Abstract
HLA class I molecules reflect the health state of cells to cytotoxic T cells by presenting a repertoire of endogenously derived peptides. However, the extent to which the proteome shapes the peptidome is still largely unknown. Here we present a high-throughput mass-spectrometry-based workflow that allows stringent and accurate identification of thousands of such peptides and direct determination of binding motifs. Applying the workflow to seven cancer cell lines and primary cells, yielded more than 22,000 unique HLA peptides across different allelic binding specificities. By computing a score representing the HLA-I sampling density, we show a strong link between protein abundance and HLA-presentation (p < 0.0001). When analyzing overpresented proteins – those with at least fivefold higher density score than expected for their abundance – we noticed that they are degraded almost 3 h faster than similar but nonpresented proteins (top 20% abundance class; median half-life 20.8h versus 23.6h, p < 0.0001). This validates protein degradation as an important factor for HLA presentation. Ribosomal, mitochondrial respiratory chain, and nucleosomal proteins are particularly well presented. Taking a set of proteins associated with cancer, we compared the predicted immunogenicity of previously validated T-cell epitopes with other peptides from these proteins in our data set. The validated epitopes indeed tend to have higher immunogenic scores than the other detected HLA peptides. Remarkably, we identified five mutated peptides from a human colon cancer cell line, which have very recently been predicted to be HLA-I binders. Altogether, we demonstrate the usefulness of combining MS-analysis with immunogenesis prediction for identifying, ranking, and selecting peptides for therapeutic use.
The highly polymorphic Human Leukocyte Antigen class I (HLA-I)1 genes are encoded by three loci (HLA-A, B, and C) in a gene-rich region on chromosome 6. They produce up to six unique cell surface receptors that bind and present the so-called HLA class I peptidome, which consists of peptides derived from proteolysis of intracellular proteins. Their function is to reflect the health state of the body's cells to CD8+ cytotoxic T cells. During thymic maturation T cells that react to self-peptides are eliminated (1), leaving T cells with the capability to recognize peptides from viruses and bacteria. This recognition is interpreted as a danger signal, leading to removal of infected cells. Transformed, preneoplastic and cancer cells also tend to display atypical self-peptides from mutated or excessively expressed self-proteins, known as tumor associated antigens (TAAs). Although HLA-I molecules are indispensable in prevention of disease, they also pose a substantial health problem by causing allergies (2), life-threatening autoimmune diseases (3), and the often fatal rejection of donor organs because of recognition of both major and minor histocompatibility antigens (4).
Finding the rules for peptide generation and selection is regarded as the most important open issue in the field of HLA-I biology by leading experts (5). Although the antigen presentation pathway is well characterized, it is still unclear how basic properties such as protein abundance, turnover, and subcellular localization influence and shape the HLA-I presented peptidome (6–10). One expectation is that protein abundance should correlate with presentation (11), but previous studies have reported conflicting and contradicting results that mostly argue against a strong link (6, 7, 10, 12, 13). It is also not fully understood why only some HLA-sampled self-peptides from cancer antigens spontaneously activate T cells, whereas others do not.
The majority of HLA-I peptides are derived from proteasomal degradation (5). Although the proteasome generates an excess of peptides, only some have the required sequence motifs for HLA binding, resulting in a selective sampling of available peptides (14). The presented peptides are typically nine amino acids long, but the length can range from eight to 15. The high degree of genetic variance of HLA-I receptors translates into allele-specific peptide-binding motifs defined by anchor positions, which are usually the second and the last positions in a peptide (15). Each cell has around 200,000 cell-surface-expressed HLA complexes, which bind about 10,000 unique peptide sequences (16). The affinity of a peptide toward the presenting HLA molecule does not correlate strongly with its immunogenicity, and neither does the number of presented HLA complexes (17). Instead, the most robust predictor of peptide immunogenicity appears to be the number of potential reactive T-cell clones (17–19).
The longer the source protein, the higher the chances it will contain sequences that fit to a certain HLA motif, which would inflate the representation of longer proteins regardless of biological role. Furthermore, some HLA-I peptide sequences can be mapped to multiple proteins, potentially causing a problem in determining the number of observed HLA peptides per protein (13). This illustrates that careful accounting of the potentially and actually presented HLA peptides is important in properly delineating trends in propensity of peptide presentation.
In cancer immunotherapy, T cells can be directed against tumors, based on the pattern of cancer associated HLA peptides. Therefore, there is great interest in determining the identity of these immunogenic peptides. Bioinformatic methods that attempt to predict HLA peptides of cancer proteins of interest are easily accessible and most commonly used. They typically score sequences with respect to proteasomal degradation, transport into the ER via the transporter associate with antigen processing (TAP) and binding to different HLA-I alleles (20). However, their precision success is modest (21, 22). The second approach is to directly capture the naturally presented peptides using mass spectrometry; however, this requires the relevant biological sample and sophisticated instruments and workflows, which have become accessible only recently for large-scale work (23–28). Although identification of cancer associated HLA peptides by MS, if performed stringently, establish the in vivo existence of the peptide, it still does not guarantee that it will elicit a potent T-cell response, which is required for further development into therapeutics (29). Therefore, like in the case of in silico predicted peptides, the immunogenicity of the peptides must in any case be tested empirically.
We here present a rich and high confidence HLA-I peptidome, established by applying state-of-the-art mass-spectrometric techniques on a collection of seven cell lines. We investigate how abundance affects the propensity of proteins to be presented as measurable HLA peptides and whether or not there are specific protein classes that are overrepresented even independent of abundance. Likewise, we explore how to use in silico immunogenicity tools on the set of identified HLA peptides from cancer-associated proteins, with a view to select vaccine candidates.
EXPERIMENTAL PROCEDURES
Cell Lines and Antibodies
JY, SupB15WT, HCC1143, HCC1937, and HB95 cells were maintained in RPMI 1640 medium and HCT116 cells in DMEM medium. SupB15RT cells were grown with the addition of 1 μm imatinib to the medium (Cayman Chemical, Ann Arbor, MI). Primary fibroblast cells were a kind gift from Dr. Stern-Ginossar (UCSF). W6/32 monoclonal antibodies were purified from the growth medium of HB95 cells that were grown in CELLine CL-350 flask (Wilson Wolf Manufacturing Corporation, New Brighton, MN) using Protein-A Sepharose (Invitrogen, Camarillo, CA). HLA-I types were determined using high-resolution genotyping (Center for Human Genetics and Laboratory Medicine, Martinsried, Germany) except for JY and SupB15 for which the information was obtained from (27) and (30), respectively (Table I).
Table I. List of cell lines and their tissue origin used for HLA-I peptidome study and their genotypic information.
| Cell line | Tissue origin | HLA-A | HLA-B | HLA-Cw |
|---|---|---|---|---|
| JY | B-cells EBV transformed | HLA-A*02:01 HLA-A*02:01 | HLA-B*07:02 HLA-B*07:02 | HLA-Cw*07:02 HLA-Cw*07:02 |
| SupB15 | B-cell leukemia | HLA-A*03 HLA-A*11 | HLA-B*51 HLA-B*52 | HLA-Cw*12:04 HLA-Cw*14:02 |
| HCC1143 | Basal like breast cancer | HLA-A*31:01 HLA-A*31:01 | HLA-B*35:08 HLA-B*37:01 | HLA-Cw*04:01 HLA-Cw*06:02 |
| HCC1937 | Basal like breast cancer | HLA-A*23:01 HLA-A*24:02 | HLA-B*07:02 HLA-B*40:01 | HLA-Cw*03:04 HLA-Cw*07:02 |
| HCT116 | Colon carcinoma | HLA-A*01:01 HLA-A*02:01 | HLA-B*45:01 HLA-B*18:01 | HLA-Cw*05:01 HLA-Cw*07:01 |
| Fibroblast | Primary fibroblast cells | HLA-A*03:01 HLA-A*23:01 | HLA-B*08:01 HLA-B*15:18 | HLA-Cw*07:02 HLA-Cw*07:04 |
Purification of HLA-I Complexes
HLA-I peptidomes were obtained from three to four biological replicates per cell line. HLA-I complexes were purified from about 5 × 108 cell pellets after lysis with 0.25% sodium deoxycholate, 0.2 mm iodoacetamide, 1 mm EDTA, 1:200 Protease Inhibitors Mixture (Sigma), 1 mm PMSF, and 1% octyl-β-D glucopyranoside (Sigma) in PBS at 4°C for 1 h. The lysates were cleared by 30 min centrifugation at 40,000 × g. We immunoaffinity purified HLA-I molecules from cleared lysate with the W6/32 antibody covalently bound to Protein-A Sepharose beads (Invitrogen, Camarillo, CA), because covalent binding of W6/32 antibody to the beads improves the purity of the eluted HLA-I complexes, diminishes co-elution of the antibodies that otherwise overload the C-18 cartridges, and enables the reuse of the column. This affinity column was washed first with 10 column volumes of 150 mm NaCl, 20 mm Tris·HCl (buffer A), 10 column volumes of 400 mm NaCl, 20 mm Tris·HCl, 10 volumes of buffer A again, and finally with seven column volumes of 20 mm Tris·HCl, pH 8.0. The HLA-I molecules were eluted at room temperature by adding 500 μl of 0.1 N acetic acid, in total seven elutions for each sample. Small aliquots of each elution fraction were analyzed by 12% SDS-PAGE to evaluate the yield and purity of the eluted HLA-I.
