Abstract
The conformational basis for reduced activity of the thermophilic ribonuclease HI enzyme from Thermus thermophilus, compared to its mesophilic homolog from Escherichia coli, is elucidated using a combination of NMR spectroscopy and molecular dynamics (MD) simulations. Explicit-solvent all-atom MD simulations of the two wild-type proteins and an E. coli mutant in which a glycine residue is inserted after position 80 to mimic the T. thermophilus protein reproduce the differences in conformational dynamics determined from 15N spin-relaxation NMR spectroscopy of three loop regions that surround the active site and contain functionally important residues: the glycine-rich region, the handle region, and the β5/αE loop. Examination of the MD trajectories indicates that the thermophilic protein samples conformations productive for substrate binding and activity less frequently than the mesophilic enzyme, although these differences may manifest as either increased or decreased relative flexibility of the different regions. Additional MD simulations indicate that mutations increasing activity of the T. thermophilus enzyme at mesophilic temperatures do so by reconfiguring the local environments of the mutated sites to more closely resemble active conformations. Taken together, the results show that both locally increased and decreased flexibility contribute to an overall reduction in activity of T. thermophilus ribonuclease H compared to its mesophilic E. coli homolog.
Keywords: chemical exchange, Michaelis–Menten enzyme kinetics, molecular dynamics simulation, order parameter, relaxation dispersion
Introduction
Ribonuclease HI (RNase H; EC 3.1.26.4) enzymes are well-conserved endonucleases that non-specifically cleave the RNA strand of RNA:DNA hybrid substrates in numerous biological processes [1]. In Escherichia coli, RNase H has been implicated in primer synthesis for replication of ColE1 plasmids, inhibition of replication from sites other than oriC by removal of R-loops [2–4], removal of Okazaki fragments during lagging strand synthesis [5,6], synthesis of multicopy single-stranded DNA [7–9], and removal of ribonucleotides misincorporated into the genome during transcription [10]. Retroviral reverse transcriptase contains a C-terminal RNase H domain that is essential for reverse transcription of the viral genome (for reviews, see Refs. [11] and [12]).RNase H belongs to a nucleotidyl-transferase superfamily with a conserved structure and mechanism; other family members include transposase, retroviral integrase, Holliday junction resolvase, and RISC nuclease Argonaute [13,14].
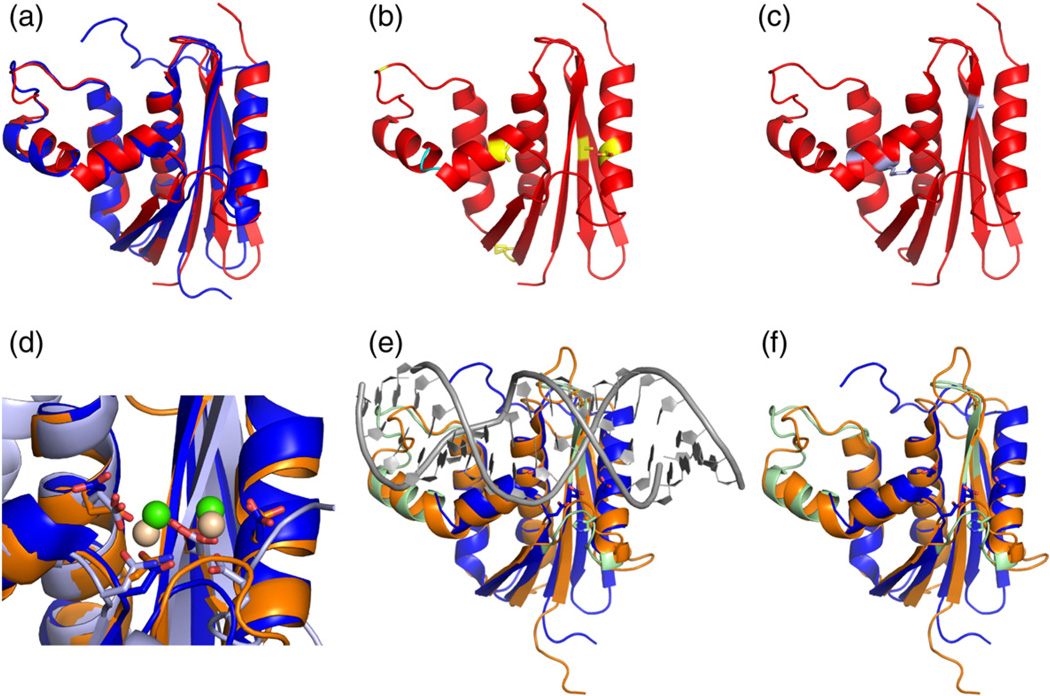
RNase H from the mesophile E. coli [E. coli RNase H (ecRNH)] and that from the thermophile Thermus thermophilus [T. thermophilus RNase H (ttRNH)] have been the subject of numerous studies of function and thermal adaptation [15–21]. Typical of other mesophile–thermophile pairs [22], the two proteins share a high level of sequence (52%) and structural similarity (<1 Å Cα RMSD in secondary structures) (Fig. 1a), but they differ significantly in stability and activity. ttRNH shows significantly higher thermal and chemical stability [23–25] and significantly lower binding affinity and catalytic activity at mesophilic temperatures compared to ecRNH [25]. Mutational studies have identified several instances of improved hydrophobic packing, more favorable electrostatic interactions, or reduced conformational strain that contribute to the increased stability of ttRNH compared to ecRNH (Fig. 1b) [26–33]. The most prominent sequence difference between the two proteins is the presence of an inserted glycine residue in ttRNH in the junction between two conserved helices (Fig. 1b), the presence of which is more common in thermophilic bacteria [34] despite conferring relatively little stability in isolation [30]. Introduction of the inserted glycine into the sequence of ecRNH (iG80b) yields a significantly less active enzyme, with a 50-fold increase in Km and a 10-fold decrease in kcat relative to wild-type (WT) ecRNH [30]. A genetic screen for mutants that enable ttRNH to complement ecRNH at mesophilic temperatures has identified three mutations— A12S, K75M, and A77P—that cooperatively increase the enzymatic activity of ttRNH without substantially compromising stability, although the mutations individually have varied effects on enzymatic activity and stability (Fig. 1c) [35]. RNase H residues are henceforth numbered according to their position in the sequence of ecRNH for simplicity.
Fig. 1.
Structural features of RNases H. (a) Superposition of ecRNH (blue, PDB code 2RN2) [70] and ttRNH (red, PDB code 1RIL) [56]. (b) Known sites of thermostabilization (yellow) and the glycine insertion (cyan). (c) Positions of the three activating mutations identified for ttRNH [35]. (d) Superposition of the active site of ecRNH (blue) with the two known substrate complexes, hsRNH (orange and green, PDB code 2QK9) [14] and RNase H domain from B. halodurans (light blue and beige, PDB code 1ZBI). (e) Superposition of ecRNH and hsRNH showing substrate (gray) and substrate-binding loops (green). (f) Superposition of ecRNH and hsRNH with substrate removed. Active-site residues from ecRNH are shown as blue sticks. Functionally important residues located in substrate-binding loops are shown as green sticks.
The RNase H active site consists of three to four conserved carboxylate residues, also referred to as the DED(D) motif, which are involved in coordinating catalytically required divalent cations and possibly in proton transfer during catalysis [36]. Activity can be supported by both Mg2+ and Mn2+ [37]. High-resolution structures of RNase H domains from Bacillus halodurans [38] and Homo sapiens [H. sapiens RNase H (hsRNH)] [14] in complex with substrate locate the RNA cleavage site between the two ions (Fig. 1d). NMR spectroscopy and molecular dynamics (MD) simulations indicate that the carboxylate side chains in the ecRNH active site are pre-organized for binding the first Mg2+ ion but may require conformational rearrangement to bind the second [39]. MD simulations of a variety of RNase H homologs suggest that the dynamics of the active-site side chains in the apo state are conserved across the RNase H family [40].
The crystal structure of hsRNH identifies three distinct regions as important for substrate binding and catalysis: the loop between β1 and β2 (termed the glycine-rich region, residues 11–22 in ecRNH), αC and the loop between αC and αD (termed the handle region, residues 81–101), and the loop between β5 and αE (termed the β5/αE loop, residues 121–127) (Fig. 1e and f). These results are consistent with mutagenesis and chemical shift mapping for ecRNH [41–43]. The glycine-rich loop and handle region form direct contacts with substrate; the handle, in particular, contains well-conserved tryptophan residues (W81 and W85) that form part of a DNA-binding channel that sterically excludes RNA [14]. A catalytic role for the β5/αE loop, located close to the active site, has been suggested, with H124 thought to be involved in product release and/or inhibition in the presence of high metal ion concentration [14,38,44]. The holo hsRNH crystal structures exhibit distinct conformations in all three regions compared to apo ecRNH and ttRNH structures (Fig. 1f), implying that conformational changes may occur upon binding substrate.
