Abstract
During the late stage of the viral life cycle, HIV-1 Gag assembles into a spherical immature capsid, and undergoes budding, release, and maturation. Here we review events involved in immature capsid assembly from the perspective of five different approaches used to study this process: mutational analysis, structural studies, assembly of purified recombinant Gag, assembly of newly-translated Gag in a cell-free system, and studies in cells using biochemical and imaging techniques. We summarize key findings obtained using each approach, point out where there is consensus, and highlight unanswered questions. Particular emphasis is placed on reconciling data suggesting that Gag assembles by two different paths, depending on the assembly environment. Specifically, in assembly systems that lack cellular proteins, high concentrations of Gag can spontaneously assemble using purified nucleic acid as a scaffold. However, in the more complex intracellular environment, barriers that limit self-assembly are present in the form of cellular proteins, organelles, host defenses, and the absence of free nucleic acid. To overcome these barriers and promote efficient immature capsid formation in an unfavorable environment, Gag appears to utilize an energy-dependent, host-catalyzed, pathway of assembly intermediates in cells. Overall, we show how data obtained using a variety of techniques has led to our current understanding of HIV assembly.
1. Introduction
1.1. Overview
One of the remarkable features of HIV-1 is its prolific ability to generate new virus particles. Estimates suggest that up to 1010 virions are produced per day in an infected individual (Chun et al., 1997), leading to levels of viremia as high as 107 virions per milliliter of blood (Perelson et al., 1996). This prodigious ability can be attributed to the impressive capacity for virus production: it is estimated that an HIV-1 infected cell produces ∼5 × 104 virions are in a single day, which is the estimated lifespan of an infected cell (Chen et al., 2007). These numbers reflect in part the remarkable efficiency of the late events in the virus life cycle within the infected cell. These numbers also speak to the importance of understanding, at a cellular and molecular level, why late events such as assembly are so efficient within cells. And yet, as described below, important puzzle pieces are missing from our picture of how HIV-1 assembles in infected cells.
Late events in the virus lifecycle can be divided into 4 main stages: 1) Gag polyprotein assembly, which leads to formation of the HIV-1 immature capsid (also called the immature lattice); 2) budding and envelopment of the immature capsid; 3) immature virus particle release; and 4) maturation into the infectious virus, which involves cleavage of the Gag polyprotein by the HIV-1 protease into its four constituent domains. This review focuses entirely on the first stage – assembly of the HIV-1 immature capsid lattice, the spherical protein shell that is located within the immature virus and encapsidates the viral genome. It is well accepted that HIV-1 has evolved sophisticated mechanisms for taking advantage of the host cell at many stages of replication in order to efficiently generate progeny virus. While mechanisms for co-opting host machinery have been described in detail for virus budding and release [reviewed in (Votteler and Sundquist, 2013)], equivalent mechanisms for co-opting host proteins during immature capsid assembly remain poorly understood. Identifying and understanding how HIV-1 utilizes cellular machinery during capsid assembly could offer novel approaches for inhibiting virus production in actively infected cells, as well as in cells reactivated out of latency.
In this section we summarize the current view of HIV-1 immature capsid assembly within cells. In subsequent sections we illustrate how different experimental systems have yielded complementary pieces of the HIV-1 assembly puzzle, while highlighting important questions that remain unanswered and concepts that could reconcile contrasting assembly models. Other topics related to HIV-1 assembly have been reviewed elsewhere and will only be mentioned here in passing, including gRNA trafficking and packaging [reviewed in (Kuzembayeva et al., 2014; Lu et al., 2011)], HIV-1 budding and release [reviewed in (Votteler and Sundquist, 2013)], HIV-1 maturation [reviewed in (Sundquist and Krausslich, 2012)], and the subcellular localization of HIV-1 assembly [reviewed in (Jouvenet et al., 2008; Klein et al., 2007)].
1.2. The current view of HIV-1 assembly in cells
The 55 kDa HIV-1 Gag polyprotein contains four domains – matrix (MA), capsid (CA), nucleocapsid (NC), and p6 – as well as two small spacer peptides, SP1 and SP2 (Fig. 1). When defined narrowly, the problem of immature capsid assembly is about how ∼1500–3000 Gag polyproteins multimerize to form a single immature capsid shell. Viewed from this limited perspective, many questions related to assembly appear resolved since the general functions of the major domains in Gag are well understood. For example, mutational analyses reveal that only the first three domains of Gag (MA, CA, and NC) are required for immature capsid assembly per se, with the fourth domain, p6, being required for budding and release. Additionally, it is well established that MA governs membrane targeting; CA contains residues that form critical Gag-Gag interactions; and NC is required for viral genomic RNA packaging as well as non-specific interactions with RNA. However, in infected cells, the process of immature capsid assembly is much more complex than would be suggested by a simple Gag-centric and domain-centric view of Gag function. To provide perspective, we start here with an overview of our current understanding of HIV-1assembly, aspects of which are discussed in more detail later.
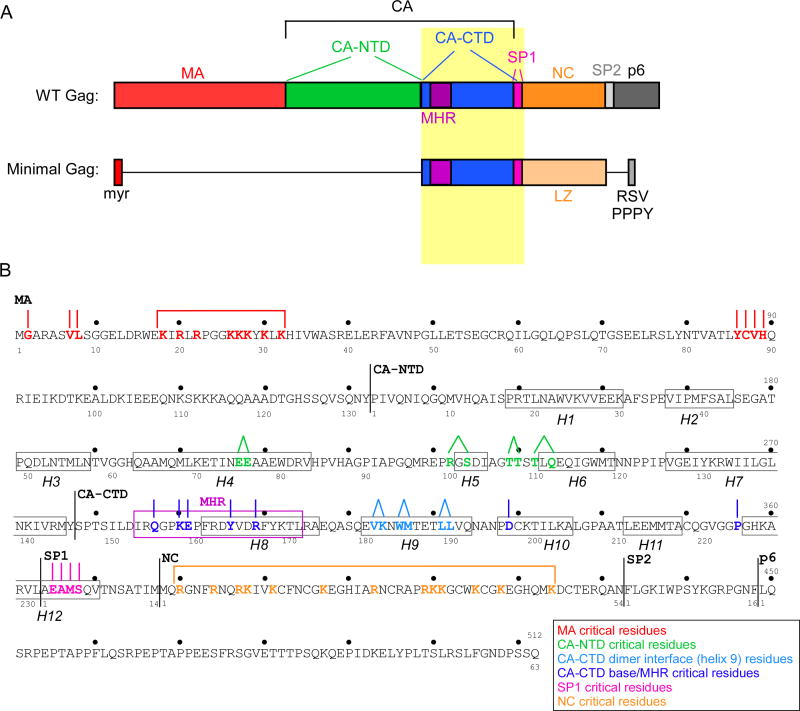
Figure 1. HIV-1 Gag domain map and amino acid sequence showing residues critical for immature capsid assembly.
(A) Schematic showing domains, subdomains, and spacer peptides in Gag [MA (red), CA-NTD (green), CA-CTD (blue), SP1 (pink), NC (orange), SP2 (light grey) and p6 (dark grey)]. The highly conserved MHR motif is highlighted in purple. Also shown is minimal Gag, a construct containing the minimal domains needed for assembly (Accola et al., 2000), in which some domains or subdomains are deleted (horizontal black lines), NC is replaced with a leucine zipper (LZ, light orange), and p6 is replaced with a short peptide containing the RSV PPPY motif (medium grey). B) Amino acid sequence of HIV-1 LAI Gag, with amino acid numbering for the complete Gag polyprotein shown above, and numbering for individual domains and spacer peptides (MA, CA, SP1, NC, and SP2) shown below. Note that the text and other figures in this review use the domain number system. Black lines indicate domain boundaries. The 12 actual or predicted α-helices in CA/SP1 are enclosed in black boxes (H1-H12). The MHR is enclosed in a purple box. Residues that are critical for immature capsid assembly are color-coded by surface (see legend). Lines above the critical residues indicate assembly-defective phenotypes produced by single (∣) or double (•) residue mutations; brackets indicate regions in which multiple basic residues are mutated to produce an assembly-defective phenotype.
Gag is the only viral protein required for assembly and release of immature virus particles in cells, although production of infectious virus requires other viral proteins. Assembly of the Gag polyprotein begins in the cytoplasm of the infected cell, where Gag is translated from a full-length capped and polyadenylated genomic RNA (gRNA), and is co-translationally myristoylated at its N-terminus. Importantly, the full-length gRNA species is both the mRNA used for translation of Gag and the genome that is packaged by assembling Gag. How the pool of full-length gRNA serves for both purposes remains poorly understood [reviewed in (Butsch and Boris-Lawrie, 2002; Kuzembayeva et al., 2014; Lu et al., 2011)]. While in the cytoplasm, newly synthesized Gag undergoes limited oligomerization (Kutluay and Bieniasz, 2010), associates with the gRNA it will package (Jouvenet et al., 2009; Kemler et al., 2010; Kutluay and Bieniasz, 2010), and also associates with several cellular factors, as described below.
Because the plasma membrane (PM) is the site of virion release in most cells (Jouvenet et al., 2006), trafficking of the initial Gag oligomer from the cytoplasm to the PM is a critical step in HIV-1 assembly. Membrane targeting of Gag requires the N-terminal myristate, as well as residues in MA that form a basic patch and interact with acidic head groups of phospholipids in the PM, including phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] found in the inner leaflet of the PM [reviewed in (Chukkapalli and Ono, 2011)]. Structural studies have shown that the myristate is initially sequestered within a cavity formed by the globular domain of MA (Tang et al., 2004) (Fig. 2A). Regulatory mechanisms control the timing of myristate exposure to ensure that membrane targeting occurs after Gag has associated with gRNA, formed an oligomer, and is in close proximity to the PM [reviewed in (Chukkapalli and Ono, 2011)]. Additionally, live imaging studies reveal that membrane targeting by Gag is responsible for anchoring gRNA at the PM (Jouvenet et al., 2009). Once at the PM, Gag continues to multimerize by a mechanism that is poorly understood; and while assembly is still in progress, the viral bud begins to form, thereby initiating the next stage in the viral life cycle.
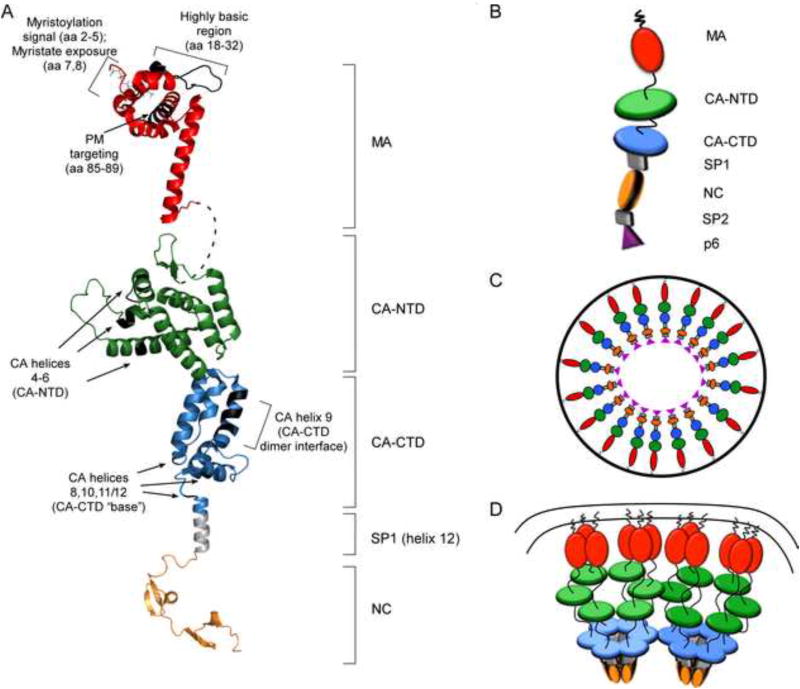
Fig. 2. Models for the structure of HIV-1 Gag domains and the architecture of the immature capsid.
(A) Ribbon diagrams of myristoylated MA in red (1UPH.pdb); full-length CA (3NTE.pdb) based on crystal structures of the individual CA-NTD and CA-CTD domains, shown in green and blue, respectively; SP1 (hypothetical α-helix) shown in grey, and NC (1mfs.pdb) shown in orange. Regions that contain residues critical for immature capsid assembly (shown in Fig. 1B) are indicated with brackets or arrows. Black highlights show locations of relevant amino acids. Dotted line shows flexible linker between MA and CA. (B) Schematic of full-length Gag as shown in Figs. 2C and D, with each domain labeled. (C) “Beads on a string model” for the arrangement of the Gag polyprotein in the immature capsid, adapted from (Fuller et al., 1997; Ganser-Pornillos et al., 2008; Yeager et al., 1998). (D) Model showing a hexameric configuration for CA-NTD, CA-CTD, and SP1 in the context of the fully assembled immature capsid lattice at the PM, adapted from (Wright et al., 2007).
