The aim of this study was to identify novel protein signatures that would predict clinical outcomes in a large cohort of patients with adrenocortical carcinoma based on data from previous gene expression microarray studies. Ki-67, TOP2A, and EZH2 were all significantly associated with poorer outcomes, whereas BARD1 was associated with improved overall survival. It is hoped that these biomarkers may help tailor additional therapy and be potential targets for directed therapy.
Keywords: Adrenocortical carcinoma, Biomarkers, Prognosis, Immunohistochemistry, Tissue microarray
Abstract
Background.
Adrenocortical carcinoma (ACC) is a rare malignancy with a poor prognosis. The aim of this study was to identify novel protein signatures that would predict clinical outcomes in a large cohort of patients with ACC based on data from previous gene expression microarray studies.
Materials and Methods.
A tissue microarray was generated from the paraffin tissue blocks of 61 patients with clinical outcomes data. Selected protein biomarkers based on previous gene expression microarray profiling studies were selected, and immunohistochemistry staining was performed. Staining patterns were correlated with clinical outcomes, and a multivariate analysis was undertaken to identify potential biomarkers of prognosis.
Results.
Median overall survival was 45 months, with a 5-year overall survival rate of 44%. Median disease-free survival was 58 months, with a 5-year disease-free survival rate of 44%. The proliferation marker Ki-67 and DNA topoisomerase TOP2A were associated with significantly poorer overall and disease-free survival. The results also showed strong correlation between the transcriptional repressor EZH2 and TOP2A expression, suggesting a novel role for EZH2 as an additional marker of prognosis. In contrast, increased expression of the BARD1 protein, with its ubiquitin ligase function, was associated with significantly improved overall and disease-free survival, which has yet to be documented for ACC.
Conclusion.
We present novel biomarkers that assist in determining prognosis for patients with ACC. Ki-67, TOP2A, and EZH2 were all significantly associated with poorer outcomes, whereas BARD1 was associated with improved overall survival. It is hoped that these biomarkers may help tailor additional therapy and be potential targets for directed therapy.
Implications for Practice:
Adrenocortical carcinoma (ACC) is a rare but highly lethal malignancy with limited treatment options. There has been an expansion of knowledge from gene expression profiling and pangenomic analyses of the genetic alterations associated with ACC. Many of the findings have yet to be validated at the tissue-specimen level to confirm their translational relevance. We report on a novel panel of protein markers that have been validated in a cohort of patients with ACC and are able to predict both poor and improved survival outcomes. Given the difficulty in envisaging outcomes with this disease, it is hoped that the identification of these new prognostic and predictive markers will aid in tailoring additional therapy and be targets of molecular therapy themselves for future research.
Introduction
Adrenocortical carcinoma (ACC) is a rare but aggressive heterogeneous malignancy with an incidence of 0.5–2.0 per 1 million population [1]. The disease is typically sporadic, with patients presenting in a bimodal age pattern, either early in the first decade of life or in the 40- to 50-year age bracket [2, 3]. At presentation, most patients have resectable disease, but disappointingly, up to 40% of patients will already have systemic disease [1, 4, 5]. Morbidity arises from aggressive local and distant disease and from dysregulated steroid hormone production. Complete surgical resection (R0) is the mainstay of therapy.
Research has been expanded to unravel the pathogenesis of ACC, and the molecular landscape of ACC has changed dramatically over the past 15 years. Significant advances in our understanding of the molecular aspects of the disease have arisen from identification of genetic changes in rare familial syndromes manifesting ACC and comparison to sporadic ACC and use of genomewide messenger RNA (mRNA) and micro-RNA (miRNA) expression studies and comparative genomic hybridization and methylation profiling studies [6–11]. In a similar fashion, genomewide gene expression profiling has identified several dysregulated genes associated with ACC [12–16] and has been able to stratify outcomes depending on the grade of ACC [12, 17].
These gene expression profiling and pangenomic analyses of the genetic and epigenetic alterations in ACC have generated a myriad of data points that will require further validation. It is important that these data be validated in tissue sections to confirm the differential expression at the RNA or protein level. Tissue microarray (TMA) technology has been gaining momentum as a unique diagnostic and research tool in the molecular analysis of cancer biology [18–22]. Our aim was to use TMA technology with immunohistochemistry (IHC) to validate previously identified dysregulated genes in ACC and to identify novel protein signatures that would predict clinical outcomes in a large cohort of patients with ACC.
Methods
Clinical Data and Patient Samples
Given the rarity of ACC, there was a need to pool resources and use a multicenter approach to gain meaningful data regarding clinical characteristics and outcomes. This multicenter study was undertaken with approval by the Northern Sydney Human Research Ethics Committee. The lead investigating center was the University of Sydney Endocrine Surgery Unit, Royal North Shore Hospital (RNSH), Sydney, Australia, with collection of clinical data from teaching and district hospitals, both public and private, across the state of New South Wales (NSW), Australia, and accruement of formalin-fixed paraffin-embedded (FFPE) tissue blocks from pathology departments across the state.
A final cohort of 71 patients diagnosed with ACC was accrued. These patients had been entered postoperatively in the New South Wales Cancer Registry from 1998 to 2013 and had available FFPE tissue samples for analysis. Relevant clinical data were obtained from various sources, including hospital records and multiple treating clinicians (surgeons, physicians, general practitioners). To ensure accuracy in data acquisition, clinical data were collected and independently recorded by four investigators (J.C.Y.I., T.C.Y.P., A.R.G., P.S.). This included basic patient demographics, clinical presentation, endocrine functional assessment, imaging, preoperative diagnosis, operative findings and technique, histopathological findings, medical management, clinical outcome, and survival data. All records were then compiled into one database using Microsoft Excel 2011 (Microsoft Corporation, Redmond, WA, http://www.microsoft.com). Data entered into the database were verified and agreed on by at least two investigators (J.C.Y.I., T.C.Y.P.).
Clinical assessment included whether the patient presented incidentally, by means of a mass effect, or with hormonal imbalance, hemorrhage, or paraneoplastic symptoms. Multiple means of presentation were also recorded. The date of diagnosis was standardized to the date of surgery. Endocrine workup of each patient was assessed by examining the medical records for results of biochemical investigations related to overproduction of cortisol, aldosterone, and/or androgens. Resection margins were determined from operative and histopathology reports and recorded in the standard fashion (R0, no evidence of tumor, complete resection; R1, microscopic evidence of residual tumor with positive margins; R2, macroscopic evidence of residual tumor).
The European Network for the Study of Adrenal Tumors (ENSAT) staging system was used because it had been shown to be superior to traditional TNM staging [23, 24]. The evaluation of medical management included an assessment of use of adjuvant radiotherapy, chemotherapy, and mitotane therapy.
Clinical outcome data were determined from the date of death as recorded by the NSW cancer registry or the Ryerson Index [25]. Survival times were determined from the date of surgery. Overall survival was defined by censoring at the date of death from any cause or the date of last follow-up. Recurrence-free survival was determined only in patients with no evidence of metastases at presentation. Time to recurrence in this case was determined by the date of death due to ACC or date of recurrence of any type (local, regional, or distant). Patients were otherwise censored at date of last follow-up.
Selection of Biomarkers
Our earlier work using microarray gene expression identified several genes known to be important in adrenal carcinogenesis [7]. These genes were noted to be significantly overexpressed in ACCs compared with adrenocortical adenomas (ACAs). From this data, we selected several candidate markers involved in cellular proliferation (Ki-67), cell-cycle control (cyclin B1, TOP2A), DNA damage repair (BARD1), cell signaling (IGF2), and transcription (GR, SF-1, EZH2) for further IHC analysis.
Creation of TMA Slides for Immunohistochemistry
A TMA was constructed from the FFPE blocks of representative ACCs using a manual tissue-arraying instrument (Chemicon ATA-100; EMD Millipore, Billerica, MA, http://www.emdmillipore.com). A 1.0-mm-diameter needle and matching stylet were used for all TMAs, and all ACC specimens were sampled in triplicate. The TMA paraffin block was placed in a rotary microtome (Leica RM2125; Leica Biosystems, Mount Waverley, Victoria, Australia, http://www.leicabiosystems.com), and multiple 4-μm sections were cut and adhered to positively charged slides (Superfrost Plus; Menzel-Glaser, Braunschweig, Germany, http://www.menzel.de).
