Abstract
Methylation of lysine residues on histone proteins is known to play an important role in chromatin structure and function. However, non-histone protein substrates of this modification remain largely unknown. An effective approach for system-wide analysis of protein lysine methylation, particularly lysine monomethylation, is lacking. Here we describe a chemical proteomics approach for global screening for monomethyllysine substrates, involving chemical propionylation of monomethylated lysine, affinity enrichment of the modified monomethylated peptides, and HPLC/MS/MS analysis. Using this approach, we identified with high confidence 446 lysine monomethylation sites in 398 proteins, including three previously unknown histone monomethylation marks, representing the largest data set of protein lysine monomethylation described to date. Our data not only confirms previously discovered lysine methylation substrates in the nucleus and spliceosome, but also reveals new substrates associated with diverse biological processes. This method hence offers a powerful approach for dynamic study of protein lysine monomethylation under diverse cellular conditions and in human diseases.
Within proteins, the lysine residue can be modified at its ε-amino side-chain by mono, di, and trimethyl groups (1, 2). Over the past few decades, study of lysine methylation (Kme) has mainly focused on core histones. Early studies demonstrated critical roles of this modification inchromatin structure and function (3–5). The methylation status of lysine is regulated by two groups of enzymes with opposing enzymatic activities: lysine methytransferases and lysine demethylases (5, 6). To date, several dozens of protein lysine methyltransferases and lysine demethylases have been identified (7–9). Dysregulation of histone methylation can result in reprogramming of gene expression networks and has been associated with diverse disease states, including cancer (4, 5, 7). Accordingly, enzymes regulating lysine methylation have emerged as a group of promising drug targets (7, 10, 11).
All post-translational modifications (PTMs)1 that have been identified in histones are also present in non-histone proteins. Identification of a large family of Kme-regulatory enzymes, the non-nuclear localization of some of these enzymes, and the recent discovery of non-histone Kme substrates clearly suggest that Kme is likely abundantly present in non-histone proteins (12–14), as has been observed for lysine acetylation. Nevertheless, non-histone methylation substrates remain largely unknown. Identification of protein substrates is a critical step for functional characterization of a PTM pathway, a principle well illustrated by the history of lysine acetylation biology. Progress in elucidating the roles of lysine acetylation in chromatin function, transcriptional regulation, and DNA-independent pathways (e.g. metabolism) followed shortly after the discovery of lysine acetylation substrates among the three groups of cognate cellular pathways (2, 15–19). Likewise, proteomic identification and quantification of Kme substrates will reveal chromatin-independent downstream protein targets and pathways of this modification, thus laying a concrete foundation for studying its functions outside the nucleus.
Nevertheless, detection of Kme substrates is not simple. The relatively low-energy radiation emitted by 3H or 14C makes it difficult to detect Kme substrates by a radio labeling approach, as is done for protein phosphorylation using 32P (20). Kme, especially monomethylation (Kme1), induces only a small structural change to the substrate lysine residue. Because of this very limited difference in physiochemical properties between monomethyllysine and unmodified lysine residues, it is difficult to use chemical methods to isolate methylated peptides from unmodified ones as is done, for example, for phosphopeptides using immobilized metal affinity chromatography (21). In addition, it has been challenging to develop a highly specific antibody against monomethyllysine with workable affinity (22). Thus, it is still a daunting challenge to develop sequence-independent (or pan) anti-Kme antibodies with adequate affinity and specificity. As a consequence, progress in identifying Kme substrates has been slow, largely because of a lack of suitable affinity enrichment technology for isolation of Kme peptides that can be used for mass spectrometry-based proteomic screening (23–26).
To overcome this technical challenge, we developed a new chemical proteomic approach for efficient enrichment and global analysis of protein lysine monomethylation. In our approach, we first chemically derivatized the monomethyl ε-amine group of the lysine residue, adding apropionyl moiety. Then, the propionyl monomethylated peptides were enriched using a pan anti-propionyl monomethyllysine antibody, followed by HPLC/MS/MS analysis of the enriched peptides for peptide identification and propionyl monomethylation site mapping. This method was clearly validated by identifying 446 monomethylation sites on 398 proteins with high accuracy, the largest monomethyllysine data set ever reported.
MATERIALS AND METHODS
Isotopic Methionine Labeling and In Vitro Propionylation
SILAC labeling medium Roswell Park Memorial Institute medium 1640 (RPMI1640) or Eagle's minimal essential medium (DMEM) (Life TechnologiesCorp., Carlsbad, CA) was reconstituted with methionine of interest (either 12CH3-methionine or 13CD3-methionine; Sigma-Aldrich), 10% dialyzed FBS (Invitrogen Corp.), and 1×penicillin (Hyclone Laboratories Inc., South Logan, UT). The cell lines were grown in either RMPI or DMEM medium at 37 °C supplied with 5% CO2. The labeling efficiency of cells cultured in medium containing 13CD3-methionine was greater than 98% as determined by mass spectrometry prior to the proteomics experiment. Cells were washed three times with ice-cold Dulbecco's PBS (Mediatech Inc., Manassas, VA) before harvesting, collected in chilled lysis buffer (8.0 m urea in two parts 0.1 m NH4HCO3 and one part 0.1 m NaHCO3 (v/v) with 1x protease inhibitor) and incubated on ice for half an hour. The debris was removed by centrifugation, and the supernatant was collected. The protein concentration of the supernatant was determined by Bradford assay. Chemical derivatization of the protein lysate with propionic anhydride was performed as described previously with slight modifications (27). Briefly, 200 μl propionic anhydride (Sigma) was added to 20 mg protein lysate and vortexed. The solution was adjusted to pH 8.0 with an adequate volume of 2.0 m NaOH. The reaction was carried out at RT for 1h. The propionic anhydride addition, pH correction, and 1h incubation were performed twice more. The reaction was terminated by adding 10 μl ethanolamine at RT for 30min. The modified protein was precipitated with trichloroacetic acid. The protein pellet was resuspended in 2 ml digestion buffer (0.1 m NH4HCO3, pH 8.5), followed by in-solution trypsin digestion (28). The tryptic peptides were centrifuged at 20,000 × g for 10min before subsequent HPLC separation.