The nature of the relationship between functionally important conformational changes and thermostability has been difficult to characterize. Conformational restriction owing to a deep and narrow global minimum in the energy landscape is a feature commonly attributed to thermostable proteins [45]. Studies of homologous mesophile–thermophile pairs have identified cases in which the thermostable protein is more rigid than its mesophilic counterpart [46]; however, examples of more flexible thermostable proteins have also been described [47–49]. Protein regions that interact with substrate are often identifiable as being particularly flexible by NMR spectroscopy [50,51]. NMR studies of the RNase H family suggest that the substrate-binding regions exhibit differences in dynamic behavior that reflect underlying energy landscapes in which the active conformations are less accessible at ambient temperature to the thermophilic homolog, resulting in its reduced activity [15,16]. Although the ttRNH protein is slightly more globally rigid, more subtle differences are observed in the functionally important dynamic regions. On the picosecond-to-nanosecond timescale, the glycine-rich region and the β5/αE loop are more rigid in ttRNH than in ecRNH, while the handle region appears somewhat more flexible [15]. On the microsecond-to-millisecond timescale, the handle region exchanges between one highly populated and one or more sparsely populated conformations [16]. The glycine insertion found in ttRNH modulates the local conformational preferences of the handle region: homologous insertion of a glycine into the sequence of ecRNH (iG80b) alters the thermodynamic equilibrium of the handle region to resemble that of ttRNH, whereas deletion of the glycine residue from ttRNH (dG80) alters the thermodynamic equilibrium to resemble that of WT ecRNH [16]. In addition, the handle region and the N-terminal hinge of the β5/αE loop, which are distal and proximal to the active site, respectively, share a common motional process in ttRNH, suggesting coupling between these two sites [16].
Previously, we performed explicit-solvent MD simulations of ecRNH, ttRNH, and other members of the RNase H family in order to characterize the temperature dependence of conformational dynamics in the handle region [34]. In the present work, simulations of ecRNH, ttRNH, and iG80b ecRNH are compared to NMR spin-relaxation data. Both simulations and experiments were carried out in the apo state and thus reflect ensembles sampled in the absence of substrate or catalytically required magnesium ions. The simulations qualitatively reproduce experimentally observed differences in conformational dynamics of the glycine-rich region, the handle region, and the β5/αE loop for the three proteins. Analysis of the simulation trajectories identifies the atomistic basis and mechanistic consequences of these differences, highlighting the ability of the iG80b mutation to confer motional properties on the ecRNH protein that closely resemble those of ttRNH. Additional simulations enable rationalization of the origins of enhanced enzymatic activity in mutant ttRNH proteins identified by a genetic screen [35]. Introduction of these mutations in silico in the ttRNH structure yields a conformational ensemble that more closely resembles ecRNH or hsRNH. Taken together, the experimental and computational results suggest that local conformational preferences in the thermophilic enzyme contribute to its reduced substrate-binding affinity.
Results and Discussion
Global backbone dynamics
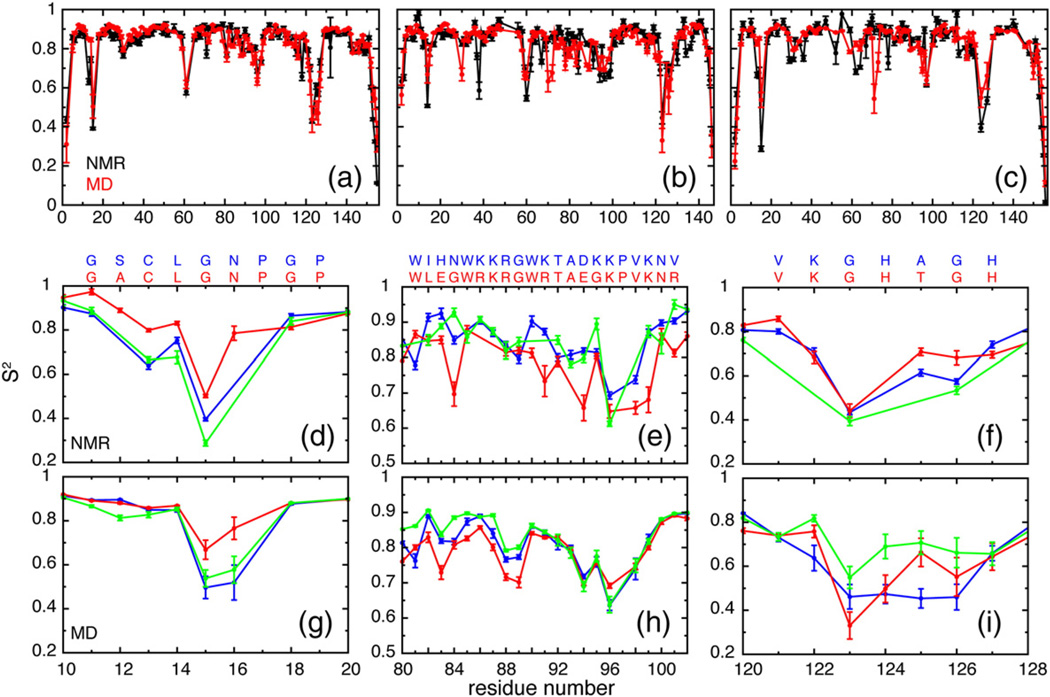
Generalized order parameters (S2) of the backbone amide N–H bond vectors, which quantify the equilibrium distributions of bond vector orientations in a molecular frame of reference, were computed from MD simulations and compared to experimentally determined values for ecRNH [19], ttRNH [15], and ecRNH iG80b (vide infra) (Fig. 2a–c). The simulations agree well with experiment, yielding correlation coefficients R = 0.89, 0.74, and 0.86 for ecRNH, ttRNH, and iG80b ecRNH, respectively; independent but comparable simulations of ecRNH and ttRNH also accurately reproduce experimental NMR chemical shifts [52]. The reduced correlation for ttRNH likely reflects subtle differences under experimental and simulation conditions: the experimental data were acquired at 310 K [15] on a cysteine-free mutant used to eliminate undesirable thiol chemistry, while simulations were performed at 300 K on the WT construct whose structure was solved by X-ray crystallography. In addition, ttRNH has relatively few sites with very low order parameters, which reduces the range of observed values. Because NMR spin-relaxation data for iG80b ecRNH are only available at a single static magnetic field, the associated order parameters may be less robust than those for WT ecRNH and ttRNH, which were derived from spin-relaxation data at three static magnetic fields [15,19]. Nonetheless, the correlation between experimental and simulated order parameters is equally good for iG80b ecRNH as the other proteins. Good agreement between simulated and experimental backbone dynamics encourages further investigation of the differences among the simulated ensembles in the substrate-binding regions of the protein.
Fig. 2.
Experimental and simulated order parameters. Full sets of (black) experimental and (red) simulated order parameters for (a) ecRNH, (b) ttRNH, and (c) iG80b ecRNH. Comparison of (d) experimental and (g) simulated order parameters in the glycine-rich region (residues 11–22). The sequence alignment between WT ecRNH (blue) and ttRNH (red) in this region is shown at the top. Comparison of (e) experimental and (h) simulated order parameters in the handle region (residues 81–101). Comparison of (f) experimental and (i) simulated order parameters in the β5/αE loop (residues 121–127). Color coding in (d–i) is (blue) ecRNH, (red) ttRNH, and (green) iG80b ecRNH. The sequence alignment between (blue) ecRNH and (red) ttRNH in each region is shown at the top of (d–f). The sequence of iG80b ecRNH differs from WT by insertion of a glycine residue N-terminal to W81.
Glycine-rich region
Experimental backbone order parameters in the glycine-rich region are higher in ttRNH than in WT and iG80b ecRNH [15], which is qualitatively reproduced in silico (Fig. 2d and g). G15 adopts an alternative left-handed α-helical backbone conformation in the MD simulations, in addition to the conformation observed in the apo-state crystal structures. This minor state is populated 39%, 3%, and 29% of the time in ecRNH, ttRNH, and iG80b ecRNH trajectories, respectively. Increased populations of the minor state manifest in lower backbone order parameters for G15 and N16 in ecRNH and iG80b ecRNH.