Immunohistochemistry
IHC was performed on the TMA slides sectioned at 4 μm onto positively charged slides (Superfrost Plus; Menzel-Glaser). The clones, dilutions, catalog numbers, and manufacturers for the different antibodies were as follows: IGF2 (clone S1F2, 1/400, 05-166; Upstate; EMD Millipore), GR (clone 4H2, 1/20, NCL-GCR; Novocastra; Leica Biosystems), SF-1 (clone N1665, 1/100, PP-N1665-00; Perseus Proteomics, Komaba, Japan, http://www.ppmx.com), cyclin B1 (clone 7A9, 1/20, NCL-cyclin B1; Novocastra; Leica Biosystems), Ki-67 (clone MIB-1, 1/50, M7240; Dako, Carpenteria, CA, http://www.dako.com), BARD1 (1/400, HPA044864; Sigma-Aldrich, St. Louis, MO, https://www.sigmaaldrich.com), EZH2 (clone D2C9, 1/100, 5246; Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com), and TOP2A (clone 3F6, 1/50, NCL-TOPIIA; Novocastra; Leica Biosystems).
All slides were processed with an automated staining system, the Leica Biosystems Bond III autostainer, used according to the manufacturer’s protocol and with the manufacturer’s retrieval solutions.
For GR, SF-1, and BARD1, heat-induced epitope retrieval was performed for 30 minutes in the manufacturers’ acidic retrieval solution ER1 (VBS part number AR9961). For cyclin B1, Ki-67, EZH2, and TOP2A, heat-induced epitope retrieval was performed for 30 minutes in the manufacturers’ alkaline retrieval solution ER2 (VBS part number AR9640). For IGF2, enzyme-based antigen retrieval was performed for 10 minutes using the manufacturer’s enzyme pretreatment kit (VBS part number AR9551). A biotin-free detection system was used (VBS part number DS9713). Appropriate external positive controls for each stain were performed.
All slides were interpreted by an experienced endocrine pathologist (A.J.G.) in conjunction with a matched hematoxylin and eosin-stained slide. At the time of examination, the pathologist was blinded to all clinical and pathological data. For Ki-67, a standard proliferative index expressed as the percentage of neoplastic cells demonstrating positive staining was determined; for all other stains, tumors were scored semiquantitatively as 0 (negative), 1+ (focally or weakly positive), 2+ (moderate staining), 3+ (diffuse strong staining), or 4+ (intense diffuse staining).
Statistical Analysis
Descriptive statistics were presented as mean ±SD, median (interquartile range), or as count (percentage), depending on distribution. Staining intensity was converted into binary values. Cutoff values were selected using regression tree analysis. Univariate and multivariate inferential survival analyses were performed using Cox proportional hazards models. A multivariate model was then fitted to the entire cohort using a stepwise approach with covariates, with p < .1 retained. The final model was assessed for validity of the proportional hazards assumption using Schoenfeld residuals and goodness of fit using Cox-Snell residuals.
A C statistic was calculated to assess the discrimination ability of the model. Internal validation of the multivariate model was performed by refitting the model using the same stepwise approach to 200 bootstrap samples, and this model’s performance was tested against the bootstrap and original data sets to assess the degree optimism of the original C statistic. An adjusted C statistic was then calculated by subtracting this optimism from the original estimate [26]. In this way, a survival model based on IHC staining was compared with a model based purely on clinical characteristics. The covariates for this clinical model were selected from analysis of the larger cohort from a previous study [27]. A combined model was also developed using a similar approach.
Univariate analysis of disease-free survival of those patients with no evidence of metastases on presentation using Cox models was performed, but no multivariate analysis was performed because of the small sample size. Association between staining characteristics and prolonged survival in advanced disease was also tested with Fisher’s exact test (univariate analysis). Data management and statistical analyses were performed using Stata SE version 11.2 for Windows (StataCorp LP, College Station, TX, http://www.stata.com).
Results
Clinical and Histopathological Characteristics
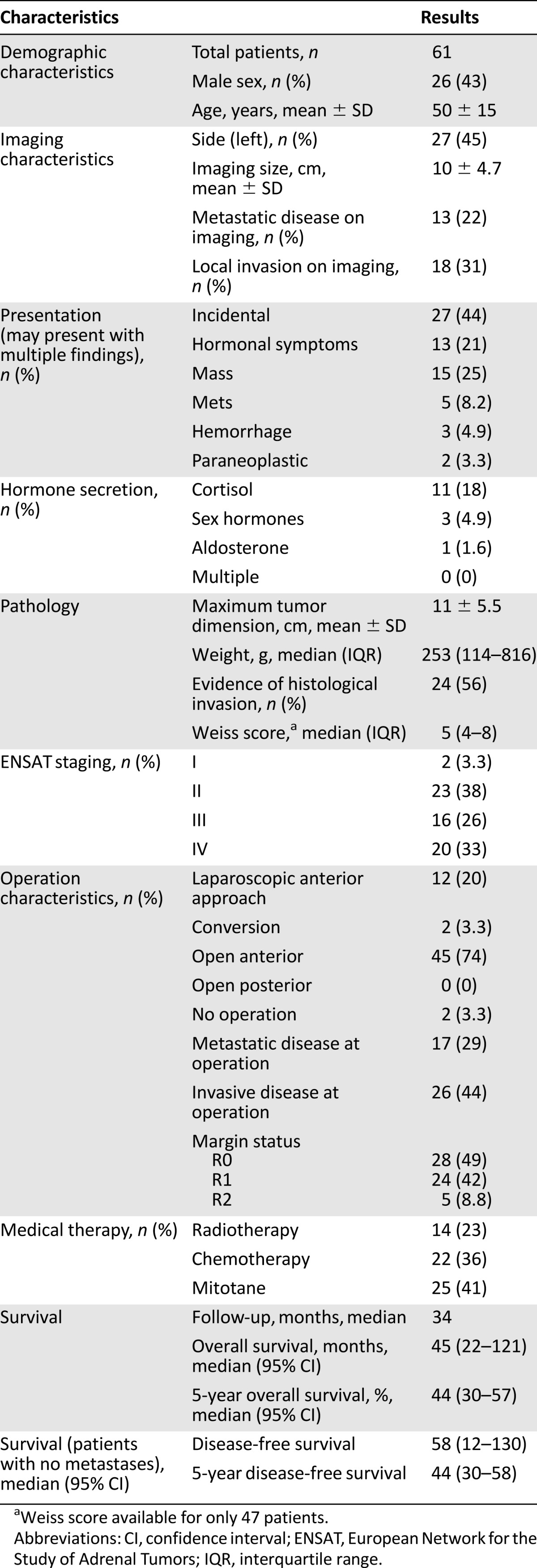
In the 15-year study period, a total of 71 patients had available tumor blocks with complete clinical data; however, 10 patients’ FFPE blocks were unsuitable for TMA construction or further analysis and subsequently were excluded from the study. This left 61 patients to be included in the study. Patient characteristics are summarized in Table 1. There was a slight female preponderance (n = 35, 57%), and the mean age was 50 ± 15 years.
Table 1.
Patient characteristics
Treatment
The majority of patients (n = 45, 74%) underwent open laparotomy (anterior approach) for their operation. Intraoperative evidence showed that adjacent organ invasion was present in 26 patients (44%) and that intraoperative metastases were present in 17 patients (29%). R0 resection was achieved in 28 patients (49%), 24 (42%) had an R1 resection, and 5 patients (8.8%) underwent an R2 resection. Four patients had missing data on resection status.
In patients who had medical therapy, 25 (41%) had mitotane therapy, 14 (23%) received radiotherapy, and 22 (36%) received various regimens of chemotherapy.
