Abstract
RAD6 is a ubiquitin E2 protein with roles in a number of different biological processes. Here, using affinity purification coupled with mass spectrometry, we identify a number of new RAD6 binding partners, including the poorly characterized ubiquitin E3 ligases KCMF1 (potassium channel modulatory factor 1) and UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4), a protein that can bind N-end rule substrates, and which was recently linked to lysosome-mediated degradation and autophagy. NMR, combined with in vivo and in vitro interaction mapping, demonstrate that the KCMF1 C terminus binds directly to RAD6, whereas N-terminal domains interact with UBR4 and other intracellular vesicle- and mitochondria-associated proteins. KCMF1 and RAD6 colocalize at late endosomes and lysosomes, and cells disrupted for KCMF1 or RAD6 function display defects in late endosome vesicle dynamics. Notably, we also find that two different RAD6A point mutants (R7W and R11Q) found in X-linked intellectual disability (XLID) patients specifically lose the interaction with KCMF1 and UBR4, but not with other previously identified RAD6 interactors. We propose that RAD6-KCMF1-UBR4 represents a unique new E2-E3 complex that targets unknown N-end rule substrates for lysosome-mediated degradation, and that disruption of this complex via RAD6A mutations could negatively affect neuronal function in XLID patients.
RAD6 is a ubiquitin E2 conjugating protein that plays a number of important roles in eukaryotes, including histone H2B ubiquitylation, postreplication DNA damage repair and degradation of proteins via the N-end rule pathway (1, 2). This wide variety of functions is mediated via interactions with at least five different ubiquitin E3 ligases, which target the multipurpose E2 to a diverse array of substrates.
In mammals, RAD6 is encoded by two genes, UBE2A (the protein product of this gene has historically been referred to as RAD6A) and UBE2B (RAD6B). The human UBE2A gene is located on the X chromosome (Xq24) and the UBE2B locus maps to 5q31.1 (3). The human RAD6 proteins share ∼95% identity at the amino acid level (supplemental Fig. S1), and the two variants appear to play redundant roles in processes such as DNA damage repair (4). Both RAD6A and RAD6B are present in all tissues and cell lines examined (albeit at varying ratios), with highest mRNA levels measured in heart, testis, and brain (5).
Previous reports have identified a variety of UBE2A coding sequence mutations in X-linked intellectual disability (XLID)1 patients (6–10), and a recent study revealed that human and Drosophila cells deficient for RAD6 function display defects in mitochondrial turnover and vesicle dynamics (6). However, the consequences of the XLID RAD6A mutations at the molecular level have not been well characterized, and the proteins that couple RAD6 to these new functions had not been identified.
Here, we used an unbiased AP-MS (affinity purification coupled with mass spectrometry) approach to identify RAD6 interacting partners in human cells. Interestingly, we identify two new RAD6-associated E3 proteins, KCMF1 and UBR4, along with several additional interactors previously linked (via genetic and/or biochemical evidence) to intracellular vesicle trafficking and mitochondrial function. Using a combination of in vivo and in vitro binding assays combined with NMR spectroscopy, our data suggest that the KCMF1 protein recruits RAD6 to UBR4, a noncanonical N-recognin recently implicated in bulk lysosomal degradation and autophagy (11, 12). Consistent with this observation, knockdown of KCMF1 or RAD6A expression alters late endosome-lysosome vesicle trafficking. Finally, we demonstrate that two different RAD6A mutant proteins (R7W, R11Q) expressed in XLID patients maintain protein-protein interactions in vivo with all previously reported RAD6-associated E3s, but specifically lose the interaction with KCMF1 and UBR4. Together our data suggest that a RAD6-KCMF1-UBR4 complex targets unknown N-end rule substrates to the lysosome, and that this function is likely to be compromised in some XLID patients.
EXPERIMENTAL PROCEDURES
Expression Constructs
XLID UBE2A point mutations were introduced using the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Using standard cloning procedures, WT and mutant UBE2A (RAD6A), and full length KCMF1, were cloned into Flag-, GFP-, and Cherry-pcDNA5/FRT/TO vectors (all generous gifts from Dr. Anne-Claude Gingras; Lunenfeld-Tanenbaum Research Institute, Toronto, ON) for mammalian expression. Full-length cDNA clones for KCMF1 and UBR2 were obtained from the Mammalian Gene Collection (National Institute of Health). Full-length KCMF1, KCMF1 truncation mutants, and UBR2 (aa 1010–1267, encompassing the Basic Rich Region and RING domains) were subcloned into the pGEX-6p-1 vector (N-terminal GST tag; GE Healthcare, Piscataway, NJ) and/or the pET15b vector (N-terminal 6xHis tag; Novagen, San Diego, CA) for bacterial expression.
Stable Cell Lines
Tetracycline-inducible, Flag-, GFP-, and Cherry-tagged proteins were expressed in human Flp-In T-REx 293 cells (Invitrogen, Carlsbad, CA). Protein expression was induced by adding 1 μg/ml tetracycline to the culture medium (DMEM + 10% fetal calf serum) for 24 h. Cells were maintained at 37 °C in DMEM supplemented with 10% fetal bovine serum, 10 mm HEPES (pH 8.0), and 1% penicillin-streptomycin.
Transfection and siRNA
HEK 293 T-REx cells were transfected with the indicated plasmids using Lipofectamine and PLUS reagents, according to manufacturer's instructions (Invitrogen). For siRNA experiments, cells were grown to 50% confluence, then transfected with siRNA (Dharmacon ON-TARGET or RNAi duplex from IDT) directed against KCMF1, UBE2A, or a nontargeting siRNA (siRNA Control) at a final concentration of 10 nm. Transfections were carried out using Dharmafect #1 or Lipofectamine RNAiMax according to manufacturer's instructions. Transfection efficiencies of >85% were achieved as quantified using dsRED control (IDT). Cells were imaged 72 h post-transfection.
Affinity Purification
For MS analysis of interacting proteins, 6 × 150 cm2 dishes of subconfluent (80%) HEK 293 T-REx cells expressing the protein of interest were scraped into PBS, pooled, washed twice in 25 ml PBS, and collected by centrifugation at 1000 × g for 5 min at 4 °C. Cell pellets were stored at −80 °C until lysis. The cell pellet was weighed, and 1:4 pellet weight/lysis buffer (by volume) was added. Lysis buffer consisted of 50 mm HEPES-NaOH (pH 8.0), 100 mm KCl, 2 mm EDTA, 0.1% Nonidet P-40, 10% glycerol, 1 mm PMSF, 1 mm DTT, and 1:500 protease inhibitor mixture (Sigma-Aldrich, St. Louis, MO). On resuspension, cells were incubated on ice for 10 min, subjected to one additional freeze-thaw cycle, then centrifuged at 27,000 × g for 20 min at 4 °C. Supernatant was transferred to a fresh 15 ml conical tube, and 1:1000 benzonase nuclease (Novagen) plus 30 μl packed, pre-equilibrated Flag-M2 agarose beads (Sigma-Aldrich) were added. The mixture was incubated for 2 h at 4 °C with end-over-end rotation. Beads were pelleted by centrifugation at 1000 × g for 1 min and transferred with 1 ml of lysis buffer to a fresh centrifuge tube. Beads were washed once with 1 ml lysis buffer and twice with 1 ml ammonium bicarbonate (ammbic) rinsing buffer (50 mm ammbic, pH 8.0, 75 mm KCl). Elution was performed by incubating the beads with 150 μl of 125 mm ammonium hydroxide (pH >11). The elution step was repeated twice, and the combined eluate centrifuged at 15,000 × g for 1 min, transferred to a fresh centrifuge tube and lyophilized.
Mass Spectrometry
One-microgram of MS-grade TPCK trypsin (Promega, Madison, WI) dissolved in 70 μl of 50 mm ammbic (pH 8.3) was added to the Flag eluate and incubated at 37 °C overnight. The sample was lyophilized and brought up in 0.1% formic acid. LC analytical columns (75 μm inner diameter) and precolumns (100 μm inner diameter) were made in-house from fused silica capillary tubing from InnovaQuartz (Phoenix, AZ) and packed with 100 Å C18-coated silica particles (Magic, Michrom Bioresources, Auburn, CA). Peptides were subjected to nanoflow liquid chromatography – electrospray ionization – tandem mass spectrometry (nLC-ESI-MS/MS), using a 90 min reversed phase (10–40% acetonitrile, 0.1% formic acid) buffer gradient running at 250 nL/min on a Proxeon EASY-nLC pump in-line with a hybrid linear quadrupole ion trap (Velos LTQ) Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A parent ion scan was performed in the Orbitrap, using a resolving power of 60,000. Simultaneously, up to the forty most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using standard CID fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a 10 ppm window, exclusion list size 500) detected three times within 45 s were excluded from analysis for 30 s.
