Abstract
Natural selection is expected to drive adaptive evolution in genes involved in host–pathogen interactions. In this study, we use molecular population genetic analyses to understand how natural selection operates on the immune system of Anopheles coluzzii (formerly A. gambiae “M form”). We analyzed patterns of intraspecific and interspecific genetic variation in 20 immune-related genes and 17 nonimmune genes from a wild population of A. coluzzii and asked if patterns of genetic variation in the immune genes are consistent with pathogen-driven selection shaping the evolution of defense. We found evidence of a balanced polymorphism in CTLMA2, which encodes a C-type lectin involved in regulation of the melanization response. The two CTLMA2 haplotypes, which are distinguished by fixed amino acid differences near the predicted peptide cleavage site, are also segregating in the sister species A. gambiae (“S form”) and A. arabiensis. Comparison of the two haplotypes between species indicates that they were not shared among the species through introgression, but rather that they arose before the species divergence and have been adaptively maintained as a balanced polymorphism in all three species. We additionally found that STAT-B, a retroduplicate of STAT-A, shows strong evidence of adaptive evolution that is consistent with neofunctionalization after duplication. In contrast to the striking patterns of adaptive evolution observed in these Anopheles-specific immune genes, we found no evidence of adaptive evolution in the Toll and Imd innate immune pathways that are orthologously conserved throughout insects. Genes encoding the Imd pathway exhibit high rates of amino acid divergence between Anopheles species but also display elevated amino acid diversity that is consistent with relaxed purifying selection. These results indicate that adaptive coevolution between A. coluzzii and its pathogens is more likely to involve novel or lineage-specific molecular mechanisms than the canonical humoral immune pathways.
Keywords: population genetics, innate immunity, balancing selection, C-type lectin, JAK-STAT, genetics of immunity
Anopheles mosquitoes live in pathogen-rich environments where survival requires an effective immune system. Population genetic studies in Anopheles gambiae have generally focused on putative antimalaria genes (Obbard et al. 2007, 2008, 2009a; Slotman et al. 2007; Cohuet et al. 2008; Parmakelis et al. 2008; White et al. 2011; Rottschaefer et al. 2011). However, mosquitos are also exposed to diverse microbes, microsporidia, and other pathogens during their larval stages when they live in septic standing water (Muspratt 1946; Bargielowski and Koella 2009; Wang et al. 2011), and these larval pathogens probably impose stronger selection on the immune system than the malaria parasite. Regardless of the proximal selective agent, any pathogen-driven evolution of the immune system is likely to shape the efficacy of resistance to parasites of public health relevance, including human malaria, through pleiotropic effects on cross-resistance (Mitri et al. 2009; White et al. 2011; Rottschaefer et al. 2011). In this study, we look broadly at the Anopheles immune system to determine targets of natural selection and identify potential instances of pathogen-driven evolution that may shape general resistance to infection.
Because host fitness depends on the ability to combat infection, pathogens are expected to impose significant selection pressure and immune system genes are often observed to evolve rapidly (Schlenke and Begun 2003; Nielsen et al. 2005; Obbard et al. 2006; Sackton et al. 2007; McTaggart et al. 2012). Comparative genomic studies show conserved gene orthology in insect innate immune systems, particularly for genes in intracellular signaling pathways, yet amino acid divergence between species is often elevated in these orthologs relative to nonimmune genes (Sackton et al. 2007; Waterhouse et al. 2007). Immunological divergence can additionally occur through lineage-specific and species-specific gene family expansions and contractions, which may reflect differences in selection pressures imposed by distinct pathogenic environments. This process is particularly pronounced in recognition and effector genes (Sackton et al. 2007; Waterhouse et al. 2007). These comparative genomic studies have provided insight into how the immune system evolves between species over longer evolutionary time, but they do not provide information about how natural selection operates to shape the immune system within a species over shorter timescales. Additionally, comparative genomic analyses are only able to reveal adaptive divergence and cannot detect the adaptive maintenance of polymorphism within species. Signatures of pathogen-driven evolution over shorter time scales can be detected more effectively from patterns of intraspecific polymorphism relative to divergence in immune system genes.
Insect immune systems can be broadly divided into categories of humoral and cellular responses. The humoral immune response involves recognition of pathogens by pattern recognition molecules that initiate intracellular signaling cascades that stimulate transcriptional activation of antimicrobial effector molecules. The Toll and Imd immune signaling pathways have been most extensively studied for their role in antibacterial and antifungal immunity in Drosophila, but these pathways have also been shown to play a role in immunity against a variety of pathogens in other species. In A. gambiae, the Imd pathway has been implicated in defense against both Gram-positive and Gram-negative bacteria (Meister et al. 2005, 2009; Garver et al. 2009) and has been shown to play an important role in mediating protection against the human malaria parasite Plasmodium falciparum (Mitri et al. 2009; Garver et al. 2009, 2012; Meister et al. 2009). The A. gambiae Toll pathway is involved in defense against bacteria and rodent malaria (Barillas-Mury et al.1996; Frolet et al. 2006, Riehle et al. 2008; Garver et al. 2009). The JAK-STAT pathway also plays a role in the response to bacterial infection in A. gambiae (Barillas-Mury et al. 1999; Gupta et al. 2009). Although most dipterans have only a single STAT gene, there are two STAT genes in A. gambiae, which appear to be the result of a retroduplication event along the Anopheline lineage. STAT-A, the ancestral gene, is most closely related to STAT genes in other insects, and STAT-B is a derived retrocopy (Gupta et al. 2009). The JAK-STAT pathway in A. gambiae is less well-studied than the Toll and Imd pathways, but studies indicate that it may play a role in killing P. falciparum parasites at the oocyst stage in A. gambiae (Gupta et al. 2009), and it has also been shown to limit Plasmodium vivax infection in Anopheles aquasalis (Bahia et al. 2011).
In the cellular immune response, pathogen recognition leads to encapsulation, phagocytosis, or melanization of the pathogen by hemocytes. The melanization response in particular has been studied in A. gambiae for its role in killing plasmodium parasites (Collins et al. 1986). Two C-type lectins, CTL4 and CTLMA2, have been shown to play a role in regulating the melanization response in A. gambiae. CTL4 and CTLMA2 primarily exist in the form of a heterodimer, which is secreted into the hemolymph (Schnitger et al. 2009). RNAi experiments indicate an important role for CTL4 and CTLMA2 in antibacterial immunity, because silencing of either gene reduces the number of Gram-negative bacteria melanized (Schnitger et al. 2009). In contrast, silencing of either CTL4 or CTLMA2 results in an increase in the number of melanized Plasmodium berghei ookinetes, suggesting that the CTL4/CTLMA2 heterodimer acts as an agonist for P. berghei parasites, preventing their melanization (Osta et al. 2004).
In this study, we examine patterns of genetic variation at 20 immune genes in a population of Anopheles coluzzii (formerly A. gambiae “M form”) (Coetzee et al. 2013) from Burkina Faso. The set of immune genes consists of genes belonging to the Toll, Imd, and JAK-STAT pathways, as well as other mosquito-specific immune factors. To control for demography and the effects of genomic location, we additionally sequenced 17 nonimmune control genes located physically nearby in the genome to our genes of interest. We found evidence that two distinct haplotypes in CTLMA2 arose before the divergence of A. coluzzii, A. gambiae (“S form”), and A. arabiensis, and they have been adaptively maintained as a balanced polymorphism in all three species. We also found evidence of rapid adaptive evolution in STAT-B, suggesting an important role for the JAK-STAT pathway in A. coluzzii. We observe higher overall rates of amino acid divergence in the immune genes relative to the control genes. Genes involved in the Imd pathway show particularly high amino acid divergence, but they also display elevated amino acid diversity that is consistent with relaxed purifying selection.
