Abstract
Chalcone synthase (CHS) catalyzes the first committed step in the flavonoid biosynthetic pathway. In this study, the cDNA (FhCHS1) encoding CHS from Freesia hybrida was successfully isolated and analyzed. Multiple sequence alignments showed that both the conserved CHS active site residues and CHS signature sequence were found in the deduced amino acid sequence of FhCHS1. Meanwhile, crystallographic analysis revealed that protein structure of FhCHS1 is highly similar to that of alfalfa CHS2, and the biochemical analysis results indicated that it has an enzymatic role in naringenin biosynthesis. Moreover, quantitative real-time PCR was performed to detect the transcript levels of FhCHS1 in flowers and different tissues, and patterns of FhCHS1 expression in flowers showed significant correlation to the accumulation patterns of anthocyanin during flower development. To further characterize the functionality of FhCHS1, its ectopic expression in Arabidopsis thaliana tt4 mutants and Petunia hybrida was performed. The results showed that overexpression of FhCHS1 in tt4 mutants fully restored the pigmentation phenotype of the seed coats, cotyledons and hypocotyls, while transgenic petunia expressing FhCHS1 showed flower color alteration from white to pink. In summary, these results suggest that FhCHS1 plays an essential role in the biosynthesis of flavonoid in Freesia hybrida and may be used to modify the components of flavonoids in other plants.
Introduction
The flower color, which is mainly determined by betalains, carotenoids and flavonoids, is one of the most important characteristics in ornamental plants [1,2]. Betalains, nitrogen-containing compounds derived from tyrosine, confer violet to red and yellow to orange coloration to flowers but are found exclusively in Caryophyllales [3,4]. Carotenoids furnish flowers with different colors ranging from yellow to red and are essential for photosynthesis. Unlike betalains and carotenoids, flavonoids are often present in the same plant tissues to create co-pigmentation phenomenon, and this combination increases the color variety [5,6]. In addition, flavonoids are the most abundant and widely distributed pigments in the plant kingdom, and they are largely responsible for diverse pigmentation from shiny orange to pink, red, violet and blue colors in the flowers and fruits. So far, more than 6,000 flavonoids have been identified [7]. Flavonoids are one of the most important secondary metabolites that comprise several different classes of compounds such as chalcones, flavones, flavonols and anthocyanins, and have a wide variety of biological functions, including protection of cells against UV radiation, resistances against phytopathogens and herbivores, signal molecules in plants and microbes interaction, pollen growth and development, root nodule organogenesis, auxin transport, and most importantly, they pigment flower to attract pollinators and disperse the fruits and seeds [8–11]. It has been suggested that flavonoids may have played a crucial role in the early evolution of land plants, first as chemical messengers and then as UV screeners [12,13]. Meanwhile, it has been reported that flavonoids also have a close connection with human health. Besides the remarkable anti-oxidant, anti-virus and anti-proliferative biological activities, they were also applied for preventative maintenance on atherosclerotic arteries and treatment of cardiovascular disease as well as many other diseases [14–16].
Over the past few decades, the flavonoid biosynthetic pathway has been well established by using petunia (Petunia hybrida), snapdragon (Anfirrhinum majus), Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) as models [17,18]. Briefly, the biosynthesis of flavonoid starts with the condensation of one molecule of p-coumaroyl-CoA (derived from the general phenylpropanoid pathway) with three molecules of malonyl-CoA, resulting in naringenin chalcone and this reaction is conducted by chalcone synthase (CHS). Then the naringenin chalcone is rapidly isomerized to naringenin by chalcone isomerase (CHI). From these central intermediates, the biosynthetic pathway diverges into side branches, each yielding a different class of flavonoids. Subsequently, flavanone 3-hydroxylase (F3H) catalyzes the hydroxylation of naringenin (the C-ring) to produce dihydrokaempferol, which is further hydroxylated at the B-ring to form dihydroquercetin and dihydromyricetin. Finally, the dihydroflavonols are converted into colored anthocyanins by the actions of dihydroflavonol reductase (DFR), anthocyanidin synthase (ANS) and flavonoid 3-O-glucosyltransferase (3GT) [19–21]. Furthermore, the action of flavonol synthase (FLS), leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) leads to branching of the flavonoid pathway to flavonols and proanthocyanidins, respectively [22–24]. In conclusion, CHS is the first key enzyme of flavonoid pathway.
CHS, the best studied plants-specific type III ployketide synthase, catalyses the first committed step of the branch of the phenylpropanoid pathway which leads to the synthesis of flavonoids [25,26]. The product of the CHS reaction is a pivotal precursor for a vast array of secondary metabolites derived from malonyl-CoA and p-coumaroyl-CoA [27]. Enzymes of the CHS superfamily share high similarity in their amino acid sequence, structure and general catalytic principles and function as a homodimer (monomer size about 43 kDa) which contains the conserved Cys-His-Asn catalytic triad in the buried active sites [28]. More recently, the crystal structure and functional studies of alfalfa CHS2 have confirmed that the conserved cysteine and histidine residues play a significant role in the catalytic function of CHS, and site-directed mutants of these residues have been investigated to elucidate the reaction mechanism of CHS [29,30]. Furthermore, a previous study also showed that the reaction catalyzed by CHS can be seen as a three-step reaction which involves transfering the p-coumaroyl moiety to the active site residue Cys164, decarboxylation of malonyl-CoA to an acetyl-CoA carbanion and extending the polyketide intermediate [31].
Because of its essential role in flavonoid biosynthesis, CHS gene has been isolated and well characterized in hundreds of plant species, especially in eudicots [32]. Previous studies have also described the phenotypic and metabolic effects of loss of CHS protein activity in different model plant species. In 1936, the first CHS mutant identified in Antirrhinum majus was the chalk white nivea mutant which lacks flavonoids and possesses the white-flowered phenotype because of the deletion of the single CHS gene [33]. White anther (wha), a petunia CHS mutant, generated by using ethylmethanesulphonate is functionally male sterile with reduced corolla pigmentation. Biochemical and transgenic complementation studies by Napoli et al. demonstrated that wha mutation was in CHS-A gene which is expressed in petunia petal and pollen tissues [34]. In Arabidopsis, tt4 mutants lacking CHS protein activities accumulate no brown condensed tannins in their seed coats and only the yellow colour of the underlying embryo is visible. Additionally, those mutants exhibited no anthocyanin pigments in the cotyledons or hypocotyls when grown in Murashige and Skoog (MS) media, while the seedlings of wild-type Arabidopsis had strong red pigmentation [35,36]. In consequence, these mutants are helpful to define the function of CHS gene and the roles of those compounds derived from flavonoid pathway.
Freesia hybrida, a monocotyledonous ornamental species, widely distributed in the world, and is becoming increasingly popular as a cut flower. The range of flower colours available in the freesias includes red, pink, yellow, white, blue, lavender, purple and bicolors. Thereby, the freesias are a good system for studying the secondary metabolism of flowers with different colors. Furthermore, the characterization of CHS gene from F. hybrida may help the studies of the duplication and evolution of the CHS gene family considering that there are only a few reports of CHS gene from Iridaceae.
In this paper, we first report the isolation of a CHS gene from Freesia hybrida and investigation of its spatial and temporal expression at different developmental stages and in different organs. Secondly, structure and enzyme activity is clarified using the purified recombinant protein from the E. coli system. Finally, functionality of this CHS gene is characterized via its ectopic expression in both Arabidopsis tt4 mutants and petunia, and detailed alteration of flavonoids in flowers of transgenic petunia lines was also assayed. In summary, these results suggest the role of FhCHS1 in biosynthesis of flavonoid during floral coloration.
