-Tic56 antiserum. A section of the Ponceau S-stained membrane is shown as a loading control. The triangles mark bands running at the expected sizes of the processed forms of the three substrates [white, eGFP; gray, E1α(62-100)eGFP; black, SSu(55-100)eGFP]. Each x indicates a nonspecific cross reaction of the anti-Tic56 antiserum with a 70-kD protein (Fig. 1B).
DISCUSSION
Protein Import in the Absence of Tic56 Disagrees with the Model of a General TIC Translocon
During precursor protein import, the TOC complexes associate with TIC complexes in the inner envelope membrane to form TOC-TIC supercomplexes that allow the translocation of protein substrates across the two membranes (Schnell et al., 1994; Kouranov et al., 1998). In this study, we provide additional evidence for the formation of TOC-TIC supercomplexes. We were able to isolate Toc159 in association with several different TIC components in the absence of precursor protein (Fig. 1). Our data, therefore, confirm that TOC-TIC supercomplexes exist independently of precursor protein translocation (Kouranov et al., 1998). The organization of the TIC complexes that associate with TOC into such supercomplexes is currently not understood. Tic20 assembles into a 1-MD translocation complex, at the inner membrane of which new subunits were recently identified (Kikuchi et al., 2013). In earlier studies, Tic20 was found in TOC-TIC supercomplexes together with Tic110, Tic40, and Tic22 (Kouranov et al., 1998). Tic110, an essential TIC component, was proposed to have a function as a translocation channel (Heins et al., 2002). However, structural data (Tsai et al., 2013) argue against this. It is more certain that Tic110 together with the cochaperone Tic40 has a role in the recruitment of stromal chaperones to the import sites (Chou et al., 2006). Initially, it was suggested that the Tic20 complex functions between the TOC complex and a Tic110 complex comprising Tic110, Tic40, and chaperones in preprotein translocation (Kikuchi et al., 2009). However, in a more recent study, Kikuchi et al. (2013) challenge a direct participation of Tic110 in the translocation process and propose the 1-MD Tic20 complex as the TIC translocon. This denomination is mainly based on the observation that Tic110 is absent in the Tic20 complex and does not comigrate in native PAGE (Kikuchi et al., 2013). Our findings support earlier data showing that Tic110 and Tic40 are part of the translocon. Our data could be explained by an association of Tic20 with Tic110 mediated by the TOC complex, which has been observed before (Kouranov et al., 1998), and point to a dynamic formation of the TOC-TIC supercomplexes (Paila et al., 2014). However, we cannot exclude the possibility that the TAP-Toc159 preparation contains different Toc159 complexes, one with Tic110 and Tic40 and a separate one with the 1-MD TIC complex. If this is the case, it would be an interesting task to determine what controls the association of the TOC complex with one or the other TIC complex.
The flexibility of the chloroplast import machinery is further reflected by our data on the protein import ability of the tic56-1 mutant. We find a remarkably high protein import capability into plastids of tic56-1 plants, despite the absence of the 1-MD Tic20 complex in this mutant (Kikuchi et al., 2013). This conclusion is based on the numerous correctly imported proteins identified by TAILS (Tables II–V). Thus, an intact 1-MD Tic20 complex is not necessarily required for chloroplast protein import, suggesting that its activity can be taken over by other constituents of the inner envelope membrane protein translocation machinery. This could be either the residual amounts of the unassembled components of the 1-MD complex in tic56-1 (e.g. Tic20-I alone or its homolog Tic20-IV). Furthermore, Tic21 or a channel protein in the Tic110-Tic40-chaperone complex might have a role in maintaining the import capability in this mutant. Taken together, these results show that plastid protein translocation can take place despite the lack of Tic56 function, thus excluding the 1-MD complex as a unique TIC translocon.
Similarly, tic56-3 plastids revealed no significant reduction in the import of different substrates in in vitro (Fig. 5) and in vivo (Fig. 6) import assays despite a shortage in Tic56. While we lack information on the state of the 1-MD TIC complex in tic56-3, our data clearly show that wild-type levels of native Tic56 are not required to allow for functional protein import, at least not in developed chloroplasts of plants grown under the given conditions (4 weeks old, short days). Presumably, the strong phenotype of tic56-1 compared with tic56-3 is caused by a default of a specific function of Tic56 during an early phase of chloroplast development that can be fulfilled by the low levels of truncated Tic56 in tic56-3.
A Large Variety of Substrates Is Imported in tic56-1 and ppi2
As mentioned above, translocon selectivity at the level of the TOC complex has been both hypothesized and demonstrated (Jarvis et al., 1998; Bauer et al., 2000; Kubis et al., 2003, 2004; Ivanova et al., 2004; Inoue et al., 2010). However, until now, a clear classification of substrates and of sequence determinants for the different TOC import routes was unsuccessful (Bischof et al., 2011). In addition, a recent yeast (Saccharomyces cerevisiae) split-ubiquitin study showed that the G domains of the TOC receptors Toc159 and Toc132 can bind both photosynthetic and nonphotosynthetic precursor proteins with overlapping specificities (Dutta et al., 2014).
