Abstract
Chromosomal translocation that results in fusion of the genes encoding RNA-binding protein EWS and transcription factor FLI1 (EWS-FLI1) is pathognomonic for Ewing sarcoma. EWS-FLI1 alters gene expression through mechanisms that are not completely understood. We performed RNA sequencing (RNAseq) analysis on primary pediatric human mesenchymal progenitor cells (pMPCs) expressing EWS-FLI1 in order to identify gene targets of this oncoprotein. We determined that long noncoding RNA-277 (Ewing sarcoma–associated transcript 1 [EWSAT1]) is upregulated by EWS-FLI1 in pMPCs. Inhibition of EWSAT1 expression diminished the ability of Ewing sarcoma cell lines to proliferate and form colonies in soft agar, whereas EWSAT1 inhibition had no effect on other cell types tested. Expression of EWS-FLI1 and EWSAT1 repressed gene expression, and a substantial fraction of targets that were repressed by EWS-FLI1 were also repressed by EWSAT1. Analysis of RNAseq data from primary human Ewing sarcoma further supported a role for EWSAT1 in mediating gene repression. We identified heterogeneous nuclear ribonucleoprotein (HNRNPK) as an RNA-binding protein that interacts with EWSAT1 and found a marked overlap in HNRNPK-repressed genes and those repressed by EWS-FLI1 and EWSAT1, suggesting that HNRNPK participates in EWSAT1-mediated gene repression. Together, our data reveal that EWSAT1 is a downstream target of EWS-FLI1 that facilitates the development of Ewing sarcoma via the repression of target genes.
Introduction
Oncoproteins that result from chromosomal translocations are key driver events in many human cancers. Many oncogenic translocations induce aberrant transcriptional activity that results in the rewiring of genetic networks. However, in most cases the precise mechanisms that lead to these changes in gene expression remain poorly characterized. In roughly 50% of sarcomas that harbor reciprocal translocations, members of the FET family of RNA-binding proteins (FUS/TLS, EWSR1, and TAF15) are involved (1). One example of this is Ewing sarcoma, a highly aggressive malignancy found primarily in children and young adults. The pathognomonic molecular lesion in the majority of Ewing sarcomas is a translocation event that fuses the FET family member EWSR1 to an ETS transcription factor, most commonly FLI1. Because these translocations always contain the transcriptional activation domain at the N terminus of the FET protein fused to the ETS DNA-binding domain, they are thought to behave as aberrant transcription factors (2, 3).
Previous work has identified genes upregulated by fusion of the genes encoding RNA-binding protein EWS and transcription factor FLI1 (EWS-FLI1) that are critical for transformation. Early efforts identified targets by expressing EWS-FLI1 in NIH3T3 cells (4, 5). However, gene expression changes found in heterologous systems may not always reflect Ewing sarcoma biology (6). As an alternative approach, microarray analysis of the transcriptome of Ewing sarcoma–derived cell lines following EWS-FLI1 knockdown has identified hundreds of potential direct and indirect targets of EWS-FLI1, a few of which have been functionally analyzed. For example, the nuclear receptor NR0B1 is upregulated by EWS-FLI1 as a consequence of direct binding to its promoter and is required for EWS-FLI1–driven oncogenesis (7). Similarly, the transcription factor NKX2.2 is an indirect EWS-FLI1 target that is also required for transformation (8).
Knockdown of EWS-FLI1 in Ewing sarcoma cell lines leads to an expression profile similar to that of bone marrow–derived mesenchymal progenitor cells (MPCs), suggesting these as the likely cell of origin for Ewing sarcoma (9, 10). Human MPCs are permissive to EWS-FLI1 expression, although EWS-FLI1 alone is insufficient to transform them (11). Microarray profiling has identified some of the transcriptional consequences of EWS-FLI1 expression in primary MPCs. For example, EZH2 is induced by EWS-FLI1 in primary adult MPCs (11, 12), and IGF1 and miRNA145 are EWS-FLI1 target genes in MPCs isolated from pediatric patients (13, 14). The majority of Ewing sarcomas arise in patients between the ages of 10 and 20 years. This age-restricted frequency suggests that pediatric MPCs may be particularly susceptible to transformation by EWS-FLI1. Consistent with this possibility, MPCs derived from pediatric patients (pMPCs) express a distinct subset of genes when induced to express EWS-FLI1 compared with adult-derived MPCs (14).
EWS-FLI1 can both activate and repress gene expression, although previous work has suggested that gene repression may be more prevalent (15). However, most well-characterized EWS-FLI1 target genes are upregulated by the translocation, and much less is known about the mechanisms involved in EWS-FLI1 repression of gene expression. One mechanism may involve upregulation of transcriptional repressors such as NKX2.2, NR0B1, and BCL11B (16–18). However, only a subset of EWS-FLI1–downregulated genes is regulated by these targets. Thus, other mechanisms accounting for gene repression likely exist and may play an important role in EWS-FLI1–driven oncogenesis.
A role for long noncoding RNAs (lncRNAs) in regulating oncogenesis is beginning to emerge. It has become clear through recent massively parallel sequencing studies that many transcribed RNAs have no protein-coding potential (19, 20), suggesting a vast network of gene regulation that is only beginning to be understood. Furthermore, recent comprehensive studies have catalogued thousands of lncRNAs, the majority of which remain to be functionally annotated (21, 22). Some lncRNAs play important roles in chromatin remodeling, RNA transport, RNA stability, and other critical functions that lead to changes in gene expression (23). Well-known examples include XIST, which acts by recruiting the PRC2 complex to initiate X-chromosome inactivation (24, 25) as well as MALAT and NEAT1, both of which play a role in mRNA processing and nuclear organization (26, 27). Some lncRNAs mediate gene expression in cis by directly recruiting or inhibiting the ability of transcription complexes to bind promoters of neighboring genes (28, 29), while others act in trans to influence the expression of distant genes (30). Given the widespread transcription of lncRNAs and their emerging role in gene regulation, they are candidate genes for participation in the complex genetic rewiring programs that occur as a consequence of oncogenic translocation events.
We used RNA sequencing (RNAseq) to elucidate the transcriptional effects of EWS-FLI1 in MPCs isolated from pediatric patients. This identified many novel EWS-FLI1 target genes. Among these was a previously uncharacterized lncRNA, lnc277 (LINC00277, herein referred to as Ewing sarcoma–associated transcript 1 [EWSAT1]). Functional and genomic studies indicate an important role of this lncRNA in the pathogenesis of Ewing sarcoma. Specifically, our studies identified EWSAT1 as a key regulator of gene repression downstream of EWS-FLI. The repressive function of EWSAT1 was mediated in part by interaction with the RNA-binding protein heterogeneous nuclear ribonucleoprotein (HNRNPK). These results suggest that further studies to identify non-protein-coding genes that contribute to oncogenesis will be fruitful for elucidating the complex genetic rewiring that is driven by EWS-FLI1 and perhaps other oncogenic translocations.
Results
Next-generation sequencing of pMPCs expressing EWS-FLI1 identifies novel targets of the oncogenic fusion.
Previous studies have suggested that a MPC may be the cell of origin of Ewing sarcoma (10). In order to identify genes regulated by EWS-FLI1 in primary human cells, we isolated pMPCs from donors (see Methods). These pMPCs expressed high levels of CD146 (Supplemental Figure 1A and data not shown; supplemental material available online with this article; doi:10.1172/JCI72124DS1), an established marker of early skeletal progenitors capable of reconstituting a bone marrow niche in vivo (31). pMPCs also expressed the mesenchymal markers CD90 and CD105 but not the hematopoietic marker CD45 (Supplemental Figure 1A). As expected, pMPCs differentiated into osteogenic and adipogenic lineages in vitro (Supplemental Figure 1, B and C) and were capable of forming colonies when plated at low density (ref. 32 and Supplemental Figure 1D). The gene expression of pMPCs was highly similar to that of Ewing sarcoma cell lines with EWS-FLI1 knockdown, supporting the possibility that pMPCs are a cell type closely related to the Ewing sarcoma cell of origin (Supplemental Figure 1E and Supplemental Table 1). Thus, pMPCs provide a system to analyze the consequences of EWS-FLI1 expression in normal cells derived from patients within the age group in which Ewing sarcoma is known to arise most frequently.
A retroviral vector containing an EWS-FLI1 cDNA under the control of a doxycycline-inducible promoter was transduced into independent pMPC isolates from 3 different patients. Exposure to doxycycline induced expression of EWS-FLI1 to levels similar to those in Ewing sarcoma cell lines (Figure 1A). EWS-FLI1 expression in pMPCs induced expression of several genes previously reported to be EWS-FLI1 targets (7, 8, 13, 33–35). In some cases, expression of these genes was below the threshold for detection in the absence of EWS-FLI1 but clearly was induced after EWS-FLI1 expression (IGF1, NKX2.2, and NR0B1), whereas, in other cases, expression was present at baseline but increased substantially after EWS-FLI1 (CAV1, ID2, and TOPK) (Figure 1B). pMPCs expressing EWS-FLI1 continued to proliferate for several passages, although their population-doubling and colony-forming ability was decreased compared with that of control uninfected pMPCs (data not shown).
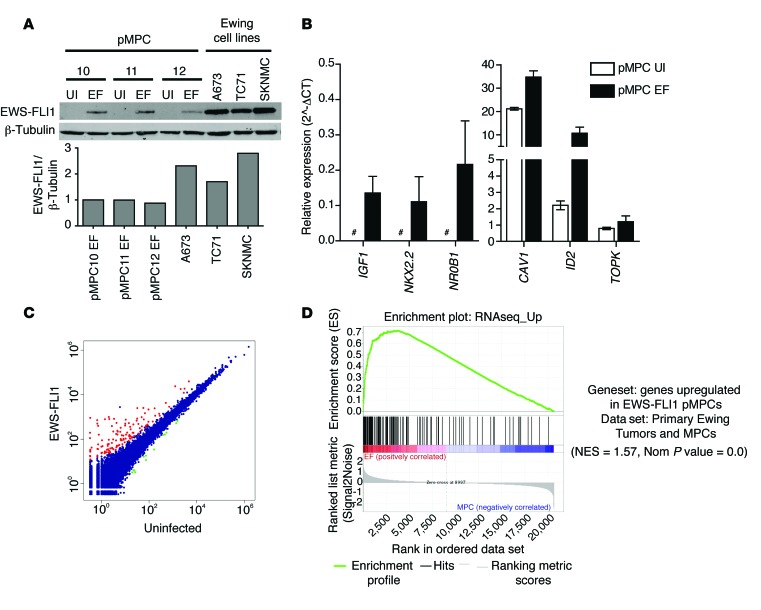
Figure 1. Expression of EWS-FLI1 in pMPCs.
