Abstract
The Syrian golden hamster has been increasingly used to study viral hemorrhagic fever (VHF) pathogenesis and countermeasure efficacy. As VHFs are a global health concern, well-characterized animal models are essential for both the development of therapeutics and vaccines as well as for increasing our understanding of the molecular events that underlie viral pathogenesis. However, the paucity of reagents or platforms that are available for studying hamsters at a molecular level limits the ability to extract biological information from this important animal model. As such, there is a need to develop platforms/technologies for characterizing host responses of hamsters at a molecular level. To this end, we developed hamster-specific kinome peptide arrays to characterize the molecular host response of the Syrian golden hamster. After validating the functionality of the arrays using immune agonists of defined signaling mechanisms (lipopolysaccharide (LPS) and tumor necrosis factor (TNF)-α), we characterized the host response in a hamster model of VHF based on Pichinde virus (PICV1) infection by performing temporal kinome analysis of lung tissue. Our analysis revealed key roles for vascular endothelial growth factor (VEGF), interleukin (IL) responses, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, and Toll-like receptor (TLR) signaling in the response to PICV infection. These findings were validated through phosphorylation-specific Western blot analysis. Overall, we have demonstrated that hamster-specific kinome arrays are a robust tool for characterizing the species-specific molecular host response in a VHF model. Further, our results provide key insights into the hamster host response to PICV infection and will inform future studies with high-consequence VHF pathogens.
Viruses of the Arenaviridae family cause persistent and asymptomatic infections of rodents throughout the world (1). However, infection with select members of the Arenaviridae can result in viral hemorrhagic fever (VHF) in humans and non-human primates. Lassa virus (LASV), a Risk Group 4 Select Agent, is responsible for severe febrile illness in 15–20% of those infected (2, 3). While pathophysiological studies for LASV are limited to high-containment laboratories, Pichinde virus (PICV), a South American arenavirus, has been investigated as a potential surrogate for LASV and can be handled under biosafety level 2 conditions (4). Further, the histopathological effects of PICV infection of guinea pigs and hamsters are similar to those induced by LASV in humans (4–6). Thus, PICV is attractive as a surrogate virus for modeling the hemorrhagic disease process of LASV.
Recently, there has been interest in the use of the Syrian golden hamster as a model for high-consequence pathogens (7–9). In contrast to mice and guinea pigs, hamsters more closely recapitulate the pathological effects of VHF infections in non-human primates, particularly with regard to coagulopathy. It has been posited that VHF infection studies in hamsters will supplant those of mouse and guinea pigs (7–9). However, such investigations are limited by the lack of available hamster-specific molecular reagents (9). Thus, there is a great need for technologies that can characterize the molecular host response of hamsters.
Despite the potential threat to global health posed by high-consequence pathogens, there is limited information regarding the role of host cell signaling network dysregulation in the molecular pathogenesis of these pathogens. As the molecular events associated with disease are essential to understanding pathogenesis and identifying drug targets, it is important to examine the dynamic cellular changes that occur during infection. From the perspective of host cellular responses, phosphorylation of host proteins is the most well-characterized post-translational modification (PTM) with regard to cell signaling regulation, as well as the most ubiquitous PTM found within host cells (10). While gene expression data are informative, defining host responses at the level of host cell kinases (the kinome) provides a unique perspective regarding host-pathogen interactions. In particular, kinase-mediated phosphorylation events provide the host with a mechanism to rapidly respond to environmental changes (stress, infection, etc.) through the modulation of cell signaling networks as compared with more long-term changes in gene or protein expression. Further, there is increasing interest in the use of kinome analyses for the identification of novel therapeutic targets, including malaria and other parasitic diseases (11). Importantly, kinases are a primary target for the development of therapeutics with 25 kinase inhibitors currently licensed for the treatment of various malignancies by the U.S. Food and Drug Administration (12). Further, the investigation of kinase inhibitors as therapeutics for infectious disease fulfills a National Institutes of Health mandate focused on the repurposing of drugs (12, 13).
Recently, there has been an increased appreciation for the role of kinase-mediated signaling events in the host response to pathogens (14–17). Members of the Arenaviridae are known to modulate normal host immune signaling function (18, 19). As many immune responses are modulated through kinase-mediated signaling cascades, it is essential to examine the host response at this level. Previous investigations have attempted to characterize these responses; however, these investigations have been limited by the lack of availability of species-specific reagents and the relative scarcity of information regarding the host-specific molecular responses (18). Thus, there has been little investigation into the molecular response of hamster to high-consequence pathogens (5, 6).
Here, we present a novel analysis for characterizing hamster-specific kinome responses that overcome these challenges. Through our analysis, we have demonstrated the utility of hamster-specific kinome arrays for assessing species-specific host responses to innate immune agonists of well-defined signaling pathways, bacterial lipopolysaccharide (LPS) and tumor necrosis factor (TNF)-α. Moreover, we have characterized temporal host responses in the lungs of PICV-infected hamsters and demonstrated for the first time that vascular endothelial growth factor (VEGF), interleukin (IL) responses, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling, and Toll-like receptor (TLR) signaling form central components of the host response to PICV infection in the lungs of the Syrian golden hamster. Further, we also demonstrated that the modulation of VEGF- and angiogenic-related cell signaling processes correlated with modulation of downstream cell adhesion molecule expression. Importantly, these events were unique to late-stage disease in PICV-infected animals as compared with sham-infected animals. Taken together, our investigation provides a methodology for the global analysis of host responses of the Syrian golden hamster to pathogen insult and illustrates the potential to use data from this method to characterize the activity state of functional signaling networks useful for identifying therapeutic targets.
EXPERIMENTAL PROCEDURES
Cell and Virus Conditions
PICV, strain An 4763, was provided by Dr. David Gangemi (Clemson University, Clemson, SC). The virus was passaged once through hamsters and the stock (3.9 × 108 pfu/ml) was prepared from pooled clarified liver homogenates. Virus stock was diluted in sterile medium and inoculated by intraperitoneal injection of 0.2 ml. BHK-21 cells (CCL-10; ATCC, Manassas, VA) were maintained in EMEM (ATCC) supplemented with 10% FCS. VeroE6 cells (CRL-1586; ATCC) were maintained in DMEM (Invitrogen) and 10% FCS (v/v). All cultures were maintained at 37 °C in a humidified 5% (v/v) CO2 incubator.
Antibodies
VE-cadherin (Invitrogen), claudin-1 (Invitrogen, Grand Island, NY), ICAM-1 (R&D Systems, Minneapolis, MN) and VCAM-1 (R&D Systems) primary antibodies were used in this study for the detection of changes in protein expression in hamster lung tissue. β-tubulin (Cell Signaling, Danvers, MA) primary antibody was used as a control for protein loading. Antibody concentrations were based off of the manufacturer's recommendations.
Viral Titration
Plaque assays were performed on VeroE6 cells. Samples were diluted in DMEM and cells were inoculated for 1 h with periodic rocking followed by addition of a 0.8% Seakem ME agarose overlay (Lonza, Walkersville, MD) and cells were incubated for 6 days. Viral titers were determined following fixation with 10% neutral buffered formalin and staining with 2% crystal violet with 10% neutral buffered formalin (Sigma Aldrich, St. Louis, MO).