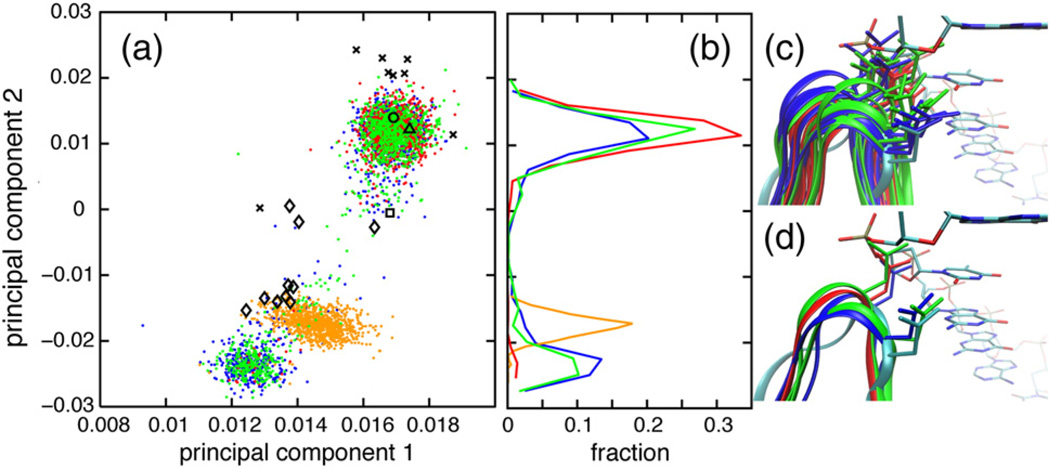
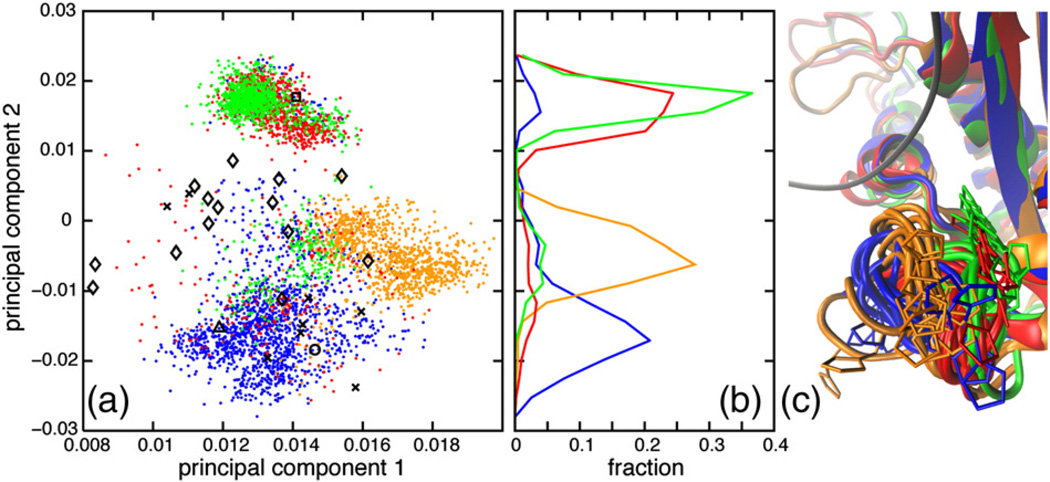
A principal components analysis (PCA) was performed using backbone dihedral angles of the N-terminal part of the glycine-rich region (residues 11–16) for WT ecRNH, ttRNH, iG80b ecRNH, and hsRNH (Fig. 3). The glycine-rich region of hsRNH contains a three-residue insertion C-terminal of N16 relative to ecRNH and ttRNH; however, residues 11–16 have a high level of sequence similarity to the bacterial proteins. The backbone conformation of G15 and the PCA indicate that the minor state resembles the conformation observed in the holo hsRNH crystal structures, which is retained in the hsRNH MD ensemble; this conformation is also observed in the structure of B. halodurans RNase H, a bacterial enzyme that lacks the handle region [38]. The dominant G15 conformation positions the N16 side chain to clash with the DNA backbone, whereas the minor conformation positions N16 for productive substrate interactions (Fig. 3c and d).
Fig. 3.
Glycine-rich region conformational dynamics. (a) PCA was conducted using the backbone dihedral angles ϕ and ψ of the N-terminal part of the glycine-rich region (residues 11–16) from (blue dots) ecRNH, (red dots) ttRNH, (green dots) iG80b ecRNH, and (orange dots) hsRNH (orange dots) MD simulations, as well as (black circle, PDB code 2RN2) ecRNH, (black triangle, PDB code 1RIL) ttRNH (black square, PDB code 1GOA) , iG80b ecRNH, and (black diamonds, PDB codes 2QK9, 2QKK, and 2QKB) hsRNH crystal structures and (black Xs, PDB code 1RCH) ecRNH NMR structures. (b) Distributions of MD conformations along the second principal component. (c and d) Conformations were extracted from MD simulations of (blue) ecRNH, (red) ttRNH, and (green) iG80b ecRNH after backbone heavy-atom superposition with respect to a (cyan, PDB code 2QK9) holo hsRNH crystal structure. The DNA strand from that hsRNH crystal structure is shown with atoms colored according to chemical identity. (c) Conformations were extracted from MD simulations in 10-ns intervals. (d) Conformations of WT and iG80b ecRNH representative of the major and minor states and a conformation of ttRNH representative of the major state were selected from MD simulations for clarity.
The only sequence difference between ecRNH and ttRNH in glycine-rich region is at position 12, where ecRNH has Ser and ttRNH has Ala. The mutation A12S in ttRNH reduces Km by a factor of 5.2 while leaving kcat almost unchanged [35]. Three independent MD trajectories of A12S ttRNH mutants (one of the double mutant A12S/K75M and two of the triple mutant A12S/K75M/A77P) yielded populations of the alternative conformation of 14%, 4%, and 9%, respectively, with an average of (9 ± 5)%. The simulated increases in the populations in the mutant proteins would account for increased affinity by a factor of 3.0 for the average population over all three trajectories of the mutants or by a factor of 4.7 for the population of the A12S/K75M simulation. Although achieving simulation convergence is difficult when sampling sparsely populated conformations, these differences in populations are in qualitative agreement with the experimental increase in substrate affinity determined for these mutants.
Handle region
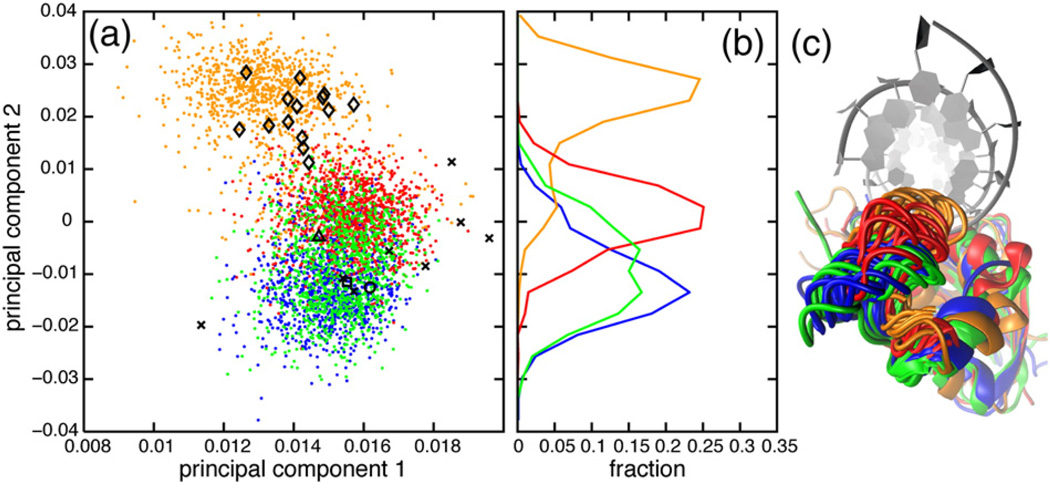
Experimental backbone order parameters in the handle region (residues 81–101) are lower in ttRNH than in ecRNH and iG80b ecRNH (Fig. 2e) [15]. The differences are relatively small, however, with average order parameters of 0.84 ± 0.01, 0.78 ± 0.02, and 0.85 ± 0.02 for ecRNH, ttRNH, and iG80b ecRNH, respectively. Simulated order parameters qualitatively reproduce the differences among the three proteins, with average order parameters of 0.81 ± 0.02, 0.78 ± 0.02, and 0.83 ± 0.02, respectively (Fig. 2h); however, the simulations do not capture fully the finer features of the conformational dynamics within the handle loop (residues 89–101). PCA was conducted using pseudo-dihedral angles among four consecutive Cα atoms of the handle region loop (αC/αD loop, residues 89–101) for ecRNH, ttRNH, iG80b ecRNH, and hsRNH (Fig. 4). Conventional backbone dihedral PCA is not informative because no transitions to alternate backbone dihedral conformations occur in this region.
Fig. 4.
Handle region conformational dynamics. (a) PCA was conducted using pseudo-dihedral angles between four consecutive Cα atoms of the handle-region loop (residues 89–101). (b) Distributions of MD conformations along the second principal component. (c) Conformations were extracted from MD simulations in 10-ns intervals, after backbone heavy-atom superposition with respect to a holo hsRNH crystal structure (PDB code 2QK9). The RNA:DNA hybrid from the hsRNH crystal structure is shown as gray ribbons. Color coding and symbolic representations are as described for Fig. 3.
The first two principal components describe the closing of the handle loop (Fig. 4c). The MD ensembles for ttRNH and hsRNH are more closed than the respective crystal structures, while the WT ecRNH crystal and NMR structures, as well as the iG80b ecRNH crystal structure, are contained within their respective MD ensembles. Consistent with previous observations [34], the handle loop of ttRNH adopts a more closed conformation than that of ecRNH. The broad distribution of iG80b ecRNH conformations along the second principal component reveals that this mutant populates both ecRNH-like and ttRNH-like conformations. The inserted glycine residue therefore modulates the conformational preferences of this region. In combination with simulations of the temperature dependence of open–closed transitions [34], these results suggest that the closed handle-loop conformation inhibits substrate binding. Transitions between open and closed conformations may underlie chemical exchange broadening on the microsecond-to-millisecond timescale previously observed experimentally [16]. This pattern in which conformational changes observed in relatively short simulations resemble those known experimentally to occur on longer timescales has previously been observed in other systems, including triose phosphate isomerase [53], dihydrofolate reductase [54], and adenylate kinase [55]. However, the timescale discrepancy precludes precise estimation of the effects of handle-loop conformational change on enzyme kinetics.