Purification and Concentration of HLA-I Peptides
Eluted HLA-I peptides and the subunits of the HLA complex were loaded on Sep-Pak tC18 (Waters, Milford, MA) cartridges that were prewashed with 80% acetonitrile (ACN) in 0.1% trifluoroacetic acid (TFA) and with 0.1% TFA only. After loading, the cartridges were washed with 0.1% TFA. The peptides were separated from the much more hydrophobic HLA-I heavy chains on the C18 cartridges by eluting them with 30% ACN in 0.1% TFA. They were further purified using a Silica C-18 column tips (Harvard Apparatus, Holliston, MA) and eluted again with 30% ACN in 0.1% TFA. The peptides were concentrated and the volume was reduced to 15 μl using vacuum centrifugation. For MS analysis, we used 5 μl of this highly enriched HLA peptides.
LC-MS/MS Analysis of HLA-I Peptides
HLA peptides were separated by a nanoflow HPLC (Proxeon Biosystems, Thermo Fisher Scientific, Bremen, Germany) and coupled on-line to a Q Exactive mass spectrometer (31) (Proxeon Biosystems, Thermo Fisher Scientific) with a nanoelectrospray ion source (Proxeon Biosystems). We packed a 20 cm long, 75 μm inner diameter column with ReproSil-Pur C18-AQ 1.9 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) in buffer A (0.5% acetic acid). Peptides were eluted with a linear gradient of 2–30% buffer B (80% ACN and 0.5% acetic acid) at a flow rate of 250 nl/min over 90 min. Data was acquired using a data-dependent “top 10” method, which isolated them and fragment them by higher energy collisional dissociation (HCD). We acquired full scan MS spectra at a resolution of 70,000 at 200 m/z with a target value of 3e6 ions. The ten most intense ions were sequentially isolated and accumulated to an AGC target value of 1e5 with a maximum injection time of generally 120 ms, except in a few cases where we used 250 ms to increase signal of the fragments. In case of unassigned precursor ion charge states, or charge states of four and above, no fragmentation was performed. The peptide match option was disabled. MS/MS resolution was 17,500 at 200 m/z. Fragmented m/z values were dynamically excluded from further selection for 15 or 20 s.
Mass Spectrometry Data Analysis of HLA Peptides
We employed the MaxQuant computational proteomics platform (32) version 1.3.10.15. Andromeda, a probabilistic search engine incorporated in the MaxQuant framework (33), was used to search the peak lists against the UniProt database (86,749 entries, June 2012) and a file containing 247 frequently observed contaminants such as human keratins, bovine serum proteins, and proteases. N-terminal acetylation (42.010565 Da) and methionine oxidation (15.994915 Da) were set as variable modifications. The second peptide identification option in Andromeda was enabled. The enzyme specificity was set as unspecific. Andromeda reports the posterior error probability and false discovery rate, which were used for statistical evaluation. A false discovery rate of 0.01 was required for peptides. As we are interested in HLA-I peptide identification rather than the protein identification common in proteomics, no protein false discovery rate was set. Likewise, as no sequence specific proteases are involved and peptides do not terminate in certain amino acids such as Arg or Lys, the special permutation rules in MaxQuant for these amino acids (32) were not used in creating the decoy database. Possible sequence matches were restricted to eight to 15 a.a., a maximum peptides mass of 1500 Da and a maximum charge states of three. The initial allowed mass deviation of the precursor ion was set to 6 ppm and the maximum fragment mass deviation was set to 20 ppm. We enabled the “match between runs” option, which allows matching of identifications across different replicates that belongs to the same cell line, in a time window of 0.5 min and an initial alignment time window of 20 min. From the “peptide.txt” output file produced by MaxQuant, hits to the reverse database and contaminants were eliminated. The resulting list of peptide is provided in supplemental Table S1.
Total Proteome Sample Preparation, MS Measurement, and Data Analysis
From the cell lines JY, HCT116, HCC1143, and HCC1937 proteins were digested by the FASP method (34). The proteomic data from SupB15WT and SupB15RT cells was provided by R. D'souza. In that data, proteins were also digested by the FASP method but separated into three SAX fractions as described (35). For the fibroblasts, we digested proteins in solution in guanidine-chloride buffer and separated them into three SCX fractions (36). For all of the above cases, we cleaned and concentrated 5 μg of the peptides on a C18 based StageTip (37), before LC-MS/MS measurement.
Peptides were separated on a 50 cm reversed phase column (75 μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 1.9 μm resin (Dr. Maisch GmbH)) over a 120 or 240-min gradient of 5–60% buffer B (0.1% (v/v) formic acid and 80% (v/v) acetonitrile) using the Proxeon Ultra EASY-nLC system (Thermo Fisher Scientific). The LC system was coupled on-line with a Q Exactive instrument (Thermo Fisher Scientific) via a nano-electrospray source. Full scans were acquired in the Orbitrap mass analyzer with resolution 70,000 at 200 m/z. For the full scans, 3e6 ions were accumulated within a maximum injection time of 20 ms and detected in the Orbitrap analyzer. The five or ten most intense ions with charge states ≥ two were sequentially isolated to a target value of 1e5 with a maximum injection time of 120 ms and in a few cases a fixed injection time of 60 ms was used. Normalized collision energy was 25% for fragmentation and resolution was 17,500 at 200 m/z (38).
Raw data analysis for proteomics data sets was performed using MaxQuant with the same version and similar settings as above, with the following alterations. Strict trypsin specificity was required allowing up to two missed cleavages. Carbamidomethylation of cysteine (57.021464) was set as fixed modification. Minimum required peptide length was set to seven amino acids. A false discovery rate of 0.01 was required for peptides and for proteins in these proteomics experiments. Calculated proteins intensities from MaxQuant “proteinGroups” output file were used for estimation of abundance and are provided for each of the cell lines in supplemental Tables S2–S8.
Predicting HLA-I Peptides from JY Cells
We observed 2874 peptides from 2368 proteins for the JY cells of which 2875 could be mapped to distinct expressed protein sequences. By default, tools for prediction of HLA-binding provide 9-mer peptides. Of the original 2875 uniquely observed HLA-binding peptides about 61% (1741) were 9-mers and we used these for comparisons to predictions. We employed the prediction tool NetMHCcon1.0 (39) to estimate the HLA-binding for A02 and B7 alleles to the associated source proteins for the 9-mer peptide set. This led to a total of 1,188,744 9-mer peptides associated with prediction values allowing the computation of ROC curve accessing the power of the prediction.
Of the observed 1741 9-mer peptides, we were able to include 1732 in the computation of the ROC curves and the derived AUC values. The inconsequential loss of 0.5% of the observe peptides was caused by incomplete matching between protein IDs from the MaxQuant output and UniProt IDs.
The suggested HLA-I alleles of the cancer peptides are indicated with their prediction binding scores in affinity Kd values, which were generated using NetMHC 3.4 for the common alleles (All A alleles, B*07:02, B*08:01, B*18:01, B*40:01, B45:01, B*51, Cw*04:01, Cw*05:01, Cw*06:02, Cw*07:01, Cw*07:02, and Cw*14:02), and for the remaining alleles using NetMHCcons 1.0 Servers (39, 40).
Gibbs Clustering of HLA-I Peptidomes
Gibbs clustering analysis was performed using the publicly available GibbsCluster-1.0 Server tool (41). We used 9-mer HLA-I peptides as input using the default settings without alignment, 1–6 number of clusters, and the default threshold score of zero for “discarding to trash” as described in (41). We compared the resulting motifs to the known and predicted motifs of the HLA-I alleles using the MHC motif viewer (42) and the SYFPEITHI database (43).
HLA-I peptide sampling density. HLA-I peptides that originate from proteins for which we had no data on expression levels were omitted from the computation of HLA-I sampling density. About 8–11% of all presented peptides can be mapped back to several expressed source protein sequences and counting such nonunique peptides could create a bias toward peptides commonly found in the human proteome and toward larger groups of homologous proteins, protein families, and genes with many isoforms. Although overall only about one in 10 HLA-I peptides have more than one protein source, we find that 18–26% of those proteins with at least one mapped HLA-I peptides contain another HLA-I peptide that is shared with other protein sequences. It is estimated that only about 20% of potential peptides are sufficiently cleaved during proteasomal degradation (44) and it is therefore reasonable to expect that only a subset of proteins will contribute to the production of peptide because of favorable downstream sequence motifs, significant expression and an adequate degradation frequency. We define a presentation density for each protein sequence based the number of observed HLA-I peptides divided by the number of 9 mers contained in the protein sequence. Then we corrected for cases in which the peptide occurred in different proteins as described in detail in Online Supplemental Experimental Procedures.