LPS and TNF-α Treatments
Baby hamster kidney (BHK)-21 cells were seeded for 100% confluency in six-well plates the day prior to treatment at a cell density of 1 × 106 cells/well. LPS (10 ng/ml) or murine TNF-α (10 ng/ml) were added to the cells for 4 h and 6 h, respectively. Cells were harvested and subject to kinome sample processing.
PICV Infection of Hamsters
Female (105–120 g) Syrian golden hamsters (Charles River, Willimantic, CT) were infected by intraperitoneal injection with 300 pfu of PICV, strain An 4763. Hamsters were euthanized in groups of three on days 1, 3, 5, and 7 postinfection. A baseline control group that did not receive a viral inoculum was euthanized at day 0. Lung tissue was collected immediately following euthanasia, flash frozen in liquid nitrogen, and stored at −80 °C until time of use. All hamster procedures used in this study complied with guidelines set by the USDA and Utah State University Animal Care and Use Committee, under IACUC protocol #2120. Hamster lung tissue was weighed and suspended in either EMEM or phosphoarray lysis buffer for 10 min [20 mm Tris-HCl, pH 7.5; 150 mm NaCl; 1 mm EDTA; 1 mm EGTA; 1% Triton-X100; 1X HALT protease/phosphatase inhibitor (Pierce, Rockford, IL)]. Samples were subsequently homogenized using the Qiagen TissueLyserII for two 3 min cycles using sterile beads and then pelleted by centrifugation at 21,000 × g for 20 min. After centrifugation, the supernatants were transferred to a fresh tube.
Selection of Hamster Kinome Array Peptides
The design of the hamster array was based on another array used by our group that contained peptides with perfect sequence conservation in the human and pig proteomes. The peptides on this array were drawn from a variety of biological pathways but with a concentration on pathways involved in immune responses. As this same focus was desired in this study, we created a hamster-specific version of this array. Specifically, each peptide on the human/pig array was used as a Basic Local Alignment Search Tool query against the hamster proteome. As the Syrian hamster genome has not yet been sequenced, we built a hamster proteome consisting of the ∼1000 Syrian hamster protein sequences that were available in the UniProt database, along with the ∼24,000 sequences available from the Chinese hamster. The sequence of the best Basic Local Alignment Search Tool match for each query peptide was used on the hamster array. Targets for our peptide kinome arrays were created as per previous kinome investigations by our group (14–16, 20, 21), and the arrays were printed by JPT Peptide Technologies (http://www.jpt.com). Briefly, designed peptides were synthesized using SPOT technology followed by immobilization to functionalized glass slides via a flexible linker through a proprietary technology. The peptide arrays were subjected to subsequent quality control testing by the manufacturer.
Kinome Sample Processing and Imaging
Samples for kinome analysis were suspended in 100 μl of phosphoarray lysis buffer for 10 min on ice. After incubation, samples were pelleted to remove cell debris at 12,000 × g for 10 min (cell culture samples) or 21,000 × g for 20 min (animal tissue samples) at 4 °C. A 10 μl aliquot of supernatant was then used to determine overall protein concentration (Pierce). The amount of protein to be spotted onto the peptide arrays was normalized, and the relevant dilutions were made in phosphoarray lysis buffer. Dilutions were then spotted onto the arrays for cell culture samples and animal tissue samples, respectively. 85 μl of supernatant and 13 μl of activation mix (50% glycerol, 50 μm ATP, 60 mm MgCl2, 0.05% Brij-35, 0.25 mg/ml BSA) were combined, and the 98 μl volume was spotted onto the arrays and incubated for 2 h at 37 °C. Following incubation, the array was washed in dH2O once and then submerged in PRO-Q Diamond Phosphoprotein Stain (Invitrogen) and gently rocked for 1 h. Arrays were subsequently destained (20% acetonitrile; 50 mm sodium acetate; pH 4) for 3 × 10 min, adding new destain each time. PRO-Q Diamond Phosphoprotein Stain is a proprietary fluorescent stain that detects phosphate groups attached to ser, thr, or tyr residues with excitation/emission maxima of ∼555/580 nm. After destain, arrays were washed with dH20 for 10 min and allowed to air dry for 10 min. To ensure the removal of all moisture, arrays were centrifuged at 300 rpm x 10 min. After drying, arrays were scanned on the TECAN Power Scanner (Morrisville, NC) to detect dye fluorescence. Images were collected using the GenePix 6.0 software (MDS, Sunnyvale, CA). Signal intensity values were measured by Array Pro (Media Cybernetics, Rockville, MD). Background values for each peptide on the arrays were subtracted from the foreground value to determine the phosphorylation states of each peptide. Peptides were printed in triplicate, and the averages of the net intensities were taken. The Platform for Integrated, Intelligent Kinome Analysis, which is a software pipeline tailored to analyze kinome array data, was used to calculate test statistics, probability values, and to perform clustering analysis of the data sets (22).
Pathway Over-Representation Analysis
InnateDB is a public bioinformatics database of the genes, gene products, and signaling pathways implicated in human, mouse, and bovine immune responses. The pathway over-representation analysis (ORA) feature can be used to identify pathways that are significantly up-regulated. Fold-change values greater than 1 and p values <.05 were considered for the ORA. In this study, the hamster kinome data sets (and the homologous human peptide target ID) were uploaded into InnateDB ORA and processed to identify over- or under-represented pathways.
Functional Network Analysis
Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood City, CA) is a bioinformatics software package used to elucidate pathways from -omics data. IPA is not limited to immune response alone; rather, it considers the “best-fit” of the kinome data sets into functional networks that have been previously well-characterized. Fold-change values greater than 1 and p values <.05 were considered for functional network analysis.
Western Blot Analysis
Hamster lung tissues were lysed in SDS loading buffer without bromphenol blue (200 mm Tris-HCl, pH 6.8; 8% SDS; 40% glycerol; 4% β-mercaptoethanol; 50 mm EDTA) and boiled for 20 min at 95 °C to inactivate remaining virus. Following inactivation, protein concentration was determined using the BCA Protein Assay kit (Pierce), according to the manufacturer's instructions. For phosphor-Western blot analysis, equal amounts of protein from lung tissue samples were loaded onto PathScan Intracellular Signaling Antibody Array membranes (Cell Signaling Technologies, Danvers, MA) and analyzed according to the manufacturer's instructions. Images were acquired using a Syngene G:Box Chemi (Syngene, Frederick, MD). For traditional Western blot analysis, equivalent amounts of total cell lysates were separated on 4–12% Bis-Tris gradient gels (Invitrogen) and wet transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). Membranes were blocked [5% (w/v) nonfat milk or 5% (w/v) BSA in TBS-Tween20 (0.5% w/v)] prior to incubation with primary antibodies and subsequent detection with appropriate horseradish peroxidase conjugated secondary antibodies (Pierce). Complexes were visualized using SuperSignal West Femto Chemiluminescent Substrate (Pierce). Images were acquired using a Syngene G:Box Chemi. Quantification of antibody spot intensities or of Western blot bands was determined through densitometry analysis using the ImageJ software suite (23).
RESULTS
Characterization of Hamster-Specific Kinome Responses to the Pro-Inflammatory Modulators LPS and TNF-α.