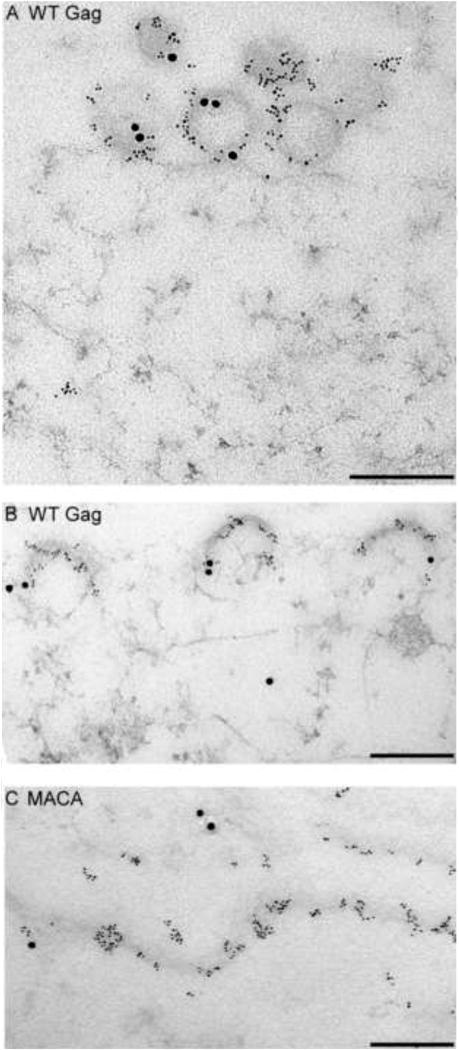
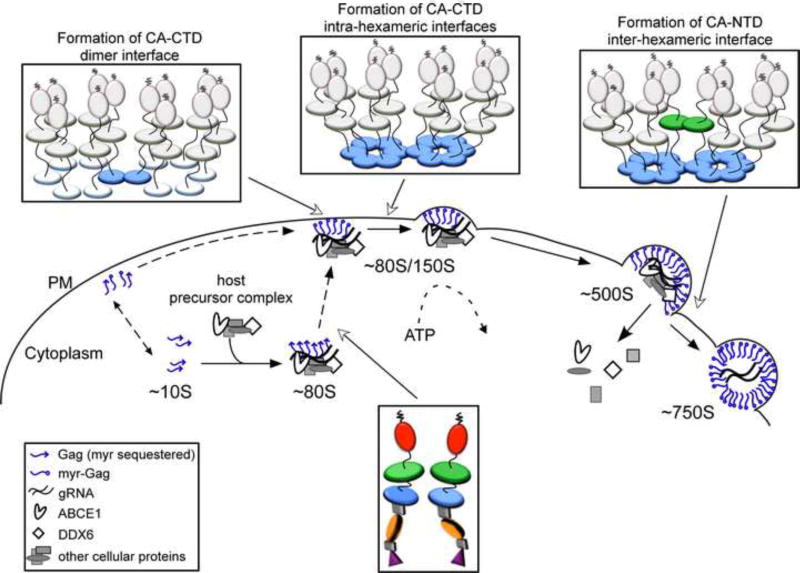
Studies reveal that Gag proteins are not alone when they oligomerize in the cytoplasm, traffic to the PM, and multimerize at the PM. Shortly after translation, Gag co-opts a cytoplasmic host complex that contains RNA granule proteins (also called processing body or P body proteins), (Reed et al., 2012). Two of the proteins in this complex are host enzymes that facilitate assembly - ABCE1 and DDX6 (Reed et al., 2012; Robinson et al., 2014; Zimmerman et al., 2002). A third co-opted host protein, Staufen, is an RNA binding protein that facilitates Gag multimerization and RNA packaging [reviewed in (Cochrane et al., 2006)]. Exactly how these cellular facilitators act remains to be determined, but studies have found that Gag remains with these proteins as it traffics to the PM and multimerizes. These studies have revealed that Gag progresses through a sequential pathway of assembly intermediates, which are described by their sedimentation values (∼10S, ∼80S, ∼150S, and ∼500S). The high-molecular-mass assembly intermediates (the ∼80S, ∼150S, and ∼500S complexes) contain gRNA and other viral proteins, as well as cellular proteins, such as ABCE1 and DDX6, that dissociate when assembly is completed (Dooher et al., 2007; Reed et al., 2012; Robinson et al., 2014; Zimmerman et al., 2002). Consistent with this model, assembly-defective Gag mutants are arrested at specific steps in this assembly pathway, resulting in accumulation of the assembly intermediates that precede the point of blockade (Fig. 3). Thus, assembly can be viewed from the perspective of Gag, or from the perspective of the cellular proteins that the virus hijacks to assist Gag in its journey through the cell. Both the assembly pathway and cellular facilitators of assembly are viewed as controversial by some. This is not surprising given the complexities of biochemical studies in cells; additionally, it may be too soon to assess the full impact of these studies. Nevertheless, the assembly pathway is included in this review because it forms the cornerstone of a fundamentally different approach to understanding assembly - in which Gag is viewed as one of many players in the poorly-understood intracellular environment - that contrasts with the Gag-centric approach that currently dominates the field.
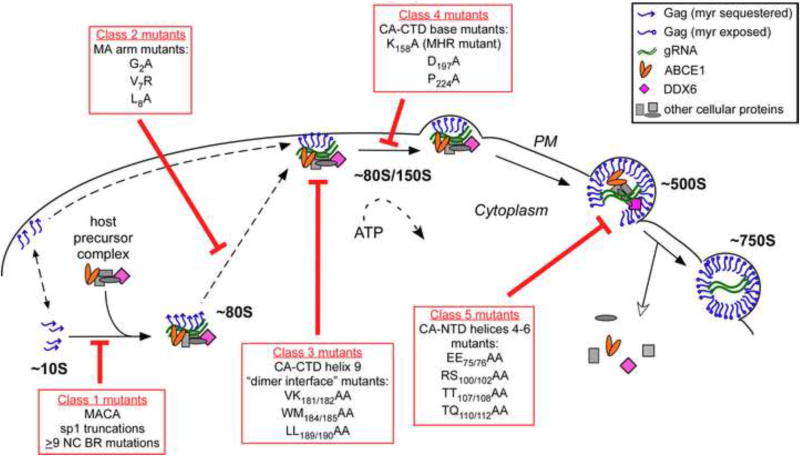
Fig. 3. Mutations of specific residues critical for immature capsid assembly cause arrest of Gag at distinct points in the assembly pathway in cells.
In cells, HIV-1 Gag progresses sequentially through an energy-dependent, stepwise pathway of assembly intermediates (∼10S, ∼80S, ∼150S, and ∼500S complexes) before forming the fully assembled ∼750S immature capsid. To form the ∼80S intermediate, Gag co-opts a precursor complex that contains the cellular facilitators of assembly, ABCE1 and DDX6. Mutations of residues critical for immature capsid assembly (shown in Fig. 1B) have been analyzed to determine the point at which they inhibit progression of Gag in the HIV-1 capsid assembly pathway. As shown here, each mutation blocks the pathway at a characteristic point, with accumulation of assembly intermediates that precede the point of inhibition. Mutations can be divided up into 5 different classes, depending on where they block the assembly pathway, as indicated in red boxes. Dotted lines indicate parts of the pathway that require further verification. For icons, see legend. References are in the text.
At the end of assembly, the completed immature capsid is released. In structural studies of the completed immature capsid, CA forms a hexameric lattice that contains irregular defects to accommodate the spherical shape (Bharat et al., 2014; Bharat et al., 2012; Briggs et al., 2009; Carlson et al.; de Marco et al., 2010; Fuller et al., 1997; Keller et al., 2011; Wright et al., 2007). Residues that form key CA-CA interfaces in the immature capsid lattice are starting to be identified with some precision in increasingly high-resolution structures (Bharat et al., 2014; Bharat et al., 2012). Such interfaces are present in the fully assembled immature capsid and likely form when Gag multimerizes. Notably, many of the residues that form CA-CA interfaces in the immature capsid are also required for efficient immature capsid assembly (Bharat et al., 2014; Bharat et al., 2012; Robinson et al., 2014). Recent biochemical studies in cells have defined when these and other critical residues act during assembly, leading to a model for the temporal sequence in when each of these critical CA-CA interfaces are formed (Robinson et al., 2014).
While many experimental approaches have contributed to our current understanding of immature capsid assembly in cells, here we focus on five approaches that have shaped our thinking about assembly in complementary ways. The first of these, mutational analysis of virus production (Section 2), was the main approach used in early studies of assembly. The second approach, structural biology (Section 3), has been used for nearly thirty years, with recent advances leading to increasingly higher resolution. The third and fourth approaches – assembly of purified recombinant Gag and assembly of newly synthesized Gag in cell extracts (Sections 4 and 5) – involve in vitro systems that were developed in the 1990's and offer two very different views of the assembly process. Finally, in the past 10-15 years, a variety of tools have been devised to study events of assembly in cells, including live imaging, as well as biochemical, ultrastructural, and siRNA-based approaches for identifying critical viral-host interactions (Section 6).
2. Effects of mutations in Gag on virus production
2.1. Overview
For over three decades, HIV-1 assembly has been studied by infecting or transfecting cells to express a mutated virus, and assessing the resulting phenotype. Because the Gag protein acts during many stages of the HIV life cycle, replication-defective Gag mutations fall into multiple categories depending on the point at which the viral life-cycle is inhibited. For example, some Gag mutations cause defects in post-entry events (e.g. uncoating or nuclear trafficking of the pre-integration complex) but do not affect the amount of virus produced. Other Gag mutations inhibit virus release or maturation but not virus assembly. Here we will focus on a category of Gag mutations that specifically inhibit immature capsid assembly (Fig. 1B, Fig. 2A). These mutations have little effect on Gag expression but cause significant reductions in particle production by inhibiting membrane targeting or multimerization of Gag at membranes. To demonstrate reductions in particle production, the amount of pelletable virus-like particles present in the cell supernatant is measured under conditions in which steady-state levels of Gag are equivalent. To demonstrate reductions in Gag targeting or multimerization at the PM, biochemical studies are utilized, ideally in conjunction with electron microscopy, allowing electron dense assembling Gag at membranes to be directly assessed. While only well-studied mutations critical for immature assembly are discussed here, other residues in Gag may also play a role.
2.2. MA residues required for immature capsid assembly in cells
The role of myristoylation and MA residues in membrane targeting has been reviewed in detail elsewhere (Chukkapalli and Ono, 2011). Briefly, myristoylation involves recognition of a specific N-terminal, six amino acid signal by a cytoplasmic enzyme, resulting in co-translational addition of myristate, a 14-carbon saturated fatty acid, to the nascent polypeptide N-terminus (Wright et al., 2010). Myristoylation of Gag is required for membrane targeting of Gag and production of immature virus particles, as revealed by the G2A mutation in MA, which abrogates the myristoylation signal in Gag (Bryant and Ratner, 1990; Gheysen et al., 1989; Gottlinger et al., 1989) (Fig. 1B; Fig. 2A). Consistent with these findings, when Gag is expressed at physiological levels, myristoylation of Gag is also required for immature capsid assembly (Dooher and Lingappa, 2004; Klein et al., 2011; Robinson et al., 2014). Since membrane targeting is thought to concentrate Gag, it is not surprising that the dependence of assembly on myristoylation observed at physiological levels of expression can be overcome by very high levels of Gag expression. For example, when the G2A Gag mutant is expressed at very high levels in insect cells, cytoplasmic capsid-like structures are observed (Gheysen et al., 1989); such structures have not been reported when G2A Gag is expressed in mammalian cells at levels comparable to those seen in HIV-1 infection.
In N-myristoylated proteins, the myristate moiety typically exists in two conformations – either sequestered within a hydrophobic pocket or exposed. The transition between these states constitutes a reversible “myristoyl switch” (McLaughlin and Aderem, 1995; Resh, 2006). In full-length HIV-1 Gag, the myristate is sequestered in a hydrophobic pocket created by α-helices in MA (Hermida-Matsumoto and Resh, 1999; Ono and Freed, 1999; Paillart and Gottlinger, 1999; Perez-Caballero et al., 2004; Saad et al., 2007; Spearman et al., 1997; Tang et al., 2004; Zhou and Resh, 1996), discussed further in Section 3.2. Mutational analyses demonstrated that a valine and leucine at positions 7 and 8 in MA (V7 and L8) are required for myristate exposure (Saad et al., 2007) and membrane targeting (Freed et al., 1994; Ono and Freed, 1999; Ono et al., 1997; Ono et al., 2000; Paillart and Gottlinger, 1999). When analyzed at a more detailed biochemical level in cells expressing the HIV-1 provirus, mutations of the myristoylation signal or the residues required for myristate exposure cause Gag to be arrested at the second step in the assembly pathway, with accumulation of the cytoplasmic ∼10S and ∼80S assembly intermediates (Robinson et al., 2014) (Class 2 mutations in Fig. 3).
In keeping with the reversible nature of myristoyl switches, myristoylation is thought to be necessary but not sufficient for stable membrane binding, with a second signal required to maintain membrane binding (McLaughlin and Aderem, 1995; Resh, 2006). In the case of Gag, the second signal is the highly basic region (HBR), a patch of basic residues (spanning amino acids 15 – 32 in MA) that form electrostatic interactions with the acidic head group of the PM phospholipid PI(4,5)P2 (Saad et al., 2006; Zhou et al., 1994; Zhou and Resh, 1996) (Fig. 1B; Fig. 2A). Additionally, mutational analyses have shown that residues around amino acids 85-89 in MA are important for directing assembling Gag to the PM, rather than to internal membranes within the cell (Freed et al., 1994; Ono et al., 2000). It is generally accepted that the PM is the main site of infectious HIV-1 assembly [reviewed in (Jouvenet et al., 2011a; Klein et al., 2007)], although there may be some situations in which targeting virus to internal compartments can lead to productive assembly and release (Joshi et al., 2009). Thus, residues that direct Gag to the PM during assembly typically ensure efficient release of infectious virus.
In summary, regions of the 132 amino acid MA domain that are critical for proper Gag assembly in cells include: i) residues 2 – 5 required for myristoylation; ii) residues 7 and 8, required for myristate exposure; iii) the MA HBR, which stabilizes membrane-targeted Gag through PI(4,5)P2 interactions; and iv) residues between amino acids 85-89, which ensure targeting of Gag specifically to the PM (Fig. 1B; Fig. 2A).