Materials and Methods
Mosquito collection and DNA isolation
The Anopheles coluzzii individuals used in this study were collected in the village of Goundry, Burkina Faso (coordinates 12°30′N, 1°20′W) in September 2008. Freshly fed females were captured indoors by manual aspirator catch and DNA was extracted from individual carcasses using DNAzol (Invitrogen). Diagnostic PCRs were performed to confirm the species (Scott et al. 1993; Favia et al. 2001), and whole genome amplification of each sample was performed using the GenomiPhi V2 DNA Amplification Kit (GE Healthcare). Anopheles merus DNA from the OPHANSI colony was obtained from Malaria Research and Reference Reagent Resource Center (MR4).
Loci analyzed
We sequenced a set of 20 immune genes including the Toll pathway genes GNBPB1, TOLL1A, TUBE, PELLE, TRAF6, CACT, and REL1, the Imd pathway genes IMD, FADD, CASPL1, TAK1, IAP2, IKK1, IKK2, and REL2, the JAK-STAT transcription factors STAT-A and STAT-B, and the Anopheles specific immune factors CTL4, CTLMA2, and LRIM1. To control for demography and background effects of genomic location, we additionally sequenced 17 nonimmune control genes located nearby our genes of interest. These controls are located within 40–100 KB of their "matched" immune genes and are similar in size and structure to the immune gene they are matched to. The names and relative chromosomal locations of all loci are shown in Supporting Information, Figure S1. All PCR primers were designed based on the published A. gambiae genome sequence (Vectorbase, A. gambiae genome, version P3). Loci were sequenced from a set of 20 A. coluzzii females. Some loci could not be amplified in all 20 individuals, but all loci were sequenced in a minimum of 18 A. coluzzii individuals as well as in A. merus.
PCR and sequencing
Each gene was amplified in a single amplicon from whole genome amplified DNA using iProof high-fidelity DNA Polymerase (BioRad). PCR products were run out on a 1% agarose gel and the product fragments were excised and purified using EZNA gel extraction kits (Omega BioTek). Adenosine tails were added to the purified products by incubating for 20 min at 72° with PCR buffer, dATP, and Taq polymerase. Products were then cloned using either TOPO or TOPO XL cloning kits (both from Invitrogen). Colonies to be sequenced were grown overnight at 37° in liquid Luria-Bertani broth supplemented with 20 mg/ml kanamycin, and the plasmids were isolated using the Qiaprep spin miniprep kit (Qiagen). The products were sequenced directly from the plasmids using the BigDye Terminator Cycle Sequencing Kit v3.1(ABI). The sequences were assembled using Sequencher (Gene Codes Corp). All sequences have been deposited in GenBank under accession numbers KP274100-KP274844.
Only one of the two alleles at each gene was sequenced from any given mosquito in the study. To correct sequencing errors, all singleton polymorphisms were verified by re-amplification and direct sequencing of heterozygous PCR products. For singleton validation, the entire gene was amplified directly from the whole genome amplified DNA using iProof high-fidelity DNA Polymerase (BioRad), and this full-length amplicon was then used as a template for a secondary PCR that used internally nested primers to robustly amplify the gene region containing the singleton to be validated. Unincorporated primers and dNTPs were inactivated from these secondary amplification products by incubation with ExoI and SAP (both manufactured by USB), and amplification products were sequenced using the BigDye Terminator Cycle Sequencing Kit v3.1 (ABI).
Population genetic analysis
Average pairwise genetic diversity (π) was calculated for all sites, and also separately for synonymous (πs) and nonsynonymous (πa) sites using DnaSP v.5 (Librado and Rozas 2009). The Tajima’s D statistic (Tajima 1989) was also calculated in DnaSP using silent sites only. The average pairwise genetic divergence at synonymous (KS) and nonsynonymous (KA) sites and their ratio (KA/KS) were calculated in DnaSP using A. merus as an outgroup. Maximum likelihoood multi-locus HKA tests were implemented using the mlhka program (Wright and Charlesworth 2004) using synonymous sites only. Multi-locus McDonald Kreitman tests were performed using the software MKtest v2.0 (Welch 2006; Obbard et al. 2009a). The multi-locus McDonald Kreitmen tests were performed on the full dataset, as well as on the Toll and Imd pathway genes separately. Three models, which varied only in the parameter α, were implemented for each dataset. In the first model (M0), α was fixed at zero for all loci. In the second model (M1), a single α value estimated from the data were shared by all loci, and in the third model (M2) α was estimated separately for the immune and control loci. For all three models, the expected neutral divergence (λ=µt) and neutral diversity (θ=4Neµ) each took a single value at all loci, and selective constraint was allowed to vary between loci. Maximum likelihood ratio tests and Akaike weighting were used to assess model fit.
To identify distinct haplotype clades in CTLMA2, neighbor-joining gene trees were constructed in MEGA5 (Tamura et al. 2011) using the maximum composite likelihood method and uniform substitution rates, with 1000 bootstrap replicates. To determine if the CTLMA2 haplotype clades were present in other species, we compared our data with published CTLMA2 sequences of A. gambiae (GenBank accession numbers EF519453–EF519450 and EF519463–EF519478) and A. arabiensis (GenBank accession numbers EF519419–EF519428), both collected from Kenyan populations, as well as the outgroup species A. quadriannulatus (GenBank accession number EF519435) (Obbard et al. 2007). To test for introgression of the CTLMA2 haplotypes among A. coluzzii, A. gambiae, and A. arabiensis, we calculated the average number of pairwise differences (Dxy) within and between each clade in DnaSP. To determine typical values of Dxy in the genomic region of CTLMA2, we calculated coluzzii-arabiensis Dxy and gambiae-arabiensis Dxy from published data for the nearby loci AGAP005540 (GenBank accession numbers EF519480–EF519501) and APL2 (GenBank accession numbers EF519504–EF519528) (Obbard et al. 2007).
No position-matched control gene was sequenced for one immune locus (TRAF6), and in two instances a pair of immune loci were located very near to each other (TAK1 and PELLE on chromosome 2R, CTL4 and CTLMA2 on chromosome 2L), so a single position control was sequenced for each pair (see Figure S1). Differences in divergence between the immune and control groups were assessed using Mann-Whitney U tests. When analyzing the full dataset, each control gene was included only once, but because TAK1 and PELLE are involved in different signaling pathways (Imd and Toll, respectively), the shared control gene was included in the control group for analysis of each individual pathway. Differences in nucleotide polymorphism between the immune and control groups were assessed using paired Wilcoxon tests. TRAF6, which lacks a position control, was excluded from these comparisons, and the shared control genes were included twice as they were paired to each immune locus. Mann Whitney U tests and paired Wilcox tests for differences in divergence and diversity were implemented in R (R Development Core Team 2011).
