Abstract
Spinal muscular atrophy is a common autosomal recessive neuromuscular disorder caused by the homozygous loss of the SMN1 gene. The absence of the SMN1 gene has been shown to occur in all types of SMA, childhood and adult forms. In rare cases, asymptomatic family members have also been found with homozygous mutations in the SMN1 gene, suggesting a role for phenotypic modifiers. We describe three unrelated asymptomatic individuals, with family histories of SMA, who were shown to have the homozygous SMN1 deletion. Quantitative studies indicated that the three individuals all had increased SMN2 copy numbers. These cases not only support the role of SMN2 in modifying the phenotype, but our data also demonstrate that expression levels consistent with five copies of the SMN2 genes maybe enough to compensate for the absence of the SMN1 gene. Lastly, in cases similar to the ones described, the measurement of the SMN2 gene copy number may provide valuable prognostic information.
Keywords: neuromuscular disorder, spinal muscular atrophy, SMN, discordant phenotype
INTRODUCTION
The autosomal recessive disorder proximal spinal muscular atrophy (SMA) is a severe neuromuscular disease characterized by degeneration of alpha motor neurons in the spinal cord, which results in progressive proximal muscle weakness and paralysis. SMA is the second most common fatal autosomal recessive disorder after cystic fibrosis, with an estimated incidence of 1 in 10,000 live births [Pearn, 1978]. Childhood SMA is subdivided into three clinical groups on the basis of age of onset and clinical course: type I SMA (Werdnig–Hoffmann) is characterized by severe, generalized muscle weakness and hypotonia at birth or within the first 3 months. Death from respiratory failure usually occurs within the first 2 years. Type II children are able to sit, although they cannot stand or walk unaided, and survive beyond four years. Type III SMA (Kugelberg–Welander) is a milder form, with onset during infancy or youth: patients learn to walk unaided. Adult-onset SMA, variably referred to as Type IIIb or Type IV, while less frequent, has also been reported. However, the large majority of SMA patients have onset of symptoms in infancy or early childhood.
The survival motor neuron (SMN) gene comprises nine exons and has been shown to be the primary SMA determining gene [Lefebvre et al., 1995]. Two almost identical SMN genes are present on 5q13: the telomeric or SMN1 gene, which is the SMA-determining gene, and centromeric or SMN2 gene. The SMN1 gene exon 7 is absent in about 95% of affected patients, while small more subtle mutations have been identified in the remaining affected patients. Although mutations of the SMN1 gene are observed in the majority of patients, no phenotype– genotype correlation was initially observed because SMN1 exon 7 is absent in the majority of patients independent of the type of SMA. However, there have been several studies which have shown that the SMN2 copy number influences the severity of the disease [Campbell et al., 1997; McAndrew et al., 1997; Wirth et al., 1999; Feldkotter et al., 2002; Mailman et al., 2002]. The copy number varies from zero to three copies in the normal population, with approximately 10–15% of normals having no SMN2. However, milder patients with type II or III have been shown to have more copies of SMN2 than type I patients. It has been proposed that the extra SMN2 in the more mildly affected patients arise through gene conversions, whereby the SMN2 gene is copied either partially or totally into the telomeric locus [Burghes, 1997].
Five base pair changes exist between SMN1 and SMN2 transcripts, and none of these differences change amino acids. Since virtually all SMA individuals have at least one SMN2 gene copy, the question that arises is why do individuals with SMN1 mutations have a SMA phenotype? It has now been shown that the SMN1 gene produces predominately full-length transcript, whereas the SMN2 copy produces predominately an alternatively transcribed (exon 7 deleted) product [Lorson et al., 1999; Lorson and Androphy, 2000]. The inclusion of exon 7 in SMN1 transcripts and exclusion of this exon in SMN2 transcripts is caused by a single nucleotide differences at +6 in SMN exon 7. Although the C to T change in SMN2 exon 7 does not change an amino acid, it does disrupt an exonic splicing enhancer ESE which results in the majority of transcripts lacking exon 7 [Lorson and Androphy, 2000]. Therefore, SMA arises because the SMN2 gene cannot fully compensate for the lack of SMN1 expression when SMN1 is mutated. However the small amount of full length transcripts generated by SMN2 are able to often produce a milder type II or III phenotype when the copy number of the SMN2 gene is increased.
The absence of detectable SMN1 in SMA patients is being utilized as a powerful diagnostic test for SMA. However, there have been reports of the occurrence of unaffected individuals with the homozygous absence of the SMN1 gene [Brahe et al., 1995; Cobben et al., 1995; Hahnen et al., 1995; Wang et al., 1996]. Although homozygosity for SMN1 deletions in unaffected persons is most likely a rare event, the finding does suggest that the phenotype is modified by other factors. Compensating factors might include alterations in the SMN2 copy number, differences in the actual extent of the SMN1 deletion, or other modifying genes.