2.3. CA and SP1 residues required for immature capsid assembly in cells
The CA domain of Gag is composed of N-terminal and C-terminal subdomains, termed CA-NTD (residues 1 – 145) and CA-CTD (residues 146-231), respectively (Fig. 1B). In the context of wild-type (WT) Gag both CA subdomains play important roles in assembly, although CA-NTD is dispensable for assembly of a minimal Gag construct (see Section 2.5). Altogether, CA contains eleven α-helices, with helices 1 – 7 in CA-NTD and helices 8 – 11 in CA-CTD (Fig. 1B; Fig. 2A). In the case of CA-NTD, alanine-scanning mutagenesis revealed that specific exposed residue pairs in helices 4, 5, and 6 (EE75/76, RS100/102, TT107/108, and TQ110/112, shown in Fig. 1B; Fig. 2A) promote efficient assembly and virus production in cells (von Schwedler et al., 2003). Subsequent studies confirmed a role for these CA-NTD residues during immature capsid assembly, and demonstrated that mutations of these residues inhibit the final step in assembly, leading to accumulation of all the intermediates in the assembly pathway with no virion release (Class 5 mutations in Fig. 3) (Robinson et al., 2014). Interestingly, a recent structural study found that residues adjacent to or near these four residue pairs form critical inter-hexameric CA-NTD interfaces in the completed immature capsid formed by CA-NC of Mason-Pfizer monkey virus (MPMV) (Bharat et al., 2012). Thus, these residues are likely required during assembly because they form inter-hexameric CA-NTD contacts that stabilize the immature capsid.
Residues in CA-CTD that are required for immature capsid assembly were initially identified through alanine-scanning mutagenesis and deletion mapping [e.g.(Borsetti et al., 1998; Dorfman et al., 1994; Joshi et al., 2006; Mammano et al., 1994; Ono et al., 2005; von Schwedler et al., 2003)]. One set of CA residues critical for immature capsid assembly are located in CA helix 9 (the second α-helix in CA-CTD), which forms the crystallographic CA-CTD dimer interface (Fig. 2A). Assembly-defective CA-CTD dimer interface mutations are exemplified by the WM184/185AA mutant (Joshi et al., 2006; Ono et al., 2005; von Schwedler et al., 2003).
A second set of CA residues critical for immature capsid assembly is located at the base of the CA-CTD crystal structure, where three non-contiguous regions of CA are in close proximity: the major homology region (MHR), CA helix 10, and the CA-CTD tail (Fig. 1B, 2A). Examples of assembly-defective CA-CTD base mutations include K158A in the MHR; D197A in CA helix 10; and P224A, in the CA-CTD tail (von Schwedler et al., 2003) (Fig. 1B; Fig. 2A). Biochemical analyses of the effect of CA-CTD mutations on the assembly pathway in cells suggest that the CA-CTD dimer interface residues stabilize the first membrane-targeted assembly intermediate, while the CA-CTD base residues are required for continued multimerization of Gag at the PM (Robinson et al., 2014) (Class 3 and 4 mutations respectively in Fig. 3).
Two of the three regions that form the CA-CTD base are of particular interest. The first is the MHR, a 20 amino acid motif that precedes and overlaps with CA helix 8 (the first α-helix in CA-CTD, Fig. 1A). This motif is unusually well conserved, especially given that the amino acid sequence of the rest of Gag, with the exception of NC, is poorly conserved. The MHR is required for replication of retroviruses (Chang et al., 2007; Craven et al., 1995; Mammano et al., 1994; Strambio-de-Castillia and Hunter, 1992; von Schwedler et al., 2003; Willems et al., 1997) as well as the Ty3 retrotransposon (Orlinsky et al., 1996). Mutations of conserved residues in the HIV-1 MHR inhibit immature capsid assembly, although when and how they act remains unclear (Chang et al., 2007; Mammano et al., 1994; von Schwedler et al., 2003). Thus, the function of the MHR during assembly is not yet understood, and is likely a critical missing piece of the assembly puzzle, given its high degree of conservation.
The second region of interest at the base of CA is the CA-CTD tail, which is disordered in the CA-CTD crystal structure (Gamble et al., 1997). Secondary structure prediction programs, NMR, and biophysical analyses reveal that the C-terminus of CA-CTD, along with the adjacent SP1 spacer, can form an α-helix (Accola et al., 1998; Datta et al., 2011; Morellet et al., 2005; Newman et al., 2004) (shown as helix 12 in Fig. 2A). These studies have led to the proposal that an intact helix 12 may be critical for assembly of full-length Gag, but is lost upon cleavage. Consistent with this hypothesis, numerous mutational analyses reveal that residues in SP1 are critical for virus particle production (Chu et al., 2006; Gross et al., 2000; Guo and Liang, 2005; Guo et al., 2005; Krausslich et al., 1995; Liang et al., 2002; Liang et al., 2003).
In summary, mutational analyses indicate that three regions of CA are required for immature capsid assembly: helices 4 – 6 in CA-NTD, the CA-CTD dimer interface in helix 9, and the three regions that form the CA-CTD crystallographic base (the MHR, helix 10, and the CA-CTD tail). Related to the third region, an α-helix that is predicted to span the CA-CTD tail and SP1 also appears to be critical for assembly of full-length Gag (Figs. 1B; Fig. 2A).
2.4. The role of nucleocapsid in immature capsid assembly in cells
The NC domain of Gag contains two Cys-X2-Cys-X4-His- X4-Cys (CCHC) motifs, also called Cys-His boxes, each of which co-ordinates a zinc ion [reviewed in (Freed, 1998)]. The NC Cys-His boxes are critical for specific gRNA encapsidation [reviewed in (Berkowitz et al., 1996; Lu et al., 2011)]. Additionally, NC contains 15 basic residues dispersed throughout the domain (Fig. 1B). In the context of WT Gag, NC is critical for Gag multimerization; this requirement has been mapped to basic residues in NC (Cimarelli et al., 2000; Dawson and Yu, 1998; Derdowski et al., 2004; Gheysen et al., 1989; Jowett et al., 1992; Sandefur et al., 1998). The basic residues at the N-terminus of NC [which form a region previously termed the interaction (I) domain] also promote non-specific association of Gag with RNA, allowing RNA to bridge, tether, or scaffold Gag; this is thought to increase the local concentration of Gag, thereby indirectly promoting Gag-Gag interactions. As discussed in Section 4, this model is supported by the finding that RNA promotes multimerization of purified recombinant Gag in vitro (Campbell and Rein, 1999; Campbell and Vogt, 1997). Thus, NC basic residues mediate indirect, RNA-mediated Gag-Gag interactions, in contrast to residues in CA helix 9 (in CA-CTD), which mediate direct Gag-Gag dimerization. In cells, mutation of nine or more of the NC basic residues results in severe defects in immature capsid assembly (Cimarelli et al., 2000), causing Gag to be arrested at the first step in the assembly pathway with accumulation of only the ∼10S assembly intermediate (Lingappa et al., 2006; Reed et al., 2012) (Class 1 mutation in Fig. 3). In contrast, the direct interactions mediated by helix 9 are required for stable membrane targeting of assembling Gag, later in the assembly pathway (Robinson et al., 2014) (Class 3 mutations in Fig. 3). Thus, these two Gag-Gag interaction domains - NC acting indirectly via RNA and CA helix 9 acting through direct protein-protein interactions - function at very different points during the assembly process.
2.5. Defining the minimal domains required for Gag assembly
As described above, point mutations and deletions within individual domains reveal that MA, CA-NTD, CA-CTD, SP1, and NC each play an important role in assembly of WT Gag. In a complementary approach, mutational analyses were used to identify Gag domains that are absolutely required for assembly (i.e. cannot be either deleted or replaced). Domain replacement was initially used to gain insights into the function of NC (Zhang et al., 1998). In this study, immature capsid assembly and release was unaffected when NC was replaced with a heterologous transcription factor leucine zipper, a canonical protein dimerization or trimerization domain (Alber, 1992). The resulting GagZip particles contained little RNA, since the leucine zipper does not bind nucleic acid, but nevertheless were morphologically similar to immature particles formed by WT Gag (Accola et al., 2000; Crist et al., 2009; Johnson et al., 2002; Zennou and Bieniasz, 2006). Moreover, like WT Gag, GagZip proteins require the critical CA-CTD dimerization residues (WM184/185) for assembly, interact with ABCE1, and form intracellular assembly intermediates unless the leucine zipper domain is mutated to inhibit dimerization (Klein et al., 2011). Thus, studies of GagZip proteins reveal that the function of NC in assembly is to promote Gag-Gag interactions; that the role of NC in Gag multimerization is independent of its role in gRNA packaging; and that replacement of NC (which promotes indirect Gag-Gag interactions via RNA) with a domain that forms direct protein-protein interactions does not affect immature capsid assembly or progression of Gag through ABCE1-containing assembly intermediates.
Another study extended this approach and demonstrated that many of the remaining regions of Gag in the assembly-competent GagZip protein could be either deleted or replaced without affecting assembly (Accola et al., 2000). Thus, immature capsids still form following removal of CA-NTD and most of MA (except for the N-terminal amino acids involved in myristoylation and myristate exposure). Additionally, the p6 domain can be replaced with a short peptide containing a budding motif from RSV Gag. In the resulting “minimal Gag” protein, the only regions of HIV-1 Gag that could not be deleted or replaced without adversely affecting assembly were CA-CTD and SP1 (Fig. 1A). Thus, these studies revealed that CA-CTD and SP1 are uniquely critical for assembly, for reasons that remain unclear. Notably, as described above, CA-CTD contains the MHR, a highly conserved and poorly understood motif required for assembly.
3. Structural biology of Gag domains
3.1. Overview of Gag structural biology
Cleavage of the Gag polyprotein by the HIV-1 protease occurs simultaneous with, or just after, release of the immature virus. Following cleavage, MA (also called p17), CA (also called p24), NC (also called p7), p6, and the spacer peptides exist as independent proteins, some of which play critical roles during the post-entry phase of the virus life cycle. X-ray structures have been solved for only three retroviral Gag domains or subdomains over the years: MA, CA-NTD, and CA-CTD [reviewed in (Ganser-Pornillos et al., 2012; Ganser-Pornillos et al., 2008)]. However, because it is the Gag polyprotein that undergoes assembly, additional insight into how Gag functions during assembly would be gained from a crystal structure of full-length Gag. Unfortunately, full-length Gag has eluded crystallization because it contains highly flexible unstructured regions (Bharat et al., 2012), including the p6 domain, the spacer peptides, and the linker regions that connect the structured MA domain and CA subdomains. The crystal structures of MA, CA-NTD, and CA-CTD have obvious relevance to events that occur following maturation, but have also helped with computationally generated structures of full-length Gag in the immature virion (Bharat et al., 2012; Briggs et al., 2009; Carlson et al.; de Marco et al., 2010; Fuller et al., 1997; Keller et al., 2011; Wright et al., 2007). Nevertheless, in the absence of a high-resolution structure of full-length Gag, we are left with a gap in our understanding of structural considerations during early events of Gag assembly. Moreover, we are also faced with the nagging question of how infected cells handle the highly flexible, and possibly unstable, newly synthesized, full-length Gag protein.
Here we will discuss structures of MA, CA-NTD, CA-CTD, the CA-SP1 boundary, and the fully assembled immature capsid. NMR structures are also available for NC complexed with individual stem loops of the HIV-1 psi-element packaging signal (Amarasinghe et al., 2000; De Guzman et al., 1998), but are not discussed here. Interestingly, the three dimensional structures of MA, CA-NTD, and CA-CTD are conserved in diverse retroviruses, despite very limited overall amino acid sequence conservation [e.g. (Bharat et al., 2012; Campos-Olivas et al., 2000; Christensen et al., 1996; de Marco et al., 2010)]. Thus, the structures of MA and CA appear to be highly constrained by their functions. Likewise, the architecture of fully assembled RSV, and MPMV immature capsids is remarkably similar to that of HIV-1 (de Marco et al., 2010), which is described in Section 3.5.
3.2. Structure of the matrix (MA) domain
In high-resolution structures, the 132 amino acid MA domain contains five α-helices and a three-stranded mixed β–sheet (Hill et al., 1996; Massiah et al., 1994) (Fig. 2A). Helices 1-3 are organized around a central, buried helix 4 to form a globular domain that is capped by the β–sheet, with helix 5 projecting away from the globular domain. MA forms a trimer when crystallized (Hill et al., 1996), and a hexamer of trimers on membranes containing PI(4,5)P2 (Alfadhli et al., 2009; Alfadhli et al., 2007). From the perspective of immature capsid assembly, the two most important features of MA are the cavity formed by the MA globular domain and the basic residues that cluster around the exposed MA β–sheet and form the aforementioned HBR.
NMR studies show that the cavity formed by the globular domain of MA can sequester the N-terminal myristate of MA (Tang et al., 2004). In these studies, MA in solution exists both in myristate-sequestered and myristate-exposed states. A shift in the equilibrium towards the myristate-exposed state occurs upon Gag oligomerization involving either direct (CA-CA-mediated) interactions or indirect (NC-mediated) interactions, thereby coupling myristate exposure to early oligomerization events in assembly. Additionally, direct binding of MA to PI(4,5)P2 in the PM favors myristate exposure as well as PM binding (Saad et al., 2006) [reviewed in (Ganser-Pornillos et al., 2012)]. In contrast, RNA binding to the MA basic residues appears to inhibit myristate exposure in the context of full-length Gag (Chukkapalli et al., 2010). Thus, myristate exposure is governed by a number of positive and negative regulators, perhaps including other cellular factors not yet described. By acting in concert, these regulators likely allow RNA binding and Gag oligomerization to precede membrane targeting (Chukkapalli and Ono, 2011).