Liver Cancer Sample Preparation
With the informed consent of the donor, liver tissue was collected from a liver cancer patient at Zhongshan Hospital (Shanghai, China). Ethical approval for the use of human subjects was obtained from the Research Ethics Committee of Zhong Shan Hospital. The fresh tissue was frozen in liquid nitrogen immediately after surgery. To extract the proteins, the liver tissue was quickly dissected using surgical scissors and washed with chilled Dulbecco's PBS (Mediatech Inc., Manassas, VA) to remove residual blood. The dissected tissue was homogenized using a Douncehomogenizer in chilled Dulbecco's PBS. Connective tissue was removed with cell strainer (70 μm size, Cell strainer, Falcon One Inc., Franklin Lakes, NJ). Hepatocytes were collected by centrifugation and lysed as described above.
HPLC Fractionation of Tryptic Peptides and Affinity Purification
Before affinity purification, the tryptic peptides were separated by HPLC (29). Briefly, the separation was performed on a Varian SD1 LC system (Agilent TechnologiesInc., Folsom, CA) with an Xbridge C18 column (19X150 cm, Waters Corp., Milford, MA) using a 70-min gradient from 2% to 40% of buffer B (10 mm ammonium formate/80%ACN, pH 8.5) at a flow rate of 10 ml/min. The sample was collected into 60 fractions based on equal time intervals (about 10 ml/tube), and combined into 20 fractions. Each fraction was dried in a SpeedVac. The peptides were redissolved in NETN buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 0.5% (w/v) Nonidet P-40) for affinity enrichment.
Affinity enrichment of the propionyl-methyl peptides was carried out using protein A-agarose beads conjugated with 20 μg of anti-propionyl-methyllysine antibody (Jingjie PTM BioLab Co. Ltd, Hangzhou, China) at 4 °C for 4 h. The beads were washed three times with 1 ml of NETN buffer and three times with ETN (20 mMTris-HCl, pH 8.0, 100 mMNaCl, and 1 mm EDTA). The bound peptides were eluted from the beads by washing three times with 100 μl of 0.1%TFA. The eluates were combined and dried in a SpeedVac.
HPLC/MS/MS Analysis and Protein Sequence Database Searching
The enriched propionyl-methyl peptide samples were dissolved in 3-μl HPLC buffer A (0.1% formic acid in water, v/v) and delivered onto the capillary RPLC trap column (2 cm length with 100 μm inner diameter (ID), packed with Luna C18 resin, 5 μm particle size, 100 Å pore size, Dikma Technologies Inc., Lake Forest, CA) through the auto-sampler at a maximum pressure of 250 bar in 100% buffer A. After loading and washing, the peptides were transferred to the analytical column (10 cm length with 75 μm ID, packed with C18resin, 3 μm particle size, 90Å pore size, Dikma Technologies Inc.) connected to an EASY-nLC 1000 HPLC system (Thermo Fisher Scientific Inc.). Peptides were eluted with a 70-min gradient of 2% to 90% HPLC buffer B (0.1% formic acid in acetonitrile, v/v) in buffer A at a flow rate of 300 nL/min. The eluted peptides were ionized and introduced into an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific Inc.) using a nanospray source. Survey full-scan MS spectra (from m/z 300 to 2000) were acquired in the Orbitrap with resolution r = 24,000 at m/z 400. The ten most intense ions were sequentially isolated in the linear ion trap and subjected to collisionally induced dissociation (CID) with a normalized energy of 35%. The exclusion duration for the data-dependent scan was 30 s, the repeat count was two, and the exclusion window was set at +2 Da and −1 Da. In addition, all HPLC/MS/MS data were analyzed by Mascot (v2.3, Matrix Science Ltd., London, UK). Peak lists were generated by Proteome Discoverer software (version 1.4) from Thermo Fisher. Precursor mass tolerance for Mascot analysis was set at ±10 ppm, and fragment mass tolerance was set at ±0.6 Da. The protease was set to trypsin/P (which accounts for in-source fragmentation at lysine or arginine residues followed by proline), allowing for a maximum of three missed cleavage sites. All data were searched against the UniProt Human database (88,817 sequences) including fixed modification of cysteine residues by carbamidomethylation as well as the following variable modifications: 13CD3-methionine, 13CD3-methionine oxidation, lysine propionylation (lysine +56.0262), lysine propionyl-13CD3-methylation (lysine +74.0640) for the heavy coded samples, and additional lysine propionyl-CH3-methylation (lysine +70.0418) for the SILAC labeled HeLa sample. While for the liver cancer, only methionine oxidation, lysine propionylation and lysine propionyl-CH3-methylation were considered as variable modifications. All spectra with a Mascot ion score of more than 20 were manually inspected using stringent criteria as previously described (30). All peptides identified with a C-terminal lysine were filtered out unless the lysine was followed by proline.
Isolation and Mass Spectrometric Analysis of CDC5L
Protein purification was performed as described previously (31). Briefly, heavy13CD3-methionine-labeled K562 cells were lysed in 1x NETN buffer. Homogenates were centrifuged at 6000 × g at 4 °C for 10 min to remove cell debris. Cleared lysates were incubated overnight at 4 °C with anti-CDC5L antibody (C20, Santa Cruz Biotechnology Inc., Dallas, TX) followed by incubation with protein A/G beads (Santa Cruz Biotechnology Inc.) for another 4h at 4 °C. After extensive washing, immunoprecipitates were boiled in loading buffer, and then separated on a NuPAGENovex 4–12% Bis-Tris Mini Gel (Invitrogen Corp). The gel bands of the target proteins were cut out and subjected to in-gel trypsin digestion. Tryptic peptides were resolubilized in 0.1% TFA buffer and analyzed by HPLC-MS/MS to map lysine methylation sites.
Subcellular Localization Analysis
Manually curated protein subcellular localization data were downloaded from the PENCE Proteome Analyst Specialized Subcellular Localization Server v2.5.
Functional Annotation Analysis
Enrichment analysis of Gene Ontology (GO) (32), KEGG pathway (33), and PFAM domain (34) databases were performed using DAVID 6.7 (35) (The Database for Annotation, Visualization, and Integrated Discovery) tools with the total Homo sapiens genome information as the background. GO biological process, molecular function, and cellular compartment categories were analyzed separately. All p values were adjusted with a Benjamini-Hochberg false discovery rate using a cutoff rate of 0.05 (36).