(A) Western blot analysis of EWS-FLI1 induction in 3 pMPC isolates following addition of doxycycline for 13 days. Levels of EWS-FLI1 protein in 3 Ewing sarcoma cell lines (A673, TC71, and SKNMC) are shown for comparison. Relative protein levels are shown (EWS-FLI1/β-tubulin), normalized to pMPC10 EWS-FLI1. EF, EWS-FLI1; UI, uninfected. (B) Known EWS-FLI1 targets are upregulated in pMPC cells expressing EWS-FLI1. Expression is shown relative to HPRT. Error bars indicate standard error of the mean for n = 2. Pound signs indicate expression below the detection threshold. (C) RNAseq identified 157 upregulated genes (red) and 13 downregulated genes (green) in 3 pMPC isolates expressing EWS-FLI1 compared with uninfected controls (blue). Data indicate average read counts for the 3 MPC lines. (D) Gene set enrichment analysis analysis using a published data set of genes expressed in Ewing sarcoma tumors compared with primary MPCs and a gene set comprised of the upregulated genes identified in pMPCs.
To identify novel EWS-FLI1–regulated transcripts and gain a genome-wide understanding of the changes induced by EWS-FLI1 expression, we performed RNAseq (3SEQ, see Methods) to compare pMPCs expressing EWS-FLI1 with control pMPCs. We identified 157 genes with substantially higher expression in cells expressing EWS-FLI1, whereas only 13 genes had reduced expression (Figure 1C; Supplemental Figure 2, A and B; and Supplemental Table 2). Genes induced by EWS-FLI1 in pMPCs were highly enriched in a previously published data set of human Ewing sarcomas (36), validating the use of pMPCs for identifying genes relevant to the pathogenesis of this disease (Figure 1D). Similarly, 5 previously published gene sets of EWS-FLI1–regulated genes were all significantly enriched in the pMPC RNAseq data set (Supplemental Figure 2, C–G, and refs. 8, 11, 14).
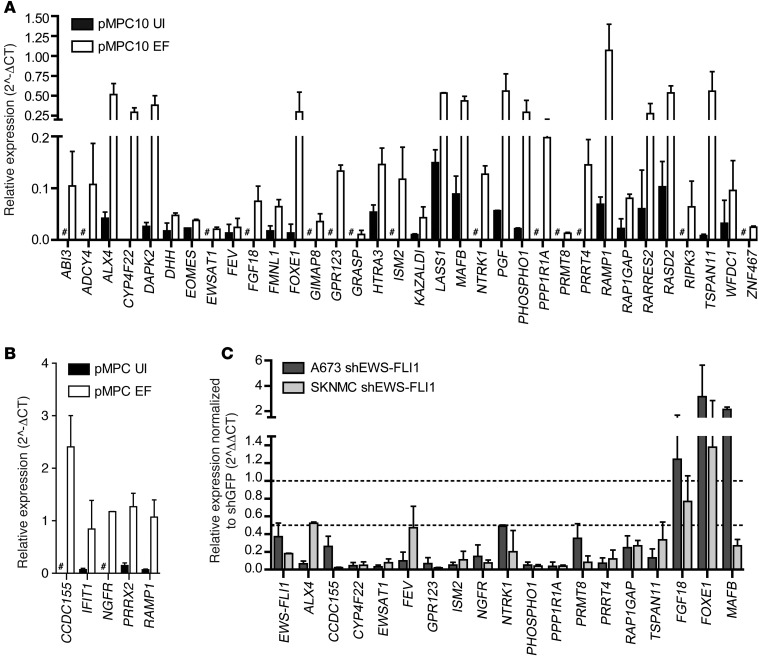
In order to maximize the potential for discovery of novel EWS-FLI1 target genes, we focused further analysis primarily on genes not previously established as EWS-FLI1 targets using other approaches. RT-PCR validation confirmed induction by EWS-FLI1 of 39 genes identified as overexpressed in pMPCs (Figure 2, A and B). Next, we tested whether these genes were dependent on EWS-FLI1 expression in the context of Ewing sarcoma by analyzing their expression in 2 Ewing sarcoma cell lines (A673 and SKNMC) after EWS-FLI1 knockdown. Of the 19 genes validated as upregulated in pMPCs, 16 also had decreased expression in both Ewing sarcoma cell lines after EWS-FLI1 knockdown (Figure 2C). Of these genes, 15 were protein coding, whereas one was a lncRNA of unknown function, EWSAT1. As a role for lncRNAs in the pathogenesis of Ewing sarcoma has not been described previously, further functional studies were focused on this novel downstream target of EWS-FLI1.
Figure 2. Validation of EWS-FLI1 targets in pMPCs and Ewing sarcoma cell lines.
(A and B) RT-PCR for genes identified as upregulated by EWS-FLI1 in pMPCs. Expression levels are relative to HPRT. Pound signs indicate expression below the threshold for detection. (C) RT-PCR analysis of a subset of target genes in A and B in 2 Ewing sarcoma cells lines after EWS-FLI1 knockdown. Expression normalized to shGFP. Error bars represent SD for n = 2.
EWSAT1 is a lncRNA induced by EWS-FLI1.
Visualization of the RNAseq reads across all controls (control pMPCs) and all induced samples (EWS-FLI1–expressing MPCs) at the EWSAT1 locus confirmed an upregulation of EWSAT1 in pMPCs expressing EWS-FLI1 compared with controls (Figure 3A). In all 3 pMPC lines expressing EWS-FLI1, multiple reads were detected at the 3′ end of the last exon of EWSAT1, as expected since the 3SEQ RNAseq protocol sequences only from the 3′ ends of transcripts, whereas few reads could be detected in the controls (Supplemental Figure 3A). Expression of EWSAT1 in pMPCs expressing EWS-FLI1 was confirmed by RT-PCR (Figure 3B). Notably, while EWSAT1 is clearly upregulated as a consequence of EWS-FLI1, expression of EWSAT1 was several-fold higher in Ewing sarcoma cell lines than in pMPCs expressing EWS-FLI1, consistent with the lower levels of expression of EWS-FLI1 in pMPCs compared with cell lines (Figure 1A).
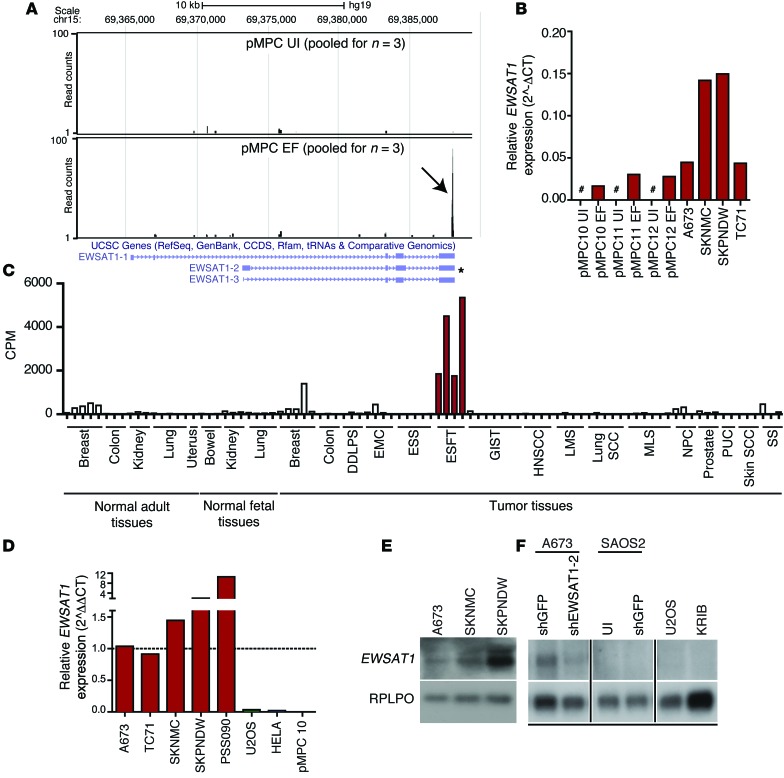
Figure 3. EWSAT1 is a lncRNA expressed in Ewing sarcoma.
(A) Visualization of reads mapped to the EWSAT1 locus in either control MPCs (pMPC UI) or EWS-FLI1–expressing MPCs (pMPC EWS-FLI1). Black arrow indicates accumulation of reads at the 3′ end of pMPCs expressing EWS-FLI1, whereas almost no reads are detected in the control. Tracks indicate pooled reads from either 3 controls or 3 pMPCs expressing EWS-FLI1. Predominant known isoforms are indicated below tracks. Asterisk indicates most abundant isoform, as determined by RT-PCR (see Results) (B) RT-PCR for EWSAT1 in uninfected pMPCs, EWS-FLI1–expressing pMPCs, and in 4 Ewing sarcoma cell lines. EWSAT1 expression is quantified relative to HPRT. Pound signs indicate expression below the threshold for detection. (C) cpm reads mapped for EWSAT1 in the RNAseq data set generated from a panel of primary human tumors and normal samples. DDLPS, dedifferentiated liposarcoma; EMC, extraskeletal myxoid chondrosarcoma; ESS, endometrial stromal sarcoma; ESFT, Ewing sarcoma family of tumors; GIST, gastrointestinal stromal tumor; HNSCC, head and neck; LMS, leiomyosarcoma; MLS, myxoid liposarcoma; NPC, nasopharyngeal carcinoma; PUC, papillary urothelial carcinoma; SS, synovial sarcoma. (D) RT-PCR for EWSAT1 in 4 Ewing cell lines and a primary patient-derived xenograft (PSS090) compared with the indicated non-EWS-FLI1–expressing cells. EWSAT1 expression is relative to HPRT and normalized to A673. (E) Northern blot detection of EWSAT1 in 3 Ewing cell lines. (F) Northern blot detection of EWSAT1 in A673 Ewing cells (control vs. knockdown of EWSAT1) and in osteosarcoma cell lines SAOS2, U20S, and KRIB. All lanes were run on the same gel but were noncontiguous.