The HBR mediates electrostatic contacts with acidic phospholipids (Zhou et al., 1994) such as PI(4,5)P2, which is found preferentially in the PM inner leaflet, and thereby helps to anchor assembling Gag to the PM. Notably, studies suggest that the HBR- PI(4,5)P2 interaction may involve other elements, besides charge alone (Chukkapalli et al., 2010; Llewellyn et al., 2013). NMR studies indicate that HBR binding to PI(4,5)P2 promotes exposure of the sequestered myristate, which in turn allows one PI(4,5)P2 acyl chain to occupy a hydrophobic groove within the MA globular head, while the other PI(4,5)P2 acyl chain remains anchored in the PM (Saad et al., 2006). Thus, two hydrophobic interactions anchor Gag to the PM: the exposed myristate of Gag inserted into the PM inner leaflet, and a PI(4,5)P2 acyl chain extending out of the lipid bilayer to sit in an MA hydrophobic pocket (reviewed in (Chukkapalli and Ono, 2011)).
3.3. Structure of the N-terminal and C-terminal domains of capsid (CA-NTD and CA-CTD)
The immature Gag lattice is stabilized by direct Gag-Gag interactions, the majority of which are in CA-NTD, CA-CTD, and SP1. CA-NTD contains seven α-helices that form an arrow shaped structure (Gamble et al., 1996; Gitti et al., 1996) (Fig. 2A). An extended loop connects helices 4 and 5 and also forms the binding site for cyclophilin A (Gamble et al., 1996), a host factor that acts post-entry, during capsid uncoating (Luban, 2007). CA-CTD is composed of a short 310 helix followed by an extended strand and four α-helices (numbered helices 8 – 11 in CA) (Gamble et al., 1997; Worthylake et al., 1999). As discussed in Section 2.3, CA-CTD contains the highly conserved 20 amino acid MHR motif, which overlaps with the strand-turn-helix 8 region at the N-terminus of HIV-1 CA-CTD. The MHR is required for immature capsid assembly of HIV-1, other retroviruses, and the Ty3 retrotransposon (Chang et al., 2007; Craven et al., 1995; Mammano et al., 1994; Orlinsky et al., 1996; Strambio-de-Castillia and Hunter, 1992; von Schwedler et al., 2003; Willems et al., 1997).
CA-CTD crystallizes as a side-by-side dimer, in which helix 9 forms the dimer interface, with hydrophobic contacts stabilizing its interaction with its symmetry mate (Byeon et al., 2009; Gamble et al., 1997; Worthylake et al., 1999). A second “domain-swapped” dimer structure has been observed in crystal structures of a CA-CTD mutant in which a single amino acid was deleted (Ivanov et al., 2007). In this structure, the MHR formed the interface between the two dimers. While the notion that Gag dimers adopt a domain-swapped conformation during assembly of the immature capsid lattice is an appealing one (Kingston and Vogt, 2005), evidence in favor of the domain-swapped conformation has not been generated by recent structural studies, including a deuterium hydrogen exchange study (Monroe et al., 2010), a 17-Å structure of the immature HIV-1 capsid (Briggs et al., 2009), and 8 to 11-Å structures of immature capsid-like tubular arrays of MPMV and HIV-1 CA-NC (Bharat et al., 2014; Bharat et al., 2012).
3.4. Structural studies of the SP1 spacer
As mentioned above, computational programs predict that the last seven residues of CA [which are disordered in the CA-CTD crystal structure (Gamble et al., 1997; Worthylake et al., 1999)] and the first seven residues of SP1 form an α-helix in the context of full-length Gag (Accola et al., 1998). The helical propensity of CA-SP1 boundary is supported by mutational approaches (Chu et al., 2006; Gross et al., 2000; Guo and Liang, 2005; Guo et al., 2005; Krausslich et al., 1995; Liang et al., 2002; Liang et al., 2003), biophysical and NMR studies (Datta et al., 2011; Morellet et al., 2005; Newman et al., 2004), and structural analyses of the fully-assembled immature capsid, all of which are consistent with this region forming a six helix bundle (Bharat et al., 2014; Wright et al., 2007). Interestingly, in studies of the purified CA-SP1 peptide in buffer an abrupt switch to a helical conformation was observed at high peptide concentrations, suggesting that this region is capable of a conformational switch that could promote assembly (Datta et al., 2011). Whether this helical conformation is present when full-length Gag assembles in cells remains to be determined.
3.5. Structure of the immature capsid
Improvements in cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) have led to increasingly higher resolution structures of the immature capsid in recent years (Bharat et al., 2012; Briggs et al., 2009; Carlson et al.; de Marco et al., 2010; Fuller et al., 1997; Keller et al., 2011; Wright et al., 2007). Others have reviewed the structure of the immature capsid revealed by cryo-ET studies in detail (Ganser-Pornillos 2012). Briefly, the immature capsid does not display icosahedral symmetry - unlike capsids of other viruses - but instead is composed of a lattice that contains areas of hexagonal order, as well as defects that allow for its spherical shape. The generally hexagonal architecture comes from the CA-NTD, CA-CTD, and SP1 regions, each of which forms closely packed hexameric structures. There is general consensus from cryoET and cryoEM studies that Gag within the completed immature capsid adopts an extended, radially arranged “beads on a string” configuration (Fig. 2 B, C). In this configuration, MA is closest to the viral membrane, with CA-NTD forming a hexameric lattice that sits below the MA layer; below that is a hexameric lattice formed by CA-CTD; and below the “hole” formed by the CA-CTD hexamer are pillars of density formed by the putative helix at the CA/SP1 boundary, which has been modeled as a six helix bundle (Fig. 2 D).
A recent 8-Å resolution cryoEM/cryoET structure of the immature-like tubular assemblies formed by MPMV CA-NC defined some of the residues in CA-CA interfaces (Bharat et al., 2012). In this study, a fitted reconstruction of the HIV-1 immature capsid revealed that CA-NTD is stabilized by inter-hexameric contacts alone, formed by residues in helices 4-7 in CA-NTD. In contrast, CA-CTD is stabilized by both intra- and inter-hexameric contacts in the immature capsid. Residues in CA helix 11 and the MHR (which overlaps with CA helix 8) contribute to intra-hexameric CA-CTD interfaces, while residues in CA helix 9 form inter-hexameric CA-CTD contacts at the hydrophobic dimer interface, in agreement with numerous previous studies. Notably, this study found that interface residues in the immature capsid differ substantially from the interface residues in the mature capsid (Bharat et al., 2012). Indeed the only interface in common was the CA-CTD dimer interface, which involves CA helix 9 in both the immature and mature capsid, albeit in different orientations (Bharat et al., 2012). A follow-up study used hybrid cryoEM/cryoET to resolve an ∼9 – 11-Å structure of immature-like tubular assemblies composed of HIV-1 CA-NC encoding a Y196 mutation that stabilizes these tubular assemblies but does not affect crystal structure of CA domains (Bharat et al., 2014). This structure confirmed that CA-CTD dimers in MPMV and HIV-1 immature assemblies are structurally similar, but found that the size of the HIV-1 CA-CTD hexamer is smaller, resulting in more extensive intra-hexameric CA-CTD contacts for HIV-1 via additional residues in the MHR and CA helix 11.
3.6. Gag structural biology: new horizons and challenges
As cryoEM/cryoET techniques improve, it is likely that increasingly high-resolution structures of the fully assembled immature capsid will be obtained in the future. Such structures will allow more accurate mapping of residues involved in Gag-Gag interfaces, thereby filling in more pieces of the puzzle. An example of the degree of resolution that can be achieved is seen in a recent 8-Å resolution cryoEM/cryoET structure of the mature HIV-1 capsid, from which large scale molecular dynamics simulation resulted in an all-atom model of an entire mature capsid (Zhao et al., 2013). Ultimately, however, the biggest challenge for structural biologists will be to obtain a structure of full-length Gag, and to better understand structural considerations underlying the interaction of Gag with cellular facilitators of assembly.
4. Assembly of recombinant Gag in vitro
4.1. Principles learned through studying the assembly of recombinant Gag in vitro
To gain insights into the process of assembly, systems have been generated that recapitulate capsid formation outside the context of the cell. One such system involves studying self-assembly of purified, isolated Gag domains (Fig. 4). In this system, purified recombinant Gag is incubated at high concentrations (∼1mg/ml, or 20 μM) in buffer. Typically a recombinant MA-CA-NC protein (GagΔp6) is used, since including the unstructured p6 domain, which contains motifs involved in budding, leads to degradation in E. coli (Campbell and Rein, 1999). Additionally, the recombinant proteins used in these systems are not myristoylated [myr(-)]. Incubation of recombinant HIV-1 myr(-)GagΔp6 with RNA or even oligonucleotides in buffer results in spherical structures that resemble HIV-1 immature capsids, except for being significantly smaller (25-30 nm vs. 100 nm in diameter) (Campbell and Rein, 1999). Thus, these studies revealed that, in a simplified environment, Gag has an intrinsic ability to assemble into spherical structures in the presence of nucleic acid alone.
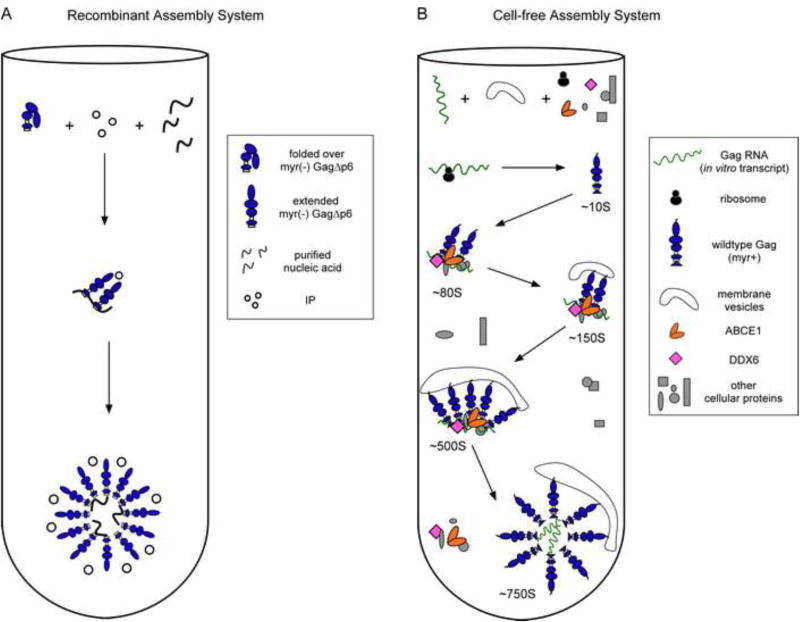
Fig. 4. Comparison of the pathways for assembly in the recombinant and cell-free assembly systems.
(A) The recombinant assembly system contains purified nucleic acids, an inositol phosphate-related compound (IP), and a purified, fully folded, recombinant myr(-)GagΔp6. In solution, myr(-)GagΔp6 is thought to exist in a folded-over form. Addition of IP and oligomerization of Gag converts Gag to an extended form resulting in formation of spherical structures that closely resemble HIV-1 immature capsids. The only intermediate in the pathway is likely a dimer. (B) In the cell-free system, as in cells, Gag progresses through an energy-dependent, host-catalyzed pathway of assembly intermediates (∼10S, ∼80S, ∼150S, and ∼500S complexes), culminating in formation of the ∼750S completed immature capsid. RNAs are found in ribonucleoprotein complexes (RNPs). A cellular extract and purified exogenous components provide factors needed for Gag translation and assembly. Gag mRNA is translated by ribosomes from the extract to produce WT myristoylated Gag de novo. Newly synthesized Gag co-opts host proteins to form the assembly intermediates. Based on studies in the cell-free system and/or cells, the assembly intermediates contain the cellular facilitators of assembly ABCE1 and DDX6. Intact membranes, likely present as vesicles, and myristoylation of Gag are required for progression past the ∼80S intermediate. References are in the text.
Subsequently it was shown that adding inositol phosphates or phosphatidylinositol phosphates to the recombinant in vitro assembly system results in capsid-like structures that are similar in size to immature capsids produced in cells (Campbell et al., 2001). This finding began with the observation that addition of a cellular extract to recombinant assembly systems resulted in particles of the correct size. Fractionation of this extract led to identification of an inositol phosphate that increased particle size when added directly to the recombinant reaction (Campbell et al., 2001). As described in Section 6.3, studies in cells later led to identification of PI(4,5)P2, a PM-associated phosphatidylinositide that binds to MA in cells, and promotes proper membrane targeting [reviewed in (Chukkapalli and Ono, 2011)].
The recombinant assembly of Gag-derived proteins has also been useful for analyzing the architecture of the immature capsid [e.g. (Bharat et al., 2014; Bharat et al., 2012; de Marco et al., 2010)], as described in Section 3.5. Additionally, studies of purified recombinant myr(-)GagΔp6 in vitro have defined the behavior of Gag in solution. Biophysical measurements reveal that Gag is in a monomer-dimer equilibrium in solution, and suggest that monomeric Gag adopts a folded over conformation that brings MA relatively close to CA (Datta et al., 2007). It remains to be determined if this folded-over structure is also found in cells. If so, then Gag must undergo a drastic conformational change via an unknown mechanism to achieve the extended conformation found in fully assembled particles. It has been suggested that such a conformational change could be achieved during oligomerization of Gag, based on in vitro studies (Datta et al., 2011). However, it is also possible that in cells Gag co-opts a host protein that either prevents Gag from adopting the folded-over form, or facilitates conversion from the folded-over to the extended form. Additionally, it is unclear how the highly unstructured p6 domain, which was not included in these recombinant studies, might affect the folded-over conformation of Gag in cells.
