Abstract
Background:
Emergence and spread of carbapenemase (blaOXA) genes in multidrug resistant Acinetobacter baumannii (MDR-AB) forming biofilm complicated treatment of the patients infected with this microorganism particularly in intensive care units (ICUs).
Objectives:
The current study aimed to determine the prevalence of molecular class-D OXA carbapenemase in biofilm and non-biofilm forming strains of MDR-AB.
Materials and Methods:
A total of 65 strains of MDR-AB were isolated from the patients hospitalized in the ICU of two hospitals in Kerman, Iran. The isolates were identified by conventional microbiological tests as well as API 20NE assay. Antibiotic susceptibility was carried out by disk diffusion method; minimum inhibitory concentration (MIC) of carbapenems was measured by E-test. The presence of blaOXA genes among the isolates were studied by duplex-polymerase chain reaction and application of appropriate primers. Biofilm formation was detected by microtiter plate method.
Results:
The isolates were highly resistant to ciprofloxacin, levofloxacin, piperacillin, nalidixic acid and third generation cephalosporins such as tigecycline (7%; n = 5) and colistin (13%; n = 8). Among the isolates, 77% (n = 50) exhibited high MIC (265μg/mL) for imipenem. Both the blaOXA-51 and blaOXA-23 like genes coexisted in all the isolates; while, blaOXA-24/40 like gene was only detected in 29 imipenem-resistant strains (P ≤ 0.05). The blaOXA-58 like gene was not detected among the isolated strains. Quantification of biofilm introduced 23 isolates (including blaOXA-24/40 strains) with efficient attachment to microtiter plate; while, those isolates without blaOXA-24/40, or imipenem-sensitive strains formed weak or no biofilm.
Conclusions:
Coexistence of the blaOXA-51, blaOXA-23 and blaOXA-24/40 like genes, along with formation of strong biofilm, in MDR-AB strains particularly with indiscriminate use of imipenem, complicated treatment of the patients infected with these bacteria in the hospitals understudy.
Keywords: Acinetobacter baumannii, Biofilms, Carbapenemase
1. Background
Acinetobacter baumannii has increasingly allocated an important situation as a hospital-acquired pathogen worldwide. The contribution of A. baumannii to nosocomial infections has increasingly raisen over the past three decades, and many outbreaks of hospital infections involving this microorganism are reported worldwide (1-3). One of the important therapeutic features of this organism is resistance to a wide range of antibiotics including the third generation cephalosporins, carbapenems, aminoglycosides, monobactams, and fluoroquinolones (4). This property, along with the ability to form biofilm, as documented by recurring outbreaks contributed significantly to the infections caused by this microorganism (5, 6).
Carbapenem hydrolyzing β-lactamases (CHDLs) belonging to molecular class-D (OXA enzymes) is one of the main enzymes contributing to the inactivation of imipenem and meropenem in A. baumannii (7, 8). The OXA carbapenemases in Acinetobacter spp. are divided into four phylogenetic subgroups: OXA-23, OXA-24, OXA-51, and OXA-58 (9), but recently some researchers have reported the presence of blaOXA-23 and blaOXA-51 in non A. baumannii species. The blaOXA-23 gene was found in A. pittii (10). Of the 174 imipenem-resistant A. baumannii isolated from clinical specimens of the ICU ward in a tertiary care hospital in Greece, from 2010 to 2011 (11), 95% carried blaOXA-23 gene. Distribution of OXA subgroups in A. baumannii strains isolated from different regions of Turkey revealed that all the isolates carried blaOXA-51; however, blaOXA-24 gene could not be found in any isolates (12). The blaOXA-23 gene was first identified in Scotland and then was increasingly reported worldwide (13, 14).
In a study carried out on 104 carbapenem-resistant A. baumannii isolated from ICU of the hospitals in Krakow, Poland, blaOXA-51 like gene was detected in all the isolates; while 46 isolates also carried blaOXA-23 gene (15). Similarly, antimicrobial susceptibility patterns and the prevalence of OXA-type carbapenemases among the clinical isolates of A. baumannii in Tehran hospitals, Iran (16), introduced 123 isolates, 100 (81.3 %) had OXA-23, 10 (8.1%) possessed OXA-24, and one (0.81%) carried OXA-58 carbapenemase.