Previously, it was demonstrated that species-specific peptide kinome arrays could be rapidly created for species in which there is a paucity of phosphorylation site data but for which genome or proteome information is available (20). To this end, we created a hamster-specific peptide array for the characterization of host kinome responses. Human peptide sequences (query) were used to identify orthologous hamster peptide sequences (hit) (supplemental Table 1). Our arrays contained 282 unique peptides representing sequences in the hamster proteome with homology to human phosphorylation sites from a broad spectrum of cell signaling pathways and processes. As an initial test of the biological validity of our approach, we characterized the cellular response of hamsters to well-annotated agonists of pro-inflammatory and innate immune responses, namely LPS and TNF-α. For these experiments, BHK-21 cells were treated with LPS or TNF-α for 4 and 6 h, respectively, followed by harvesting of cells for kinome analysis. Kinome analysis with peptide arrays relies on the phosphorylation of specific kinase targets (peptides) on the array by active kinases in a cell lysate, allowing the subsequent identification and quantitation of these phosphorylation events. Following image analysis, the kinome array data sets were analyzed using Platform for Intelligent, Integrated Kinome Analysis 2 (16).
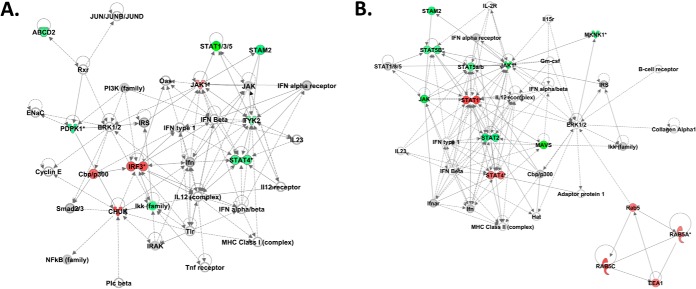
To provide biological insight into the specific cell signaling events that were modulated in the hamster cells, we performed functional network and pathway over-representation analysis. Functional network analysis of the kinome data from LPS-treated BHK-21 cells demonstrated that the top functional network modulated by LPS stimulation was related to antimicrobial response, inflammatory response, and cell death and survival (Fig. 1A). Importantly, our data corresponded well to characterized host response to LPS, including the modulation of NF-κB, TLR, and Akt signaling (24–27). Further, IL-12 occupied a central position in this functional network and the induction of IL-12 by LPS has been well characterized (28–31).
Fig. 1.
Characterization of cellular responses to LPS or TNF-α treatment validates the functionality of the hamster-specific kinome arrays. IPA generated functional network for the BHK-21 cellular response to known immune agonists LPS and murine TNF-α. A Top functional network for LPS-treated cells. Biological responses associated with this network include: cell death and survival, cell-mediated immune response, cellular development. B Top functional network for murine TNF-α treated cells. Biological responses associated with this network include: infectious disease, cellular assembly and organization, cell signaling.
As expected, pathway over-representation analysis of the LPS kinome data sets using the online software InnateDB (www.innateDB.com) demonstrated a strong up-regulation of cell signaling networks related to pro-inflammatory responses. These included up-regulation of TNF/stress-related signaling pathways, endogenous TLR signaling, IL-1 signaling, and modulation of I kappa B kinase (IKK)/ NF-κB signaling responses (Table I). Stimulation of BHK-21 cells with TNF-α resulted in a similar pattern of up-regulation of pro-inflammatory-related signaling networks (Fig. 1B). Analysis with InnateDB revealed the over-representation of pathways related to TNF and TLR signaling as well as the activation of c-Jun N-terminal kinase and Fas signaling, for which TNF-α is a known agonist (32, 33). In addition, activation of both NOD- and RIG-1-mediated signaling was noted following TNF-α treatment as has been described previously (Table I) (34, 35). Taken together, these results demonstrated the ability of the hamster-specific kinome arrays to accurately detail the activation state of hamster innate immune responses at the level of cell signaling networks.
Table I. Pathway over-representation analysis of kinome data from LPS and TNF-α treated BHK-21 cells.
| Pathway name | Uploaded proteins | Upregulated p value |
|---|---|---|
| Pro-inflammatory canonical pathways | ||
| TNF/stress related signaling | 6 | .04 |
| Endogenous TLR signaling | 7 | .05 |
| Fas (CD95) signaling pathway | 9 | .06 |
| NOD1/2 signaling pathway | 9 | .06 |
| IL-1 signaling | 10 | .07 |
| TNF receptor signaling pathway | 10 | .07 |
| Ceramide signaling pathway | 11 | .08 |
| IL-1 signaling (through IKK- NF-κB cascade)(canonical) | 11 | .08 |
| Signal transduction through IL-1 receptor | 11 | .08 |
| TNFR1 signaling pathway (TNFR1 signaling pathway) | 11 | .08 |
| TLR signaling pathway (through LPS, TLR4, MyD88, IRAK, TAK1 and IKK- NF-κB cascade) | 11 | .08 |
| IL1-mediated signaling events | 12 | .08 |
| TLR pathway | 12 | .08 |
| IL-1 signaling | 13 | .09 |
| NOD-like receptor signaling pathway | ||
| Alternative NF-κB pathway | 3 | .02 |
| Activation of NF-κB in B Cells | 4 | .03 |
| IKK complex recruitment mediated by RIP1 | 4 | .03 |
| IKK- NF-κB cascade (CD4 T cell receptor signaling) | 5 | .03 |
| Canonical NF-κB pathway | 6 | .04 |
| IKK- NF-κB cascade (TNFR1 signaling pathway) | 6 | .04 |
| IRAK1 recruits IKK complex | 6 | .04 |
| NF-κB signaling (TLR pathway (through NF-κB)) | 6 | .04 |
| TAK1 activates NF-κB by phosphorylation and activation of IKKs complex | 7 | .05 |
| RIP-mediated NF-κB activation via DAI | 8 | .06 |
| NF-κB signaling pathway | 11 | .08 |
| NF-κB activation by nontypeable hemophilus influenzae | 11 | .08 |
| Canonical signaling pathways | ||
| Fas signaling pathway | 7 | .00 |
| TGF-beta super family signaling pathway (canonical) | 10 | .02 |
| TNFR1 signaling pathway | 11 | .02 |
| NOD-like receptor signaling pathway | 13 | .04 |
| RIG-I-like receptor signaling pathway | 16 | .06 |
| TGF beta signaling pathway (through TAK1) | 9 | .10 |
| TGF-beta signaling pathway | 9 | .10 |
| TLR signaling pathway (through ECSIT, MEKK1, MKKs, JNK cascade) | 9 | .10 |
| JNK-mediated signaling | ||
| Activation of the AP-1 family of transcription factors | 3 | .01 |
| JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 | 3 | .01 |
| JNK cascade (TNFR1 signaling pathway) | 3 | .01 |
| JNK cascade (TGF-beta signaling(through TAK1)) | 4 | .02 |
| JNK cascade (CD4 T cell receptor signaling) | 5 | .03 |
| JNK cascade (TLR signaling pathway (through ECSIT, MEKK1, MKKs, JNK cascade)) | 5 | .03 |
| JNK cascade ( IL-1 signaling pathway (through JNK cascade)) | 6 | .05 |
| JNK cascade (TLR signaling pathway (through JNK cascade)) | 6 | .05 |
Infection of Syrian Hamsters with PICV
Given the success of the validation experiments described above, we next applied the hamster-specific kinome arrays to characterize the temporal host response to PICV infection in vivo in Syrian golden hamsters (Fig. 2A). Previous investigations regarding the molecular pathogenesis of PICV in hamster host tissues although previous investigations of PICV infection in hamsters have demonstrated that vascular permeability was significantly increased on day 7 post-infection in the lung tissue of PICV-infected hamsters (4–6). However, the molecular events underlying this phenomenon have not been characterized. Thus, we characterized the hamster host response in lung samples in naïve and PICV-infected hamsters (D0, D1, D3, D5, and D7 post-infection). Prior investigations have demonstrated that PICV-infection of hamsters results in terminal disease 7–9 days post-infection (Gowen references) that is characterized by the presence of virus in multiple organs (liver, kidney, spleen, and lung) by D2, decreased serum albumin levels beginning on D4, systemic PICV burden by D4–5, elevated levels of circulating white blood cells and modulation of serum cytokine levels (D6), the onset of vascular leakage and peak virus titers by D7, and the terminal stage of disease (D7-D9) (6). As prior investigations have demonstrated that the lungs are a target organ for PICV during infection in hamsters and that peak viral titers within the lungs correlate with the peak of vascular leakage, we focused on the characterization of kinome responses within the lungs of PICV-infected hamsters for our investigation. Animals in our study displayed general signs of late-stage disease (hunched posture, lethargy, etc.) on D7 as previously described. Quantifiable viral titers were found in the lungs of our infected hamsters beginning on day 3 (4.97 × 103 pfu/g) and steadily increased over the course of the infection and peaked at D7 at 8.02 × 105 pfu/g (Fig. 2B) (6).