4.2. The recombinant Gag assembly system recapitulates a simplified environment, not an intracellular environment
The in vitro capsid assembly system reconstitutes the process of assembly in a very specific environment - one in which purified Gag is present at very high concentrations (1 mg/ml or ∼20 μM) and purified RNA is available to nucleate assembly. This approach established two concepts that have dominated the field: that RNA nucleates the assembly of Gag (in conjunction with Gag-Gag and Gag-lipid interactions); and that in the presence of nucleic acid Gag can assemble spontaneously, i.e. uncatalyzed and without the input of energy. To put these concepts into context, however, it must be recognized that there are a number of key differences between the recombinant assembly system and the intracellular environment. First, in cells the concentration of Gag is likely much lower than in the recombinant assembly system, and the concentration of total cellular protein is estimated to be very high - up to 200 – 400 mg/ml (Ellis, 2001). Both of these features would be expected to reduce the frequency of spontaneous Gag-Gag interactions, making spontaneous assembly of Gag less likely in cells. Secondly, because RNA is such a potent activator of the innate immune response [reviewed in (Gurtler and Bowie, 2013)], viral RNA would likely not be present in a purified form in cells, but instead would be sequestered within host ribonucleoprotein (RNP) complexes where RNA is shielded by proteins. Thirdly, in cells HIV-1 must co-ordinate the many events needed to produce an infectious virus with great efficiency. For example, in cells Gag must find the gRNA sequestered within a host RNP complex, remove the RNPs that coat the gRNA and replace them with Gag to promote encapsidation of gRNA - and succeed in these tasks rapidly with limited detection by the host.
Given that the cytoplasm of infected cells is best described as a hostile environment full of obstacles, Gag is unlikely to assemble spontaneously in cells the way it does in the simplified environment of the recombinant assembly system. Instead, retroviral Gag proteins are likely to have developed specialized mechanisms to avoid detection and enhance assembly efficiency in cells. A parallel can be seen in the study of protein folding. Specifically, Nobel prize-winning experiments in the 1960's demonstrated that the primary amino acid sequence of a protein contains all the information needed for proper protein folding (Anfinsen, 1973). And yet, we now know that because of the vectorial nature of protein synthesis and the detrimental consequences of protein aggregation, most protein folding in cells is facilitated by a diverse array of molecular chaperones - cellular proteins that assist protein folding in cells without becoming part of the protein's final structure (Hartl, 1996; Kim et al., 2013). If the same is true for Gag assembly, then the principle of “Gag self-assembly” revealed by the recombinant assembly system would best be viewed as demonstrating that Gag can self-assemble, not that Gag does self-assemble. As discussed in Sections 5 and 6 below, systems that recapitulate HIV-1 assembly in the context of the cellular environment strongly favor a model in which assembly in cells is energy-dependent, host-catalyzed, and sequestered. Nevertheless, the recombinant assembly system has proven very useful for understanding Gag's intrinsic properties.
4.3. Questions that remain to be answered in the recombinant Gag self-assembly system
In the future, it would be of interest to determine how “spontaneous” assembly of recombinant Gag is affected when the system is altered to more closely approximate the intracellular environment. For example, how is the efficiency of assembly affected by reducing the concentration of recombinant Gag? Additionally, how does inclusion of other proteins or RNP complexes (rather than purified RNA) affect the efficiency of recombinant Gag assembly? Given that facilitators of assembly are absent in the recombinant system, one might expect spontaneous assembly to occur only at extremely high concentrations of recombinant Gag, and that addition of RNP complexes as a source of RNA or addition of high concentrations of other proteins would reduce or eliminate spontaneous assembly.
Another question that arises is how recombinant Gag self-assembly is affected by mutations found to be critical for immature capsid assembly in cells. As discussed in Section 2, the requirement for specific residues in MA, CA, and NC is an important hallmark of Gag assembly under physiologically relevant conditions. An example of the discrepancy between assembly in cells and recombinant assembly is seen with the non-myristoylated Gag. In cells, mutations that inhibit myristoylation of Gag also inhibit assembly (Bryant and Ratner, 1990; Gottlinger et al., 1989), except in cases of significant overexpression (Gelderblom et al., 1987). However, in the recombinant assembly system non-myristoylated GagΔp6 is assembly competent, most likely because the concentration-dependence that is a hallmark of assembly in cells is not recapitulated by the in vitro assembly system. Thus, these findings support the hypothesis that assembly takes different pathways to a similar endpoint in these two systems. It would be useful to examine whether other assembly-defective Gag mutants assemble in the recombinant system. If they do, this would suggest that other barriers to assembly in cells, besides low Gag concentration, are not recapitulated in the recombinant assembly system. Differences in assembly phenotype between cells and the recombinant assembly system could be useful for understanding the role of cellular facilitators of assembly, which HIV-1 may have evolved to utilize in order to overcome barriers present only in the more challenging intracellular environment.
5. Assembly of newly synthesized Gag using cell extracts
5.1. Cell-free assembly systems recapitulate the intracellular environment
Like the recombinant assembly system, the cell-free system is well suited for studying how biochemical manipulations affect Gag assembly; however, unlike the recombinant assembly system, the cell-free system recapitulates key features of the intracellular environment. This is achieved through the use of cell extracts, such as rabbit reticulocyte extract (Sakalian et al., 1996; Spearman and Ratner, 1996) or wheat germ extract (Lingappa et al., 1997), and by linking Gag assembly to translation. When comparing the cell-free and recombinant assembly systems, five major differences should be noted (Fig. 4). First, the cell-free assembly system begins with translation of Gag from an mRNA (de novo Gag synthesis), while purified Gag proteins in the recombinant assembly system are fully folded at the start of the assembly reaction. Secondly, Gag proteins translated in the cell-free system are full-length and myristoylated, rather than myr(-) and Δp6. Thirdly, the cell-free system contains cellular proteins while the recombinant system does not; in fact the concentration of cellular protein in the cell-free system is as high as 30 – 40 mg/ml. Additionally, the cell-free system contains much lower concentrations of Gag, in the range of 10 ng/ml or 0.5 micromolar, which is 40 fold lower than the concentration of Gag in the recombinant assembly system (Lingappa et al., 2005). This sub-micromolar Gag concentration is more likely to approximate steady-state levels of Gag found in infected cells. Fourthly, membranes are present in cell-free systems, allowing recapitulation of membrane targeting of Gag, a critical feature of assembly in vivo. Finally, in cell extracts as in cells, non-translating RNAs are likely to be in RNP complexes within RNA granules, as discussed further in Section 6.5.
5.2. Assembly in cell-free systems reproduces key features of assembly in cells
In cell-free systems, the cellular extract provides all the cellular factors needed for protein synthesis and post-translational events, as well as membrane vesicles. Also added to the reaction are an in vitro transcribed Gag mRNA that serves as the template for translation, unlabeled and radiolabeled amino acids, an energy regenerating system, and any critical cofactors that are removed during preparation of the extract, such as myristoyl CoA, which is needed for myristoylation. In a typical retroviral cell-free reaction, Gag, the only viral protein present, is also the only protein that is translated de novo; thus, newly synthesized Gag is radiolabeled, while the cellular proteins in the reaction are unlabeled.
During the 2h cell-free reaction, Gag synthesis is followed by post-translational events of assembly. Three approaches have demonstrated that cell-free assembly of Gag closely mimics Gag assembly in cells (Lingappa et al., 1997). First, the Gag-containing particles formed at the end of the reaction closely resemble HIV-1 immature capsids from cells in their size, shape, and density, as judged by biochemical and ultrastructural analyses. Secondly, key features of the HIV-1 capsid assembly process in cells are also faithfully reconstituted in this system. For example, assembly is inhibited at the same step by addition of detergents expected to solubilize membrane vesicles at the start of the reaction, in reactions that lack myristoyl coA, and in reactions programmed with the G2A Gag mutant that fails to undergo myristoylation. Together, these findings suggest that assembly in the cell-free system requires myristate-dependent targeting of Gag to membranes, as observed in cells. Thirdly, analysis of Gag mutants has revealed that phenotypes of assembly-competent and assembly-defective Gag mutants, originally identified in cells (see Section 2 above), are faithfully reconstituted in the cell-free system (Lingappa et al., 1997; Singh et al., 2001). Thus, analyses of the end product, the assembly process, and phenotypes of Gag mutants confirm that the cell-free assembly system closely recapitulates the process of assembly that occurs in cells.
5.3. Assembly in cell-free systems proceeds through an energy-dependent, host-catalyzed pathway of assembly intermediates
One notable feature of the cell-free system is its inefficiency: at best ∼30% of the newly synthesized Gag ends up as completed capsids. This is likely attributable to the use of cell extracts, in which cellular proteins undergo progressive denaturation and are not replaced. Additionally, the heterologous nature of the system may be a contributing factor, since the cell extracts that are used are from cells with a high synthetic capacity and few proteases, rather than cells that HIV-1 evolved to infect efficiently. This inefficiency turns out to be a strength of the cell-free system, since it allows intermediates in the assembly process to be trapped, identified, isolated, and analyzed. Such intermediates are found in extremely small quantities in human cells expressing Gag, where the assembly process occurs much more efficiently. Indeed, their existence was not even hypothesized until they were first revealed by studies in the cell-free assembly system (Lingappa et al., 1997).
When cell-free assembly reactions were analyzed by velocity sedimentation, the ∼750S completed immature capsid was observed along with discrete Gag-containing complexes of smaller sizes: ∼10S, ∼80S, ∼150S, and ∼500S (Lingappa et al., 1997). While the cell-free assembly reaction is typically performed in the absence of detergents to allow newly synthesized Gag to target to membrane vesicles, velocity sedimentation is performed after treating the reaction products with non-ionic detergents that solubilize membranes, thereby allowing analysis of Gag-containing protein complexes that are free of membranes. Together pulse-chase and mutational analysis established that the smaller complexes observed in the cell-free system represent true intermediates in the assembly process (Lingappa et al., 1997). Specifically, pulse-chase experiments revealed that Gag proteins labeled during a short radiolabeling “pulse” subsequently chase out of the ∼10S complex, first into the ∼80S and ∼150S complexes and later into the ∼500S and ∼750S complexes. These experiments demonstrated that the Gag-containing complexes were not dead-end “off pathway” side products but instead represent intermediates in a pathway that culminates in formation of the ∼750S completed immature capsid. Additionally, in the cell-free system, assembly-defective Gag mutants identified in cellular systems were arrested at specific points in the assembly pathway, accumulating only those intermediates that precede the point of blockade (Lingappa et al., 1997; Singh et al., 2001), thereby confirming the order of intermediates in the pathway. Notably, after the approach for identifying these complexes was developed in the cell-free system, it was adapted to allow identification of comparable assembly intermediates in transfected and HIV-1 infected cells expressing Gag [e.g. (Dooher and Lingappa, 2004; Dooher et al., 2007; Lingappa et al., 1997; Reed et al., 2012)].
Studies also revealed that capsid assembly in the cell-free system is energy-dependent, that ATP hydrolysis and not just ATP binding was likely required, and that the energy requirement is post-translational and therefore separate from the well-understood requirement for energy during translation (Lingappa et al., 1997). This initiated a shift in thinking about Gag assembly. The “self-assembly” model had been dominant in the field (and still is, as revealed in textbooks, e.g. Fields' Virology Sixth edition p. 136, 2013); but the finding that assembly is energy-dependent in cells implies that the mechanism of assembly in cells differs from the spontaneous assembly observed with recombinant Gag in vitro. This point was subsequently underscored by additional studies demonstrating the energy-dependence of retroviral capsid assembly in the cell-free system and in cells (Dooher and Lingappa, 2004; Weldon et al., 1998), and by the identification of an ATP-binding protein, ABCE1, that facilitates assembly in cells (Zimmerman et al., 2002). Likewise, the finding that Gag assembles via a pathway of intermediates, in the cell-free system and in cells, is also at odds with results of recombinant assembly systems, in which no intermediates larger than dimers have been identified (Ma and Vogt, 2004). These contrasting models could be reconciled if Gag evolved the ability to form assembly intermediates in cells to evade host innate immune defenses and access cellular facilitators of assembly, as discussed in Section 7.
5.4. Identification of ABCE1, a cellular ATPase that facilitates HIV-1 capsid assembly
Initial studies revealed that immature capsid assembly in the cell-free system is dependent on one or more large host cell complexes that could be removed from the system by ultracentrifugation of the cell extract (Lingappa et al., 1997). Like the finding that immature capsid assembly is energy-dependent, the suggestion that cellular factors are critical for proper assembly of Gag did not conform to expectations at the time. The subsequent identification of ABCE1, a cellular ATPase that facilitates assembly (Zimmerman et al., 2002), helped to explain the dependence of assembly on energy and cellular factors. ABCE1 is a cytoplasmic member of the ATP-binding cassette (ABC) family of proteins (E1 subfamily) and contains two nucleotide binding domains as well as two N-terminal iron sulfur binding domains (Kerr, 2004). ABCE1 is associated with Gag in the ∼80S, ∼150S, and ∼500S assembly intermediates (collectively termed the high-molecular-mass assembly intermediates) in both the cell-free system and in cells (Fig. 3) (Zimmerman et al., 2002), but is not associated with Gag in the ∼10S fraction even though ABCE1 and Gag are abundant in this fraction.
Importantly, capsid formation in the cell-free system was inhibited when reactions were programmed with extracts immunodepleted of ABCE1. Conversely, when recombinant ABCE1 was added back to the depleted extracts, capsid assembly was restored (Zimmerman et al., 2002). In addition, a truncated ABCE1 mutant containing only one of the two ATPase domains acted as a dominant negative inhibitor of immature capsid assembly in cells (Zimmerman et al., 2002). Together these studies provided strong support for ABCE1 being a cellular facilitator of assembly. However, because ABCE1 is an essential protein, ABCE1 knockdown leads to cell death within hours (Chen et al., 2006; Dooher et al., 2007); thus, it has not been possible to test whether ABCE1 is critical for assembly in cells using siRNA knockdown approaches. Other studies performed in cells have provided considerable information about ABCE1-containing intermediates (see Section 6.7) and identified an additional ATP-dependent cellular facilitator of assembly, DDX6 (see Section 6.5).