For protein identification, Thermo .RAW files were converted to the .mzXML format using Proteowizard (13), then searched against Human RefSeq Version 45 (containing 36,113 entries, appended with a reversed decoy database of equal size based on RefSeq v45) using the Mascot (14) and Comet (15) search engines. Search parameters specified a parent MS tolerance of 15 ppm and an MS/MS fragment ion tolerance of 0.4 Da, with up to two missed cleavages allowed for trypsin. Oxidation of methionine was allowed as a variable modification. Each AP was analyzed using at least two technical replicates. Statistical validation of peptide and protein identifications was performed using iProphet (16) as part of the Trans-Proteomic Pipeline (17). For each search, the iProphet probability at 1% error rate was used as a cutoff value to generate a matrix input file to upload to the CRAPome (v1.1). Thirty additional control datasets were selected from within the CRAPome (all FLAG-AP/MS data in HEK 293 cells). SAINTexpress options were; LowMode = 0, MinFold = 0, Normalize = 1, Virtual controls = 4, and Controls = All Controls. All raw data files have been uploaded to the Mass spectrometry Interactive Virtual Environment (MassIVE) repository (massive.ucsd.edu). Accession number: MSV000078734.
Protein Expression and Purification
GST- and His-tagged proteins were expressed in E. coli BL21 (DE3) cells (Invitrogen, Carlsbad, CA) grown in TB at 37 °C to an OD600 of ∼1.0, then induced with 0.5–1 mm IPTG at 4 °C overnight. Cells were harvested by centrifugation and stored at −80 °C until processing. For His-tagged proteins, cell pellets were thawed briefly on ice and resuspended in 50 mm Tris (pH 7.5), 500 mm NaCl, 10% glycerol, 5 mm imidazole, 1 mm benzamidine, and 1 mm PMSF. Proteins were purified using Talon beads (BD Biosciences, San Jose, CA) and eluted with buffer containing 500 mm imidazole. For GST-tagged proteins, thawed cell pellets were resuspended in phosphate buffered saline (PBS, pH 7.3), 0.1% Triton X-100, 10% glycerol, 2 mm DTT, 1 mm PMSF, and 1 mm benzamidine. Proteins were purified using MagneGSTTM glutathione beads (Promega, Madison, WI), and stored at 4 °C, or purified using Glutathione-Sepharose 4B beads (GE Healthcare) and eluted with 10 mm reduced glutathione, pH 8.0 in native buffer.
In Vitro Binding Assays
Two to four-micrograms GST-KCMF1 protein on MagneGSTTM glutathione beads were incubated with 2 μg purified His-tagged RAD6A for 1 h at 4 °C in 50 μl binding buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 2 mm DTT, 25 μm ZnCl2, and 0.05% Triton). Magnetic beads were washed six times with binding buffer, and bound proteins eluted with SDS sample buffer. Western blot analysis using monoclonal anti-Penta His antibody (Qiagen, Valencia, CA) was used to detect RAD6A. Primary antibody was visualized with the aid of secondary horseradish peroxidase (HRP) anti-mouse antibody (Bio-Rad, Hercules, CA) using enhanced chemiluminescence (ECL) (Immuno-Star HRP, Bio-Rad).
Autoubiquitylation Reactions
Autoubiquitylation reactions were performed in a volume of 30 μl in a buffer of 50 mm Tris, pH 8.0, 50 mm NaCl, 50 mm KCl, 10 mm MgCl2, 5 mm ATP, 0.1 mm DTT, with 5 μg ubiquitin (human recombinant; Boston Biochem), 3 μm E2 and E3, and 90 nm E1 (Human Recombinant, UBE1; Boston Biochem). After incubation at 30 °C for 3 h, reactions were stopped by the addition of SDS sample buffer and resolved on Criterion TGX Precast Gels 4–20% (BioRad). Ubiquitylated proteins were evaluated by Western blotting using a mouse monoclonal antibody directed against ubiquitin (P4G7, equivalent to P4D1; Covance, Emeryville, CA), HRP-conjugated goat anti-mouse secondary antibody (Bio-Rad) and Immuno-Star HRP ECL.
Immunofluorescence and Image Acquisition and Processing
Stable cell lines expressing tagged proteins or cultured HEK 293 cells were grown on coverslips, fixed with 4% formaldehyde for 15 min, and washed in PBS with 0.25% Triton X-100. In some cases, 10 μm MG132 (Calpain inhibitor IV, Z-Leu-Leu-Leu-CHO; American Peptide Company, Sunnyvale, CA) was added to cells for 12 h before fixation with 100% methanol at −20 °C for 10 min. Fixed cells were blocked in 5% bovine serum albumin (BSA) in PBS for 30 min before incubating in the indicated primary antibodies for 1 h at RT. Primary antibodies were used at the following concentrations: anti-KMCF1 polyclonal antibody (rabbit, 1:450; Sigma prestige antibodies), anti-Flag M2 antibody (1:500; Sigma-Aldrich), anti-EEA1 (1:200; Developmental Studies Hybridoma Bank (DSHB), Univ. Iowa), anti-GM130 (1:500; DSHB), anti-LAMP1 (1:100; DSHB). Secondary antibodies conjugated to Alexa 488 or Texas Red were used at 1:500, and incubated at RT for 1 h. After removing the antibody solution, cells were incubated with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min. After washing with PBS three times for 5 min each, coverslips were mounted with ProLong Gold Antifade (Thermo Fischer Scientific). Cells were imaged using PlanApo 60× oil lens, NA 1.40 on an Olympus FV1000 confocal microscope (zoom factor between 3–5; Olympus America, Melville, NY). Images were processed using the Volocity Viewer v.6 and assembled using Adobe Illustrator CS5 (Adobe Systems Inc.).
Live-cell Confocal Microscopy and Distance Measurements
For live-cell studies, cells were seeded in labtek chambers (Thermo Fisher), and stained with 1 μg/ml of Hoechst 33342 dye (Fisher Scientific) for 15 min at 37 °C. Cells were washed twice with warm PBS before incubating in phenol red-free DMEM media (GIBCO, Invitrogen). Cells were incubated 30 min in 50 nm Lysotracker Red DND-99 (Molecular Probes) in phenol red-free DMEM media, in a Chamlide TC stage incubator (LCI, Seoul, Korea) during acquisition (37 °C; 5% CO2). Minimum distance from Lysotracker-positive vesicles to the nucleus (see below) was calculated with the measurement tool of Volocity 6, and corresponds to the shortest distance between the centroid of the vesicles and the closest nuclear edge (Hoechst positive). To avoid inter-cellular measurement cells were seeded at low density and measurements performed with constant acquisition and calculation parameters (Confocal Cy3 channel: 300 ms exposure, fixed sensitivity and gain; constant size and intensity thresholds for both nuclei and vesicles). Data were analyzed using Student's t test.
For time-lapse live-cell video microscopy, one frame every ∼2 s was acquired, at maximum speed (confocal Cy3 channel exposure set to 100 ms; confocal GFP channel exposure set to 300 ms; auto-contrast on for both channels) for 2 mins. Movies are played at five frames per second.
NMR Spectroscopy and Data Analysis
E. coli BL21 (DE3, NEB) expressing His-tagged KCMF1 protein (aa 302–381) or His-tagged RAD6A protein (full length, aa 1–152) were grown at 37 °C to an A600 of ∼1.0, then induced with 0.5 mm IPTG at 4 °C overnight. For RAD6A-expressing cells, bacteria were grown in M9-defined medium supplemented with 15N-ammonium chloride (0.8 g/L) and/or 13C6-d-glucose (0.4 g/L) for 15N or 15N/13C-labeled RAD6A samples. Proteins were purified using HisPur Cobalt Resin (Fisher) affinity chromatography under native conditions and eluted with buffer containing 500 mm imidazole. The proteins were further purified by size exclusion chromatography using a HiLoad 26/60 Superdex-75 column (GE Healthcare). Proteins were monomeric in solution as determined by size exclusion chromatography. Final NMR samples were prepared in buffer containing 50 mm KH2PO4, pH 8.0, 200 mm NaCl, 5 mm β-mercaptoethanol, 1 mm benzamide, 0.5 mm PMSF, and 5% D2O. The RAD6A-KCMF1 complex was prepared for NMR by titrating aliquots of unlabeled KCMF1 into the 15N labeled RAD6A (500 μm) in molar ratios from 1:1 to 1:20, until no further changes in chemical shifts were detected in the 1H-15N HSQC spectrum. Weighted chemical shift displacements were calculated using: Δppm = ((ΔδHN)2 +(ΔδN/5)2)1/2. Assignment of RAD6A was conducted using the ABACUS approach (18) from NMR data collected at high resolution from nonlinearly sampled spectra, and processed using multidimensional decomposition (19, 20). All NMR spectra were recorded at 310K on Bruker Avance 600 and 800-MHz spectrometers equipped with cryoprobes. 1H, 13C, and 15N resonances were assigned using the ABACUS protocol (18) from peak lists derived from manually peak picked spectra using SPARKY. Figs. were prepared using PyMOL (DeLano Scientific).