Staining Characteristics and Survival
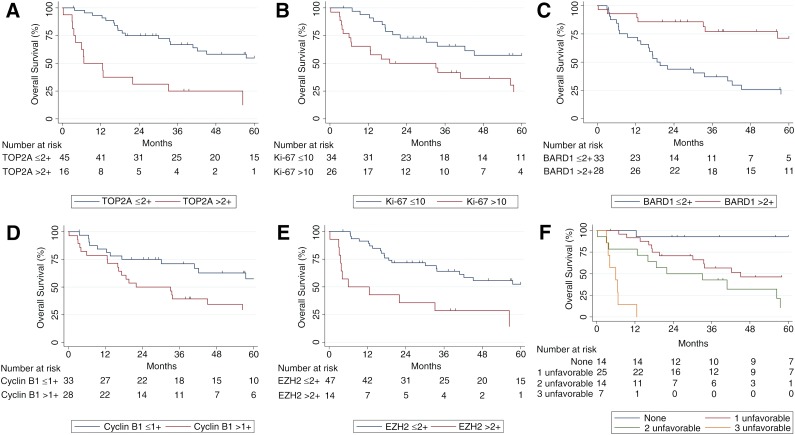
Survival data were available for all patients. Representative IHC images for various stains can be seen in Figure 1. The univariate and multivariate models for patients’ overall survival are presented in Tables 2 and 3. On both univariate and multivariate analysis, staining for TOP2A (staining score >2) (Fig. 2A) and Ki-67 (staining score >10%) (Fig. 2B) were found to be significant factors associated with poor prognosis. In contrast, staining for BARD1 (staining score >2) (Fig. 2C) was associated with significantly better overall survival on both univariate analysis (hazard ratio [HR]: 0.33; 95% confidence interval [CI]: 0.14–0.76; p = .009) and multivariate analysis (HR: 0.11; 95% CI: 0.043–0.28; p < .001). Cyclin B1 was also significantly associated with poorer overall survival on univariate analysis (HR: 2.4; 95% CI: 1.0–5.2) (Fig. 2D).
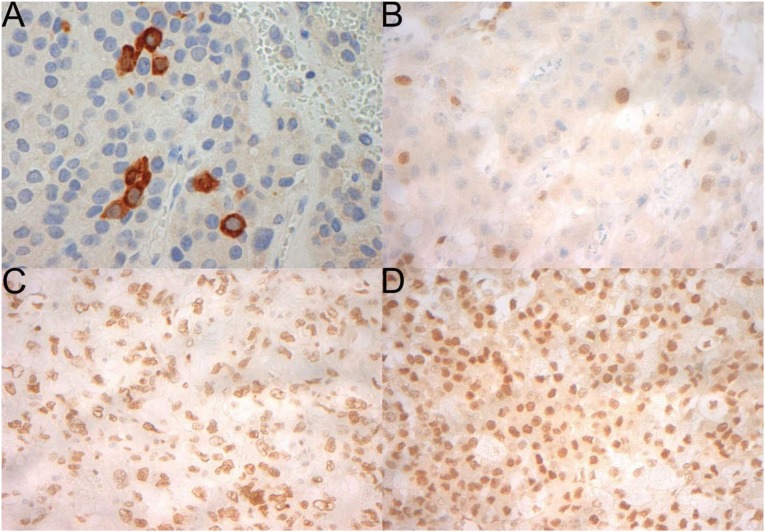
Figure 1.
Representative immunohistochemistry for cyclin B1, which demonstrates a cytoplasmic pattern of staining (A); TOP2A, which is nuclear (B); EZH2, which is nuclear (C); and BARD1, which is nuclear (D). All magnification ×400.
Table 2.
Univariate models for staining (overall survival)
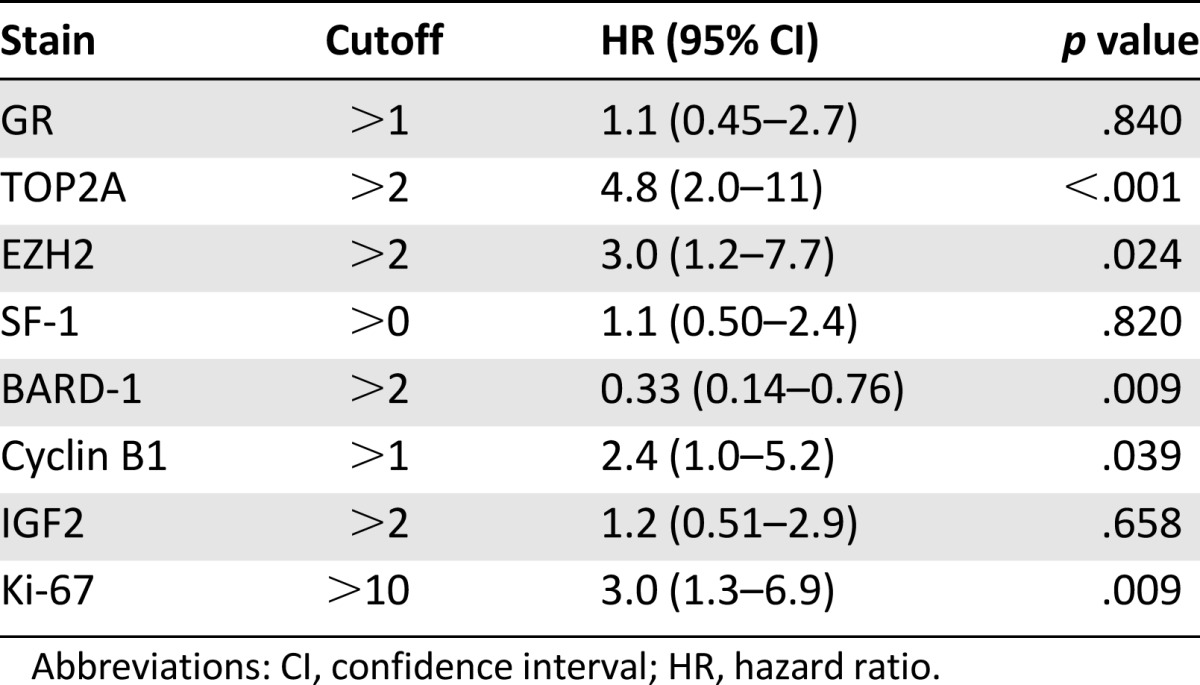
Table 3.
Multivariate models (overall survival) with and without clinical data incorporated
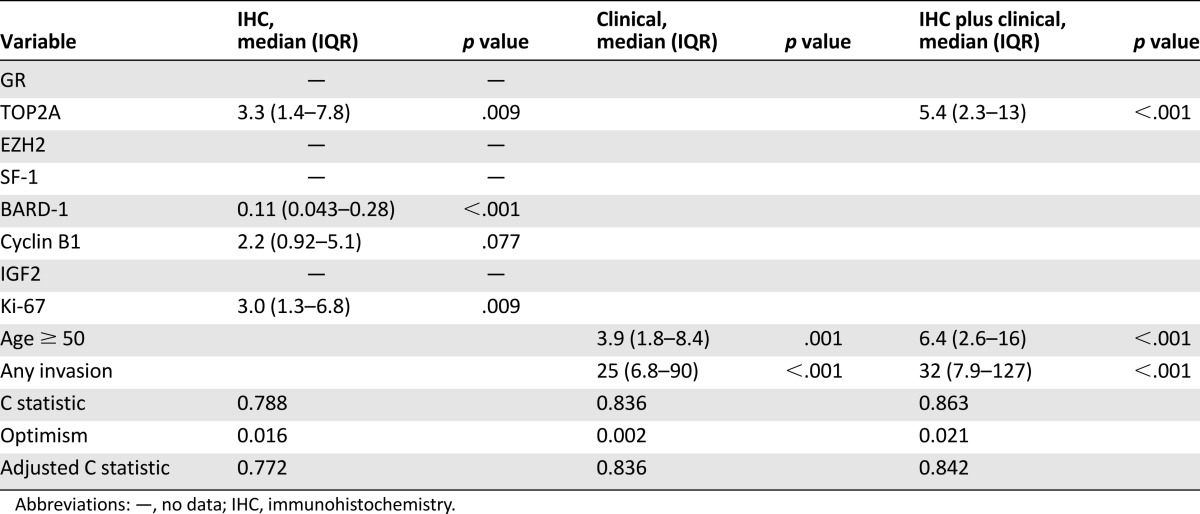
Figure 2.
Kaplan Meier charts summarizing overall survival of the cohort according to biomarker over- or underexpression (A–E). (F): Kaplan Meier chart describing overall survival in patients with zero, one, two, or three unfavorable staining characteristics (TOP2A >2, BARD1 ≤2+, and Ki-67 >10%).
