Abstract
The presence of large genomic regions with suppressed recombination (SR) is a key shared property of some sex- and mating-type determining (mat) chromosomes identified to date in animals, plants, and fungi. Why such regions form and how they evolve remain central questions in evolutionary genetics. The smut fungus Microbotryum lychnis-dioicae is a basidiomycete fungus in which dimorphic mat chromosomes have been reported, but the size, age, and evolutionary dynamics of the SR region remains unresolved. To identify the SR region in M. lychnis-dioicae and to study its evolution, we sequenced 12 genomes (6 per mating type) of this species and identified the genomic contigs that show fixed sequence differences between the mating types. We report that the SR region spans more than half of the mat chromosome (>2.3 Mbp) and that it is of very recent origin (∼2 × 106 years) as the average sequence divergence between mating types was only 2% in the SR region. This contrasts with a much higher divergence in and around the mating-type determining pheromone receptor locus in the SR, suggesting a recent and massive expansion of the SR region. Our results comprise the first reported case of recent massive SR expansion documented in a basidiomycete fungus.
Keywords: Microbotryum, evolution, fungi, mating type, sex chromosome
SEX chromosomes play a fundamental role in evolutionary biology, and are thought to contribute toward key phenomena such as dichotomous sexes, adaptation, and speciation (Rice 1984; Presgraves 2008; Bergero and Charlesworth 2009; Bachtrog 2013). A key feature shared in sex chromosomes from animals and plants, and some mating-type (mat) chromosomes in algae and fungi, is the presence of regions with suppressed recombination (SR) (Merino et al. 1996; Charlesworth and Charlesworth 2005; Bergero and Charlesworth 2009; Ferris et al. 2010; Bachtrog 2013; Charlesworth 2013; Vicoso et al. 2013). SR regions on fungal mat chromosomes have been reported for various fungi including the ascomycete Neurospora tetrasperma and the basidiomycete fungi Cryptococcus neoformans and Microbotryum lychnis-dioicae (previously named Microbotryum violaceum and Ustilago violacea) (Merino et al. 1996; Lengeler et al. 2002; Hood 2002; Fraser et al. 2004; Menkis et al. 2008; Coelho et al. 2010; Ellison et al. 2011; Whittle et al. 2011a).
SR regions of many organisms, including humans, contain a signature of stepwise expansion over time, termed “evolutionary strata,” with older SR regions showing higher divergence between the X- and Y-linked homologous genes compared to SR regions that stopped recombining more recently (Lahn and Page 1999; Handley et al. 2004; Menkis et al. 2008; Bergero and Charlesworth 2009). The causes of SR expansion are not well understood and remain the subject of active research (e.g., Qiu et al. 2013). According to the prevailing hypothesis, the formation and expansion of sex-specific SR regions in animals and plants is driven by sexual antagonism—mutations advantageous in one sex and harmful in the other (Charlesworth and Charlesworth 1980; Rice 1987; Bergero and Charlesworth 2009). Indeed, the SR regions in species that lack sexual antagonism, such as self-incompatibility loci in plants, are usually relatively small compared to the chromosome size (e.g., Goubet et al. 2012). In certain fungi, it has been proposed that SR may arise and expand via a different mechanism—selection acting to promote linkage between the mat locus and the centromere (Antonovics and Abrams 2004; Abbate and Hood 2010; Ellison et al. 2011).
The smut fungus M. lychnis-dioicae is a well-studied biotrophic parasite of the plant Silene latifolia (Day and Cummings 1981; Bernasconi et al. 2009). M. lychnis-dioicae has a unifactorial mating system (also referred to as bipolar) with two mating types, A1 and A2. Sexual reproduction and mating are regulated by a biallelic mating-type locus (Day and Cummings 1981), located on heteromorphic mat chromosomes (Hood 2002), making the species a useful model for studying evolution of SR in fungi. In basidiomycete fungi, the unifactorial mating system is considered to be a derived state from the ancestral bifactorial (tetrapolar) system, which allows mating only when alleles at two independently segregating mating-type loci are different (reviewed in Nieuwenhuis et al. 2013). For example, the closest (but still very distant) relative of M. lychnis-dioicae with a fully sequenced genome, Ustilago maydis, has a tetrapolar system (Kämper et al. 2006), while its close relative U. hordei has evolved bipolarity, where the two mating-type factors (pheromone/pheromone receptor and homeodomain genes) are linked together into a single “mat locus” (Lee et al. 1999). The evolution of linkage between the two mating-type loci, leading to a switch from tetrapolar (bifactorial) to bipolar (unifactorial) system, has occurred on multiple occasions in Basidiomycetes, including the progenitor of M. lychnis-dioicae (Nieuwenhuis et al. 2013). Such linkage should lead to the evolution of an SR region between the mating-type loci. However, the physical distance between the two component mating-type factors in M. lychnis-dioicae is not clear from the current genome assembly, because these factors are on disconnected genomic contigs (Petit et al. 2013).