RESULTS
RAD6 AP-MS Identifies a Set of Novel Interacting Partners
To more fully characterize the human RAD6 interactome, we employed affinity purification coupled with mass spectrometry (AP-MS). Tetracycline-inducible HEK 293 cell lines (T-REx) stably expressing Flag-tagged RAD6A or RAD6B were established. Following induction (1 μg/ml tetracycline, 24 h), the Flag-RAD6 proteins were immunopurified under nondenaturing conditions from two independent cell pools, and RAD6-interacting proteins were identified using nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry (nLC-ESI-MS/MS). Using the ProHits (21) system, MS data were analyzed with the Mascot (14) and Comet (15) database search algorithms, and the resulting peptide identifications subjected to iProphet (16) analysis. Finally, data were subjected to Significance Analysis of INTeractomes (SAINT) (22), using our own control runs (comprising untransfected cells, cells expressing the Flag-tag alone, and several unrelated Flag-tagged bait proteins) and the CRAPome database (23) to identify bona fide RAD6 interacting partners. Polypeptides identified in Flag-RAD6A or Flag-RAD6B AP-MS with an iProphet confidence value ≥0.95 and assigned a SAINT score ≥0.90 are reported in Table I (all MS data in supplemental Table S1).
Table I. A human RAD6 interactome. N-terminally Flag-tagged RAD6A, RAD6B, KCMF1 and UBR2 were expressed in Flp-In 293 T-REx cells. Cells were lysed under non-denaturing conditions and Flag-tagged proteins isolated using Flag-M2 agarose beads. Immunopurified proteins were eluted, digested with trypsin, and analyzed by nLC-ESI-MS/MS. Each cell pool represents a biological replicate. Two technical replicates were conducted for each pool. Spectral counts for each protein identification are indicated. All proteins highlighted here were detected with a >95% confidence level (iProphet) and assigned a SAINT score >0.95. Previously reported and new RAD6 interactors are indicated at left. KCMF1 interactions shared with RAD6A/B, and UBR2-specific interactions are indicated at right. *Most of the identified RAD6 peptides do not distinguish between RAD6A and RAD6B; for clarity all RAD6 peptides were pooled (and listed under RAD6A) for the Flag-KCMF1 and Flag-UBR2 analyses.
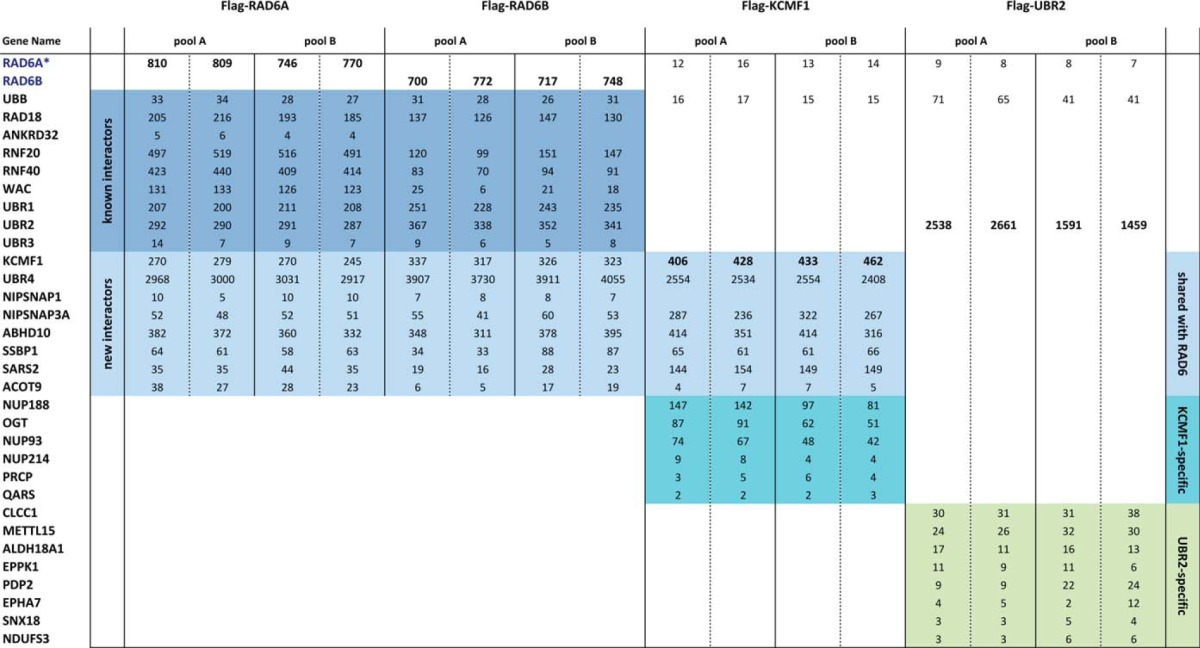
All of the previously reported RAD6-interacting E3 ligases (RAD18, RNF20/RNF40, UBR1, UBR2, and UBR3 (1, 2)) were detected in this analysis, highlighting the sensitivity of the approach. Also identified were the RNF20/RNF40 binding partner WW domain-containing adaptor with coiled-coil (WAC) (24), and the RAD18 interactor ANKRD32 (also known as BRCTx) (25).
Several previously unreported high-confidence interacting partners were also identified in this analysis, including: the putative ubiquitin E3 ligase KCMF1 (potassium channel modulatory factor 1); the ubiquitin N-recognin containing protein UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4); NIPSNAPs (4-nitrophenylphosphatase domain and non-neuronal SNAP25-like proteins) 1 and 3A; and several mitochondrial proteins, including the abhydrolase domain-containing protein ABHD10, the mitochondrial seryl-tRNA synthetase 2 (SARS2) and mitochondrial single stranded DNA binding protein 1 (SSBP1) (Table I and supplemental Table S1).
KCMF1 is an evolutionarily conserved 42kDa zinc-finger protein previously reported to function as a ubiquitin E3 ligase in vitro (26) with the E2 UBCH5 (UBE2D1). In vivo E2 partners for this protein had not been identified. KCMF1 mRNA expression is up-regulated in gastric cancer cells and epithelial tumors (26, 27), and previous reports have linked KCMF1 to cellular motility, invasion, and cancer progression (26–28). UBR4 (also known as p600) contains a ubiquitin recognin box but no HECT or RING domains (and is thus unlikely to act as an E3 ligase in isolation), and has been linked to membrane and cytoskeleton dynamics, neuronal migration (29) and survival (30), and brain, liver, and cardiac development (31). UBR4 interacts with the Dengue virus protein NS5, and both KCMF1 and UBR4 interact with the Human Papilloma Virus (HPV) E7 proteins across a wide range of HPV isotypes (32–34). Notably, like the other N-recognins, UBR4 can bind to N-end rule substrates, and was recently linked to lysosome-mediated degradation and autophagy (11, 12). Consistent with these findings, UBR4 is also a candidate gene for episodic ataxia (EA), a group of rare human neurological channelopathies characterized by a lack of balance and coordination (35), and D. melanogaster strains mutant for the UBR4 ortholog pushover display sluggishness, a lack of coordination, a defective escape response and deregulated synaptic vesicle fusion (36).
NIPSNAP1 is predominantly expressed in brain, spinal cord, liver, and kidney, is localized to mitochondria and postsynaptic densities (37), and appears to play a role in pain transmission (38). NIPSNAP3A is associated with phagosomes (39), mitochondria (40) and other cellular membrane components (41), and is a host cell target protein for the Salmonella virulence protein SpiC (39). SpiC-mediated inactivation of NIPSNAP3A function is required to prevent the fusion of Salmonella-containing vesicles with lysosomes.