Materials and Methods
Plant materials
Wild-type, Arabidopsis thaliana tt4 mutant, and T2 transgenic seedlings of Arabidopsis were germinated in the greenhouse (16 h of light/8 h of dark cycle, 22°C). Seven-day-old seedlings were collected and stored at -80°C. Petunia hybrida plants (Xinhui Horticulture Co., China) for transformation were grown aseptically on Murashige and Skoog medium supplemented with 3% sucrose, and transgenic flowers were harvested at the full-bloom stage. Tissues and flowers at different stages of development from Freesia hybrida were collected and stored at -80°C until required.
Cloning of the full-length cDNA of FhCHS1
A cDNA library was constructed from petals of red Freesia hybrida using a pBluescript II XR Predigested Vector Kit (Invitrogen). Colonies were transferred onto Hybond-N+ nylon membranes, and then denatured and fixed. The polymerase chain reaction (PCR) fragment previously isolated from Iris germanica was used as a probe. The membranes were prehybridized for 1 h at 42°C before the probe was added and hybridized in the manufacturer’s hybridization buffer at 42°C overnight. Labeling of the probe was carried out according to the manufacturer’s instructions using an ECL direct nucleic acid labeling and detection system (Amersham). Positively hybridized plaques were isolated and sequenced, one of which was identified to be a CHS homologue with the 3′ end sequence of FhCHS1. Total RNA from petals of red Freesia hybrida was isolated using the RNAiso Plus (TaKaRa), and then was treated with RNase-free DNase I (TaKaRa), which was heat inactivated at 65°C for 10 min. SMARTer RACE cDNA Amplification Kit (Clontech-Takara, Catalogue no. 634923) was used to synthesise the first-strand cDNA from Freesia hybrida for 5′ RACE, according to the manufacturer’s protocol. After dilution, the first-strand reaction product was subjected to PCR for amplification. The gene-specific primers GSP (5′-CTCCGGTCGTACCGTTCTTCTCCT-3′) as reverse primer in combination with the universal primer A mix (UPM, provided in the kit) as the forward primer were used with the following cycling program: initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 68°C for 30 s and 72°C for 2 min, and a final extension at 72°C for 5 min. The amplified PCR products were analyzed by electrophoresis on a 1% agarose gel, and then cloned into pGEM-T Easy Vector (Promega) to sequence. Once the 5′ sequences were determined, the full-length FhCHS1 was amplified from the cDNA and genomic DNA with the following primer sequences, forward primer 5′-GCAAGAAAAATGGTGAATG-3′ and reverse primer 5′-TGGGAATGATATATAGGGAGTC-3′, in order to determine the accurate nucleotide sequence of cDNA and the presence of intron.
CHS enzyme assays
The full-length open reading frame of FhCHS1 cDNA was amplified by PCR using Premix Taq (TaKaRa) with the following primers: forward primer 5′-CGGGATCCAGAAAAATGGTGAATG-3′ (the BamHI site is underlined) and reverse primer 5′-CGAGCTCTGGGAATGATATATAGGGAGTC-3′ (the SacI site is underlined). The amplified fragment was then introduced into BamHI/SacI-digested pET-28a (+) expression vector, and the construct was sequenced to ensure that the gene was in the correct reading frame. After transformation into the Escherichia coli strain BL-21 (DE3), colonies were selected on LB medium containing 100 mg/L kanamycin (KAN) plates. Individual colonies were grown overnight in 4 mL of LB-KAN medium at 37°C, and 2 mL of the culture was used to inoculate 400 mL of LB-KAN fresh medium. Cells were grown to OD600 of 0.6 at 37°C and induced for 8 h with 0.55 mM isopropyl thio-β-D-galactoside at 25°C, followed by harvest by centrifugation at 4,000 g for 10 min and resuspended in 20 mL of phosphate-buffered saline. After sonication on ice, the supernatant and pellet were tested by SDS-PAGE for solubility of the fusion protein by Coomassie blue stain. Finally, the soluble proteins were purified using Ni Sepharose High Performance following the manufacturer’s instructions (GE Healthcare).
The purified recombinant FhCHS1 was used to examine enzyme activity with p-Coumaroyl-CoA (MicroCombiChem e.K., Wiesbaden, Germany) and Malonyl-CoA (Sigma) as substrates. The standard assay contained 80 μM starter CoA, 160 μM malonyl-CoA, 0.1 M potassium phosphate (pH 7.2), 30 μg of protein, and was incubated at 30°C for 1 h. Subsequently, reactions were extracted twice with 300 μl ethyl acetate and centrifuged at 10,000 g for 20 min. After drying under vacuum, the samples were dissolved in 100 μl of methanol. Meanwhile, BL-21 (DE3) cells transformed with an empty pET-28a (+) vector were used as a control and the resulting protein extract was assayed under the same conditions. For analysis of the product, the mixture was filtered with a 0.22 μm filter and analyzed by high-performance liquid chromatography (HPLC). The mobile phases were water (A) and methanol (B) at a flow rate of 0.6 ml min-1. Detection of the enzymatic products was carried out in the detection range of 289 nm by means of a linear gradient elution for 30% B for 3 min, 30–70% B for 22 min, 70–80% B for 2 min, 80–95% B for 3 min, and 95% B for 5 min.
Crystallization and data collection
Recombinant FhCHS1 protein was affinity purified and concentrated to 7.4 mg/ml, then vapour diffusion crystallization drops was then set up at 2 μl drop size with 1:1 ratio of mixture between protein and crystallization mother liquor (0.2 M di-ammonium hydrogen citrate, 20% w/v PEG 3350). Bar shaped crystals were observed after up to 7 days of incubation at 4°C. Crystals were cryo-protected with cryo buffer (0.2 M di-ammonium hydrogen citrate, 20% w/v PEG 3350, 25% glycerol) before flash frozen at 105 K by liquid nitrogen.
Diffraction data was collected at energy of 14.2080 keV from beamline ID23–2 in European Synchrotron Radiation Facility, Grenoble, France. Typically, the obtained crystals diffracted to 2.6 Å of resolution, with a best case of 1.6 Å resolution. The CHS crystals belonged to the space group P212121 with unit cell dimensions of a = 233.76 Å, b = 63.45 Å, c = 108.66 Å, α = β = γ = 90°. Diffraction data was processed by xds [37]. Phase was obtained by carrying out Molecular Replacement in the program Phaser [38] by using PDB ID of 1BI5 as the search model. The generated model was further refined by the programme Phenix [39] with start Rwork = 0.2193, Rfree = 0.2957 and final Rwork = 0.2260, Rfree = 0.2645.
Phylogenetic analysis
To construct the phylogenetic tree, the full-length amino acid sequences of CHS protein were aligned with the ClustalW2 using default parameters (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The alignments were saved and executed by MEGA version 5.1 to generate a neighbor-joining tree with bootstrapping (2,000 replicates) analysis and handling gaps with pairwise deletion.
Expression profile analysis
For real-time quantitative PCR, total RNA was extracted from different tissues and flowers of Freesia hybrida. This was done by grinding fresh tissue in liquid nitrogen to a fine powder and extracting in RNAiso Plus (TaKaRa), according to manufacturer’s protocol. Then total RNA extracted from the above samples was treated with RNase-free DNase I (TaKaRa) before cDNA synthesis.