The lncRNA EWSAT1 is found on chromosome 15 between 2 protein-coding genes, NOX5 and GLCE (Supplemental Figure 3B). Several alternatively spliced isoforms for this gene have been reported. Northern blot analysis using several different probes as well as isoform-specific RT-PCR confirmed that one isoform (EWSAT1-2, indicated in Figure 3A) was predominantly expressed in Ewing sarcoma cells and dependent on EWS-FLI1 expression (Supplemental Figure 3, C–F). To confirm the annotation of EWSAT1 as a noncoding RNA, we used the coding potential calculator (CPC) algorithm and found a negative value (–0.626559) for EWSAT1, making it highly unlikely that this gene codes for a protein (37).
To confirm expression of EWSAT1 in primary Ewing sarcomas and to assess the expression of this gene in other cancer types, we analyzed a large RNAseq data set of paraffin-embedded primary human tumor samples that included several Ewing sarcoma samples (38). Among the 17 cancer types in this data set, the highest expression of EWSAT1 was found in the 4 Ewing sarcoma tumors (Figure 3C). Lower but detectable levels of expression of EWSAT1 were seen in breast tumors and normal breast samples (Figure 3C). However, expression of EWSAT1 was low in all of the 7 other sarcomas analyzed. Of these, myxoid liposarcoma and extraskeletal myxoid chondrosarcoma harbored other translocations involving members of the FET family of RNA-binding proteins, suggesting that EWSAT1 expression is specific to EWS-FLI1. We also confirmed higher levels of expression of EWSAT1 in a first-passage patient-derived xenograft compared with Ewing cell lines (Figure 3D). Expression of EWSAT1 was undetectable in several non-Ewing sarcoma cancer cell lines analyzed, including osteosarcoma and medulloblastoma (Figure 3D and data not shown). Two prostate cancer cell lines (VCaP and NCI-H660) that harbor the TMPRSS2-ERG translocation were also negative for EWSAT1 expression. Northern blot analysis further confirmed expression of EWSAT1 in Ewing sarcoma cell lines and the lack of EWSAT1 expression in primary osteosarcoma tumors and cell lines (Figure 3, D–F). The specificity of the detected band was confirmed using an shRNA against EWSAT1 (Figure 3F). Taken together, the analysis of archival and fresh primary tumor samples as well as that of tumor cell lines strongly suggests that EWSAT1 is a lncRNA specifically upregulated as a consequence of the driving oncogenic event in Ewing sarcoma.
EWSAT1 expression is dependent on EWS-FLI1 and can be induced in heterologous cell types.
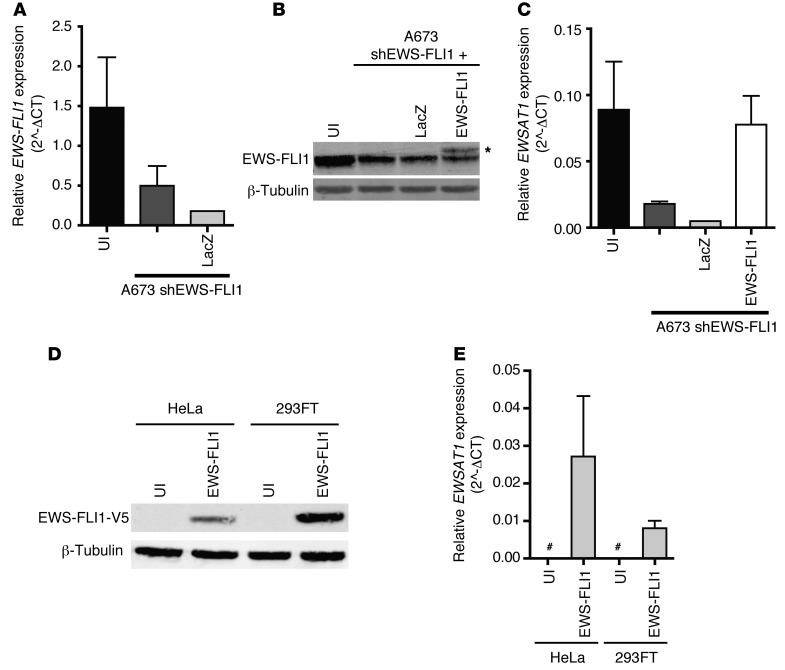
To further analyze the role of EWS-FLI1 in regulating EWSAT1 expression, we used an shRNA targeting EWS-FLI1 via the 3′UTR of FLI1 (Figure 2B). To rescue the knockdown of EWS-FLI1, cells were infected with a lentivirus expressing an EWS-FLI1 cDNA construct not targeted by this shRNA (Figure 4, A and B). EWSAT1 expression levels were rescued upon reexpression of EWS-FLI1, confirming that the observed effect of EWS-FLI1 knockdown on EWSAT1 expression is mediated via EWS-FLI1 and is not off target (Figure 4C).
Figure 4. Expression of EWSAT1 is dependent on EWS-FLI1 and independent of the cell of origin.
(A) RT-PCR for EWS-FLI1 expression in the Ewing sarcoma cell line A673 transduced with a hairpin targeting EWS-FLI1 and the indicated cDNA constructs. EWS-FLI1 expression is relative to HPRT. Error bars represent SD for n = 2. (B) Western blot for EWS-FLI1–expressing cDNA constructs. Asterisk indicates V5-tagged EWS-FLI1. (C) RT-PCR for EWSAT1 expression in shEWS-FLI1 A673 cells transduced with indicated cDNA constructs. EWSAT1 expression is relative to HPRT. Error bars represent SD for n = 2. (D) Western blot analysis for EWS-FLI1 using an antibody against the V5 epitope tag in transduced HeLa and 293FT cells. (E) RT-PCR for EWSAT1 expression in transduced HeLa and 293FT cells expressing ectopic EWS-FLI1. EWSAT1 expression is relative to HPRT expression. Pound signs indicate expression below the threshold for detection. Error bars indicate SD.
The close correlation between EWS-FLI1 expression and EWSAT1 could be a consequence of the underlying epigenetic landscape present in the cell of origin of Ewing sarcoma. Alternatively, previous work has demonstrated that EWS-FLI1 has chromatin-remodeling activity and leads to nucleosome depletion at sites near upregulated genes (39). If the epigenetic state of the cell of origin is the key determinant of EWS-FLI1–mediated EWSAT1 expression, it would be expected that expression of EWS-FLI1 in other cell types would not induce EWSAT1. To determine whether EWS-FLI1 expression in heterologous cell types is sufficient to induce expression of EWSAT1, HeLa and 293FT cells were transduced with a lentivirus containing a V5-tagged EWS-FLI1 cDNA (Figure 4D). Expression of EWS-FLI1 led to an increase in the expression of EWSAT1 in both cell types (Figure 4E). These results suggest that EWS-FLI1 is sufficient to activate EWSAT1 independent of the epigenetic landscape of the cell in which is it expressed. Notably, we were unable to confirm EWS-FLI1–regulated expression of a luciferase reporter construct containing a 4-kb fragment of the EWSAT1 promoter upstream of luciferase, suggesting that regulation of EWSAT1 by EWS-FLI1 either does not occur due to proximal promoter binding or involves more distal enhancer elements (data not shown).
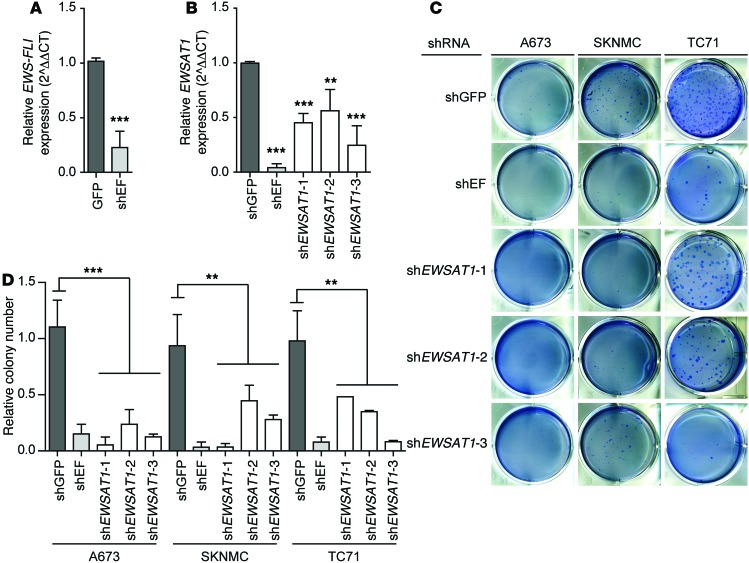
EWSAT1 is required for EWS-FLI1–induced soft agar colony formation but is dispensable in normal cells.