In view of the existence of different, separate TIC complexes, one cannot avoid the question of whether translocon selectivity also exists at the level of the TIC complex. In fact, substrate specificity at the TIC level has already been discussed for Tic20-I and Tic20-IV (Hirabayashi et al., 2011; Kasmati et al., 2011) as well as Tic21 (Teng et al., 2012), and a preference for photosynthetic precursors was attributed to Tic20-I and Tic21 (Kikuchi et al., 2009).
In the course of this work, we employed an N-terminal enrichment strategy combined with proteomics (TAILS experiment) to identify correctly imported substrates in the tic56-1 mutant. Remarkably, many different substrates that function in various metabolic pathways were found to be imported, including several photosynthesis-related proteins (Tables II–V). Considering that Tic56 is a central component of the Tic20-I complex and is required for its integrity (Kikuchi et al., 2013), one can exclude a clear-cut preference of this complex for photosynthetic precursor proteins. This is also supported by the in vitro import studies with the tic56-3 mutants with different classes of import substrates (Figs. 5 and 6). A similar observation was made for ppi2 here (Tables II–V) and in a previous report (Bischof et al., 2011), demonstrating that Toc159 contributes to the import of a broad variety of proteins and is not restricted to photosynthetic substrates.
We also found a remarkable overlap of imported proteins in the two albino mutants blocked at the TOC (ppi2) and TIC (tic56-1) levels of the translocation machinery (Fig. 4B; Tables II–V). This might suggest a common function of Toc159 and Tic56 in the same import pathway, which fits well with the occurrence of the two proteins in one TOC-TIC supercomplex as isolated from Toc159 TAP-tagged plants (Fig. 1). However, one cannot exclude that the high amount of common imported substrates detected in the two mutants is partially due to specific characteristics of the experimental system. For instance, the abundance or properties of some (N-terminal) peptides could render them more easily detectable by TAILS than others. A comparative quantitative proteomics approach with the two mutants could be helpful to complement the TAILS data.
The detection of a nucleus-encoded chloroplast peptidase (AT5G56730.1) as unprocessed precursor protein in both mutants (Table I) further points to a common function of Toc159 and Tic56. Such an accumulation of unprocessed precursor proteins in the cytosol was reported previously for Toc159-deficient ppi2 plants (Bischof et al., 2011), and one of the precursors in that study, FERREDOXIN2 (AT1G60950.1), also was identified here in ppi2 (Table I). A second precursor protein we found in ppi2 only, ribosomal S5 family protein (AT2G33800.1), was classified as a Toc159-dependent substrate by Bischof et al. (2011). This agreement of the data further demonstrates the power of the TAILS approach, which identified chloroplast-localized β-amylase (AT4G17090.1) as another unprocessed precursor in ppi2.
The second protein identified in its unprocessed form by TAILS in tic56-1 was a plastidic 3-phosphoglycerate dehydrogenase designated EMBRYO SAC DEVELOPMENTAL ARREST9 (EDA9; Toujani et al., 2013). The fact that the phenotype of tic56-1 is milder than the phenotype of the eda9 mutant (Toujani et al., 2013) indicates that the import of EDA9 is not blocked completely in tic56-1. EDA9 is a good candidate for an import substrate that is imported by a pathway comprising Tic56 but that is independent on Toc159. However, further experimentation is required to confirm this hypothesis.
TAILS Reveals Correct Targeting to the Thylakoid Lumen in tic56-1 and ppi2 and Points to a Second Proteolytic Event after Transit Peptide Removal
The power of the TAILS approach lies in its functional implications, because N termini identified in vivo provide a direct read out of either protein import with correct processing or precursor protein accumulation. We found several imported proteins in tic56-1 and ppi2 that were correctly processed at the predicted processing site by the stromal processing protease (SPP; Tables II–V). Furthermore, in both mutants, known thylakoid lumen-localized proteins were found whose luminal targeting signal was correctly removed (Tables II and IV). While most of these are substrates of the Tat pathway, one (AT1G54780; thylakoid lumen 18.3-kD protein) turned out to be a substrate of the Sec pathway. Therefore, our results suggest that the two albino mutants ppi2 and tic56-1 have functional Tat and Sec translocation pathways as well as an active thylakoid processing peptidase (plastidic type I signal peptidase1). These findings provide indirect evidence for the identity of the membrane structures observed in tic56-1 plastids by transmission electron microscopy (Fig. 2B). The indirect evidence for the existence of a thylakoid lumen in tic56-1 and the occurrence of plastoglobules in close proximity to the observed membrane structures support their identification as (pro)thylakoid structures. This means that tic56-1 and ppi2 chloroplasts, despite their severe developmental defect (Fig. 2; Supplemental Fig. S2), have the basic compartments that make up a chloroplast.