On univariate analysis, EZH2 (staining score >2) was associated with poorer overall survival (HR: 3.0; 95% CI: 1.2–7.7) (Fig. 2E). It is interesting to note that EZH2 and TOP2A staining correlated highly with each other. As a binary variable, the correlation coefficient was 0.83, and Fisher’s exact test strongly suggested an association (p < .001), with only 4 of 61 patients classified in different groups on the two tests. Consequently, EZH2 was not included in the multivariate model to prevent collinearity issues.
When examining IHC as a model compared with clinical factors, IHC staining results in isolation gave a C statistic of 0.788, which was adjusted for optimism to 0.772 on internal bootstrap validation (Table 3). Figure 2F summarizes overall survival depending on the number of unfavorable staining characteristics (TOP2A >2, BARD1 ≤2+, and Ki-67 >10%). When examining clinical factors in isolation, age ≥50 years and any invasion were significant risk factors for poor prognosis. After accounting for these two clinical factors, TOP2A remained a significant predictor of poor prognosis. The combined model (TOP2A IHC and clinical factors) demonstrated a C statistic (adjusted for optimism) of 0.842 (strong) (Table 3).
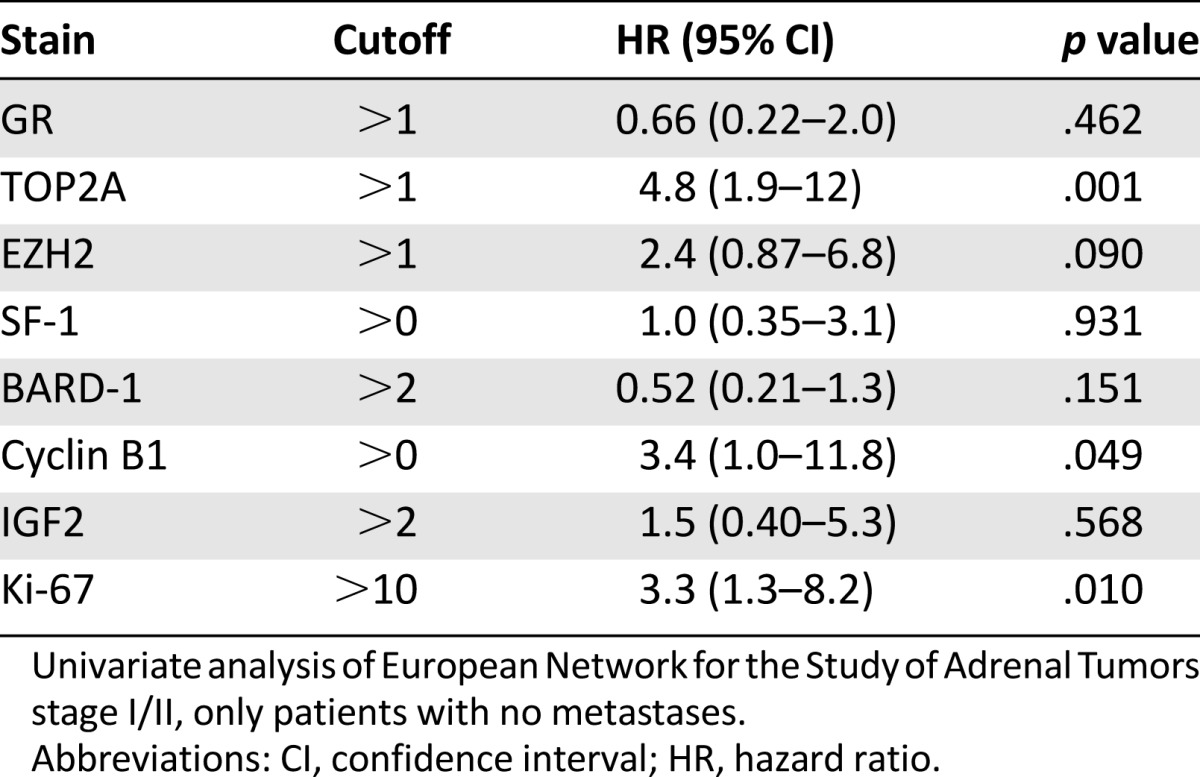
In an attempt to gain further insight into whether protein marker expression correlated with prognosis depending on the stage of presentation, grouped stage-specific analysis of outcomes of patients with ENSAT stage I or II and ENSAT stage III or IV disease was undertaken. When disease-free survival was examined with patients presenting with ENSAT stage I or II disease, TOP2A, cyclin B1, and Ki-67 were associated with poor prognosis on univariate analysis (Table 4). Multivariate analysis was not performed because of the small sample size.
Table 4.
Univariate analysis of disease-free survival
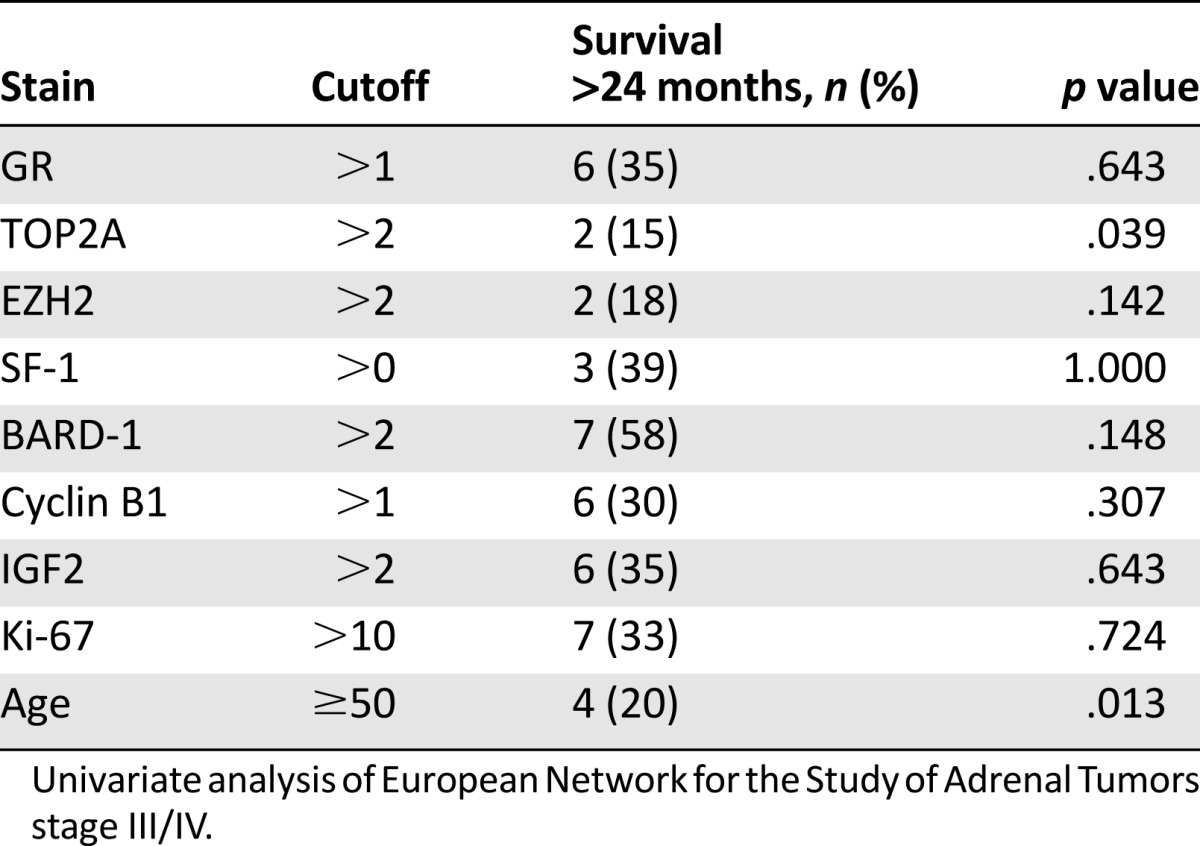
For patients with advanced disease (ENSAT stage III or IV), analysis revealed that increased TOP2A expression (p = .039) and age ≥50 years (p = .013) were found to be associated with a significantly lower likelihood of survival beyond 2 years (Table 5). Again, multivariate analysis was not performed because of the small sample size; however, the effect of TOP2A did not seem to be related to the effect of age because correlation between age category and TOP2A staining (binary variable) was poor (0.15). No patients aged >50 years and who had high TOP2A staining survived >24 months, although this result failed to achieve statistical significance (p = .117).