To determine whether EWSAT1 plays a specific role in maintaining oncogenesis in Ewing sarcoma cell lines, we used 3 independent shRNAs targeting EWSAT1 and 2 control shRNAs, one targeting GFP as a negative control and one targeting EWS-FLI1 as a positive control. Reduced expression of EWS-FLI1 and EWSAT1 was confirmed by RT-PCR (Figure 5, A and B). Knockdown of EWSAT1 with these shRNAs impaired the ability of Ewing cell lines to proliferate (Supplemental Figure 4A). EWSAT1 knockdown also had a dramatic effect on soft agar colony formation in all 3 Ewing sarcoma cell lines tested (Figure 5, C and D). These data strongly support a requirement for EWSAT1 in the proliferation and soft agar colony formation of Ewing sarcoma cell lines. In contrast, knockdown of EWSAT1 had no significant effect on survival of IMR90 cells (Supplemental Figure 4B) or BJTERT cells (data not shown). The effect of EWSAT1 on proliferation was further supported by analysis of BrDU incorporation in the A673 Ewing sarcoma cell line (Supplemental Figure 4, C and D). These data suggest that EWSAT1 is specifically required for proliferation and soft-agar growth in Ewing cells but is dispensable in other cell types. Given these results, we hypothesized that EWSAT1 expression may increase the proliferative capacity of Ewing cell lines or modulate other oncogenic properties such as their metastatic capacity. However, EWSAT1 overexpression in A673 cells (which express the lowest level of the lncRNA among all cell lines tested) did not result in increased proliferation and had no discernible effects in a scratch assay (data not shown).
Figure 5. Knockdown of EWSAT1 decreases soft agar colony growth of Ewing sarcoma cell lines.
(A) RT-PCR for EWS-FLI1 expression in the A673 Ewing cell line transduced with shRNAs for GFP or EWS-FLI1. RT-PCR normalized to shGFP. (B) RT-PCR for EWSAT1 expression in the A673 Ewing cell line transduced with shRNAs for GFP, EWS-FLI, or EWSAT1. RT-PCR normalized to shGFP. (C) Representative images of soft agar colony assay in cells infected with indicated shRNAs. (D) Quantification of soft agar colonies normalized to uninfected cells. Error bars represent SD for n = 2–5. t test was performed comparing colony numbers for shGFP and colony numbers for all 3 shRNAs together. **P < 0.01, ***P < 0.001 by 2-tailed t tests.
EWSAT1 functions through regulating gene expression downstream of EWS-FLI1.
Many functionally characterized lncRNAs have been shown to play a role in gene expression, primarily acting through interactions with chromatin modifying complexes (40). Notably, EWSAT1 is expressed both in the nucleus and in the cytoplasm, suggesting that it may have complex roles in gene regulation (Supplemental Figure 4, E and F). To ascertain the mechanism of action of EWSAT1 in EWS-FLI1–driven oncogenesis and to determine whether the lncRNA plays a role in regulating gene expression changes downstream of EWS-FLI1, we evaluated the global effects of EWSAT1 knockdown compared with those of EWS-FLI1 knockdown using RNAseq. The A673 Ewing sarcoma cell line was transduced with shRNAs targeting EWS-FLI1, EWSAT1, or a control shRNA (shGFP) (Supplemental Figure 5A). Effective knockdown of the intended targets was validated by RT-PCR and Northern blot analysis (Supplemental Figure 5, B–F) prior to library construction and sequencing (see Methods). Principal component analysis indicated a closer relationship between shEWS-FLI1 and shEWSAT1 gene expression profiles than either of these two compared with the shGFP controls (Supplemental Figure 6A).
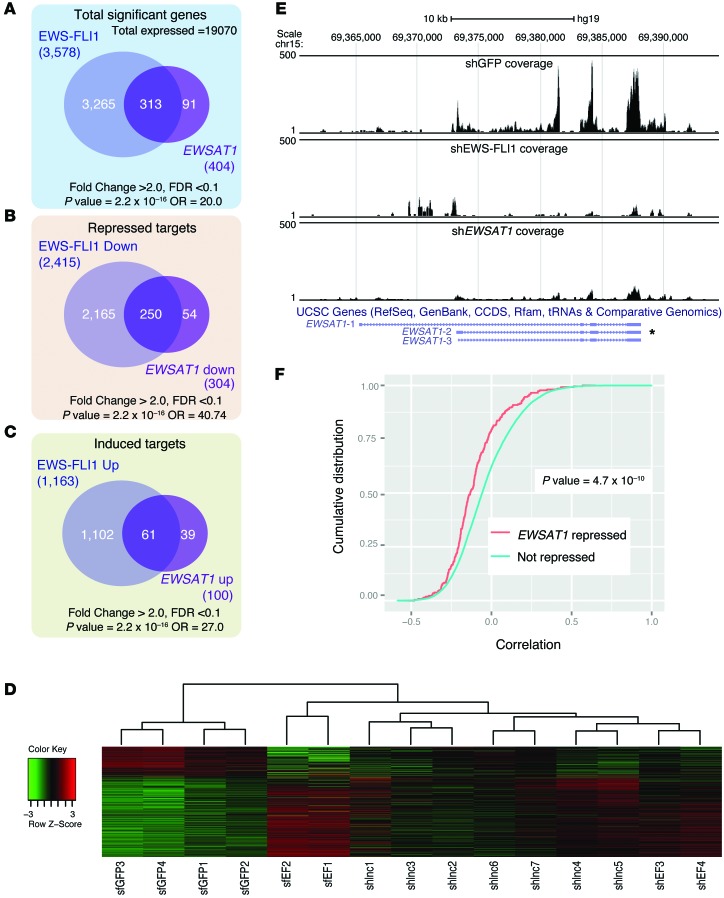
A total of 3,578 genes were significantly upregulated or downregulated in A673 cells as a consequence of EWS-FLI1 knockdown (FDR <0.1, fold change >2.0; see Methods and Supplemental Table 3). In contrast, a much smaller set of 404 genes were upregulated or downregulated by EWSAT1 knockdown (FDR <0.1, fold change >2.0; Supplemental Table 4). Surprisingly, the majority of EWSAT1-regulated genes (313 of 404, 77%) were within the set of EWS-FLI1–regulated genes, vastly more than expected by chance alone (P = 2.2 × 10–16) (Figure 6A and Supplemental Figure 6, B and C). While EWS-FLI1 causes both aberrant gene activation and gene repression (1,163 genes induced and 2,415 genes repressed), EWSAT1 repressed a much larger number of genes than it induced (100 genes induced and 304 genes repressed). Most importantly, the majority of the genes repressed by EWSAT1 (increased expression after EWSAT1 knockdown) overlapped with the genes repressed by EWS-FLI1 (Figure 6B). While a smaller number of genes was induced by EWSAT1 (repressed by EWSAT1 knockdown), most of these also overlapped with genes induced by EWS-FLI1 (Figure 6C and Supplemental Table 5). Clustering of all 3 data sets in the set of genes repressed by EWSAT1 is shown in Figure 6D.
Figure 6. EWSAT1 mediates repression of genes downstream of EWS-FLI1.
(A) Venn diagram of all genes regulated by EWS-FLI1 compared with all genes regulated by EWSAT1. (B) Venn diagram of all EWS-FLI1–repressed genes compared with all EWSAT1-repressed genes. (C) Venn diagram of all EWS-FLI1–upregulated genes compared with all EWSAT1-upregulated genes. P values and odds ratios (OR) for overlaps in A–C were calculated using Fisher’s exact test. (D) Heat map of genes differentially regulated by EWSAT1 in the indicated samples. (E) UCSC genome browser view of reads mapping to the EWSAT1 locus in shGFP, shEWS-FLI1, and shEWSAT1 samples. The predominant EWSAT1 isoform expressed in Ewing cells is indicated with an asterisk. (F) Cumulative distribution function of Pearson correlations for genes repressed by EWSAT1 and that for genes not repressed. Left shift (anticorrelation, indicating repression) of EWSAT1-repressed genes (red) is seen. Correlations calculated from a data set of Ewing sarcoma primary tumor RNAseq. Two-sided KS test was used to calculate P value.
A reduction of reads mapping to the EWSAT1 locus by either knockdown of EWS-FLI1 or knockdown of EWSAT1 compared with the control was confirmed by visualization on a genome browser (Figure 6E). The overlap of repressed targets between EWS-FLI1 knockdown and EWSAT1 knockdown in a Ewing cell line suggests that EWSAT1 may be an important driver of EWS-FLI1 target gene expression in vivo. To test this hypothesis, we analyzed an available RNAseq data set of 24 primary Ewing tumors (41) (see Methods). We computed the correlation of expression of each mRNA with that of EWSAT1 across all of these primary Ewing tumor samples. Next, we identified the mRNAs that we had found to be repressed by EWSAT1 in vitro (Supplemental Table 6). Notably, the cumulative distribution of the expression correlations for these EWSAT1-repressed genes was shifted to the left (more anticorrelated) compared with the overall list of genes expressed in Ewing tumors. The difference in the cumulative distributions of these 2 gene sets was highly statistically significant (Figure 6F). This suggests that the repression of these genes via EWSAT1 represents an important regulatory mechanism present in primary Ewing patient tumors.