An unbiased RAD6A and RAD6B AP-MS analysis thus identified two ubiquitin E3 ligases not previously linked with this E2 protein, along with several novel interacting partners previously linked to mitochondrial function, vesicle trafficking, and lysosome-mediated degradation.
KCMF1 Links RAD6 to UBR4 and Vesicle- and Mitochondria-Associated Proteins
To begin to characterize these novel RAD6 interactions, we stably expressed Flag-tagged KCMF1 in 293 T-REx cells and conducted AP-MS, as above. Notably, Flag-KCMF1 AP-MS isolated RAD6, UBR4, and all of the other newly identified RAD6 interacting partners (in addition to O-linked N-acetyl glucosamine transferase (OGT) and several nucleoporins not observed in RAD6 AP-MS), but did not co-isolate any of the previously reported RAD6 binding proteins (Table I and supplemental Table S1). By comparison, Flag-UBR2 AP-MS isolated RAD6 and several other high-confidence interactors (representing putative N-end rule targets), but did not co-isolate KCMF1, UBR4, the NIPSNAPs, or mitochondrial proteins (Table I and supplemental Table 1). KCMF1 and UBR4 thus appear to be components of a new, unique RAD6-containing protein complex.
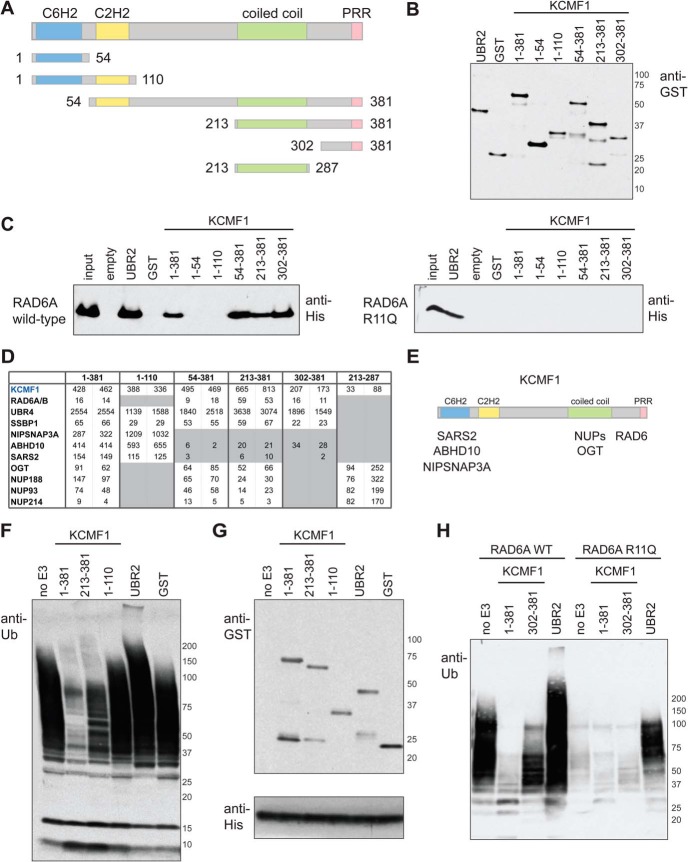
The previously reported functions of the KCMF1 interacting proteins suggested that KCMF1 could play a role in recruiting RAD6 to proteins associated with intracellular membrane-bound structures. To test this hypothesis, glutathione-S-transferase (GST)-tagged KCMF1 protein fragments (Fig. 1A, 1B), along with the previously characterized UBR2 RAD6-binding region (aa 1010–1267; the basic residue-rich domain, or BRR (42)) and GST alone (as positive and negative controls, respectively) were tested for RAD6 binding. Both UBR2 BRR and the full-length (aa 1–381) KCMF1 proteins bound to purified RAD6A in vitro (Fig. 1C, left panel). Although the N-terminal region of KCMF1 (fragments consisting of aa 1–54 and 1–110) displayed no RAD6 binding activity, KCMF1 fragments containing the C terminus (aa 54–381, 213–381, and 302–381) bound to RAD6 as efficiently as the full-length polypeptide (Fig. 1C, left panel).
Fig. 1.
In vitro and in vivo interaction mapping of KCMF1. A, KCMF1 domain structure map, and deletion mutants used here for in vitro and in vivo binding assays. Amino acid residues assigned to each domain based on the Jpred3 secondary structure prediction program (49). PRR, proline-rich region. B, Recombinant GST alone, GST-UBR2 (BRR + RING domain fragment) and GST-tagged KCMF1 proteins (as indicated) were expressed in E. coli, purified, subjected to SDS-PAGE and analyzed via anti-GST Western blotting. C, In vitro RAD6A binding assay. Recombinant GST-KCMF1 (full-length and truncation mutants, as indicated), GST-UBR2 (BRR+RING domain) and GST alone were incubated with recombinant full-length His-RAD6A for 1h at 4 °C, followed by extensive washing. Proteins were eluted with Laemmli buffer, resolved by SDS-PAGE and analyzed by immunoblotting with an antibody directed against 6xHis; (left) wild type RAD6A, (right) RAD6A R11Q mutant. D, In vivo KCMF1 interaction mapping. Flag-tagged full-length KCMF1 and truncation mutants were expressed in 293 T-REx cells and subjected to AP-MS. Two cell pools were generated independently. For each pool, two technical runs were conducted, and maximum spectral counts for each protein identification are indicated. All proteins highlighted here were detected with a >95% iProphet confidence level and SAINT score >0.95. Spectral count values highlighted in gray are ≤ 10% of those observed for full length (1–381) KCMF1. E, Binding regions for the indicated KCMF1 interactors, as determined by AP-MS. F, Autoubiquitylation reactions, performed with recombinant human E1 (90 nm), ubiquitin (5 ug) and His-RAD6A (3 μm) alone, or in the presence of equimolar amounts of GST alone, GST-UBR2 (BRR+RING), or GST-tagged KCMF1 protein fragments (as indicated), for 3 h at 30 °C. Reactions were subjected to SDS-PAGE and anti-ubiquitin Western blotting. G,, anti-GST (top panel) and anti-His (bottom panel) Western blotting of the autoubiquitylation reactions shown in F. H, Autoubiquitylation assays as in F, conducted with wild type and R11Q RAD6A proteins.
The same set of KCMF1 protein domains (with the exception of fragment 1–54, which did not express well in our cell system) was also stably expressed as N-terminal Flag fusions in HEK 293 T-REx cells, and subjected to AP-MS (Fig. 1D and supplemental Table S2). Consistent with our in vitro data, an N-terminal KCMF1 fragment (aa 1–110) displayed no RAD6 binding in vivo, whereas Flag-fusion proteins containing the KCMF1 C terminus (fragments 54–381, 213–381, and 302–381) retained RAD6 binding. The KCMF1 C terminus is thus necessary and sufficient for RAD6 binding both in vitro and in vivo.
Notably, the KCMF1 N-terminal fragment (aa 1–110) retained interactions with UBR4, NIPSNAP3A, ABHD10, and SARS2 (Fig. 1D). Deletion of the N-terminal C6H2 (ZZ) domain (aa 54–381) resulted in a specific loss of ABHD10, SARS2, and NIPSNAP binding, and a Flag-tagged KCMF1 coiled-coil (aa 213–287) fragment coprecipitated only OGT and nucleoporins. The interactions with NIPSNAP3A, ABHD10, and SARS2 are thus dependent on the KCMF1 N terminus, and the coiled-coil region is required to mediate interactions with OGT and nucleoporins. UBR4 and SSBP1 appear to interact with multiple regions of KCMF1, as these proteins were detected with both N- and C-terminal fragments. Together, these data delimit interaction domains for a number of KCMF1 binding proteins, and suggest that KCMF1 is required to recruit vesicle- and mitochondria-associated proteins to RAD6 (Fig. 1E).
KCMF1 Modulates RAD6 Activity
To determine whether KCMF1 affects RAD6 activity, we conducted in vitro autoubiquitylation reactions, as in (43). As previously demonstrated (44), RAD6 can synthesize polyubiquitin oligomers in vitro even in the absence of an E3 (Fig. 1F–1H). Nevertheless, the addition of the recombinant UBR2 BRR fragment to the reactions further stimulated RAD6 ubiquitin chain-building activity under these conditions (Fig. 1F–H). In contrast, the addition of full-length (1–381) KCMF1 had a clear inhibitory effect (Fig. 1F–H). Inclusion of GST alone or the KCMF1 N terminus (aa 1–110) had no effect on RAD6 activity. The addition of equimolar amounts of the KCMF1 C terminus (aa 213–381 or 302–381) also inhibited RAD6A autoubiquitylation activity, albeit not to the same extent as the full-length protein, suggesting that other regions of KCMF1 probably also play a role in this process.