The handle-loop conformations of hsRNH are distinct from the three bacterial proteins throughout the MD trajectories. Although this difference could arise because the trajectory was initiated from the substrate-bound conformation, reciprocal in silico mutations have attributed this difference to the identity of the residue at position 88, which is conserved as Arg in ecRNH and ttRNH but is an Asn in hsRNH [34].
Tryptophan 81
Tryptophan side-chain Nε order parameters for ecRNH, ttRNH, and iG80b ecRNH are presented in Table 1 and show good agreement between experiment and simulation. Both methods identify W81 in ttRNH as the only flexible tryptophan side chain. Located at the N-terminus of helix C, W81 is highly conserved among handle-region-containing bacterial RNases H and is the C-terminal neighbor of the inserted glycine residue in ttRNH and iG80b ecRNH. The inserted Gly residue, in addition to its role in modulating handle-loop closure, reconfigures the energy landscape sampled by its neighbor W81. In the holo hsRNH crystal structures, the side chain of W81 forms part of a channel important for recognition of the DNA strand [14]. The side-chain conformation adopted in the holo hsRNH structure is likely to be the binding-competent conformation and is the dominant state in ecRNH. However, the side chain adopts different conformations in MD simulations of WT ecRNH, characterized by χ2 = trans, and iG80b ecRNH, characterized by χ2 = gauche−. Both states are populated in ttRNH, resulting in a reduced side-chain order parameter for this protein (Fig. 5). The crystallographic conformations of W81 in ttRNH and iG80b ecRNH correspond to neither of the two states visited in silico, likely due to the presence of distorting crystal contacts [56].
Table 1.
Order parameters for tryptophan ring Nε1
| Residue | WT ecRNH |
ttRNH |
iG80b ecRNH |
|||
|---|---|---|---|---|---|---|
| S2NMR | S2MD | S2NMR | S2MD | S2NMR | S2MD | |
| W81 | 0.83 ± 0.02 | 0.83 ± 0.01 | 0.67 ± 0.02 | 0.66 ± 0.06 | 0.82 ± 0.01 | 0.79 ± 0.01 |
| W85 | 0.85 ± 0.02 | 0.75 ± 0.01 | N/A | N/A | 0.86 ± 0.01 | 0.84 ± 0.01 |
| W90 | 0.84 ± 0.01 | 0.85 ± 0.01 | 0.82 ± 0.01 | 0.81 ± 0.01 | 0.86 ± 0.01 | 0.86 ± 0.01 |
| W104 | 0.92 ± 0.08 | 0.87 ± 0.01 | 0.96 ± 0.01 | 0.87 ± 0.01 | 0.87 ± 0.02 | 0.88 ± 0.01 |
| W118 | 0.86 ± 0.01 | 0.85 ± 0.02 | — | — | 0.85 ± 0.01 | 0.87 ± 0.01 |
| W120 | 0.89 ± 0.01 | 0.84 ± 0.01 | — | — | 0.80 ± 0.01 | 0.87 ± 0.01 |
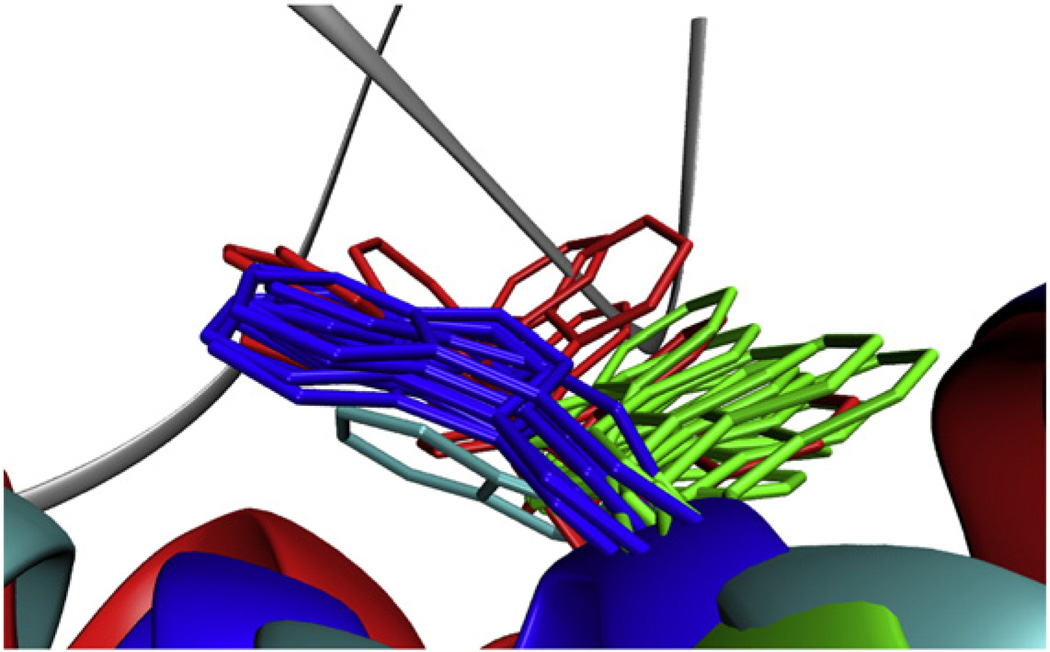
Fig. 5.
W81 conformational space. Representative side-chain conformations of W81 were extracted from (blue) ecRNH, (red) ttRNH, and (green) iG80b ecRNH MD simulations. The side chain of the homologous tryptophan from a holo hsRNH crystal structure (PDB code 2QK9) is shown in cyan. The backbone of the RNA:DNA hybrid from that crystal structure is shown as a gray ribbon.
Superposition of the ttRNH MD ensemble with the holo hsRNH crystal structures suggests that the side chain of W81 would clash with the DNA strand in the alternative conformation. This conformation is thus considered binding incompetent (Fig. 5). In iG80b ecRNH, W81 predominantly populates this binding-incompetent conformation, resulting in a high sidechain order parameter comparable to that observed in WT ecRNH, in good agreement with experiment. The NMR spin-relaxation data alone are insensitive to conformational differences between WT and iG80b ecRNH in this case; the atomistic interpretation afforded by MD simulations reveals distinct conformational ensembles with distinct functional consequences.
Cooperativity has been observed between the presence of the glycine insertion and the identity of the residue located at position 77 [30]. In ttRNH, the kink introduced by the glycine insertion orients A77 in close spatial proximity to W81. The A77P mutation in ttRNH reduces Km by a factor of 2.3 and increases kcat by 50% [35]. Table 2 reports simulated rotamer populations and order parameters for W81. Although the observed reduction in the population of the binding-incompetent rotamer—from 7% in the WT ttRNH simulation to 0% in the triple activating mutant—does not fully account for the reported change in Km, the fact that this rotamer is accessible only in the presence of the glycine insertion and is reduced or eliminated by changes in local packing suggests that this is at least one mechanism through which the A77P mutation influences substrate affinity.
Table 2.
MD simulated order parameters for W81 ring Nε1
| Protein | Rotamers | Population | S2MD |
|---|---|---|---|
| ttRNH WT | t/g – | 0.93/0.07 | 0.66 ± 0.06 |
| ttRNH A12S/K75M | t/g – | 0.98/0.02 | 0.72 ± 0.14 |
| ttRNH A12S/K75M/A77P | t | 1.00 | 0.84 ± 0.01 |
| ttRNH dG80 | t | 1.00 | 0.80 ± 0.01 |
| ecRNH WT | t | 1.00 | 0.83 ± 0.01 |
| ecRNH iG80b | g – | 1.00 | 0.79 ± 0.01 |
| ecRNH iG80b Q80L | t | 0.97 | 0.67 ± 0.13 |
| ecRNH iG80b V101R | t | 0.48 | 0.85 ± 0.01 |
| g – | 0.52 | 0.69 ± 0.04 | |
| ecRNH iG80b G77A | t/ | 0.66 | 0.82 ± 0.01 |
| g – | 0.34 | 0.76 ± 0.02 |
S2MD values are calculated in blocks corresponding to the expected global tumbling time of approximately 10 ns; therefore, rotamer-specific order parameters are calculated only in cases where the persistence time for each state exceeds 10 ns.