Results
Reduced purifying selection in the IMD pathway
To test the hypothesis that A. coluzzii immune genes might evolve under positive selection, we analyzed patterns of intraspecific and interspecific genetic variation in 20 immune genes and 17 control genes from a single population in Burkina Faso. Population genetic statistics for each locus are listed in Table S1. Across all 37 genes, the average per-site nucleotide diversity was 1.3% at all sites (π), 2.6% at synonymous sites (πs), and 0.3% at nonsynonymous sites (πa). Nucleotide diversity was lower for genes on the X chromosome compared with the autosomes (X mean: π = 0.4%, πs = 0.7%, πa = 0.1%; autosome mean: π = 1.5%, πs = 2.9%, πa = 0.4%), in keeping with the lower effective population size on the X (Cohuet et al. 2008). Nucleotide divergence was measured using A. merus as an outgroup. The average per-site nucleotide divergence was 3.3% at all sites (K), 6% at synonymous sites (KS), and 0.9% at nonsynonymous sites (KA). These estimates for diversity and divergence are comparable with previously published estimates in A. coluzzii (Cohuet et al. 2008).
Genes that are the target of recurrent positive selection are expected to have elevated rates of amino acid evolution, which can be measured using the ratio of nonsynonymous to synonymous divergence (KA/KS). The average KA/KS ratio of the immunity genes is significantly higher than that of the position-matched control genes (immune KA/KS = 0.175; control KA/KS = 0.077; Mann-Whitney U test P = 0.001) (Table 1). This difference is driven by divergence at nonsynonymous sites (KA), which is three-times higher in the immune genes than controls, whereas divergence at synonymous sites (KS) is not significantly different between the immune and control groups (Table 1). When we looked at genes involved in the Toll and Imd pathways separately, we found that genes involved in the Imd pathway have higher average KA/KS values than their controls (Imd immune KA/KS = 0.197; Imd control KA/KS = 0.068; Mann-Whitney U-test P = 0.003) (Table 1). This difference is not driven by one or a few outliers, because the difference remains significant even after removing the highest three KA/KS ratios. The average KA/KS value in Toll pathway genes is also higher than their controls, although the difference is not statistically significant (Toll immune KA/KS = 0.109; Toll control KA/KS = 0.056; Mann-Whitney U-test P = 0.063) (Table 1). In both the Toll and Imd pathways, divergence at synonymous sites is not different between the immune and control groups, so the higher KA/KS ratio in immune genes is attributable to elevated replacement divergence in the immune genes (Table 1). When all genes in the Imd pathway are excluded from the analysis, immune genes still have marginally higher average KA/KS relative to the control genes (0.16 vs. 0.084; Mann-Whitney U-test P = 0.048) (Table 1), indicating that the Imd pathway is not the only driver of high amino acid divergence found in the immune genes.
Table 1. Average pairwise divergence in immune and control groups.
| na | Ktotalb | KSc | KAd | KA/KSe | |
|---|---|---|---|---|---|
| All data | |||||
| Immune | 20 | 0.0335 | 0.0652 | 0.0120 | 0.1747 |
| Control | 17 | 0.0319 | 0.0545 | 0.0043 | 0.0772 |
| U = 149, P = 0.537 | U =117.5, P = 0.113 | U = 64.5, P = 0.001 | U = 62.5, P = 0.001 | ||
| IMD pathway | |||||
| Immune | 8 | 0.0336 | 0.0613 | 0.0121 | 0.1968 |
| Control | 8 | 0.0302 | 0.0526 | 0.0041 | 0.0675 |
| U = 24, P = 0.442 | U = 21, P = 0.279 | U = 8, P = 0.010 | U = 5, P = 0.003 | ||
| TOLL pathway | |||||
| Immune | 7 | 0.0332 | 0.0713 | 0.0087 | 0.1090 |
| Control | 6 | 0.0335 | 0.0565 | 0.0032 | 0.0555 |
| U = 22, P = 0.945 | U = 14, P = 0.366 | U = 8, P = 0.073 | U = 7.5, P = 0.063 | ||
| Exclude IMD pathway | |||||
| Immune | 12 | 0.0335 | 0.0679 | 0.0119 | 0.1600 |
| Control | 10 | 0.0322 | 0.0557 | 0.0043 | 0.0835 |
| U = 58, P = 0.923 | U = 44, P = 0.314 | U = 28.5, P = 0.041 | U = 29.5, P = 0.048 | ||
Number of genes considered.
Average per-gene divergence at all sites between A. coluzzii and A. merus
Average per-gene silent divergence between A. coluzzii and A. merus
Average per-gene replacement divergence between A. coluzzii and A. merus
Average per-gene KA/KS ratio between A. coluzzii and A. merus.
If the increased rates of amino acid evolution in the immune genes were due to recurrent fixation of beneficial alleles, then we would also expect to see reduced diversity at physically linked sites and a shift in the allele frequency spectrum (Nielsen et al. 2005). We found that diversity at synonymous sites was similar in the immune and control genes overall (immune πs = 2.83%; control πs = 2.52%; paired Wilcox V = 128; P = 0.196) (Table 3), but elevated in Imd pathway genes relative their controls (Imd immune πs = 3.85%; Imd control πs = 2.97%; paired Wilcox V = 34; P = 0.023) (Table 3). We found no difference in Tajima’s D, which measures shifts in the allele frequency spectrum, between the immune and control groups overall or when split by immune pathway (Table 3). We did, however, see significantly higher average polymorphisms at nonsynonymous sites in the immune genes relative to controls (immune πa = 0.48%; control πa =0.16%; paired Wilcox V = 172; P = 0.001) (Table 2), driven primarily by elevated πa in the Imd pathway (Imd immune πa = 0.66%; control πa =0.18%; paired Wilcox V = 36; P = 0.008) (Table 3). The Toll genes do not have significantly higher average πa than their controls (Toll immune πa = 0.31%; control πa = 0.14%; paired Wilcox V = 19; P = 0.219) (Table 3).
Table 3. Average pairwise genetic diversity in immune and control groups.
| na | πtotalb | πsc | πad | De | |
|---|---|---|---|---|---|
| All data | |||||
| Immune | 19 | 0.0140 | 0.0283 | 0.0048 | −0.434 |
| Control | 19 | 0.0127 | 0.0252 | 0.0016 | −0.764 |
| V = 102, P = 0.798 | V = 128, P = 0.196 | V = 172, P = 0.001 | V = 141, P = 0.066 | ||
| IMD pathway | |||||
| Immune | 8 | 0.0182 | 0.0385 | 0.0066 | −0.512 |
| Control | 8 | 0.0134 | 0.0297 | 0.0018 | −0.601 |
| V = 27, P = 0.25 | V = 34, P = 0.023 | V = 36, P = 0.008 | V = 20, P = 0.844 | ||
| TOLL pathway | |||||
| Immune | 6 | 0.0116 | 0.0231 | 0.0031 | −0.353 |
| Control | 6 | 0.0141 | 0.0239 | 0.0014 | −0.834 |
| V = 4, P = 0.219 | V = 9, P = 0.844 | V = 17, P = 0.219 | V = 19, P = 0.094 |
Number of genes considered, 18–20 alleles sampled per gene.
Average per-gene genetic diversity calculated for all sites.
Average per-gene genetic diversity calculated for synonymous sites.
Average per-gene genetic diversity calculated for nonsynonymous sites.
Average per-gene Tajima’s D calculated using silent sites.