The SR region of M. lychnis-dioicae is therefore of unknown size and age: the region spans at least 1 Mbp (estimated from strains Sl-405 and Sl-127; Votintseva and Filatov 2009), and the A1 region might be as large as 3.3 Mbp, and 4.0 Mbp for the A2 chromosome (estimated from the Lamole strain)—89 and 91% of the estimated length of mat A1 and A2 chromosomes, respectively (Hood et al. 2013). However, only three M. lychnis-dioicae strains have been examined so far and other strains may have larger or smaller SR regions.
Based on optical mapping, which failed to align the A1 and A2 maps, Hood et al. (2013) concluded that A1–A2 divergence was high and that the SR region in M. lychnis-dioicae is ancient. This conflicts with results of Votintseva and Filatov (2009) that indicated a low divergence between mat A1- and A2-linked alleles, for a small number of short DNA sequences (ranging from 0 to 8.6% across all sites), suggesting that the SR region stopped recombining recently. It was also reported that the SR region had evolutionary strata (Votintseva and Filatov 2009), similar to many animal systems (Lahn and Page 1999). However, the presence of evolutionary strata in M. lychnis-dioicae was disputed by Hood et al. (2013). Until now, adequate genome-wide sequence data were not available to confirm the size and age of the SR region in M. lychnis-dioicae. Specifically, sequence data were limited only to the A1 mating type, whose assembly is fragmented, consisting of >1000 disconnected contigs (http://www.broadinstitute.org/).
In this study, we report complete genome sequences for 12 independent M. lychnis-dioicae strains, including both mating types (Table 1). By assessing sequence divergence between mat-A1 and -A2 chromosomes, we identify the SR region that is mating-type specific and includes the pheromone receptor and homeodomain genes. Furthermore, we demonstrate that, contrary to the recent optical mapping results (Hood et al. 2013), the most of this region is of very recent origin.
Table 1. Strains of M. lychnis-dioicae used in the study.
| Strain name (with mating type) | Collection location |
|---|---|
| Lamole (reference) | Lamole, Italy |
| 405A1 | Balsta, Sweden |
| 1088A1 | Lausanne, Switzerland |
| 562A1 | Krakow, Poland |
| 769A1 | Galicia, Spain |
| 1069A1 | Cotswolds, England |
| 1075A1 | Sardinia, Italy |
| 1089A2 | Lausanne, Switzerland |
| 1090A2 | Pyrenees, France |
| 563A2 | Krakow, Poland |
| 920A2 | Valencia, Spain |
| 1076A2 | Sardinia, Italy |
| 1103A2 | Woodstock, England |
Materials and Methods
Data
Twelve M. lychnis-dioicae samples (six per mating type) were collected across Europe (Table 1), haploidized, and grown in axenic culture in the lab, as described previously (Votintseva and Filatov 2009). The genomic DNA for each sample was extracted using Qiagen DNeasy Plant Kit. The DNA was sequenced by the Oxford Wellcome Trust Centre for Human Genetics (WTCHG) using Illumina sequencing technology.
A de novo assembly of the A2 genome was conducted using the CLC genomics workbench 6 (CLCgw) with k-mer size 41. Scaffolding was conducted in the same program as a part of the de novo assembly. The de novo-assembled A2 genome was aligned to the genome sequence of the Lamole A1 strain of this species produced by the Broad Institute (26-Mb genome in 1231 scaffolds; N50 = 185,029) (hereafter referred to as lam1). We used a progressive mauve algorithm implemented in Mauve v. 2.3.1 (Darling et al. 2010).
To assemble sequence alignments for downstream analyses, Illumina reads for all 12 sequenced M. lychnis-dioicae strains were mapped to the lam1 genomic scaffolds, using the CLCgw software with a high stringency (mismatch cost 3, insertion cost 3, deletion cost 3, minimum identity 0.95, nonuniquely mapping reads excluded). Less stringent mapping parameters (mismatch cost 2, insertion cost 2, deletion cost 2, minimum identity 0.9, nonuniquely mapping reads excluded) did not affect our conclusions. All results were robust to the choice of read mapping parameters. Excluding the nonuniquely mapping reads effectively masks all repetitive regions, as no reads map to such regions. Read coverage values for each scaffold in each strain are listed in Supporting Information, Table S1. The mapped reads were exported as BAM files, and a consensus was called for each strain with samtools and vcf/bcf tools (Danecek et al. 2011). The consensus sequences from the 12 M. lychnis-dioicae strains were combined into alignments for each reference contig using ProSeq v. 3.6 (Filatov 2009). The same program was used to annotate coding regions in these alignments by importing information about protein coding regions (CDS) from annotation files for the lam1 reference downloaded from the Broad Institute website.
Scaffolding of the lam1 reference was conducted with sspace v. 1.1 using paired-end sequences of A1-mating-type samples. The matA1 scaffold containing the pheromone receptor and homeodomain genes was identified with blastn using published sequences of these genes (e.g., Petit et al. 2013). Nine lam1 supercontigs that were included in the matA1 scaffold were identified and verified by aligning these lam1 supercontigs with the matA1 scaffold (Figure S1). The links between the nine supercontigs were verified by remapping the paired-end sequence data to the scaffolds using CLCgw with high stringency and manually checking every junction between the lam1 supecontigs against the matA1 scaffold (Figure S2).