Table IV. Correctly processed plastid proteins identified with TAILS in tic56-1 (thylakoid proteins).
Chloroplast proteins are listed that were identified in their mature (i.e. correctly processed) form on the basis of their N-terminal peptides in tic56-1. Most processing events of nucleus-encoded plastid proteins are consistent with the ChloroP prediction or the UniProt entry for thylakoid lumenal proteins, arguing for functional import and functional processing peptidases. Thylakoid proteins were classified according to AT_CHLORO (Ferro et al., 2010). Proteins were classified into pathways using MapMan (Thimm et al., 2004).
| Identifier | Description | Start, TAILS | Start, ChloroP/UniProt | MapMan Classification/Thylakoid Import Pathway |
|---|---|---|---|---|
| AT1G03130.1 | PSAD-2/PSI subunit D-2 | 45 | 44/45 | PS.lightreaction.photosystem I.PSI polypeptide subunits/ not assigned |
| AT1G03600.1 | PSB27/PSII family protein | 69 | 68/69 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT1G54780.1 | TLP18.3/thylakoid lumen 18.3-kD protein | 85 | 80/85 | not assigned.no ontology/ Sec |
| AT1G71500.1 | Rieske (2Fe-2S) domain-containing protein | 64 | 63/– | misc. other Ferredoxins and Rieske domain/ not assigned |
| AT2G01140.1 | Aldolase superfamily protein | 41 | 41/40 | PS.calvin cycle.aldolase/ not assigned |
| AT2G17630.1 | Pyridoxal phosphate-dependent transferases superfamily protein | 51 | 50/– | amino acid metabolism.synthesis.Ser-Gly-Cys group.Ser.phospho-Ser aminotransferase/ not assigned |
| AT3G44880.1 | ACD1, LLS1, PAO/pheophorbide a oxygenase family protein with Rieske [2Fe-2S] domain | 50 | 50/50 | cell.cell death.plants/ not assigned |
| AT3G56650.1 | Mog1/PsbP/DUF1795-like PSII reaction center PsbP family protein | 68 | 66/68 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT3G57560.1 | NAGK/N-acetyl-l-Glu kinase | 50 | 50/51 | nucleotide metabolism.phosphotransfer and pyrophosphatases.misc/ not assigned |
| AT4G02530.1 | Chloroplast thylakoid lumen protein | 74 | 39/74 | not assigned.no ontology/ Tat |
| AT4G02770.1 | PSAD-1/PSI subunit D-1 | 46 | 45/46 | PS.lightreaction.photosystem I.PSI polypeptide subunits/ not assigned |
| AT4G04020.1 | FIB/fibrillin | 56 | 56/56 | cell.organization/ not assigned |
| AT4G05180.1 | PSBQ, PSBQ-2, PSII-Q/PSII subunit Q-2 | 83 | 49/85 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT4G09650.1 | ATPD/ATP synthase δ-subunit gene | 48 | 49/– | PS.lightreaction.ATP synthase.delta chain/ not assigned |
| AT4G21280.1 | PSBQ, PSBQA, PSBQ-1/PSII subunit QA | 76 | 45/78 | PS.lightreaction.photosystem II.PSII polypeptide subunits/ Tat |
| AT5G08740.1 | NDC1/NAD(P)H dehydrogenase C1 | 53 | 53/53 | mitochondrial electron transport / ATP synthesis.NADH-DH.type II.mitochondrial/ not assigned |
| AT5G23120.1 | HCF136/PSII stability/assembly factor, chloroplast (HCF136) | 79 | 61/79 | PS.lightreaction.photosystem II.biogenesis/ Tat |
An important observation that we made with the TAILS experiment is the divergence between predicted (ChloroP) and established in vivo N termini (Fig. 4C; Supplemental Fig. S3). A similar observation was made during the development of ChloroP (Emanuelsson et al., 1999) and in proteome analyses probing the N termini of plastid proteins (Gómez et al., 2003; Zybailov et al., 2008; Huesgen et al., 2013). This interesting finding was interpreted in two different ways: on the one hand, it was attributed to the inaccuracy of the ChloroP prediction tool (Zybailov et al., 2008), and on the other hand, it was suggested that most imported chloroplast proteins undergo additional N-terminal proteolysis after transit peptide removal by SPP (Emanuelsson et al., 1999; Huesgen et al., 2013). Our data show that the starting amino acid aligns with the preferential exposure of stabilizing amino acids at the N terminus. Cys residues are removed from the N terminus, and the number of proteins starting with Ala as the first amino acid is significantly reduced compared with the ChloroP prediction (Supplemental Fig. S3B). As a result, we observed a shift in the starting amino acids from known stabilizing to destabilizing residues in plastids according to Apel et al. (2010). The one-amino acid shift results in the change of the N-terminal amino acid from a strong instability-conferring amino acid (Cys) to residues that confer intermediate stability (Ser and Gly) or to the stabilizing amino acid Glu (Supplemental Fig. S3B; Apel et al., 2010). Such a second proteolytic event after SPP cleavage would be very much reminiscent of the presequence trimming of mitochondrial (pre)proteins by Icp55 (for intermediate cleaving peptidase of 55 kD) in yeast (Naamati et al., 2009; Vögtle et al., 2009). This protease exposes stabilizing residues at the N terminus of intermediate processing products generated by the mitochondrial processing peptidase, thereby determining their half-lives (Vögtle et al., 2009; Venne et al., 2013). It is conceivable that a similar proteolytic system exists in chloroplasts; however, despite the existence of Icp55 homologs in plants (Kwasniak et al., 2012), a plastid-localized Icp55-like protease remains to be discovered.