A SYBR Green-based real-time PCR assay was carried out in a total volume of 10 μl of reaction mixture containing 5 μl of 2 × Master Mix (TOYOBO), 0.5 μM of each primer, and 1 μl cDNA. Cycling conditions were 95°C for 60 s, followed by 40 cycles of 95°C for 5 s and 60°C for 60 s. The primers FhCHS1F1 (5′-CACCAA CAGCGAGGATAA-3′) and FhCHS1R1 (5′-AGACATTCGGATTTTCCT-3′) were used for real-time PCR to amplify a 122-bp fragment. The FhCHS1 expression data was normalized against the expression level of an internal control 18s rRNA gene (forward primer 5′-TCCTGATACGGGGAGGTAGTGACA-3′, reverse primer 5′-ACTTGCCCTCCAATGGATCCTCG-3′) fragment with 111-bp long. The fidelity of the PCR products was confirmed by using agarose gel electrophoresis and sequencing. All analyses were repeated three times using biological replicates. Differences in cycle thresholds between target and 18s rRNA genes corresponded to relative expression level of the target gene.
Expression of FhCHS1 in Arabidopsis mutants
A pair of primers, 5′- CGGTCTAGAGCAAGAAAAATGGTGAAT-3′ (the XbaI site is underlined) and 5′-CGAGGATCCGTGCTACTTAAGCTTCA-3′ (the BamHI site is underlined), was designed to amplify the whole coding regions of FhCHS1 gene. The PCR products were digested with XbaI and BamHI and then was subcloned into the vector pBI-121containing the constitutive cauliflower mosaic virus 35S promoter. Arabidopsis thaliana tt4 mutants (CHS; At5g13930) were transformed using Agrobacterium tumefaciens strain GV3101 according to the floral dip techniques [40]. For transgenic plant selection, transformants were germinated on 1/2 MS medium containing 50 mg/L kanamycin. After selection, kanamycin-resistant plants with red cotyledons were transplanted to soil and grown in the greenhouse under long days. Total RNA extracted from T2 transgenic seedlings was used for reverse transcription PCR (RT-PCR) to confirm the expression of FhCHS1 gene in the transformed mutant lines, and the actin-1 gene was used as an internal control gene [41].
Transformation of petunia by Agrobacterium tumefaciens
The vector used for Arabidopsis transformation was also used for petunia transformation by using an Agrobacterium tumefaciens-mediated leaf transformation protocol. Two-month-old petunia plants were used for transformation by following a previously described protocol, and selected with 50 mg/L of kanamycin [42]. After rooting, kanamycin-resistant plants were transferred to soil and grown in a greenhouse. To confirm the expression of FhCHS1 gene in the transformed plants, total RNA was isolated from flowers of T0 transgenic plants and used in RT-PCR. The 18s rRNA gene was used as a constitutive control with the following primer sequences: 5′-TACCACATCCAAGGAAGGCA-3′ and 5′-ACCCAAAGTCCAACTACGAG-3′.
Anthocyanins and flavonols analysis
Anthocyanins and flavonols were extracted from 100 mg of finely ground tissues in 1 ml of extraction solvent mixture containing H2O:MeOH:HCl (75/24/1,v/v/v) at 4°C for 12 h in the dark. The extract was centrifuged at 10,000 rpm for 10 min at room temperature, and then supernatant was filtered with 0.22 μm filter and used for HPLC analyses. HPLC assay was performed on a Shimadzu HPLC system equipped with an ACCHROM XUnion 5 μm C18 column (250 × 4.6mm). The HPLC flow rate was set at 1 ml min-1 and the mobile phases are composed of A (5% formic acid in H2O) and B (methanol). The elution gradient was as follows: 0–10 min, 14–17% B; 10–35 min, 17–23% B; 35–60min, 23–47% B; 60–67 min, 47–14% B; 67–70 min, 14% B. HPLC separations were monitored at 520 and 360 nm. The HPLC-ESI-MS system adopted in our study consisted of an API2000 mass spectrometer (AB Sciex) and a SPD-20AV UV/VIS Dectector (Shimadzu, Kyoto, Japan). An Analyst (version 1.5, Applied Biosystems) was used for data acquisition and processing. The optimal settings of the mass spectrometer were as follows: positive mode, ion spray voltage 4,500 V, scan range of 100 to 1,000 m/z, declustering potential +80 V and entrance potential 10 V.
Quantification analysis was performed by comparison with commercial standards, including cyanindin-3-glucoside chloride and quercetin-3-glucoside (Sigma) [43]. Data were represented as mean values from at least three independent experiments with three replicates each. Statistical significances of the differences were determined using Student’s t-test. Differences between wild-type and transgenic lines were considered significant when P< 0.01.
Results
Sequences and structure analysis of FhCHS1
Through heterologous screening of a cDNA library and PCR amplication, cDNA (deposited in GenBank, accession no. JF732898) and genomic sequence of FhCHS1 gene were obtained. The gene has one intron with an open reading frame of 1,170 bp encoding 389 amino acid residues with a calculated molecular mass of 42.58 kDa and an isoelectric point of 5.74 (Fig. 1A). The intron of FhCHS1 begins with the nucleotides ‘GT’ and ends with the nucleotides ‘AG’ (Fig. 1B), and the intron length was 129 nt (S1 Fig.), following the GT-AG rule. Multiple sequence alignments revealed that FhCHS1 shared 83.55% to 85.86% identity with AtCHS, PhCHS and MsCHS2 (Fig. 2A). Consistent with previous results with other CHS proteins, the FhCHS1 protein also contained the three important catalytic residues of Cys164, His303, and Asn336 (marked with pink background) and two highly conserved Phe residues, Phe215 and Phe265 (marked with blue background), which were identified to be important for the formation of the active site as well as for the substrate specificity of the CHS. Additionally, the malonyl-CoA binding motif (marked with yellow background) and a highly conserved CHS signature sequence (marked with red background) were also found in the FhCHS1 sequence (Fig. 2A). As shown in Fig. 2B&C, the crystallographic symmetry of FhCHS1 is a dimer of dimer, with the homodimer as the biological functional unit. The individual monomer consists of roughly two domains with the active site located at the clef between the two domains and the dimerization interface, therefore sharing the accessible substrate binding area with the dimer partner. On the other hand, FhCHS1 belonged to the cluster of CHSs expressed in flower tissue and was most closely related to CHS from Iris germanica (Fig. 3) in the phylogenetic analysis.
Fig 1. Schematic diagram of the generalized structure of FhCHS1 gene.
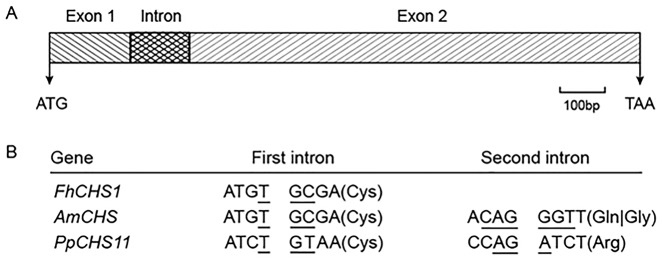
A, Exons-intron architecture of FhCHS1. exons, intron, initiation codon (ATG) and terminator codon (TAA) are labeled. B, Nucleotide sequences surrounding splice sites in FhCHS1, AmCHS (Antirrhinum majus CHS, X03710) and PpCHS11 (Physcomitrella patens CHS, ABU87504). Amino acid residues and codons (underlined) split by introns are shown.
Fig 2. Sequence and crystal structural analysis of FhCHS1.