Identification of recombinationally suppressed regions on the mat chromosomes
Mating-type chromosomes from the mat A1 and mat A2 genomes do not recombine and are expected to exhibit elevated sequence divergence in the SR region as compared to recombining genome regions (cf. Menkis et al. 2008). Accordingly, the SR region was identified on the basis of sequence divergence between strains of the two mating types sampled from multiple natural populations. For this, we identified the 5000-bp segments where the mating types differed by at least five fixed sites (FS) including sites with gaps/missing data (including single sites with missing information, state N) specific to one mating type, and shared no more than five polymorphisms (SP). This allowed us to identify the SR regions specific to the A1 mating type and regions with high A1-A2 divergence. Supercontigs matching these criteria were classified as candidates for inclusion in the SR and all others as non-SR (NSR).
A randomly chosen subset of these supercontigs (listed in Table S1) was tested for linkage to mating type using segregation analysis in a previously described genetic cross (Votintseva and Filatov 2009). Sequence differences identified in genomic sequences of the parental strains 405A1 and 1090A2 (the same as strains Sl-405 and Sl-127 in Votintseva and Filatov 2009), were used as genetic markers in our segregation analysis and were genotyped in progeny of the cross, by PCR and sequencing as described previously (Votintseva and Filatov 2009). Single nucleotide polymorphism (SNP) variants were genotyped in 30 supercontigs listed in Table S1, which also provides the numbers of SNPs that were used. Complete cosegregation with the mating was found in all of an initial set of 48 progeny scored for all 30 contigs (with two missing data points where we failed to determine the variant in a progeny for a single marker). A further set of 51 progeny was scored for 15 of the same 30 contigs, and again perfect cosegregation was observed.
Analyses of sequence divergence
For each between-group comparison (i.e., six A1 vs. six A2 genomes), we measured genetic differentiation using net divergence (Da; Nei 1987), by subtracting intragroup polymorphism from average pairwise sequence divergence between the groups (Dxy; Nei 1987). Our sequence divergence estimate between the lam1 sequence and our de novo assembled A2 genome used Jukes–Cantor corrected measure of sequence divergence (K; Nei 1987). Divergence was estimated for all positions as well as for silent (fourfold degenerate coding) sites only. Analyses were conducted for nonoverlapping windows of 5000 bp (unless stated otherwise) along the genome using ProSeq v. 3.6 (Filatov 2009).
Results
Comparison of the genome sequences of the two mating types
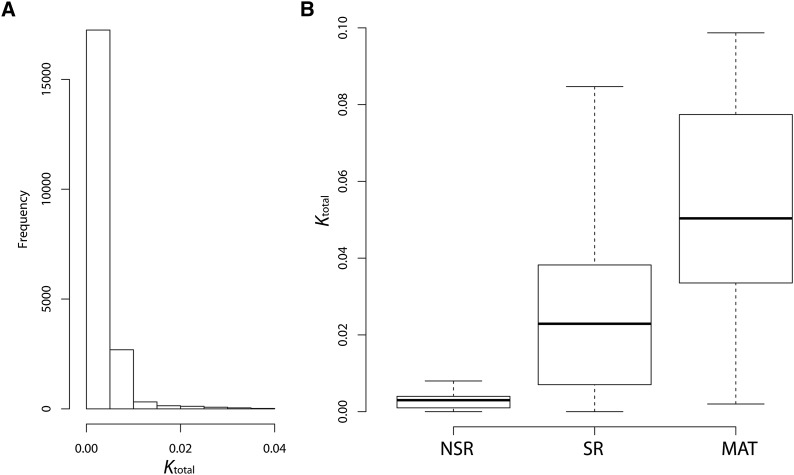
Currently only the A1 reference sequence is available (see above). To compare the genomes of the two mating types, we sequenced one of our A2 strains (1090A2) and performed de novo assembly, resulting in 2801 contigs with total length of 24.4 Mb (N50 = 70,658). Aligning to the A1 lam1 genome identified 191 local homology blocks with total alignment length of 21.2 Mb. A1–A2 divergence was very low based on all site types (Ktotal) (Figure 1A), but ranged up to Ktotal = 0.2. Similarly, the median for A1–A2 divergence at fourfold degenerate sites was only K4deg = 0.002, but ranged up to K4deg = 0.175. The 4.5-kb region around the A1 pheromone receptor locus (hereafter prA1, a gene involved in mating-type determination), is highly diverged from its A2 homolog and the DNA sequences are not alignable (Figure 2A).
Figure 1.