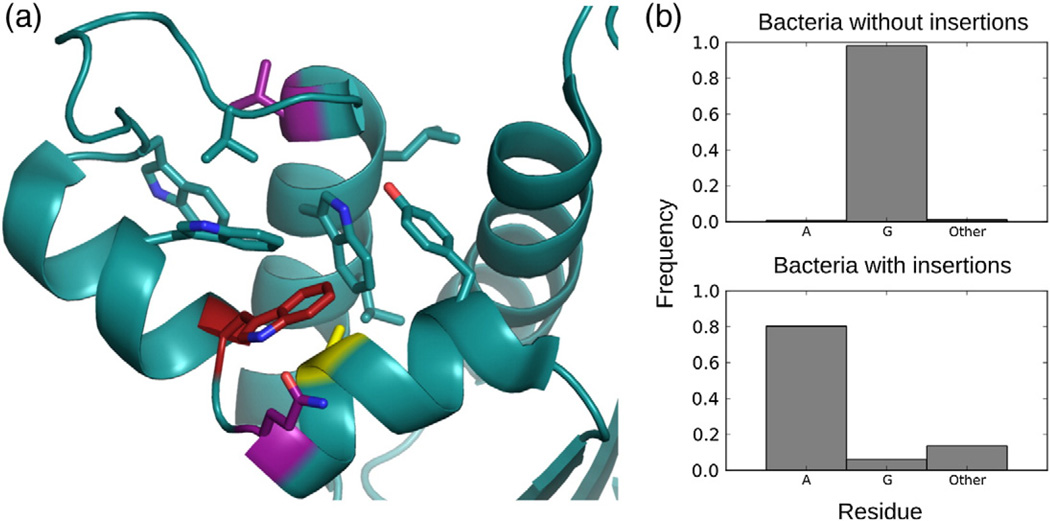
To further explore the effects of local hydrophobic packing on W81 conformational preferences, we calculated additional trajectories for mutations to iG80b ecRNH, which favors the binding-incompetent rotamer (Table 2). Mutations that alter local hydrophobic packing or provide hydrophobic surfaces adjacent to the W81 side chain increase the population of the productive rotamer relative to ecRNH iG80b. Sites at which mutations can affect W81 conformational preferences are shown in Fig. 6a. Interestingly, examination of bacterial RNase H sequences containing handle regions reveals strong evolutionary coupling between the Gly insertion and position 77 (Fig. 6b). In aggregate, the results show that the inserted glycine residue affects the conformational ensemble of the handle region in two ways—by reducing the population of the open handle loop and by increasing the population of the binding-incompetent W81 rotamer—both of which result in lower binding affinity toward substrate in ttRNH.
Fig. 6.
Hydrophobic environment surrounding W81. (a) Hydrophobic and aromatic residues surrounding W81 (red) are shown as sticks in the crystal structure of the ecRNH double mutant iG80b/G77A (PDB code 1GOC). Mutations at sites shown in purple (Q80 and V101) and yellow (A77) promote the trans conformation of W81. (b) Sequence analysis of bacterial RNase H sequences [34] containing handle regions show a strong coupling between presence of insertion and residue identity at position 77.
The β5/αE loop and the αB/β5 interface
Experimental backbone order parameters for residues 125 and 126 at the C-terminal end of the β5/αE loop are somewhat higher in ttRNH than in WT ecRNH (Fig. 2f). The limited spin-relaxation data for iG80b ecRNH yield order parameters for only two of the seven backbone amides in the loop, preventing quantitative comparison. Trends observed experimentally are not well reproduced in silico for this region, likely because limited sampling is exacerbated by the presence of distorted initial conformations resulting from crystal contacts. A well-documented crystal contact common in ecRNH crystal structures and present in both WT and iG80b is known to distort the conformation of this loop [57]. Nevertheless, comparisons between even unconverged simulations may prove instructive, given that the purpose of the analysis is to explore the possibility of differential conformational restriction among the RNases H tested.
The β5/αE loop occupies a more closed conformation in ttRNH than in ecRNH, characterized by increased backbone order parameters in qualitative agreement with experiment (Fig. 2f and i). The first two components of a PCA conducted using backbone dihedral angles in the β5/αE loop are plotted in Fig. 7. The results indicate that conformational preferences of the β5/αE loop in iG80b ecRNH and ttRNH are very similar and preferentially populate a relatively restricted state largely unique to those two proteins (Fig. 7c). Notably, the β5/αE-loop conformation in the ttRNH crystal structure more closely resembles the WT ecRNH and hsRNH ensembles than the simulated ttRNH ensemble. The conformational space accessible to the catalytically important H124 residue contained in this loop is substantially more limited in ttRNH and iG80b ecRNH than in WT ecRNH and hsRNH. Comparison of multiple holo hsRNH crystal structures reveals that the β5/αE loop retains substantial flexibility in complex with substrate. The closed conformation and more restricted flexibility in ttRNH may contribute to decreased activity through the reported effects of H124 on metal ion binding or product release. The conformational preferences of the β5/αE loop in iG80b ecRNH mirror those in ttRNH, indicating a dynamic coupling to the glycine insertion in the handle region on the picosecond-to-nanosecond timescale. This coupling may also be responsible for the similarities in the microsecond-to-millisecond dynamics observed between the two proteins experimentally [16].
Fig. 7.
β5/αE-loop conformational dynamics. (a) PCA was conducted using the backbone dihedral angles ϕ and ψ of the β5/αE loop (residues 121–127). (b) Distributions of MD conformations along the second principal component are also shown. (c) Conformations were extracted from MD simulations in 10-ns intervals, after backbone heavy-atom superposition with respect to a holo hsRNH crystal structure (PDB code 2QK9). The RNA strand from that hsRNH crystal structure is shown as a gray ribbon. Color coding and symbolic representations are as described for Fig. 3.
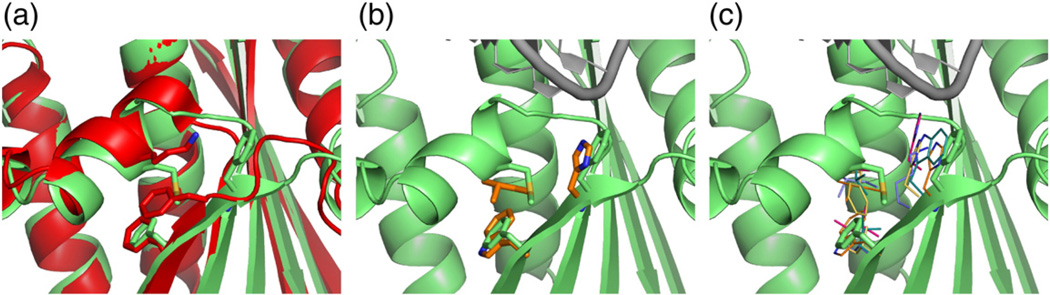
The activating mutations of ttRNH partially reorganize the packing of the hydrophobic interface between αB and β5, thereby suggesting a candidate pathway through which this coupling occurs. This interface is significantly different in crystal structures of RNases H in the apo state compared to the hsRNH complex. In particular, aromatic groups (Phe in ttRNH and Trp in ecRNH) at position 120 are oriented “down”, pointing away from the substrate-binding site, in both ecRNH and ttRNH; however, a 180° χ2 rotation orients the equivalent H120 residue in hsRNH “up” so that its ring nitrogen interacts with the backbone of the RNA strand of the bound substrate. In the A12S/K75M and A12S/K75M/A77P ttRNH trajectories, a transition of F120 to the “up” state occurs consistently and reproducibly, thus reconfiguring the interface to resemble that present in the bound state (Fig. 8). The effect of the F120 “up” and “down” conformational equilibrium on Km cannot be estimated because only single transitions are observed in these simulations. However, structures of other, more distantly related RNases H, including the subdomain found in HIV (human immunodeficiency virus) reverse transcriptase and the atypical RNase H, occupy the “up” conformation even in the absence of substrate (Fig. 8c). In fact, ttRNH is unusual among family members for the absence of a polar or charged residue at position 120. Both K75M/A77P and A12S/K75M/A77P ttRNH mutants exhibit small differences in near- and far-UV circular dichroism spectra compared to either WT ttRNH or any of the single mutants, implying a subtle difference in the packing of aromatic groups despite little change in secondary structure [35], which is consistent with the reorganization of local packing near F120 and W81 observed in simulation.
Fig. 8.
Changes at the αB/β5 interface due to the activating mutation K75M. (a) Superposition of ttRNH crystal structure (red) with a representative MD conformation from the A12S/K75M trajectory (green). (b) Superposition of a representative A12S/K75M conformation as in (a) with equivalent residues from hsRNH (orange). (c) Superposition of equivalent residues from additional members of the RNase H family: H. sapiens (orange, 2QK9), B. halodurans (purple, 1ZBI), HIV (cyan, 1HRH), prototype foamy virus (pink, 2LSN), and XMRV (yellow, 3V1O).
Conclusion
The results presented herein indicate that complex changes in the conformational preferences of three key loop regions contribute to reduced activity of ttRNH relative to ecRNH at mesophilic temperatures. The differences in dynamics of the three regions are summarized in Fig. 9. Examples of both overall increase [47] and overall decrease [58] in conformational flexibility have been reported for thermostable proteins, as well as examples of broader native-state ensembles coupled to reduced longer-timescale fluctuations far from the native state [59,60]. In the present work, we describe a case in which increased and decreased flexibility coexist in the same protein on similar timescales, with the common effect of reduced substrate affinity. Notably, several well-studied examples of mesophile–thermophile pairs are enzymes with small-molecule substrates where differences in activity can be correlated with differences in conformational dynamics of a loop involved in substrate binding [61–63], whereas RNase H has a large substrate-binding surface and thus can accommodate multiple forms of adaptation through subtle changes in the relative energies of local conformational states. Proteins with large, dynamic binding surfaces and whose substrates exhibit complex internal dynamics of their own complicate the emerging picture of flux through conformational selection and induced-fit binding pathways as determined primarily by conformational exchange kinetics and ligand concentration [64]. The behavior observed in the RNase H family, in which partially binding-competent conformations are sampled in the apo state but require additional induced-fit accommodation after binding substrate, is consistent with experimental observations of interdomain protein–protein interactions [65] and with simulations of RNA-binding U1A protein [66]. The present work extends these observations to account for reduced substrate affinity in a thermostable protein.