5.5. Strengths and limitations of the cell-free assembly system
The cell-free system provided the first evidence that HIV-1 Gag assembly in cells proceeds through a stepwise, energy-dependent, and host-catalyzed pathway of intermediates. Assembly intermediates were readily identified in the cell-free system because it is a closed system that does not recapitulate envelopment and virion release, its inefficiency results in accumulation of Gag in assembly intermediates, and it is easy to manipulate biochemically. Thus, the cell-free system provided a “road map” for detection of assembly intermediates in cells, which are transient and present in very small quantities. Additionally, the cell-free system is currently being used as a screen for inhibitors of capsid assembly of a variety of viruses (Lingappa et al., 2013), and therefore holds promise for identifying novel antiviral small molecules in the future (see Section 6.4).
Notably, the heterologous nature of the cell-free system – in which the plant extracts support a virus that infects human cells - is both a strength and weakness. Initially, the fact that HIV-1 capsids could be assembled utilizing proteins from a plant extract was puzzling to some since HIV-1 does not infect plants. However, it had long been known that immature capsids can be generated in lower eukaryotes (Gheysen et al., 1989). Moreover, cellular facilitators of assembly identified to date, e.g. ABCE1 and DDX6 (Reed et al., 2012; Zimmerman et al., 2002), are well conserved and likely function quite similarly to their homologues in human cells. A downside to the cell-free system is that fewer tools are available for studying proteins in wheat germ compared to human cells. For this reason - and to settle questions regarding the presence of the host-catalzyed assembly pathway in infected human cells - studies of the assembly pathway in the past decade have been largely performed using human or non-human primate cell lines.
6. Imaging, biochemical, and siRNA knockdown analyses of assembly in cells
6.1. Live imaging of assembly in cells
In recent years, live imaging has been used to study many aspects of the HIV-1 life cycle, including genome packaging [reviewed in (Moore and Hu, 2009)], budding and release [reviewed in (Baumgartel et al., 2012)], and cell-to-cell transmission [reviewed in (Campbell et al., 2008)]. Here we will confine our discussion to how live cell imaging has been used to study assembly of Gag in cells. Live imaging studies established that HIV-1 assembly occurs primarily at the PM, with Gag going from undetectable to fully assembled in less than 10 min (Ivanchenko et al., 2009; Jouvenet et al., 2008; Jouvenet et al., 2009). Fluorescence resonance energy transfer studies confirmed that Gag tagged with a fluorescent protein undergoes multimerization at the PM (Jouvenet et al., 2008). Additionally, fluorescent recovery after photobleaching allowed fully assembled sites to be distinguished from assembling sites. Many of the highest resolution live imaging studies use total internal reflection fluorescent microscopy (TIR-FM), a method that only excites fluorescent proteins that are in close proximity to the coverslip, i.e. near the PM. TIR-FM analysis demonstrated that gRNA expressed alone moves rapidly, appearing only transiently at the PM, while gRNA expressed with Gag was able to anchor at the PM (Jouvenet et al., 2009). In this study, Gag fluorescence was initially observed ∼4.5 min after gRNA appeared at the PM, after which Gag fluorescence increased, consistent with multimerization. Another live imaging study confirmed this conclusion by showing that accumulation of gRNA at the PM requires Gag (Kemler et al., 2010). Both studies proposed that the Gag-gRNA association is likely initiated in the cytosol, and that the number of Gag molecules in each membrane targeting event is very small, below the level that can be detected using live imaging (Jouvenet et al., 2009; Kemler et al., 2010). Subsequently, the initial packaging complex inferred from live imaging studies was identified by immunoprecipitation of complexes containing Gag and gRNA from cell lysates (Kutluay and Bieniasz, 2010). Thus, based on live imaging and biochemical studies of cells, it appears that gRNA first associates in the cytoplasm with a small number of Gag proteins, possibly a dimer given results of crosslinking experiments (Kutluay and Bieniasz, 2010).
While live imaging has greatly advanced our understanding of the behavior of HIV-1 genomes near the PM and assembling Gag in the late stages of multimerization, it is limited when proteins of interest are expressed at concentrations that are below the threshold for detection using this approach. For example, while a study successfully detected fluorescently tagged ALIX, a host protein involved in HIV-1 budding, by TIR-FM, the same study was unable to detect fluorescently tagged TSG101, a well-established cellular factor critical for HIV-1 budding (Jouvenet et al., 2011b). Additionally, it is difficult to use live imaging to study early events of assembly because diffuse background fluorescence in the cytosol reduces imaging resolution (Jouvenet et al., 2011a). In contrast, biochemical and ultrastructural studies of assembly in cells expressing HIV-1 proviruses are able to detect small amounts of Gag, both in the cytoplasm and at the PM (Kutluay and Bieniasz, 2010; Reed et al., 2012; Robinson et al., 2014); thus, these techniques nicely complement live imaging studies.
6.2. Overview of cellular facilitators of assembly studied in cells
Broadly speaking, cellular factors critical for late events in the HIV-1 life cycle act either during assembly, RNA encapsidation, budding, or release. The majority of such factors identified to date function during budding and release. These include TSG101, ALIX, ESCRTIII proteins, and VPS4 [reviewed in (Votteler and Sundquist, 2013)]. Here we focus on the smaller number of factors shown to facilitate immature capsid assembly per se. The best understood among them is PI(4,5)P2, which acts during membrane targeting of assembling Gag and will be described first. Next we will discuss cellular studies of ABCE1 and DDX6, both of which facilitate Gag assembly. Finally, we will briefly discuss Staufen, an RNA binding protein that has been implicated in Gag translation, multimerization, and gRNA packaging (Chatel-Chaix et al., 2007; Chatel-Chaix et al., 2008; Chatel-Chaix et al., 2004; Dugre-Brisson et al., 2005; Milev et al., 2010); and AP-3, a cellular factor involved in endosome recycling that influences Gag trafficking (Dong et al., 2005; Garcia et al., 2005; Kyere et al., 2012; Liu et al., 2012).
6.3. PI(4,5)P2 facilitates proper membrane targeting and myristate exposure
Assembling Gag targets primarily to the PM rather than to other cellular membranes (Jouvenet et al., 2006) because of its interaction with PI(4,5)P2, an anionic lipid that is found primarily in the cytoplasmic leaflet of the PM. PI(4,5)P2 plays two roles during assembly, acting to target assembling Gag specifically to the PM and also facilitating myristate exposure [reviewed in (Chukkapalli and Ono, 2011)]. While a role in assembly for compounds related to phosphoinositides was first suggested by studies in the recombinant Gag assembly system (Campbell et al., 2001), definitive evidence for this role came from knockdown studies in cells. When PI(4,5)P2 was depleted from cells using siRNA directed against a kinase critical for PI(4,5)P2 biogenesis, virus particle release was significantly reduced (Ono and Freed, 2004). This study also demonstrated that redirecting PI(4,5)P2 to internal compartments rather than the PM caused assembling Gag to be mislocalized, with resulting reductions in virus release. Thus, it is clear that PI(4,5)P2 is critical for targeting assembling Gag to the PM through the results of cellular studies (Chukkapalli et al., 2008; Monde et al., 2011; Ono et al., 2004), as well as structural studies that delineated the binding sites for PI(4,5)P2 in MA (Saad et al., 2006; Shkriabai et al., 2006).
6.4. ABCE1, the first cellular facilitator of assembly to be identified
As described in Section 5.4, ABCE1, the first host protein found to facilitate Gag assembly was initially identified in the cell-free system [(Zimmerman et al., 2002); reviewed in (Stevenson, 2003)]. Because siRNA depletion of ABCE1 causes rapid cell death, knockdowns have not been useful for understanding the function of ABCE1 during assembly. However, other studies - including depletion-reconstitution studies in the cell-free system and dominant negative studies in cells - point strongly to an important role for ABCE1 during Gag assembly (Zimmerman et al., 2002). Interestingly, two findings suggest that ABCE1 binds Gag directly, in an RNA-independent manner, to a region in Gag other than NC. First, binding of Gag to endogenous ABCE1 in cells is resistant to RNase A treatment (Lingappa et al., 2006). Secondly, in coimmunoprecipitation experiments, ABCE1 binds to GagZip chimeras (Klein et al., 2011), in which NC has been replaced with a leucine zipper that promotes protein-protein interactions but does not bind to nucleic acid. Thus, ABCE1 differs from many Gag binding factors, including APOBEC3G [e.g. (Schafer et al., 2004; Zennou et al., 2004)] and Staufen (Milev et al., 2012), which bind indirectly to the Gag NC domain via RNA.
Recently, the lethality of ABCE1 knockdowns has been explained by the finding that ABCE1 is critical for coupling translation termination to ribosome recycling (Barthelme et al., 2011; Becker et al., 2012; Khoshnevis et al., 2010; Pisarev et al., 2010; Shoemaker and Green, 2011), and likely promotes splitting the 40S and 60S ribosomal subunits [reviewed in (Hopfner, 2012; Nurenberg and Tampe, 2013)]. The critical role for ABCE1 in ribosome disassembly also explains why ABCE1 is exceptionally well conserved, present in all eukaryotes and all archaebacteria studied to date (Chen et al., 2006). ABCE1 was initially identified as an inhibitor of RNase L (Bisbal et al., 1995), however RNase L inhibition is not a conserved function of ABCE1 given that the phylogenetic distribution of ABCE1 is much broader than that of RNase L (Kerr, 2004). Crystal structure of archaebacterial ABCE1 reveals it to be a clamp-shaped protein that undergoes an impressive degree of tweezer-like movement (Karcher et al., 2005; Karcher et al., 2008). To date, the function of ABCE1 in HIV-1 capsid assembly has remained elusive; however, studies in cells have added considerably to our understanding of how Gag progresses through ABCE1-containing assembly intermediates (see Section 6.7).
Studies also suggest a possible role for ABCE1 in replication of other viruses. A small molecule screen based on a cell-free system for assembly of rabies virus capsids identified a compound that inhibits replication of infectious rabies virus. Interestingly, this relatively non-toxic inhibitor targets a subfraction of ABCE1 (Lingappa et al., 2013). Thus, ABCE1 may play a broad role in viral replication, and could be a target for antiviral compounds that selectively inhibit its action in viral replication. Additionally, studies suggest that ABCE1 may be under positive selection (Crawford et al., 2009). While the sample size of HIV-1 infected vs. uninfected individuals in this study was insufficient to conclude whether rare variation in ABCE1 influences outcomes, a separate study found that HIV-1 replication in CD4+ T cells was markedly reduced in three heterozygotes carrying a deletion in the ABCE1 3′ UTR (Bleiber et al., 2005).
6.5. DDX6 promotes immature capsid formation in an ATP-dependent manner
The DEAD-box RNA helicase DDX6 first came to the attention of retrovirologists because of the yeast retrotransposon Ty3 (Beliakova-Bethell et al., 2006), which shares a common ancestor with retroviruses (Pelisson et al., 1997). Studies in yeast demonstrated that P body proteins, including the yeast DDX6 homologue Dhh1, are required for Ty3 retrotransposition (Aye et al., 2004; Irwin et al., 2005) and are associated with assembling Ty3 Gag (Beliakova-Bethell et al., 2006). P bodies are a subclass of RNA granules, which are sites of RNA metabolism to which cellular mRNAs are sent for silencing, storage, and degradation in healthy cells - i.e. for everything besides translation [reviewed in (Anderson and Kedersha, 2006)]. Cellular proteins in RNA granules include enzymes that facilitate events in RNA metabolism, for example RNA helicases like DDX6 that enable RNP remodeling, a process in which one set of proteins coating an RNA are replaced with another set of proteins. Since the fate of RNA is typically dictated by associated proteins in the RNP complex [reviewed in (Lee and Lykke-Andersen, 2013)], enzymes like RNA helicases could have a profound influence on the efficiency of Gag assembly and encapsidation in cells.
Using siRNA knockdown approaches, DDX6 was found to facilitate events in HIV-1 assembly independent of RNA packaging (Reed et al., 2012). Like ABCE1, DDX6 was associated with all the high-molecular-mass assembly intermediates, as were other P body proteins, including AGO2 and DCP2. In this study, quantitative EM analyses found that DDX6 acts at or after targeting of Gag to the PM (Fig. 3). Moreover, DDX6 knockdowns were rescued with WT DDX6 but not with a DDX6 mutant incapable of ATP hydrolysis, indicating that DDX6 acts enzymatically during assembly. Additionally, this study also showed that in uninfected cells, DDX6 is present in a complex with ABCE1 and AGO2, termed the precursor complex [Fig. 4 in (Reed et al., 2012)]. Thus, it appears that newly synthesized Gag co-opts the precursor complex to form the ∼80S assembly intermediate. Notably, the precursor complex is smaller than a typical P body, and P bodies have not yet been found to contain ABCE1. Hence, the precursor complex present in uninfected cells may represent a unique subclass of RNA granules that are significantly smaller than P bodies, but similar to P bodies in composition except for the presence of ABCE1 (Reed et al., 2012).
Why HIV-1 Gag co-opts this unique subclass of host RNA granules remains to be determined, but two possible explanations make intuitive sense. First, because RNA helicases remodel RNPs, they liberate RNAs from one set of proteins, allowing them to be replaced by a new set of proteins that dictate a different function [reviewed in (Lee and Lykke-Andersen, 2013)]. Thus, co-opting host RNA helicases during assembly could allow the virus to reprogram gRNA and/or cellular mRNAs involved in translation to function as nucleators of assembly. A second reason for HIV-1 Gag to co-opt host RNA granules is to access gRNA that is sequestered within RNA granules to evade innate immune defenses. Thus, sequestration of gRNA and assembling Gag in RNA granule-like assembly intermediates could serve the dual purpose of allowing access to enzymatic facilitators of capsid assembly, while at the same time concealing viral components from the host immune response. In support of this, recent studies in chronically infected T cells reveal that HIV-1 gRNA is present in ABCE1- and DDX6-containing high-molecular-mass assembly intermediates (Robinson et al., 2014), along with Gag.