CONCLUSION
Altogether, our results provide further evidence for a role of Tic56 in chloroplast protein translocation (Kikuchi et al., 2013). This is supported by its interaction with Toc159 and the intersection of substrates that are imported or not in tic56-1 and the protein import mutant ppi2. However, the tic56-1 TAILS data and the import studies with tic56-3 argue against a function of Tic56 as an essential component of a unique general TIC translocon, because considerable import was observed despite Tic56 deficiency. Our results also demonstrate that TAILS is a powerful tool to study plastid or mitochondrial protein import in mutants whose severe phenotype (e.g. albino, dwarf) hamper classical approaches such as in vitro protein import assays.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The following Arabidopsis (Arabidopsis thaliana) mutants were used: tic56-1 (SAIL_810_G07), tic56-3 (FLAG_579H12), and ppi2 (toc159; CS11072 introgressed into the Columbia-0 ecotype; Kubis et al., 2004). As control, wild-type plants of the ecotypes Wassilewskija or Columbia-0 were used. Plants were grown at 21°C under short-day conditions (8-/16-h photoperiod) on agar plates. Agar plates contained 1% (w/v) plant agar, one-half-strength MS medium, and 0.8% (w/v) Suc.
TAP Tag Purification
The transgenic plants (TAP-Toc159:ppi2) used for the isolation of TAP-Toc159-containing complexes have been described before, as has the procedure of purification and mass spectrometry analysis (Agne et al., 2010).
DNA Constructs
To obtain a vector for Tic56 antigen production, the coding sequence for Tic56 without transit peptide was amplified with forward primer 5′-GTATAGGATCCTTCTCAAAGAAGTCCC-3′ and reverse primer 5′-GAGTGCGGCCGCTCAATCTTTTTTGGAGTTGC-3′ from clone RAFL05-19-G19 (pda02782; RIKEN Bioresource Center). The insert was subsequently cloned using BamHI/NotI into pGEX-4T-2. The vector backbone of all plasmids for Arabidopsis protoplast transformation was pRT100Ω/Not/Asc (Uberlacker and Werr, 1996) containing the coding sequence of eGFP (Clontech). The plasmid pRT100Ω/Not/Asc-eGFP as well as the construct pRT100Ω/Not/Asc pRT100 encoding the first 55 amino acids of spinach (Spinacia oleracea) FNR fused to eGFP was kindly provided by Ralf Bernd Klösgen. The coding sequences for the first 100 residues of the SSu and E1α were cloned into pRT100Ω/Not/Asc-eGFP between the cauliflower mosaic virus 35S promotor region and the eGFP coding sequence. For in vitro transcription/translation of chloroplast precursor proteins, the complementary DNAs of E1α and SSu were cloned using NcoI/SalI into pET21d. An internal NcoI site in the complementary DNA of the small subunit was mutated by site-directed mutagenesis with forward primer 5′-CCGGCTCAAGCCACGATGGTCGCTCCATTCACTGG-3′ and reverse primer 5′-CCAGTGAATGGAGCGACCATCGTGGCTTGAGCCGG-3′.
Anti-Tic56 Antiserum Production
A fusion protein of glutathione S-transferase (GST) and Tic56 without transit peptide (amino acids 48–527) was overexpressed in Escherichia coli BL21(DE3). Inclusion bodies were prepared according to the method described by Palmer and Wingfield (2012). Proteins of the inclusion bodies were separated on a gradient SDS-PAGE (5%–12% acrylamide), and the most intensive bands after Coomassie Blue staining were cut out. The presence of the GST-Tic56 antigen in these slices was confirmed by mass spectrometry. The gel slices were sent to Eurogentec for polyclonal antibody production.