Pairwise sequence divergence between the genomes of the two mating types. (A) Genome-wide divergence between lam1 and de novo assembled A2 genome. (B) A comparison of mating-type sequence divergence in NSR, entire SR, and matA1 scaffolds. The box and whiskers plot shows quantiles of the distribution, while the horizontal line inside the box indicates the median. The analysis of divergence (with Jukes–Cantor correction) was conducted in 1-kb windows across the specified regions; only windows with at least 500 gap-free alignment positions are shown.
Figure 2.
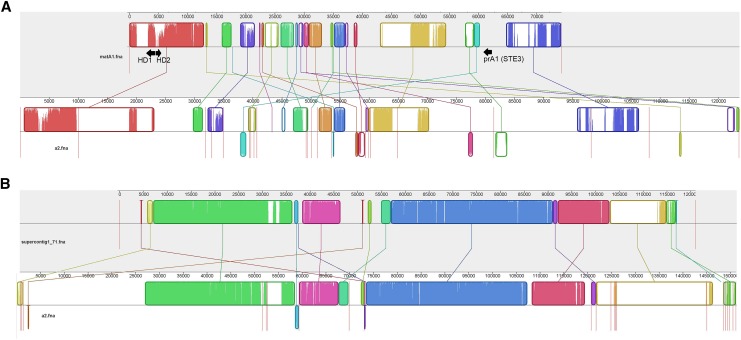
Alignment of selected mating-type A1-linked scaffolds to the A2 genome. Curved colored blocks represent collinear regions of homology between the genomes (connected by lines) and regions outside such blocks lack detectable homology between the aligned genome sequences. Inside each block are local sequence similarity profiles, with the heights of the color bars being inversely proportional to average entropy of alignment columns in the region. The color in the blocks is arbitrary. Red vertical lines indicate the boundaries of genomic scaffolds. (A) The alignment of the matA1 scaffold to homologous regions in the A2 genome; arrows indicate homeodomain and pheromone receptor loci. (B) The alignment of lam1 supercontig1.71 (longest SR scaffold in lam1 genome) with homologous regions in the A2 genome. The extent of colinearity and sequence similarity between the two mating types is much higher for this region compared to the matA1 region (A). For alignments of the full genomes and the NSR contigs see Figure S3.
The prA1 locus is located on supercontig_1.215 of the current lam1 reference assembly, which is only 15 kb long. Using paired-end sequence reads to “step” across the gaps and locate adjacent genomic contigs we extended the sequence of this region to 74 kb (Figure S1 and Figure S2). According to the Broad Institue annotation of the lam1 genome assembly, this extended scaffold contains 39 exons in 12 genes (0.16 genes/kb). This gene density is nearly 43% lower than the genomic average of 0.28 genes/kb. The extended scaffold includes both mating-type determining components—the pheromone receptor and the homeodomain genes (Figure 2A). Thus we refer to it as the mating-type A1 (matA1) scaffold. The distance between the prA1 locus and the homeodomain genes is ∼56 kb (Figure 2A and Figure S1).
An attempt to identify and align regions in the A2 genome homologous to the matA1 scaffold resulted in a very fragmented alignment (Figure 2A), in striking contrast to other parts of the genome (Figure 2B and Figure S3). Only 36.5 kb of the matA1 scaffold aligned to the A2 genome sequence, most of it in short blocks of homology on different A2 scaffolds. A1–A2 divergence in these aligned blocks was much higher than that for the rest of the genome (Figure 1B). The region around prA1 (located on an A2 genomic contig1636, of only 4 kb long) shows no homology to the A2 pheromone receptor. Our attempts to extend this contig were unsuccessful with the available paired-end data, perhaps because the de novo assembly of the A2 genome already included a scaffolding step, so no new information was available to extend the scaffolds further.
A large proportion (4.9 Mb; nearly one-fifth of the total genome) of the A1 lam1 genome sequence had no homologous sequence in the A2 genome, and 3.2 Mb (13.1%) of the A2 sequence had no homologous sequence in the A1 sequence. These sequences may be mating-type specific; however, neither genome assembly is complete, which could account for a portion of the genome-specific sequence. Furthermore, given the above small sample of strains, some of these sequences could be specific to certain strains, rather than mating-type-specific. To better describe the sequence differences between mat-A1 and A2 genomes, we sequenced the genomes from additional strains of both mating types.
Identification of the mating-type-specific regions
To locate the nonrecombining mating-type-specific region and study its evolution in M. lychnis-dioicae we resequenced the genomes of 12 haploid strains, 6 of each mating type (Table 1). The genome sequences had high coverage (median coverage = 62.47; see Table S1). Using these data, we scanned for regions with SNPs present in all strains of one mating type, but were absent from the other type (see Materials and Methods). Such fixed sequence differences should be more common in the SR than the NSR regions of mating-type chromosomes, as SR regions do not recombine and DNA sequences diverge between the mating types. We aligned the 12 newly sequenced M. lychnis-dioicae genomes against the genome assembly of the lam1 reference strain and found a total of 179 of the 1231 scaffolds with SNPs fixed between the two mating types (listed in Table S1), suggesting linkage to the mating type (mat linked). All nine supercontigs that were joined into the matA1 scaffold (Figure S1) were among these putatively mat-linked regions. To confirm linkage to mating type we used segregation analysis in a genetic cross described previously (Votintseva and Filatov 2009). We tested segregation of markers from 30 of these 179 scaffolds (Table S1) and all showed complete cosegregation with the mating type, confirming the accuracy of our assignment of scaffolds to the mat-specific SR region. Hereafter we refer to these as SR scaffolds and scaffolds in NSR regions as NSR scaffolds.