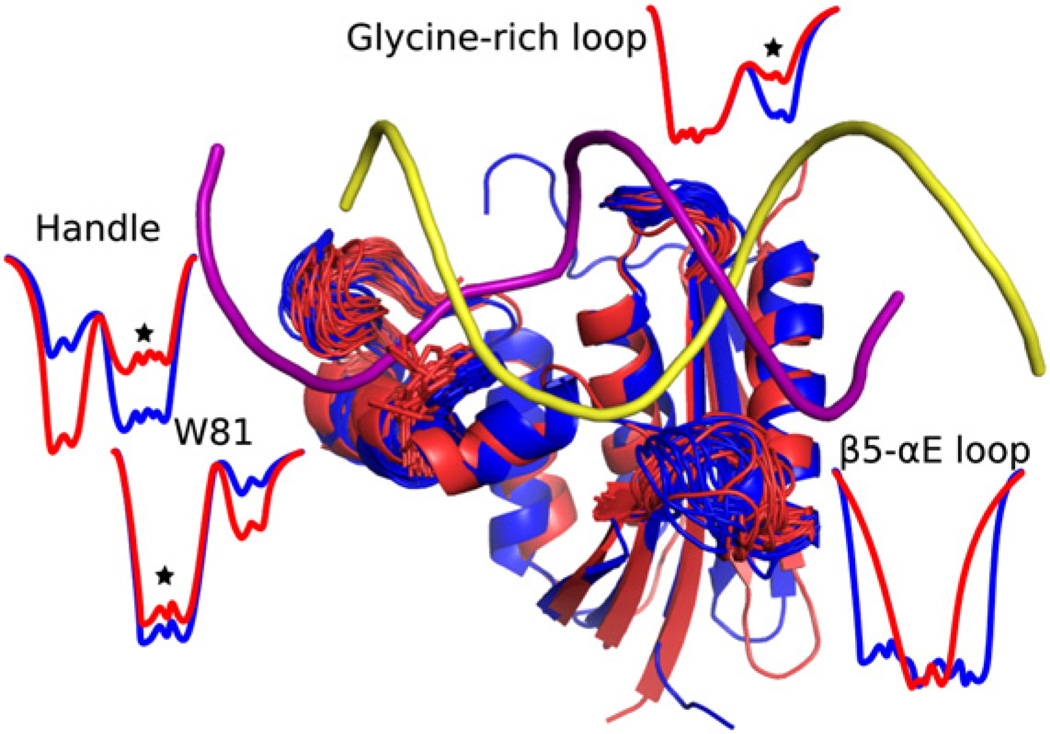
Fig. 9.
Summary of dynamic differences between (blue) ecRNH and (red) ttRNH in regions neighboring the RNA: DNA (yellow and purple, respectively) hybrid substrate. Superposed snapshots from the simulations are shown with energy landscape cartoons to illustrate differences in local conformational preferences for each region. Stars indicate the presumptively binding-competent state. The glycine-rich region is more restricted in ttRNH than in ecRNH and populates a minor binding-competent state substantially less in the thermophile enzyme, the handle loop is more closed in ttRNH than in ecRNH, W81 populates a binding-incompetent conformation in ttRNH in addition to the binding-competent conformation populated in ecRNH, and the β5/αE loop is more restricted in ttRNH than in ecRNH.
Although NMR experiments and MD simulations reported herein were conducted in the absence of catalytically required divalent metal ions, independent NMR spin-relaxation data for the side-chain carboxylates of the catalytic Asp and Glu residues in apo ecRNH suggest that the active site is rigid and pre-organized for binding the first Mg2+ ion [39]. Furthermore, active-site dynamics are conserved among RNases H [40] and their positions are conserved in crystal structures of the endonuclease members of the broader nuclease family [67]. The active site is therefore likely to be relatively insensitive to changes in the conformational dynamics of the substrate-binding loops on the picosecond-to-nanosecond timescale.
Interpretation of NMR spin-relaxation data through MD simulations provides a structure-based explanation for activity differences between mesophilic and thermophilic RNase H enzymes. Differences in the underlying conformational ensembles were correlated to function through structural comparison to holo hsRNH crystal structures for three flexible regions implicated in substrate binding and/or product release: the glycine-rich region, the handle region, and the β5/αE loop. Each of these regions is spatially close to sites of mutation in ttRNH that positively affect activity and that are shown in simulation to reconfigure their local environments to more closely resemble active conformations. Taken together, these findings suggest that differences in the conformational ensembles of dynamic regions not reflected in the corresponding crystal structures contribute to the difference in activity between ecRNH and ttRNH at mesophilic temperatures.
Methods
NMR spectroscopy
[U-2H, U-15N]iG80b ecRNH was prepared as described elsewhere [16]. NMR data were recorded for a sample of 1.0 mM [pH 5.5; 100 mM d3-sodium acetate at pH 5.5, 10% (v/v) deuterium oxide, 0.02% (w/v) sodium azide, and 1 mM d10-dithiothreitol] at 300 K on a Bruker DRX500 NMR spectrometer. 15N R1, 15N R2, and 1H-15N nuclear Overhauser enhancement values were measured using standard pulse sequences [68], essentially as described for the WT ecRNH protein [15]. Relaxation parameters were analyzed using FAST-Modelfree [69] and ModelFree [20].
MD simulations
Hydrogens were added to the crystal structures of WT ecRNH (1.5 Å resolution, PDB code 2RN2) [70], ttRNH (2.8 Å resolution, PDB code 1RIL) [56], iG80b ecRNH (1.9 Å resolution, PDB code 1GOA) [30], and a holo hsRNH crystal structure with the substrate removed (2.55 Å resolution, PDB code 2QK9) [14] using the Maestro program [71]. The hsRNH sequence incorporates a D210N inactivating mutation. Protonation states of ionizable groups in WT ecRNH under experimental conditions (pH 5.5) [19] were assigned based on previous NMR studies [72,73]. Protonation states of ionizable groups in ttRNH and iG80b ecRNH under experimental conditions (pH 5.5) [15] were assigned by estimating pKa values using the H++ Web server‡, indicating protonation of D10, protonation of H114 at the Nε2 position, protonation of H29 and H127 at the Nδ1 position, and double protonation of H31, H72, H119, and H124 in ttRNH; protonation of D10, protonation of H83 and H114 at the Nε2 position, and double protonation of H62, H124, and H127 in iG80b ecRNH; and protonation of D10, protonation of H31 at the Nε2 position, and double protonation of the N-terminal histidine residue, H50, H120, and H124 in hsRNH. H++ predictions for WT ecRNH agree well with the experimental data. Although the experimental pH optimum for the RNase H reaction is 7.5–8.5 [25], previous simulations of protonation states consistent with higher pH showed only small changes in dynamics, particularly of the active site [34,39]. The protonated structures were solvated in cubic boxes of 8505, 8207, and 8809 TIP3P [74] water molecules, respectively, to accommodate a minimum water shell thickness of 1 nm. Seven, thirteen, and six chloride ions, respectively, were added to the systems to maintain electric neutrality. The proteins were described by the AMBER ff99SB force field [75]. Each system was energy minimized and subsequently equilibrated to 300 K over 5 ns of NPT simulation with Desmond Academic Release 3 [76] using periodic boundary conditions and a cutoff of 0.9 nm for both particle mesh Ewald [77–79] and Lennard–Jones interactions. The SHAKE [80] algorithm was applied for constraining vibrations of bonds involving hydrogen atoms. A RESPA integrator was used [81] with a time step of 2.5 fs for bonded and short-range non-bonded interactions, as well as 7.5 fs for long-range electrostatics. For each protein, a structure with a box volume close to the average box volume over the last 4 ns of NPT equilibration was extracted from the last 1 ns of equilibration (volume and temperature reached their equilibrium values in less than 100 ps in all simulations). This structure was used as the starting structure for a 100-ns constant volume and constant temperature (NVT) production run, using identical simulation parameters. Coordinate sets were saved every 4.5 ps in the production runs. Activating mutations and other point mutants were introduced using MODELLER 9v5 [82] prior to protonation-state prediction using H++. No mutation caused changes in the predicted protonation states of titratable residues. One trajectory was calculated for the double mutant ttRNH A12S/K75M; two independent trajectories were calculated from different initial models of the triple mutant ttRNH A12S/K75M/A77P due to the relative difficulty of introducing a proline into a helix. All mutant simulations were performed identically, except for the use of Desmond 2.4.2.1 in place of Academic Release 3.