Motif Analysis
An iceLogo standalone version (version 1.2) was used for consensus flanking sequence analysis of methylation sites. Six neighboring amino acids residues on each side of the methylation site were selected as the positive set for analysis. The embedded Swiss-Prot “Homo sapiens” data set was used as the negative set.
Protein–Protein Interaction Network Analysis
All interactions for Homo sapiens with high confidence (0.7) in the STRING 9.05 database were used for protein–protein interaction analysis. The network was visualized in Cytoscape (v.3.0.1 beta) (37) and highly connected clusters were identified by the MCODE plug-in (38).
Protein Complex Analysis
The manually curated CORUM protein complex database for Homo sapiens was used for protein complex analysis (39). Overrepresented complexes were identified using Fisher's exact test with a cutoff of<0.05.
RESULTS
Affinity Enrichment of Lysine Monomethylated Peptides
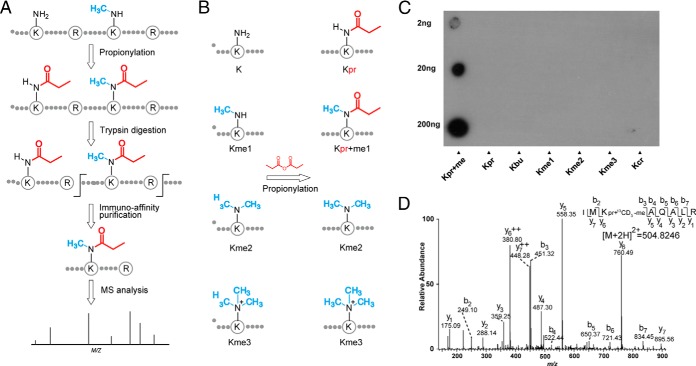
Affinity enrichment is a key step toward efficient global analysis of a PTM, as has been well demonstrated in large-scale studies of protein phosphorylation and lysine acetylation (40–42). To this end, we developed a novel strategy to purify peptides containing monomethyllysine, involving three main steps (Fig. 1A): 1) chemical derivatization of the monomethylated ε-amine group of the lysine residue with propionyl anhydride to form propionyl-methyl ε-amine; 2) affinity enrichment of the propionyl-methyllysine-containing peptides using a pan antibody against propionyl-methyllysine; and 3) HPLC/MS/MS analysis of the resulting peptides to identify the modified peptides and map the modification sites. For the proposed procedure for proteomic screening to be efficient, the propionylation reaction should be as complete as possible. The chemical propionylation of lysine residues was first introduced by Garcia et al. to enhance the hydrophobicity of tryptic peptides and increase sequence coverage during mass spectrometric analysis of histone modifications (27). However, this chemistry has not yet been applied to proteomics analysis of lysine methylation. To determine whether this chemistry works efficiently in the more complex environment of a protein lysate, we subjected HeLa whole-cell lysate to the chemical reaction using propionyl anhydride in NH4CO3-NaHCO3 buffer (pH 8.5). After the reaction, we digested the proteins with trypsin and then analyzed the proteolytic peptides by HPLC/MS/MS. The propionylation reaction occurs at unmodified lysine and monomethyllysine residues and the N-terminal amine, but not at di or trimethylated lysine residues (Fig. 1B) (27). If the propionylation reaction goes to near completion, the percentage of tryptic peptides with unmodified lysine should be low. Thus, the intensity ratio of modified to unmodified lysine-containing peaks provides a relevant measure of the efficiency of the propionylation reaction. Our analysis showed that after treatment with propionyl anhydride, less than 2% of the detected peptides contained unmodified lysine, suggesting that our procedure for lysine propionylation is efficient (supplemental Fig. S1).
Fig. 1.
Strategy for monomethylome profiling. A, Schematic diagram of the experimental workflow for identifying monomethyllysine-containing peptides by chemical propionylation and immuno-affinity enrichment followed by MS analysis. B, The propionylation reaction occurs at monomethylated lysine but not di or trimethylated lysine. C, Specificity of the anti-propionyl monomethyllysineantibody by dot-blot assay. Kpr+me, Kpr, Kbu, Kme1, Kme2, Kme3, and Kcr denote peptide libraries of sequence xxxxxKxxxxx, where K represents a fixed lysine residue modified by propionyl-methyl, propionyl, butuyryl, monomethyl, dimethyl, trimethyl, and crotonyl groups, respectively. D, A typical annotated ion trap CID spectrum of a propionyl-13CD3-methyllysine-bearing peptide from protein HSP90.
Antibody Generation
The pan antibody against propionyl monomethyllysine was developed in Jingjie PTM BioLab (Hangzhou, China) Co. Ltd. Briefly, propionyl-methyllysine-conjugated keyhole limpet hemocyanin (KLH) was used to immunize rabbits. The anti-propionyl monomethyllysine antibody was isolated from the resulting serum. The specificity of the pan antibody was then tested by dot-blot assay with peptide libraries containing propionyllysine (Kpr), butyryllysine (Kbu), crotonyllysine (Kcr), or mono, di, or trimethyllysine residues (Kme1, Kme2, and Kme3) in a fixed position (Fig. 1C). The antibody showed more than 100-fold specificity for propionyl monomethyllysine relative to the other modified lysine residues. Because propionyl monomethyllysine is structurally similar to propionyllysine and butyryllysine, this result clearly suggests that the anti-propionyl monomethyllysine antibody has good affinity and specificity. We therefore used the antibody for enriching propionyl monomethyllysine peptides in our subsequent proteomic screenings.