Table 2. Multilocus MK-test model comparison.
| Model | Description | Par | log(L) | 2∆log(L) | χ2 P value | AICca | Akaike Weightb | αac | αbd |
|---|---|---|---|---|---|---|---|---|---|
| All loci | |||||||||
| M0 | α = 0 | 38 | −615.83 | 1335.9 | 0.806 | [0] | [0] | ||
| M1 | α ∼ (all loci) | 39 | −615.51 | 0.64 | 0.4237 | 1339.0 | 0.169 | [0.07] | [0.07] |
| M2 | α ∼ (control, immune) | 40 | −615.48 | 0.06 | 0.8065 | 1342.8 | 0.025 | 0.04 | 0.09 |
| Imd Pathway | |||||||||
| M0 | α = 0 | 18 | −235.44 | 522.1 | 0.003 | [0] | [0] | ||
| M1 | α ∼ (all loci) | 19 | −228.01 | 14.86 | 0.0001 | 511.3 | 0.695 | [−0.64] | [−0.64] |
| M2 | α ∼ (control, immune) | 20 | −226.72 | 2.58 | 0.1069 | 513.0 | 0.302 | −0.20 | −0.91 |
| Toll pathway | |||||||||
| M0 | α = 0 | 14 | −181.22 | 403.2 | 0.305 | [0] | [0] | ||
| M1 | α ∼ (all loci) | 15 | −178.55 | 5.34 | 0.0208 | 402.1 | 0.520 | [−0.5] | [−0.5] |
| M2 | α ∼ (control, immune) | 16 | −177.37 | 2.36 | 0.1245 | 404.3 | 0.175 | −1.09 | −0.24 |
The Akaike information criterion corrected for sample size.
The likelihood of the model, given the relative support for each of the models tested.
Estimate of the proportion of adaptive substitutions in the control genes. Square brackets indicate where α is constrained by the model.
Estimate of the proportion of adaptive substitutions in the immune genes. Square brackets indicate where α is constrained by the model.
The elevated Ka/Ks observed in immune genes could be consistent with positive selection driving adaptive evolution in these genes, but could also arise if immune genes are generally less constrained and therefore free to accumulate amino acid substitutions. Under a model of neutral evolution, the ratio of nonsynonymous to synonymous divergence is expected to equal the ratio of nonsynonymous to synonymous polymorphism. The McDonald-Kreitman test compares the number of polymorphisms and fixed differences at synonymous and nonsynonymous sites to detect deviation from the neutral expectation (McDonald and Kreitman 1991) and can be used to estimate the proportion of nonsynonymous fixations attributed to positive selection (α) (Smith and Eyre-Walker 2002). To determine whether the immune genes as a class show a higher proportion of adaptive substitutions than the control genes, we implemented a multi-locus MK test using the software MKtest v2.0 (Welch 2006; Obbard et al. 2009a). We first implemented the test on the full dataset. Using a likelihood ratio test, the fit of a model allowing α to take the maximum likelihood value showed no significant improvement over the null model where α is fixed at zero [M1 vs. M0 2∆log(L)=0.64; P = 0.4] (Table 2). We then implemented the test on genes in the Toll and Imd pathways separately. In both cases, the model that allowed a single value of α estimated from the data showed a significant improvement over the null model where α is fixed at zero [Imd M1 vs. M0 2∆log(L)=14.8, P = 0.0001; Toll M1 vs. M0 2∆log(L)=5.34, P = 0.0208] (Table 2). A model that allowed a separate α value to be estimated for the immune and control genes did not provide any additional improvement in model fit for either pathway [Imd M2 vs. M1 2∆log(L)=2.6, P = 0.1069; Toll M2 vs. M1 2∆log(L)=2.36, P = 0.1245] (Table 2). We thus see a proportional increase in both nonsynonymous polymorphism and nonsynonymous divergence in immune genes, providing no support for the hypothesis that the immune genes experience more positive selection than the control genes.
The elevated nucleotide diversity at both synonymous and nonsynonymous sites in the Imd pathway genes could be consistent with adaptive maintenance of polymorphism, or alternatively could indicate that purifying selection on these genes is weak relative to control genes, allowing deleterious nonsynonymous mutations to persist in the population as effectively neutral polymorphisms. The ratio of polymorphism to divergence is predicted to be equivalent across neutrally evolving loci, whereas elevated polymorphism relative to divergence is predicted under adaptive maintenance of polymorphisms. The HKA test (Hudson et al. 1987), which compares the ratio of polymorphism relative to divergence across multiple loci, can help distinguish between these hypotheses. We used a multilocus HKA test in a maximum-likelihood framework to compare the polymorphism to divergence ratios of the eight genes in Imd pathway genes with the other 29 genes in the dataset. Under this framework, a model allowing selection on Imd genes as a class did not show a significant improvement over a model that assumed all genes evolve neutrally [χ2(8)=6.52; P = 0.6]. These results indicate the elevated amino acid diversity observed in Imd pathway genes cannot be explained by a model of adaptive maintenance of polymorphism, but rather that these genes experience weakened purifying selection that allows deleterious nonsynonymous mutations to persist in the population as effectively neutral polymorphisms that may drift to fixation and contribute to divergence between species.
Adaptive evolution in STAT-B
STAT-B is an intronless duplicate of STAT-A that arose through retrotransposition of a STAT-A mRNA. The duplication event occurred more recently than the divergence of the Anopheline and Culicine lineages (145-200MYA; Krzywinski 2006), yet the two STAT copies are remarkably divergent. STAT-A shows only 43% amino acid identity with STAT-B, whereas it retains 74% identity with Aedes aegypti STAT and 63% identity with Culex quinquefasciatus STAT. Despite the rapid divergence of STAT-B, there is no evidence of pseudogenization, and both STAT-A and STAT-B appear to play a role in immunity. STAT-A and STAT-B are differentially expressed in various developmental stages, with STAT-A being absent from the pupal stage when STAT-B is highly expressed and STAT-A being expressed at higher levels than STAT-B in adults (Gupta et al. 2009). Both STAT-A and STAT-B appear to be involved in resistance to bacteria and Plasmodium parasites (Gupta et al. 2009). Gene duplicates are often maintained in the genome because they acquire a function that is distinct from that of the ancestral gene. In this case, we might predict evidence of adaptive evolution in STAT-B, particularly when compared with STAT-A.
Synonymous site divergence between A. coluzzii and A. merus in STAT-B is typical of genes in our dataset (STAT-B KS = 6.3%; mean KS = 6%). Amino acid divergence, however, is remarkably high at this locus (STAT-B KA= 3%; mean KA= 0.9%), giving it the highest KA/KS ratio in the dataset (STAT-B KA/KS= 0.46; mean KA/KS= 0.13). Such an excess of replacement divergence, along with a deficit of intraspecific nucleotide diversity (STAT-B π = 0.18%; mean π = 1.3%), is consistent with a model of recent adaptive evolution at this locus. To test the hypothesis that positive selection is driving the rapid divergence of STAT-B, we used the maximum likelihood multi-locus HKA test to test for a departure from neutrality in STAT-B as compared with all other genes in the dataset. A model that hypothesized that STAT-B was evolving by directional selection fit the empirical data significantly better than the null model that assumed neutral evolution in all genes [χ2(1) = 9.1; P = 0.0026]. In contrast to the high amino acid divergence in STAT-B, STAT-A exhibits a complete lack of replacement divergence or polymorphism, consistent with a model of purifying selection. Estimates of total and synonymous polymorphism are low in STAT-A, but they are consistent with the estimates from other genes on the X chromosome. (X mean π = 0.42%, πS = 0.7%; STAT-A π = 0.42%, πS = 0.64%). This pattern of purifying selection in STAT-A, the ancestral gene, and rapid evolution in the derived duplicate STAT-B, is consistent with a model of neofunctionalization of STAT-B.