Fig. 2.
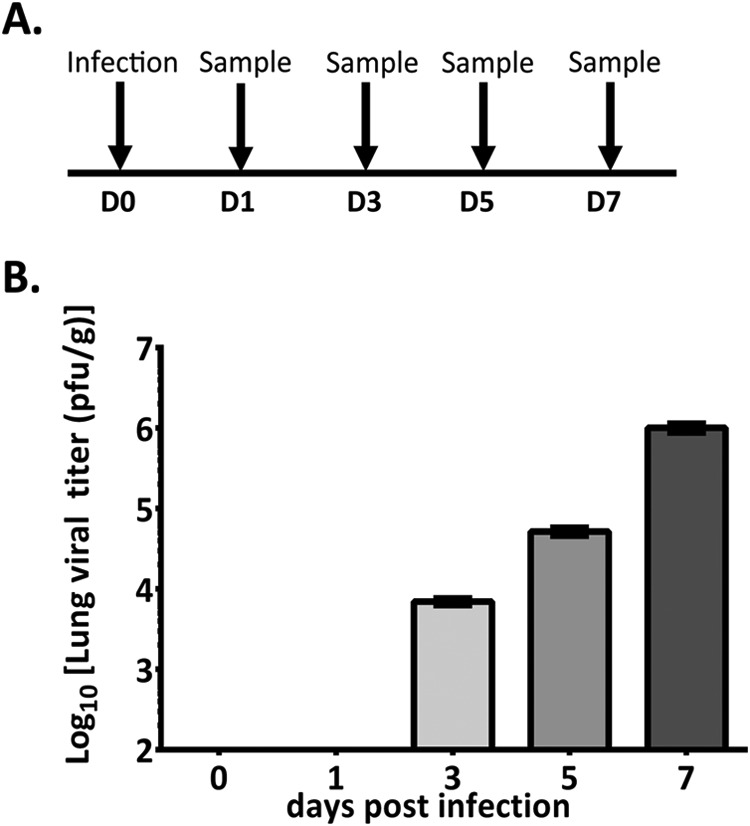
Study design for PICV infection of Syrian golden hamsters and lung viral titers. A At D0, three uninfected hamsters were euthanized to serve as baseline controls. The remaining hamsters were challenged with 300 PFU of PICV and euthanized in groups of three at D1, D3, D5, and D7. B Lung tissue was processed and titrated for PICV as described in Materials and Methods. Samples were titrated three times on three different days. Results are plotted as the average ± one standard deviation.
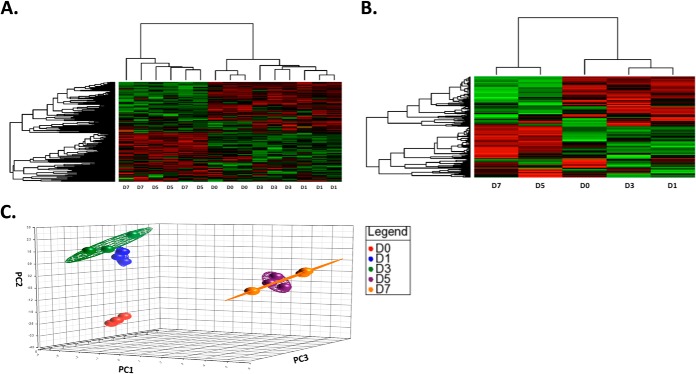
Hierarchal Clustering and Principal Component Analysis of Temporal Kinome Data from PICV-Infected Lung Samples
As the PICV infection progressed in the lungs, we observed differential phosphorylation of peptide targets compared with D0 animals. Hierarchal analysis of individual animals showed that the D5 and D7 kinome data sets clustered with each other but separately from the D0, D1, and D3 animals (Fig. 3A). Next, data from each group of three animals were averaged together to create combined data sets. A similar clustering pattern was observed between averaged data sets and for each individual animal (Fig. 3B). Principal component analysis demonstrated clear separation of the D0, D1/D3, and D5/D7 data sets (Fig. 3C).The distinct separation of D1/D3 from D5/D7 suggested that important signaling events might distinguish these time points and may be important to viral pathogenesis.
Fig. 3.
Hierarchal clustering and principal component analysis (PCA) of temporal kinome data from PICV-infected hamster lung tissue. A Hierarchal clustering analysis of kinome profile of individual PICV-infected hamster lungs. B Hierarchal clustering analysis of PICV-infected hamster groups based on day of euthanasia. The letter and number combination designate the day of euthanasia. The distance metric used was (1 − Pearson correlation), while McQuitty linkage was used as the linkage method. Rows correspond to probes (phosphorylation targets), and columns correspond to samples. Colors indicate the averaged (over three intra-array replicates) phosphorylation intensity of each target, with red indicating greater amounts of phosphorylation and green indicating lesser amounts of phosphorylation. The intensity of the color corresponds to the measured level of phosphorylation. C Three-dimensional principal component analysis of PICV-infected hamster data sets across the time course of the study.
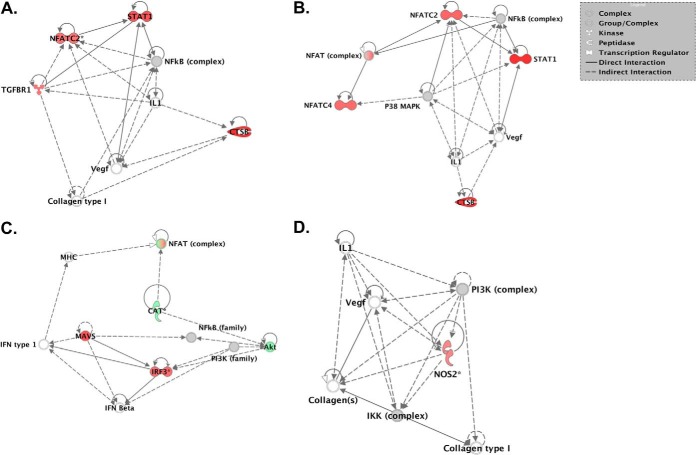
Fig. 4.