Confident Detection of Lysine Propionyl Monomethylated Peptides
When identifying monomethylated peptides, false positives can arise for two reasons. Lysine monomethylation induces a mass shift of 14.0157 Da, which is the same as the shift observed for several amino acid substitutions or mutations: Ser to Thr, Asp to Glu, Val to Leu or Ile, and Asn to Glu (22). In addition, propionyl monomethylation has the same mass shift as lysine butyrylation, a protein modification that occurs in vivo (43). Therefore, our strategy should include a proper control to prevent false identification of propionyl monomethylated peptides. To address these concerns, we used the well-known method of isotopic labeling, using 13CD3-methionine. The isotopic label results in a mass shift of 18.037 at the lysine residue. Therefore, propionyl-13CD3-methyllysine induces a unique mass shift of 74.0640 localized at the lysine residue, which can be distinguished by MS from the mass shift caused by any known PTM (including butyrylation), or any amino acid mutation (44). As an example, we identified a propionyl monomethylated peptide from HSP90 (Fig. 1D), where the propionyl monomethylation can be unambiguously localized by the mass shift of 74.0640 Da at residue K3 within the peptide.
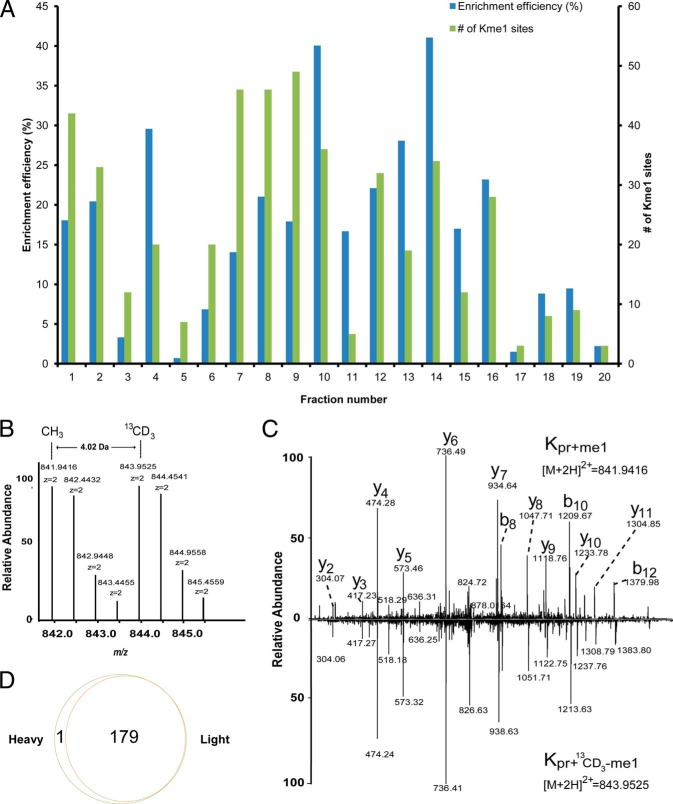
To test reliability of this approach, we cultured two groups of HeLa cells in parallel in DMEM media containing either 12CH3-methionine or 13CD3-methionine. After more than six doublings, we mixed the two pools of cells and lysed them. The resulting protein lysate was propionylated, digested with trypsin and resolved into fractions by preparative HPLC. In each fraction, the propionyl monomethylated peptides were enriched using the pan anti-propionyl monomethyllysine antibody and then subjected to nano-HPLC/MS/MS analysis. The MS/MS spectra of the enriched peptides were searched against the human Uniprot proteins database with the Mascot algorithm, and positive identifications were manually inspected using stringent criteria to verify each identification as previously described (30). Using this method, we identified 180 propionyl monomethylated peptides from HeLa cells, with the enrichment specificity in the fractions ranging from 3 to 41% (Fig. 2A). A truly propionyl monomethylated peptide will have twin parent ions with a mass difference of 4.02 Da derived from 13CD3-SAM and 12CH3-SAM, and the fragment ions will have an identical pattern if both light and heavy peptides are selected for MS/MS (see Fig. 2B and 2C for characteristic spectra). When manually checking the raw data, 179 of the 180 putative propionyl monomethylated peptides were found with paired precursor ions, corresponding to a total of 185 methylation sites on 163 proteins. Only one “heavy” peptide was identified without a counterpart ion (Fig. 2D), suggesting that our procedure is reliable for mapping monomethylation sites, and filtering out false positives. In the proteomic screening experiments reported below, we used only 13CD3-methionine-labeled cells.
Fig. 2.
Proteome-wide analysis of lysine monomethylation using an isotopic labeling approach. A, Affinity enrichment efficiency and the number of methylated peptides identified in each fraction. B, MS spectrum of a “Light” and “Heavy” methyl-coded monomethylated peptide of protein EZH2from HeLa cells, showing twin peaks with a mass difference of 4.02 Da induced by isotopic labeling using 13CD3. C, Ion trap CID spectra from the “Light” and “Heavy” methylated peptide YSQADALKme1YVGIER of protein EZH2 from HeLa cells. The 4-Da mass shift of the b- and y-ions of the “Heavy” peptide relative to its “Light” counterpart was induced by the isotopic methyl group. D, The overlap of the methylated peptides from “Light” and “Heavy” methionine-labeled HeLa cells.
Landscape of Protein Lysine Monomethylome
We next extended the method of using 13CD3-methionine labeling to profile Kme1 in four other human cancer cell lines: K562 (chronic myelogenous leukemia), SW620 (colon cancer), A549 (lung cancer), and SMM7721 cells (liver cancer). Using the strategy described above, we identified 434 nonredundant monomethylated peptides corresponding to 446 sites on 398 proteins. Most of the monomethylation sites have not been reported previously (24, 25, 45, 46). A complete list of the methyllysine peptides and proteins identified in this study is provided in supplemental Table S1 and supplemental Fig. S2.