Protein Extraction and Immunoblotting
The plant material was shock frozen in liquid nitrogen and ground to a fine powder. Protein extraction buffer (100 mm sodium chloride, 50 mm Tris, pH 7.5, 4% [w/v] SDS, 0.1% [v/v] plant protease inhibitor cocktail [Sigma-Aldrich], and 1 mm phenylmethylsulfonyl fluoride) was added, and the samples were incubated for 10 min at 70°C while shaking. The samples were clarified by centrifugation at maximum speed in a tabletop centrifuge for 10 min at room temperature. The protein content was determined using the bicinchoninic acid assay (Serva), and proteins were chloroform/methanol precipitated (Wessel and Flügge, 1984). For SDS-PAGE, the protein pellets were dissolved in SDS sample buffer and incubated at 95°C for 5 min. Subsequent to separation by SDS-PAGE, the proteins were transferred to polyvinylidene difluoride membranes by semidry blotting (Kyhse-Andersen, 1984) or to nitrocellulose by tank blotting using Dunn carbonate buffer. Anti-Lhcb2, anti-Lhcb4, anti-RbcS, anti-PsbA, and anti-Tic40 sera were purchased from Agrisera, and monoclonal anti-actin (A0480) and monoclonal anti-GFP (G6795) sera were from Sigma-Aldrich. Other primary antibodies and antisera used in this study include anti-atTic56 (this study), anti-atTic20 (Teng et al., 2006), anti-atToc33 (Agne et al., 2009), anti-atToc75 (Hiltbrunner et al., 2001b), anti-atToc132 (from D.J. Schnell, University of Massachusetts), anti-atToc159 (Bauer et al., 2000), anti-atPgl35 (Vidi et al., 2006), and anti-atTic110 (Bauer et al., 2000). Band intensities of blots were quantified using ImageJ software (National Institutes of Health).
Transmission Electron Microscopy
Leaf segments were fixed with 3% (v/v) glutaraldehyde in sodium cacodylate buffer (SCB), pH 7.2, for 4 h at room temperature, washed with SCB, postfixed with 1% (w/v) osmium tetroxide in SCB for 1 h, dehydrated in a graded ethanol series, and embedded in epoxy resin (Spurr, 1969). After polymerization, the material was sectioned with an ultramicrotome S (Leica). Semithin sections (1 µm) were stained with 1% (w/v) Toluidine Blue and observed with an Axioskop 20 light microscope (Carl Zeiss Microscopy). Ultrathin sections (80 nm) were transferred to coated copper grids and poststained with uranyl acetate and lead citrate. The sections were observed using a LIBRA 120 device (Carl Zeiss Microscopy) operating at 120 kV. Images were taken applying a Dual-Speed on axis SSCCD camera (BM-2k-120; TRS).
Transient Expression of eGFP Fusion Proteins in Arabidopsis Protoplasts
Protoplast isolation and transformation were done using a polyethylene glycol-based method adapted from the protocols of Jin et al. (2001) and Yoo et al. (2007). Twenty-six-day-old wild-type (Wassilewskija) and tic56-3 plants grown on agar plates were harvested in enzyme buffer without enzymes (400 mm sorbitol, 5 mm MES, and 8 mm calcium chloride, pH 5.6). After cutting the leaves with a razor blade, the buffer was replaced by the same buffer containing 1.5% (w/v) Cellulase Onozuka R-10 (Serva) and 0.375% (w/v) Macerozyme R-10 (Serva). After two vacuum infiltration steps (5 min, −800 mbar), digestion was allowed to proceed for 4 h at room temperature. Protoplasts were collected by gentle shaking, filtering through a 50-µm mesh, and centrifugation for 5 min at 100g. Protoplasts were washed once in 5 to 15 mL of W5 solution (154 mm sodium chloride, 125 mm calcium chloride, 5 mm potassium chloride, 5 mm Glc, and 1.5 mm MES, pH 5.6) and resuspended in cold W5 solution to a final concentration of 2 × 106 protoplasts mL−1. After incubation for 30 min on ice, the protoplasts were sedimented by centrifugation and resuspended in the same volume of MaMg solution (400 mm sorbitol, 15 mm magnesium chloride, and 5 mm MES, pH 5.6). For each transformation, 400 µL of protoplast suspension was combined with 40 µg of plasmid and subsequently carefully mixed with 440 µL of PEG-CMS (1 g of polyethylene glycol 4000, 375 µL of water, 1 mL of 500 mm sorbitol, and 250 µL of 1 m calcium nitrate). After incubation for 20 min at room temperature, the protoplasts were washed by stepwise addition of W5 solution and centrifugation for 2 min with 100g. The protoplasts were washed in 4 mL of protoplast culture medium (4.4 g L−1 MS medium, 350 mm sorbitol, 50 mm Glc, 3 mm calcium chloride, and 50 µg mL−1 ampicillin, pH 5.8) and finally resuspended in 2 mL of protoplast culture medium. After an incubation of approximately 20 h, the samples were analyzed using confocal laser scanning microscopy. Twenty-four hours after transformation, protoplast were collected by centrifugation for 5 min at 500g and resuspended in 50 mm Tris-HCl, pH 6.8, 10 mm dithiothreitol, and 0.2% (v/v) plant protease inhibitor cocktail P9599 (Sigma-Aldrich) prior to chloroform/methanol precipitation (Wessel and Flügge, 1984). Fifty micrograms of protoplast protein was used for anti-GFP and anti-Tic56 western-blot analyses.