The total length of SR scaffolds detected in the lam1 assembly was 2323 kb, implying that the A1-specific SR region is at least 2.3 Mb long. According to the current annotation of the lam1 genome assembly, the SR scaffolds include 2104 exons in 402 genes. The average gene density in the matA1 scaffold (0.16 genes/kb) is similar to that in the rest of the SR scaffolds (0.17 genes/kb) and approximately half the genome-wide average (0.28 genes/kb).
Sequence divergence between the mat A1- and A2-specific SR regions
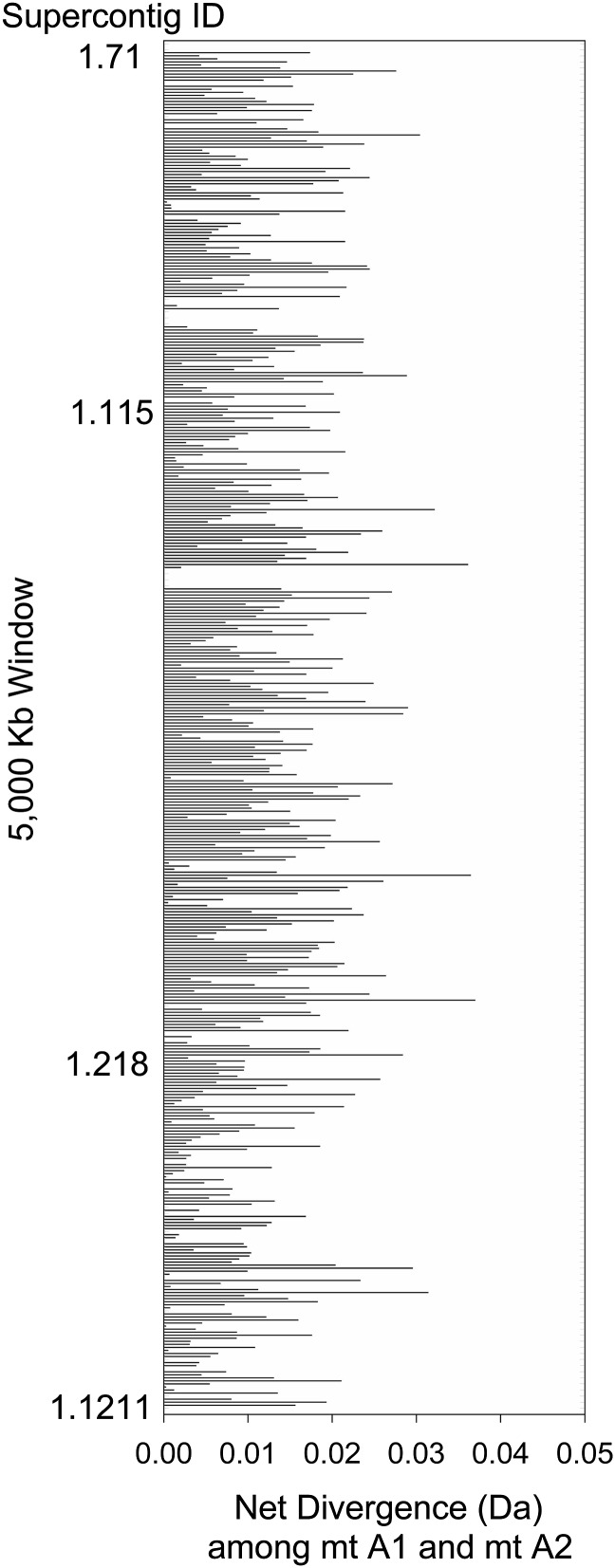
Raw divergence between the SR regions of the lam1 sequence and the de novo assembled A2 genome was relatively low (median Ktotal= 0.023, Figure 1B), indicating that much of this region has likely been recombining relatively recently. Most of the alignable SR region sequence (1618 kb out of 1890 kb, 85.6%) had A1–A2 divergence of Ktotal <3%. DNA polymorphism within each mating type could inflate the divergence, if K is low and polymorphism is high. However, SR net sequence divergence (Da) between the A1 and A2 sequences, using our samples from the natural populations, which corrects for any intragroup polymorphism (Nei 1987), varied from 0 to 0.0389 for all sites (Figure 3), with an average value of only 0.0110 (±3.5 × 10−4). The Da values for the SR were 55-fold higher than that for NSR regions, which had a mean value of 0.0002 (±1.4 × 10−5; t-test P = 3.9 × 10−120). Fewer than 5.2% of windows from the SR region had Da lower than the mean for the NSR regions. The results for silent sites give similar conclusions: average Da = 0.0099 (± 3.37 × 10−3) and 0.00035 (±1.84 × 10−4) for the SR and NSR, respectively. The low Da values between the matA1 and matA2 sequences in the SR indicate that this region stopped recombining recently. The estimates of fungal molecular clock rate vary by an order of magnitude (Kasuga et al. 2002; Berbee and Taylor 2010), which does not allow us to be more precise with an estimate of the age of the SR region.