Representative core nodes of functional networks from temporal kinome data of PICV-infected hamster lung tissue. Kinome data sets were uploaded to IPA for functional network analysis. These nodes are derived from the full functional networks identified for each data comparison. A D1 versus D0; B D3 versus D0; C D5 versus D0; D D7 versus D0.
Functional Network Analysis of Temporal Kinome Data from PICV-Infected Lung Samples
To provide biological insight into our kinome analysis at the level of cellular responses, we performed systems analysis at the level of both functional networks and individual cell signaling pathways. In order to characterize the host response in a temporal manner, we compared the kinome data sets to those from the baseline lung samples (D0). We first examined our kinome data sets for associated biological responses at the level of functional networks. Biological responses related to infectious disease or immune-related responses formed central components of the functional networks at D1 and D3 (supplemental Fig. 1A and B) as compared with baseline whereas cell death and survival responses dominated the biological responses within the functional groups for the D5 and D7 data sets (supplemental Fig. 1C and D). Analysis of the individual nodes within the top functional networks for each group demonstrated that nodes related to VEGF-mediated cellular responses formed the cores of each network. In particular, nuclear factor of activated T cells (NFAT) or VEGF were found within the core of each network. Previously, it has been demonstrated that NFAT can regulate the expression of VEGF (36). In addition, nodes related to cell morphology (collagen) or extracellular matrix stability (cathepsin B) were also found within these networks. Interleukin-related nodes, in particular IL-1, were found within the D1, D3, and D7 functional networks but were absent at D5. These observations suggested that VEGF/NFAT-related cellular responses formed central components of the host response to PICV infection and may contribute to the associated disease pathology.
To gain greater insight into the particular cell signaling networks that were related to these responses, we next performed pathway ORA. The complete pathway analysis data demonstrated that the total number of up-regulated pathways was greater at D5 and D7 post-infection than at D1 and D3 post-infection (supplemental Table 2). These observations correlated with our PCA results as the D5 and D7 clusters were further separated in three-dimensional space from the D0 cluster as compared with the D1 and D3 data sets suggesting a greater degree of difference associated with the D5 and D7 data sets. We next assessed the over-representation of specific signaling pathways related to our functional network analysis throughout the course of infection (Table II). The full list of signaling pathways and associated p values identified from InnateDB are presented in supplemental Table 2. Our pathway analysis demonstrated that VEGF-mediated signaling responses were predicted to be up-regulated at all of the post-infection time points examined. These results suggested that VEGF and/or angiogenesis-related signaling pathways played a central role in the host response within the lungs throughout the course of PICV infection. In addition, our pathway analysis demonstrated that multiple signaling pathways related to angiogenic responses were up-regulated at D5 and D7 post-infection, including Tie2-, Rac1-, Wnt-, BCR-, and NFAT-mediated signaling responses. Interestingly, Tie2-mediated responses were predicted to be downregulated at the D1 time point suggesting that angiogenesis-related signaling responses may be differentially modulated during the early course of infection as compared with later time points. Also of note was the down-regulation of signaling pathways related to integrin signaling at D1. Further, our data also suggested that G-protein coupled receptors formed central components of signaling pathways modulated throughout all of the post-infection time points suggesting a critical role for G-protein coupled receptors in the response to PICV infection in hamster lung tissue (supplemental Table 2). Innate immune- and NF-κB-related signaling responses were largely found to be downregulated in our pathway analysis data and had also comprised central nodes in our functional networks. Overall, our data suggested that interleukin responses were largely downregulated. IL-6 and IL-7 signaling responses were downregulated at D1; IL-2 was predicted to be activated at D5 and then repressed at D7 along with IL-1-related responses. This is consistent with a previous investigation of PICV in hamsters that identified significantly lower levels of IL-1 in the serum at D7 post-infection (6). Pattern recognition receptor signaling pathways were also predicted to be downregulated at D7, including TLR and RIG/MD5 signaling responses as well as NF-κB-related signaling responses. The general down-regulation of immune signaling is consistent with previous investigations of PICV infection in guinea pigs (18).
Table II. Pathways identified to be related to vascular, cell adhesion and immune/inflammatory/stress processes as identified from temporal kinome data sets of PICV-infected hamsters.
| D1 vs D0 | D3 vs D0 | D5 vs D0 | D7 vs D0 | |
|---|---|---|---|---|
| VEGF/NFAT/Tie2-related pathways | ||||
| Up-regulated pathways | VEGF signaling pathway | VEGF signaling Pathway | NFAT and hypertrophy of the heart | VEGF signaling pathway |
| Role of calcineurin-dependent NFAT signaling in lymphocytes | Signaling events mediated by VEGFR1 and VEGFR2 | |||
| VEGF signaling pathway | ||||
| BCR | ||||
| Tie2 Signaling | ||||
| Angiopoietin receptor Tie2-mediated signaling | ||||
| Down-regulated pathways | Tie2 signaling | |||
| Angiopoietin receptor Tie2-mediated signaling | ||||
| Tie2 signaling | ||||
| Angiopoietin receptor Tie2-mediated signaling | ||||
| Cell junction/adhesion/motility-related pathways | ||||
| Up-regulated pathways | Wnt signaling | Rac1 cell motility signaling pathway | Rac1 cell motility signaling pathway | |
| Wnt signaling pathway | Leukocyte transendothelial migration | |||
| Down-regulated pathways | Focal adhesion | |||
| A6b1 and a6b4 integrin signaling | ||||
| Alpha6Beta4 Integrin | ||||
| Integrins in angiogenesis | ||||
| Immune/inflammatory/stress-related pathways | ||||
| Up-regulated pathways | Fmlp induced chemokine expression in hmc-1 cells | Fmlp induced chemokine gene expression in hmc-1 cells | Negative regulators of RIG-I/MDA5 signaling | |
| P38 mapk signaling pathway | IL2 signaling events mediated by PI3K | |||
| Interleukin-7 signaling | ||||
| Downregulated pathways | Interferon alpha/beta signaling | Interleukin-1 signaling | Interleukin-1 signaling | |
| TNFR1 signaling pathway | TNFR1 signaling pathway | |||
| FAS (CD95) signaling pathway | Toll-like receptor pathway | |||
| IRAK1 recruits IKK complex | IL-7 signaling | |||
| IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation | Il 2 signaling pathway | |||
| Fas signaling pathway | Interleukin-2 signaling | |||
| NF-κB signaling pathway | Signal transduction through il1r | |||
| CD4 T cell receptor signaling | NF-κB signaling pathway | |||
| CD40/CD40L signaling |
Phospho-Western blot array analysis was also performed to examine the phosphorylation state of various intracellular molecules that were found to be significantly modulated (p < .1) in our kinome data. Lung samples from each animal within the D0 through D7 groups were subjected to Western blot array analysis and spot intensity quantification. The fold-change values for phosphorylation events that were found to be significantly modulated in our kinome analysis comparatives (comparison of post-infection samples to those from D0 animals) were largely conserved between the kinome and Western blot array analyses (supplemental Table 3). Twenty-four proteins were selected for phosphor-Western analysis. Of the 24 selected protein phosphorylations examined by phosphor-Western blot, 19 of the phosphorylations were significantly up- or downregulated as compared with the D0 animals (p < 0.1). Of these, 18/19 were completely conserved between the kinome arrays and the phosphor-Western blot arrays in terms of the direction of the change in phosphorylation (upregulated or downregulated) as compared with the D0 samples (supplemental Table 3).