Table 5.
Univariate analysis of survival >24 months
Discussion
Long-term outcome and survival of malignant ACC is difficult to predict, given the heterogeneity of the disease, both clinically and pathologically. The modified Weiss score is the most widely accepted scoring system for classification of an adrenocortical tumor as a carcinoma or an adenoma [28, 29]. Further clinical intervention, whether it be surgery, radiotherapy, and/or chemotherapy, is predicated on the diagnosis; however, the system is not infallible, with benign tumors giving rise to metastases and with a tumor identified as malignant behaving in a benign fashion [30].
One of the more significant clinical advances in predicting prognosis arose from the ENSAT classification system in 2009 [24]. Nevertheless, given the variability in outcomes regardless of grading or staging of disease, there has been a pressing need for more clinicopathological markers to aid in the prediction of outcomes in ACC.
Modern molecular analysis has made a significant impact on the understanding of cancer biology. It has also highlighted the usefulness of these molecules as potential prognostic and predictive transcriptomic, epigenomic, and genomic markers. The usefulness of RNA-based global gene expression profiling techniques in cancers, for example, has been well established [31–33]. For ACC, several landmark series have highlighted distinct gene expression profiles between ACCs and benign adrenal disease [12, 17, 34]. These have paved the way for further studies attempting to correlate these profiles with clinical outcomes [17, 35, 36]. Notably, more recent integrated genomic analyses involving exome sequencing, DNA methylation analysis, mRNA expression arrays, and miRNA sequencing have suggested that aggressive and indolent ACCs belong to two distinct molecular entities, each spearheaded by separate oncogenic mutations [16]. Despite these advances in knowledge of the underlying molecular pathology of ACCs, there has been a paucity of data on immunohistologically validated markers. In this study, we characterized a panel of established and novel IHC markers, based on global expression microarrays, that are able to provide insight into survival outcomes for patients after surgery for ACC.
In the present study, we were able to demonstrate that overexpression of Ki-67 (>10%) is associated with significantly worse overall survival on multivariate analysis of all patients (HR: 3.0; 95% CI: 1.3–6.8). It was also associated with worse disease-free survival (HR: 3.3; 95% CI: 1.3–8.2) for patients without metastatic disease at presentation. Ki-67 is a well-established marker of proliferation in ACC and has been shown to be able to differentiate ACCs from ACAs [12, 37, 38]. It has been shown to be a powerful prognostic marker for localized, advanced, and recurrent ACC [39–41]. Earlier work in our unit using microarray gene expression and IHC analysis has also shown that Ki-67, together with IGF2, is a useful marker in differentiating ACC from benign adrenal disease [7]. In addition, Ki-67 has been demonstrated to be of importance in determining a patient’s likelihood of recurrence after surgery, with a high index (>10%) associated with poorer disease-free and overall survival [42]. Given the mounting body of evidence showing Ki-67 as a useful marker of prognosis and outcome, this IHC stain has become routine for all adrenal specimens processed at the laboratory facilities of RNSH (Pacific Laboratory Medicine Services).
We have shown that overexpression of TOP2A on IHC (score >1) correlates with significantly poorer overall on both univariate (p < .001) and multivariate (p = .009) analyses. For patients without metastases, TOP2A was also associated with poorer disease-free survival on univariate analysis (p = .001). We noted that age ≥50 years and any evidence of invasion were significant risk factors for poor prognosis. Even after accounting for these two clinical factors, TOP2A remained a significant predictor of poor prognosis. The combined model demonstrated a C statistic (adjusted) of 0.842 (strong). TOP2A staining (p = .039) and age ≥50 years (p = .013) were found to be associated with a significantly lower likelihood of survival beyond 2 years in patients who had advanced disease (ENSAT stage III or IV) on presentation, with TOP2A staining being independent of age, as shown by a low correlation coefficient (0.15).
TOP2A has been previously shown to be overexpressed in ACC compared with ACAs in genomewide expression profiling studies [12, 14, 43]. The TOP2A gene encodes for TOP2A, a DNA topoisomerase that plays a role in maintaining DNA structure by reducing DNA torsional stress, chromosomal condensation, and segregation during mitosis [44]. TOP2A has been implicated as a marker of poor prognosis in numerous malignancies, including breast, renal cell, hepatocellular, prostate, and colorectal carcinomas [43, 45–51]. In addition, TOP2A has been reported to be a sensitive and specific marker for actively proliferating cells in colorectal carcinoma [44].
More recent work has shown that TOP2A is increased in ACC compared with normal adrenal cortex and plays a role in ACC cellular proliferation, cell invasion, and anchorage-independent growth [52]. Our data support the increasing evidence linking TOP2A to tumorigenesis, and its overexpression is believed to be linked with mutations of p53 and the retinoblastoma protein pRB, both of which have been postulated to be negative regulators of TOP2A [37, 51]. Mutations in the TP53 gene have been well documented in both inherited and sporadic forms of ACC [53–57], leading to increased expression of TOP2A.
Our IHC data regarding TOP2A overexpression and its correlation with poorer survival outcomes reinforce its role in tumorigenesis. Functional work in H295R and SW13 ACC cell lines by Jain et al. have shown promising results with TOP2A inhibitors; as such, it represents a potential therapeutic target [52].
Interestingly, we were also able to demonstrate a strong correlation between EZH2 and TOP2A expression (correlation coefficient: 0.83; p < .001). On univariate analysis, a staining score >2 was associated with significantly worse overall survival (p = .024). Although multivariate analysis of EZH2 was not performed because of potential multicollinearity, given the high degree of correlation with TOP2A expression, staining patterns for EZH2 would also carry potential prognostic value, with higher levels of staining implying poorer disease-free and overall survival patterns. The data in our series demonstrating EZH2 as a novel and independent prognostic biomarker in ACC are unique, and we believe this report is the first of its kind.
EZH2 is a polycomb group (PcG) protein homologous to Drosophila enhancer of zeste [58]. As a PcG protein, EZH2 functions as a transcriptional repressor by forming a complex with EED and the resultant EED-EZH2 complex methylating nucleosomal H3 at lysine 27 (H3-K27), resulting in chromatin condensation [59]. This mechanism of gene silencing has downstream effects on target genes that are involved in cell fate decisions, cell-cycle regulation, cellular differentiation, and senescence. Moreover, dysregulation of this epigenetic control over transcriptional memory and gene silencing has implied a role for EZH2 in tumorigenesis [60, 61]. Epigenetic silencing resulting in the inactivation of tumor suppressor genes is believed to be the underlying mechanism through which overexpression of EZH2 results in the transcriptional repression of tumor suppressor genes. EZH2 overexpression has been associated with aggressive carcinoma of the breast, colon, prostate, kidney, and endometrium and sarcomas and lymphomas [62–67].
Given the increasing evidence of the oncogenic role of EZH2 overexpression in other tumors, this seemed to be a logical marker to pursue. Moreover, we also chose EZH2 as a potential novel target because it was 1 of the 100 genes that were found to be significantly differentially expressed between ACCs and ACAs in previous work from our unit [7]. In our microarray, EZH2 carried an odds of differential expression (B statistic) of 2.70 in ACC compared with ACAs, with a log2 fold change of 2.31.
Similarly, from our previous microarray data, BARD1 carried an odds of differential expression (B statistic) of 5.95 in ACC compared with ACAs, with a log2 fold change of 2.05. In the present study, we demonstrated that BARD1 protein overexpression was associated with significantly better overall survival in both univariate and multivariate analyses. BARD1 is commonly found in many tissues, including the breast, colon, ovary, and uterus [68]. It plays an important tumor-suppressor function in conjunction with BRCA1 by heterodimerizing with it. This BARD1-BRCA1 heterodimer has a ubiquitin ligase function that is believed to be of importance in DNA damage repair, transcription regulation, RNA processing, and cell-cycle regulation [69–71]. BARD1 also has a BRCA1-independent function in mediating p53-dependent apoptosis; by binding to p53, BARD1 facilitates phosphorylation and stabilization of p53 [72].