A, Comparison of amino acid sequences between FhCHS1, AtCHS (At5913930), PhCHSA (X14591) and MsCHS2 (L02902). Functionally important conserved residues are highlighted with a colored background: pink, the three conserved catalytic residues in all CHS; blue, two important residues determining the substrate specificity of CHS; yellow, the malonyl-CoA binding motif; red, highly conserved CHS signature sequence. B, Functional unit of CHS. The homo-dimer with one of the monomer highlighting secondary structure components: cyan for α-helix, magenta for β-strand, pink for loop. C, Structure comparison of the current CHS of freesia (blue, PDB ID 4WUM) and previous structure model CHS2 of alfalfa (yellow, PDB ID 1BI5), indicating strong structural conservation of the CHS protein.
Fig 3. Phylogenetic tree based on the amino acid sequences of plant CHSs.
The tree was constructed using the MEGA 5.1 and neighbor-joining method with 2000 bootstrap replicates. Freesia hybrida CHS is boxed. GenBank accession numbers used are: Nelumbo nucifera (NnCHS, FJ999628.1), Abelmoschus manihot (AmCHS, EU573212.1), Nicotiana tabacum (NtCHS, AF311783.1), petunia hybrida (PhCHSA, X14591; PhCHSB, X14592; PhCHSD, X14593; PhCHSF, X14594; PhCHSG, X14595; PhCHSJ, X14597), Saussurea medusa (SmCHS, DQ350888.1), Humulus lupulus (HlCHS, CAK19317.1), Anthurium andraeanum (AaCHS, DQ421809.1), Freesia hybrida (FhCHS1, JF732897.1), Iris germanica (IgCHS, AB219147.1), Camellia chekiangoleosa (CcCHS1, JN944573.1; CcCHS2, JN944574.1), Rosa chinensis (RcCHS, HQ423171.1), Melastoma malabathricum (MmCHS, KF234569.1), Rhododendron simsii (RsCHS, AJ413277.1), Camellia nitidissima (CnCHS, HQ269804.1).
Enzymatic activity of freesia CHS proteins
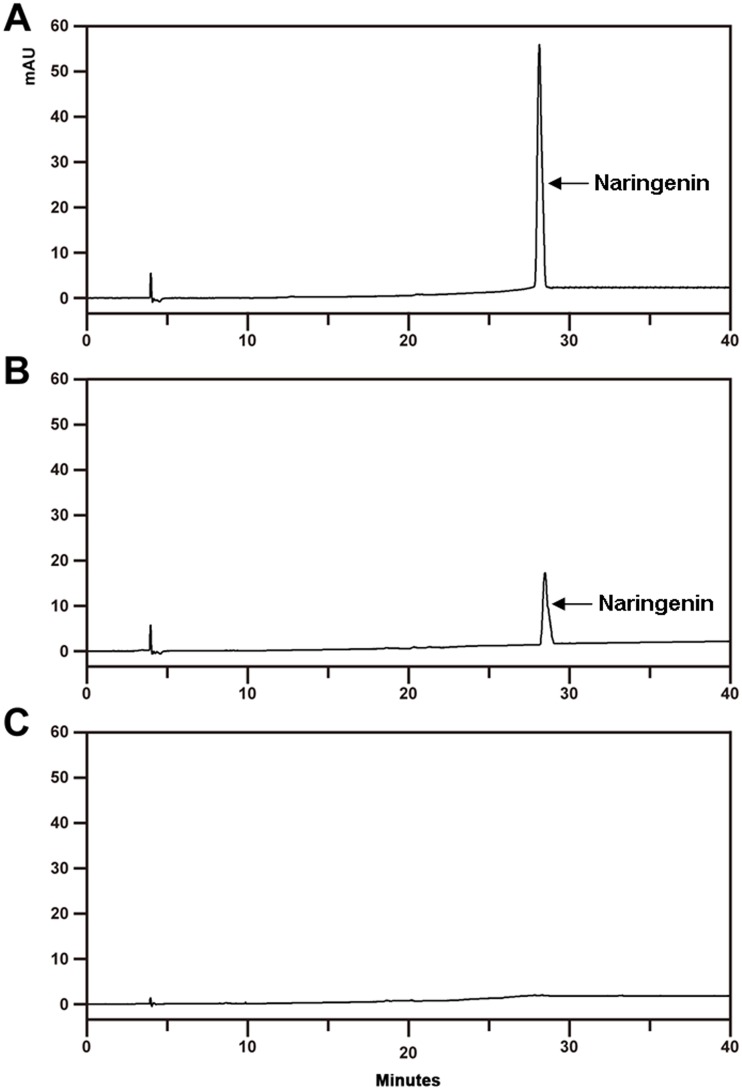
In order to examine the catalytic properties of FhCHS1, it was heterologously expressed in E. coli with an additional his-tag at the N terminus. Enzyme activity of FhCHS1 was tested with p-coumaroyl-CoA, malonyl-CoA and purified protein, and the reaction products were analyzed by HPLC in comparison to authentic standards. As well known, naringenin chalcone is converted non-enzymatically to naringenin in the course of the reactions performed in vitro, and therefore naringenin is the main product in the analysis of HPLC. Compared with control, the reaction product generated a new peak at a retention time (28.5 min) that matched the authentic naringenin (Fig. 4). This indicates that FhCHS1 has a typical CHS enzymatic function, catalyzing the conversion of p-coumaroyl-CoA and malonyl-CoA substrates to naringenin chalcone.
Fig 4. Enzyme activity of freesia (Freesia hybrida) chalcone synthase (FhCHS1).
A, The sample of naringenin standard. B, The reaction products catalyzed by FhCHS1. C, Control.
Expression patterns of FhCHS1 in developing flowers and other tissues
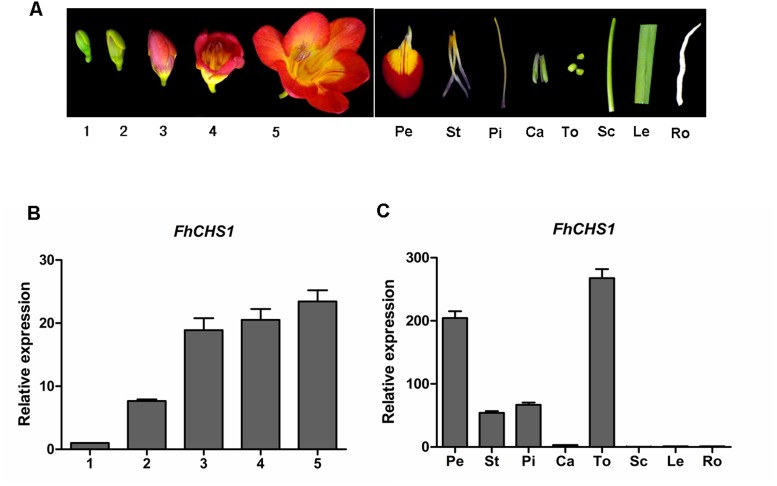
Expression pattern of FhCHS1 was examined using quantitive real-time qPCR. Total RNA was extracted from different freesia tissues including flowers at five developmental stages (stage 1, <10 mm long with unpigmented buds; stage 2, 10–20 mm long with slightly pigmented buds; stage 3, 20–30 mm long with pigmented buds; stage 4, fully pigmented flowers before complete opening; stage 5, fully opened flowers), and different tissues including petals, stamens, pistils, calyxes, toruses, scapes, leaves and roots (Fig. 5A). Overall, transcripts of FhCHS1 were detected in all examined samples at different expression levels. The expression level of FhCHS1 gradually increased during flower development and peaked at stage 5 (Fig. 5B). And this expression profile of FhCHS1 during developmental stages corresponded to the accumulation of total anthocyanin during flower development. Furthermore, the transcript level of FhCHS1 was also found significantly higher in flower organs than those in vegetative organs as shown in Fig. 5C. The experiment data showed that FhCHS1 reached its highest expression level in torus and then decreased to a very low level in leaves and roots. Taken together, these findings indicated that FhCHS1 was involved in the development of pigmentation in flower and biosynthesis of flavonoid in other tissues.