Balancing selection in CTLMA2
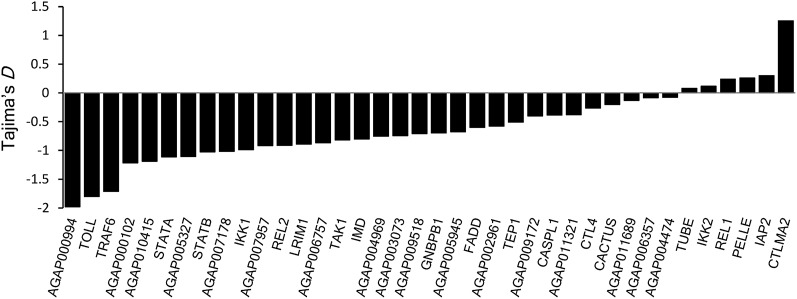
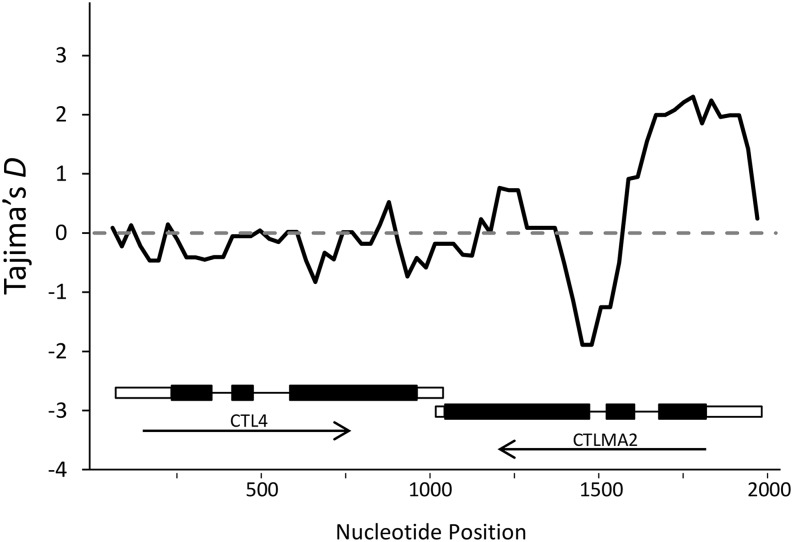
CTL4 and CTLMA2 play a role in regulating the melanization response in A. gambiae (Osta et al. 2004). They primarily exist in the form of a heterodimer, which is secreted into the hemolymph (Schnitger et al. 2009). CTL4 and CTLMA2 are located directly adjacent on the chromosome and, in our study, were PCR amplified and cloned as a single fragment. Although all but three of the genes examined in this dataset have Tajima’s D values that are negative or nearly zero, CTLMA2 stands out with a Tajima’s D value of positive 1.25 (Figure 1). A sliding window analysis of the entire CTL4/CTLMA2 region shows that the high positive Tajima’s D values are limited to the 5′ end of CTLMA2, and that Tajima’s D throughout the rest of the gene is similar to that of CTL4 (Figure 2). Inspection of the sequences reveals the presence of two distinct haplotype clades, hereafter referred to as clades A and B. There are 11 fixed SNP differences between clades A and B; all are located in the 5′ 350 bp of the gene. This block of fixed differences, which spans part of the 5′ UTR, the first exon, and the first intron, includes four SNPs that result in three amino acid changes near the predicted signal cleavage site (Figure 3). We considered that the divergent haplotype could have been introduced through paralogous gene conversion, but we were unable to find significant sequence matches to any CTL paralogs in either the A. gambiae genome or the entire NCBI nr database.
Figure 1.
Distribution of Tajima’s D across all 37 genes. Tajima’s D was calculated for each gene using silent sites and plotted as a histogram showing deviation from neutral expectations. Loci are ordered based on the value of D to draw attention to the contrast between CTLMA2 and the rest of the dataset.
Figure 2.
Sliding window analysis of Tajima’s D in CTL4 and CTLMA2. CTL4 and CTLMA2 are located directly adjacent on the chromosome and were PCR amplified and cloned as a single fragment. Tajima’s D was calculated using silent sites in a sliding window along the entire sequenced region using a 200-bp window with a 25-bp step size. The dashed line indicates the expected value of D under a neutral equilibrium model. The schematic below the plot shows the exon structure and direction of transcription for CTL4 and CTLMA2. Positive values of D in the 5′ region of CTLMA2 indicate an excess of intermediate frequency polymorphism in this region.
Figure 3.
Amino acid alignment of CTLMA2 sequences of A. coluzzii collected in Burkina Faso, along with previously published sequences from A. gambiae and A. arabiensis collected in Kenya (Obbard et al. 2007). Sites matching the top sequence are indicated with a dot (•). Dashes (–) note unavailable sequence because we sequenced a different amplicon than was available for A. gambiae or A. arabiensis
To determine if the presence of the two CTLMA2 haplotypes could be consistent with a partial selective sweep, a sweep with recombination, or a sweep from standing variation, we compared the patterns of genetic variation within each haplotype clade to genetic variation in CTL4 and AGAP005327. If the presence of the haplotypes were due to a partial selective sweep, where the frequency of a selected allele increases in the population but does not or has not yet become fixed in the population, then we would expect to see evidence of selection in one clade, whereas variation in the other clade should be consistent with surrounding regions (Hudson et al. 1997). If the haplotypes are the result of a sweep from standing variation or a sweep with recombination, then we would expect both clades to show signatures of selection (Pritchard et al. 2010). It is important to note that the expected values of neutrality statistics like those used here are not the same for both a random sample of chromosomes and a partitioned subset of chromosomes. Most notably, the effective population size, which is a central parameter in these statistics, will be smaller in clade-wise estimates, because we are specifically conditioning on a specific subset of chromosomes in the population. In our sample, the clades are segregating at 50%, so we should expect an approximately 50% reduction in within-clade nucleotide diversity under neutral equilibrium conditions, assuming these haplotypes have segregated at this frequency for many generations. Tajima’s D is calculated as the normalized difference between two neutrality statistics that both depend on effective population size (Tajima 1989), so we do not expect the reduction in effective population size to affect this statistic. We found that estimates of nucleotide diversity and the site frequency spectra (Tajima’s D) of clade A are consistent with those observed in the adjacent CTL4, as well as the position-matched control gene AGAP005327 (Table 4). Nucleotide diversity is slightly lower in clade B, which also shows a nonsignificant shift in the site frequency spectrum, but on the whole there is little evidence of positive selection on either clade, so we cannot attribute the presence of the two CTLMA2 haplotype clades to partial or soft selective sweep.
Table 4. Population genetic statistics for CTLMA2 haplotype clades and surrounding loci.
| na | Sb | πc | Dd | |
|---|---|---|---|---|
| CTLMA2 all | 20 | 55 | 0.022 | 1.254 |
| Clade A | 10 | 30 | 0.010 | −0.465 |
| Clade B | 10 | 25 | 0.007 | −1.737 |
| CTL4 | 20 | 44 | 0.013 | −0.263 |
| AGAP005327 | 19 | 54 | 0.010 | −1.107 |
Number of alleles sampled.
Number of segregating sites.
Average number of pairwise differences per site.
Tajima’s D calculated using silent sites.