Several studies reported the relationship between biofilm and antibiotic resistance biofilm-based infections. In this regard, A. baumannii is among the leading nosocomial pathogens capable of producing severe biofilm-related infections such as colonization on venous catheters (CVCs) and lower respiratory tract infections (due to contaminated ventilators) (17). Furthermore, biofilm formation is important in the chronic infections, and colonizing bacteria can resist phagocytosis and evade the immune system.
2. Objectives
Due to overwhelming increase in carbapenem-resistant A. baumannii (CR-AB) in ICUs of the hospitals in Kerman, Iran, the present study was carried-out to evaluate the prevalence of carbapenem-resistant genes belonging to molecular class-D (OXA) in biofilm and non-biofilm forming A. baumannii by duplex–polymerase chain reaction.
3. Materials and Methods
3.1. Patients and Bacterial Isolates
A total of 65 non-duplicated MDR-AB strains were isolated from 266 patients hospitalized in the ICU wards of two main hospitals affiliated to the Kerman University of Medical Sciences (Afzali Pour and Shahid-Bahonar) in Kerman, Iran, from February to August 2013. From these isolates, 42% (n = 28) were collected from lung of the patients assisted by ventilator, 27.5% (n = 18) from UTI patients, and the remaining were collected from blood and soft tissue of burnt patients (P ≤ 0.05). The length of hospitalization among these patients was between 15 and 20 days. Samples were collected by an expert laboratory technician and inoculated into 5 mL sterile Stuart Transport (ST) medium (Hi-Media, India) broth and transferred to the department of microbiology laboratory within 24 hours of the collection. Prior to collection of the samples, demographic information including gender, age, clinical conditions and previous antibiotic exposure were considered.
3.2. Bacterial Identification
The isolates were inoculated into blood and Luria-Bertani (LB) agar (Merck, Germany) medium and incubated overnight at 37°C. The individual colonies were identified by conventional microbiological tests such as colony characteristic on cysteine lactose electrolyte-deficient agar (CLED) (Hi-Media, India), Gram staining, oxidation/fermentation of various sugars (OF), motility, oxidase and catalase tests (18), and further the species were confirmed by API 20NE assay (BioMerieux, Marcy l’Etoile, France). The reference strains of A. baumannii 19606 and Escherichia coli K12 DH5α were used as positive and negative controls.
3.3. Antibiotic Susceptibility Testing
Antimicrobial susceptibility of the isolates to the following antibiotics were carried out by the Kirby-Bauer (KB) disk diffusion method according to the Clinical Laboratory Standards Institute (CLSI 2012) guide lines (19); tetracycline (TE) [30 μg], amikacin (AN) [30 μg], tigecycline (TIG) [15 μg], colistin (CL) [10 μg], gentamicin (GM) [10 μg], piperacillin (PIP) [100 μg], ciprofloxacin (CIP) [5 μg], ceftazidime (CAZ) [30 μg], cefotaxime (CTX) [30 μg], tobramycin (TOB) [10 μg], amoxicillin + clavulanic acid (AMC) [30 μg], rifampin (Rif) [30 μg], cefixime (CFM) [5 μg], nalidixic acid (NA) [30 μg], imipenem (IMP) [10 μg] and meropenem (MEM) [10 μg] (Hi-media-India). Susceptibility to tigecycline was classified based on EUCAST criteria (20) (MIC ≤ 0.5 μg/mL; inhibition zone ≤ 17 mm). Minimum inhibitory concentrations (MICs) of the IMP and MEM were determined by E-test (Hi-Media, India) as described by the manufacturer’s instructions and interpreted according to CLSI guidelines (19). E. coli ATCC 25922 was used as the quality control strain.
3.4. Genomic DNA Isolation
Genomic DNA was isolated from individual colonies grown on TSA medium with the Genomic isolation kit (Thermo scientific, Lithuania) (21). DNA quantification was performed by spectrophotometry at 260 nm. The purity of DNA was evaluated by the ratio of the absorbance at 260 and 280 nm (A260/A280).