Fig 5. FhCHS1 gene expression profiles in Freesia hybrida.
A, The phenotypes of different samples. 1–5, represent the flowers of different developmental stages; Pe, petals; St, stamens; Pi, pistils; Ca, calyxes; To, toruses; Sc, scapes; Le, leaves; Ro, roots. B, Expression profile of FhCHS1 in flowers at different developmental stages. C, Expression levels of FhCHS1 in different tissues. Data represent means ± SD of three biological replicates.
Complementation of CHS function in Arabidopsis tt4 mutants
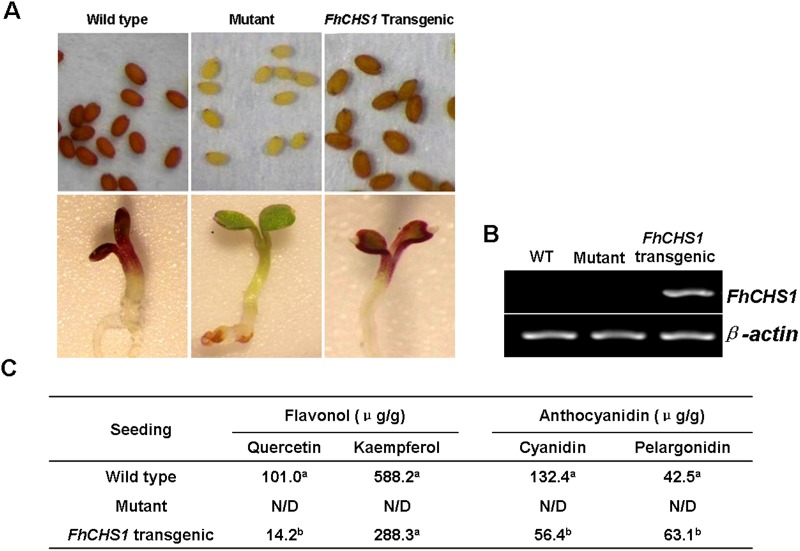
In order to examine the FhCHS1 function in flavonoid biosynthesis, the cDNA was transferred into the Arabidopsis tt4 mutants under the control of the cauliflower mosaic virus 35S promoter. The Arabidopsis tt4 mutants cannot synthesize tannins in their seed coats as well as anthocyanins in cotyledons and hypocotyls (Fig. 6A). After transformation, several T1 kanamycin-resistant plants showed pigmentation at the rosette base similar to the wild type Arabidopsis, whereas Arabidopsis tt4 mutant lacked the visible pigments (S2 Fig.). Additionally, seeds collected from these T1 plants exhibited pigmentation characteristic of the wild-type Arabidopsis. When the T2 seeds were grown on 1/2 MS medium containing 3% sucrose, germinating seedlings regained the red cotyledons compared with the green cotyledons in Arabidopsis tt4 mutants (Fig. 6A), and this could be caused by the expression of FhCHS1 (Fig. 6B). Furthermore, 100 mg of T2 seedlings were extracted with extraction solvent mixture and analyzed by HPLC to determine the amounts of individual anthocyanins and flavonols (Table A in S1 File). The results showed that tt4 mutant had reduced peak area for all the peaks of anthocyanin (monitored at 520) and some peaks of flavonol (monitored at 360) relative to the wild type control, as anticipated, T2 seedlings carrying FhCHS1 cDNA could restore these peaks (S3 Fig.). As for specific components, T2 seedlings had higher levels of pelargonidin but lower levels of cyanidin, kaempferol and quercetin than wild-type Arabidopsis (Fig. 6C). Overall, these results clearly demonstrated that CHS1 gene from Freesia hybrida was fully functional in flavonoid biosynthesis in Arabidopsis.
Fig 6. Complementation of the pigmentation of Arabidopsis tt4 mutant seedlings with FhCHS1 gene.
A, Phenotypes of wild-type, tt4 mutant, and transgenic Arabidopsis seeds and seedlings. B, Expressional analysis of the FhCHS1 gene by reverse transcription polymerase chain reaction in the wild-type, tt4 mutant and transgenic lines. C, Contents of anthocyanidins and flavonols in Arabidopsis seedlings. Data correspond to means of three biological replicates. Means with different letters within the same column are significantly different at the 0.01 level of probability. N/D, not detected.
Overexpression of FhCHS1 gene in petunia
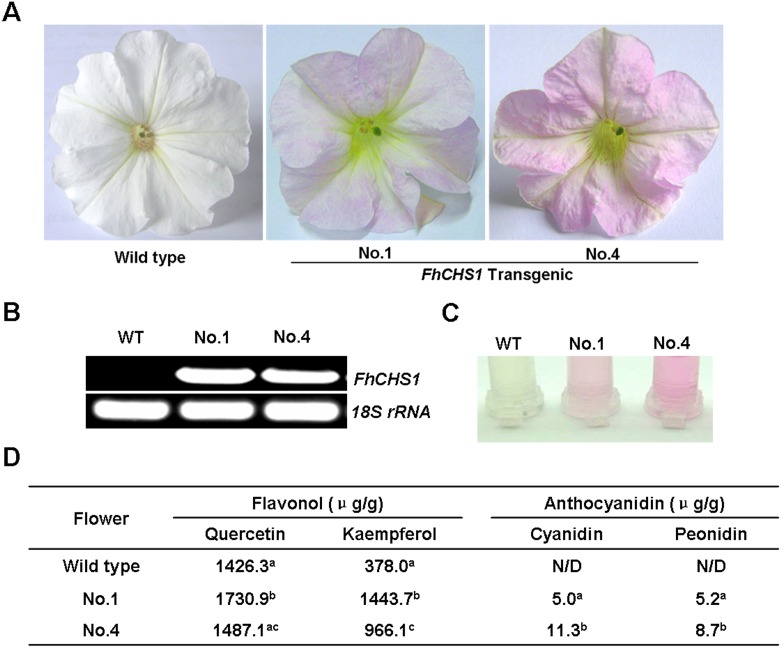
The coding region of FhCHS1 was also transferred into petunia with white flowers, and a total of five transgenic lines were obtained. Among them, two lines (clone No. 1 and 4) showed significant changes of flower color (Fig. 7A). Thus, these two lines were subjected to further analyses, and the results showed that ectopic expression of FhCHS1 (Fig. 7B) in petunia could alter the flower color from white to pink. Furthermore, the anthocyanin and flavonol composition in flowers of transgenic lines was also analyzed in detail (Table B in S1 File and S4 Fig.). The transgenic flowers showed markedly increased accumulation of cyanidin and peonidin derivatives reflecting by the color intensities of the extracted solutions (Fig. 7C). In contrast, the level of anthocyanidin pigments in wild type flowers was undetectable. On the other hand, flowers of those transgenic lines also exhibited notably higher levels of kaempferol derivatives than those of wild type petunia, while the content of quercetin derivatives was scarcely increased (Fig. 7D). Taken together, these results demonstrated that ectopic expression of FhCHS1 gene promoted the biosynthesis of anthocyanins and flavonols in petunia flowers.
Fig 7. Functional characterization of FhCHS1 gene following its overexpression in transgenic petunia lines.