Confocal Laser Scanning Microscopy
Protoplasts were observed using a Plan-Apochromat 40×/0.95 objective on an LSM-780 confocal microscope (Zeiss). The chlorophyll fluorescence was excited with a helium-neon laser (633 nm) and the eGFP with an argon laser (488 nm). Emission wavelengths were 647 to 721 nm for chlorophyll and 493 to 598 nm for eGFP. Data from both channels were collected simultaneously in one scan event. After acquisition, images were processed using Carl Zeiss ZEN lite 2012 software.
TAILS
TAILS was done in two experiments with two biological replicates for tic56-1 compared with the wild type. In the second experiment, the ppi2 mutant was analyzed in addition. First, total protein was extracted from 8-week-old plant material. Plant material was frozen in liquid nitrogen and ground with a pestle. In the first experiment, HEPES/NaOH buffer (pH 7) was added; in the second experiment, the buffer contained additionally 0.1% (v/v) plant protease inhibitor cocktail from Sigma-Aldrich, 1 mm phenylmethylsulfonyl fluoride, and 4% (w/v) SDS to solubilize membrane proteins. The extract was clarified from the remaining plant material by centrifugation. From each plant line, 100 µg of total protein was used for further steps. The N-terminal blocking, protein digestion, peptide filtering, and sample desalting were done as described (Doucet et al., 2011). In the first experimental setup, the protein samples from both plant lines were blocked using different isotopes of formaldehyde (wild type, 12C1H2O; tic56-1, 13C2H2O). Afterward, both samples were combined and treated together in further steps. In the second experimental setup, all three protein samples were blocked using normal formaldehyde and treated separately. The samples were analyzed on an LTQ Orbitrap Velos device (Thermo Scientific). The desalted and completely dried samples were resuspended in water with 2% (v/v) acetonitrile and 0.1% (v/v) formic acid. Samples were separated by liquid chromatography using C18 columns from Proxeon (guard column, 2 cm, with i.d. of 100 µm, 5 µm; analytical column, 10 cm, with i.d. of 75 µm, 3 µm). For separation, a gradient consisting of water with 0.1% (v/v) formic acid (A) and acetonitrile with 0.1% (v/v) formic acid (B) was used (0–150 min, 5%–40% B; 150–160 min, 40%–80% B; 160–170 min, 80% B). Precursor detection was done in a mass-to-charge ratio range from 400 to 1,850, and the 20 most intensive signals were used for tandem mass spectrometry.
TAILS Data Processing
The raw files were analyzed using Proteome Discoverer 1.2 (Thermo Scientific) with the search engine SEQUEST and with MaxQuant 1.3.0.5 (Cox and Mann, 2008) using the search engine Andromeda (Cox et al., 2011). In both cases, The Arabidopsis Information Resource 10 database was used, and the false discovery rate of the data set was set to 5%. As variable modifications, N-terminal acetylation and Met oxidation, and as fixed modifications, carbamidomethylation of Cys, dimethylation of N termini or on Lys side chains, originating from the blocking procedure, were allowed. In Proteome Discoverer, the precursor mass tolerance was set to 7 ppm and the fragment mass tolerance to 0.8 D. We accepted semitryptic peptides with a maximum number of one missed cleavage. Using MaxQuant, Lys and N-terminal dimethylation were accepted in the group-specific parameters table as labels. The precursor mass was set to an accuracy of 10 ppm and the fragment mass to 0.5 D. Here, we also accepted semitryptic peptides with a maximal number of three missed cleavages. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the data set identifier PXD000660. For further analyses, the identified peptides must have had blocked N termini by dimethylation or acetylation and had to be unique for one protein. Peptides without a C-terminal Arg or Lys as evidence for a tryptic origin were filtered out. The lists of determined peptides (Supplemental Table S1) from both experiments and all data analysis were combined for each plant line. The minimal starting position of the assigned proteins was determined using a program that detects the position of the peptides in the proteins (from Katja Bärenfaller). The mapping of the observed proteins to the organelles was done using a chloroplast reference proteome (van Wijk and Baginsky, 2011) and SUBAIII (Tanz et al., 2013). The theoretical starting position was predicted with ChloroP (Emanuelsson et al., 1999). For proteins that were assigned as thylakoid localized by AT_CHLORO (Ferro et al., 2010), the start position of the mature protein listed in the UniProt database was taken. Sequences were analyzed using WebLogo (Crooks et al., 2004).