BARD1 has been implicated in a variety of common cancers, including breast, ovarian, colorectal, and lung [73–75]. The BARD1 protein is known to exist as 19 different splice variations [76]. Zhang et al. demonstrated that expression of certain BARD1 isoforms predicted improved survival in colon cancer, whereas other isoforms expressing the truncated isoforms of BARD1 correlated with decreased survival [73]. In the present study, the antibody used was directed at the full length of BARD1, and as such, strong staining for the full length of BARD1 in IHC (staining score >2) was associated with improved disease-free and overall survival. This is in keeping with other series. Sporn et al. discovered that loss of the full-length BARD1 protein is associated with a poor prognosis in colon cancer [76]. To the best of our knowledge, this report is the first to show that the full-length BARD1 protein has been identified and validated in ACC tumor specimens using IHC. Consequently, it represents a novel marker to improve outcome stratification in patients with the disease.
Cyclin B1 has also been previously shown to be upregulated in ACCs [7, 12, 34]. We were able to demonstrate this at a protein level with IHC and show that the presence of positive staining for cyclin B1 (>1) was associated with poorer overall survival (HR: 2.4; 95% CI: 1.0–5.2; p = .039) and disease-free survival (HR: 3.4; 95% CI: 1.0–11.8; p = .049; patients with no metastases) in univariate analysis. Cyclin B1 is a common marker of cellular proliferation. By forming a complex with cyclin-dependent kinase 1 (Cdk1), cyclin B1 plays a key role in the transition from G2 to M phase during cell division. The complex acts as a switch at the G2-M checkpoint of the cell cycle, determining whether the cell commits to mitosis [77]. In various malignancies such as lung, bladder, colorectal, prostate, and breast cancers and glioma, it becomes dysregulated and overexpressed, resulting in uncontrolled cell growth [78]. The downstream ramifications of unchecked cyclin B1 binding to cyclin-dependent kinases is also believed to be responsible for the inactivation of the tumor suppressor protein p53, which has been demonstrated to suppress cyclin B1 expression in the nonmalignant state [79, 80]. Its use as a potential marker of prognosis has been entertained in breast and esophageal carcinoma [81, 82].
When studying a rare cancer such as ACC, adequate sample size is always a challenge, and this is one of the limitations of our study. Our results demonstrate that, along with the established proliferation marker Ki-67, the increased expression of TOP2A and EZH2 proteins was associated with a poorer prognosis with decreased overall and disease-free survival. In contrast, the increased expression of BARD1 demonstrated significantly improved overall survival.
Conclusion
Despite the many advances in the understanding of the disease, ACC remains a diagnostic and prognostic dilemma, with disappointing long-term rates of survival. Molecular markers predicting prognosis that would allow for rapid stratification of further surgery and adjuvant therapies would likely improve outcomes and quality of life for patients.
With the rapid identification of genes associated with prognosis in ACC through global gene expression profiling technology, there has been limited IHC data validating the protein products of these genes at the tissue level. For the first time in the literature, we report on EZH2 as a potential novel biomarker of tumor aggressiveness, correlating closely with the recently established TOP2A as a marker of poor prognosis in patients with ACC. Overexpression of these markers on IHC is associated with significantly poorer disease-free and overall survival, with strong TOP2A staining being associated with a significantly lower likelihood of survival beyond 2 years. Both EZH2 and TOP2A have shown potential as therapeutic targets in a variety of other cancers. In particular, EZH2 has been shown previously to be a marker of tumor aggressiveness in various other cancers, and further work is required to assess the feasibility of PcG proteins as targets for therapy.
Our findings also highlight the full-length BARD1 isoform as a novel marker for improved survival in ACC. Although BARD1 and its various isoforms have been shown to predict outcomes in other malignancies, to the best of our knowledge, this is the first time this has been reported in the literature for ACC.
Because of the limitations of a small sample size, further stage-specific analysis with a larger cohort may provide a more definitive picture of the true prognostic value of these potential markers. Nevertheless, as a whole, the data presented have exciting prognostic and predictive implications for patients with ACC. Further work is required to elucidate any potential role for these biomarkers as targets for personalized therapy that may be useful as an adjunct to the limited treatment options currently available for ACC.
Acknowledgments
We thank the clinicians who have kindly provided access to their patients’ medical records and thus made this study possible. Julian C.Y. Ip is supported by a Royal Australasian College of Surgeons Foundation for Surgery Richard Jepson Research Scholarship. Stan B. Sidhu is a University of Sydney Medical School Foundation fellow.
Footnotes
For Further Reading: Lyndal J. Tacon, Ruth S. Prichard, Patsy S. H. Soon et al. Current and Emerging Therapies for Advanced Adrenocortical Carcinoma. The Oncologist 2011;16:36–48.
Abstract: Adrenocortical carcinoma (ACC) is a rare but aggressive malignancy with a poor prognosis. Complete surgical resection offers the only potential for cure; however, even after apparently successful excision, local or metastatic recurrence is frequent. Treatment options for advanced ACC are severely limited. Mitotane is the only recognized adrenolytic therapy available; however, response rates are modest and unpredictable whereas systemic toxicities are significant. Reported responses to conventional cytotoxic chemotherapy have also been disappointing, and the rarity of ACC had hampered the ability to undertake randomized clinical studies until the establishment of the First International Randomized Trial in Locally Advanced and Metastatic Adrenocortical Carcinoma. This yet-to-be reported study seeks to identify the most effective first- and second-line cytotoxic regimens. The past decade has also seen increasing research into the molecular pathogenesis of ACCs, with particular interest in the insulin-like growth factor signaling pathway. The widespread development of small molecule tyrosine kinase inhibitors in broader oncological practice is now allowing for the rational selection of targeted therapies to study in ACC. In this review, we discuss the currently available therapeutic options for patients with advanced ACC and detail the molecular rationale behind, and clinical evidence for, novel and emerging therapies.
Author Contributions
Conception/Design: Julian C.Y. Ip, Jing Ting Zhao, Bruce G. Robinson, Anthony J. Gill, Stan B. Sidhu
Provision of study material or patients: Julian C.Y. Ip, Patsy Soon, Bruce G. Robinson, Anthony J. Gill, Stan B. Sidhu
Collection and/or assembly of data: Julian C.Y. Ip, Tony C.Y. Pang, Anthony R. Glover, Patsy Soon, Anthony J. Gill, Stan B. Sidhu
Data analysis and interpretation: Julian C.Y. Ip, Tony C.Y. Pang, Anthony R. Glover, Jing Ting Zhao, Stephen Clarke, Anthony J. Gill, Stan B. Sidhu
Manuscript writing: Julian C.Y. Ip, Tony C.Y. Pang, Patsy Soon, Jing Ting Zhao, Stephen Clarke, Anthony J. Gill, Stan B. Sidhu
Final approval of manuscript: Julian C.Y. Ip, Tony C.Y. Pang, Anthony R. Glover, Patsy Soon, Jing Ting Zhao, Stephen Clarke, Bruce G. Robinson, Anthony J. Gill, Stan B. Sidhu
Disclosures
Bruce G. Robinson: Bayer (C/A); Mayne Pharma (OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Else T, Kim AC, Sabolch A, et al. Adrenocortical carcinoma. Endocr Rev. 2014;35:282–326. doi: 10.1210/er.2013-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McAteer JP, Huaco JA, Gow KW. Predictors of survival in pediatric adrenocortical carcinoma: A Surveillance, Epidemiology, and End Results (SEER) program study. J Pediatr Surg. 2013;48:1025–1031. doi: 10.1016/j.jpedsurg.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Fassnacht M, Kroiss M, Allolio B. Update in adrenocortical carcinoma. J Clin Endocrinol Metab. 2013;98:4551–4564. doi: 10.1210/jc.2013-3020. [DOI] [PubMed] [Google Scholar]