Calculation of order parameters
Simulated order parameters were calculated from
| (1) |
in which μ1, μ2, and μ3 are the x, y, and z components of the relevant bond vector scaled to unit magnitude, μ, respectively [83]. The experimental values were scaled by the respective slope of a linear regression against the simulated values for visual representation (0.91 for ecRNH, 0.92 for ttRNH, and 1.08 for iG80b ecRNH.).
Angular brackets in Eq. (1) indicate averaging over the snapshots in a given analysis block, after superposition of backbone heavy atoms to remove the effects of overall tumbling. The block length was chosen to mimic the timescale limitation of NMR spin relaxation [84], yielding ten 10-ns blocks because ecRNH has an overall tumbling time of 9.7 ns. To assess the separability of internal and overall protein motion, the rotationally invariant iRED approach [85] was also used to calculate order parameters from the appropriate blocks for the WT ecRNH and ttRNH simulations. The results obtained from the two methods agree within errors (data not shown).
Acknowledgments
This work was supported by the National Institutes of Health grant GM50291 (A.G.P.), a National Science Foundation predoctoral fellowship (K.A.S.), and a Boehringer Ingelheim Fonds Ph.D. scholarship (N.T.). We thank Columbia University's Center for Computational Biology and Bioinformatics for computational resources. We thank Dr. Paul Robustelli (Columbia University) for helpful discussions.
Abbreviations used
- RNase H
ribonuclease HI
- ecRNH
E. coli RNase H
- ttRNH
T. thermophilus RNase H
- hsRNH
H. sapiens RNase H
- MD
molecular dynamics
- WT
wild type
- PCA
principal components analysis
Footnotes
References
- 1.Hostomsky Z, Hostomska Z, Mathews DA. Ribonucleases H. In: Linn SM, Roberts RJ, editors. Nucleases. 2nd edit. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 341–376. [Google Scholar]
- 2.de Massy B, Fayet O, Kogoma T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. J Mol Biol. 1984;178:227–236. doi: 10.1016/0022-2836(84)90141-4. [DOI] [PubMed] [Google Scholar]
- 3.von Meyenburg K, Boye E, Skarstad K, Koppes L, Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K-12. J Bacteriol. 1987;169:2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horiuchi T, Maki H, Sekiguchi M. RNase H-defective mutants of Escherichia coli: a possible discriminatory role of RNase H in initiation of DNA replication. Mol Gen Genet. 1984;195:17–22. doi: 10.1007/BF00332717. [DOI] [PubMed] [Google Scholar]
- 5.Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J Mol Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa T, Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193:231–237. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- 7.Inouye M, Shimamoto T, Shimada M, Inouye S. Role of ribonuclease H in the biosynthesis of msDNA in Escherichia coli. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. Paris: INSERM; 1998. pp. 67–78. [Google Scholar]
- 8.Lima TM, Lim D. Isolation and characterization of host mutants defective in msDNA synthesis: role of ribonuclease H in msDNA synthesis. Plasmid. 1995;33:235–238. doi: 10.1006/plas.1995.1026. [DOI] [PubMed] [Google Scholar]
- 9.Shimamoto T, Shimada M, Inouye M, Inouye S. The role of ribonuclease H in multicopy single-stranded DNA synthesis in retron-Ec73 and retron-Ec107 of Escherichia coli. J Bacteriol. 1995;177:264–267. doi: 10.1128/jb.177.1.264-267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, Koh KD, Weiss B, Storici F. Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nat Struct Mol Biol. 2012;19:98–104. doi: 10.1038/nsmb.2176. [DOI] [PubMed] [Google Scholar]
- 11.Skalka AM, Goff SP, editors. Reverse transcriptase. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [PubMed] [Google Scholar]
- 12.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Steitz TA. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 14.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell. 2007;28:264–276. doi: 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Butterwick JA, Loria JP, Astrof NS, Kroenke CD, Cole R, Rance M, et al. Multiple time scale backbone dynamics of homologous thermophilic and mesophilic ribonuclease HI enzymes. J Mol Biol. 2004;339:855–871. doi: 10.1016/j.jmb.2004.03.055. [DOI] [PubMed] [Google Scholar]
- 16.Butterwick JA, Palmer AG. An inserted Gly residue fine tunes dynamics between mesophilic and thermophilic ribonucleases H. Protein Sci. 2006;15:2697–2707. doi: 10.1110/ps.062398606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanaya S. Enzymic activity and protein stability of E. coli ribonuclease HI. In: Crouch RJ, Toulmé JJ, editors. Ribonucleases H. Paris: INSERM; 1998. pp. 1–38. [Google Scholar]
- 18.Kroenke CD, Loria JP, Lee LK, Rance M, Palmer AG. Longitudinal and transverse 1H-15N dipolar/15N chemical shift anisotropy relaxation interference: unambiguous determination of rotational diffusion tensors and chemical exchange effects in biological macromolecules. J Am Chem Soc. 1998;120:7905–7915. [Google Scholar]
- 19.Kroenke CD, Rance M, Palmer AG. Variability of the 15N chemical shift anisotropy in Escherichia coli ribonuclease H in solution. J Am Chem Soc. 1999;121:10119–10125. [Google Scholar]
- 20.Mandel AM, Akke M, Palmer AG. Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 21.Mandel AM, Akke M, Palmer AG. Dynamics of ribonuclease H: temperature dependence of motions on multiple time scales. Biochemistry. 1996;35:16009–16023. doi: 10.1021/bi962089k. [DOI] [PubMed] [Google Scholar]
- 22.Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- 23.Hollien J, Marqusee S. Structural distribution of stability in a thermophilic enzyme. Proc Natl Acad Sci USA. 1999;96:13674–13678. doi: 10.1073/pnas.96.24.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollien J, Marqusee S. A thermodynamic comparison of mesophilic and thermophilic ribonucleases H. Biochemistry. 1999;38:3831–3836. doi: 10.1021/bi982684h. [DOI] [PubMed] [Google Scholar]
- 25.Kanaya S, Itaya M. Expression, purification, and characterization of a recombinant ribonuclease H from Thermus thermophilus HB8. J Biol Chem. 1992;267:10184–10192. [PubMed] [Google Scholar]
- 26.Akasako A, Haruki M, Oobatake M, Kanaya S. Conformational stabilities of Escherichia coli RNase HI variants with a series of amino acid substitutions at a cavity within the hydrophobic core. J Biol Chem. 1997;272:18686–18693. doi: 10.1074/jbc.272.30.18686. [DOI] [PubMed] [Google Scholar]
- 27.Haruki M, Noguchi E, Akasako A, Oobatake M, Itaya M, Kanaya S. A novel strategy for stabilization of Escherichia coli ribonuclease HI involving a screen for an intragenic suppressor of carboxyl-terminal deletions. J Biol Chem. 1994;269:26904–26911. [PubMed] [Google Scholar]
- 28.Ishikawa K, Kimura S, Kanaya S, Morikawa K, Nakamura H. Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability. Protein Eng. 1993;6:85–91. doi: 10.1093/protein/6.1.85. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa K, Nakamura H, Morikawa K, Kanaya S. Stabilization of Escherichia coli ribonuclease HI by cavity-filling mutations within a hydrophobic core. Biochemistry. 1993;32:6171–6178. [PubMed] [Google Scholar]
- 30.Ishikawa K, Nakamura H, Morikawa K, Kimura S, Kanaya S. Cooperative stabilization of Escherichia coli ribonuclease HI by insertion of Gly-80b and Gly-77 → Ala substitution. Biochemistry. 1993;32:7136–7142. doi: 10.1021/bi00079a010. [DOI] [PubMed] [Google Scholar]
- 31.Kimura S, Kanaya S, Nakamura H. Thermostabilization of Escherichia coli ribonuclease HI by replacing left-handed helical Lys95 with Gly or Asn. J Biol Chem. 1992;267:22014–22017. [PubMed] [Google Scholar]
- 32.Kimura S, Nakamura H, Hashimoto T, Oobatake M, Kanaya S. Stabilization of Escherichia coli ribonuclease HI by strategic replacement of amino acid residues with those from the thermophilic counterpart. J Biol Chem. 1992;267:21535–21542. [PubMed] [Google Scholar]
- 33.Kimura S, Oda Y, Nakai T, Katayanagi K, Kitakuni E, Nakai C, et al. Effect of cavity-modulating mutations on the stability of Escherichia coli ribonuclease HI. Eur J Biochem. 1992;206:337–343. doi: 10.1111/j.1432-1033.1992.tb16932.x. [DOI] [PubMed] [Google Scholar]
- 34.Stafford KA, Robustelli P, Friesner RA, Palmer AG. Thermal adaptation of conformational dynamics in ribonuclease H. PLoS Comput Biol. 2013;9:e1003218. doi: 10.1371/journal.pcbi.1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano N, Haruki M, Morikawa M, Kanaya S. Enhancement of the enzymatic activity of ribonuclease HI from Thermus thermophilus HB8 with a suppressor mutation method. Biochemistry. 2000;39:13285–13294. doi: 10.1021/bi0015008. [DOI] [PubMed] [Google Scholar]
- 36.Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- 37.Huang H-W, Cowan JA. Metallobiochemistry of the magnesium ion. Eur J Biochem. 1994;219:253–260. doi: 10.1111/j.1432-1033.1994.tb19936.x. [DOI] [PubMed] [Google Scholar]