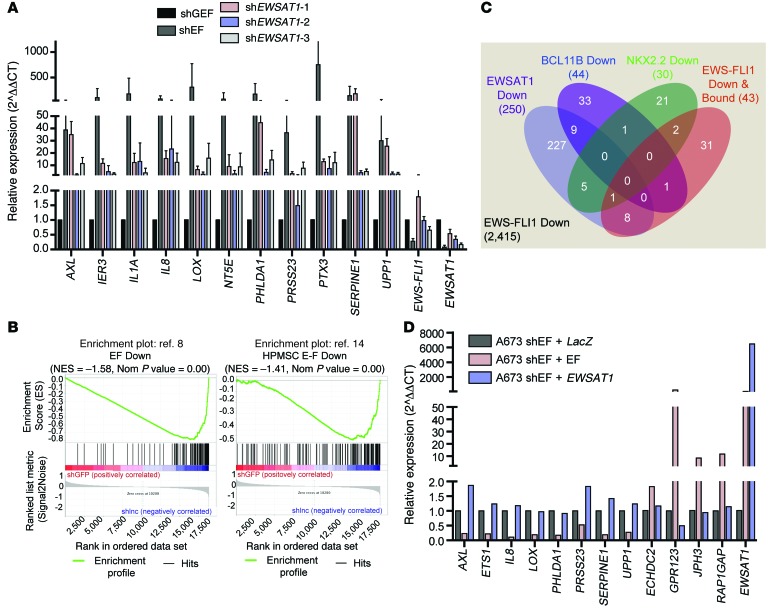
Regulation of a subset of the overlapping target genes was validated by RT-PCR in EWSAT1 knockdown experiments independent of those used to generate the RNAseq data. Twenty-two of twenty-eight of the genes tested validated with at least 2 shRNAs against EWSAT1 (Figure 7A and Supplemental Figure 6, C and D). Next, we compared the EWSAT1 knockdown data set with published gene lists of EWS-FLI1–downregulated targets (8, 14). A previously published gene set of EWS-FLI1–downregulated genes identified by knockdown of EWS-FLI1 was highly enriched in the shGFP versus shEWSAT1 data set (normalized enrichment score [NES] = –2.72, P = 0) (ref. 8 and Figure 7B). Similarly, a gene set of targets downregulated by expression of EWS-FLI1 in pMPCs was also highly enriched in the shGFP versus shEWSAT1 comparison (NES = –2.07, P = 0) (ref. 14 and Figure 7B). The overlap of these independent gene sets with the EWSAT1 knockdown data further supports a role for EWSAT1 as a critical negative regulator of gene expression involved in the pathogenesis of Ewing sarcoma.
Figure 7. EWSAT1 mediates a mechanism of gene repression downstream of EWS-FLI1.
(A) RT-PCR validation of genes identified as repressed by both EWS-FLI1 and EWSAT1. RT-PCR normalized to shGFP. Error bars represent SD for n = 3. (B) Gene set enrichment analysis of 2 published EWS-FLI1 gene sets in the data set of EWSAT1-regulated genes in A673 cells. (C) Venn diagram of the overlap between the indicated data sets within the set of genes repressed by EWS-FLI1, as defined by RNAseq in A673 cells. See Methods for source of each indicated gene set. (D) RT-PCR on A673 Ewing sarcoma cells with knockdown of EWS-FLI1 and overexpression of cDNAs for LacZ, EWS-FLI1, or EWSAT1. LacZ was used as a control cDNA. RT-PCR normalized to shEWS-FLI1 + LacZ.
Recent studies have identified mechanisms through which EWS-FLI1 represses gene expression, either directly or through other transcriptional repressors. To determine whether EWSAT1 functions as part of previously known mechanisms of repression downstream of EWS-FLI1, we compared the repressed gene expression signature of EWSAT1 to 3 previously described subsets of EWS-FLI1–repressed targets. The first gene set consisted of genes identified by ChIPseq and gene expression analysis as directly repressed targets of EWS-FLI1 (42). A second gene set contained genes downregulated through the EWS-FLI1 target gene BCL11B (18). Last, we included genes downregulated by the EWS-FLI1 target gene NKX2.2 (16). Strikingly, these 3 gene sets had little overlap with the set of genes identified as repressed by EWSAT1. Furthermore, EWSAT1 regulated a substantially larger subset (~10%) of the total number of EWS-FLI1–downregulated genes in A673 cells (Figure 7C). Taken together, these results identify EWSAT1 as a key target of EWS-FLI1 that is involved in establishing a repressive gene expression program that plays a critical role in maintaining the oncogenicity of Ewing sarcoma through a mechanism likely to be distinct from those of previously described repressed targets of EWS-FLI1.
These results raised the possibility that expression of EWSAT1 may be sufficient to drive gene repression in Ewing cells. To test this possibility, we overexpressed the main isoform of EWSAT1 found in Ewing sarcoma cells (Supplemental Figure 3C) after knockdown of EWS-FLI1. Notably, the overexpression of EWSAT1 alone did not rescue the repression or induction of genes in EWS-FLI1–depleted A673 Ewing sarcoma cells. As a control, overexpression of a shRNA-resistant EWS-FLI1 cDNA both upregulated EWSAT1 and led to the expected change in levels of these mRNAs. These results suggest that other targets downstream of EWS-FLI1 are required for the ability of EWSAT1 to regulate gene expression (Figure 7D). In addition, we found that overexpression of EWSAT1 in pMPCs did not dramatically alter the growth of these cells or the ability of the pMPCs to form fibroblast-like colonies (CFU-Fs) or differentiate into mesenchymal lineages (Supplemental Figure 7). Moreover, overexpression of EWSAT1 in the A673 Ewing sarcoma cell line did not result in an increased capacity of the cells to form tumors upon injection into immunocompromised mice (data not shown). Thus, EWSAT1 appears to be necessary, but not sufficient, to induce gene repression and drive the oncogenic transformation that occurs in the context of EWS-FLI1.
To identify biological pathways regulated by EWSAT1, repressed target genes with known gene identifiers (415 of 430) were queried to determine whether they could be classified into specific annotation clusters using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (refs. 43, 44, and Supplemental Table 7). The top 10% of annotation clusters with the highest enrichment scores was analyzed (Supplemental Figure 6E). The largest annotation cluster (“extracellular region part,” 189 genes) included genes coding for secreted proteins, such as LOX, SERPINE1, PGF, AXL, IGFBP5, and the interleukins IL1A, IL6R, and IL8. Nine of the eleven genes shown in Figure 7A validated by RT-PCR as EWSAT1-repressed targets are in this annotation cluster. In addition, the annotation cluster “skeletal system development” includes genes involved in mesenchymal differentiation into bone (BMP1, MGP, COL1A1, PTGS2, PDGFRA, MMP2, RUNX2, and IGFBP5). Thus, EWSAT1 may play a role in oncogenesis by inhibiting genes involved in a variety of processes such as extracellular signaling and differentiation.
EWSAT1 controls gene expression partially through an interaction with HNRNPK.
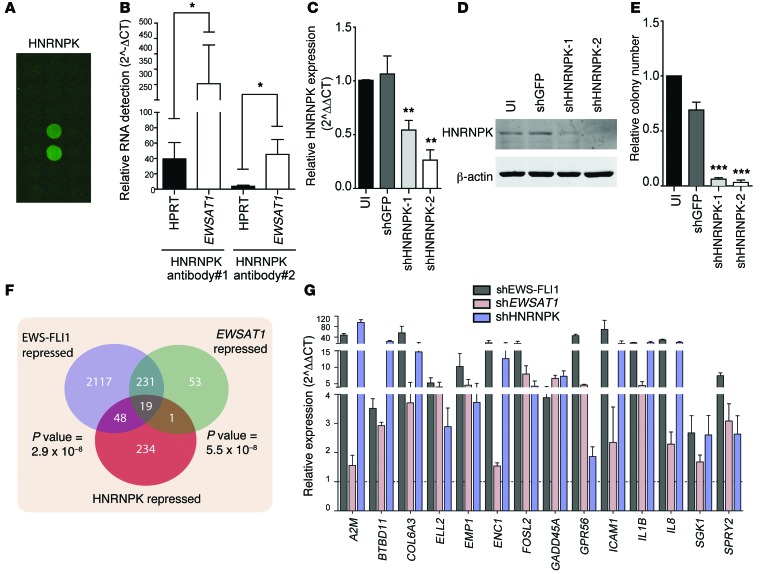
lncRNAs generally act through interactions with proteins. To understand the mechanism by which EWSAT1 regulates gene expression downstream of EWS-FLI1, we used a previously described protein array method to identify lncRNA-binding proteins (45). EWSAT1 bound consistently to 22 proteins, out of >9,000 proteins, on the protein array (see Methods and Supplemental Table 8). Two of these twenty-two proteins, HNRNPK and STAU1, have previously been described to alter gene expression through interactions with lncRNAs (refs. 46, 47; Figure 8A; and data not shown). We confirmed an interaction between HNRNPK and EWSAT1 using endogenous RNA immunoprecipitation (RIP) with 2 different HNRNPK antibodies (Figure 8B).
Figure 8. EWSAT1 interacts with HNRNPK to regulate gene expression.
(A) Image of protein array with replicate spots indicating binding of EWSAT1 to HNRNPK. (B) RIP using 2 different HNRNPK antibodies followed by RT-PCR for EWSAT1. HPRT binding to HNRNPK is shown as control. Expression levels are relative to IgG RIP. Error bars indicate SD for 3 independent experiments. (C) RT-PCR for HNRNPK knockdown using 2 different shRNAs. Error bars indicate SD for 3 experiments. PCR is normalized to uninfected cells. (D) Western blot analysis for HNRNPK. β-Actin was used as a loading control. (E) Soft agar colony assay on A673 cells with knockdown of HNRNPK. Data are presented as mean + SD for 3 independent experiments. ***P < 0.001 by 2-tailed t test compared with shGFP. (F) Venn diagram indicating overlap between genes repressed by EWS-FLI1, EWSAT1, and HNRNPK. P value for overlap between each pairwise comparison is indicated. (G) RT-PCR for subset of genes repressed by EWS-FLI1, EWSAT1, and HNRNPK. Fourteen of nineteen genes in overlap were validated by RT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001 by 2-tailed t test. Data are presented as mean + SD.
Knockdown of HNRNPK using 2 independent shRNAs substantially decreased cell growth (Supplemental Figure 8A) as well as soft agar colony formation (Figure 8, C–E). In addition, loss of HNRNPK led to upregulation of a subset of genes regulated by EWSAT1 by RT-PCR, whereas knockdown of STAU1 had no appreciable effect on EWSAT1-regulated genes (Supplemental Figure 8, B–F). To analyze the genome-wide transcriptional consequences of knockdown of HNRNPK on Ewing cells, we used RNAseq to compare Ewing cells after HNRNPK knockdown and control cells. Using the same analysis pipeline and the same significance cutoffs as used for the equivalent EWS-FLI1 and EWSAT1 RNAseq experiments, we identified 502 genes upregulated and 302 genes repressed by HNRNPK (Supplemental Table 9). Notably, there was no overlap in the HNRNPK-upregulated gene list with genes upregulated by EWS-FLI1 or EWSAT1. However, a highly statistically significant overlap (P < 5.5 × 10–8, Figure 8F) was noted for the 302 genes repressed by HNRNPK (i.e., upregulated after knockdown) and the list of genes repressed by both EWS-FLI1 and EWSAT1 knockdown. RT-PCR analysis validated differential expression of a subset of these genes (Figure 8G). Thus, EWSAT1 and HNRNPK participate in gene repression downstream of EWS-FLI1.