Characterization of the KCMF1-RAD6 Interaction
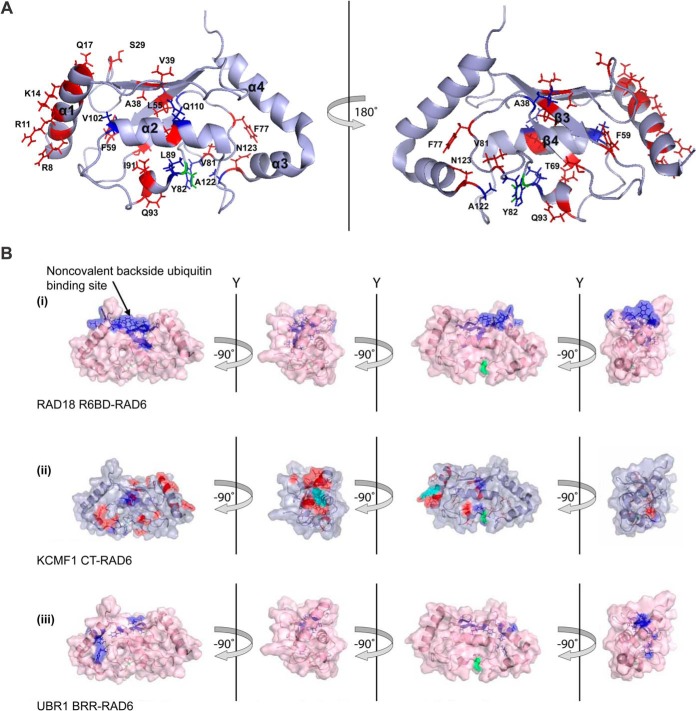
To better understand the KCMF1-RAD6 interaction, a series of 1H-15N heteronuclear single quantum coherence (HSQC) spectra of 15N-labeled RAD6A was recorded, to which increasing amounts of an unlabeled KCMF1 C-terminal fragment (aa 302–381) were added (Fig. 2A and supplemental Fig. S2). Addition of the KCMF1 C terminus effected chemical shift perturbations (CSPs) at: 1) the RAD6A N-terminal α1 helix, primarily at residues R7, R8, R11, F13, K14, and Q17 (all located at the canonical E3-binding surface), and; 2) a region near the catalytic Cys residue (C88), on both the α2 and 310 helices. In this region, L89, V102, Q110, and A122 displayed a marked decrease in peak intensity (>90%) in response to the addition of KCMF1.
Fig. 2.
Further analysis of the KCMF1-RAD6 interaction. A, Significant (>0.04) chemical shift perturbations (red) and resonances that disappear (blue) upon KCMF1 CT addition mapped onto the RAD6 structure PDB 2Y4W (50). Catalytic residue Cys88 highlighted in green. B, Binding sites for RAD18, UBR1 and KCMF1 protein fragments on RAD6. (i) RAD6 residues implicated in the RAD18 interaction (44). Residues that displayed significant CSPs when 15N-RAD6 was titrated with a RAD6-binding domain (R6BD) synthetic peptide derived from RAD18 are indicated in blue. Cys88 in green. (ii) RAD6 residues involved in the KCMF1 CT interaction. Red highlighted residues display significant CSPs, residues with decreasing intensity in blue. (iii) RAD6 residues implicated in the interaction with UBR1. Residues displaying significant CSPs when 15N-RAD6 was titrated with a basic residue-rich region (BRR) synthetic peptide derived from UBR1 (44) are shown in blue. CSPs for RAD6A were mapped onto the RAD6B structure (PDB 2Y4W).
Interestingly, KCMF1 is not the first example of an “inhibitory” RAD6-associated protein. The E3 ligase RAD18 (involved in DNA damage repair) similarly affects RAD6 ubiquitin chain building activity, biasing it toward monoubiquitylation of the substrate protein PCNA via an interaction with the noncovalent ubiquitin-binding “backside” of the E2 to inhibit ubiquitin chain formation (44). However, because the KCMF1 C terminus makes no detectable contacts with the RAD6 backside, it appears to inhibit RAD6 polyubiquitylation activity via a mechanism that differs from that of RAD18 (Fig. 2B).
Several proteins involved in intracellular vesicle trafficking possess ubiquitin interacting motifs, and are thought to bind monoubiquitylated substrates to regulate intracellular vesicle trafficking (45). Like RAD18, KCMF1 may thus bias RAD6 toward monoubiquitylation of cytoplasmic substrates to target them to lysosome-directed vesicles. Further study will be required to test this model.
RAD6A XLID Mutants Lose the Interaction with KCMF1 and UBR4
Several different UBE2A (RAD6A) coding sequence mutations have been identified in patients with X-linked intellectual disability (XLID) (6–10). To explore the role of these mutations in RAD6 protein–protein interactions, we expressed four different RAD6A point mutants in the HEK 293 T-REx system and conducted AP-MS, as above (Table II; Supplemental Table S3). A RAD6A Q128X nonsense mutant appeared to be unstable in our cell system (supplemental Table S3). A RAD6A G23R missense mutant was expressed at WT RAD6A levels, and displayed no apparent change in protein–protein interactions (supplemental Table S3; n.b. this mutation was previously shown to negatively affect RAD6 activity (44)). Notably, however, although both the RAD6A R7W and R11Q proteins were expressed at levels comparable to the WT polypeptide, and maintained interactions with RAD18, RNF20, RNF40, UBR1, and UBR2 in vivo, both of these mutant polypeptides displayed a dramatic decrease in the interactions with KCMF1 and UBR4 (Table II).
Table II. RAD6A XLID mutations disrupt specific protein-protein interactions. Flag-tagged RAD6A WT and mutant proteins (R7W, R11Q) were expressed in 293 T-REx cells, and interacting proteins identified via AP-MS, as above. Each column lists maximum spectral counts from two technical runs for each cell pool. Interactions maintained (blue) or lost (grey) in the mutant proteins are indicated.
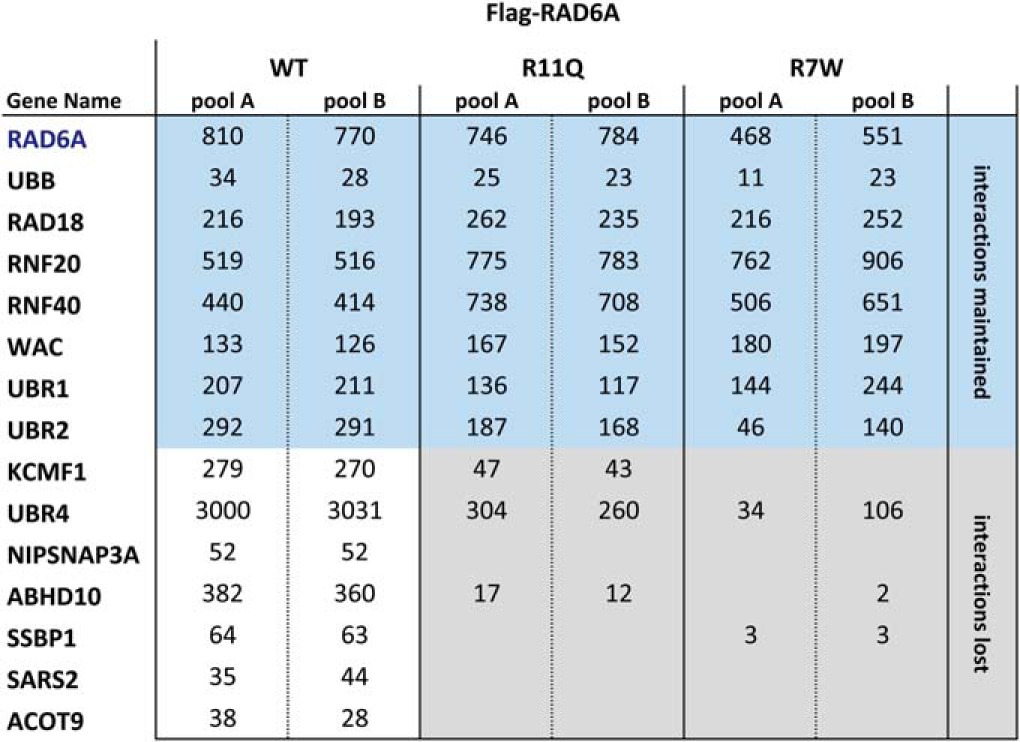
Consistent with these data, the UBR2 BRR fragment, but not KCMF1, bound to a recombinant RAD6A R11Q protein in vitro (Fig. 1C, right panel). The RAD6A R11Q mutant also displayed lower intrinsic polyubiquitylation activity in vitro, as compared with the wild type protein (Fig. 1H). However, addition of the UBR2 BRR was able to elicit a substantial increase in RAD6 R11Q ubiquitin chain-building activity (Fig. 1H), whereas the KCMF1 protein had no apparent effect. Together, these data indicate that two different UBE2A (RAD6A) mutations found in XLID patients (R7W and R11Q), and which map to the KCMF1 binding site as determined by NMR (Fig. 2A), negatively affect binding to KCMF1 and UBR4.