- 38.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Stafford KA, Ferrage F, Cho JH, Palmer AG. Side chain dynamics of carboxyl and carbonyl groups in the catalytic function of Escherichia coli ribonuclease H. J Am Chem Soc. 2013;135:18024–18027. doi: 10.1021/ja409479y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stafford KA, Palmer AG. Evidence from molecular dynamics simulations of conformational preorganization in the ribonuclease H active site. F1000 Research. 2014;3:67. doi: 10.12688/f1000research.3605.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanaya S, Katsuda-Nakai C, Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J Biol Chem. 1991;266:11621–11627. [PubMed] [Google Scholar]
- 42.Nakamura H, Oda Y, Iwai S, Inoue H, Ohtsuka E, Kanaya S, et al. How does RNase H recognize a DNA-RNA hybrid? Proc Natl Acad Sci USA. 1991;88:11535–11539. doi: 10.1073/pnas.88.24.11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oda Y, Iwai S, Ohtsuka E, Ishikawa M, Ikehara M, Nakamura H. Binding of nucleic acids to E. coli RNase HI observed by NMR and CD spectroscopy. Nucleic Acids Res. 1993;21:4690–4695. doi: 10.1093/nar/21.20.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keck JL, Goedken ER, Marqusee S. Activation/attenuation model for RNase H. A one-metal mechanism with second-metal inhibition. J Biol Chem. 1998;273:34128–34133. doi: 10.1074/jbc.273.51.34128. [DOI] [PubMed] [Google Scholar]
- 45.D'Amico S, Marx J-C, Gerday C, Feller G. Activity–stability relationships in extremophilic enzymes. J Biol Chem. 2003;278:7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 46.Zavodszky P, Kardos J, Svingor A, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA. 1998;95:7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fitter J, Herrmann R, Dencher NA, Blume A, Hauss T. Activity and stability of a thermostable α-amylase compared to its mesophilic homologue: mechanisms of thermal adaptation. Biochemistry. 2001;40:10723–10731. doi: 10.1021/bi010808b. [DOI] [PubMed] [Google Scholar]
- 48.Hernández G, LeMaster DM. Reduced temperature dependence of collective conformational opening in a hyperthermophile rubredoxin. Biochemistry. 2001;40:14384–14391. doi: 10.1021/bi0112560. [DOI] [PubMed] [Google Scholar]
- 49.Meinhold L, Clement D, Tehei M, Daniel R, Finney JL, Smith JC. Protein dynamics and stability: the distribution of atomic fluctuations in thermophilic and mesophilic dihydrofolate reductase derived using elastic incoherent neutron scattering. Biophys J. 2008;94:4812–4818. doi: 10.1529/biophysj.107.121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 51.Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, et al. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robustelli P, Stafford KA, Palmer AG. Interpreting protein structural dynamics from NMR chemical shifts. J Am Chem Soc. 2012;134:6365–6374. doi: 10.1021/ja300265w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Massi F, Wang C, Palmer AG. Solution NMR and computer simulation studies of active site loop motion in triosephosphate isomerase. Biochemistry. 2006;45:10787–10794. doi: 10.1021/bi060764c. [DOI] [PubMed] [Google Scholar]
- 54.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 55.Pontiggia F, Zen A, Micheletti C. Small- and large-scale conformational changes of adenylate kinase: a molecular dynamics study of the subdomain motion and mechanics. Biophys J. 2008;95:5901–5912. doi: 10.1529/biophysj.108.135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishikawa K, Okumura M, Katayanagi K, Kimura S, Kanaya S, Nakamura H, et al. Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 Å resolution. J Mol Biol. 1993;230:529–542. doi: 10.1006/jmbi.1993.1169. [DOI] [PubMed] [Google Scholar]
- 57.Tsunaka Y, Takano K, Matsumura H, Yamagata Y, Kanaya S. Identification of single Mn2+ binding sites required for activation of the mutant proteins of E. coli RNase HI at Glu48 and/or Asp134 by X-ray crystallography. J Mol Biol. 2005;345:1171–1183. doi: 10.1016/j.jmb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Bae E, Phillips GN. Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 59.Wintrode PL, Zhang D, Vaidehi N, Arnold FH, Goddard WA. Protein dynamics in a family of laboratory evolved thermophilic enzymes. J Mol Biol. 2003;327:745–757. doi: 10.1016/s0022-2836(03)00147-5. [DOI] [PubMed] [Google Scholar]
- 60.Niesen MJM, Bhattacharya S, Grisshammer R, Tate CG, Vaidehi N. Thermostabilization of the β1-adrenergic receptor correlates with increased entropy of the inactive state. J Phys Chem B. 2013;117:7283–7291. doi: 10.1021/jp403207c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic–mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 62.Toth K, Amyes TL, Wood BM, Chan KK, Gerlt JA, Richard JP. An examination of the relationship between active site loop size and thermodynamic activation parameters for orotidine 5′-monophosphate decarboxylase from mesophilic and thermophilic organisms. Biochemistry. 2009;48:8006–8013. doi: 10.1021/bi901064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu CT, Hanoian P, French JB, Pringle TH, Hammes-Schiffer S, Benkovic SJ. Functional significance of evolving protein sequence in dihydrofolate reductase from bacteria to humans. Proc Natl Acad Sci USA. 2013;110:10159–10164. doi: 10.1073/pnas.1307130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greives N, Zhou H-X. Both protein dynamics and ligand concentration can shift the binding mechanism between conformational selection and induced fit. Proc Natl Acad Sci USA. 2014;111:10197–10202. doi: 10.1073/pnas.1407545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clore GM. Interplay between conformational selection and induced fit in multidomain protein–ligand binding probed by paramagnetic relaxation enhancement. Biophys Chem. 2014;186:3–12. doi: 10.1016/j.bpc.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurisaki I, Takayanagi M, Nagaoka M. Combined mechanism of conformational selection and induced fit in U1A–RNA molecular recognition. Biochemistry. 2014;53:3646–3657. doi: 10.1021/bi401708q. [DOI] [PubMed] [Google Scholar]
- 67.Majorek KA, Dunin-Horkawicz S, Steczkiewicz K, Muszewska A, Nowotny M, Ginalski K, et al. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014;42:4160–4179. doi: 10.1093/nar/gkt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy. 2nd edit. San Diego, CA: Academic Press; 2007. [Google Scholar]
- 69.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 70.Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Nakamura H, et al. Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution. J Mol Biol. 1992;223:1029–1052. doi: 10.1016/0022-2836(92)90260-q. [DOI] [PubMed] [Google Scholar]
- 71.Banks JL, Beard HS, Cao YX, Cho AE, Damm W, Farid R, et al. Integrated modeling program, applied chemical theory (IMPACT) J Comput Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oda Y, Yamazaki T, Nagayama K, Kanaya S, Kuroda Y, Nakamura H. Individual ionization-constants of all the carboxyl groups in ribonuclease HI from Escherichia coli determined by NMR. Biochemistry. 1994;33:5275–5284. doi: 10.1021/bi00183a034. [DOI] [PubMed] [Google Scholar]
- 73.Yamazaki T, Yoshida M, Nagayama K. Complete assignments of magnetic resonances of ribonuclease-H from Escherichia coli by double-resonance and triple-resonance 2D and 3D NMR spectroscopies. Biochemistry. 1993;32:5656–5669. doi: 10.1021/bi00072a023. [DOI] [PubMed] [Google Scholar]
- 74.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 75.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (SC06) Tampa, FL: ACM Press; 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. [Google Scholar]
- 77.Cheatham TE, Miller JL, Fox T, Darden TA, Kollman PA. Molecular-dynamics simulations on solvated biomolecular systems—the particle mesh Ewald method leads to stable trajectories of DNA, RNA, and proteins. J Am Chem Soc. 1995;117:4193–4194. [Google Scholar]
- 78.Darden T, York D, Pedersen L. Particle mesh Ewald—an N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 79.York DM, Darden TA, Pedersen LG. The effect of long-range electrostatic interactions in simulations of macromolecular crystals—a comparison of the Ewald and truncated list methods. J Chem Phys. 1993;99:8345–8348. [Google Scholar]
- 80.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 81.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- 82.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 83.Chandrasekhar I, Clore GM, Szabo A, Gronenborn AM, Brooks BR. A 500-ps molecular-dynamics simulation study of interleukin-1-beta in water—correlation with nuclear-magnetic-resonance spectroscopy and crystallography. J Mol Biol. 1992;226:239–250. doi: 10.1016/0022-2836(92)90136-8. [DOI] [PubMed] [Google Scholar]
- 84.Maragakis P, Lindorff-Larsen K, Eastwood MP, Dror RO, Klepeis JL, Arkin IT, et al. Microsecond molecular dynamics simulation shows effect of slow loop dynamics on backbone amide order parameters of proteins. J Phys Chem B. 2008;112:6155–6158. doi: 10.1021/jp077018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prompers JJ, Brüschweiler R. General framework for studying the dynamics of folded and nonfolded proteins by NMR relaxation spectroscopy and MD simulation. J Am Chem Soc. 2002;124:4522–4534. doi: 10.1021/ja012750u. [DOI] [PubMed] [Google Scholar]