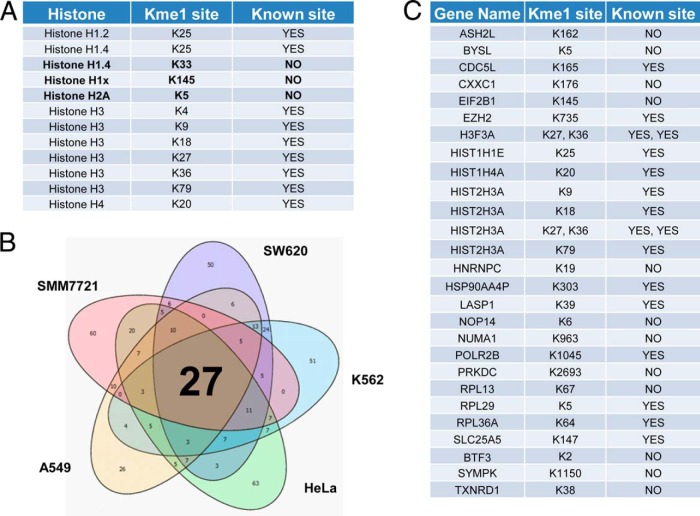
The core histone proteins have been extensively analyzed for lysine methylation, thus providing a good positive control. As expected, we identified 12 histoneKme1 sites. In addition to the well-studied methylation sites at H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20 (47), we identified the little-studied H3K18 monomethylation (48). Most of these sites were identified in all five of the cell lines, indicating that our method is robust for detecting Kme1 peptides. Intriguingly, we identified three previously unknown monomethylation sites: H2AK5, H1K33, and H1K145 (Fig. 3A). In addition to the histone sites, we identified new Kme1 sites on proteins previously known to be monomethylated. For instance, we found two new monomethylated sites (K1321 and K1463) on the protein WIZ, in addition to the previously reported site at K967 (26).
Fig. 3.
A landscape of the protein lysine monomethylome. A, List of the monomethylation sites identified from histone proteins in this study, three of which were not previously described: H1.4K33me1, H1.xK145me1, and H2AK5me1. B, Venn diagram comparison of the methylated peptides identified from five cell lines: HeLa (cervical cancer), K562 (chronic myelogenous leukemia), SW620 (colon cancer), A549 (lung cancer), and SMM7721 (liver cancer). C, List of the methylated proteins identified inall five cell lines, including 20 novel methylation sites.
Next, we applied our method to analyze the scope of Kme1 substrates in human liver cancer tissue taken directly from a patient. With our stringent criteria, we identified 59 monomethylation sites located on 49 proteins. We compared the Kme1 data sets among the five types of cells. Only 29 monomethylation sites among 27 monomethylated peptides were found in all five cell types, including 20 novel methylation sites and five histone H3 Kme1 sites (Fig. 3B and 3C).
Validation of the Novel Kme1 Sites
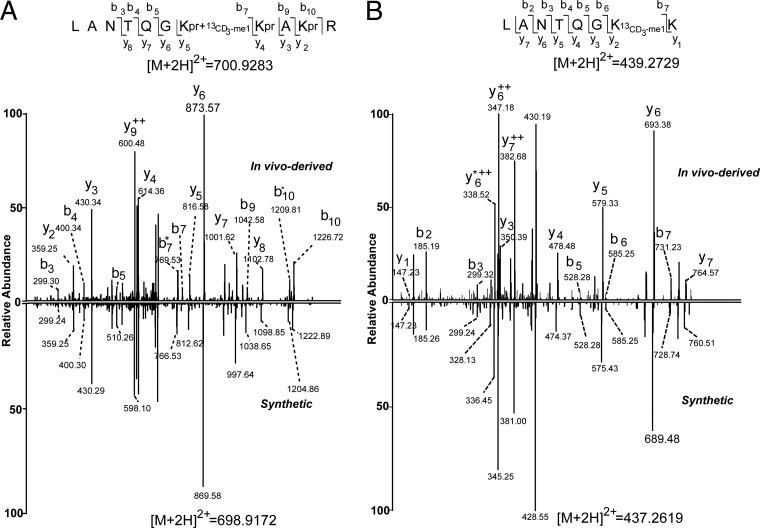
Because site-specific anti-Kme1 antibodies for non-histone proteins are not available, we validated the identified Kme1-containing peptides by analyzing corresponding synthetic peptides. Because synthetic peptides and their cell-derived counterparts will generate identical MS/MS patterns under the same mass spectrometric conditions, this approach is the gold standard for peptide validation. To this end, we randomly selected and synthesized 10 propionyl-monomethylated peptides identified with Mascot ion scores ranging from 20 to 60. The MS/MS spectra of these synthetic propionyl-methyllysine peptides matched perfectly with those of their cell-derived counterparts (Fig. 4A and supplemental Fig. S3), confirming the identification of all of these propionyl-methyllysine peptides. These results suggest that our approach for identifying Kme1 peptides is reliable and highly accurate.
Fig. 4.
Validation of the K114 methylated site on protein CDC5Lfrom K562 cells. A, The tandem mass spectrum of the synthetic propionyl-methylated peptide LANTQGKpr+me1KprAKprR (bottom) showed a pattern that was almost identical to the pattern derived from its in vivo counterpart from K562 cells (top). B, Tandem mass spectra of in vivo-derived peptide LANTQGKme1K (top) and its synthetic counterpart peptide (bottom). Some y-ions and b-ions have a 4-Da difference between the in vivo-derived and synthetic peptides because of the isotopic labeling.
In addition to testing our method using synthetic peptides, we also validated the monomethylation sites in vivo for CDC5L, a protein involved in cell cycle control (49). To this end, we first metabolically labeled the methyl group using 13CD3-methionine as described above, and then isolated the protein from cultured K562 cells by immunoprecipitation for MS analysis. We successfully identified the K164 13CD3-methylated peptide, LANTQGK13CD3-me1K, from the in vivo-derived protein CDC5L (Fig. 4B). To further confirm this peptide, we analyzed the MS/MS spectrum of its corresponding synthetic counterpart, which showed an almost identical MS/MS spectra, except for the 4-Da difference caused by isotopic labeling (Fig. 4B). Taken together, our data clearly confirmed monomethylation at the K165 residue of CDC5L.