KCMF1 and RAD6 are Localized to Late Endosomes and Lysosomes
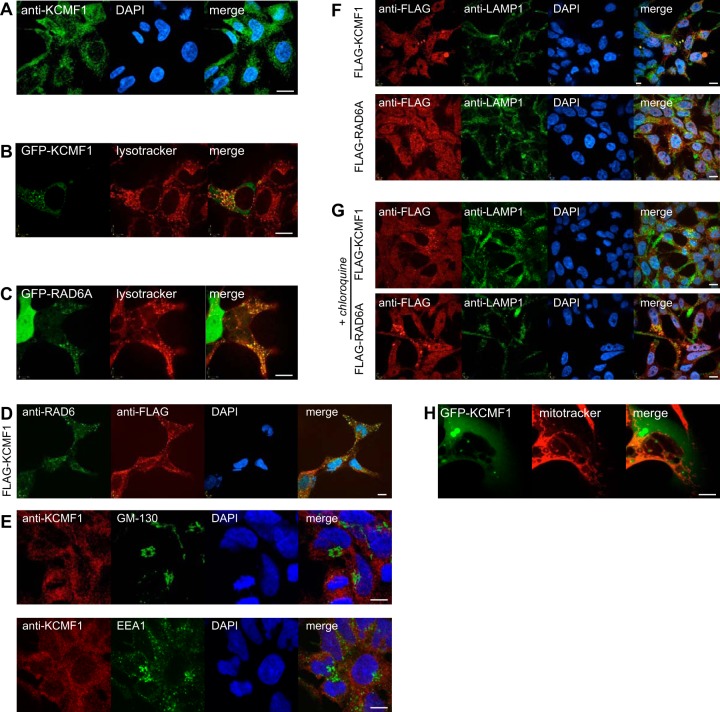
Previous immunofluorescence studies of human pancreas slices localized KCMF1 to the cytoplasm in normal cells (27). Consistent with this report, immunofluorescence using a commercially available antibody (Sigma Aldrich) directed against endogenous KCMF1 revealed cytoplasmic puncta in 293 T-REx cells (Fig. 3A). Flag- and GFP-tagged KCMF1 proteins displayed a similar localization (e.g. Fig. 3B, 3F). GFP-KCMF1 also partially colocalized with the late endosome/lysosome marker LysoTracker Red in live cells (Fig. 3B), and when GFP-KCMF1 localization was followed in real time, KCMF1-associated vesicles were observed to fuse with structures marked with LysoTracker Red (supplemental Video S1).
Fig. 3.
KCMF1 links RAD6 to vesicle dynamics. A, Immunofluorescence was conducted in untransfected HEK 293 cells using an antibody directed against KCMF1. DNA stained with DAPI (blue). All scale bars 10 μm. B, Colocalization of GFP-KCMF1 with Lysotracker red. C, Colocalization of GFP-RAD6A with Lysotracker red. D, Colocalization of endogenous RAD6 (green) with Flag-KCMF1 (red) in 293 T-REx cells. E, Endogenous KCMF1 does not colocalize with GM-130 (top) or EEA1 (bottom). F, Colocalization of Flag-KCMF1 (top panel, red) and Flag-RAD6A (bottom panel, red) with the late endosome/lysosome marker LAMP1 (green). G, cells treated with the lysosome inhibitor chloroquine were analyzed as in F. H, GFP-KCMF1 does not colocalize with the mitochondrial marker Mitotracker (red).
Consistent with previous reports (e.g. (46)), GFP-tagged RAD6A displayed both nuclear and cytoplasmic localization (Fig. 3C). GFP-RAD6A also partially colocalized with Lysotracker-marked vesicles (Fig. 3C) and Flag-KCMF1 (Fig. 3D). No obvious differences in localization were observed between the wild type RAD6A protein and the GFP-RAD6A R11Q mutant, suggesting that the interaction with KCMF1-UBR4 may not be strictly required for its presence at late endosomes (data not shown). As observed with KCMF1, in a real-time live cell analysis GFP-RAD6A-positive vesicles also fused with LysoTracker-marked structures (supplemental Video S2).
Immunofluorescence microscopy revealed no colocalization of KCMF1 or RAD6A with markers of the Golgi apparatus (GM-130) or early endosomes (EEA1; Fig. 3E), but both KCMF1 and RAD6A partially colocalized with the late endosome and lysosome marker LAMP1 (Fig. 3F), and like LAMP1 displayed increased cytoplasmic puncta in the presence of the lysosome inhibitor chloroquine (Fig. 3G).
GFP-KCMF1 did not colocalize with the mitochondrial marker MitoTracker Red CMXRos (Fig. 3H). Thus, even though KCMF1 and RAD6 clearly interact with polypeptides previously assigned a function in mitochondria, they do not appear to localize to mitochondria themselves (at least to steady-state levels that we can detect under normal growing conditions). These observations are not inconsistent with RAD6-KCMF1 targeting some mitochondrial proteins for lysosome-mediated degradation, but further study will be required to understand the role of RAD6-KCMF1 in mitochondrial function.
KCMF1 and RAD6 Inactivation Affect Vesicle Dynamics
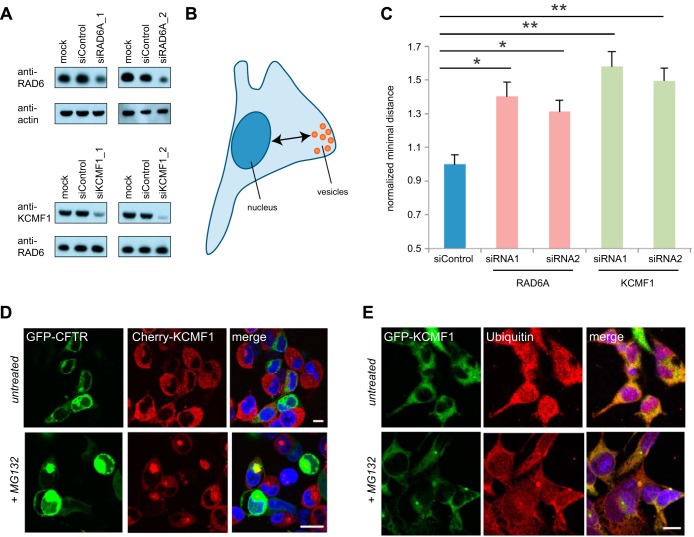
To begin to explore the role of KCMF1 in vesicle function, HEK 293 cells were transfected with siRNAs directed against KCMF1 or RAD6A (Fig. 4A; n.b. this siRNA is not expected to target the UBE2B mRNA. The RAD6B protein could thus make up all or part of the residual RAD6 signal in these Western blots). At 72 h post-transfection, Lysotracker-marked structures were analyzed for size distribution and intracellular localization. Although no significant differences in vesicle size were detected, cells deficient for either KCMF1 or RAD6A expression displayed a significant increase in the average distance of vesicles from the nucleus, as compared with untreated cells or cells transfected with control siRNA (p < 0.02, Fig 4B, C, and supplemental Table S4). Knockdown of either KCMF1 or RAD6A thus has a striking effect on vesicle trafficking dynamics in human cells.
Fig. 4.