In 1996, we developed a competitive quantitative PCR assay, using alternative internal controls, for the identification of SMA carriers and for the measurement of the SMN2 gene copy number [McAndrew et al., 1997]. Over the last 7 years, our laboratory has performed 1,080 SMA carrier test and have identified 407 SMA carriers. Two unaffected and unrelated cases with SMN1 homozygous deletions were identified through our carrier screening program. A diagnostic prenatal case also revealed a homozygous deletion in a young asymptomatic sibling, which showed that siblings with the same SMN1 locus may have different levels of the SMN2 gene. In this report, we present the clinical and molecular findings of these three cases.
CLINICAL REPORTS
Case 1
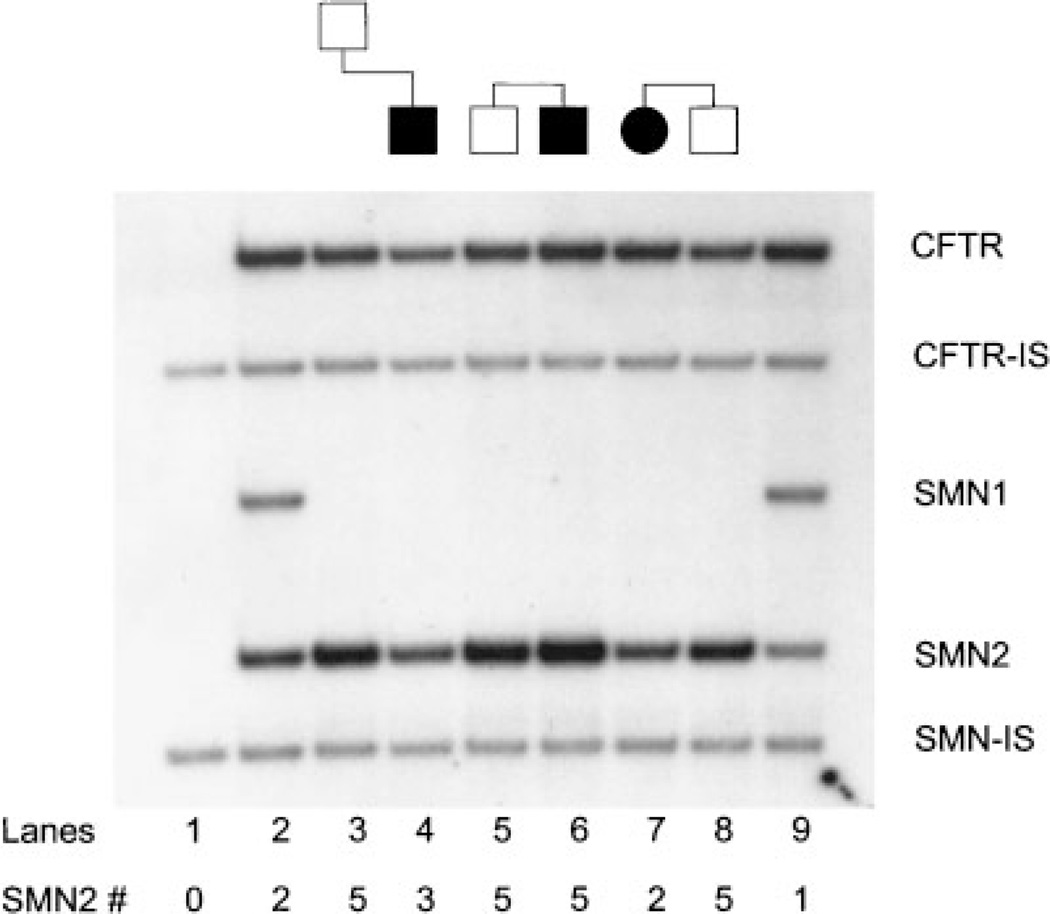
The affected is a SMA type 3 patient who was evaluated at 42 months of age as part of a natural history study. Prenatal, postnatal, and early developmental motor milestones were normal. He was able to walk unassisted by 13 months, but could never arise from a squatting position on his own. Maximum motor function relative to peers was achieved at approximately 12 months. Gowers’ sign in arising from the floor was noted at 14 months. Currently, the patient still walks independently, but with some hip girdle weakness. Compound motor action potential amplitude (CMAP) maximum was 5.92 mV (normal >5 mV). Motor unit number estimation (MUNE) value was 165 (normal >200) based on collection of 18 representative single motor unit potentials (SMUPs). DNA testing was performed on the patient and an absence of the SMN1 gene (exons 7 and 8) was found, confirming the diagnosis of type III SMA. SMN2 copy number was determined and indicated three copies of the SMN2 gene (Fig. 1, lane 4). Carrier testing was performed on the 35-year-old asymptomatic father and a homozygous deletion of the SMN1 gene was present. Quantitative testing revealed five SMN2 copies in the father (Fig. 1, lane 3). The father shows no clinical weakness on thorough neurological examination, with absent tongue fasiculation, normal deep tendon reflexes, and normal lean/fat mass and bone density on DEXA scan. He has an active lifestyle which includes running 3 to 6 miles regularly several times per week and he is extremely well muscled and in excellent physical health. CMAP maximum was 18.5 mV (normal >5 mV). MUNE value was 228, which is normal for his age, based on collection of 15 representative SMUPs.