Bioinformatics Analysis of the Methylated Proteins
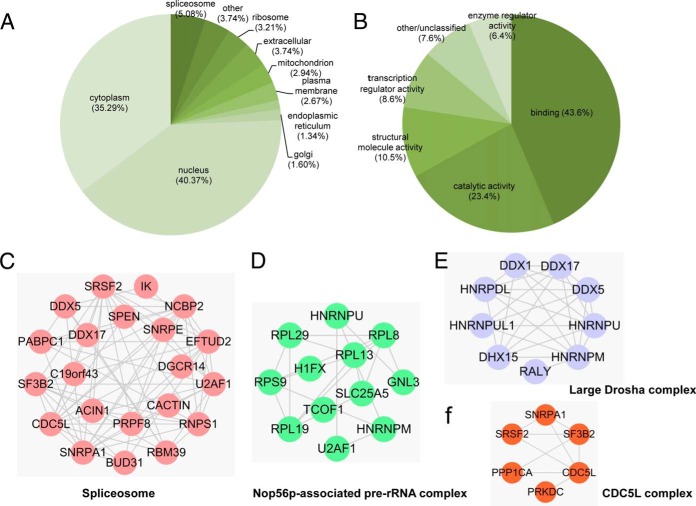
We next performed bioinformatics analysis to investigate the possible roles of protein lysine monomethylation in cellular regulation. We first analyzed the subcellular localizations of the Kme1 substrates. Earlier studies suggest that most arginine-methylated proteins are associated with RNA processing and are mainly located in the nucleus or ribosome (12). In our study, nuclear proteins accounted for the largest subgroup (∼40.37%) of the identified lysine monomethylated proteins. Thirty-four percent of the Kme1 proteins were predicted to localize to the cytoplasm, with an additional 8% of proteins being found in the ribosome or spliceosome. This result suggests that lysine monomethylation may have diverse previously unknown functions in the cytosolic compartment. Intriguingly, we also identified 11 mitochondrial Kme1 proteins. Most of these proteins play roles in regulating various aspects of mitochondrial function (supplemental Table S2). For example, SLC25A4, SLC25A5, and SLC25A6, three members of the SLC25 protein family, catalyzes the exchange of cytoplasmic ADP for mitochondrial ATP across the mitochondrial inner membrane (50). Intriguingly, the monomethylation site of these three proteins is located at lys147, an evolutionary conserved domain. The data suggest that mitochondrial functions may be regulated by protein methylation.
We then carried out analysis of the Kme1-containing proteins using the Gene Ontology (GO) categories. Within the molecular function (MF) domain, the largest group of proteins belong to the “Binding” category, and the majority of these carry RNA-binding activity. We additionally found that the second-largest category of Kme1 proteins has the function of catalytic activity (Fig. 5B). Analysis of the Biological Process (BP) domain suggested that the methylated proteins have roles in diverse biological processes, of which metabolic process, cellular process, cell cycle, and cell communication comprised the largest part (supplemental Fig. S4).
Fig. 5.
Bioinformatic analysis of the methylated proteins. A, Pie chart of predicted subcellular localization of the methylated proteins according to the PENCE Proteome Analyst databases. Proteins predicted to be present in the “ribosome” and “spliceosome” were differentiated from those predicted to be in the “cytoplasm.” Proteins were designated “other” if their localizations or functions were not annotated in the database. B, Pie chart of Molecular Function analysis of the methylated proteins analyzed with DAVID (The Database for Annotation, Visualization, and Integrated Discovery). C–F, Representative protein complexes enriched in monomethyllysine proteome by querying CORUM database (39).
Further assessment revealed considerable enrichment for RNA-related GO terms and Pfam database categories, such as chromosome organization and RNA processing (BP), RNA binding (MF), nucleolus (cellular component (CC) domain of GO), and RNA recognition motif (i.e. RRM, RBD, or RNP domain, in Pfam). Additional KEGG pathway analysis underscored the significant overrepresentation of the methylated proteins in RNA-related spliceosome and ribosome pathways (supplemental Fig. S5 and supplemental Table S3).
We further used the STRING protein interaction database (51) and Comprehensive Resource of Mammalian protein complex (CORUM) database (39) to visualize the protein–protein interaction networks and identify overrepresented protein complexes. The interaction network showed that the methylated proteins were highly connected to each other and formed a number of overrepresented subclusters, the largest of which were proteins involved in the cellular functional categories of spliceosomes, RNA splicing and cell cycle(supplemental Fig. S6 and supplemental Table S4). By querying the CORUM database, we found 30 enriched protein complexes among the lysine monomethylated proteins (p < 0.05). Among these are the spliceosome, the Nop56p-associated pre-rRNA complex, Large Drosha complex, and CDC5L complex (Fig. 5C–5F and supplemental Table S5), consistent with the study reported previously (24).
To examine whether any of the monomethylated lysine residues could be alternatively modified by acetylation or ubiquitination, we compared our data set with the public database of known PTMs from UniProt. In total, 37 Kme1 sites, including the known H3K9 and H3K27 sites (52) and 30 non-histone proteins, could be alternatively modified with either methyl or acetyl groups (supplemental Table S6). In addition, when we compared the methylation sites with our recently published lysine succinylome in mice (53), we identified seven monomethylated sites that overlapped with succinylation sites (supplemental Table S7). Importantly, we found that two sites, from the proteins vimentin and histone H3, could be alternatively modified by all three different types of modifications.
Finally, we asked whether any disease-related genetic mutations are located around the Kme1 sites. We compared the sequences of our methylated peptides with the data set of inherited amino acid substitutions in humans from UniProt. We identified 39 mutations (amino acid substitutions) located in a window of ±10 amino acids surrounding the methylated lysine (supplemental Fig. S7 and supplemental Table S7). Eight of the mutations are known to be associated with certain human diseases, including Rett syndrome, colorectal cancer, cirrhosis (CIRRH), argininosuccinicaciduria (ARGINSA), Diamond-Blackfan anemia 6 (DBA6), and focal segmental glomerulosclerosis 5 (FSGS5) (supplemental Table S8).
DISCUSSION
Although affinity-based proteomic strategy has been described for protein lysine methylation (22, 25, 26, 54–56), the proteome-wide survey of protein lysine monomethylation remains challenging, mainly because of the negligible physicochemical difference induced by the methyl group (57). The 446 methylation sites found on 398 proteins identified in this study represent the largest data set of lysine monomethylated protein ever reported. The overlap between the three independent studies with different strategies was around 10% suggested that there approaches were complement with each other (supplemental Fig. S8) (24, 25).
Identification of the substrates is a critical step for functional characterization of protein lysine methylation and remains the most challenging for large-scale study. Despite the biological significance of lysine di and trimethylation, our method was designed to focus on the proteomic screening of monomethylated lysine because di and trimethylated lysine residues cannot react with propionyl anhydride. We also had to use heavy isotope-coded methionine to improve the accuracy of detection of Kme1 peptides (22). Although this method is not directly applicable to clinical tissue samples, there liability of peptide identification can be enhanced by alternative methods, such as stringent manual verification and MS/MS analysis of synthetic peptides.