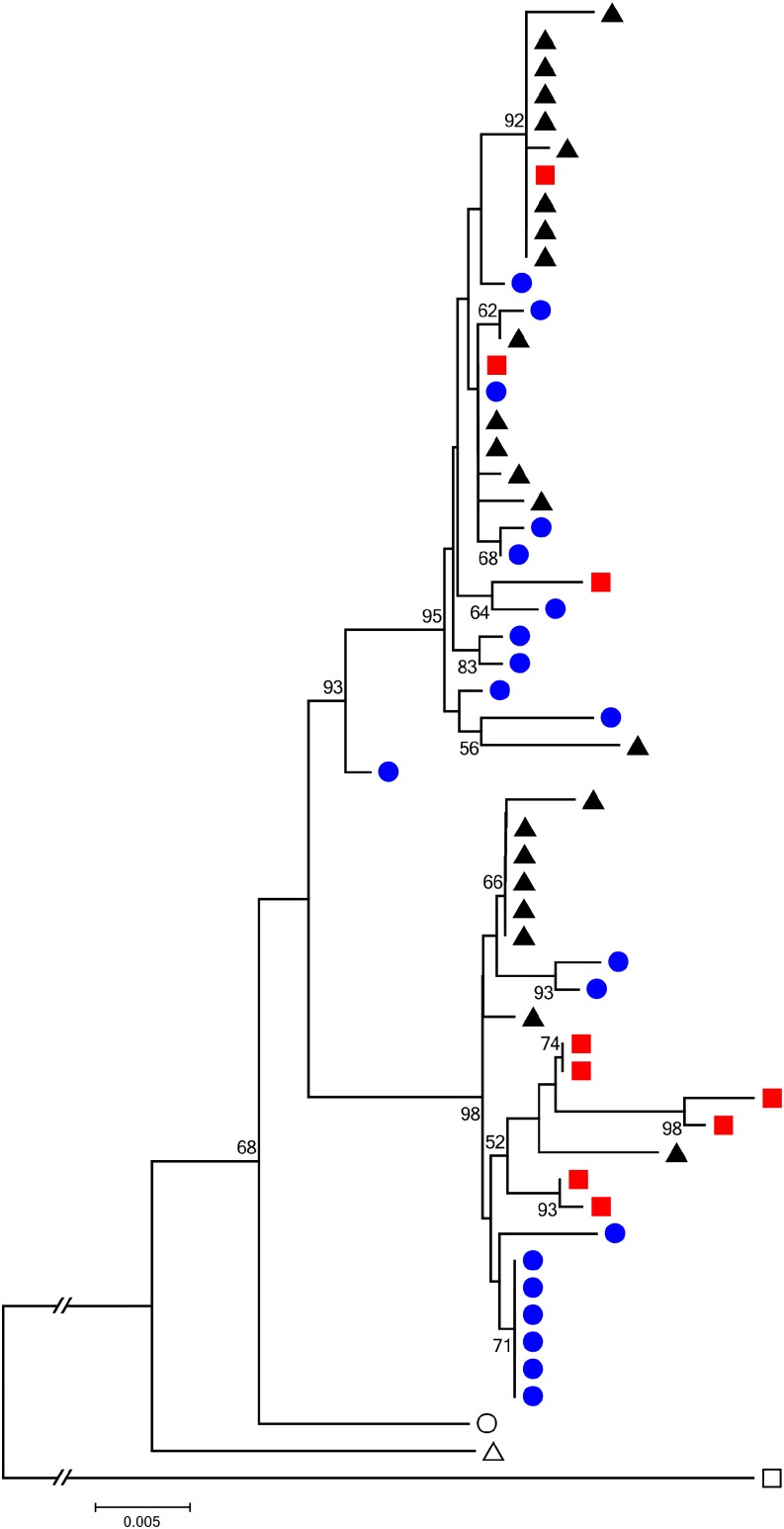
Alternatively, these two divergent haplotypes may be balanced polymorphisms maintained by frequency-dependent selection, spatially varying selection pressures, or overdominance. We might then predict that these haplotypes have been segregating for evolutionarily long times and thus might also be found in closely related species. We tested this hypothesis by comparing our sequences to a published dataset (Obbard et al. 2007) and found that both CTLMA2 haplotypes are also segregating in A. gambiae (“S form”) as well as in A. arabiensis (Figure 3, Figure 4). The presence of both haplotypes in all three species could be consistent with an origin that predates the divergence of the species or, alternatively, they could have been shared among species through adaptive introgression. Introgressed loci are expected to be less diverged between species compared with adjacent chromosomal regions. Therefore, if the CTLMA2 haplotypes arose after the species diverged and were shared by introgression, then we would expect the introgressed haplotype to show relatively low divergence between species and divergence in the ancestral haplotype to be typical for the genomic region. If the CTLMA2 haplotypes predate the divergence of the species, then divergence within each haplotype clade should be similar to the genome average, whereas divergence between haplotype clades should be elevated, reflecting a longer coalescence time. To determine which of these models best fits the data for CTLMA2, we calculated the average number of pairwise differences, Dxy, among species within and between the CTLMA2 haplotype clades, and also in two nearby loci from a published dataset (Obbard et al. 2007). Because Dxy depends on effective population size (Nei 1987), which is lower within clades as noted above, we would expect Dxy to be smaller relative to nearby regions without similar long-term haplotype structure. We found that Dxy among species ranged from 0.008 to 0.012 within each haplotype clade (Table 5), and from 0.030 to 0.035 between haplotype clades (Table 5). The within-clade values are consistent with Dxy among species calculated for two nearby loci (Dxy = 0.016–0.018, Table 5) and estimated genome-wide gambiae-arabiensis Dxy of 0.011 (O’Loughlin et al. 2014), whereas the between-clade values are approximately three-times higher, and even higher than the estimated genome-wide gambiae-merus Dxy of 0.0235, most likely reflecting an ancient origin. Although we can only compare our values with the mean and SD among individual locus values of Dxy in the O’Loughlin et al. (2014) dataset, our between-clade values are greater than the mean plus 1 SD of even the gambiae-merus comparison, suggesting that the CTLMA2 values exceed most of the many independent genealogies in their study. Moreover, such high values of Dxy across clades are exceptional, given the expectation that clade-wise comparisons should be scaled downward. These results support the hypothesis that the two divergent CTLMA2 haplotypes arose before the species divergence and have been adaptively maintained as a balanced polymorphism in all three species.
Figure 4.
Neighbor-joining tree of CTLMA2 sequences from A. coluzzii (blue circles), A. gambiae (black triangle), A. arabiensis (red squares), A. quadriannulatus (open circle), A. merus (open triangle), and A. christyi (open square). Bootstrap support was determined using 1000 bootstrap replicates, and nodes with >50% bootstrap support are shown.
Table 5. Dxy among species within and between CTLMA2 haplotype clades and in two nearby loci.
| A. coluzzii | A. arabiensisa | Dxy |
|---|---|---|
| Clade A | Clade A | 0.009 |
| Clade B | Clade B | 0.011 |
| Clade A | Clade B | 0.034 |
| Clade B | Clade A | 0.031 |
| AGAP005540a | AGAP005540a | 0.016 |
| APL2a | APL2a | 0.017 |
| A. gambiaea | A. arabiensisa | Dxy |
| Clade A | Clade A | 0.008 |
| Clade B | Clade B | 0.012 |
| Clade A | Clade B | 0.035 |
| Clade B | Clade A | 0.030 |
| AGAP005540a | AGAP005540a | 0.017 |
| APL2a | APL2a | 0.018 |
Sequences from published data (Obbard et al. 2007).