To estimate which proteins are actually more presented than expected, we modeled the expected value of presented HLA-p densities from the length of the protein (L) and the measured protein abundance (A) as: D′ = D(protein A) = F(L, A). To determine F, we first divide the proteins into bins of proteins with similar abundance, calculate the average HLA-p density for each bin, and then fit a sigmoid function to this. The resulting function is used to calculate the expected HLA-p density given A and L for each protein.
RESULTS
Mass Spectrometry-Based Capturing of the HLA Class I Peptidome
Elucidating global properties of the presented proteome requires analysis of HLA-I peptidomes across different cell types and multiple alleles. To this end, we developed a workflow employing a robust protocol for the purification of HLA-I peptides and their identification using a state-of-the-art quadrupole Orbitrap mass spectrometer (Q Exactive) and the MaxQuant environment (Fig. 1). In contrast to earlier studies that focused on one or few selected alleles across a single cell type (27, 28, 45), we analyzed seven primary and cancer cell lines expressing a total of seven different HLA-A alleles, ten HLA-B alleles, and nine HLA-Cw alleles (Table I). Remarkably, just 5 × 108 cells per biological replicate turned out to be sufficient to obtain high coverage of HLA peptides in a relatively short measurement time of 2 h per replicate. Although recently published studies applied a permissive FDR threshold of 5% or more (26, 27), we wished to restrict the analysis to peptides identified with high certainty. Using a stringent FDR of 1%, we reached a high median Andromeda identification score of 123. With a more permissive 5% FDR threshold, the median Andromeda scores is reduced to 98, whereas the total number of identified peptides increases to 49,011. (For comparison, Andromeda scores are generally about threefold larger than Mascot scores (33); the median identification score of our peptidomics data set is thus equivalent to a Mascot score of about 41.) We identified from 2758 to 5324 peptides sequences in HCC1143 and fibroblast cells, respectively, and a total of 22,244 sequence-distinct peptides from all seven cell lines. More than 93% of the peptides have the typical length of HLA-I peptides, that is nine to 11 amino acids. The measured MS signals of the peptides span four orders of magnitude. Although most of the peptides are doubly charged, we were also able to accurately identify singly charged ions, which constitute about 15% of the peptidome (supplemental Table S1 and Fig. 2).
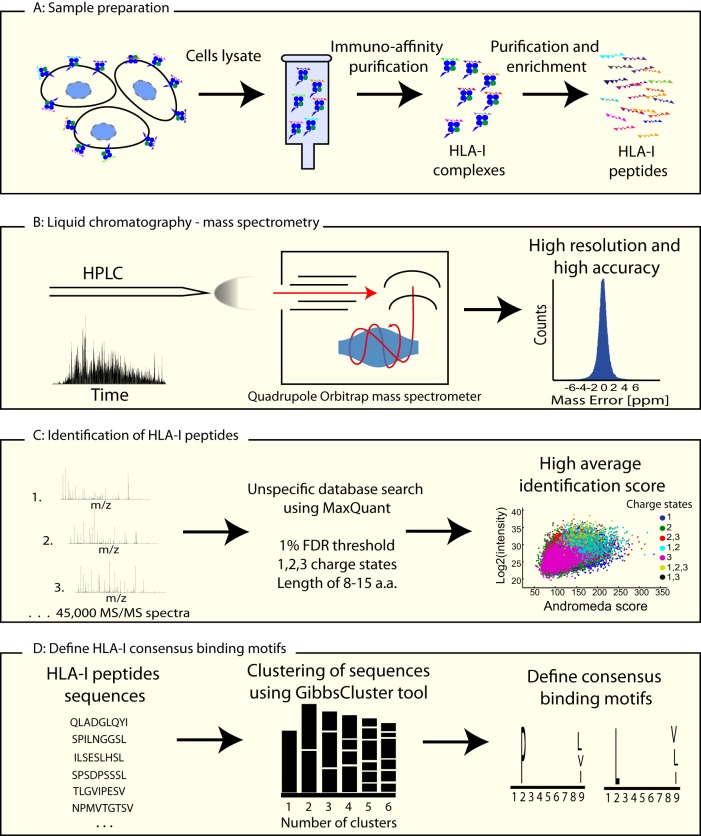
Fig. 1.
Schematic overview of HLA-I peptidomics. A, Sample preparation. HLA-I complexes were immunoaffinity purified from cells lysates using anti-HLA-I (W6–32) antibody cross-linked to Protein-A Sepharose beads. HLA-I peptides were purified from the heavy chain based on their hydrophobicity using a C-18 column. B, Liquid chromatography and mass spectrometry. The enriched mixtures of HLA-I peptides were measured on a quadrupole Orbitrap mass spectrometer (Q Exactive), resulting in high resolution and high mass accuracy at the MS and MS/MS levels. C, Identification of HLA-I peptides. HLA-I peptides were analyzed with MaxQuant software, using an unspecific search, allowing the identification of peptides with one, two, and three charge states. In total, 22,244 unique HLA-I peptides were identified with a median identification score of 123 using a threshold of 1% FDR at the peptide level. D, HLA-I consensus binding motifs. Using the GibbsCluster tool the consensus binding motifs were defined from the identified peptides sequences.
Fig. 2.
Properties of the HLA-I peptidomes data set obtained from seven cell lines. A, Number of HLA-I peptides identified in each peptidome sample. B, Length distribution of HLA-I peptides. C, Intensities of HLA peptides span over four orders of magnitudes. D, Identification score distribution of HLA-I peptides for each of the charge states. E, Number of HLA-I peptides that were identified with the different charge states.
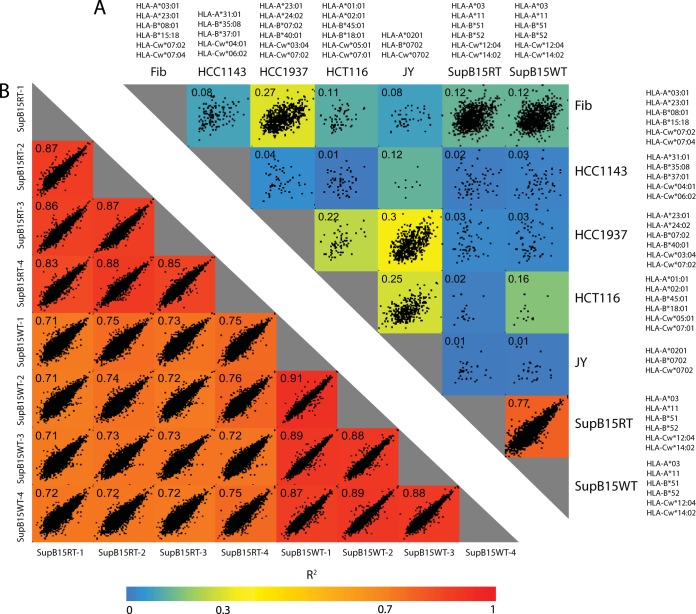
Unique repertoires of peptides are presented on the surface of cells that express different sets of HLA alleles according to the specificity of their binding motifs. As expected, we observed that cells expressing similar alleles, such as fibroblast and HCC1937 cells, shared drastically more peptide sequences than cells expressing different set of alleles, like fibroblast and HCT116 cells (Fig. 3A). We also found a higher correlation between the peptidome intensities from cells that share HLA-I alleles, as highlighted by the analysis of the isogenic cells SupB15WT and SupB15RT (resistance to imatinib treatment). The quantitative reproducibility of the peptidomes was excellent (R2 = 0.83–0.91) between biological replicates and very good (R2 = 0.71–0.76) between these two isogenic lines (Fig. 3B). This demonstrates both the robustness of our protocol and the stability of the cellular presentation machinery, which sampled the same peptides sequences and therefore the same source proteins in cells with similar genetic background.
Fig. 3.
HLA-I peptidomes are highly reproducible. A, Cells expressing similar alleles shared more peptide sequences than cells expressing different set of alleles. Very low quantitative reproducibility of peptidomes isolated from cells expressing different set of alleles. B, Quantitative reproducibility of the peptidomes isolated from SupB15WT and SupB15RT. The reproducibility of the peptidomes was excellent (R2 = 0.83 –0.91) between biological replicates and very good (R2 = 0.71 – 0.76) between the two isogenic lines.