Among the Kme1substrates we identified, a noteworthy subgroup of protein methyltransferases. In this study, we identified lysine monomethylation among eight protein lysine methyltransferases (SETDB1, EZH2, CAMKMT, EHMT1, EHMT2, SETD2, MLL4, and NSD1), and one methyltransferase interaction partner (ASH2L). Previous research showed that automethylation of G9a methyltransferase at K239 led to its interaction with HP1, consequently exerting its biological functions (58). The automethylation motif of G9a flanking K239 (ARKT) resembles the sequence around histone H3 lysine 9 (ARKS), which is the specific substrate of G9a (58). Similarly, we found a novel methylation site (K490) on methyltransferase SETDB1 with a flanking motif of AKKST, which resembles the ARKST motif of its specific substrate histone H3K9 (59). Hence, it is likely that modification of K490 of SETDB1 resulted from auto-methylation, which could consequently lead to the recruitment of HP1 to form an HP1a-CAF1-SETDB1 complex (58, 60). One question worthy of future study is whether lysine automethylation among lysine methyltransferaseshas regulatory roles similar to those of autophosphorylation of receptor tyrosine kinases (61) and autoacetylation of acetyltransferases (e.g. p300 and CBP) (62).
Previous studies have identified arginine methylated protein substrates and revealed the likely role of this modification in RNA-related biological events such as gene transcription, mRNA splicing and regulation of small RNAs (63). Our study showed that proteins in these pathways are also lysine methylated. Given the high abundance of both arginine methylation and lysine methylation, it is very likely that protein methylation has an important role in the regulation of these processes.
Cross-talk among lysine modifications, such as between methylation and acetylation, has been reported (64). For example, H3K9 acetylation (H3K9ac) is generally associated with active gene transcription, whereas methylation of H3K9 (H3K9me) correlates with inactivation of gene expression and could compete with H3K9ac (65). In this study, we found overlap of methylation sites with sites of acetylation and succinylation, including two sites which could be alternately modified with all three types of modification (supplemental Table S8). Given that methylation retains the positive charge on lysine, acetylation neutralizes the charge, and succinylation converts the positive charge to a negative charge, these three different types of modification provide a mechanism to produce distinct physicochemical states in the substrate proteins.
In recent years, the biological importance of protein methylation has attracted extensive attention in the research community. Enzymes regulating lysine methylation have been proposed as drug targets for diverse diseases (11). In addition, some inhibitors of lysine methylation regulatory enzymes, such as inhibitors of EZH2 and DOTL1, are currently under clinical evaluation (66). However, current knowledge of lysine methylation is restricted to a limited number of proteins, such as histones and p53 (4, 67). As a consequence, non-histone substrates of the enzymes and methylation substrate-based biomarkers for compounds under clinical investigation remain unknown, hindering our understanding of the mode of action for these enzymes and their inhibitors. The proteomic technology described here offers a new approach to addressing these issues by identifying methylation substrates in response to either the enzymes or their inhibitors.
The study presented here not only provides a robust methodology for global screening of lysine monomethylation substrates, but also dramatically extends the inventory of lysine monomethylated proteins. A combination of this proteomic technology with genetic manipulation of expression of methylation regulatory enzymes can be used to identify substrates for methytransferases (for Kme1) and demethylases (for Kme1 and Kme2), and for dynamic analysis of Kme substrates under disease conditions. In addition, our data set is a valuable starting point for further biological research to study the biological functions of lysine-methylated proteins and the anatomy of lysine methylation pathways specific to human diseases.
Supplementary Material
Acknowledgments
We thank the China Postdoctoral Science Foundation (No. 2013M541567) for their support.
Footnotes
Author contributions: Z.W., Z.C., M.T., and Y.Z. designed research; Z.W., M.S., and P.L. performed research; Z.C. and T.H. contributed new reagents or analytic tools; Z.W., M.S., and X.W. analyzed data; Z.W., M.T., and Y.Z. wrote the paper.
* This work is supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2012ZX09301001-007), the National Basic Research Program of China (973 Program) (No. 2014CBA02004), the Natural Science Foundation of China (No. 31370814), the National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China (2014ZX09507-002), and the Shanghai Pujiang Program (No. 13PJ1410300) to M.T. This work was also supported by NIH grants GM105933, CA160036, RR020839, and American Cancer Society (# RSG-13-198-01-DDC) to Y.Z.
 This article contains supplemental Figs. S1 to S8 and Tables S1 to S8.
This article contains supplemental Figs. S1 to S8 and Tables S1 to S8.
1 The abbreviations used are:
- PTM(s)
- Post-translational modification(s)
- Kme
- Lysine methylation
- ACN
- Acetonitrile
- CID
- Collisionally induced dissociation
- EDTA
- Ethylenediaminetetraacetic acid disodium salt dehydrate
- HPLC
- High Performance Liquid Chromatography
- Kbu
- Butyryllysine
- Kcr
- Crotonyllysine
- Kme1
- Monomethyllysine
- Kme2
- Dimethyllysine
- Kme3
- Trimethyllysine
- Kpr
- Propionyllysine
- PBS
- Phosphate buffered saline
- RPLC
- Reversed-phase liquid chromatography
- RT
- Room temperature
- SILAC
- Stable isotope labeling by amino acids in cell culture
- SAM
- S-adenosyl methionine
- TFA
- Trifluoroacetic acid.