Discussion
We examined patterns of genetic variation and divergence in 20 immune-related genes and 17 control genes. We found that, on average, immune genes have elevated rates of amino acid divergence compared to nonimmune genes, and this was particularly true for genes involved in the Imd pathway. Our findings are consistent with studies of other insects, which show that, as a group, immune genes are rapidly evolving (Schlenke and Begun 2003; Sackton et al. 2007; Viljakainen et al. 2009). The rapid divergence of immune system genes is often hypothesized to be the result of positive selection driving adaptive evolution of the immune system in response to pathogen pressure (Sackton et al. 2007). However, when we examined patterns of nucleotide diversity in the genes in our dataset, we found no evidence of recent positive selection driving the evolution of most of the immune genes. For example, although genes in the Imd pathway exhibit elevated amino acid divergence, they also tended to have higher levels of amino acid diversity than the control genes, which is parsimoniously consistent with relaxed purifying selection on this pathway.
Our data are in contrast to those in studies of Drosophila and termites that conclude adaptive evolution is particularly common in the Imd pathway. Rapid amino acid evolution has been detected in termites in the terminal NF-κB transcription factor of the Imd pathway (Bulmer and Crozier 2006). In D. melanogaster, positively selected sites appear to cluster in the interacting domains of Relish, IKKβ, and Dredd, suggesting that the entire Relish cleavage complex is evolving by positive selection (Sackton et al. 2007). Likewise, a survey of genes throughout the D. melanogaster immune system found that the trend of elevated rates of adaptive evolution observed in immune genes overall is primarily driven by genes in the Imd and RNAi pathways (Obbard et al. 2009b), although adaptation is not uniform throughout the pathways. It has been hypothesized that pathogen interaction with signaling molecules is driving the rapid evolution of the Imd pathway in D. melanogaster (Begun and Whitley 2000). Our findings of relaxed purifying selection in the Imd pathway may seem surprising, because signaling through the Imd transcription factor REL2 has been implicated in antibacterial and antiplasmodium immunity in A. gambiae, and given that the Relish cleavage complex is thought to be a site of pathogen-driven evolution in Drosophila. However, the specific target or mode of pathogen-driven evolution is likely to be different among insects, shaped by the unique suite of pathogens to which they are exposed, and A. gambiae is exposed to a distinct set of pathogens due to its aquatic larval environment as well as its obligate blood feeding and potential exposure to human and animal pathogens. Furthermore, unlike Drosophila Relish, the NF-κB transcription factor for the A. gambiae Imd pathway, REL2, exists in two isoforms (Meister et al. 2005). The full-length isoform (REL2-F) contains an inhibitory domain that must be cleaved off prior to nuclear translocation, whereas the short isoform (REL2-S) that lacks the inhibitory domains is constitutively active (Meister et al. 2005). Additionally, some immune responses involving REL2 signaling have been shown to occur independent of the IMD protein (Garver et al. 2012), suggesting that novel mechanisms may play a role in REL2 signaling. Our observation of relaxed purifying selection on Imd pathway genes could be consistent with IMD-independent regulation of REL2 signaling in A. gambiae.
Evolution in lineage-specific genes or gene families may reflect adaptation to novel pathogens. We found that STAT-B, a retrotransposed duplicate of the STAT transcription factor STAT-A, which is specific to the Anopheles lineage, shows a strong signature of adaptive evolution, whereas the ancestral copy STAT-A is highly conserved. This pattern of selective constraint in STAT-A and rapid evolution in STAT-B is consistent with a model of neofunctionalization after duplication (Hahn 2009; Lynch and Force 2000). There are seven members of the STAT family in vertebrates, of which different copies have adopted specialized roles in the endocrine or immune system. Interestingly, the vertebrate STAT genes, which are predominantly involved in the endocrine system, evolve more slowly than those that are predominantly involved in immunity (Gorissen et al. 2011). The A. gambiae JAK-STAT pathway plays a role in regulating nitric oxide synthase in response to bacterial and Plasmodium infections, and may additionally regulate TEP1 expression during late-phase Plasmodium infections (Gupta et al. 2009). A recent study reported that STAT-B, along with various other detoxification and immune genes, is expressed at higher levels in A. coluzzii larvae than in A. gambiae larvae (Cassone et al. 2014), presumably reflecting the shift to the more biotically and abiotically complex larval habitats preferred by A. coluzzii, and suggesting that adaptive evolution in STAT-B could be important to adaptation to novel ecological or pathogen environments.
Balancing selection can act to maintain functionally important polymorphisms in a population. Genes involved in host–pathogen interactions are considered likely targets of balancing selection, although instances of balancing selection are rarely observed (Leffler et al. 2013). We found two distinct haplotype clades in the 5′ end of CTLMA2, with 11 fixed SNPs between the clades that result in three amino acid changes near the predicted signal cleavage site. We identified both clades in previously published A. gambiae and A. arabiensis sequences. Patterns of divergence within and between the CTLMA2 haplotype clades suggest that they arose before the species divergence and have been adaptively maintained as a balanced polymorphism in all three species, although further study will be required to determine what functional effect the amino acid variants between the two clades might have. CTLMA2 is primarily secreted into the hemolymph in the form of a heterodimer with CTL4 (Schnitger et al. 2009); because most of the amino acid substitutions are located in the signal peptide, the different haplotypes might lead to changes in peptide secretion. CTLs are a family of carbohydrate binding proteins that are involved in the immune response through pathogen recognition and as modulators of melanization. The CTL4/CTLMA2 heterodimer is required for successful melanization of Gram-negative bacteria (Schnitger et al. 2009), so it is tempting to speculate that the amino acid variants in CTLMA2 might change the configuration of the heterodimer to create a unique carbohydrate recognition profile. In contrast to the protective role against bacterial pathogens, the CTL4/CTLMA2 heterodimer prevents melanization of rodent malaria (Osta et al. 2004), suggesting that this heterodimer has conflicting pleiotropic functions in the mosquito immune response that might contribute to the evolutionary pattern observed in CTLMA2.
Conclusion
In this study, we used population genetic analysis to understand how natural selection operates on the A. coluzzii immune system. We found evidence of two distinct haplotypes in the Anopheles-specific C-type lectin CTLMA2, which have been adaptively maintained as a balanced polymorphism in A. coluzzii and the sister species A. gambiae and A. coluzzii. We also found strong evidence of adaptive evolution in the Anopheles-specific JAK-STAT transcription factor STAT-B, consistent with neofunctionalization after duplication. In contrast to these Anopheles-specific immune genes, we found no evidence of adaptive evolution in genes involved in the canonical immune signal transduction pathways in A. coluzzii. As a group, genes in the Imd pathway exhibit patterns of elevated amino acid diversity and accelerated rates of protein evolution, consistent with relaxed purifying selection, relative to nonimmune control loci. Taken together, these results suggest that host–pathogen interactions involving novel or lineage-specific molecular mechanisms likely play a larger role than canonical immune pathways in adaptively evolving resistance to infection in A. coluzzii.