Unbiased and Direct Definition of HLA-I Binding Motifs
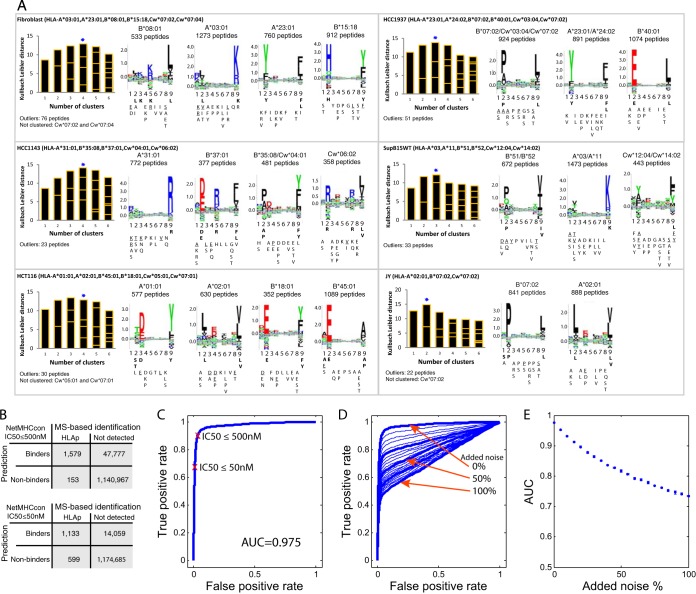
HLA-I peptides are characterized by their typical binding motifs. In an attempt to obtain the motifs directly from the peptidome data, without the need for genotyping information, we clustered the peptides by sequence similarity. We expected to find up to six motifs and fewer in case of motif redundancy, homozygosity, or abolished expression of one or more alleles. Using the Gibbs clustering algorithm (41) we resolved the motifs and their relative contribution to the total presentation from cells expressing only three alleles such as JY cells, and more alleles like HCC1143 and HCC1937 (Fig. 4A, and supplemental Data). In most cases the motifs of HLA-Cw alleles could not be resolved, probably because of their low expression levels, and because of redundancy in their specificities to HLA-A and B alleles. For cell types that share one of their alleles, we should detect the corresponding binding motives in both and this was indeed the case. For example, similar motifs were obtained for HLA-A*23:01 in HCC1937 and the fibroblast cells, for HLA-A*03:01 in the fibroblast and SupB15 cells, and for HLA-A*02:01 in JY and HCT116 cells. Moreover, we could assign the motif for the poorly defined HLA-Cw*06:02 allele (42, 43, 46) based on 358 peptides sequences identified from HCC1143 cells.
Fig. 4.
High confident identification of purified HLA-I peptides. A, Defining motifs directly from the mixture of identified peptides. Gibbs clustering analysis was performed for the purified 9-mer HLA-I peptides from the different cell lines. The motifs of the isogenic cell lines SupB15WT and SupB15RT cells were identical; therefore the results are shown only for SupB15WT. For each initial number of clusters the information content of the alignment is shown as a bar plot, where the size of each block within a bar is proportional to the size of a given cluster. The blue star marks the number of clusters that were selected based on the optimal fitness (higher KLD values) and minimum outliers, and their sequence logo plots are shown with the number of HLA-I peptides in each cluster and the assigned HLA-I alleles that fit each cluster. Binding motifs were calculated for each cluster from the frequency of the amino acids (AA) in positions P1 to P9 in the peptides sequences (see Supplemental Data). Frequency of more than 30% was classified as a dominant anchor motif (bold), more than 20% as a strong motif (underline), and more that 10% as a weak motif. B, Confirming the accurate identification of the observed peptides by predicting their affinity to the expressed alleles. We predicted using NetMHCcon (39) the binding affinity (maximal predicted binding affinity; HLA-A*02:01 and HLA-B*07:02) of the peptidome data set of 9-mer peptides from JY cells, and estimated the performance of the predictor using the expressed proteins as the set of input sequences. We compared the default affinity score <500 nm to include weak binders and the high affinity score of <50 nm to restrict to strong binders only. C, The computed Receiver Operating Characteristic (ROC) curve for the binding affinity to the HLA-I based on the predicted 9-mer epitopes from JY cells. The AUC (area under the curve) value is 0.975. D, Evaluating the deterioration in the ROC analysis when introducing noise of randomly selected 9-mer sequences from the expressed proteins. 9-mer peptides were added from 0 to100%, in steps of 5%, to the list of observed 9-mer peptides from JY cells. E, AUC values calculated from ten iterations of noise introduction.
Accurate Identification of the Naturally Presented Peptidomes
To quantitatively assess the ability of our protocol to enrich for HLA-I binders and discriminate against potential co-eluting contaminants, we predicted the HLA binding for the presented peptides of the two HLA-I alleles expressed in JY cells using the proteins expressed in this cell line as a set of input sequences) (Fig. 4B, and detailed description in Experimental Procedures). JY cells specifically express A*02:01 and B*07:02. (They also express very low levels of HLA-Cw*07:02, but because the binding motif of HLA-Cw*07:02 overlaps considerably with the motifs of A*02:01 and B*07:02, only these motifs where qualitatively observed in the Gibbs clustering analysis.) We evaluate the agreement between the observed peptidome of JY cells and the predicted peptidome as a receiver operating characteristic (ROC) curve (Fig. 4C). We summarize the ROC curve as the area under the curve (AUC) value, which ranges from 0.5 for random agreement to 1.0 for perfect agreement. The resulting AUC value of 0.975 reflects excellent agreement, meaning that the predictor confirmed that the vast majority of isolated peptides fit the HLA-A*02:01 and HLA-B*07:02 motifs. A threshold of predicted affinity <500 nm is commonly applied for retrieving weak binders, and yielded 1579 out of the 1732 identified peptides (91.2%). A total of 1133 (65%) of the peptides are predicted to be strong binders (<50 nm) (Fig. 4B).
To estimate the purity of the isolated peptidomes further, we introduced noise in silico, which will imitate the presence of impurities in our peptidomes. As the most likely contaminating components are peptides originating from nonspecific degradation of co-eluting proteins, we randomly added 9-mer peptides from the source proteins of the HLA-I peptides to the list of observed 9-mer peptides from JY cells. We did this from 0 to 100%, in steps of 5%, and repeated the computation of the resulting ROC curve and AUC values. The addition of 10%, 25%, and 50% random peptides reduced the AUC values to 0.931, 0.877, and 0.816, respectively (Fig. 4D and 4E). These analyses show quantitatively how AUC values are affected by impurities and here they revealed that this impurity must be minimal in our data set (much less than 5%).
Computing Presentation of Expressed Proteins from HLA-I Peptide Counts
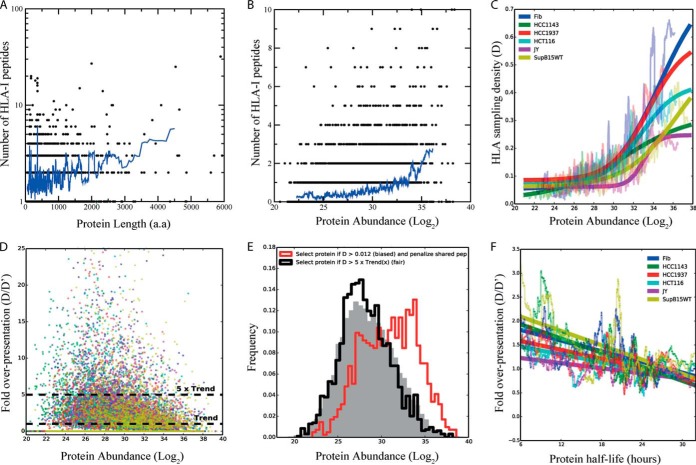
Because cell lines that express unique sets of HLA-I alleles have few presented peptides in common, it is difficult to directly compare peptide intensities and estimate HLA-I sampling. The measured intensities of HLA-I peptides reflect a combination of their intracellular processing, their binding affinities, and their compatibility with MS detection (ionization and fragmentation). Therefore, even HLA-I peptides that originate from the same protein within one sample can differ significantly in intensity. Our data set contains hundreds of proteins, which are a source for several detected HLA-I peptides (Fig. 5A and 5B and supplemental Tables S2–S8). This allowed us to investigate the correlation between the number of HLA-I peptides (rather than their MS-intensities) and expression level of their source proteins, which was measured in an independent proteomic analysis for each of the cell lines.
Fig. 5.
HLA-I sampling for presentation correlates with proteins length, abundance, and half-life. A, HLA-I peptides in proteins as a function of proteins length. The plot represents the comparison between the number of presented peptides and the length of the source proteins as detected in proteomic analysis of total cell lysates. The blue line is a running average calculation of the data points. B, HLA-I peptides in proteins as a function of proteins abundance. The plot represents the comparison between the number of presented peptides and protein abundance, in Log2(intensity) of their source proteins as detected in proteomic analysis of total cell lysates. Every protein that was detected in the proteomics analysis in each of the cell lines is presented in the plot according to the number of resulting detected HLA-I. Therefore, the same protein can be represented in the plot several times in case that it was detected in different intensities and gave rise to different number of epitopes in each of the cell lines. The blue line is a running average calculation of the data points. C, HLA-I sampling density (D) correlates with protein abundance. Using a running average, HLA-I sampling density is significantly correlated with the abundance of the source proteins (p < 0.0001). The data was fitted with a trend line (solid line) for each cell line. D, Fold HLA-I sampling density over the expected sampling (D′). For each protein the ratio D/D′ is represented as a function of protein abundance. The criteria for the selection of overpresented proteins was set to D>5 times larger than the expected HLA-I sampling density (D′). E, Unbiased selection of overpresented proteins. An explanatory plot showing how without correcting for the bias that originates from preferential presentation of highly expressed proteins the selection of overpresented proteins will result in selecting mainly the abundant proteins. The histogram represents protein abundance. The emphasized black histogram (5 x trend) shows the protein abundance for the subset of proteins with D>5 times larger than the expected HLA-I sampling density (D′), and it has the same shape as the proteome (in gray). The red histogram (biased) illustrates what would happen if D′ was a constant (D′ = 0.012), resulting in a biased selection toward highly expressed but not overpresented proteins. F, Presentation efficiency (D/D′) in relation to proteins half-life. Regardless of expression levels, turnover rates measures as half-life values, statistically significant correlate with presentation, in all cell lines (p < 0.001).