PICV Infection Results in the Modulation of Expression of Proteins Related to Cellular Adhesion or Vascular Permeability
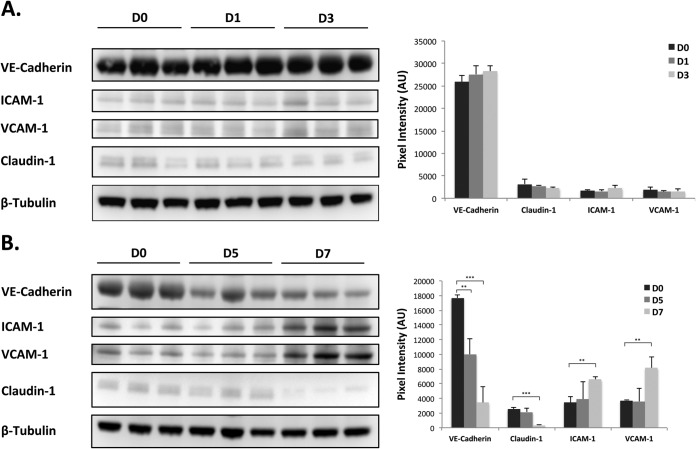
In order to provide insight into the modulation of VEGF and/or angiogenesis-related signaling responses as identified from our kinome analysis, we performed Western blot analysis and assessed the expression of various proteins related to cell adhesion or vascular permeability. Modulation of protein expression or cellular redistribution of claudin-1 and/or VE-cadherin have been associated with elevated levels of VEGF and hyperpermeability of hantavirus-infected endothelial cells and in the breakdown of paraendothelial barrier function in response to Marburg virus (37, 38). Similarly, we observed a significant temporal decrease in the levels of both claudin-1 and VE-cadherin as infection progressed, which correlated with the up-regulation of VEGF-mediated signaling networks (Fig. 5B) (37, 39). In addition, we also analyzed the expression of intracellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1). It has been demonstrated that the expression of ICAM-1 and VCAM-1 are induced during hantavirus infection of endothelial cells and Gowen et al. demonstrated increased levels of VCAM in the serum of PICV-infected hamsters at late time points during infection (6, 40). Our data were in good agreement with these observations as we found a significant increase in ICAM-1 and VCAM-1 when comparing the lung tissues from D7 to those from D0 (Fig. 5B). Taken together, these data provide biological confirmation for the up-regulation of VEGF and other angiogenic-related signaling in our pathway analysis and suggest a role for these responses in the pathogenesis of PICV in hamsters.
Fig. 5.
Temporal Western blot analysis of claudin-1, VE-cadherin, ICAM-1, and VCAM-1 expression. Lung samples were processed and Western blot analysis was performed as described in Materials and Methods. Spot intensities were quantified and normalized to the loading control. Data were averaged for each group (D0, D1, D3, D5, and D7) and the plotted pixel intensities represent the average intensity of the expressed marker for each animal from the respective group (n = 3 for each group). Standard deviations were calculated using the Student's t test.
DISCUSSION
The feasibility of performing species-specific investigations of host responses has been fostered by the increasing availability of sequence information from genome or proteome investigations for a broad range of animal species (16, 20). To this end, we created hamster kinome arrays that were comprised of kinase peptide targets that encompass a broad range of cell-related responses, including innate immunity. Hamsters are being increasingly employed for pathogenesis investigations for a diverse range of human diseases, including viral infections, prion disease, cardiomyopathy, pancreatic cancer, alcoholism, and sleep deprivation (41–46). Insight into the molecular events associated with these diseases may provide valuable information concerning pathogenesis and novel therapeutic targets or treatment strategies.
Phosphorylation events are often the first cellular modifications in response to cell stimulation or perturbation as nearly 1/3 of intracellular proteins are thought to be phosphorylated (10). Thus, information about changes in phosphorylation may provide key insights into viral pathogenesis and host response. We have previously utilized systems kinomics to identify differential host responses to Congo basin and West African monkeypox virus infection as well as cellular responses to Ebola virus (14, 15). Here, we have characterized important components of the host response to PICV infection in the Syrian golden hamster.
We have identified several signaling pathways and functional cellular responses important for hamster host response to PICV infection. Functional network analysis using IPA demonstrated that the lower viral titers found at D1 and D3 were associated with functional networks comprised of multiple interleukin and/or interferon family members. Further, IFN signaling was central in D1 to D5 animals but absent in D7. It is tempting to postulate that the increase in viral load modulated IFN signaling in the immune response, and this is reflected in the differential clustering and functional analysis of D1 and D3 versus D5 and D7 animals. The large down-regulation of interleukin-responses, NF-κB signaling, and TLR signals, especially at later time points, may also be indicative of PICV-mediated down-regulation of the host immune response. The later time points also correlate with the peak in viral titer, thus the down-regulation in IL- and IFN-mediated responses, as well as other innate immune signaling networks, could also be associated with cellular damage and apoptotic events either from the virus or host-mediated responses, as evident in the functional network analysis for D5 and D7 animals. These results may also be indicative of cellular responses to host-mediated tissue damage such as those associated with the infiltration of natural killer cells at the site of infection. Interestingly, natural killer cells are known to accumulate in the livers of mice infected with lymphocytic choriomeningitis virus, a member of the Arenaviridae (47). The up-regulation of the leukocyte transendothelial migration signaling pathway found in the analysis of the kinome data from the D5 animals further supports this idea. Interestingly, aldosterone-regulated sodium reabsorption was up-regulated in D7 animals. Given the natural history of disease in the hamster, this is also likely part of the molecular host response to the vascular leak characteristic of PICV infection in a hamster model (6).
Interestingly, our kinome analysis demonstrated that VEGF-mediated signaling responses were up-regulated in all infected animals and angiogenic responses were modulated at D5 and D7. These results are in agreement with a prior investigation of PICV infection in hamsters that demonstrated that both vascular permeability and serum concentrations of VEGF were increased in the late stage of infection (6). Here, we elucidated the specific signaling pathways that are responsible for this phenotype of vascular permeability late in infection. Further, we confirmed the involvement of these pathways by assessing the expression of molecules that are known to be downstream of VEGF and other angiogenic signaling activity (21, 36, 38, 48). Specifically, the expression of claudin-1 and VE-cadherin decreased as infection progressed, whereas the expression of VCAM-1 and ICAM-1 was upregulated late in infection when vascular permeability is known to occur (6). Interestingly, the down-regulation of claudin-1 and VE-cadherin is thought to be associated with the hyperpermeability of endothelial cells in Hantavirus infection (39). Additionally, the significant up-regulation of VCAM-1 in lung tissue is in agreement with a previous investigation of PICV in hamsters that identified increases in systemic VCAM-1 expression at D7 post infection (6). Further, the up-regulation of ICAM-1 has been associated with vascular permeability in the retina (48). Taken together, these data provide evidence for the modulation of specific molecules known to be associated with the maintenance of cell-cell adhesion in late-stage PICV infection.
As the recognition sites for kinases within humans and hamsters do not share 100% homology, it is important that molecular analyses such as kinome analysis incorporate species-specific targets. We have demonstrated that the fold-changes associated with phosphorylation events that met threshold significance criteria in our kinome analysis correlated with those found using phosphor-Western blot array analysis. The overall similarity between the kinome and Western blot results provide validity to our investigational approach and although small fluctuations in the overall fold-change were present, the specificity of phospho-specific human antibodies for hamster protein phosphorylations could contribute to fluctuations in this correlation, In addition, phosphor-specific antibody arrays provide a quantitative measure of phosphoproteins as compared with a relative measure of kinase activity, as with peptide kinome arrays and may therefore further contribute to differences in phosphorylation fold-changes between the two methodologies (49).