Chloroplast Preparation and in Vitro Chloroplast Import Studies
The preparation of intact Arabidopsis chloroplasts and in vitro import were performed as described before (Agne et al., 2009). For western-blot analysis, chloroplasts corresponding to 50 µg of chlorophyll were treated or not with 50 µg mL−1 thermolysin in 100 µL of HS buffer (50 mm HEPES-KOH, pH 8, and 330 mm sorbitol) supplemented with 0.5 mm CaCl2 for 30 min on ice. The reaction was stopped with 20 mm EDTA in HS buffer for 5 min on ice. Chloroplasts were reisolated, washed with HS buffer, and denatured in SDS-PAGE loading buffer. For fractionation into soluble and membrane proteins, chloroplasts were resuspended in 10 mm Tris-HCl, pH 8, 1 mm EDTA, 5 mm dithiothreitol, and 0.1% (v/v) plant protease inhibitor cocktail P9599 (Sigma-Aldrich) to a final chlorophyll concentration of 0.5 mg mL−1. After lysis for 30 min on ice, soluble and membrane fractions were obtained by centrifugation for 30 min at 15,000g and 4°C. For in vitro import, chloroplasts isolated from 27-d-old wild-type and tic56-3 mutant seedlings grown under an 8-h photoperiod were adjusted to equal amounts of protein (corresponding to 17.5 µg of chlorophyll of the wild-type chloroplasts per time point). The samples of the import reactions were resolved by SDS-PAGE and analyzed by phosphorimaging (Fujifilm; BAS1500 or BAS1800) and quantification with the Tina 2.10i software (Raytest).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Rubisco (At5g38430), Lhcb4.1 (At5g01530), E1α (At1g01090), Toc159 (At4g02510), and Tic56 (At5g01590).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotyping and gene expression analysis of the tic56-3 mutant.
Supplemental Figure S2. Phenotypes of the mutants ppi2 and tic56-1 compared with the wild type.
Supplemental Figure S3. Comparison of experimental and theoretical processing sites and resulting N-terminal amino acids.
Supplemental Figure S4. Proteins of tic56-1 plastids are protected during thermolysin treatment.
Supplemental Table S1. Summary of all proteins identified in the TAILS experiment with the minimal starting positions.
Supplemental Table S2. Summary of all peptides identified in the TAILS experiments used for further analyses.
Supplemental Table S3. Summary of unique peptides identified in the proteome analysis of tic56-1 and wild-type plastids (thermolysin treatment).
Supplemental Methods S1. Experimental approaches for Supplemental Figures S1 and S4.
Supplementary Material
Acknowledgments
We thank Paul Jarvis (University of Leicester) for providing ppi2 in the Columbia-0 background; Julia Grimmer, Anja Rödiger, and Ralf Bernd Klösgen (Martin-Luther-University Halle-Wittenberg) for providing plasmid constructs; Hsou-min Li (Institute of Molecular Biology, Academia Sinica) for donating the anti-atTic20 antibody; the ProteomeXchange Consortium for the possibility to deposit the proteomics data; and Dr. Katja Bärenfaller (Eidgenössisch Technische Hochschule) and Dr. Dirk Dobritzsch and Nina Pötzsch (Martin-Luther-University Halle-Wittenberg) for providing tools for the determination of the minimal peptide positions and peptide positions in protein sequences, respectively.
Glossary
- TAILS
terminal amine isotopic labeling of substrates
- T-DNA
transfer DNA
- Tat
twin arginine translocase
- SPP
stromal processing protease
- MS
Murashige and Skoog
- SCB
sodium cacodylate buffer
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Ba1902/3–1 to S.B.).
References
- Agne B, Andrès C, Montandon C, Christ B, Ertan A, Jung F, Infanger S, Bischof S, Baginsky S, Kessler F (2010) The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol 153: 1016–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agne B, Infanger S, Wang F, Hofstetter V, Rahim G, Martin M, Lee DW, Hwang I, Schnell D, Kessler F (2009) A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J Biol Chem 284: 8670–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M, Goetze TA, Kovács-Bogdán E, Schürmann P, Wagner R, Buchanan BB, Soll J, Bölter B (2009) Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J Biol Chem 284: 2603–2616 [DOI] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, Troesch R, Vidi PA, Roschitzki B, Hirsch-Hoffmann M, Hennig L, Kessler F, Gruissem W, et al. (2011) Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Reddick LE, McWilliams DR, Campbell IM, Bruce BD (2012) Differential transit peptide recognition during preprotein binding and translocation into flowering plant plastids. Plant Cell 24: 3040–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Chu CC, Chen LJ, Akita M, Li HM (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J Cell Biol 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K (1984) Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol 75: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet A, Kleifeld O, Kizhakkedathu JN, Overall CM (2011) Identification of proteolytic products and natural protein N-termini by terminal amine isotopic labeling of substrates (TAILS). Methods Mol Biol 753: 273–287 [DOI] [PubMed] [Google Scholar]
- Dutta S, Teresinski HJ, Smith MD (2014) A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors. PLoS ONE 9: e95026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez SM, Bil’ KY, Aguilera R, Nishio JN, Faull KF, Whitelegge JP (2003) Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol Cell Proteomics 2: 1068–1085 [DOI] [PubMed] [Google Scholar]
- Heins L, Mehrle A, Hemmler R, Wagner R, Küchler M, Hörmann F, Sveshnikov D, Soll J (2002) The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J 21: 2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A, Bauer J, Alvarez-Huerta M, Kessler F (2001a) Protein translocon at the Arabidopsis outer chloroplast membrane. Biochem Cell Biol 79: 629–635 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Bauer J, Vidi PA, Infanger S, Weibel P, Hohwy M, Kessler F (2001b) Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J Cell Biol 154: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Kikuchi S, Oishi M, Nakai M (2011) In vivo studies on the roles of two closely related Arabidopsis Tic20 proteins, AtTic20-I and AtTic20-IV. Plant Cell Physiol 52: 469–478 [DOI] [PubMed] [Google Scholar]