Figure 3.
The values for net divergence (Da) between the two mating types in the SR region. The value of Da is shown for each of the 5000-bp windows based on six strains of M. lychnis-dioicae sequenced for each of the mating types. Each bar represents the value for one window. The order of supercontigs from top to bottom of this figure is listed in Table S1 (ranked by supercontig size—the largest on top).
Discussion
Our data demonstrate that the SR region in M. lychnis-dioicae is larger than our prior estimate of ∼1 Mb (Votintseva and Filatov 2009) and is probably closer to 3.3 Mbp (Hood et al. 2013). It may be even larger than our new estimate, given that SR regions tend to accumulate repetitive DNA, which is difficult to assemble into contigs. Indeed, most SR scaffolds in the lam1 genome are short (N50: SR = 44,175; ALL = 185,029); the longest is only 121 kb (Figure 2B), much shorter than the longest NSR scaffold (868 kb).
Importantly, our large-scale sequence data sets herein revealed that A1 vs. A2 average sequence divergence in the SR regions is very low for M. lychnis-dioicae. This finding disagrees fundamentally with the conclusion by Hood et al. (2013) that most of the SR region is highly divergent and, hence, ancient. Their analysis, however, was based on optical mapping rather than on DNA sequence data and the conclusion of high A1–A2 divergence was based on a failure to align A1 and A2 optical maps. However, an inability to align optical maps might result from rearrangements within the SR region, rather than high divergence at the sequence level. Our alignment of the lam1 SR scaffolds against the A2 genome revealed that these regions are indeed not collinear (Figure 2). Furthermore, comparative DNA sequence analysis among the mat chromosomes, as conducted here, quantifies divergence, allowing us to conclude that a large proportion of the SR region is of very recent origin, as suggested previously (Votintseva and Filatov 2009).
Evolutionary strata
There is little doubt that the divergence between the A2 and A1 pheromone receptor alleles is ancient (Devier et al. 2009). The sequences of these homologs are excessively challenging to align, so recombination between the two mating types in this region must have stopped a very long time ago, perhaps as long as 4 × 108 years ago (Devier et al. 2009). If we define “strata” as SR regions with different times since cessation of homologous meiotic recombination, following Lahn and Page (1999), the 4.5-kb region adjacent to the pheromone receptor (Figure 2A) exhibiting poor alignment with the opposite mating type can be regarded as a remnant of an ancient stratum, similar to the Y chromosome region carrying the mammalian sex-determining SRY locus. A1–A2 DNA sequence divergence in this region cannot be calculated due to inability to align the sequences from the two mating types, and divergence at the protein level in the pheromone receptor locus is very high (A1–A2 protein identity = 28.4%; Devier et al. 2009). Suppression of recombination in this ancient stratum around the pheromone receptor is necessary to prevent recombination between the pheromone and pheromone receptor genes to ensure recognition of the opposite mating type (Kahman and Schirawski 2007; Bakkeren et al. 2008). This stratum has probably been maintained by balancing selection ever since early in fungal history (Devier et al. 2009). The physical sizes of the strata in M. lychnis-dioicae are not yet clear, particularly for this region, as intrachromosomal rearrangements could have fragmentated the ancient stratum, and other parts of this highly diverged region may be scattered over different SR scaffolds. Indeed, the alignment of the matA1 scaffold with homologous regions in the A2 genome appears highly fragmented and rearranged (Figure 2A).
The ∼56-kb region of the matA1 lam1 scaffold between the two mating-type factors appears to be less divergent than the pheromone receptor region, as parts of it can be aligned with the sequences from individuals with the A2 mating type (Figure 2A). A1–A2 sequence differences in this region (median Ktotal= 0.052; Figure 1B), and possibly a wider region, may have been accumulating since the transition from a tetrapolar to bipolar mating system in the ancestor of M. lychnis-dioicae.
However, our divergence estimates suggest that much of the SR region stopped recombining very recently, representing a young evolutionary stratum, perhaps due to inversions, consistent with the lack of collinearity between the A1 and A2 mating-type chromosomes based on optical maps (Hood et al. 2013). Some rearrangements are detectable in the younger part of the SR region (Figure 2B) and may contribute to suppression of recombination in the region.
While it is evident that the SR region of M. lychnis-dioicae contains at least two, or possibly three, strata (depending on whether the pheromone receptor locus is regarded as a separate stratum from the rest of the matA1 scaffold), even more strata could exist. The differences between strata can be blurred by variation in the underlying mutation rate, and strata can be intermingled by intrachromosomal rearrangements. Thus, lack of clear discontinuity in A1–A2 divergence in the SR region does not exclude the presence of more evolutionary strata in the SR region of this species.