A, Differences in color between wild-type and transgenic petunia flowers. B, Expressional analysis of the FhCHS1 gene by reverse transcription polymerase chain reaction in the wild-type and transgenic lines. C, Extracted solutions from flowers of wild-type and transgenic lines. D, Contents of anthocyanidins and flavonols in the wild-type and transgenic petunia flowers. Data correspond to means of three biological replicates. Means with different letters within the same column are significantly different at the 0.01 level of probability. N/D, not detected.
Discussion
In addition to several other biological functions, flavonoids are important secondary metabolites that contribute to the appearance and quality of many flowers and fruits. The biosynthetic pathway of flavonoid has been extensively investigated in different model and economically important plant species, such as potato [44], grape [45] and apple [46]. In many plant species, core enzymes in the flavonoid pathway, such as CHS and DFR, are encoded by a gene family with up to 10 members [47], and it is deduced that each gene family was derived as a result of gene duplication events and subsequent functional divergence drove by positive selection [48]. Deng et al. found that an ancient duplication occurred in a common ancestor of monocots and eudicots, and gene families of the species tested in phylogenetic analysis are subject to various evolutionary fates [49]. However, in their study, the monocots they selected are exclusively from Poales only. In order to further trace the origination and evolution of the CHS gene family in angiosperm, it is crucial to isolate and characterize related gene families from other monocotyledonous species. The Iridaceae, one of the largest families of the monocots, is an ideal target, because the family comprises plants with large, brightly colored flowers, and is one of the most important plant families in horticulture [50]. Indeed, we have found additional CHS genes in Freesia hybrida through transcriptome analysis (unpublished data, available when requested), and FhCHS1 is the one that is expressed dominantly in the red pigmented flowers. Here, we extensively studied the FhCHS1, and found that it is significantly responsible for biosynthesis of flavonoid in Freesia hybrida.
Firstly, an FhCHS1 cDNA which had one intron was successfully isolated (Fig. 1A). Numerous studies have reported that most plant CHS superfamily genes contain a single intron that splits the conserved Cys in the consensus sequence of (K/Q)R(M/I)C(D/E)KS, while the second intron is not shared by all [13,51]. Furthermore, CHS genes which contain two introns also have the first intron at the conserved Cys and position of the second intron is situated within the codons for conserved amino acid residues such as Gly in AmCHS [52] and Arg in PpCHS11 [13]. In agreement with the previous studies, the CHS gene from Freesia hybrida contained one intron and two exons, and the intron was inserted in the first cysteine codon as in almost all the CHS genes reported (Fig. 1B). Protein sequence alignment and structure analysis indicated the strong conservation of the sequence and structure of CHS protein, and these results provided some evidence for its enzymatic function [28].
The catalytic activity of the CHS enzyme has been detected in cell suspension cultures of parsley (Petroselinum hortense) by Kreuzaler et al. in 1972 [53]. In our study, molecular weight of the purified enzyme was about 43 KDa which is similar to CHS from Hypericum androsaemum and Polygonum cuspidatum [51,54]. Then enzyme assay using p-coumaroyl-CoA and malonyl-CoA as substrates demonstrated that FhCHS1 performed a typical CHS function (Fig. 4). Recently, Jiang et al. reported that PpCHS was able to convert p-coumaroyl-CoA and malonyl-CoA to naringenin chalcone as a major product together with two derailment products (p-coumaroyltriacetic acid lactone and bisnoryangonin) [55]. However, the two derailment products were not detected in our study, likely due to that the enzyme reaction mixture was not acidified prior to extraction. Next, further experiments are needed to examine whether FhCHS1 can convert p-coumaroyl-CoA and malonyl-CoA to other products such as p-coumaroyltriacetic and bisnoryangonin. In addition, it has been reported that CHS has broad substrate preference toward aromatic and aliphatic CoA esters [56]. For example, CHS from Physcomitrella patens has the ability to convert cinnamoyl-CoA and dihydro-p-coumaroyl-CoA to the corresponding chalcones. Among the CoA esters investigated, however, the most preferred substrate for the cyclization reaction of PpCHS is still p-coumaroyl-CoA [55]. Similarly, substrate specificity analysis of CHS from Scutellaria baicalensis also revealed that it could accept both aromatic and aliphatic CoA esters such as benzoyl-CoA, phenylacetyl-CoA, isovaleryl-CoA and isobutyryl-CoA to produce various reaction products including the unnatural aromatic polyketide [57]. In light of foregoing findings, further studies are needed to investigate the catalytic efficiency of FhCHS1 toward different substrates so as to confirm its in vivo substrate. Moreover, enzymatic analysis is also a vital tool to investigate the functional divergence of a gene family after duplication [58].
Real-time PCR results indicated that transcript level of FhCHS1 was related to the stage of floral development with gradually increased expression pattern during flower blossoming, and this pattern showed significant correlation with the flower anthocyanin accumulation (Fig. 5B). This result is in line with the previous studies of wheat, in which the CHS expression in purple pericarp of caryopsis exhibited a growing trend during maturation [59]. Expression analysis in different developmental stages illustrates the fact that FhCHS1 is a key enzyme in the anthocyanin biosynthetic pathway as described by Moore et al. [60]. Subsequent analysis demonstrated that relatively low FhCHS1 expression was observed in non-pigmented organs with the exception of torus (Fig. 5C). This suggests that FhCHS1 is not flower specific and can be involved in the biosynthesis of other flavonoids.
Transfer of FhCHS1 into the Arabidopsis tt4 mutant successfully restored the pigmentation of their seed coats and purple coloration in the cotyledons and hypocotyls, confirming the in vivo function of FhCHS1 as a CHS (Fig. 6A). In the past years, complementation of Arabidopsis flavonoid mutants has revealed the functional conservation of enzymes involved in biosynthesis of flavonoid such as dihydroflavonol 4-reductase from sweet potato [61] and flavonoid 3-O-glucosyltransferase from black soybean [62]. Recently, the maize C2 gene encoding CHS was expressed in tt4 plants under the control of cauliflower mosaic virus 35S promoter, and transgenic seedlings showed similar patterns of flavonoid accumulation as those of wild-type Arabidopsis [63]. In accordance with above result, both the pigmentation phenotype and accumulation of anthocyanins and flavonols in FhCHS1 transgenic seedlings were similar to the wild-type Arabidopsis (Fig. 6). Taken as a whole, these findings provide evidence that CHS proteins in the flavonoid biosynthesis are functionally exchangeable among different plant species.
Comparing to wild-type petunia, transgenic flowers carrying FhCHS1 abundantly accumulated flavonol derivatives together with colored anthocyanins. As shown in Fig. 7D, flowers of transgenic petunia only accumulated cyanidin-based pigments, and the proposed pathway in transgenic petunia flowers was presented in S5 Fig.. The content of kaempferol derivatives in flowers expressing FhCHS1 was enhanced approximately two to threefold. Meanwhile, a slight increase of quercetin derivatives was also observed. Dihydroflavonols, the common precursors for anthocyanins and flavonols, are substrates for both DFR and FLS [22]. As well known, DFR from petunia cannot accept the dihydrokaempferol as a substrate [64]. Therefore, the formation of pelargonidin-based pigments in transgenic petunia flowers was blocked, and the increased conversion of dihydrokaempferol to kaempferol derivatives by FLS might have occurred. By contrast, when the biosynthesis of cyanidin-based pigments was activated, accordingly, the increase in the contents of quercetin derivatives would be relatively lower than those of kaempferol derivatives. In Arabidopsis, it has been reported that disruption of DFR could enhance accumulation of flavonol [65], while fls1 mutants showed increased anthocyanin levels [66,67]. Our results are consistent with these findings and reveal that substrate competition between FLS and DFR controls metabolic flux in the flavonoid pathway. In addition, transgenic flowers showed color alteration from white to pink and accumulated higher levels of cyanidin-based pigments than those of wild-type petunia. The results suggest that manipulation of the FhCHS1 gene will contribute to modification of coloration in other plants.