Discussion
Many large-scale sequencing studies have demonstrated pervasive transcription in mammalian genomes (48). Recent estimates suggest that >9,000 genomic loci express lncRNAs, and yet the vast majority of these remain functionally uncharacterized (49). Many lncRNAs appear to act by regulating gene expression either in cis (50) or in trans (30). Increasing evidence suggests that lncRNAs play important roles in oncogenesis. For example, several lncRNAs have been demonstrated to play precise roles in regulating genes expression in prostate cancer (51). The lncRNAs PCGEM1 and PRNCR1/PCAT8 increase loading of the androgen receptor (AR) to specific enhancers, even in the absence of AR ligand, thus playing a critical role in driving AR-regulated gene expression (52). Another prostate cancer–associated lncRNA, schLAP, acts by antagonizing the genome-wide localization of the SWI-SNF chromatin remodeling complex (53). In melanoma, the lncRNA BANCR is upregulated by oncogenic BRAF or NRAS and plays an important role in pathogenesis (54). In breast cancer, the lncRNA HOTAIR is upregulated and regulates the association of the polycomb complex 2 (PRC2) and LSD1/REST to target loci (55, 56). These and other examples suggest that lncRNAs are critical to many oncogenic processes.
To our knowledge, specific regulation of a lncRNA by an oncogenic translocation has not been described. Here, we identified EWSAT1 as a EWS-FLI1–upregulated lncRNA that plays an important role in the pathogenesis of Ewing sarcoma. This lncRNA is expressed in primary mesenchymal cells in response to EWS-FLI1, indicating that its upregulation is an early event in oncogenesis. In addition, EWSAT1 is upregulated in Ewing sarcoma tumor samples and downregulated in Ewing cell lines after EWS-FLI1 knockdown, indicating that its expression remains linked to EWS-FLI1 during the process of tumor progression. Functionally, knockdown of EWSAT1 in Ewing sarcoma cell lines significantly limits proliferation and soft agar colony growth, indicating that loss of function of this lncRNA is deleterious to the pro-oncogenic program downstream of EWS-FLI1. Notably, while loss of function of EWSAT1 suggested that this lncRNA may have oncogenic properties by itself, we were unable to detect a reproducible effect of EWSAT1 overexpression in either Ewing tumor cells or fibroblasts. Indeed, overexpression of EWSAT1 was poorly tolerated in some cell types, suggesting that EWSAT1 levels are tightly controlled or titrated to the presence of other components of the genetic network in which it participates. Thus, EWSAT1 is necessary but not sufficient to increase the oncogenic capacity of Ewing cells.
lncRNAs have been demonstrated to play complex roles in many biological processes. To elucidate the molecular mechanisms through which EWSAT1 is involved in Ewing pathogenesis, we analyzed the transcriptional consequences of EWSAT1 loss and compared them to those of EWS-FLI1 loss. Consistent with previous microarray experiments, RNAseq analysis identified a large number of genes repressed by EWS-FLI1 (8, 16). Direct activation of gene expression by EWS-FLI1 occurs via binding to a consensus motif near promoters or by binding to a distal microsatellite repeat (57, 58). To date, the mechanisms responsible for EWS-FLI1–mediated repression remain unclear. EWS-FLI1 directly binds to promoters of only a small subset (~5%) of repressed targets (42). Thus, EWS-FLI1 likely represses genes primarily through indirect mechanisms. One possible mechanism may involve regulation of microRNAs (14, 59). Another mechanism may involve specific EWS-FLI1 target genes that can act as transcriptional repressors downstream of EWS-FLI1, such as BCL11B, NR0B1, and NKX2.2 (7, 16, 18). Gene repression may also involve chromatin remodeling, since both EWS-FLI1 and BCL11B have been shown to interact with components of the NuRD chromatin-modifying complex (18, 42, 60).
Here, we identified a mechanism through which EWS-FLI1 regulates gene expression, namely through the upregulation of EWSAT1. Our analysis suggests that this lncRNA regulates approximately 10% of EWS-FLI1–repressed genes. This is markedly higher than the number of targets that have been demonstrated to be regulated by specific transcriptional repressors or direct binding to promoters (see Figure 7C). Thus, EWSAT1 expression leads to gene repression downstream of EWS-FLI1 and accounts for a substantial number of the repressive gene expression changes seen as a consequence of the EWS-FLI1 translocation. We identified EWSAT1 by analyzing early transcriptional events that occur after EWS-FLI1 expression in primary human MPCs, the likely cell of origin of Ewing sarcoma. Interestingly, expression of EWS-FLI1 in these cells did not lead to full activation of the EWS-FLI1 expression program seen in fully transformed cells. Specifically, EWS-FLI1 expression in pMPCs did not result in repression of genes previously shown to be repressed by EWS-FLI1 in Ewing sarcoma cell lines (Figure 1), despite expression of EWSAT1. This suggests that other genetic or epigenetic events are needed in order for EWSAT1 to regulate the repressive program downstream of EWS-FLI1.
Using protein arrays and RIP, we identified HNRNPK and an RNA-binding protein that binds to EWSAT1. Notably, loss of HNRNPK in Ewing sarcoma cells recapitulated the phenotype noted after EWSAT1 knockdown. In addition, a significant fraction of the genes regulated by EWSAT1 loss were similarly regulated by loss of HNRNPK. Thus, EWSAT1 may act in part by regulating the interaction of HNRNPK with target genes, although EWSAT1 probably interacts with other RNA-binding proteins that will need to be identified to fully understand its molecular function. HNRNPK has been reported to play a role in regulating cell signaling, transcription, and translation (61). Previous work supports a role for HNRPNK-lncRNA interactions as mediators of gene repression (43, 62). HNRNPK may mediate gene repression through disrupting DNA-protein interactions at specific cytosine/thymidine-rich regions within promoters without direct binding to DNA (63). Our studies suggest that EWSAT1 and HNRNPK act together to regulate gene repression of a subset of genes in the context of Ewing sarcoma. However, as many EWSAT1-repressed genes were not altered by HNRNPK downregulation, it is likely that EWSAT1 interacts with other yet to be identified proteins.
In summary, our study describes a pro-oncogenic function for a previously uncharacterized lncRNA. To our knowledge, these results provide one of the first indications that the function of an oncogenic translocation can be regulated by expression of a lncRNA. Indeed, unpublished data suggest that EWS-FLI1 regulates expression of hundreds of other lncRNAS. Our results also suggest that lncRNAs may play a larger role in mediating the pathogenesis of Ewing sarcoma than has been appreciated up to now. Our findings warrant careful analysis of other sarcoma translocations that may also function at least in part by regulating lncRNAs. Thus, these studies may have wide implications beyond the relatively rare Ewing sarcoma.
Methods
Cell lines.
pMPCs were isolated as previously described (64). Clinical characteristics of the isolated samples are shown in Supplemental Table 8. pMPCs were cultured in aMEM supplemented with 20% FBS (Omega Scientific) and 100 U pen/100 μg strep/2 mM glutamine (Invitrogen). Ewing sarcoma cell lines and osteosarcoma were purchased from ATCC and cultured in DMEM supplemented with 10% bovine growth serum (BGS) (HyClone) and 100 U pen/100 μg strep/2 mM glutamine. TC71 was a gift from Timothy Triche (University of Southern California, Los Angeles, California, USA). Prostate cancer cell lines were a gift from Jonathan Pollack (Stanford University, Stanford, California, USA). Breast cancer cell lines were a gift from Michael Clark (Stanford University). Patient-derived xenograft PSS090 was obtained from a sample obtained directly from a patient with Ewing sarcoma and propagated in mice. IMR90 cells were purchased from ATCC and cultured in DMEM containing 10% BGS with pen/strep/glutamine.
FACS analysis.
Freshly trypsinized cells were spun down at 100 g at 4°C and washed with 1 ml 2% FBS in PBS. Cells were resuspended in the appropriate antibody dilution and left in the dark on ice for 30 minutes, washed with 2 ml 2% FBS in PBS, and resuspended in 200 μl 2% FBS in PBS. All analysis was done on a FACSCalibur apparatus (BD Biosciences). Antibodies used and their dilutions are as follows: FITC-CD146 (Millipore MAB16985F) used at 1:100, FITC-CD90 (BD Pharmingen 555595) used at 1:100, FITC CD45 (BD 340664) used at 1:100, FITC-IgG1 (Chemicon CBL600F) used at 1:100, PE-CD105 (Abcam AB53321) used at 1:20, and PE-IgG2a (eBiosciences 12-4321-81) used at 1:20.
Differentiation assays.
For osteogenic differentiation, 1.2 × 104 pMPCs per well were plated on a 12-well plate. After 24 hours, cells were induced with osteogenic differentiation media (Lonza, PT-3002). Media were changed every 3 to 4 days. After 3 weeks in culture, wells were washed 3 times with PBS and fixed in 100% ethanol for 15 minutes at room temperature. Wells were stained with 0.2% Alizarin Red S (Sigma-Aldrich 130-22-3, diluted in water) for 30 to 60 minutes at room temperature, rinsed with water, and then left to dry overnight at room temperature. For adipogenic differentiation, 8.0 × 104 pMPCs per well were plated on a 12-well plate. Upon confluency (2–3 days later), cells were induced with adipogenic induction media (Lonza PT-3004). Three days later, the media were replaced with adipogenic maintenance media and then changed every 3 days thereafter, alternating between adipogenic induction media and adipogenic maintenance media. After 3 rounds of induction and maintenance (~24 days after induction), the wells were washed with PBS and then fixed in 10% phosphate-buffered formalin for 20 to 30 minutes at room temperature. Wells were stained with Oil Red O (0.5% Oil Red O diluted in methanol, Sigma-Aldrich 00625) for 30 minutes at 37°C. Wells were washed with PBS and allowed to dry overnight at room temperature.