Although our streamlined workflow enabled analysis of a very large number of HLA peptides, about 70% of the expressed proteins were not presented at all in our peptidomics data set (supplemental Tables S2–S8). Most likely, many of them are still presented to some extent on the cells surface; however, in peptide copy numbers that made them inaccessible to detection. These nonrepresented proteins are nevertheless distinguished from the others by the fact that they present no or undetectable HLA peptides and we set out to further characterize these proteins in the following sections.
HLA-I Presentation Correlates Significantly with Protein Abundance
Because HLA-I peptides originate primarily from proteasomal cleavage of proteins, a possible expectation would be that highly abundant proteins lead to more proteins being degraded per unit of time than less abundant proteins. Moreover, longer proteins contain more 9-mers and should therefore give rise to more potential HLA-I binders. Thus, one might expect a positive correlation between the number of measured HLA-I peptides and the abundance and length of their source proteins, although establishing such a relationship has so far been proven difficult.
First we investigated if there was a relationship between the length of the protein and the number of presented peptides in our data set. This clearly the case as can be seen in Fig. 5A and supplemental Fig. S1A. Similarly, Fig. 5B and supplemental Fig. S1B establish an unambiguous relationship between protein abundance and resulting HLA peptides. Next, to quantify the correlation between presentation and protein abundance in an unbiased manner, we normalized for protein length by calculating the HLA-I sampling density (D). For each source protein we computed D from its count of HLA-I peptides, weighting down shared peptides to eliminate bias (supplemental Experimental Procedures). This analysis revealed a highly significant correlation between HLA-presentation and the abundance of the source proteins (p < 0.0001) (Fig. 5C). The figure also reveals that in the cell lines tested, the mean sampling density of the 20% most abundant proteins was 3.04- to 5.17-fold higher than the mean density of the 20% least abundant proteins (supplemental Tables S2–S8). Thus as expected, low abundant proteins will on average be the source of fewer HLA-I peptides than more abundant proteins.
We further wanted to explore which proteins are more presented than should be expected from their abundance. To this end we identified a set of “overpresented” proteins, which we defined as the proteins with HLA-I sampling density at least fivefold higher than their expected predicted sampling density (termed D′) given their abundance (D/D′>5) (Fig. 5D). This strategy ensured that the protein abundance distribution of overpresented proteins resembles that of other proteins (black line in Fig. 5E), whereas this is not the case without this correction (red line).
Protein Half-Life Correlates with Efficient HLA-I Presentation
Although we find a clear correlation between protein abundance and HLA-I sampling rates, about half of the 20% most abundant proteins are not represented at all in our peptidomics data set. We focused on these “hidden” proteins (abundant but not represented) because their absence in the HLA peptidome is less likely to be caused by the finite detection capability of our MS-setup. They can only to small extent be explained by a generally shorter median protein length (311 for the hidden proteins versus 441 for the presented ones). In search of other factors we noticed that hidden proteins have longer half-lives. We obtained the half-life for 4066 of the proteins in our data set from (47). Among these, we identified 154 overpresented proteins for which we were able to find a corresponding hidden protein with a closely matched abundance and length (154 matched hidden proteins). This matching ensures that any observed difference between the two sets will not be caused by the small difference in protein length between hidden and presented proteins or difference in their abundance. We found that hidden proteins are significantly longer-lived (median half-life 23.6h) than overpresented proteins (20.8 h, p = 4.8E-5) (Fig. 6A).
Fig. 6.
Characterization of overpresented proteins. A, Protein degradation and HLA-I presentation. Hidden proteins, with the same length and same abundance level, are longer lived than overpresented but also in general longer lived than other highly abundant proteins. From the top 20% abundant proteins we compared 154 proteins, which were overpresented proteins (D/D′>5) to balanced set of similar size of hidden proteins (D′ = 0) with matched expression and protein length. B, Predicting immunogenicity of HLA-I peptides from cancer antigens and from overpresented proteins. The 229 epitopes from the set of validated cancer antigens are significantly more immunogenic than the 82 HLA-I peptides from the same proteins which we identified in our peptidomics data set. Overpresented proteins (D/D′>5) are not more immunogenic than the rest of the presented peptidome (0<D<5) both in primary and cancer cell lines.
Extending this analysis, we next correlated the presentation efficiency (D/D′) in relation to turnover rate for the entire data set. This revealed a statistically significant correlation (p < 0.001) in all cell lines (Fig. 5F). Clearly, regardless of expression level, turnover rates influence the level of presentation; however, without normalizing for protein length and abundance this correlation could not have been resolved (supplemental Fig. S1C).
Faster turnover for otherwise matched protein properties should lead to a higher concentration of HLA peptide precursors, therefore, our statistical association are biologically plausible.
Preferentially Presented Biological Processes and Cellular Compartments
We investigated whether overpresented proteins have a shared biological function or cellular localization that favors their presentation. For each of the cell lines, we selected the overpresented proteins and used the proteome expressed in these cells as a customized background set, and performed gene ontology enrichment analysis using the DAVID functional annotation tool (48, 49). The significantly enriched terms (Benjamini-Hochberg corrected p value of 5% or less) in most of the cell lines regardless of their alleles and binding motifs are the mitochondrion (all cells except JY and SupB15RT), ribosome (all cells except JY cells), and the nucleosome in Supb15WT and SupB15RT cells (supplemental Tables S2–S8). Specifically, the overpresented mitochondrial proteins belong to the NADH dehydrogenase, ATP synthase, and cytochrome oxidase complexes as well as the mitochondrial ribosomes. The mitochondrial and cytoplasmic ribosomes were both represented with the large and small subunits as well as associated proteins.
Prioritizing HLA-I Peptides Via Immunogenicity Prediction
HLA peptides originating from cancer-associated proteins are used clinically in turning the immune system toward attacking the tumor (29, 50–55). The presentation of peptides from overexpressed, cancer-associated self-proteins is known to be elevated on cancer cells compared with healthy cell (56, 57). This elevated presentation can break the inherited tolerance against self and can lead to antitumor response. A key challenge in the field is to select the most promising HLA epitopes even from one candidate protein. Given the large number of high quality hits from reported cancer-associated proteins resulting from our workflow, we explored ways to prioritize them via in-silico methods.
Using a tool that is able to predict immunogenicity of viral HLA-I peptides (58), we first evaluated if immunogenicity of cancer and viral peptides follows the same trends employing the list of 229 immunogenic epitopes from cancer-associated proteins published in ref (59). The T-cell-defined epitopes were selected based on their good performance in activating T cells, hence, we expect them to be the most immunogenic HLA-I peptides from these proteins. We name them “epitopes” to distinguish them from other HLA-I peptides we detected in our study. Our data set contains four known cancer epitopes from this list, among them well characterized epitopes from the melanoma-associated antigens family (MAGE-A1 and MAGE-A3) and 82 more HLA-I peptides from 25 cancer-associated proteins (59) (supplemental Table S9). We predicted the immunogenicity of the 229 empirically validated immunogenic T-cell epitopes associated with cancer and compared their scores to the 82 HLA peptides we identified in our data set (Fig. 6B). We find that T-cell-defined cancer epitopes are predicted to be significantly more immunogenic than other MS-identified HLA-I peptides that originate from the same proteins (p < 5 × 10−6). Based on these findings, we suggest ranking the experimentally identified HLA peptides based on their immunogenicity score. For example, in our data set we found that peptides that score higher than 0.15 are the top 20% most immunogenic peptides (supplemental Table S9).
Serving as a control, HLA-I peptides that originate from the overpresented proteins (D>5) turn out to be less immunogenic compared with the cancer epitopes. They also seem to be slightly less immunogenic than HLA-I peptides from other presented proteins (0<D<5), although this difference is not statistically significant. We conclude that the immunogenicity of cancer-associated HLA-I peptides can to some extent be predicted and utilize this to suggest potential new epitopes (supplemental Table S9).
Direct Identification of Mutated HLA-I Peptides
During revision of this manuscript, a resource of cell line-specific HLA type and somatic mutations has been published, including predictions of HLA-I binding peptide sequences that harbor these mutations (60). In this catalog, 152, 167, and 1903 mutated sequences were predicted to be presented in HCC1143, HCC1937, and HCT116 cell lines, respectively. We added these sequences to the list of UniProt proteins used for the database search, and reanalyzed the peptidomics data with MaxQuant, again applying the stringent 1% FDR for peptide identification. We clearly and unambiguously identified five mutated HLA-I peptides from HCT116 cell line, bearing mutations that were reported to be presented in this particular cell line (60) (Table II; and supplemental Data 2 for annotated spectra). All peptides were predicted to be strong binders with Kd values of less than 150 nm.