3.5. Detection of Carbapenemase (OXA) Genes by Duplex-PCR
Presence of blaOXA-like genes in A. baumannii strains were detected by duplex-PCR. The primers sequence (forward and reverse) specific to the above genes are shown in Table 1. PCR conditions were as follows: 10 μL master mix containing, 0.9 μL primer for each blaOXA like gene, OXA-51 (324 base pair), OXA-23 (501 bp), OXA-24 (246 bp), OXA-58 (599 bp), 2 μL genomic DNA, 5.5 μL DDW, 1U Taq polymerase, 1 × PCR buffer containing, 1.5 μM MgCl2 (25 mM) (QIAGEN Inc., Valencia, CA, USA) and 0.7 μM of each deoxynucleoside triphosphate (dNTPs). The duplex-PCR amplification was conducted in the gradient thermal cycler (Biometra-T gradient, Australia) and the initial denaturation temperature was 94°C for five minutes, 30 cycles of 94°C for 35 seconds, annealing 60°C for 35 seconds followed by 72°C for 40 seconds with the final extension step at 72°C for six minutes (21). DNA ladder consisted of a plasmid double digest with the size range 100-1200 bp purchased from Cinnagen Co. (Tehran, Iran). Simultaneously, positive and negative controls (E. coli K12 DH5α provided from institute pasture, Iran) were run along with the tested samples.
Table 1. Primers Used to Detect blaOXA Genes .
| blaOXA Gene | Sequence (5´→3´) | Size, bp | Reference |
|---|---|---|---|
| OXA-23 | F:GAT CGG ATT GGA GAACCAGA, R:ATT TCT GAC CGC ATT TCC AT | 501 | (11) |
| OXA-24 | F:GGT TAG TTG GCC CCC TTA AA, R:AGTTGAGCGAAA AGGGGATT | 246 | (20) |
| OXA-58 | F:AAG TAT TGG GGC TTG TGC TG, R:CCCCTCTGCGCTCTACATAC | 599 | (7) |
| OXA-51 | F: 5-TAA TGC TTT GAT CGG CCT TG-3, R: 5-TGG ATT GCA CTTCATCTT GG-3 | 324 | (22) |
3.6. Primary Attachment Assay
The initial attachment to surfaces is an important step in biofilm formation, and the subsequent pathogenesis of biofilm-associated infections. Therefore, the current study tested the capability of the strains to attach to a polystyrene surface. Briefly, overnight growth of A. baumannii cultures were adjusted to an OD 600 nm of 0.5 (1 × 108 CFU/mL) and diluted to 103 CFU/mL. Aliquots (100 μL) of the suspension were inoculated on microtiter plate wells. After 30 minutes of incubation at 37°C, the plate was gently rinsed three times with sterile 0.1 M phosphate-buffered saline (PBS) and then covered with 150 μL of Tryptic Soy Broth (TSB) (Merck, Germany). Primary attachment was expressed as the mean percentage of CFU (± SD) remaining on the microtiter plate compared to the initial inoculum.
3.7. Biofilm Formation Under Static Condition
Formation of the biofilm in each A. baumannii strain was quantified by microtiter plate method (22) with some modifications. Briefly, one loopful from each colony grown on LB-agar was inoculated into a sterile TSB medium (2 mL) containing glucose (1% W/V) to optimize biofilm production. The optical density (OD 650 nm) was then adjusted to 0.13 to reach 0.5 McFarland standard (1.5 × 108 CFU/mL) followed by further dilution of the prepared bacterial suspension to reach ~106 CFU/mL. One hundred microliter of each prepared inoculum was added into 96-well polystyrene flat bottom tissue culture microplate. Similarly, 100 μL of the TSB medium free of bacteria (negative control) was added to the related well and the microtiter plate was then incubated at 37°C under static condition. The TSB medium free of bacteria was used as the no biofilm-forming control.
After 24 hours, non-adherent cell suspensions were aseptically aspirated, washed and replaced with 10 μL of sterile phosphate buffered solution (pH = 7.2) to remove any remaining suspended cells. In order to fix the biofilm, 150 μL of methanol was added to each well and kept at room temperature (25°C) for 20 minutes. The methanol was then removed and replaced with 200 μL of crystal violet solution (1% W/V). The wells containing biofilm matrix were slowly washed with sterile deionized water and kept at room temperature till dried. Thereafter, 200 μL of glacial acetic acid (33% V/V) was added to each well and the optical density was measured at OD570 nm by S2 multi-mode microplate reader (BioTek, USA). The isolates were classified into strong, moderate, weak, and no biofilm based on attachment to microtiter plate using the formula given by Stepanovic et al. (21). Simultaneously, CFU/mL of each isolate in the wells were determined and subtracted from CFU/mL control. All the mentioned experiments were performed in triplicate and the most potent biofilm producer isolates were selected for further investigations.