Fig. 1.
Molecular analysis of the SMN gene. The protocol is described in detail in McAndrew et al. [1997]. SMN2 copy number is determined by densitometry using the SMN2/CFTR dosage ratio, which accounts for differences in the amount of DNA in each lane. The use of two internal standards (SMN-IS and CFTR-IS) allows one to monitor the efficiency of the PCR reactions. Lane 1: blank consisting of only the two internal standards. Lane 2: carrier control with 1 SMN1 and two SMN2 copies. Lanes 3 and 4: case 1. Lanes 5 and 6: case 2. Lanes 7 and 8: case 3. Lane 9: carrier control with one SMN1 and one SMN2.
Case 2
The forty-year-old individual came to the clinic in order to be tested for SMA carrier status, since he has an affected brother with a mild SMA type III phenotype. He has experienced no neurological problems and no muscle weakness. He is compulsive about his physical fitness and regularly does weight training and runs 5 miles every other day. On physical exam; he is well developed, of normal weight, and has no evidence of muscle weakness, atrophy, or any other neurological abnormalities. Carrier testing indicated a homozygous deletion of SMN1 and quantitative testing for SMN2 revealed five copies (Fig. 1, lane 5). The affected brother was diagnosed at the age of 14, when he first exhibited muscle weakness and cramping. He was very active in sports and had been playing football, baseball and tennis. He is now 42 years old, still ambulant, and uses a cane mainly for balance. Molecular studies showed the homozygous SMN1 deletion and five copies of the SMN2 gene were also detected (Fig. 1, lane 6).
Case 3
The affected is a 3½-year-old female patient with SMA type 1. Pregnancy and delivery were unremarkable. Hypotonia and diminished leg movement were apparent by 2 months of age. By 4 months old, she was quadriparetic, with absent head control and limited antigravity upper extremity movement. She has been g-tube fed since 6 months of age, and has been dependent on noninvasive respiratory support including nocturnal BiPAP and cough inexsufflator and other respiratory therapies on a daily basis since about 12 months of age. She has significant facial diplegia and antigravity limb movement is no longer possible. CMAP maximum was 0.24 mV (normal >5 mV). Molecular testing at 4 months of age revealed a SMN1 homozygous deletion and two copies of SMN2 (Fig. 1, lane 7). The patient’s brother was diagnosed prenatally to have a homozygous SMN1 deletion. The parents elected to continue with the pregnancy. SMN2 quantitation was then performed postnally and five copies of SMN2 were identified (Fig. 1, lane 8). He remains completely asymptomatic at 6 months of age. He has normal tone and head control, no evident muscle weakness, no tongue or muscle fasiculations, and is able to roll and sit independently on his own. His phenotype is in marked contrast to his sibling’s clinical picture at that age. Routine electromyography and nerve conduction testing demonstrates normal compound motor action potential amplitudes for age (6.1 mV), and excellent recruitment of normal appearing motor units.
DISCUSSION
Although the absence of the SMN1 gene is responsible for the majority of SMA cases, the clinical phenotype is quite variable ranging from the severe type I to the milder type III. This report also confirms the rare occurrence of asymptomatic persons with SMN1 homozygous deletions. From a total of 1,081 carrier test performed in our molecular pathology laboratory, 408 asymptomatic carriers have been identified and two of the carriers (in cases 1 and 2) were shown to have SMN1 homozygous deletions. One of the individuals was the parent of a type III patient and the other individual was the brother of a type III patient. A diagnostic test performed prenatally on the brother of an SMA type I child (case III) also revealed an absence of SMN1, yet he remains asymptomatic at six months of age. The homozygous deletion has not been reported in individuals without family histories of SMA. In our experience, the homozygous deletion has not been observed in greater than 500 normal control individuals with no history of SMA.