Table II. List of MS-identified mutated HLA-I peptides purified from HCT116 cell line. Potential mutation bearing peptide sequences were obtained from (60) and they were added to the UniProt database for MaxQuant search. The mutated positions are marked in bold.
| Protein | Peptide | AA change | Length | Identification score | HLA Allele best fit (NetMHC 3.4 predicted affinity, Kd values in nM) |
|---|---|---|---|---|---|
| CHMP7 | QTDQMVFNTY | p.A324T | 10 | 166 | A*01:01 (39), Cw*05:01 (23) |
| BCL2L13 | EEEYPGVTA | p.I216V | 9 | 148 | B*45:01 (20) |
| NR1D1 | YSDNSNDSF | p.G39D | 9 | 134 | A*01:01 (36), Cw*05:01 (2) |
| RBBP7 | EERVIDEEY | p.N17D | 9 | 116 | B*18:01 (149) |
| UQCRB | EEEKFYLEP | p.N88K | 9 | 117 | B*45:01 (54) |
DISCUSSION
Although much is known about the antigen presentation pathway of intracellular proteins, this knowledge has mostly been derived over several decades from small-scale studies involving the processing of relatively few proteins, typically of microbial origin (5). The advent of modern, high performance proteomics technologies, which has already revolutionized our ability to identify and quantify complete proteomes, now promises to allow global, systems-wide investigation of peptides presented to the immune system.
Here we have developed a powerful and streamlined workflow for the detection of HLA-I peptides presented by cells. We took advantage of the new generation of improved mass spectrometers, specifically the quadrupole-Orbitrap instrument (31), and better approaches to immune purification of HLA-I peptides (23). The high resolution of the Q Exactive, in combination with the HCD fragmentation, enabled an in-depth and accurate identification of the HLA-I peptidomes, whereas our purification protocol minimizes unnecessary steps and is optimized for sensitive, robust, and fast measurements. We used at least 20 times less sample compared with recently published studies (27, 28); therefore, our workflow makes it possible to identify HLA-I peptidomes from small amounts of sample such as tumors. Furthermore, we achieve high numbers of HLA-I peptide identifications even in a single-shot format, drastically reducing measurement time compared with protocols involving extensive fractionations (i.e. up to 40 fractions (27, 28, 61)).
A recently published study combines two fragmentation modes and therefore generates particularly information rich fragment spectra (EThcD fragmentation method), leading to a high identification percentage of HLA peptides (28). However, by its nature, that method is restricted to at least doubly charged precursors, while we also target singly charged peptides for fragmentation. They comprise about 15% of the data set reported here, but in our hands they can reach to 40% of total identified peptides, depending on the amino acids that restrict the binding motifs.
It in principle possible that our HLA peptide enrichment protocol would also copurify non-HLA peptides, such as co-eluting degradation products of cellular proteins. To assess the presence or absence of such peptides we employed NetMHCcon (39), a widely used tool to score peptides by their predicted affinity for selected alleles. We performed this analysis for the JY cell line because 1) this is the only cell line in our set that is homozygous for the main alleles (A*02:01 and B*07:02), 2) the expressed alleles are the best studied one and predictors have been optimally trained for them, and 3) as a consequence of the fact that the available receptors only bind two motifs, the intensity of the HLA peptides is increased. When we predicted the binding affinities of the experimentally identified peptides from the JY cells to these alleles, we found that 91% of them had immunologically relevant affinities of better than 500 nm. Of the remaining peptides, many also have appreciable predicted affinities; therefore, the level of possible contaminations is likely to be much less than 10%. Additionally, we modeled the effect of possible contaminant peptides, which also predicted absence or very low level of contaminating peptides.
This analysis also implies a robust performance of the predictor. However, prediction alone cannot yield the in vivo presented HLA peptidome. For instance, by its nature, the prediction algorithm does not account for the abundance of the source proteins; they predict sequences that could not be presented under physiological conditions, for instance proteins that are expressed at too low levels. Furthermore, these algorithms generally overpredict: Only about 1:30 predictions were observed in our data at the default threshold that includes weak binders. In principle, we could change this ratio to the still very large ratio of 1:12 by increasing the threshold to include only the strong binders. However, this is already too stringent because we then lose 35% of the observed peptides. Conversely, HLA peptides may be present but go undetected because of low abundance or unusual peptide properties or because of the stringent peptide identification criteria. Therefore the performance of the HLA predictors is somewhat higher than what we calculate here, which, however, does not solve the overprediction or false-positive problem.
Having established that our data reflect the actual peptidome well, we proceeded to estimate HLA-I presentation in relation to the expressed proteome. We computed the HLA-I sampling density by counting observed peptides regardless of their reported intensities. Intensity based computation of the HLA-I sampling would be biased by the MS compatibility of the individual peptides because about 61% of source proteins are singletons, 22% have two epitopes, and the rest three and more. Therefore, we count the peptides, and then normalize the count by the length of the source protein, as longer proteins give rise to more epitopes.
HLA peptide sequences may occur in more than one protein in the database, complicating analysis and the correct computation of the HLA-I sampling density. Hoof et al. excluded these peptides from their analysis (13), yet this may lead to a bias because abundant proteins have more epitopes, and therefore they have higher chance to share epitopes. The solution is to down-weight the contribution of multiple-target HLA-I peptides in a proper way assuming a posterior model of cleavage given the peptide is known to be cleaved at least by one potential sequence.
There have been conflicting reports on how protein abundance influences the degree of presentation. Marginal correlations as low as 6% (R2) have been reported (10), whereas a recent publication found that presented proteins are generally more abundant than nonsampled (13). On the basis of the fact that MS signals of the HLA-I ligandome varied greatly even in the same protein, it has been concluded that it is not possible to predict the ligand copy number from the overall proteins copy number, and hence, that there is no clear selection on the basis of protein properties during antigen processing (27). A comparison of mRNA levels and peptidome found a weak link (r = 0.32) (8), and another noted that HLA-I binding peptides are preferentially derived from highly abundant mRNAs (6). In contrast, it has also been reported that that only a minority of differentially expressed HLA-peptides originate from differentially expressed genes (7).
These disagreements have led to speculations about the involvement of novel biological effects that contribute to the production of HLA-I binding peptides, such as the DRiPs hypothesis and the pioneer round of mRNA translation. These theories suggest that a major source of HLA peptides derived from premature translation termination or downstream initiation (62, 63), hence there can be effective presentation even in the near absence of protein expression. However, only few studies have attempted to rigorously quantify the contribution of such alternative pathways to the generation of the peptidomes (10, 63–65). No positional bias was found in recently published studies among DRiPs toward the N terminus or the C terminus (64, 65). DRiPs were described to originate from the rapid degradation of “normal” proteins and subunits of complexes that are produced in excess (64). Our results indicate that high turnover of proteins, many of which are subunits of large complexes, enables their efficient presentation.
Here, we propose that the previously reported lack of correlation between presentation and proteins abundance was largely the result of insufficient statistical and data processing methods. Using our in-depth and accurate data set, we now demonstrate a highly statistically significant correlation between protein abundance and HLA-I sampling density (p<0.0001) indicating a direct mechanistic relationship between the protein copy number and the tendency to present peptides derived from them. Two factors enabled us to detect the clear significant correlation between presentation and expression of source proteins: the improved computation of HLA-I sampling density instead of peptide intensities and employing a method of moving average, which is better suited to analyze trends in data that are both sparse and dominated by single numerical values like the many nonpresented proteins, which results in a majority of zeroes.
We further characterized the two extreme states of presentation efficiencies: hidden proteins, which have no detectable HLA-I peptides in our experimental conditions and proteins with higher than expected sampling densities. We showed that abundant proteins without any detectable peptides have relatively lower turnover rates than the overpresented abundant proteins. Hidden proteins are relatively stable and long-lived and may not give rise to enough degradation products to be further processed and presented at a level that we could detect.
Previous efforts to characterize the cellular localization and functional properties of the source proteins did not account for the expression levels of the source proteins (9, 66). Here we removed this bias and investigated the functional properties of proteins with higher than expected sampling densities. In most of the cell lines investigated, mitochondrial proteins, histones, and ribosomes were presented even in excess of the degree expected from their abundance. Mitochondrial proteins are susceptive to oxidative stress-related damage caused by free radicals, and are routinely replaced to maintain proper organelle function. Ribosomal proteins are expressed in excess and a significant fraction of ribosomal proteins, which are imported into the nucleus, is degraded by the proteasome (67), which helps to explain their substantial contribution to the HLA-I peptidome (64). Nucleosomes were shown to turnover multiple times during each cell cycle (68) in a proteasome mediated degradation (69, 70). In these cases of high protein turnover, the increased HLA presentation can most simply be explained by their increased flux through the proteasome and subsequent parts of the antigen processing machinery.