3.8. Statistical Analyses
Pearson χ2 or Fisher’s exact tests were performed to compare the number of isolates resistant and sensitive to antibiotics. Statistical analyses were performed using SPSS 17.0 (SPSS, Chicago, IL, USA); P ≤ 0.05 was considered as level of significance for the two-tailed tests.
4. Results
4.1. Susceptibility to Antibiotics
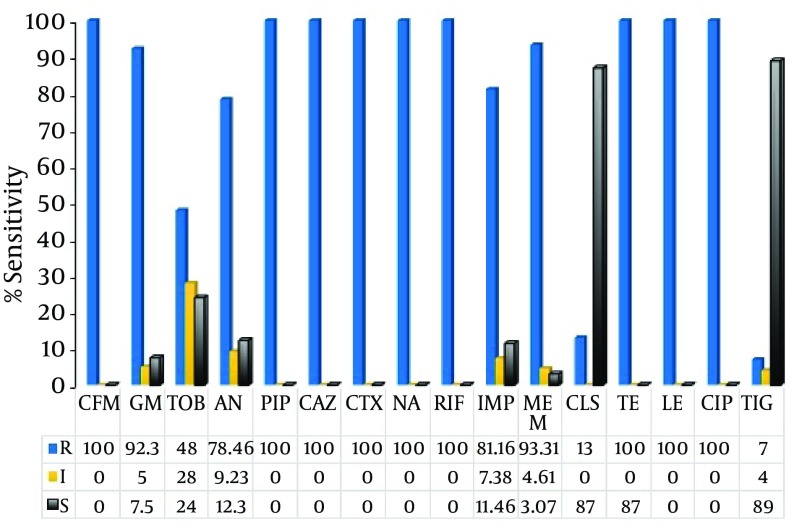
The susceptibility of the A. baumannii isolates to various antibiotics is shown in Figure 1. Approximately, all of the isolates were fully resistant to CIP, LE, CAZ, CTX, PI, TE, followed by AN (78.5%; n = 51) and TOB (48%; n = 31); whereas, 93% (n = 61) of the isolates were susceptible to tigecycline, a derivative of minocycline (Figure 1). In case of colistin, 13% of the population was resistant to this antibiotic. The isolates exhibited high MIC to the imipenem and meropenem as illustrated in Table 2. Eighty percent of the A. baumannii exhibited an MIC of 256 μg/mL and 15.4% showed an MIC of 128 μg/mL to both the IMP and MEM (P ≤ 0.05).
Figure 1. Antibiotic Susceptibility of Acinetobacterbaumannii Strains Isolated From ICU Patients.
Abbreviations; CFM, Cefixime; GM, gentamicin; TOB, tobramycin; AN, amikacin; PIP, piperacillin; LE, levofloxacin; CAZ, ceftazidime; CTX, cefotaxime; NA, nalidixic acid; RIF, rifampin; IMP, imipenem; MEM, meropenem; CL, colistin; TIG, tigecycline. Figures in the table indicate percent in bacterial population; R, resistance; I, intermediate; S, sensitive; Muller-Hinton agar was used for susceptibility test. The inoculum concentration was 10 × 108.
Table 2. Minimum Inhibitory Concentrations of Imipenem and Meropenem for Acinetobacterbaumannii.
| Antibiotic | MIC , μg/mL a | MIC 50 | MIC 90 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤ 0.5 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | |||
| IMP b | 0 (0) | 3.1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1.5 (1) | 3.1 (2) | 15.4 (10) | 76.9 (50) | 64 | 256 |
| MEM | 0 (0) | 1.5 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1.5 (1) | 1.5 (1) | 4.2 (4) | 12.1 (8) | 76.7 (51) | 64 | 256 |
a MIC was determined by E-strip test and the result was interpreted after 24 hours of incubation at 37ºC. Figures in the brackets indicate the number of isolates and the ones out of brackets indicate the percent in the population. The inoculum concentration was 10 × 108. The break point for each antibiotic was calculated according to the CLSI procedure.
b IMP, imipenem; MEM, meropenem.