The causes of SR formation and expansion
While suppression of recombination between the pheromone and pheromone receptor loci (stratum 1) is ancient, the second evolutionary stratum, which comprises the matA1 scaffold and possibly a wider region, may have arisen more recently as a result of the switch from tetrapolar to bipolar mating-type system to ensure linkage between the two mating-type factors—the pheromone/pheromone receptor and homeodomain genes. Such linkages have also evolved in other basidiomycete lineages (e.g., Lee et al. 1999), and the driving force behind this selection for close linkage is to ensure that selfing is possible. For instance, in tetrapolar systems such as U. maydis, different alleles are required at both mating-type loci, such that each meiotic segregant can mate with only 25% of the other progeny, thereby promoting outcrossing (Fraser et al. 2004). Linkage of the homeodomain and pheromone/pheromone receptor into a single locus in U. hordei and M. lychnis-dioicae allows mating success among 50% of the products of the same meiosis, which is advantageous when finding a genetically nonidentical mate is difficult (Giraud et al. 2008). Bipolarity is associated to selfing in some fungi, particularly those with a parasitic lifestyle (Fraser et al. 2004; Whittle et al. 2011b; Laurie et al. 2013; Nieuwenhuis et al. 2013).
The most recent stratum, apparently comprising most of the SR region in M. lychnis-dioicae, appears to have formed only a few million years ago, perhaps under selection for linkage of the mating-type locus with the centromere (Antonovics and Abrams 2004; Zakharov 2005; Giraud et al. 2008; Abbate and Hood 2010). Selection for linkage to the centromere was hypothesized to be responsible for evolution of large SR regions on mat chromosomes of N. tetrasperma (Ellison et al. 2011). Further work is needed to test whether selection for centromere linkage was responsible for recent SR expansion in M. lychnis-dioicae. In particular, establishing the location of the centromere on the mating-type determining chromosome and assembling a less fragmented genome sequence for both mating types will help to reconstruct the genomic rearrangements associated with recent SR expansion. A comparative genomic analysis of other Microbotryum species that diverged before the SR expansion will also be informative about the evolution of the SR region in these fungi.
Conclusions
Based on evolutionary analyses of genome-wide sequence data from multiple M. lychnis-dioicae populations, this study demonstrates that the SR region of the mat chromosomes spans at least 2.3 Mbp and it has undergone a major expansion. The low divergence between the mating types observed in the largest stratum of the SR region indicates that this expansion has occurred recently. On the other hand, around the mating-type-determining factors, sequence divergence between the mating types is much higher than elsewhere in the SR region, providing support for the presence of previously debated (Votintseva and Filatov 2009; Hood et al. 2013) evolutionary strata in M. lychnis-dioicae. Future studies should assess what evolutionary forces have driven the recent massive SR expansion of this species. Furthermore, it will be of interest to test whether genetic degeneration has occurred in the SR region of M. lychnis-dioicae, as has been reported for SR regions on other eukaryotic sex and mat chromosomes (Charlesworth and Charlesworth 2005; Nicolas et al. 2005; Bergero and Charlesworth 2009; Whittle et al. 2011a; Charlesworth 2013).
Supplementary Material
Acknowledgments
We are grateful to staff of the Wellcome Trust Centre for Human Genetics Oxford for generating high-throughput sequence data used in this study, to Mike Chester for checking the text of the manuscript, and to Deborah Charlesworth for many helpful suggestions and corrections. The study was funded by Oxford John Fell fund.
Footnotes
Whole-genome shotgun sequencing FASTQ files have been deposited with the NCBI sequence read archive (SRA) under accession no. SRP050602.
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171702/-/DC1.