CFU fibroblasts.
100 pMPCs cells were plated per 10-cm plate and then left to grow for 2 weeks. Plates were then washed with PBS and fixed for 15 to 20 minutes in 10% phosphate-buffered formalin. Colonies were stained using 0.1% crystal violet for 30 minutes at room temperature, rinsed in water, and allowed to dry overnight at room temperature.
Viral infections.
EWS-FLI1 (fusion type 1) cDNA cloned into the pLRT retroviral vector was a gift from Fernando Lecanda (Centro de Investigación Médica Aplicada, Pamplona, Spain) (65). Virus was made using calcium phosphate transfection into 293FT cells. Virus was filtered, spun for 2 hours at 50,000 g at 4°C, resuspended overnight in PBS with 40 mg/ml lactate, and then applied to pMPCs with polybrene for 24 hours. Cells were selected with 7.5 μg/ml Blasticidin (Invitrogen). Doxycycline at 0.1 μg/ml was used to induce EWS-FLI1 expression. The pLKO.1 shRNA lentiviral system was used to knockdown genes of interest. pLKO.1-shGFP, pLKO.1-shEWSAT1-1, and pLKO.1-shEWSAT1-2 were purchased from Open Biosystems. pLKO.1-shEWS-FLI1 and pLKO.1-shEWSAT1-8 were generated by cloning oligos into pLKO.1. Target sequences for shRNAs can be found in the Supplemental Methods. Puromycin at 2.5 μg/ml was used to select transduced cells. cDNAs for EWS-FLI1 and EWSAT1 were cloned into pLenti6/V5-DEST vector (Invitrogen).
RNA isolation.
RNA was isolated using TRIzol (Invitrogen) following the manufacturer’s specifications. cDNA was synthesized using either the DyNAmo cDNA Synthesis Kit (New England Biolabs, F470) or the Maxima First Strand cDNA Synthesis Kit (Thermo, K1642). RT-PCR was performed using SYBR Green (Applied Biosystems or Quanta Biosciences). RT-PCR primer sequences can be found in the Supplemental Methods. Northern blotting was carried out with standard protocols using a probe against the first exon of EWSAT1. Nuclear/cytoplasmic fractionation was performed using the PARIS Kit (Ambion) following the manufacturer’s instructions.
Western blot.
Cell pellets were lysed in RIPA buffer supplemented with protease inhibitor cocktail tablets (Roche) and phosphatase inhibitor cocktails 2 and 3 (Sigma-Aldrich). Proteins were quantified using the BCA kit (Thermo Scientific). 25 to 50 μg of total protein was resolved by SDS-PAGE and transferred onto Immobilon-FL membranes (Millipore). Membranes were blocked in Li-Cor blocking buffer for 1 hour and then probed in primary antibody overnight. Membranes were probed for 1 hour at room temperature using the appropriate Li-Cor IRDye 680 and IRDye 800 secondary antibodies, depending on the species. Membranes were then scanned on the Odyssey Imaging Scanner (Li-Cor). Anti-FLI1 antibody was used at 1:500 (BD Pharmingen 554266). Anti–β-Tubulin was used at 1:1,000 (Li-Cor, 926-42211). Anti-V5 antibody was used at 1:5,000 (Invitrogen R96025). Anti-HNRPK antibody was used at 1:1,000 (Santa Cruz Biotechnology SC-25373). Anti–β-Actin was used at 1:5,000 (Sigma-Aldrich A5316). Quantification was done by using Li-Cor Odyssey Scanner and Li-Cor Odyssey Software and dividing the levels of EWS-FLI1 by the levels of β-tubulin and then normalized to pMPC10.
Microarray analysis.
RNA from pMPCs was isolated using TRIzol (Invitrogen) following the manufacturer’s specifications and then purified using a RNAeasy column (Qiagen) followed by hybridization to Affymetrix arrays following the manufacturer’s instructions. Microarray data were normalized with Expression Console software (Affymetrix) using the RMA algorithm (66). Low signals (below 50) were filtered out using the Preprocess Data Set module in GenePattern (http://broad.mit.edu/cancer/software/genepattern/). Differentially expressed genes in published data sets of Ewing sarcoma cell lines with knockdown of EWS-FLI1 compared with control knockdown were identified using Significance Analysis of Microarrays software, with the cutoff FDR of 5% (8, 67). To compare the arrays from different sources, array probe data were collapsed to gene symbols and filtered for a minimum fold change of 1.5, followed by standardization (SDs from the mean) of each gene symbol in each data set (list of arrays in Supplemental Table 1). Hierarchical clustering was used to compare standardized array data, using Pearson correlation to measure sample distance (68).
Analysis of published human 3SEQ data.
Raw data were downloaded from the NCBI Sequence Read Archive (SRA accession SRP006575) (69). Reads were converted to FASTQ format and mapped to hg19 using BWA (70). Using the Broad Institute lincRNA annotations (71), raw read counts and cpm values for each annotated lincRNA were obtained using NGSUtils (72). cpm values were normalized to the millions of reads mapped to hg19.
RNAseq library preparation and analysis.
For RNAseq of pMPCs, sequencing was carried out using the 3SEQ method following published protocols (73). Total RNA was extracted by TRIzol (Invitrogen) and subjected to poly-A selection with the Dynabeads mRNA Selection Kit (Invitrogen catalog no. 610-06). The mRNA was then heat sheared, and the 3′ ends of the transcripts were transcribed into cDNA using an oligo-dT_P7 primer. After cDNA synthesis of the first and second strands, the P5 linker was ligated to the end opposite the P7 linker. The linker-ligated product was selected for 220- to 320-bp fragments on a Nusieve GTG Agarose Gel (Lonza, 50080) and subsequently amplified by 15-cycle PCR to generate the directional sequencing library. Sequencing was performed from the P5 primer to generate 36-bp reads using an Illumina Genome Analyzer II. Processing was carried out as described previously (74). Briefly, reads were mapped to the human genome (hg19) using DNAnexus. The analysis module “3SEQ/Transcriptome-based quantification” was used for each sample using the parameters “sense read” for mapping direction and the ENSEMBL transcriptome as the reference. To identify differentially expressed genes, read counts were rounded to the nearest integer and the DESeq R script was used with an adjusted P value (padj) cutoff of 0.1.
For RNA sequencing of shRNA-infected A673 cells, RNA from A673 cells with shRNA-mediated knockdown of GFP (4 libraries), EWS-FLI1 (4 libraries), or EWSAT1 (7 libraries) or HNRNPK (2 libraries) was isolated with TRIzol (Invitrogen) according to the manufacturer’s specifications. Each sample was cleaned up on an RNeasy Mini Column (Qiagen), treated with DNase, and analyzed for quality on an Agilent 2100 Bioanalyzer. 100–150 ng RNA was synthesized into cDNA and amplified using the NuGen Encore Complete Kit to produce strand-specific and rRNA-depleted libraries. Samples were multiplexed (4 per lane) for 2 × 100-bp paired-end sequencing on an Illumina HiSeq 2000. Reads were mapped to hg19 using STAR with default options (75). A splice-junction aware STAR index was constructed using Ensembl gene annotations. SAM output was then sorted and converted to BAM files using Picard (http://picard.sourceforge.net). Read counts for each gene were obtained with NGSUtils using Ensembl gene annotation (72). Differential expression analysis was performed using edgeR (76) for each comparison (EWS-FLI1 vs. GFP, lncRNA vs. GFP, and HNRNPK vs. GFP). Only genes with a mean read count of 10 in at least one condition were retained for further analysis. Genes with a FDR less than 0.1 and a fold count greater than 2.0 were considered to be significant. Analysis was performed using the R statistical environment. All data for RNAseq experiments are available through the Gene Expression Omnibus (GSE60891 and GSE60949). For analysis of human Ewing sarcoma RNAseq, aligned RNASeq data from Ewing sarcoma tumors was generated as previously described (41). We calculated the reads per kilobase per million mapped reads values for coding and noncoding transcripts by applying the RNA-SeQC tool (Firehose RNASeqMetrics v35), with a modified Genecode (v12) annotation file(http://www.broadinstitute.org/cancer/cga/tools/rnaseqc/examples/gencode.v12.annotation.patched_contigs.gtf.gz) (77).
Visualization of RNAseq reads.
For the uninfected pMPCs and pMPCs expressing EWS-FLI1, reads from each replicate were combined and overall coverage was calculated. Coverage amounts were normalized using the total number of reads in each sample. BigWig files were generated from the coverage data and used for visualization. For shEWS-FLI1, shGFP, and shEWSAT1 samples, reads from each replicate were combined and overall coverage for the sample was calculated. Coverage amounts were normalized using the total number of reads in each sample. BigWig files were generated from the coverage data and used for visualization with the UCSC genome browser.
Gene set enrichment analysis.
Gene set enrichment analysis was performed as described previously (78). Either the pMPC RNAseq list or published gene sets were used, as indicated. Data sets were generated from either the pMPC RNAseq or the A673 shGFP and shEWSAT1 knockdown experiments; otherwise, published data sets were used: GSE12102 (36), GSE9520 (79), and GSE12274 (80).
Gene ontology analysis.
To identify functional categories of genes regulated by EWSAT1, we used DAVID for gene annotation enrichment analysis (http://david.abcc.ncifcrf.gov/).
In vitro assays.