In order to elucidate the role of potential phenotypic modifiers, we determined the SMN2 gene copy number and all of the unaffected individuals were shown to have five copies of the SMN2 gene. The existence of these rare asymptomatic individuals lacking SMN1 and possessing five copies of SMN2 has only been observed in SMA families, and maybe the consequence of an already unstable loci prone to either unequal cross overs between the two genes or conversion events whereby SMN1 is replaced by multiple copies of SMN2. In our previous studies on type III patients; 76% have three, 24% have four, and only 1% have demonstrated five copies of the SMN2 gene [Mailman et al., 2002]. Our results in the nonaffected cases are therefore consistent with the previous genotype/phenotype associations established in SMA patients, again supporting the role of SMN2 in ameliorating the disease severity and providing prognostic information. These results are also in agreement with the mouse model, in which elevated SMN2 copies have been shown to rescue the disease phenotype [Hsieh-Li et al., 2000; Monani et al., 2000]. However, SMN2 cannot be the sole modifier, since we have observed rare type III patients with five copies. As was shown in case 2, both the mildly affected type III patient and the asymptomatic brother both demonstrated five SMN2 copies. Differences in splicing factors may allow more full-length expression from the SMN2 gene and account for some of the variability observed between discordant sibs [Hoffman et al., 2000]. Still it remains unclear whether these are always fully intact copies of the SMN2 gene in these patients. Due to the nature of the SMN loci, deletion breakpoints have been difficult to identify. If the SMN2 genes are truncated, they may not produce full length transcript and therefore be nonfunctional. Also, SMN2 was not increased in several of the unaffected individuals recently described by Helmken et al. [2003]. However in the families described by Helmken et al. [2003], all family members, regardless of the phenotype, were shown to have the same SMN2 copy number and less than five copies. In this report, we described sibs with homozygous SMN1 deletions with different SMN2 levels (case 3) and discordant phenotypes. It is possible for recombinational events to occur between the two genes and therefore family members with the same SMN1 locus may have different levels of SMN2. Therefore SMN2 differences may contribute to intrafamilial variability observed in some families. This is a potential limitation of the prenatal test, for it is often assumed that the fetus will have a similar SMA phenotype as the previously affected family member when both are shown to be positive for the SMN1 deletion. Furthermore, asymptomatic individuals with homozygous SMN1 deletions would be a source of false positives if a screening program for the diagnosis of SMA was to be developed.
Although these cases are rare, the clinical and counseling implications are significant. If an asymptomatic individual is still young and found to have an absence of the SMN1 gene, there still remains the possibility of late onset manifestations. This may be the situation in case III, since the unaffected sib is only 6 months of age. However, in these cases, the measurement of the SMN2 copy number may provide additional and valuable prognostic information. If the SMN2 copy number is higher in the unaffected family member (case 1 and 3), the disease may be milder or late onset and in some cases no manifestations may occur. It is also important to correctly distinguish the rare homozygous from the standard heterozygous SMN1 carriers, since there is an increased recurrence risk of future affected offspring from homozygous carriers and all of their offspring will be carriers. Lastly, the cases presented support the potentially therapeutic benefit in increasing the SMN2 gene expression in order to decrease the severity of the disorder. If the SMN2 expression can equal or exceed the five copy level, and can occur early in the diseased state, the increase maybe able to correct or partially compensate for the loss of SMN1 function.
Acknowledgments
Grant sponsor: Families of SMA, MDA of America; Grant sponsor: NIH; Grant number: NS41634.
REFERENCES
- Brahe C, Servidei S, Zappatp S, Ricci E, Tonali P, Neri G. Genetic homogeneity between childhood-onset and adult-onset autosomal recessive spinal muscular atrophy. Lancet. 1995;346:741–742. doi: 10.1016/s0140-6736(95)91507-9. [DOI] [PubMed] [Google Scholar]
- Burghes AHM. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Genomic variation and gene conversion in spinal muscular atrophy: Implications for disease process and clinical phenotype. Am J Hum Genet. 1997;61:40–50. doi: 10.1086/513886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobben JM, van der Steege G, Grootscholten P, de Visser M, Scheffer H, Buys CH. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet. 1995;57:805–808. [PMC free article] [PubMed] [Google Scholar]
- Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analysis of SMN1 and SMN2 based on real-time LightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, Wirth B. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: Evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- Helmken C, Hofmann Y, Schoenen F, Oprea G, Raschke H, Rudnik-Schoneborn S, Zerres K, Wirth B. Evidence for a modifying pathway in SMA discordant families: Reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat Genet. 2000;26:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Burghes AHM, Wirth B, Prior TW. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AHM. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Coovert DD, Burghes AHM. Animal models of spinal muscular atrophy. Hum Mol Genet. 2000;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Xu J, Carter TA, Ross BM, Dominski MK, Bellcross CA, Penchaszadeh GK, Munsat TL, Gilliam TC. Characterization of survival motor neuron (SMNT) gene deletions in asymptomatic carriers of spinal muscular atrophy. Hum Mol Genet. 1996;5:359–365. doi: 10.1093/hmg/5.3.359. [DOI] [PubMed] [Google Scholar]
- Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schoneborn S, Wienker T, Zerres K. Quantitative analysis of survival motor neuron copies: Identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype–phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64:1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]