In conclusion, we have successfully isolated FhCHS1, a CHS gene from Freesia hybrida, and examined its biological function in flavonoid biosynthesis. In addition, overexpression of FhCHS1 in petunia has increased the total amount of flavonoids in flowers. This clearly demonstrates that FhCHS1 plays a crucial role in flavonoid biosynthesis and can be used to alter the composition of flavonoids of flowers to improve their aesthetic values. Furthermore, the characterized gene will probably contribute to the investigation of the origination and evolution of CHS gene families in angiosperm.
Supporting Information
The start condon (ATG) and the stop condon (TAA) are highlighted with red background, and the intron is underline.
(TIF)
(TIF)
(A-F) HPLC chromatograms of the samples from seedlings of WT, mutant and transgenic lines. (A, D) WT, (B, E) Mutant, (C, F) Transgenic lines. (A-C) Absorbance at 520 nm for analysis of anthocyanins. (D-F) Absorbance at 360 nm for analysis of flavonols.
(TIF)
(A-F) HPLC chromatograms of the samples from flowers of WT and transgenic lines. (A, D) WT, (B, E) NO.1, (C, F) NO.4. (A-C) Absorbance at 520 nm for analysis of anthocyanins. (D-F) Absorbance at 360 nm for analysis of flavonols.
(TIF)
Hoary areas indicate biosynthesis of pelargonidin-based pigments is blocked.
(TIF)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31170276, 31300271) and Jilin provincial Government of the People’s Republic of China (20130102061JC, 201201074, 20125035). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chandler SF, Brugliera F (2011) Genetic modification in floriculture. Biotechnol Lett 33: 207–214. 10.1007/s10529-010-0424-4 [DOI] [PubMed] [Google Scholar]
- 2. Maleka MF, Albertyn J, Spies JJ (2013) The floriculture industry and flower pigmentation. Philos Trans Genet 2: 55–110. [Google Scholar]
- 3. Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749. 10.1111/j.1365-313X.2008.03447.x [DOI] [PubMed] [Google Scholar]
- 4. Zhou XW, Fan ZQ, Chen Y, Zhu YL, Li JY, Yin HF (2013) Functional analyses of a flavonol synthase—like gene from Camellia nitidissima reveal its roles in flavonoid metabolism during floral pigmentation. J Biosci 38: 593–604. [DOI] [PubMed] [Google Scholar]
- 5. Niyogi KK (2000) Safety valves for photosynthesis. Curr Opin Plant Biol 3: 455–460. [DOI] [PubMed] [Google Scholar]
- 6. Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780. [DOI] [PubMed] [Google Scholar]
- 7. Ferrer JL, Austin M, Stewart C Jr, Noel J (2008) Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Bioch 46: 356–370. 10.1016/j.plaphy.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217. [Google Scholar]
- 9. Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223. [DOI] [PubMed] [Google Scholar]
- 10. Buer CS, Imin N, Djordjevic MA (2010) Flavonoids: new roles for old molecules. J Intergr Plant Biol 52: 98–111. 10.1111/j.1744-7909.2010.00905.x [DOI] [PubMed] [Google Scholar]
- 11. Ferreyra MLF, Rius SP, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3: 1–15. 10.3389/fpls.2012.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stafford HA (1991) Flavonoid evolution: an enzymic approach. Plant Physiol 96: 680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koduri PH, Gordon GS, Barker EI, Colpitts CC, Ashton NW, Suh DY (2010) Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens . Plant Mol Biol 72: 247–263. 10.1007/s11103-009-9565-z [DOI] [PubMed] [Google Scholar]
- 14. Commenges D, Scotet V, Renaud S, Jacqmin-Gadda H, Barberger-Gateau P, Dartigues JF (2000) Intake of flavonoids and risk of dementia. Eur J Epidemiol 16: 357–363. [DOI] [PubMed] [Google Scholar]
- 15. Knekt P, Kumpulainen J, Järvinen R, Rissanen H, Heliövaara M, Reunanen A, et al. (2002) Flavonoid intake and risk of chronic diseases. Am J Clin Nutr 76: 560–568. [DOI] [PubMed] [Google Scholar]
- 16. Teixeira S, Siquet C, Alves C, Boal I, Marques MP, Borges F, et al. (2005) Structure—property studies on the antioxidant activity of flavonoids present in diet. Free Radical Bio Med 39: 1099–1108. [DOI] [PubMed] [Google Scholar]
- 17. Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181: 219–229. 10.1016/j.plantsci.2011.05.009 [DOI] [PubMed] [Google Scholar]
- 18. Pourcel L, Bohorquez-Restrepo A, Irani NG, Grotewold E (2012) Anthocyanin biosynthesis, regulation, and transport: New insights from model species. Rec Adv Polyphenol Res 3: 143–160. [Google Scholar]
- 19. Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forkmann G, Martens S (2001) Metabolic engineering and applications of flavonoids. Curr Opin Biotech 12: 155–160. [DOI] [PubMed] [Google Scholar]
- 21. Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18: 477–483. 10.1016/j.tplants.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 22. Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, et al. (2003) Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131: 259–268. [Google Scholar]
- 23. Martens S, Mithöfer A (2005) Flavones and flavone synthases. Phytochemistry 66: 2399–2407. [DOI] [PubMed] [Google Scholar]
- 24. Yoshida K, Iwasaka R, Shimada N, Ayabe S-i, Aoki T, Sakuta M (2010) Transcriptional control of the dihydroflavonol 4-reductase multigene family in Lotus japonicus . J Plant Res 123: 801–805. 10.1007/s10265-010-0325-6 [DOI] [PubMed] [Google Scholar]
- 25. Winkel-Shirley B (2001) It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 127: 1399–1404. [PMC free article] [PubMed] [Google Scholar]
- 26. Pang Y, Shen G, Wu W, Liu X, Lin J, Tan F, et al. (2005) Characterization and expression of chalcone synthase gene from Ginkgo biloba . Plant Sci 168: 1525–1531. [Google Scholar]
- 27. Austin MB, Noel JP (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep 20: 79–110. [DOI] [PubMed] [Google Scholar]
- 28. Abe I, Morita H (2010) Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep 27: 809–838. 10.1039/b909988n [DOI] [PubMed] [Google Scholar]
- 29. Lanz T, Tropf S, Marner F, Schröder J, Schröder G (1991) The role of cysteines in polyketide synthases. Site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J Biol Chem 266: 9971–9976. [PubMed] [Google Scholar]
- 30. Suh D-Y, Kagami J, Fukuma K, Sankawa U (2000) Evidence for catalytic cysteine—histidine dyad in chalcone synthase. Biochem Bioph Res Co 275: 725–730. [DOI] [PubMed] [Google Scholar]
- 31. Ferrer J-L, Jez JM, Bowman ME, Dixon RA, Noel JP (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Mol Bio 6: 775–784. [DOI] [PubMed] [Google Scholar]
- 32. Raja Abdul Rahman RNZ, Zakaria II, Salleh AB, Basri M (2012) Enzymatic properties and mutational studies of chalcone synthase from physcomitrella patens . Int J Mol Sci 13: 9673–9691. 10.3390/ijms13089673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuckuck H (1936) Über vier neue Serien multipler Allele bei Antirrhinum majus . Mol Gen Genet 71: 429–440. [Google Scholar]