Supplementary Material
Acknowledgments
We thank Mark Jandricic for his help collecting the data. We also thank Rob Unkless and Moria Chambers for helpful discussion and comments on the manuscript. This work was supported by NIH grants R01 AI062995 and R01 AI042361.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014845/-/DC1
Communicating editor: S. A. Tishkoff
Literature Cited
- Bahia A. C., Kubota M. S., Tempone A. J., H. R. C. Arau´ jo, B. A. M. Guedes et al, 2011. The JAK-STAT pathway controls Plasmodium vivax load in early stages of Anopheles aquasalis infection. PLoS Negl. Trop. Dis. 5: e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C., Charlesworth A., Gross I., Richman A., Hoffmann J. A., et al. , 1996. Immune factor Gambif1, a new rel family member from the human malaria vector, Anopheles gambiae. EMBO J. 15: 4691–4701. [PMC free article] [PubMed] [Google Scholar]
- Bargielowski I., Koella J. C., 2009. A possible mechanism for the suppression of Plasmodium berghei development in the mosquito Anopheles gambiae by the microsporidian Vavraia culicis. PLoS ONE 4: e4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillas-Mury C., Han Y. S., Seeley D., Kafatos F. C., 1999. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J. 18: 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun D. J., Whitley P., 2000. Adaptive evolution of Relish, a Drosophila NF-κB/IκB protein. Genetics 154: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. S., Crozier R. H., 2006. Variation in positive selection in termite GNBPs and Relish. Mol. Biol. Evol. 23: 317–326. [DOI] [PubMed] [Google Scholar]
- Cassone B. J., Kamdem C., Cheng C., Tan J. C., Hahn M. W., et al. , 2014. Gene expression divergence between malaria vector sibling species Anopheles gambiae and An. coluzzii from rural and urban Yaounde Cameroon. Mol. Ecol. 23: 2242–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M., Hunt R. H., Wilkerson R., Della Torre A., Coulibaly M. B., et al. , 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619: 246–274. [PubMed] [Google Scholar]
- Cohuet A., Krishnakumar S., Simard F., Morlais I., Koutsos A., et al. , 2008. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics 9: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. H., Sakai R. K., Vernick K. D., Paskewitz S., Seeley D. C., et al. , 1986. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science 234: 607–610. [DOI] [PubMed] [Google Scholar]
- Favia G., Lanfrancotti A., Spanos L., Siden-Kiamos I., Louis C., 2001. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol. Biol. 10: 19–23. [DOI] [PubMed] [Google Scholar]
- Frolet C., Thoma M., Blandin S., Hoffmann J. A., Levashina E. A., 2006. Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 25: 677–685. [DOI] [PubMed] [Google Scholar]
- Garver L. S., Y. Dong Y, and G. Dimopoulos, 2009. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 5: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver L. S., Bahia A. C., Das S., Souza-Neto J. A., Shiao J., et al. , 2012. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 8: e1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorissen M., de Vrieze E., Flik G., Huising M. O., 2011. STAT genes display differential evolutionary rates that correlate with their roles in the endocrine and immune system. J. Endocrinol. 209: 175–184. [DOI] [PubMed] [Google Scholar]
- Gupta L., Molina-Cruz A., Kumar S., Rodrigues J., Dixit R., et al. , 2009. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 5: 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100: 605–617. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguade M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Sáez A. G., Ayala F. J., 1997. DNA variation at the Sod locus of Drosophila melanogaster: an unfolding story of natural selection. Proc. Natl. Acad. Sci. USA 94: 7725–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski J., Grushko O. G., Besansky N. J., 2006. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol. Phylogenet. Evol. 39: 417–423. [DOI] [PubMed] [Google Scholar]
- Leffler E. M., Gao Z., Pfeifer S., Segurel L., Auton A., et al. , 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339: 1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate-gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart S. J., Obbard D. J., Conlon C., Little T. J., 2012. Immune genes undergo more adaptive evolution than non-immune system genes in Daphnia pulex. BMC Evol. Biol. 12: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Meister S., Kanzok S. M., Zheng X. L., Luna C., Li T. R., et al. , 2005. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102: 11420–11425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S., Agianian B., Turlure F., Relogio A., Morlais I., et al. , 2009. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 5: e1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri C., Jacques J. C., Thiery I., Riehle M. M., Xu J., et al. , 2009. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 5: e1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muspratt J., 1946. On Coelomomyces fungi causing high mortality of Anopheles gambiae larvae in Rhodesia. Ann. Trop. Med. Parasitol. 40: 10–17. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., et al. , 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular evolutionary genetics, Columbia University Press, New York, NY. [Google Scholar]
- Obbard D. J., Jiggins F. M., Halligan D. L., Little T. J., 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16: 580. [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Linton Y. M., Jiggins F. M., Yan G., Little T. J., 2007. Population genetics of Plasmodium resistance genes in Anopheles gambiae: no evidence for strong selection. Mol. Ecol. 16: 3497–3510. [DOI] [PubMed] [Google Scholar]
- Obbard D. J., Callister D. M., Jiggins F. M., Soares D. C., Yan G., et al. , 2008. The evolution of TEP1, an exceptionally polymorphic immunity gene in Anopheles gambiae. BMC Evol. Biol. 8: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Welch J. J., Little T. J., 2009a Inferring selection in the Anopheles gambiae species complex: an example from immune-related serine protease inhibitors. Malar. J. 8: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Welch J. J., Kim K. W., Jiggins F. M., 2009b Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5: e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin S. M., Magesa S., Mbogo C., Mosha F., Midega J., et al. , 2014. Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Mol. Biol. Evol. 31: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osta M. A., Christophides G. K., Kafatos F. C., 2004. Effects of mosquito genes on Plasmodium development. Science 303: 2030–2032. [DOI] [PubMed] [Google Scholar]
- Parmakelis A., Slotman M. A., Marshall J. C., Awono-Ambene P. H., Antonio-Nkondjio C., et al. , 2008. The molecular evolution of four anti-malarial immune genes in the Anopheles gambiae species complex. BMC Evol. Biol. 8: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle M. M., Xu J., Lazzaro B. P., Rottschaefer S. M., Coulibaly B., et al. , 2008. Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE 3: e3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottschaefer S. M., Riehle M. M., Coulibaly B., Sacko M., Niaré O., et al. , 2011. Exceptional diversity, maintenance of polymorphism, and recent directional selection on the APL1 malaria resistance genes of Anopheles gambiae. PLoS Biol. 9: e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T. B., Lazzaro B. P., Schlenke T. A., Evans J. D., Hultmark D., et al. , 2007. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39: 1461–1468. [DOI] [PubMed] [Google Scholar]
- Smith N. G. C., Eyre-Walker A., 2002. Adaptive protein evolution in Drosophila. Nature 415: 1022–1024. [DOI] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2003. Natural selection drives Drosophila immune system evolution. Genetics 164: 1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitger A. K., Yassine H., Kafatos F. C., Osta M. A., 2009. Two C-type lectins cooperate to defend Anopheles gambiae against gram-negative bacteria. J. Biol. Chem. 284: 17616–17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. A., Brogdon W. G., Collins F. H., 1993. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 49: 520–529. [DOI] [PubMed] [Google Scholar]
- Slotman M. A., Parmakelis A., Marshall J. C., Awono-Ambene P. H., Antonio-Nkondjo C., et al. , 2007. Patterns of selection in anti-malarial immune genes in malaria vectors: evidence for adaptive evolution in LRIM1 in Anopheles arabiensis. PLoS ONE 2: e793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljakainen L., Evans J. D., Hasselmann M., Rueppell O., Tingek S., et al. , 2009. Rapid evolution of immune proteins in social insects. Mol. Biol. Evol. 26: 1791–1801. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gilbreath T. M., III, Kukutla P., Yan G., Xu J., 2011. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 6: e24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Kriventseva E. V., Meister S., Xi Z., Alvarez K. S., et al. , 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316: 1738–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. J., 2006. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics 173: 821–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Lawniczak M. K., Cheng C., Coulibaly M. B., Wilson M. D., et al. , 2011. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc. Natl. Acad. Sci. USA 108: 244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. I., Charlesworth B., 2004. The HKA test revisited: a maximum likelihood-ratio test of the standard neutral model. Genetics 168: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.