- Huang S, Shingaki-Wells RN, Taylor NL, Millar AH (2013) The rice mitochondria proteome and its response during development and to the environment. Front Plant Sci 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Alami M, Lange PF, Foster LJ, Schröder WP, Overall CM, Green BR (2013) Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS ONE 8: e74483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infanger S, Bischof S, Hiltbrunner A, Agne B, Baginsky S, Kessler F (2011) The chloroplast import receptor Toc90 partially restores the accumulation of Toc159 client proteins in the Arabidopsis thaliana ppi2 mutant. Mol Plant 4: 252–263 [DOI] [PubMed] [Google Scholar]
- Inoue H, Rounds C, Schnell DJ (2010) The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova Y, Smith MD, Chen K, Schnell DJ (2004) Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell 15: 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey RA III, Subramanian C, Bruce BD (2000) Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol 122: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmati AR, Töpel M, Patel R, Murtaza G, Jarvis P (2011) Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J 66: 877–889 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Oishi M, Hirabayashi Y, Lee DW, Hwang I, Nakai M (2009) A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21: 1781–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol 28: 281–288 [DOI] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol 143: 991–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács-Bogdán E, Benz JP, Soll J, Bölter B (2011) Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Baldwin A, Patel R, Razzaq A, Dupree P, Lilley K, Kurth J, Leister D, Jarvis P (2003) The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S, Patel R, Combe J, Bédard J, Kovacheva S, Lilley K, Biehl A, Leister D, Ríos G, Koncz C, et al. (2004) Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwasniak M, Pogorzelec L, Migdal I, Smakowska E, Janska H (2012) Proteolytic system of plant mitochondria. Physiol Plant 145: 187–195 [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. (1984) Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods 10: 203–209 [DOI] [PubMed] [Google Scholar]
- Lange PF, Overall CM (2013) Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol 17: 73–82 [DOI] [PubMed] [Google Scholar]
- Li HM, Teng YS (2013) Transit peptide design and plastid import regulation. Trends Plant Sci 18: 360–366 [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Motohashi R, Rödiger A, Agne B, Baerenfaller K, Baginsky S (2012) Common and specific protein accumulation patterns in different albino/pale-green mutants reveals regulon organization at the proteome level. Plant Physiol 160: 2189–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O (2009) Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem 284: 30200–30208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Schnell DJ (2014) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer I, Wingfield PT (2012) Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr Protoc Protein Sci Chapter 6: Unit 6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Wienk H, Sips W, de Swaaf M, Talboom I, van’t Hof R, de Korte-Kool G, Demel R, Weisbeek P, de Kruijff B (1995) Functional domains of the ferredoxin transit sequence involved in chloroplast import. J Biol Chem 270: 3882–3893 [DOI] [PubMed] [Google Scholar]
- Richly E, Dietzmann A, Biehl A, Kurth J, Laloi C, Apel K, Salamini F, Leister D (2003) Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep 4: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol 7: 303–304 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G (1994) Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta 1833: 314–331 [DOI] [PubMed] [Google Scholar]
- Smith MD, Rounds CM, Wang F, Chen K, Afitlhile M, Schnell DJ (2004) atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J Cell Biol 165: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–43 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Chan PT, Li HM (2012) Differential age-dependent import regulation by signal peptides. PLoS Biol 10: e1001416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng YS, Su YS, Chen LJ, Lee YJ, Hwang I, Li HM (2006) Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18: 2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toujani W, Muñoz-Bertomeu J, Flores-Tornero M, Rosa-Téllez S, Anoman AD, Alseekh S, Fernie AR, Ros R (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol 163: 1164–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JY, Chu CC, Yeh YH, Chen LJ, Li HM, Hsiao CD (2013) Structural characterizations of the chloroplast translocon protein Tic110. Plant J 75: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberlacker B, Werr W (1996) Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol Breed 2: 293–295 [Google Scholar]
- van Wijk KJ, Baginsky S (2011) Plastid proteomics in higher plants: current state and future goals. Plant Physiol 155: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venne AS, Vögtle FN, Meisinger C, Sickmann A, Zahedi RP (2013) Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J Proteome Res 12: 3823–3830 [DOI] [PubMed] [Google Scholar]
- Vidi PA, Kanwischer M, Baginsky S, Austin JR, Csucs G, Dörmann P, Kessler F, Bréhélin C (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem 281: 11225–11234 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, et al. (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439 [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.