- 34. Napoli CA, Fahy D, Wang H-Y, Taylor LP (1999) White anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 120: 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM (1988) Restriction fragment length polymorphism linkage map for Arabidopsis thaliana . PNAS 85: 6856–6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, et al. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671. [DOI] [PubMed] [Google Scholar]
- 37. Kabsch W (2010) Xds. Acta Crystallogr Sect D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clough SJ, Bent AF (1998) Floral dip: a simplified method forAgrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 41. Penninckx I, Eggermont K, Terras F, Thomma B, De Samblanx GW, Buchala A, et al. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sui X, Gao X, Ao M, Wang Q, Yang D, Wang M, et al. (2011) cDNA cloning and characterization of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in Freesia hybrida . Plant Cell Rep 30: 1209–1218. 10.1007/s00299-011-1029-7 [DOI] [PubMed] [Google Scholar]
- 43. Fanali C, Dugo L, D’Orazio G, Lirangi M, Dachà M, Dugo P, et al. (2011) Analysis of anthocyanins in commercial fruit juices by using nano-liquid chromatography-electrospray-mass spectrometry and high-performance liquid chromatography with UV-vis detector. J Sep Sci 34: 150–159. 10.1002/jssc.201000665 [DOI] [PubMed] [Google Scholar]
- 44. Jung CS, Griffiths HM, De Jong DM, Cheng S, Bodis M, Kim TS, et al. (2009) The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor Appl Genet 120: 45–57. 10.1007/s00122-009-1158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis G, Ananga A, Krastanova S, Sutton S, Ochieng JW, Leong S, et al. (2012) Elevated gene expression in chalcone synthase enzyme suggests an increased production of flavonoids in skin and synchronized red cell cultures of North American native grape berries. DNA Cell Biol 31: 939–945. 10.1089/dna.2011.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, et al. (2013) Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus× domestica). Plant J 74: 398–410. 10.1111/tpj.12140 [DOI] [PubMed] [Google Scholar]
- 47. Thomas B, Ryder SAH, Bell JN, Liang X, Clouse SD, Lamb CJ (1987) Organization and differential activation of a gene family encoding the plant defense enzyme chalcone synthase in Phaseolus vulgaris . Mol Gen Genet 210: 219–233. [DOI] [PubMed] [Google Scholar]
- 48. Yrjo Helariuta MK, Elomaa P, Kalkkinen N, Bremer K, Teeri TH, Albert VIA (1996) Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: Evolution with substrate change and catalytic simplification. PNAS 93: 9033–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deng X, Bashandy H, Ainasoja M, Kontturi J, Pietiainen M, Laitinen RA, et al. (2014) Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida . New Phytol 201: 1469–1483. 10.1111/nph.12610 [DOI] [PubMed] [Google Scholar]
- 50. Peter Goldbiatt JCM (2008) The Iris family: natural history and classification. Portland: Timber Press; 9 p. [Google Scholar]
- 51. Ma LQ, Pang XB, Shen HY, Pu GB, Wang HH, Lei CY, et al. (2009) A novel type III polyketide synthase encoded by a three-intron gene from Polygonum cuspidatum . Planta 229: 457–469. 10.1007/s00425-008-0845-7 [DOI] [PubMed] [Google Scholar]
- 52. Sommer H, Saedler H (1986) Structure of the chalcone synthase gene of Antirrhinum majus . Mol Gen Genet 202: 429–434. [DOI] [PubMed] [Google Scholar]
- 53. Kreuzaler F, Hahlbrock K (1972) Enzymatic synthesis of aromatic compounds in higher plants: formation of naringenin (5, 7, 4′-trihydroxyflavanone) from p-coumaroyl coenzyme A and malonyl coenzyme A. FEBS Lett 28: 69–72. [DOI] [PubMed] [Google Scholar]
- 54. Liu B, Falkenstein‐Paul H, Schmidt W, Beerhues L (2003) Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site‐directed mutagenesis of two polyketide synthases. Plant J 34: 847–855. [DOI] [PubMed] [Google Scholar]
- 55. Jiang C, Schommer CK, Kim SY, Suh DY (2006) Cloning and characterization of chalcone synthase from the moss, Physcomitrella patens . Phytochemistry 67: 2531–2540. [DOI] [PubMed] [Google Scholar]
- 56. Abe I, Takahashi Y, Noguchi H (2002) Enzymatic formation of an unnatural C6-C5 aromatic polyketide by plant type III polyketide synthases. Org lett 4: 3623–3626. [DOI] [PubMed] [Google Scholar]
- 57. Morita H, Takahashi Y, Noguchi H, Abe I (2000) Enzymatic formation of unnatural aromatic polyketides by chalcone synthase. Biochem Bioph Res Co 279: 190–195. [DOI] [PubMed] [Google Scholar]
- 58. Des Marais DL, Rausher MD (2008) Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765. 10.1038/nature07092 [DOI] [PubMed] [Google Scholar]
- 59. Trojan V, Musilová M, Vyhnánek T, Klejdus B, Hanáček P, Havel L (2014) Chalcone synthase expression and pigments deposition in wheat with purple and blue colored caryopsis. J Cereal Sci 59: 48–55. [Google Scholar]
- 60. Moore S, Vrebalov J, Payton P, Giovannoni J (2002) Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J Exp Bot 53: 2023–2030. [DOI] [PubMed] [Google Scholar]
- 61. Wang H, Fan W, Li H, Yang J, Huang J, Zhang P (2013) Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PloS One 8: e78484 10.1371/journal.pone.0078484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kovinich N, Saleem A, Arnason JT, Miki B (2010) Functional characterization of a UDP-glucose: flavonoid 3-O-glucosyltransferase from the seed coat of black soybean (Glycine max(L.) Merr.). Phytochemistry 71: 1253–1263. 10.1016/j.phytochem.2010.05.009 [DOI] [PubMed] [Google Scholar]
- 63. Dong X, Braun EL, Grotewold E (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meyer P, Heidmann I, Forkmann G, Saedler H (1987) A new petunia flower colour generated by transformation of a mutant with a maize gene. Nature 330: 677–678. [DOI] [PubMed] [Google Scholar]
- 65. Pelletier MK, Burbulis IE, Winkel-Shirley B (1999) Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol Biol 40: 45–54. [DOI] [PubMed] [Google Scholar]
- 66. Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel Brenda SJ (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis . Plant Physiol 147: 1046–1061. 10.1104/pp.108.117457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stracke R, De Vos RC, Bartelniewoehner L, Ishihara H, Sagasser M, Martens S, et al. (2009) Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229: 427–445. 10.1007/s00425-008-0841-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The start condon (ATG) and the stop condon (TAA) are highlighted with red background, and the intron is underline.
(TIF)
(TIF)
(A-F) HPLC chromatograms of the samples from seedlings of WT, mutant and transgenic lines. (A, D) WT, (B, E) Mutant, (C, F) Transgenic lines. (A-C) Absorbance at 520 nm for analysis of anthocyanins. (D-F) Absorbance at 360 nm for analysis of flavonols.
(TIF)
(A-F) HPLC chromatograms of the samples from flowers of WT and transgenic lines. (A, D) WT, (B, E) NO.1, (C, F) NO.4. (A-C) Absorbance at 520 nm for analysis of anthocyanins. (D-F) Absorbance at 360 nm for analysis of flavonols.
(TIF)
Hoary areas indicate biosynthesis of pelargonidin-based pigments is blocked.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.