Knockdown of KCMF1 or RAD6A alters late endosomal vesicle dynamics. A, siRNA-mediated knockdown of RAD6A (top) or KCMF1 (bottom), followed by Western blotting. Loading controls as indicated. B, C, KCMF1 or RAD6A knockdown leads to vesicular trafficking defects. Cells were transfected with the indicated siRNA (or siControl). Forty-eight hours post-transfection, cells were stained with Lysotracker red and minimum distance to the nucleus measured (as in diagram, B). Distances normalized to siControl ± S.E. n = 12,000–17,000 vesicles (from 100–130 cells) in each of three experiments. * p < 0.025; ** p < 0.01 D, KCMF1 colocalizes with GFP-CFTR ΔF508. T-REx 293 cells stably expressing cherry-KMCF1 were transfected with GFP-CFTR ΔF508. Twenty four-hours after transfection, cells were treated with 10 μm MG132 for 12 h, then imaged using live-cell confocal microscopy. E, KCMF1 colocalizes with ubiquitin upon MG132 treatment. T-REx 293 cells stably expressing GFP-KMCF1 were treated with 10 μm MG132 for 12 h, then fixed and stained with antibodies directed against GFP and ubiquitin.
KCMF1, but not RAD6, is Associated with Aggresomes
Finally, we noted that under standard culture conditions GFP-KCMF1 and Flag-KCMF1 were found in perinuclear inclusions in a small percentage of cells in culture, and that addition of the proteasome inhibitor MG132 significantly increased these KCMF1-containing inclusions (data not shown). The intracellular location and size of these KCMF1-positive structures suggested that they could be aggresomes. Consistent with this hypothesis, KCMF1 colocalized with the aggresome marker GFP-CFTRΔF508, and with ubiquitin, in these structures (Fig. 4D, E). Unlike KCMF1, RAD6 did not appear to localize to aggresomes (data not shown), suggesting that the RAD6-KCMF1 interaction is disrupted prior to aggresome delivery. These data are consistent with an earlier report indicating that UBR4 is also targeted to autophagic vesicles, presumably escorting its N-end rule targets for lysosome-mediated degradation (11).
DISCUSSION
Here, using standard AP-MS, NMR spectroscopy and in vivo and in vitro interaction mapping, we demonstrate that the poorly characterized ubiquitin E3 ligase KCMF1 binds directly to RAD6, bridging this multifunctional E2 protein with another E3 protein, UBR4, which was recently implicated in bulk lysosome-mediated degradation and autophagy (11, 12). KCMF1 and RAD6 colocalize at late endosomes and lysosomes, and cells disrupted for KCMF1 or RAD6 function display defects in vesicle dynamics. Notably, we also find that two different RAD6A point mutants (R7W and R11Q) found in X-linked intellectual disability (XLID) patients specifically lose the interactions with KCMF1 and UBR4, but not with other previously identified RAD6 interactors. We thus identify a new set of RAD6 interacting partners linked to lysosome-mediated degradation, and highlight specific protein–protein interactions that are lost in some RAD6 XLID mutant proteins.
A New RAD6-Containing Protein Complex
RAD6 was previously demonstrated to interact with the ubiquitin N-recognin domain-containing E3 proteins UBR1, UBR2 and UBR3, to effect the ubiquitylation of N-end rule substrates (2, 12). As expected, these interactions were successfully recapitulated in both our Flag-RAD6A and Flag-RAD6B AP-MS analyses (Table I). Here we report for the first time that RAD6 also interacts with UBR4. Like the other N-recognins, UBR4 can bind to N-end rule substrates, and was recently linked to autophagy and lysosome-mediated degradation (11, 47). Notably, however, the molecular details of this link were not previously known.
Despite its large size, UBR4 does not contain recognizable RING or HECT domains, and thus, is not likely to function as a typical E3 ligase in isolation. Here we find that, unlike the other UBR family members, the UBR4 interaction with RAD6 appears to be dependent on an unrelated protein, the RING-containing polypeptide KCMF1. Taking all of these data into account, we propose that RAD6, KCMF1, and UBR4 comprise a unique E2-E3 protein complex that targets unknown N-end rule substrates for lysosomal degradation.
Defects in lysosomal function and autophagy have been linked to neurodevelopmental and neurodegenerative disorders (48). Disruption of a RAD6-KCMF1-UBR4 complex via RAD6A mutations (such as R7W and R11Q) could thus lead to a buildup of toxic proteins and/or organelles to negatively affect neuronal function in XLID patients. Further study will be required to test this hypothesis.
Supplementary Material
Acknowledgments
We thank Dr. Anne-Claude Gingras for assistance with MS data analysis and critical reading of the manuscript.
Footnotes
Author contributions: J.H.H., L.K., C.A., and B.R. designed research; J.H.H., L.K., E.C., T.S., and J.W. performed research; L.K., T.S., C.A., and B.R. analyzed data; B.R. wrote the paper.
* This research was funded by Canadian Institutes of Health Research (CIHR) grant MOP 119289 to BR, and a Natural Sciences and Engineering Research Council (NSERC) and Canadian Cancer Society Research Institute (CCSRI) grant 2011 700932 to CA. JH and TS were funded by CIHR student fellowships. CA holds the Canada Research Chair in Structural Genomics, and BR holds the Canada Research Chair in Proteomics and Molecular Medicine.
 This article contains supplemental Tables S1 to S4, Figs. S1 and S2, and Videos S1 and S2.
This article contains supplemental Tables S1 to S4, Figs. S1 and S2, and Videos S1 and S2.
1 The abbreviations used are:
- XLID
- X-linked intellectual disability
- ABHD10
- abhydrolase domain-containing protein 10
- AP-MS
- affinity purification coupled with mass spectrometry
- BRR
- basic residue-rich domain
- EA
- episodic ataxia
- HECT
- Homologous to E6-AP Carboxyl Terminus
- HSQC
- 1H-15N heteronuclear single quantum coherence
- KCMF1
- potassium channel modulatory factor 1
- NIPSNAP
- 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein
- OGT
- O-linked N-acetyl glucosamine transferase
- RING
- really interesting new gene
- SAINT
- Significance Analysis of INTeractomes
- SARS2
- mitochondrial seryl-tRNA synthetase 2
- SSBP1
- mitochondrial single stranded DNA binding protein 1
- UBR4
- ubiquitin N-recognin domain-containing E3 ligase 4
- WAC
- WW domain-containing adaptor with coiled-coil.