4.2. Detection of blaOXA Like Genes by Duplex-PCR
The results of OXA genes detection among the isolates by duplex-PCR are presented in Figure 2 A and Figure 2 B. All of the isolates carried blaOXA-23 as well as blaOXA-51 like genes both in the imipenem resistant and imipenem sensitive strains, while blaOXA-24/40 was detected in 29 isolates with high MIC to imipenem and meropenem (256 μg/mL) (P ≤ 0.5). The blaOXA-58 like gene was absent in the entire population.
Figure 2. A) Duplex–PCR for blaOXA-23 and blaOXA-24/40 Like Genes; B) Multiplex-PCR for blaOXA-51 and blaOXA-58 genes for A. baumannii isolated from ICU patients; OXA-58 was absent in all the tested isolates.
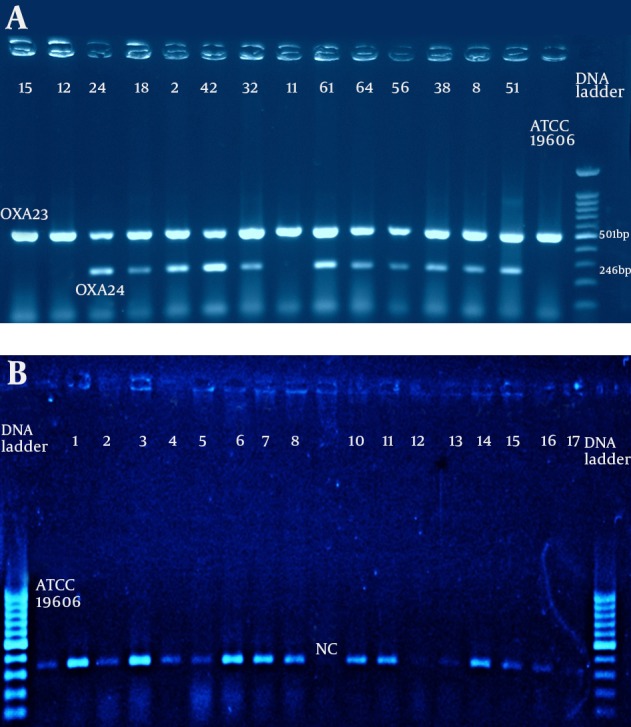
Positive control: A. baumannii ATCC19606. Negative control: E. coli DH5α. Electrophoresis conduct in 2% agarose gel concentration for one hour and 85V and the DNA bands were observed on UV-gel documentation system (UVITEC-Cambridge) attached to a camera.
4.3. Primary Attachment Assay
Primary attachment assays revealed significant initial binding differences between the isolates with strong biofilm, and moderate or weak biofilm forming A. baumannii to polystyrene surface. Those with both blaOXA-23 and blaOXA-24 genes had stronger initial attachment to microtiter wells compared to imipenem sensitive isolates.
4.4. Biofilm Assay Under Static Condition
Quantification of biofilm formation by microtiter plate method introduced 35.3% (n = 23) of the isolates as strong biofilm producer, furthermore these isolates had stronger initial attachment ability to microtiter wells. About 16.9% (n = 11) exhibited moderate biofilm formation, and also showed initial binding activity; while, in 23 % (n = 15) no biofilm was observed. Those isolates with both blaOXA-23 and blaOXA-24 genes had strong biofilm capability.
5. Discussion
Acinetobacter baumannii is one of the six most important multidrug-resistant microorganisms in hospitals worldwide. A. baumannii antimicrobial resistance has progressively increased since the 1970s, when the vast majority of strains were sensitive to the commonly used antibiotics. By 2007, up to 70% of the isolates in certain settings were MDR, including resistance to carbapenems, which were once considered the mainstay against MDR A. baumannii infections. The current study found 65 strains of A. baumannii recovered from the ICU of two main hospitals in Kerman, Iran, which were highly resistant to different classes of antibiotics. These clinical isolates formed various degrees of attachments and biofilms in microtiter plates contributed to their outstanding antibiotic resistance properties to a wide range of antibiotics (23). Thirty percent of the isolates were resistant to colistin.