Here, we have confirmed several previously identified and identified several novel signaling pathways and functional host responses to PICV infection in the Syrian golden hamster. This, in combination with our initial validation of the novel hamster-specific kinome arrays, provides strong evidence for the effectiveness of this technology for characterizing host responses in the hamster in response to high-consequence pathogen insult. Insights gained from these analyses provide valuable information that will inform future studies on host response, viral pathogenesis, and drug targets for therapeutic intervention.
Supplementary Material
Acknowledgments
B. B. G is grateful to Jonna Westover (Utah State University) for facilitating the in vivo study.
Footnotes
Author contributions: S.F., B.B.G., R.F.J., V.W., P.B.J., and J.K. designed the research; S.F., B.B.G., B.T., S.N., A.K., V.W., and J.K. performed the research; B.T., S.N., A.K., D.S., J.P., and J.K. contributed new reagents or analytic tools; S.F., B.B.G., B.T., S.N., A.K., R.F.J., D.S., J.P., V.W., P.B.J., and J.K. analyzed data; S.F. and J.K. wrote the paper.
* This study was supported, in part, by the NIAID Division of Intramural Research. B. B. G. was supported by NIH grant U54 AI-065357 (Rocky Mountain RCE).
 This article contains supplemental Tables 1–3 and supplemental Fig. 1.
This article contains supplemental Tables 1–3 and supplemental Fig. 1.
1 The abbreviations used are:
- PICV
- Pichinde virus
- BHK-21
- baby hamster kidney
- ICAM-1
- intracellular adhesion molecule
- IL
- interleukin
- IPA
- Ingenuity Pathway Analysis
- JNK
- c-Jun N-terminal kinase
- LASV
- Lassa virus
- LPS
- lipopolysaccharide
- NF-κB
- nuclear factor kappa-light-chain-enhancer of activated B cells
- ORA
- over-representation analysis
- TLR
- Toll-like receptor
- TNF
- tumor necrosis factor
- VCAM-1
- vascular cell adhesion molecule 1
- VEGF
- vascular endothelial growth factor
- VHF
- viral hemorrhagic fever.
REFERENCES
- 1. Albariño C. G., Posik D. M., Ghiringhelli P. D., Lozano M. E., Romanowski V. (1998) Arenavirus phylogeny: a new insight. Virus Genes 16, 39–46 [DOI] [PubMed] [Google Scholar]
- 2. McCormick J. B. (1986) Clinical, epidemiologic, and therapeutic aspects of Lassa fever. Med. Microbiol. Immunol. 175, 153–155 [DOI] [PubMed] [Google Scholar]
- 3. Richmond J. K., Baglole D. J. (2003) Lassa fever: Epidemiology, clinical features, and social consequences. BMJ 327, 1271–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jahrling P. B., Hesse R. A., Rhoderick J. B., Elwell M. A., Moe J. B. (1981) Pathogenesis of a Pichinde virus strain adapted to produce lethal infections in guinea pigs. Infect. Immun. 32, 872–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lukashevich I. S. (2013) The search for animal models for Lassa fever vaccine development. Expert Rev. Vacc. 12, 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gowen B. B., Julander J. G., London N. R., Wong M. H., Larson D., Morrey J. D., Li D. Y., Bray M. (2010) Assessing changes in vascular permeability in a hamster model of viral hemorrhagic fever. Virol. J. 7, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ebihara H., Zivcec M., Gardner D., Falzarano D., LaCasse R., Rosenke R., Long D., Haddock E., Fischer E., Kawaoka Y., Feldmann H. (2013) A Syrian golden hamster model recapitulating Ebola hemorrhagic fever. J. Infect. Dis. 207, 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wahl-Jensen V., Bollinger L., Safronetz D., de Kok-Mercado F., Scott D. P., Ebihara H. (2012) Use of the Syrian hamster as a new model of Ebola virus disease and other viral hemorrhagic fevers. Viruses 4, 3754–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zivcec M., Safronetz D., Haddock E., Feldmann H., Ebihara H. (2011) Validation of assays to monitor immune responses in the Syrian golden hamster (Mesocricetus auratus). J. Immun. Meth. 368, 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson S. A., Hunter T. (2005) Kinomics: methods for deciphering the kinome. Nature Meth. 2, 17–25 [DOI] [PubMed] [Google Scholar]
- 11. Doerig C., Billker O., Pratt D., Endicott J. (2005) Protein kinases as targets for antimalarial intervention: Kinomics, structure-based design, transmission-blockade, and targeting host cell enzymes. Biochim. Biophys. Acta 1754, 132–150 [DOI] [PubMed] [Google Scholar]
- 12. Fabbro D., Cowan-Jacob S. W., Möbitz H., Martiny-Baron G. (2012) Targeting cancer with small-molecular-weight kinase inhibitors. Meth. Mol. Biol. 795, 1–34 [DOI] [PubMed] [Google Scholar]
- 13. Huang R., Southall N., Wang Y., Yasgar A., Shinn P., Jadhav A., Nguyen D. T., Austin C. P. (2011) The NCGC pharmaceutical collection: A comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Trans. Med. 3, 80ps16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kindrachuk J., Wahl-Jensen V., Safronetz D., Trost B., Hoenen T., Arsenault R., Feldmann F., Traynor D., Postnikova E., Kusalik A., Napper S., Blaney J. E., Feldmann H., Jahrling P. B. (2014) Ebola virus modulates transforming growth factor-beta signaling and cellular markers of mesenchymal-like transition in hepatocytes. J. Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kindrachuk J., Arsenault R., Kusalik A., Kindrachuk K. N., Trost B., Napper S., Jahrling P. B., Blaney J. E. (2012) Systems kinomics demonstrates Congo Basin monkeypox virus infection selectively modulates host cell signaling responses as compared to West African monkeypox virus. Mol. Cell Proteomics 11, M111.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arsenault R. J., Li Y., Maattanen P., Scruten E., Doig K., Potter A., Griebel P., Kusalik A., Napper S. (2013) Altered Toll-like receptor 9 signaling in mycobacterium avium subsp. paratuberculosis-infected bovine monocytes reveals potential therapeutic targets. Infect. Immun. 81, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kindrachuk J., Napper S. (2013) Probing the kinome for biomarkers and therapeutic targets: Peptide arrays for global phosphorylation-mediated signal transduction. In: Horvatovich P., Bischoff R, ed., Comprehensive biomarker discovery and validation for clinical application, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 18. Bowick G. C., Fennewald S. M., Scott E. P., Zhang L., Elsom B. L., Aronson J. F., Spratt H. M., Luxon B. A., Gorenstein D. G., Herzog N. K. (2007) Identification of differentially activated cell-signaling networks associated with Pichinde virus pathogenesis by using systems kinomics. J. Virol. 81, 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fennewald S. M., Aronson J. F., Zhang L., Herzog N. K. (2002) Alterations in NF-kappaB and RBP-Jkappa by arenavirus infection of macrophages in vitro and in vivo. J. Virol. 76, 1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jalal S., Arsenault R., Potter A. A., Babiuk L. A., Griebel P. J., Napper S. (2009) Genome to kinome: Species-specific peptide arrays for kinome analysis. Sci. Signal. 2, pl1. [DOI] [PubMed] [Google Scholar]