Communicating editor: D. Charlesworth
Literature Cited
- Abbate J. L., Hood M. E., 2010. Dynamic linkage relationships to the mating-type locus in automictic fungi of the genus Microbotryum. J. Evol. Biol. 23: 1800–1805. [DOI] [PubMed] [Google Scholar]
- Antonovics J., Abrams J. Y., 2004. Intratetrad mating and the evolution of linkage relationships. Evolution 58: 702–709. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2004. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36: 518–522. [DOI] [PubMed] [Google Scholar]
- Bachtrog D., 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren G., Kämper J., Schirawski J., 2008. Sex in smut fungi: structure, function and evolution of mating-type complexes. Fungal Genet. Biol. 45: S15–S21. [DOI] [PubMed] [Google Scholar]
- Berbee M. L., Taylor J. W., 2010. Dating the molecular clock in fungi: How close are we? Fungal Biol. Rev. 24: 1–16. [Google Scholar]
- Bergero R., Charlesworth D., 2009. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 24: 94–102. [DOI] [PubMed] [Google Scholar]
- Bernasconi G., Antonovics J., Biere A., Charlesworth D., Delph L. F., et al. , 2009. Silene as a model system in ecology and evolution. Heredity 103: 5–14. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., 2013. Plant sex chromosome evolution. J. Exp. Bot. 64: 405–420. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet. Res. 35: 205–214. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 2005. Sex chromosomes: evolution of the weird and wonderful. Curr. Biol. 15: R129–R131. [DOI] [PubMed] [Google Scholar]
- Coelho M. A., Sampaio J. P., Goncalves P., 2010. A deviation from the bipolar-tetrapolar mating paradigm in an early diverged basidiomycete. PLoS Genet. 6: e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCF tools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T., 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. W., Cummings J. E., 1981. The genetics and cellular biology of sexual development in Ustilago violacea, pp. 379–401 in Sexual Interactions in Eukaryotic Microbes, edited by O’Day D. H., Horgen P. A. Academic Press, New York. [Google Scholar]
- Devier B., Aguileta G., Hood M. E., Giraud T., 2009. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics 181: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison C. E., Stajich J. E., Jacobson D. J., Natvig D. O., Lapidus A., et al. , 2011. Massive changes in genome architecture accompany the transition to self-fertility in the filamentous fungus Neurospora tetrasperma. Genetics 189: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P., Olson B., De Hoff P. L., Douglass S., Casero D., et al. , 2010. Evolution of an expanded sex-determining locus in Volvox. Science 328: 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov D. A., 2009. Processing and population genetic analysis of multigenic datasets with ProSeq3 software. Bioinformatics 25: 3189–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Diezmann S., Subaran R. L., Allen A., Lengeler K. B., et al. , 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T., Yockteng R., Lopez-Villavicencio M., Refregier G., Hood M. E., 2008. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot. Cell 7: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet P. M., Bergès H., Bellec A., Prat E., Helmstetter N., et al. , 2012. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 8:e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero, R. F., M. Kirkpatrick, and N. Perrin, 2012 Cryptic recombination in the ever-young sex chromosomes of Hylid frogs. J. Evol. Biol. 25: 1947–1954. [DOI] [PubMed] [Google Scholar]
- Handley L. J., Ceplitis H., Ellegren H., 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., 2002. Dimorphic mating-type chromosomes in the fungus Microbotryum violaceum. Genetics 160: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. E., Petit E., Giraud T., 2013. Extensive divergence between mating-type chromosomes of the anther-smut fungus. Genetics 193: 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahman R., Schirawski J., 2007. Mating in the smut fungi: from a to b to the downstream cascades, pp. 377–387 in Sex in Fungi, edited by Heitman J., Kronstad J., Taylor J., Casselton L. American Society of Microbiology, Washington, DC. [Google Scholar]
- Kämper J., Kahmann R., Bölker M., Ma L. J., Brefort T., et al. , 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kasuga T., White T. J., Taylor J. W., 2002. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol. Biol. Evol. 19: 2318–2324. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999 Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Laurie J. D., Linning R., Wong P., Bakkeren G., 2013. Do TE activity and counteracting genome defenses, RNAi and methylation, shape the sex lives of smut fungi? Plant Signal. Behav. 4: e23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Bakkeren G., Wong K., Sherwood J. E., Kronstad J. W., 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl. Acad. Sci. USA 96: 15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler K. B., Fox D. S., Fraser J. A., Allen A., Forrester K., et al. , 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1: 704–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkis A., Jacobson D. J., Gustafsson T., Johannesson H., 2008. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genet. 4: e1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino S. T., Nelson M. A., Jacobson D. J., Natvig D. O., 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York. [Google Scholar]
- Nicolas M., Marais G., Hykelova V., Janousek B., Laporte V., et al. , 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis B. P. S., Billiard S., Vuilleumier S., Petit E., Hood M. E., et al. , 2013. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity 11: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E., Giraud T., de Vienne D. M., Coelho M. A., Aguileta G., et al. , 2013. Linkage to the mating-type locus across the genus Microbotryum: insights into nonrecombining chromosomes. Evolution 66: 3519–3533. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S., Bergero R., Charlesworth D., 2013. Testing for the footprint of sexually antagonistic polymorphisms in the pseudoautosomal region of a plant sex chromosome pair. Genetics 194: 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 28: 911–914. [DOI] [PubMed] [Google Scholar]
- Rice W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Vicoso B., Kaiser V. B., Bachtrog D., 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl. Acad. Sci. USA 110: 6453–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votintseva A. A., Filatov D. A., 2009. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics 182: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Sun Y., Johannesson H., 2011a Degeneration in codon usage within the region of suppressed recombination in the mating-type chromosomes of Neurospora tetrasperma. Eukaryot. Cell 10: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle C. A., Nygren K., Johannesson H., 2011b Consequences of reproductive mode on genome evolution in fungi. Fungal Genet. Biol. 48: 661–667. [DOI] [PubMed] [Google Scholar]
- Zakharov I. A., 2005. Intratetrad mating and its genetic and evolutionary consequences. Russ. J. Genet. 41: 402–411. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.