REFERENCES
- 1. Game J. C., Chernikova S. B. (2009) The role of RAD6 in recombinational repair, checkpoints, and meiosis via histone modification. DNA Repair 8, 470–482 [DOI] [PubMed] [Google Scholar]
- 2. Tasaki T., Sriram S. M., Park K. S., Kwon Y. T. (2012) The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koken M. H., Smit E. M., Jaspers-Dekker I., Oostra B. A., Hagemeijer A., Bootsma D., Hoeijmakers J. H. (1992) Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24-q25 and 5q23-q31. Genomics 12, 447–453 [DOI] [PubMed] [Google Scholar]
- 4. Roest H. P., Baarends W. M., de Wit J., van Klaveren J. W., Wassenaar E., Hoogerbrugge J. W., van Cappellen W. A., Hoeijmakers J. H., Grootegoed J. A. (2004) The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24, 5485–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koken M. H., Hoogerbrugge J. W., Jasper-Dekker I., de Wit J., Willemsen R., Roest H. P., Grootegoed J. A., Hoeijmakers J. H. (1996) Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173, 119–132 [DOI] [PubMed] [Google Scholar]
- 6. Haddad D. M., Vilain S., Vos M., Esposito G., Matta S., Kalscheuer V. M., Craessaerts K., Leyssen M., Nascimento R. M., Vianna-Morgante A. M., De Strooper B., Van Esch H., Morais V. A., Verstreken P. (2013) Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol. Cell 50, 831–843 [DOI] [PubMed] [Google Scholar]
- 7. de Leeuw N., Bulk S., Green A., Jaeckle-Santos L., Baker L. A., Zinn A. R., Kleefstra T., van der Smagt J. J., Vianne Morgante A. M., de Vries B. B., van Bokhoven H., de Brouwer A. P. (2010) UBE2A deficiency syndrome: mild to severe intellectual disability accompanied by seizures, absent speech, urogenital, and skin anomalies in male patients. Am. J. Med. Genet. A. 152A, 3084–3090 [DOI] [PubMed] [Google Scholar]
- 8. Budny B., Badura-Stronka M., Materna-Kiryluk A., Tzschach A., Raynaud M., Latos-Bielenska A., Ropers H. H. (2010) Novel missense mutations in the ubiquitination-related gene UBE2A cause a recognizable X-linked mental retardation syndrome. Clin. Genet. 77, 541–551 [DOI] [PubMed] [Google Scholar]
- 9. Honda S., Orii K. O., Kobayashi J., Hayashi S., Imamura A., Imoto I., Nakagawa E., Goto Y., Inazawa J. (2010) Novel deletion at Xq24 including the UBE2A gene in a patient with X-linked mental retardation. J. Hum. Genet. 55, 244–247 [DOI] [PubMed] [Google Scholar]
- 10. Nascimento R. M., Otto P. A., de Brouwer A. P., Vianna-Morgante A. M. (2006) UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am. J. Hum. Genet. 79, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tasaki T., Kim S. T., Zakrzewska A., Lee B. E., Kang M. J., Yoo Y. D., Cha-Molstad H. J., Hwang J., Soung N. K., Sung K. S., Kim S. H., Nguyen M. D., Sun M., Yi E. C., Kim B. Y., Kwon Y. T. (2013) UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc. Natl. Acad. Sci. U.S.A. 110, 3800–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tasaki T., Mulder L. C., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25, 7120–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 15. Eng J. K., Jahan T. A., Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 16. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111 007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller A., Shteynberg D. (2011) Software pipeline and data analysis for MS/MS proteomics: the trans-proteomic pipeline. Methods Mol. Biol. 694, 169–189 [DOI] [PubMed] [Google Scholar]
- 18. Lemak A., Gutmanas A., Chitayat S., Karra M., Fares C., Sunnerhagen M., Arrowsmith C. H. (2011) A novel strategy for NMR resonance assignment and protein structure determination. J. Biomol. NMR 49, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gutmanas A., Jarvoll P., Orekhov V. Y., Billeter M. (2002) Three-way decomposition of a complete 3D 15N-NOESY-HSQC. J. Biomol. NMR 24, 191–201 [DOI] [PubMed] [Google Scholar]
- 20. Orekhov V. Y., Ibraghimov I., Billeter M. (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J. Biomol. NMR 27, 165–173 [DOI] [PubMed] [Google Scholar]
- 21. Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z. Y., Breitkreutz B. J., Ding Y., Colwill K., Pasculescu A., Pawson T., Wrana J. L., Nesvizhskii A. I., Raught B., Tyers M., Gingras A. C. (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28, 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu G., Zhang J., Choi H., Lambert J. P., Srikumar T., Larsen B., Nesvizhskii A. I., Raught B., Tyers M., Gingras A. C. (2012) Using ProHits to store, annotate, and analyze affinity purification-mass spectrometry (AP-MS) data. Curr. Protoc. Bioinformatics Chapter 8, Unit 8 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang F., Yu X. (2011) WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol. Cell 41, 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams D. J., van der Weyden L., Gergely F. V., Arends M. J., Ng B. L., Tannahill D., Kanaar R., Markus A., Morris B. J., Bradley A. (2005) BRCTx is a novel, highly conserved RAD18-interacting protein. Mol. Cell. Biol. 25, 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jang J. H. (2004) FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett. 578, 21–25 [DOI] [PubMed] [Google Scholar]
- 27. Beilke S., Oswald F., Genze F., Wirth T., Adler G., Wagner M. (2010) The zinc-finger protein KCMF1 is overexpressed during pancreatic cancer development and downregulation of KCMF1 inhibits pancreatic cancer development in mice. Oncogene 29, 4058–4067 [DOI] [PubMed] [Google Scholar]
- 28. Kreppel M., Aryee D. N., Schaefer K. L., Amann G., Kofler R., Poremba C., Kovar H. (2006) Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing's sarcoma cells. Oncogene 25, 2795–2800 [DOI] [PubMed] [Google Scholar]
- 29. Shim S. Y., Wang J., Asada N., Neumayer G., Tran H. C., Ishiguro K., Sanada K., Nakatani Y., Nguyen M. D. (2008) Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J. Neurosci. 28, 3604–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belzil C., Neumayer G., Vassilev A. P., Yap K. L., Konishi H., Rivest S., Sanada K., Ikura M., Nakatani Y., Nguyen M. D. (2013) A Ca2+-dependent mechanism of neuronal survival mediated by the microtubule-associated protein p600. J. Biol. Chem. 288, 24452–24464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakaya T., Ishiguro K. I., Belzil C., Rietsch A. M., Yu Q., Mizuno S. I., Bronson R. T., Geng Y., Nguyen M. D., Akashi K., Sicinski P., Nakatani Y. (2013) p600 plays essential roles in fetal development. PLoS One 8, e66269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. White E. A., Sowa M. E., Tan M. J., Jeudy S., Hayes S. D., Santha S., Munger K., Harper J. W., Howley P. M. (2012) Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. U.S.A. 109, E260–E267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huh K. W., DeMasi J., Ogawa H., Nakatani Y., Howley P. M., Munger K. (2005) Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. U.S.A. 102, 11492–11497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeMasi J., Huh K. W., Nakatani Y., Munger K., Howley P. M. (2005) Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc. Natl. Acad. Sci. U.S.A. 102, 11486–11491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conroy J., McGettigan P., Murphy R., Webb D., Murphy S. M., McCoy B., Albertyn C., McCreary D., McDonagh C., Walsh O., Lynch S., Ennis S. (2013) A novel locus for episodic ataxia:UBR4 the likely candidate. Eur. J. Hum. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richards S., Hillman T., Stern M. (1996) Mutations in the Drosophila pushover gene confer increased neuronal excitability and spontaneous synaptic vesicle fusion. Genetics 142, 1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nautiyal M., Sweatt A. J., MacKenzie J. A., Mark Payne R., Szucs S., Matalon R., Wallin R., Hutson S. M. (2010) Neuronal localization of the mitochondrial protein NIPSNAP1 in rat nervous system. Eur. J. Neurosci. 32, 560–569 [DOI] [PubMed] [Google Scholar]
- 38. Okuda-Ashitaka E., Minami T., Tsubouchi S., Kiyonari H., Iwamatsu A., Noda T., Handa H., Ito S. (2012) Identification of NIPSNAP1 as a nocistatin-interacting protein involving pain transmission. J. Biol. Chem. 287, 10403–10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee A. H., Zareei M. P., Daefler S. (2002) Identification of a NIPSNAP homolog as host cell target for Salmonella virulence protein SpiC. Cell Microbiol. 4, 739–750 [DOI] [PubMed] [Google Scholar]
- 40. Verhagen A. M., Kratina T. K., Hawkins C. J., Silke J., Ekert P. G., Vaux D. L. (2007) Identification of mammalian mitochondrial proteins that interact with IAPs via N-terminal IAP binding motifs. Cell Death Differ. 14, 348–357 [DOI] [PubMed] [Google Scholar]
- 41. Buechler C., Bodzioch M., Bared S. M., Sigruener A., Boettcher A., Lapicka-Bodzioch K., Aslanidis C., Duong C. Q., Grandl M., Langmann T., Dembinska-Kiec A., Schmitz G. (2004) Expression pattern and raft association of NIPSNAP3 and NIPSNAP4, highly homologous proteins encoded by genes in close proximity to the ATP-binding cassette transporter A1. Genomics 83, 1116–1124 [DOI] [PubMed] [Google Scholar]
- 42. Xie Y., Varshavsky A. (1999) The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18, 6832–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheng Y., Hong J. H., Doherty R., Srikumar T., Shloush J., Avvakumov G. V., Walker J. R., Xue S., Neculai D., Wan J. W., Kim S. K., Arrowsmith C. H., Raught B., Dhe-Paganon S. (2012) A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell, Proteomics 11, 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hibbert R. G., Huang A., Boelens R., Sixma T. K. (2011) E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. U.S.A. 108, 5590–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Husnjak K., Dikic I. (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81, 291–322 [DOI] [PubMed] [Google Scholar]
- 46. Shekhar M. P., Lyakhovich A., Visscher D. W., Heng H., Kondrat N. (2002) Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 62, 2115–2124 [PubMed] [Google Scholar]
- 47. Kim S. T., Tasaki T., Zakrzewska A., Yoo Y. D., Sa Sung K., Kim S. H., Cha-Molstad H., Hwang J., Kim K. A., Kim B. Y., Kwon Y. T. (2013) The N-end rule proteolytic system in autophagy. Autophagy 9, 1100–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee K. M., Hwang S. K., Lee J. A. (2013) Neuronal autophagy and neurodevelopmental disorders. Exp. Neurobiol. 22, 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang A., Hibbert R. G., de Jong R. N., Das D., Sixma T. K., Boelens R. (2011) Symmetry and asymmetry of the RING-RING dimer of Rad18. J. Mol. Biol. 410, 424–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.