- 21. Kim I., Moon S. O., Kim S. H., Kim H. J., Koh Y. S., Koh G. Y. (2001) Vascular endothelial growth factor expression of intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin through nuclear factor-kappa B activation in endothelial cells. J. Biol. Chem. 276, 7614–7620 [DOI] [PubMed] [Google Scholar]
- 22. Trost B., Kindrachuk J., Määttänen P., Napper S., Kusalik A. (2013) PIIKA 2: An expanded, web-based platform for analysis of kinome microarray data. PloS ONE 8, e80837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nature Meth. 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grilli M., Chiu J. J., Lenardo M. J. (1993) NF-kappa B and Rel: Participants in a multiform transcriptional regulatory system. Int. Rev. Cytol. 143, 1–62 [DOI] [PubMed] [Google Scholar]
- 25. Guha M., Mackman N. (2002) The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J. Biol. Chem. 277, 32124–32132 [DOI] [PubMed] [Google Scholar]
- 26. Triantafilou M., Triantafilou K. (2002) Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol. 23, 301–304 [DOI] [PubMed] [Google Scholar]
- 27. Xie Q. W., Kashiwabara Y., Nathan C. (1994) Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J. Biol. Chem. 269, 4705–4708 [PubMed] [Google Scholar]
- 28. Delgado M., Munoz-Elias E. J., Gomariz R. P., Ganea D. (1999) VIP and PACAP inhibit IL-12 production in LPS-stimulated macrophages. Subsequent effect on IFNgamma synthesis by T cells. J. Neuroimmunol. 96, 167–181 [DOI] [PubMed] [Google Scholar]
- 29. Happel K. I., Dubin P. J., Zheng M., Ghilardi N., Lockhart C., Quinton L. J., Odden A. R., Shellito J. E., Bagby G. J., Nelson S., Kolls J. K. (2005) Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skokos D., Nussenzweig M. C. (2007) CD8- DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J. Exp. Med. 204, 1525–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Snijders A., Hilkens C. M., van der Pouw Kraan T. C., Engel M., Aarden L. A., Kapsenberg M. L. (1996) Regulation of bioactive IL-12 production in lipopolysaccharide-stimulated human monocytes is determined by the expression of the p35 subunit. J. Immunol. 156, 1207–1212 [PubMed] [Google Scholar]
- 32. Chen Y. R., Tan T. H. (1998) Inhibition of the c-Jun N-terminal kinase (JNK) signaling pathway by curcumin. Oncogene 17, 173–178 [DOI] [PubMed] [Google Scholar]
- 33. Wallach D., Varfolomeev E. E., Malinin N. L., Goltsev Y. V., Kovalenko A. V., Boldin M. P. (1999) Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17, 331–367 [DOI] [PubMed] [Google Scholar]
- 34. Takahashi Y., Isuzugawa K., Murase Y., Imai M., Yamamoto S., Iizuka M., Akira S., Bahr G. M., Momotani E., Hori M., Ozaki H., Imakawa K. (2006) Up-regulation of NOD1 and NOD2 through TLR4 and TNF-alpha in LPS-treated murine macrophages. J. Vet. Med. Sci./JSVS 68, 471–478 [DOI] [PubMed] [Google Scholar]
- 35. Matsumiya T., Prescott S. M., Stafforini D. M. (2007) IFN-epsilon mediates TNF-alpha-induced STAT1 phosphorylation and induction of retinoic acid-inducible gene-I in human cervical cancer cells. J. Immunol. 179, 4542–4549 [DOI] [PubMed] [Google Scholar]
- 36. Armesilla A. L., Lorenzo E., Gómez del Arco P., Martínez-Martínez S., Alfranca A., Redondo J. M. (1999) Vascular endothelial growth factor activates nuclear factor of activated T cells in human endothelial cells: a role for tissue factor gene expression. Mol. Cell. Biol. 19, 2032–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Wang W., Wang J. P., Pan L., Zhang Y., Yu H. T., Jiang W., Wang P. Z., Bai X. F. (2012) Elevated vascular endothelial growth factor levels induce hyperpermeability of endothelial cells in hantavirus infection. J. Int. Med. Res. 40, 1812–1821 [DOI] [PubMed] [Google Scholar]
- 38. Bockeler M., Stroher U., Seebach J., Afanasieva T., Suttorp N., Feldmann H., Schnittler H. J. (2007) Breakdown of paraendothelial barrier function during Marburg virus infection is associated with early tyrosine phosphorylation of platelet endothelial cell adhesion molecule-1. J. Infect. Dis. 196 Suppl 2, S337–S346 [DOI] [PubMed] [Google Scholar]
- 39. Deissler H. L., Deissler H., Lang G. E. (2010) Inhibition of protein kinase C is not sufficient to prevent or reverse effects of VEGF165 on claudin-1 and permeability in microvascular retinal endothelial cells. Invest. Ophthalmol. Visual Sci. 51, 535–542 [DOI] [PubMed] [Google Scholar]
- 40. Geimonen E., Neff S., Raymond T., Kocer S. S., Gavrilovskaya I. N., Mackow E. R. (2002) Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. U.S.A. 99, 13837–13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Antle M. C., Mistlberger R. E. (2000) Circadian clock resetting by sleep deprivation without exercise in the Syrian hamster. J. Neurosci. 20, 9326–9332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chau D. T., Gulick D., Xie H., Dawson R., Green A. I. (2010) Clozapine chronically suppresses alcohol drinking in Syrian golden hamsters. Neuropharmacology 58, 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crespo M. J., Cruz N., Altieri P. I., Escobales N. (2011) Chronic treatment with N-acetylcysteine improves cardiac function but does not prevent progression of cardiomyopathy in Syrian cardiomyopathic hamsters. JCPT 16, 197–204 [DOI] [PubMed] [Google Scholar]
- 44. Mancinelli R., Vargiu R., Cappai A., Floris G., Fraschini M., Faa G. (2005) A metabolic approach to the treatment of dilated cardiomyopathy in BIO T0–2 cardiomyopathic Syrian hamsters. BioFactors 25, 127–135 [DOI] [PubMed] [Google Scholar]
- 45. Nicot S., Bencsik A., Morignat E., Mestre-Francés N., Perret-Liaudet A., Baron T. (2012) Differentiation of prions from L-type BSE versus sporadic Creutzfeldt-Jakob disease. Emerg. Infect. Dis. 18, 2028–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pour P., Krüger F. W., Althoff J., Cardesa A., Mohr U. (1974) Cancer of the pancreas induced in the Syrian golden hamster. Am. J. Pathol. 76, 349–358 [PMC free article] [PubMed] [Google Scholar]
- 47. McIntyre K. W., Welsh R. M. (1986) Accumulation of natural killer and cytotoxic T large granular lymphocytes in the liver during virus infection. J. Exp. Med. 164, 1667–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miyamoto K., Khosrof S., Bursell S. E., Moromizato Y., Aiello L. P., Ogura Y., Adamis A. P. (2000) Vascular endothelial growth factor (VEGF)-induced retinal vascular permeability is mediated by intercellular adhesion molecule-1 (ICAM-1). Am. J. Pathol. 156, 1733–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arsenault R. J., Li Y., Potter A., Griebel P. J., Kusalik A., Napper S. (2012) Induction of ligand-specific PrP (C) signaling in human neuronal cells. Prion 6, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.