REFERENCES
- 1. Ambler R. P., Rees M. W. (1959) Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature 184, 56–57 [DOI] [PubMed] [Google Scholar]
- 2. Allfrey V. G., Faulkner R., Mirsky A. E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rice J. C., Allis C. D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- 4. Martin C., Zhang Y. (2005) The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 5. Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 [DOI] [PubMed] [Google Scholar]
- 6. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 7. Copeland R. A., Solomon M. E., Richon V. M. (2009) Protein methyltransferases as a target class for drug discovery. Nat. Rev. Drug Discov. 8, 724–732 [DOI] [PubMed] [Google Scholar]
- 8. Schapira M. (2011) Structural chemistry of human SET domain protein methyltransferases. Curr. Chem. Genomics 5, 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pedersen M. T., Helin K. (2010) Histone demethylases in development and disease. Trends Cell Biol. 20, 662–671 [DOI] [PubMed] [Google Scholar]
- 10. Copeland R. A., Moyer M. P., Richon V. M. (2013) Targeting genetic alterations in protein methyltransferases for personalized cancer therapeutics. Oncogene 32, 939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arrowsmith C. H., Bountra C., Fish P. V., Lee K., Schapira M. (2012) Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 [DOI] [PubMed] [Google Scholar]
- 12. Clarke S. G. (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38, 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang J., Berger S. L. (2008) The emerging field of dynamic lysine methylation of non-histone proteins. Curr. Opin. Genet. Dev. 18, 152–158 [DOI] [PubMed] [Google Scholar]
- 14. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., Falnes P. O. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gu W., Roeder R. G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 16. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 17. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 18. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y., Zhao W., Yang J. S., Cheng Z., Luo H., Lu Z., Tan M., Gu W., Zhao Y. (2012) Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol. Cell. Proteomics 11, 1048–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelman Z., Naktinis V., O'Donnell M. (1995) Radiolabeling of proteins for biochemical studies. Methods Enzymol. 262, 430–442 [DOI] [PubMed] [Google Scholar]
- 21. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 22. Ong S. E., Mittler G., Mann M. (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 [DOI] [PubMed] [Google Scholar]
- 23. Carlson S. M., Moore K. E., Green E. M., Martin G. M., Gozani O. (2014) Proteome-wide enrichment of proteins modified by lysine methylation. Nat. Protoc. 9, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bremang M., Cuomo A., Agresta A. M., Stugiewicz M., Spadotto V., Bonaldi T. (2013) Mass spectrometry-based identification and characterization of lysine and arginine methylation in the human proteome. Mol. Biosyst. 9, 2231–2247 [DOI] [PubMed] [Google Scholar]
- 25. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moore K. E., Carlson S. M., Camp N. D., Cheung P., James R. G., Chua K. F., Wolf-Yadlin A., Gozani O. (2013) A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol. Cell 50, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., Hunt D. F. (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S. C., Chen Y., Mirza S., Xu Y., Lee J., Liu P., Zhao Y. (2006) A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J. Proteome Res. 5, 3446–3452 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Yang F., Gritsenko M. A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M. E., Lopez-Ferrer D., Reno T., Moore R. J., Klemke R. L., Camp D. G., 2nd, Smith R. D. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11, 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y., Kwon S. W., Kim S. C., Zhao Y. (2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 31. Wu Z., Moghaddas Gholami A., Kuster B. (2012) Systematic identification of the HSP90 candidate regulated proteome. Mol. Cell. Proteomics 11, M111 016675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M., Goto S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) The Pfam protein families database. Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hochberg Y. B. a. Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 12 [Google Scholar]
- 37. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bader G. D., Hogue C. W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruepp A., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Stransky M., Waegele B., Schmidt T., Doudieu O. N., Stumpflen V., Mewes H. W. (2008) CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 36, D646–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Macek B., Mann M., Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 41. Olsen J. V., Mann M. (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao Y., Jensen O. N. (2009) Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. http://www.unimod.org/modifications_list.php?
- 45. http://www.uniprot.org/.
- 46. Liu M., Liu Y., Cheng J., Zhang S. L., Wang L., Shao Q., Zhang J., Yang Q. (2004) Transactivating effect of hepatitis C virus core protein: a suppression subtractive hybridization study. World J. Gastroenterol. 10, 1746–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greer E. L., Shi Y. (2012) Histone methylation: a dynamic mark in health, disease, and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem. 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 49. Boudrez A., Beullens M., Groenen P., Van Eynde A., Vulsteke V., Jagiello I., Murray M., Krainer A. R., Stalmans W., Bollen M. (2000) NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem. 275, 25411–25417 [DOI] [PubMed] [Google Scholar]
- 50. Palmieri F. (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 51. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Musselman C. A., Lalonde M. E., Cote J., Kutateladze T. G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park J., Chen Y., Tishkoff D. X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B. M., Skinner M. E., Lombard D. B., Zhao Y. (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bothwell I. R., Islam K., Chen Y., Zheng W., Blum G., Deng H., Luo M. (2012) Se-adenosyl-L-selenomethionine cofactor analog as a reporter of protein methylation. J. Am. Chem. Soc. 134, 14905–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu H., Galka M., Mori E., Liu X., Lin Y. F., Wei R., Pittock P., Voss C., Dhami G., Li X., Miyaji M., Lajoie G., Chen B., Li S. S. (2013) A method for systematic mapping of protein lysine methylation identifies functions for HP1beta in DNA damage response. Mol. Cell 50, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cao X. J., Arnaudo A. M., Garcia B. A. (2013) Large-scale global identification of protein lysine methylation in vivo. Epigenetics 8, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 58. Chin H. G., Esteve P. O., Pradhan M., Benner J., Patnaik D., Carey M. F., Pradhan S. (2007) Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 35, 7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarraf S. A., Stancheva I. (2004) Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 [DOI] [PubMed] [Google Scholar]
- 60. Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J. P., Imhof A., Nakatani Y., Dent S. Y., Almouzni G. (2009) The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10, 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Downward J., Parker P., Waterfield M. D. (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311, 483–485 [DOI] [PubMed] [Google Scholar]
- 62. Jain S., Wei J., Mitrani L. R., Bishopric N. H. (2012) Auto-acetylation stabilizes p300 in cardiac myocytes during acute oxidative stress, promoting STAT3 accumulation and cell survival. Breast Cancer Res. Treat. 135, 103–114 [DOI] [PubMed] [Google Scholar]
- 63. Yang Y., Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 [DOI] [PubMed] [Google Scholar]
- 64. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 65. Guillemette B., Drogaris P., Lin H. H., Armstrong H., Hiragami-Hamada K., Imhof A., Bonneil E., Thibault P., Verreault A., Festenstein R. J. (2011) H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 7, e1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wagner T., Jung M. (2012) New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 30, 622–623 [DOI] [PubMed] [Google Scholar]
- 67. Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., Reinberg D. (2004) Regulation of p53 activity through lysine methylation. Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.