- 4.Fassnacht M, Allolio B. Clinical management of adrenocortical carcinoma. Best Pract Res Clin Endocrinol Metab. 2009;23:273–289. doi: 10.1016/j.beem.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Stojadinovic A, Ghossein RA, Hoos A, et al. Adrenocortical carcinoma: Clinical, morphologic, and molecular characterization. J Clin Oncol. 2002;20:941–950. doi: 10.1200/JCO.2002.20.4.941. [DOI] [PubMed] [Google Scholar]
- 6.Soon PSH, Tacon LJ, Gill AJ, et al. miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clin Cancer Res. 2009;15:7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- 7.Soon PSH, Gill AJ, Benn DE, et al. Microarray gene expression and immunohistochemistry analyses of adrenocortical tumors identify IGF2 and Ki-67 as useful in differentiating carcinomas from adenomas. Endocr Relat Cancer. 2009;16:573–583. doi: 10.1677/ERC-08-0237. [DOI] [PubMed] [Google Scholar]
- 8.Sidhu S, Marsh DJ, Theodosopoulos G, et al. Comparative genomic hybridization analysis of adrenocortical tumors. J Clin Endocrinol Metab. 2002;87:3467–3474. doi: 10.1210/jcem.87.7.8697. [DOI] [PubMed] [Google Scholar]
- 9.Patterson EE, Holloway AK, Weng J, et al. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer. 2011;117:1630–1639. doi: 10.1002/cncr.25724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertherat J, Bertagna X. Pathogenesis of adrenocortical cancer. Best Pract Res Clin Endocrinol Metab. 2009;23:261–271. doi: 10.1016/j.beem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca AL, Kugelberg J, Starker LF, et al. Comprehensive DNA methylation analysis of benign and malignant adrenocortical tumors. Genes Chromosomes Cancer. 2012;51:949–960. doi: 10.1002/gcc.21978. [DOI] [PubMed] [Google Scholar]
- 12.Giordano TJ, Thomas DG, Kuick R, et al. Distinct transcriptional profiles of adrenocortical tumors uncovered by DNA microarray analysis. Am J Pathol. 2003;162:521–531. doi: 10.1016/S0002-9440(10)63846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slater EP, Diehl SM, Langer P, et al. Analysis by cDNA microarrays of gene expression patterns of human adrenocortical tumors. Eur J Endocrinol. 2006;154:587–598. doi: 10.1530/eje.1.02116. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Ranvier GG, Weng J, Yeh R-F, et al. Identification of biomarkers of adrenocortical carcinoma using genomewide gene expression profiling. Arch Surg. 2008;143:841–846; discussion 846. doi: 10.1001/archsurg.143.9.841. [DOI] [PubMed] [Google Scholar]
- 15.Velázquez-Fernández D, Laurell C, Geli J, et al. Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery. 2005;138:1087–1094. doi: 10.1016/j.surg.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Assié G, Letouzé E, Fassnacht M, et al. Integrated genomic characterization of adrenocortical carcinoma. Nat Genet. 2014;46:607–612. doi: 10.1038/ng.2953. [DOI] [PubMed] [Google Scholar]
- 17.de Reyniès A, Assié G, Rickman DS, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27:1108–1115. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- 18.Jawhar NMT. Tissue Microarray: A rapidly evolving diagnostic and research tool. Ann Saudi Med. 2009;29:123–127. doi: 10.4103/0256-4947.51806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zlobec I, Koelzer VH, Dawson H, et al. Next-generation tissue microarray (ngTMA) increases the quality of biomarker studies: An example using CD3, CD8, and CD45RO in the tumor microenvironment of six different solid tumor types. J Transl Med. 2013;11:104. doi: 10.1186/1479-5876-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radhakrishnan R, Solomon M, Satyamoorthy K, et al. Tissue microarray - a high-throughput molecular analysis in head and neck cancer. J Oral Pathol Med. 2008;37:166–176. doi: 10.1111/j.1600-0714.2007.00606.x. [DOI] [PubMed] [Google Scholar]
- 21.Makretsov NA, Huntsman DG, Nielsen TO, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin Cancer Res. 2004;10:6143–6151. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- 22.Klevesath MB, Pantel K, Agbaje O, et al. Patterns of metastatic spread in early breast cancer. Breast. 2013;22:449–454. doi: 10.1016/j.breast.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Lughezzani G, Sun M, Perrotte P, et al. The European Network for the Study of Adrenal Tumors staging system is prognostically superior to the International Union Against Cancer-staging system: A North American validation. Eur J Cancer. 2010;46:713–719. doi: 10.1016/j.ejca.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Fassnacht M, Johanssen S, Quinkler M, et al. Limited prognostic value of the 2004 International Union Against Cancer staging classification for adrenocortical carcinoma: Proposal for a revised TNM classification. Cancer. 2009;115:243–250. doi: 10.1002/cncr.24030. [DOI] [PubMed] [Google Scholar]
- 25.The Ryerson Index to death notices and obituaries in Australian newspapers. Available at http://www.ryersonindex.org. Accessed July 10, 2014.
- 26.Liu X, Minin V, Huang Y, et al. Statistical methods for analyzing tissue microarray data. J Biopharm Stat. 2004;14:671–685. doi: 10.1081/BIP-200025657. [DOI] [PubMed] [Google Scholar]
- 27.Ip JCY, Pang TCY, Glover AR, et al. Improving outcomes in adrenocortical cancer: An Australian perspective. Ann Surg Oncol. 2014 doi: 10.1245/s10434-014-4133-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Aubert S, Wacrenier A, Leroy X, et al. Weiss system revisited: A clinicopathologic and immunohistochemical study of 49 adrenocortical tumors. Am J Surg Pathol. 2002;26:1612–1619. doi: 10.1097/00000478-200212000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Weiss LM. Comparative histologic study of 43 metastasizing and nonmetastasizing adrenocortical tumors. Am J Surg Pathol. 1984;8:163–169. doi: 10.1097/00000478-198403000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Medeiros LJ, Weiss LM. New developments in the pathologic diagnosis of adrenal cortical neoplasms. A review. Am J Clin Pathol. 1992;97:73–83. doi: 10.1093/ajcp/97.1.73. [DOI] [PubMed] [Google Scholar]
- 31.Argani P, Rosty C, Reiter RE, et al. Discovery of new markers of cancer through serial analysis of gene expression: Prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4324. [PubMed] [Google Scholar]
- 32.Hough CD, Sherman-Baust CA, Pizer ES, et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 33.Nacht M, Ferguson ATA, Zhang W, et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59:5464–5470. [PubMed] [Google Scholar]
- 34.de Fraipont F, El Atifi M, Cherradi N, et al. Gene expression profiling of human adrenocortical tumors using complementary deoxyribonucleic acid microarrays identifies several candidate genes as markers of malignancy. J Clin Endocrinol Metab. 2005;90:1819–1829. doi: 10.1210/jc.2004-1075. [DOI] [PubMed] [Google Scholar]
- 35.Giordano TJ, Kuick R, Else T, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15:668–676. doi: 10.1158/1078-0432.CCR-08-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fragoso MCBV, Almeida MQ, Mazzuco TL, et al. Combined expression of BUB1B, DLGAP5, and PINK1 as predictors of poor outcome in adrenocortical tumors: Validation in a Brazilian cohort of adult and pediatric patients. Eur J Endocrinol. 2012;166:61–67. doi: 10.1530/EJE-11-0806. [DOI] [PubMed] [Google Scholar]
- 37.Iino K, Sasano H, Yabuki N, et al. DNA topoisomerase II alpha and Ki-67 in human adrenocortical neoplasms: A possible marker of differentiation between adenomas and carcinomas. Mod Pathol. 1997;10:901–907. [PubMed] [Google Scholar]
- 38.Gupta D, Shidham V, Holden J, et al. Value of topoisomerase II alpha, MIB-1, p53, E-cadherin, retinoblastoma gene protein product, and HER-2/neu immunohistochemical expression for the prediction of biologic behavior in adrenocortical neoplasms. Appl Immunohistochem Mol Morphol. 2001;9:215–221. doi: 10.1097/00129039-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Libè R, Fratticci A, Bertherat J. Adrenocortical cancer: Pathophysiology and clinical management. Endocr Relat Cancer. 2007;14:13–28. doi: 10.1677/erc.1.01130. [DOI] [PubMed] [Google Scholar]