Many studies, and especially immunotherapeutic interventions, have for the sake of predictability and simplicity focused on a limited set of frequently expressed HLA-I alleles with exceptionally well-characterized motifs (25, 27, 55). Indeed, this formed a strict inclusion criterion for patients eligible for receiving experimental cancer vaccines. With the limited performance of MS-based technology that was available when immunotherapy was emerging into clinical settings, it was necessary to predict potential cancer epitopes in silico rather than measure them directly. Even today, HLA-I binding predictors are used almost exclusively to discover cancer epitopes. Yet the inevitable high number of false positives, which we estimated to be around nine out of 10, often carries a high risk of investing resources on the wrong targets. As we show here, MS-based workflows now enable accurate detection of the true and often relatively well-presented epitopes in great detail. We can resolve binding motifs directly from our data regardless of genotyping information, a capability that can be applied to explore alleles with yet unknown motifs. As a result, new cancer epitope can be identified across different HLA-binding specificities, as there is no need to only select donors with particular HLA subtypes. Furthermore, in combination with prior knowledge of the repertoire of somatic mutations, we identified five HLA-I peptides bearing mutations with binding specificities to five HLA-I alleles expressed in the human colon carcinoma HCT116 cell line. Such peptides are especially prized for developing immunotherapeutics because they do not occur naturally in the noncancer tissues.
Applying MS-based workflows to clinical tumor samples will result in similar rich peptidomics data set and hence the straightforward identifications of cancer-associated HLA-I peptides and mutated peptides. Moreover, de novo sequencing algorithms will enable identification of such peptides without prior knowledge of somatic mutation. However, because there is no correlation between affinities to the HLA-I allele or copy number of peptides with immunogenicity (17), neither it nor HLA-I binding prediction can highlight the most immunogenic epitopes to elicit an efficient potent antitumor response. Instead, in silico methods can efficiently prioritize the peptides shown to be presented by mass spectrometry. We propose to select these candidate epitopes based on: (1) the individual's six possible HLA binding motifs and the relative expression levels of the corresponding receptor in the investigated tumor sample, (2) the abundance and turnover rate of the source protein, and (3) the predicted immunogenicity score. In case no cancer epitopes can be identified from the patient's tumor, the resulting peptidome can be used to predict potential epitopes that fit the more dominantly expressed alleles. We envision that better immunotherapeutics could be developed based on wider and more accurate repertoires of HLA-I peptides and that such treatments will be available to a larger cohort of patients.
Supplementary Material
Acknowledgments
We thank the members of the Department of Proteomics and Signal Transduction for fruitful discussions.
Data availability – The mass spectrometry data, MaxQuant version 1.3.10.15 and the search results output files have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (71) with the data set identifier PXD000394. All peptides spectral matches and their MS/MS spectra can be viewed in the freely available MaxQuant viewer.
Footnotes
Author contributions: M.B. and M.M. designed research; M.B. performed research; S.P. contributed new reagents or analytic tools; M.B., S.P., L.J.J., and M.M. analyzed data; M.B., S.P., L.J.J., and M.M. wrote the paper.
* M.B.S is supported by the Alexander von Humboldt Foundation.
 This article contains supplemental Fig. S1, Tables S1 to S9, Data, and Experimental Procedures.
This article contains supplemental Fig. S1, Tables S1 to S9, Data, and Experimental Procedures.
1 The abbreviations used are:
- HLA-I
- Human leukocyte antigen class I
- TAAs
- Tumor associated antigens
- ER
- Endoplasmic reticulum
- FDR
- False discovery rate
- HCD
- Higher energy collision dissociation
- ROC
- Receiver operating characteristic
- AUC
- Area under the curve
- DRiPs
- Defective ribosomal products
- mRNA
- Messenger RNA.
REFERENCES
- 1. Mouchess M. L., Anderson M. (2014) Central tolerance induction. Curr. Top. Microbiol. Immunol. 373, 69–86 [DOI] [PubMed] [Google Scholar]
- 2. Yun J., Adam J., Yerly D., Pichler W. J. (2012) Human leukocyte antigens (HLA) associated drug hypersensitivity: consequences of drug binding to HLA. Allergy 67, 1338–1346 [DOI] [PubMed] [Google Scholar]
- 3. Zehn D., Bevan M. J. (2006) T cells with low avidity for a tissue-restricted antigen routinely evade central and peripheral tolerance and cause autoimmunity. Immunity 25, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dierselhuis M., Goulmy E. (2009) The relevance of minor histocompatibility antigens in solid organ transplantation. Curr. Opin. Organ Transplant. 14, 419–425 [DOI] [PubMed] [Google Scholar]
- 5. Neefjes J., Jongsma M. L., Paul P., Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 [DOI] [PubMed] [Google Scholar]
- 6. Granados D. P., Yahyaoui W., Laumont C. M., Daouda T., Muratore-Schroeder T. L., Cote C., Laverdure J. P., Lemieux S., Thibault P., Perreault C. (2012) MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood 119, e181–e191 [DOI] [PubMed] [Google Scholar]
- 7. Caron E., Vincent K., Fortier M. H., Laverdure J. P., Bramoulle A., Hardy M. P., Voisin G., Roux P. P., Lemieux S., Thibault P., Perreault C. (2011) The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol. Syst. Biol. 7, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weinzierl A. O., Lemmel C., Schoor O., Muller M., Kruger T., Wernet D., Hennenlotter J., Stenzl A., Klingel K., Rammensee H. G., Stevanovic S. (2007) Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol. Cell. Proteomics 6, 102–113 [DOI] [PubMed] [Google Scholar]
- 9. Hickman H. D., Luis A. D., Buchli R., Few S. R., Sathiamurthy M., VanGundy R. S., Giberson C. F., Hildebrand W. H. (2004) Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J. Immunol. 172, 2944–2952 [DOI] [PubMed] [Google Scholar]
- 10. Milner E., Barnea E., Beer I., Admon A. (2006) The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteomics 5, 357–365 [DOI] [PubMed] [Google Scholar]
- 11. Rock K. L., Farfan-Arribas D. J., Colbert J. D., Goldberg A. L. (2014) Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 35, 144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Croft N. P., Smith S. A., Wong Y. C., Tan C. T., Dudek N. L., Flesch I. E., Lin L. C., Tscharke D. C., Purcell A. W. (2013) Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 9, e1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoof I., van Baarle D., Hildebrand W. H., Kesmir C. (2012) Proteome sampling by the HLA class I antigen processing pathway. PLoS Comput. Biol. 8, e1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yewdell J. W., Reits E., Neefjes J. (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 [DOI] [PubMed] [Google Scholar]
- 15. Falk K., Rotzschke O., Stevanovic S., Jung G., Rammensee H. G. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 [DOI] [PubMed] [Google Scholar]
- 16. Engelhard V. H. (1994) Structure of peptides associated with class I and class II MHC molecules. Annu. Rev. Immunol. 12, 181–207 [DOI] [PubMed] [Google Scholar]
- 17. Regner M., Mullbacher A., Blanden R. V., Lobigs M. (2001) Immunogenicity of two peptide determinants in the cytolytic T-cell response to flavivirus infection: inverse correlation between peptide affinity for MHC class I and T-cell precursor frequency. Viral Immunol. 14, 135–149 [DOI] [PubMed] [Google Scholar]
- 18. Kotturi M. F., Scott I., Wolfe T., Peters B., Sidney J., Cheroutre H., von Herrath M. G., Buchmeier M. J., Grey H., Sette A. (2008) Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 181, 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Engelhard V. H., Brickner A. G., Zarling A. L. (2002) Insights into antigen processing gained by direct analysis of the naturally processed class I MHC associated peptide repertoire. Mol. Immunol. 39, 127–137 [DOI] [PubMed] [Google Scholar]
- 20. Zhang G. L., Ansari H. R., Bradley P., Cawley G. C., Hertz T., Hu X., Jojic N., Kim Y., Kohlbacher O., Lund O., Lundegaard C., Magaret C. A., Nielsen M., Papadopoulos H., Raghava G. P., Tal V. S., Xue L. C., Yanover C., Zhu S., Rock M. T., Crowe J. E., Panayiotou C., Polycarpou M. M., Duch W., Brusic V. (2011) Machine learning competition in immunology – Prediction of HLA class I binding peptides. J. Immunol. Methods 374, 1–4 [DOI] [PubMed] [Google Scholar]
- 21. Larsen M. V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. (2007) Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H., Lundegaard C., Nielsen M. (2009) Pan-specific MHC class I predictors: a benchmark of HLA class I pan-specific prediction methods. Bioinformatics 25, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bassani-Sternberg M., Barnea E., Beer I., Avivi I., Katz T., Admon A. (2010) Feature Article: soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl. Acad. Sci. U.S.A. 107, 18769–18776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berlin C., Kowalewski D. J., Schuster H., Mirza N., Walz S., Handel M., Schmid-Horch B., Salih H. R., Kanz L., Rammensee H. G., Stevanovic S., Stickel J. S. (2014) Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25. Dutoit V., Herold-Mende C., Hilf N., Schoor O., Beckhove P., Bucher J., Dorsch K., Flohr S., Fritsche J., Lewandrowski P., Lohr J., Rammensee H. G., Stevanovic S., Trautwein C., Vass V., Walter S., Walker P. R., Weinschenk T., Singh-Jasuja H., Dietrich P. Y. (2012) Exploiting the glioblastoma peptidome to discover novel tumor-associated antigens for immunotherapy. Brain 135, 1042–1054 [DOI] [PubMed] [Google Scholar]