6.6. Staufen and AP-3δ: Additional cellular factors implicated in late events
Another cellular factor that is implicated in late events is Staufen 1, a cellular double-stranded RNA binding protein. Unlike ABCE1, Staufen binds to Gag indirectly, via RNA (Abrahamyan et al., 2010; Chatel-Chaix et al., 2007; Chatel-Chaix et al., 2008; Chatel-Chaix et al., 2004; Dugre-Brisson et al., 2005; Milev et al., 2010; Milev et al., 2012; Mouland et al., 2000). While overexpression of Staufen inhibits virus infectivity and enhances gRNA packaging (Mouland et al., 2000), Staufen 1 knockdown also inhibits virus infectivity (Chatel-Chaix et al., 2004). Additionally, Staufen has been reported to play a role in Gag translation (Dugre-Brisson et al., 2005) and multimerization (Chatel-Chaix, 2007 #772;Chatel-Chaix, 2008 #1078}. Thus, Staufen's role appears to be complex, and exactly how it acts during assembly remains to be determined. Notably, Staufen is found in association with ABCE1 (Milev et al., 2012), raising the possibility that Staufen is part of the aforementioned precursor complex, as well as the high-molecular-mass assembly intermediates.
The AP-3δ subunit, which is part of the AP-3 complex involved in endosomal-lysosomal sorting (Dell'Angelica, 2009), has been reported to bind to Gag in an MA-dependent manner and facilitate HIV-1 particle production. These studies used knockdown and dominant-negative experiments in 293T cells (Dong et al., 2005), as well as analysis of cells that are genetically deficient in AP-3 (Liu et al., 2012). However, studies have shown that released virus can be endocytosed in HIV-1 expressing 293T cells (Jouvenet et al., 2006). Since AP-3δ acts during endosomal-lysosomal sorting, the possibility that AP-3δ affects virus production by modulating endocytosis of released virus will need to be explored further. These questions are underscored by a recent study reporting that AP-3δ does not bind directly to myr(-)MA, and concluding that the effects of AP-3δ in virus assembly remain unclear (Kyere et al., 2012).
6.7. Biochemical approaches to studying capsid assembly intermediates in cells
Biochemical studies of assembly intermediates have led to insights into how assembly proceeds in cells. Cells that are HIV-1 infected or transfected to express HIV-1 or other primate lentiviral Gag proteins (SIVmac, SIVagm, or HIV-2) contain complexes similar in size and composition to the assembly intermediates initially identified in the cell-free system (Dooher and Lingappa, 2004). In cells, as in the cell-free system, enzymatic depletion of ATP results in accumulation of the ∼80S complex (Dooher and Lingappa, 2004). Confirmation that the Gag-containing complexes are assembly intermediates has come from pulse-chase experiments in cells expressing the HIV-1 provirus (Dooher et al., 2007). Additionally, critical Gag mutations have been examined for their effects on the assembly pathway, and each has been found to inhibit the assembly at one of five specific points in the pathway in human 293T cells, with accumulation of only the assembly intermediates that precede the point of blockade (Class 1 – 5 mutants in Fig. 3) (Dooher and Lingappa, 2004; Klein et al., 2011; Lingappa et al., 1997; Robinson et al., 2014). Thus, a wealth of data support the notion that Gag assembles through a stepwise pathway of assembly intermediates in cells. Studies that employed other biochemical approaches have also detected similar Gag-containing complexes in cells (Holm et al., 2003; Lee et al., 1999; Lee and Yu, 1998; Morikawa et al., 2004; Tritel and Resh, 2000). Despite extensive evidence for their existence, assembly intermediates in cells are understudied, in large part because they are technically difficult to identify for a number of reasons. First, they are present in small quantities because they rapidly progress to the next step in the pathway; secondly, Gag epitopes are known to be masked as Gag multimerizes at the PM (Ono et al., 2005) making it difficult to isolate assembly intermediates using Gag antibodies; and thirdly, endocytosis of released virus in many commonly used cell lines (e.g. 293T cells) results in large ∼750S signals from intracellular completed capsids that overwhelm smaller signals from the assembly intermediates in velocity sedimentation gradients (Robinson et al., 2014).
The high-molecular-mass assembly intermediates in cells contain Gag, gRNA, and the viral proteins GagPol and Vif (Dooher et al., 2007; Robinson et al., 2014; Zimmerman et al., 2002). Notably, GagPol is present in a 1:20 ratio relative to Gag in the assembly intermediates (Dooher et al., 2007), as would be expected from the efficiency of ribosomal frameshifting (Jacks et al., 1988). In addition, immunoprecipitation with ABCE1 antibody reveals that these complexes contain cellular factors, including ABCE1 and the RNA granule proteins DDX6, AGO2, and DCP-2 (Dooher and Lingappa, 2004; Dooher et al., 2007; Klein et al., 2011; Lingappa et al., 2006; Reed et al., 2012; Robinson et al., 2014 Zimmerman, 2002 #81). These findings were confirmed by quantitative immunogold labeling, which demonstrated that ABCE1 and DDX6, the two cellular proteins in the intermediates that facilitate assembly (Reed et al., 2012; Zimmerman et al., 2002), are enriched in early and late sites of Gag assembly at the PM (Dooher et al., 2007; Reed et al., 2012) (Fig. 5). The subcellular localization of specific assembly intermediates has also been determined using membrane flotation analysis of cells expressing the HIV-1 provirus (Robinson et al., 2014). While the ∼10S and ∼80S assembly intermediates are found in the cytoplasm, the ∼80S, ∼500S, and ∼750S intermediates are found primarily at the PM ((Robinson et al., 2014); Fig. 3). The presence of the ∼80S intermediate in both subcellular locations suggests that this may be the intermediate that takes assembling Gag from the cytoplasm to the PM. Consistent with this possibility, non-myristoylated Gag mutants, which fail to target to the PM, are arrested as cytoplasmic ∼80S assembly intermediates. However, both EM and membrane flotation experiments suggest that some ∼10S Gag is also present at the PM, but not in association with cellular factors. Thus, the data argue for a model in which two populations of Gag are found at the PM: one that is anchored at the PM, initiates assembly, and is associated with gRNA, ABCE1, and DDX6; and another consisting of ∼10S Gag proteins that are not associated with ABCE1 or DDX6 and could move laterally in the PM to add subunits to the anchored site of assembly during multimerization ((Robinson et al., 2014); Fig. 3). This model is consistent with conclusions from live imaging studies (Jouvenet et al., 2008; Jouvenet et al., 2009) but will require further testing to be validated. Notably, unlike other cellular proteins (Chertova et al., 2006), ABCE1 and DDX6 are not found in released virions, suggesting that these cellular facilitators of assembly dissociate from the immature capsid after assembly is completed (Dooher et al., 2007; Reed et al., 2012).
Fig. 5. Double immunogold labeling shows enrichment of ABCE1 at sites of WT Gag assembly at the PM of HIV-1 infected cells.
HIV-1-infected COS-1 cells expressing WT Gag or an assembly-incompetent MACA mutant were analyzed by immunogold double-label EM, using antibodies to Gag (small gold) and ABCE1 (large gold). Images were chosen to reflect previous quantitative immunolabeling studies that show significant recruitment of ABCE1 to WT Gag assembly sites at the PM, versus low levels of background ABCE1-Gag colocalization with the MACA mutant (Reed et al., 2012). (A) Six late WT assembly sites, half of which colocalize with ABCE1. (B) Three early WT assembly sites, all of which colocalize with ABCE1. (C) PMs of two adjacent cells expressing assembly-incompetent, membrane targeted MACA, displaying background levels of colocalization with ABCE1. Scale bars, 200 nm.
6.8. Correlating biochemical studies of assembly in cells to advances in structural biology
Because pulse-chase and mutational analyses have identified the temporal sequence in which capsid assembly intermediates are formed in cells, the assembly pathway can also be used to map the point of action in the pathway of residues critical for Gag assembly (Fig. 3). Studies show that the NC basic residues are required at the first step of the assembly pathway, enabling progression from the unassembled ∼10S intermediate to the ∼80S intermediate (Lingappa et al., 2006), while residues required for myristoylation and myristate exposure (at the extreme N-terminus of MA) act next, being required for membrane targeting of the ∼80S intermediate (Robinson et al., 2014). After that, residues in the CA-CTD dimer interface (in CA-CTD helix 9) are required for formation of a stable membrane-bound ∼80S intermediate, while critical residues in the CA-CTD base are required for progression past the membrane-bound ∼80S intermediate. Finally, this study demonstrated that critical residues in the CA-NTD H4-6 region act at the end of the pathway, being required for progression beyond the membrane-associated ∼500S intermediate (Fig. 3).
Interestingly, many of the CA residues found to be critical for progression of Gag through each step of the assembly pathway are also found at CA-CA interfaces in the fully assembled immature capsid (Bharat et al., 2014; Bharat et al., 2012; Robinson et al., 2014). Given that the main function of CA is thought to be formation of CA-CA interfaces, mutation of residues at each interface might be expected to block assembly at the specific step where that critical interface is formed. If one makes this assumption, then a model can be proposed for when specific CA-CA interfaces form based on the point of arrest of these mutants in the assembly pathway (Robinson et al., 2014). In this model, inter-hexameric CA-CTD contacts are required for formation of a stable membrane-associated ∼80S intermediate; intra-hexameric CA-CTD contacts must be formed for progression past the membrane-associated ∼80S intermediate; and CA-NTD contacts must be formed for progression past the membrane-associated ∼500S intermediate ((Robinson et al., 2014); Fig. 6). While this model is speculative, it is based on reasonable assumptions and provides a framework for understanding events that may be occurring at each step of the assembly pathway.
Fig. 6. A model for the temporal sequence of CA-CA interactions during assembly of HIV-1 Gag in cells.
Critical residues in CA that are required for assembly (Class 3, 4, and 5 mutants in Fig. 3) are identical to, or in close proximity with, residues that form CA-CA interfaces in a recent high-resolution structure of the immature capsid formed by MPMV CA-NC (Bharat et al., 2012; Robinson et al., 2014). If one assumes that each interface is formed at the step when the relevant residues are required during assembly, the blockade data from Fig. 3 can be used to generate a map of when key CA-CA interfaces might form during cells. The diagram shows such a model, adapted from (Robinson et al., 2014), which proposes that formation of the inter-hexameric CA-CTD dimer interface (involving residues in CA helix 9) is required for the ∼80S intermediate to stably target to membranes; and that formation of the CA-CTD intra-hexameric interface (involving residues in the CA-CTD crystallographic base) may be required for Gag to progress past the ∼80S membrane-associated intermediate. These inter- and intra-hexameric CA-CTD contacts may be formed in repeated rounds, as additional Gag is added to the growing capsid during multimerization at the PM. Finally, for a stable ∼500S assembly intermediate to be produced, formation of the inter-hexameric CA-NTD interface may be required (using residues in CA helices 4 – 6).
7. Concluding remarks and future directions: Reconciling different models of assembly
In this review, we summarized conclusions regarding HIV-1 assembly and described the experimental approaches that led to them. There is general consensus for conclusions obtained using many of these approaches, such as mutational analysis and live imaging. Likewise, in the area of structural biology, there is considerable agreement. In other areas of the assembly field, consensus has not yet been reached. Perhaps the most striking example of controversy is the model of host-catalyzed assembly supported by studies in the cell-free system and in cells, which contrasts starkly with the well-established model of spontaneous assembly of purified recombinant Gag. On the surface, these two models appear to be at odds, but a more careful analysis of the underlying concepts suggests ways of reconciling them. Specifically, both models make sense if the ability to self-assemble is necessary for assembly, but not sufficient for assembly in the intracellular environment where the virus encounters innate immune defenses and energetic barriers not present in the simplified environment of the recombinant assembly system. In the intracellular environment, the underlying principles revealed by the recombinant assembly system still hold: i.e. assembly is initiated by dimerization of Gag, oligomerization of Gag is nucleated by RNA, and Gag has an intrinsic ability to form Gag-Gag interactions. However, the intracellular environment likely complicates each of these basic features of assembly, potentially leading to the evolution of viral mechanisms for overcoming these intracellular barriers. For example, studies suggest that to find nucleating RNA in an environment where RNAs are coated with RNPs, the virus may have evolved mechanisms for co-opting helicases that remodel RNPs; likewise, to evade host immune defenses, the virus may sequester gRNA within host RNA granules, which Gag must enter to access RNA and RNA helicases. Additionally, we speculate that to avoid the unfavorable energetics of unfolding “folded-over” monomeric Gag, the virus may have evolved to utilize cellular chaperones to retain Gag in an extended conformation and promote dimerization.
Note that assembly in cells is considerably more complex than assembly in the simplified recombinant assembly system. Thus, more work will need to be done to advance our primitive understanding of how cellular facilitators function during assembly – and in the process controversial ideas and technically challenging approaches will need to be considered, explored, and patiently pursued. Importantly, as our knowledge of intracellular events in assembly advances, more pieces of the puzzle are falling into place. As an example, findings in a recent temporal analysis of events that occur at each step of the host-catalyzed assembly pathway corresponded surprisingly well with results of previous structural, mutational, and live imaging studies (Robinson et al., 2014). Future studies coordinating data from the various experimental approaches discussed herein will likely lead to even more integration of different models of HIV-1 assembly.
Highlights.
Various experimental approaches have shaped our understanding of HIV-1 assembly.