- 40.Beuschlein F, Obracay J, Saeger W. Prognostic value of histological markers in localized adrenocortical carcinoma after complete resection. Endocr Rev. 2013:29–33. [Google Scholar]
- 41.Terzolo M, Boccuzzi A, Bovio S, et al. Immunohistochemical assessment of Ki-67 in the differential diagnosis of adrenocortical tumors. Urology. 2001;57:176–182. doi: 10.1016/s0090-4295(00)00852-9. [DOI] [PubMed] [Google Scholar]
- 42.Morimoto R, Satoh F, Murakami O, et al. Immunohistochemistry of a proliferation marker Ki67/MIB1 in adrenocortical carcinomas: Ki67/MIB1 labeling index is a predictor for recurrence of adrenocortical carcinomas. Endocr J. 2008;55:49–55. doi: 10.1507/endocrj.k07-079. [DOI] [PubMed] [Google Scholar]
- 43.Dawany NB, Dampier WN, Tozeren A. Large-scale integration of microarray data reveals genes and pathways common to multiple cancer types. Int J Cancer. 2011;128:2881–2891. doi: 10.1002/ijc.25854. [DOI] [PubMed] [Google Scholar]
- 44.Tsavaris N, Lazaris A, Kosmas C, et al. Topoisomerase I and IIalpha protein expression in primary colorectal cancer and recurrences following 5-fluorouracil-based adjuvant chemotherapy. Cancer Chemother Pharmacol. 2009;64:391–398. doi: 10.1007/s00280-008-0886-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coss A, Tosetto M, Fox EJ, et al. Increased topoisomerase IIalpha expression in colorectal cancer is associated with advanced disease and chemotherapeutic resistance via inhibition of apoptosis. Cancer Lett. 2009;276:228–238. doi: 10.1016/j.canlet.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 46.Wong N, Yeo W, Wong W-L, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 47.Albadine R, Wang W, Brownlee NA, et al. Topoisomerase II alpha status in renal medullary carcinoma: Immuno-expression and gene copy alterations of a potential target of therapy. J Urol. 2009;182:735–740. doi: 10.1016/j.juro.2009.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosari F, Munz J, Savci-Heijink CD et al. Identification of prognostic biomarkers for prostate cancer. AACR Meeting Abstracts 2008:219a. [DOI] [PubMed] [Google Scholar]
- 49.Tretiakova M, Turkyilmaz M, Grushko T. Topoisomerase IIα in Wilms’ tumour: Gene alterations and immunoexpression. J Clin Pathol 2006. J Clin Pathol. 2006;59:1272–1277. doi: 10.1136/jcp.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faggad A, Darb-Esfahani S, Wirtz R, et al. Topoisomerase IIalpha mRNA and protein expression in ovarian carcinoma: Correlation with clinicopathological factors and prognosis. Mod Pathol. 2009;22:579–588. doi: 10.1038/modpathol.2009.14. [DOI] [PubMed] [Google Scholar]
- 51.O’Connor JK, Hazard LJ, Avent JM, et al. Topoisomerase II alpha expression correlates with diminished disease-free survival in invasive breast cancer. Int J Radiat Oncol Biol Phys. 2006;65:1411–1415. doi: 10.1016/j.ijrobp.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 52.Jain M, Zhang L, He M, et al. TOP2A is overexpressed and is a therapeutic target for adrenocortical carcinoma. Endocr Relat Cancer. 2013;20:361–370. doi: 10.1530/ERC-12-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levine AJ. The evolution of the p53 family of genes. Cell Cycle. 2012;11:214–215. doi: 10.4161/cc.11.2.18899. [DOI] [PubMed] [Google Scholar]
- 54.Waldmann J, Patsalis N, Fendrich V, et al. Clinical impact of TP53 alterations in adrenocortical carcinomas. Langenbecks Arch Surg. 2012;397:209–216. doi: 10.1007/s00423-011-0868-6. [DOI] [PubMed] [Google Scholar]
- 55.Ragazzon B, Libé R, Gaujoux S, et al. Transcriptome analysis reveals that p53 and beta-catenin alterations occur in a group of aggressive adrenocortical cancers. Cancer Res. 2010;70:8276–8281. doi: 10.1158/0008-5472.CAN-10-2014. [DOI] [PubMed] [Google Scholar]
- 56.Raymond VM, Else T, Everett JN, et al. Prevalence of germline TP53 mutations in a prospective series of unselected patients with adrenocortical carcinoma. J Clin Endocrinol Metab. 2013;98:E119–E125. doi: 10.1210/jc.2012-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hisada M, Garber JE, Fung CY, et al. Multiple primary cancers in families with Li-Fraumeni syndrome. J Natl Cancer Inst. 1998;90:606–611. doi: 10.1093/jnci/90.8.606. [DOI] [PubMed] [Google Scholar]
- 58.Laible G, Wolf A, Dorn R, et al. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao W, Ribeiro RO, Liu D, et al. EZH2 promotes malignant behaviors via cell cycle dysregulation and its mRNA level associates with prognosis of patient with non-small cell lung cancer. PLoS One. 2012;7:e52984. doi: 10.1371/journal.pone.0052984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacobs JJ, Scheijen B, Voncken JW, et al. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacobs JJ, Kieboom K, Marino S, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 62.Zeidler M, Varambally S, Cao Q, et al. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bachmann IM, Halvorsen OJ, Collett K, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 64.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu ZQ, Zhang L, Gao BS, et al. EZH2 promotes tumor progression by increasing VEGF expression in clear cell renal cell carcinoma. Clin Transl Oncol. 2015;17:41–49. doi: 10.1007/s12094-014-1195-5. [DOI] [PubMed] [Google Scholar]
- 66.Panousis D, Patsouris E, Lagoudianakis E, et al. The value of TOP2A, EZH2 and paxillin expression as markers of aggressive breast cancer: Relationship with other prognostic factors. Eur J Gynaecol Oncol. 2011;32:156–159. [PubMed] [Google Scholar]
- 67.Chang C-J, Hung M-C. The role of EZH2 in tumour progression. Br J Cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irminger-Finger I, Jefford CE. Is there more to BARD1 than BRCA1? Nat Rev Cancer. 2006;6:382–391. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 69.Wu LC, Wang ZW, Tsan JT, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 70.Baer R, Ludwig T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev. 2002;12:86–91. doi: 10.1016/s0959-437x(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 71.Hashizume R, Fukuda M, Maeda I, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 72.Feki A, Jefford CE, Berardi P, et al. BARD1 induces apoptosis by catalysing phosphorylation of p53 by DNA-damage response kinase. Oncogene. 2005;24:3726–3736. doi: 10.1038/sj.onc.1208491. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y-Q, Bianco A, Malkinson AM, et al. BARD1: An independent predictor of survival in non-small cell lung cancer. Int J Cancer. 2012;131:83–94. doi: 10.1002/ijc.26346. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y-Q, Pilyugin M, Kuester D, et al. Expression of oncogenic BARD1 isoforms affects colon cancer progression and correlates with clinical outcome. Br J Cancer. 2012;107:675–683. doi: 10.1038/bjc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu J-Y, Vlastos A-T, Pelte M-F, et al. Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Int J Cancer. 2006;118:1215–1226. doi: 10.1002/ijc.21428. [DOI] [PubMed] [Google Scholar]
- 76.Sporn JC, Hothorn T, Jung B. BARD1 expression predicts outcome in colon cancer. Clin Cancer Res. 2011;17:5451–5462. doi: 10.1158/1078-0432.CCR-11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Song H, Walsh S, et al. Combinatorial control of cyclin B1 nuclear trafficking through phosphorylation at multiple sites. J Biol Chem. 2001;276:3604–3609. doi: 10.1074/jbc.M008151200. [DOI] [PubMed] [Google Scholar]
- 78.Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu M, Zhan Q, Finn OJ. Immune recognition of cyclin B1 as a tumor antigen is a result of its overexpression in human tumors that is caused by non-functional p53. Mol Immunol. 2002;38:981–987. doi: 10.1016/s0161-5890(02)00026-3. [DOI] [PubMed] [Google Scholar]
- 80.Innocente SAS, Abrahamson JLJ, Cogswell JPJ, et al. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nozoe T, Korenaga D, Kabashima A, et al. Significance of cyclin B1 expression as an independent prognostic indicator of patients with squamous cell carcinoma of the esophagus. Clin Cancer Res. 2002;8:817–822. [PubMed] [Google Scholar]
- 82.Winters ZE, Hunt NC, Bradburn MJ, et al. Subcellular localisation of cyclin B, Cdc2 and p21(WAF1/CIP1) in breast cancer. association with prognosis. Eur J Cancer. 2001;37:2405–2412. doi: 10.1016/s0959-8049(01)00327-6. [DOI] [PubMed] [Google Scholar]