This is very interesting, since the majority of the evaluated isolates in the present study showed high MICs to imipenem. Resistance to colistin in non-fermenters such as A. baumannii is rarely reported. In general, few studies have shown a relationship between possession of distinct antibiotic resistant genes and the resulted biofilm formation in the treatment failure infected patients with such strains in ICU (24). Experimental data in the current case showed three different subtypes of OXA genes in the strains showing strong biofilm. The study found that blaOXA-51 along with blaOXA-23 were the sole carbapenemase genes detected in all the isolates (including imipenem sensitive and resistant); while, blaOXA24/40 was only observed in the isolates showing high MIC value to imipenem and meropenem. The study also found that, those A. baumannii strains containing both blaOXA-23 and blaOXA-24/40-like genes exhibited strong biofilm activity. This is very interesting, since till now no other research studied the relationship between the presence of carbapenemase genes and biofilm quantity in this bacterium.
The OXA carbapenemase genes (CHDLs) are well distributed among the species of Acinetobacter spp. and are documented by various authors (22, 25, 26). These genes play an important role in the antibiotic resistant phenomenon among nosocomial bacteria and are reported on transposons, integrons or plasmid. In conclusion, the experimental data obtained in the current investigation revealed that many A. baumannii isolated in the ICU wards under study were resistant to high concentrations of imipenem and meropenem. The blaOXA-51, and blaOXA-23 like genes were presented in imipenem resistant and susceptible strains. Coexistence of the blaOXA-51, blaOXA-23 and blaOXA-24/40 like genes suggest the existence of integrons such as elements that encoded these genes and conferred resistance to carbapenems along with aminoglycosides. The blaOXA-58 carbapenemase gene was not found in the present study. The isolates were sensitive to tigecycline and colistin. The current research was limited to OXA genes; however, further research should be conducted on sequencing and molecular typing of the isolates (27).
Acknowledgments
The authors would like to thank the staff members of the Research Center for Infectious Diseases and Tropical Medicine, Department of Microbiology and Virology, Kerman University of Medical Sciences (Kerman, Iran) and Bacteriology Unit of the Pasture Institute of Iran for their help with this investigation. The current research was supported by the research council of Kerman University of Medical Sciences (Grant number 124/92 as part of Ph.D. research provided to Mr. Azizi). Authors also wish to thank the staff members of the microbiology laboratory and ICU ward of the Afzalipoor Hospital in Kerman for their cooperation.
Footnotes
Authors’ Contributions:Dr. Shakibaie supervised the project, wrote the manuscript and helped in analysis and interpretation of data: Azizi and Modarresi performed experimental data and contributed to the development of the protocol; Dr. Shahcherghi contributed to the Critical reading of the manuscript.
Funding/Support:This research was financially supported as grant no.18/92 provided to Dr. M.R. Shakibaie by Research Council of Kerman University of Medical Sciences, Kerman, Iran.