Here we describe complementary findings obtained with these approaches.
Major contributions of each approach are discussed, along with their limitations.
Key unanswered questions about how HIV-1 assembles in cells are discussed.
We also explain why Gag assembly path differs depending on the environment.
Acknowledgments
We would like to thank O. Pornillos and B. Torbett for comments on the manuscript, and B. Schneider, S. MacFarlane, S. Knecht, and the Fred Hutchinson Cancer Research Center Electron Microscopy Shared Resources for assistance with transmission EM imaging. This work was supported by NIH R01AI106397 to J.R.L and an NIH T32 AI750913 fellowship award to B.A.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jonathan C Reed, Email: reedjon@u.washington.edu.
Motoko Tanaka, Email: mt1982@u.washington.edu.
Kasana Chutiraka, Email: jadukdik@uw.edu.
Bridget A Robinson, Email: robinsob@u.washington.edu.
References
- Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- Accola MA, Hoglund S, Gottlinger HG. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accola MA, Strack B, Gottlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- Alfadhli A, Barklis RL, Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology. 2009;387:466–472. doi: 10.1016/j.virol.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A, Huseby D, Kapit E, Colman D, Barklis E. Human immunodeficiency virus type 1 matrix protein assembles on membranes as a hexamer. J Virol. 2007;81:1472–1478. doi: 10.1128/JVI.02122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, Sandmeyer S. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics. 2004;168:1159–1176. doi: 10.1534/genetics.104.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci U S A. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartel V, Muller B, Lamb DC. Quantitative live-cell imaging of human immunodeficiency virus (HIV-1) assembly. Viruses. 2012;4:777–799. doi: 10.3390/v4050777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, Thomm M, Hopfner KP, Green R, Beckmann R. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N, Beckham C, Giddings TH, Jr, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA (New York, NY. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- Bharat TA, Castillo Menendez LR, Hagen WJ, Lux V, Igonet S, Schorb M, Schur FK, Krausslich HG, Briggs JA. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc Natl Acad Sci U S A. 2014;111:8233–8238. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. Structure of the immature retroviral capsid at 8 A resolution by cryo-electron microscopy. Nature. 2012;487:385–389. doi: 10.1038/nature11169. [DOI] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- Bleiber G, May M, Martinez R, Meylan P, Ott J, Beckmann JS, Telenti A, Swiss HIVCS. Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J Virol. 2005;79:12674–12680. doi: 10.1128/JVI.79.20.12674-12680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsetti A, Ohagen A, Gottlinger HG. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci U S A. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Vogt VM. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- Carlson LA, de Marco A, Oberwinkler H, Habermann A, Briggs JA, Krausslich HG, Grunewald K. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Wang SM, Huang KJ, Wang CT. Mutations in capsid major homology region affect assembly and membrane affinity of HIV-1 Gag. J Mol Biol. 2007;370:585–597. doi: 10.1016/j.jmb.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J Virol. 2007;81:6216–6230. doi: 10.1128/JVI.00284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 2008;5:41. doi: 10.1186/1742-4690-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Di Mascio M, Perelson AS, Ho DD, Zhang L. Determination of virus burst size in vivo using a single-cycle SIV in rhesus macaques. Proc Natl Acad Sci U S A. 2007;104:19079–19084. doi: 10.1073/pnas.0707449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The Essential Vertebrate ABCE1 Protein Interacts with Eukaryotic Initiation Factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AM, Massiah MA, Turner BG, Sundquist WI, Summers MF. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J Mol Biol. 1996;264:1117–1131. doi: 10.1006/jmbi.1996.0700. [DOI] [PubMed] [Google Scholar]
- Chu HH, Chang YF, Wang CT. Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production. Journal of biomedical science. 2006;13:645–656. doi: 10.1007/s11373-006-9094-6. [DOI] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. Journal of virology. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Ono A. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol. 2011;410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RC, Leure-duPree AE, Weldon RA, Jr, Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Zheng N, Speelmon EC, Stanaway I, Rieder MJ, Nickerson DA, McElrath MJ, Lingappa J. An excess of rare genetic variation in ABCE1 among Yorubans and African-American individuals with HIV-1. Genes and immunity. 2009;10:715–721. doi: 10.1038/gene.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J Virol. 2009;83:2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. Conformation of the HIV-1 Gag protein in solution. J Mol Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SA, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J Virol. 2011;85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson L, Yu XF. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- de Marco A, Davey NE, Ulbrich P, Phillips JM, Lux V, Riches JD, Fuzik T, Ruml T, Krausslich HG, Vogt VM, Briggs JA. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J Virol. 2010;84:11729–11736. doi: 10.1128/JVI.01423-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21:552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Derdowski A, Ding L, Spearman P. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J Virol. 2004;78:1230–1242. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, Spearman P. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120:663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Dooher JE, Lingappa JR. Conservation of a step-wise, energy-sensitive pathway involving HP68 for assembly of primate lentiviral capsids in cells. J Virol. 2004;78:1645–1656. doi: 10.1128/JVI.78.4.1645-1656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at Plasma Membrane HIV Assembly Sites and Its Dissociation from Gag is Linked to Subsequent Events of Virus Production. Traffic. 2007;8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger HG. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Current opinion in structural biology. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SD, Wilk T, Gowen BE, Krausslich HG, Vogt VM. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Current biology : CB. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Advances in experimental medicine and biology. 2012;726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Current opinion in structural biology. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E, Pion M, Pelchen-Matthews A, Collinson L, Arrighi JF, Blot G, Leuba F, Escola JM, Demaurex N, Marsh M, Piguet V. HIV-1 trafficking to the dendritic cell-T-cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic. 2005;6:488–501. doi: 10.1111/j.1600-0854.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- Gelderblom HR, Hausmann EH, Ozel M, Pauli G, Koch MA. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich HG. A conformational switch controlling HIV-1 morphogenesis. Embo J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liang C. Opposing effects of the M368A point mutation and deletion of the SP1 region on membrane binding of human immunodeficiency virus type 1 Gag. Virology. 2005;335:232–241. doi: 10.1016/j.virol.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Guo X, Roldan A, Hu J, Wainberg MA, Liang C. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J Virol. 2005;79:1803–1812. doi: 10.1128/JVI.79.3.1803-1812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends in microbiology. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hermida-Matsumoto L, Resh MD. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J Virol. 1999;73:1902–1908. doi: 10.1128/jvi.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K, Weclewicz K, Hewson R, Suomalainen M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55(gag) associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J Virol. 2003;77:4805–4817. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP. Rustless translation. Biological chemistry. 2012;393:1079–1088. doi: 10.1515/hsz-2012-0196. [DOI] [PubMed] [Google Scholar]
- Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome research. 2005;15:641–654. doi: 10.1101/gr.3739005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci U S A. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Johnson MC, Scobie HM, Ma YM, Vogt VM. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J Virol. 2002;76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. J Virol. 2009;83:5375–5387. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Nagashima K, Freed EO. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, gag-gag interactions, gag-membrane binding, and virion maturation. J Virol. 2006;80:7939–7951. doi: 10.1128/JVI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Simon SM, Bieniasz PD. Visualizing HIV-1 Assembly. J Mol Biol. 2011a;410:501–511. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nature cell biology. 2011b;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowett JB, Hockley DJ, Nermut MV, Jones IM. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. The Journal of general virology. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- Karcher A, Buttner K, Martens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure (Camb) 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol. 2011;85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol. 2010;84:6352–6366. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr ID. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun. 2004;315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annual review of biochemistry. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kingston RL, Vogt VM. Domain swapping and retroviral assembly. Mol Cell. 2005;17:166–167. doi: 10.1016/j.molcel.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Klein KC, Reed JC, Lingappa JR. Intracellular destinies: degradation, targeting, assembly, and endocytosis of HIV Gag. AIDS Rev. 2007;9:150–161. [PubMed] [Google Scholar]
- Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J Virol. 2011;85:7419–7435. doi: 10.1128/JVI.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich HG, Facke M, Heuser AM, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6:e1001200. doi: 10.1371/journal.ppat.1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzembayeva M, Dilley K, Sardo L, Hu WS. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014 doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyere SK, Mercredi PY, Dong X, Spearman P, Summers MF. The HIV-1 matrix protein does not interact directly with the protein interactive domain of AP-3delta. Virus research. 2012;169:411–414. doi: 10.1016/j.virusres.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Lykke-Andersen J. Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends in cell biology. 2013;23:504–510. doi: 10.1016/j.tcb.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Liu B, Yu XF. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Yu XF. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- Liang C, Hu J, Russell RS, Roldan A, Kleiman L, Wainberg MA. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J Virol. 2002;76:11729–11737. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Hu J, Whitney JB, Kleiman L, Wainberg MA. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1. J Virol. 2003;77:1772–1783. doi: 10.1128/JVI.77.3.1772-1783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J Biol Chem. 2006;281:3773–3784. doi: 10.1074/jbc.M507255200. [DOI] [PubMed] [Google Scholar]
- Lingappa JR, Hill RL, Wong ML, Hegde RS. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappa JR, Newman MA, Klein KC, Dooher JE. Comparing capsid assembly of primate lentiviruses and hepatitis B virus using cell-free systems. Virology. 2005;333:114–123. doi: 10.1016/j.virol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Lingappa UF, Wu X, Macieik A, Yu SF, Atuegbu A, Corpuz M, Francis J, Nichols C, Calayag A, Shi H, Ellison JA, Harrell EK, Asundi V, Lingappa JR, Prasad MD, Lipkin WI, Dey D, Hurt CR, Lingappa VR, Hansen WJ, Rupprecht CE. Host-rabies virus protein-protein interactions as druggable antiviral targets. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E861–868. doi: 10.1073/pnas.1210198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Sutton J, Woodruff E, Villalta F, Spearman P, Dong X. Defective HIV-1 particle assembly in AP-3-deficient cells derived from patients with Hermansky-Pudlak syndrome type 2. Journal of virology. 2012;86:11242–11253. doi: 10.1128/JVI.00544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn GN, Grover JR, Olety B, Ono A. HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region. J Virol. 2013;87:6441–6454. doi: 10.1128/JVI.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YM, Vogt VM. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J Virol. 2004;78:52–60. doi: 10.1128/JVI.78.1.52-60.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammano F, Ohagen A, Hoglund S, Gottlinger HG. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends in biochemical sciences. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- Milev MP, Brown CM, Mouland AJ. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Ravichandran M, Khan MF, Schriemer DC, Mouland AJ. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Frontiers in microbiology. 2012;3:367. doi: 10.3389/fmicb.2012.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde K, Chukkapalli V, Ono A. Assembly and replication of HIV-1 in T cells with low levels of phosphatidylinositol-4,5-bisphosphate. Journal of virology. 2011;85:3584–3595. doi: 10.1128/JVI.02266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE., Jr Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure. 2010;18:1483–1491. doi: 10.1016/j.str.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Hu WS. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009;11:91–102. [PMC free article] [PubMed] [Google Scholar]
- Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. Helical structure determined by NMR of the HIV-1 345-392Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein science : a publication of the Protein Society. 2005;14:375–386. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Goto T, Momose F. Human immunodeficiency virus type 1 Gag assembly through assembly intermediates. J Biol Chem. 2004;279:31964–31972. doi: 10.1074/jbc.M313432200. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JL, Butcher EW, Patel DT, Mikhaylenko Y, Summers MF. Flexibility in the P2 domain of the HIV-1 Gag polyprotein. Protein science : a publication of the Protein Society. 2004;13:2101–2107. doi: 10.1110/ps.04614804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurenberg E, Tampe R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends in biochemical sciences. 2013;38:64–74. doi: 10.1016/j.tibs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol 4,5 bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Huang M, Freed EO. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J Virol. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Orenstein JM, Freed EO. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Joshi A, Freed EO. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlinsky KJ, Gu J, Hoyt M, Sandmeyer S, Menees TM. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillart JC, Gottlinger HG. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Teysset L, Chalvet F, Kim A, Prud'homme N, Terzian C, Bucheton A. About the origin of retroviruses and the co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica. 1997;100:29–37. [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Perez-Caballero D, Hatziioannou T, Martin-Serrano J, Bieniasz PD. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on gag precursor-membrane interactions. J Virol. 2004;78:9560–9563. doi: 10.1128/JVI.78.17.9560-9563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nature chemical biology. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- Robinson BA, Reed JC, Geary CD, Swain JV, Lingappa JR. A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J Virol. 2014;88:5718–5741. doi: 10.1128/JVI.03609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. Point Mutations in the HIV-1 Matrix Protein Turn Off the Myristyl Switch. J Mol Biol. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakalian M, Parker SD, Weldon RA, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AR, Hill RL, Lingappa JR. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology. 2001;279:257–270. doi: 10.1006/viro.2000.0706. [DOI] [PubMed] [Google Scholar]
- Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M. HIV-1 pathogenesis. Nat Med. 2003;9:853–860. doi: 10.1038/nm0703-853. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb Perspect Med. 2012;2:a015420. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc Natl Acad Sci U S A. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritel M, Resh MD. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J Virol. 2000;74:5845–5855. doi: 10.1128/jvi.74.13.5845-5855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon RA, Jr, Parker WB, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L, Kerkhofs P, Attenelle L, Burny A, Portetelle D, Kettmann R. The major homology region of bovine leukaemia virus p24gag is required for virus infectivity in vivo. The Journal of general virology. 1997;78(Pt 3):637–640. doi: 10.1099/0022-1317-78-3-637. [DOI] [PubMed] [Google Scholar]
- Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta crystallographica Section D, Biological crystallography. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. Embo J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. Journal of chemical biology. 2010;3:19–35. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006 doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman C, Klein KC, Kiser PK, Singh ARS, Firestein BL, Riba SC, Lingappa JR. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]