Cellular growth was assessed by plating cells into a 12-well plate after selection at a specific density and then counting in triplicate 24 hours after plating and then again 72 to 96 hours after plating. Growth was assessed by dividing the average number of cells at the final time point by the average number of cells at the first time point and then normalized to control cells. Soft agar assays were performed following standard protocols, seeding the cells into 0.35% agar and allowing them to grow for 2 weeks. Plates were then refrigerated for an hour and liquid media were removed. Colonies were stained with 0.02% Giemsa diluted in PBS for 10 minutes at room temperature and then rinsed with water and scanned 24 hours later. Relative cell viability was measured using MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; thiazolyl blue) tetrazolium salt and and plate reader (Synergy H1 Hybrid Reader, Biotek). Viability was measured by dividing the readout from day n (n = day 2 or onward) by the value from day 1 (e.g., day 3 = 0.75, day 1 = 0.5, relative proliferation/growth = 0.75/0.5 = 1.5).
CPC analysis.
The CPC analysis was performed using the sequence information for RefSeq accession no. NR_026949.1 using the online tool (http://cpc.cbi.pku.edu.cn).
Protein array analyses.
pGEMT-EWSAT1 plasmid was digested with SacII, and 4 μg linear DNA was used for in vitro transcription in a 50-μl total volume consisting of 1x Transcription buffer (Promega), 10 mM DTT (Promega), 1 mM NTP (Invitrogen), 40 units RNAseOUT (Invitrogen), and 60 units RNA polymerases SP6. The reaction was carried out at 37°C for 4 hours, after which DNA was digested by addition of 2 U DNase I at 37°C for 15 minutes. RNA was phenol-chloroform extracted, measured after ethanol precipitation using a NanoDrop 1000 spectrophotometer (Thermo Scientific), and visualized using denaturing agarose gel-electrophoreses. RNA labeling for microarray incubations was performed using a Label IT μArray Cy5 labeling Kit (Mirus) using 5 μg RNA and 3 μl Label IT Cy5, diluted 1:10 in water in a final volume of 25 μl. Reaction was incubated for 30 minutes at 37°C, terminated with 10x STOP buffer (Mirus), and purified via ethanol precipitation. RNA labeling density was evaluated using a NanoDrop 1000 spectrophotometer (Thermo Scientific), resulting in 1 molecule of Cy5 dye per 1,898 bp of EWSAT1, and visualized using denaturing agarose gel-electrophoreses. For RNA incubation, ProtoArray Human Protein Microarray v5.0 (Invitrogen) was used. 4 mg Cy5-labeled EWSAT1 was hybridized to protein arrays as described previously (45), slides were scanned at 635 nm (Cy5) using a GenePix 4000B Microarray scanner (Molecular Devices), and the intensity of the wavelength signal at each spotted protein location was determined with GenePix Pro 6.1 software (Molecular Devices). Data were filtered based on signal above the background for each of the duplicate feature to be greater than 2.5-fold from the global mean signal from all of the spotted proteins.
RIP analysis.
HNRPK RIP was performed as described previously (46). Briefly, A673 Ewing sarcoma cells were harvested by trypsinization and washed with PBS. Cells were resuspended in 4 ml ice-cold PBS and 4 ml C1 buffer (1.28 M sucrose, 40 mM Tris HCl [pH 7.4], 20 mM MgCl2, 4% Triton X-100) followed by 12 ml ice-cold water. After incubation on ice, cells were spun at 500 g for 15 minutes at 4°C, and the nuclear pellet was resuspended in 3 ml RIP buffer (150 mM KCl, 25 mM Tris [pH 7.4], 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, protease inhibitors, and RNAse inhibitors). The nuclei were dounced, and debris was collected by centrifugation. The supernatant was collected and diluted with RIP buffer. Antibody was added and incubated overnight with rotation at 4°C. 60 μl prewashed protein G dynabeads (Invitrogen 10003D) were then added and incubated for 2 hours with rotation at 4°C. After, 3 washes with RIP buffer and 1 with PBS, beads were resuspended in TRIzol for RNA extraction. IgG (Abcam 37415) and HNRPK (Abcam AB70492 and AB39975) antibodies were used.
Statistics.
P values were calculated using either 2-tailed Student’s t test (2-sided) or Fisher’s exact test (for overlaps between gene sets). A P value of less than 0.05 was considered significant.
Study approval.
Bone fragments used for establishment of pMPC lines were obtained with parental permission following IRB guidelines from Lucile Packard Children’s Hospital Stanford, Palo Alto, California, USA. PSS090 is a xenograft established directly from a Ewing tumor isolated from a patient with informed consent and IRB approval from both the University of Washington and Stanford University. Mice used for propagation of xenografts were housed in the Stanford Animal Care Facility with approval of the Animal Care Committee.
Supplementary Material
Acknowledgments
We thank the members of the Sweet-Cordero lab and the labs of Laura Attardi, Steven Artandi, and Julien Sage for useful comments and advice. We thank Howard Chang, Arend Sidow, Javed Khan, and Maite Huarte for protocols and useful discussions. We thank Fernando Lecanda for the inducible EWS-FLI1 vector. We thank Jonathan Pollack, Jae Cho, and Michael Clark for their generous gift of cancer cell lines. E.A. Sweet-Cordero received grant support from the Doris Duke Clinical Scientist Development Fund, the St. Baldrick’s Foundation, the Kavner Family Fund, the Hope Street Kids Foundation, the Sunbeam Foundation, the Elsa U. Pardee Foundation, the Bear Necessities Foundation, and Hyundai Hope on Wheels. M.M. Howarth and D. Simpson were funded in part by the National Cancer Institute of the NIH under award number T32CA009302. R. Chen was funded by a Stanford Graduate Fellowship.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(12):5275–5290. doi:10.1172/JCI72124.
References
- 1.Riggi N, Cironi L, Suva ML, Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins. Part 1: The fellowship of TET. J Pathol. 2007;213(1):4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- 2.May WA, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13(12):7393–7398. doi: 10.1128/mcb.13.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8(21):2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 4.Deneen B, Hamidi H, Denny CT. Functional analysis of the EWS/ETS target gene uridine phosphorylase. Cancer Res. 2003;63(14):4268–4274. [PubMed] [Google Scholar]
- 5.Thompson AD, et al. EAT-2 is a novel SH2 domain containing protein that is up regulated by Ewing’s sarcoma EWS/FLI1 fusion gene. Oncogene. 1996;13(12):2649–2658. [PubMed] [Google Scholar]
- 6.Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5(23):2753–2759. doi: 10.4161/cc.5.23.3505. [DOI] [PubMed] [Google Scholar]
- 7.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol Cancer Res. 2006;4(11):851–859. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 8.Smith R, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9(5):405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Kauer M, et al. A molecular function map of Ewing’s sarcoma. PLoS One. 2009;4(4):e5415. doi: 10.1371/journal.pone.0005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 11.Riggi N, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68(7):2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 12.Richter GH, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106(13):5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cironi L, et al. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One. 2008;3(7):e2634. doi: 10.1371/journal.pone.0002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riggi N, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sankar S, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32(42):5089–5100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS One. 2008;3(4):e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinsey M, Smith R, Iyer AK, McCabe ER, Lessnick SL. EWS/FLI and its downstream target NR0B1 interact directly to modulate transcription and oncogenesis in Ewing’s sarcoma. Cancer Res. 2009;69(23):9047–9055. doi: 10.1158/0008-5472.CAN-09-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiles ET, Lui-Sargent B, Bell R, Lessnick SL. BCL11B is up-regulated by EWS/FLI and contributes to the transformed phenotype in Ewing sarcoma. PLoS One. 2013;8(3):e59369. doi: 10.1371/journal.pone.0059369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 20.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 21.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derrien T, Guigo R, Johnson R. The long non-coding RNAs: A New (P)layer in the “Dark Matter”. Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351(6324):325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21(4):400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 26.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacchetti B, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuma M, Okita H, Hata J, Umezawa A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene. 2003;22(1):1–9. doi: 10.1038/sj.onc.1206055. [DOI] [PubMed] [Google Scholar]
- 34.Herrero-Martin D, et al. Stable interference of EWS-FLI1 in an Ewing sarcoma cell line impairs IGF-1/IGF-1R signalling and reveals TOPK as a new target. Br J Cancer. 2009;101(1):80–90. doi: 10.1038/sj.bjc.6605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tirado OM, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006;66(20):9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 36.Scotlandi K, et al. Overcoming resistance to conventional drugs in Ewing sarcoma and identification of molecular predictors of outcome. J Clin Oncol. 2009;27(13):2209–2216. doi: 10.1200/JCO.2008.19.2542. [DOI] [PubMed] [Google Scholar]
- 37.Kong L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunner AL, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13(8):R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel M, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22(2):259–270. doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crompton BD, et al. The Genomic Landscape of Pediatric Ewing Sarcoma. Cancer Discov. doi: 10.1158/2159-8290.CD-13-1037. [published online ahead of print September 3, 2014]. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 42.Sankar S, et al. Mechanism relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2012;32(42):5089–5100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Huang da W, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siprashvili Z, et al. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012;13(1):633. doi: 10.1186/1471-2164-13-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470(7333):284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 51.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flockhart RJ, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012;22(6):1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One. 2009;4(3):e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gangwal K, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105(29):10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Vito C, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21(6):807–821. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 60.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24(45):6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 61.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26(6):629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 62.Moumen A, Masterson P, O’Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123(6):1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 63.Stains JP, Lecanda F, Towler DA, Civitelli R. Heterogeneous nuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter. Biochem J. 2005;385(pt 2):613–623. doi: 10.1042/BJ20040680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuznetsov SA, et al. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J Bone Miner Res. 1997;12(9):1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- 65.Gonzalez I, Vicent S, de Alava E, Lecanda F. EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. J Mol Med (Berl). 2007;85(9):1015–1029. doi: 10.1007/s00109-007-0202-5. [DOI] [PubMed] [Google Scholar]
- 66.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 67.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brunner AL, et al. Transcriptional profiling of lncRNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13(8):R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics. 2013;29(4):494–496. doi: 10.1093/bioinformatics/bts731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beck AH, et al. 3’-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5(1):e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larson BL, Ylostalo J, Prockop DJ. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26(1):193–201. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 80.Wagner W, et al. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS One. 2009;4(6):e5846. doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.