References
- 1.Cisneros JM, Rodriguez-Bano J. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin Microbiol Infect. 2002;8(11):687–93. doi: 10.1046/j.1469-0691.2002.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Giamarellou H, Antoniadou A, Kanellakopoulou K. Acinetobacter baumannii: a universal threat to public health? Int J Antimicrob Agents. 2008;32(2):106–19. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–51. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 4.Navon-Venezia S, Ben-Ami R, Carmeli Y. Update on Pseudomonas aeruginosa and Acinetobacter baumannii infections in the healthcare setting. Curr Opin Infect Dis. 2005;18(4):306–13. doi: 10.1097/01.qco.0000171920.44809.f0. [DOI] [PubMed] [Google Scholar]
- 5.Kuo HY, Chang KC, Kuo JW, Yueh HW, Liou ML. Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 2012;39(1):33–8. doi: 10.1016/j.ijantimicag.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12(9):826–36. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 7.Sohrabi N, Farajnia S, Akhi MT, Nahaei MR, Naghili B, Peymani A, et al. Prevalence of OXA-type beta-lactamases among Acinetobacter baumannii isolates from Northwest of Iran. Microb Drug Resist. 2012;18(4):385–9. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 8.Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006;57(1):1–3. doi: 10.1093/jac/dki425. [DOI] [PubMed] [Google Scholar]
- 9.Dalla-Costa LM, Coelho JM, Souza HA, Castro ME, Stier CJ, Bragagnolo KL, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J Clin Microbiol. 2003;41(7):3403–6. doi: 10.1128/JCM.41.7.3403-3406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boo TW, Walsh F, Crowley B. First report of OXA-23 carbapenemase in clinical isolates of Acinetobacter species in the Irish Republic. J Antimicrob Chemother. 2006;58(5):1101–2. doi: 10.1093/jac/dkl345. [DOI] [PubMed] [Google Scholar]
- 11.Liakopoulos A, Miriagou V, Katsifas EA, Karagouni AD, Daikos GL, Tzouvelekis LS, et al. Identification of OXA-23-producing Acinetobacter baumannii in Greece, 2010 to 2011. Euro Surveill. 2012;17(11) [PubMed] [Google Scholar]
- 12.Ciftci IH, Asik G, Karakece E, Oksuz L, Yagci S, Sesli Cetin E, et al. [Distribution of blaOXA genes in Acinetobacter baumannii strains: a multicenter study]. Mikrobiyol Bul. 2013;47(4):592–602. doi: 10.5578/mb.6388. [DOI] [PubMed] [Google Scholar]
- 13.Heritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49(8):3198–202. doi: 10.1128/AAC.49.8.3198-3202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J Antimicrob Chemother. 2009;63(1):55–9. doi: 10.1093/jac/dkn434. [DOI] [PubMed] [Google Scholar]
- 15.Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35(3):317–25. [PubMed] [Google Scholar]
- 16.Karmostaji A, Peerayeh SN, Salmanian AH. Distribution of OXA-Type Class D β-Lactamase Genes Among Nosocomial Multi Drug Resistant Acinetobacter baumanniiIsolated in Tehran Hospitals. Jundishapur J Microbiol. 2013;6(5) [Google Scholar]
- 17.Merkier AK, Catalano M, Ramirez MS, Quiroga C, Orman B, Ratier L, et al. Polyclonal spread of bla(OXA-23) and bla(OXA-58) in Acinetobacter baumannii isolates from Argentina. J Infect Dev Ctries. 2008;2(3):235–40. doi: 10.3855/jidc.269. [DOI] [PubMed] [Google Scholar]
- 18.Bouvet PJ, Grimont PA. Identification and biotyping of clinical isolates of Acinetobacter. Ann Inst Pasteur Microbiol. 1987;138(5):569–78. doi: 10.1016/0769-2609(87)90042-1. [DOI] [PubMed] [Google Scholar]
- 19.Cockerill FR, Clinical Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement;[... provides updated tables for... M02-A11 and M07-A9]. National Committee for Clinical Laboratory Standards; 2012. [Google Scholar]
- 20.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–3. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–9. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 22.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974–6. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: A study in western Iran. Iran J Microbiol. 2013;5(3):195–202. [PMC free article] [PubMed] [Google Scholar]
- 24.de Breij A, Dijkshoorn L, Lagendijk E, van der Meer J, Koster A, Bloemberg G, et al. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grosso F, Quinteira S, Poirel L, Novais A, Peixe L. Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob Agents Chemother. 2012;56(7):3969–72. doi: 10.1128/AAC.06255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shacheraghi F, Shakibaie MR, Noveiri H. Molecular Identification of ESBL genes blaGES-1, blaVEB-1, blaCTX-M blaOXA-1, blaOXA-4, blaOXA-10 and blaPER-1 in Pseudomonas aeruginosa strains isolated from burn patients by PCR, RFLP and sequencing techniques. Int J Biol life Sci. 2010;3(6):138–42. [Google Scholar]
- 27.Shakibaie MR, Adeli S, Salehi MH. Antibiotic resistance patterns and extended-spectrum beta-lactamase production among Acinetobacter spp. isolated from an intensive care Unit of a hospital in Kerman, Iran. Antimicrob Resist Infect Control. 2012;1(1):1. doi: 10.1186/2047-2994-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]