- 26. Granados D. P., Sriranganadane D., Daouda T., Zieger A., Laumont C. M., Caron-Lizotte O., Boucher G., Hardy M. P., Gendron P., Cote C., Lemieux S., Thibault P., Perreault C. (2014) Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat. Commun. 5, 3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hassan C., Kester M. G., Ru A. H., Hombrink P., Drijfhout J. W., Nijveen H., Leunissen J. A., Heemskerk M. H., Falkenburg J. H., Veelen P. A. (2013) The human leukocyte antigen-presented ligandome of B lymphocytes. Mol. Cell. Proteomics 12, 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mommen G. P., Frese C. K., Meiring H. D., van Gaans-van den Brink J., de Jong A. P., van Els C. A., Heck A. J. (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc. Natl. Acad. Sci. U.S.A. 111, 4507–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haen S. P., Rammensee H. G. (2013) The repertoire of human tumor-associated epitopes–identification and selection of antigens and their application in clinical trials. Curr. Opin. Immunol. 25, 277–283 [DOI] [PubMed] [Google Scholar]
- 30. Hasenkamp J., Borgerding A., Wulf G., Uhrberg M., Jung W., Dingeldein S., Truemper L., Glass B. (2006) Resistance against natural killer cell cytotoxicity: analysis of mechanisms. Scand. J. Immunol. 64, 444–449 [DOI] [PubMed] [Google Scholar]
- 31. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111 011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 33. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 34. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, U359–U360 [DOI] [PubMed] [Google Scholar]
- 35. Wisniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 36. Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 37. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, prefractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 38. Kelstrup C. D., Young C., Lavallee R., Nielsen M. L., Olsen J. V. (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole Orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497 [DOI] [PubMed] [Google Scholar]
- 39. Karosiene E., Lundegaard C., Lund O., Nielsen M. (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64, 177–186 [DOI] [PubMed] [Google Scholar]
- 40. Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. (2008) NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 36, W509–W512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andreatta M., Lund O., Nielsen M. (2013) Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics 29, 8–14 [DOI] [PubMed] [Google Scholar]
- 42. Rapin N., Hoof I., Lund O., Nielsen M. (2010) The MHC motif viewer: a visualization tool for MHC binding motifs. Current Protocols in Immunology, John E. Coligan. [et al.] (Eds.). Chapter 18, Unit 18 17 [DOI] [PubMed] [Google Scholar]
- 43. Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanovic S. (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 [DOI] [PubMed] [Google Scholar]
- 44. Yewdell J. W., Bennink J. R. (1999) Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17, 51–88 [DOI] [PubMed] [Google Scholar]
- 45. de Verteuil D., Granados D. P., Thibault P., Perreault C. (2012) Origin and plasticity of MHC I-associated self peptides. Autoimmun. Rev. 11, 627–635 [DOI] [PubMed] [Google Scholar]
- 46. Falk K., Rotzschke O., Grahovac B., Schendel D., Stevanovic S., Gnau V., Jung G., Strominger J. L., Rammensee H. G. (1993) Allele-specific peptide ligand motifs of HLA-C molecules. Proc. Natl. Acad. Sci. U.S.A. 90, 12005–12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boisvert F. M., Ahmad Y., Gierlinski M., Charriere F., Lamont D., Scott M., Barton G., Lamond A. I. (2012) A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics 11, M111 011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 49. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamada A., Sasada T., Noguchi M., Itoh K. (2013) Next-generation peptide vaccines for advanced cancer. Cancer Science 104, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rammensee H. G., Singh-Jasuja H. (2013) HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev. Vaccines 12, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Overwijk W. W., Wang E., Marincola F. M., Rammensee H. G., Restifo N. P. (2013) Mining the mutanome: developing highly personalized Immunotherapies based on mutational analysis of tumors. J. Immunother. Cancer 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noguchi M., Sasada T., Itoh K. (2013) Personalized peptide vaccination: a new approach for advanced cancer as therapeutic cancer vaccine. Cancer Immunol. Immunother. 62, 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoshiyama K., Terazaki Y., Matsueda S., Shichijo S., Noguchi M., Yamada A., Mine T., Ioji T., Itoh K., Shirouzu K., Sasada T., Takamori S. (2012) Personalized peptide vaccination in patients with refractory nonsmall cell lung cancer. Int. J. Oncol. 40, 1492–1500 [DOI] [PubMed] [Google Scholar]
- 55. Walter S., Weinschenk T., Stenzl A., Zdrojowy R., Pluzanska A., Szczylik C., Staehler M., Brugger W., Dietrich P. Y., Mendrzyk R., Hilf N., Schoor O., Fritsche J., Mahr A., Maurer D., Vass V., Trautwein C., Lewandrowski P., Flohr C., Pohla H., Stanczak J. J., Bronte V., Mandruzzato S., Biedermann T., Pawelec G., Derhovanessian E., Yamagishi H., Miki T., Hongo F., Takaha N., Hirakawa K., Tanaka H., Stevanovic S., Frisch J., Mayer-Mokler A., Kirner A., Rammensee H. G., Reinhardt C., Singh-Jasuja H. (2012) Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 18, 1254–1261 [DOI] [PubMed] [Google Scholar]
- 56. Davitt K., Babcock B. D., Fenelus M., Poon C. K., Sarkar A., Trivigno V., Zolkind P. A., Matthew S. M., Grin'kina N., Orynbayeva Z., Shaikh M. F., Adler V., Michl J., Sarafraz-Yazdi E., Pincus M. R., Bowne W. B. (2014) The anti-cancer peptide, PNC-27, induces tumor cell necrosis of a poorly differentiated nonsolid tissue human leukemia cell line that depends on expression of HDM-2 in the plasma membrane of these cells. Ann. Clin. Lab. Sci. 44, 241–248 [PubMed] [Google Scholar]
- 57. Singh-Jasuja H., Emmerich N. P., Rammensee H. G. (2004) The Tubingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol. Immunother. 53, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Calis J. J., Maybeno M., Greenbaum J. A., Weiskopf D., De Silva A. D., Sette A., Kesmir C., Peters B. (2013) Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 9, e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vigneron N., Stroobant V., Van den Eynde B. J., van der Bruggen P. (2013) Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immun. 13, 15. [PMC free article] [PubMed] [Google Scholar]
- 60. Boegel S., Löwer M., Bukur T., Sahin U., Castle J. C. (2014) A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. Oncoimmunology 3, e954893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yaciuk J. C., Skaley M., Bardet W., Schafer F., Mojsilovic D., Cate S., Stewart C. J., McMurtrey C., Jackson K. W., Buchli R., Olvera A., Cedeno S., Plana M., Mothe B., Brander C., West J. T., Hildebrand W. H. (2014) Direct Interrogation of Viral Peptides Presented by the Class I HLA of HIV-Infected T Cells. J. Virol. 88, 12992–13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yewdell J. W. (2011) DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 32, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Apcher S., Daskalogianni C., Lejeune F., Manoury B., Imhoos G., Heslop L., Fahraeus R. (2011) Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc. Natl. Acad. Sci. U.S.A. 108, 11572–11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paul P., van den Hoorn T., Jongsma M. L., Bakker M. J., Hengeveld R., Janssen L., Cresswell P., Egan D. A., van Ham M., Ten Brinke A., Ovaa H., Beijersbergen R. L., Kuijl C., Neefjes J. (2011) A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell 145, 268–283 [DOI] [PubMed] [Google Scholar]
- 65. Kim Y., Yewdell J. W., Sette A., Peters B. (2013) Positional bias of MHC class I restricted T-cell epitopes in viral antigens is likely due to a bias in conservation. PLoS Comput. Biol. 9, e1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Juncker A. S., Larsen M. V., Weinhold N., Nielsen M., Brunak S., Lund O. (2009) Systematic characterization of cellular localisation and expression profiles of proteins containing MHC ligands. PloS One 4, e7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Deal R. B., Henikoff J. G., Henikoff S. (2010) Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qian M. X., Pang Y., Liu C. H., Haratake K., Du B. Y., Ji D. Y., Wang G. F., Zhu Q. Q., Song W., Yu Y., Zhang X. X., Huang H. T., Miao S., Chen L. B., Zhang Z. H., Liang Y. N., Liu S., Cha H., Yang D., Zhai Y., Komatsu T., Tsuruta F., Li H., Cao C., Li W., Li G. H., Cheng Y., Chiba T., Wang L., Goldberg A. L., Shen Y., Qiu X. B. (2013) Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 153, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Singh R. K., Kabbaj M. H., Paik J., Gunjan A. (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.