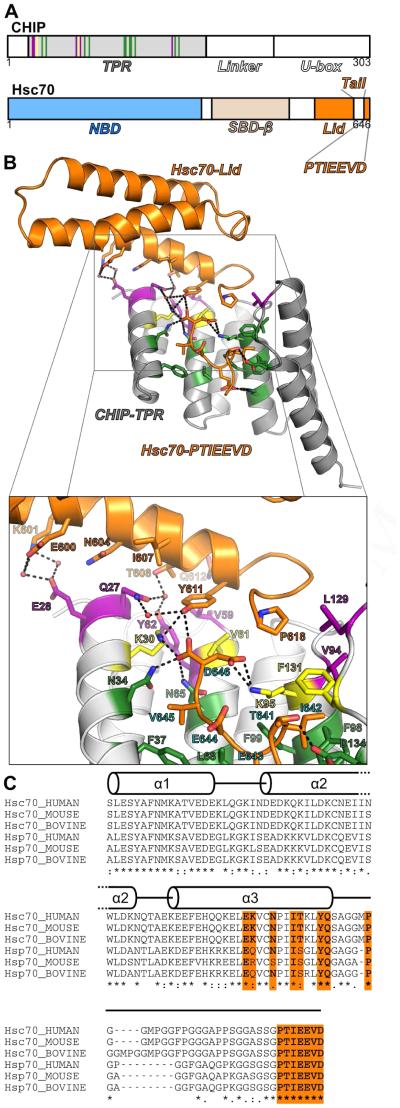
Summary
The ubiquitin ligase CHIP plays an important role in cytosolic protein quality control by ubiquitinating proteins chaperoned by Hsp70/Hsc70 and Hsp90, thereby targeting such substrate proteins for degradation. We present a 2.91 Å resolution structure of the TPR domain of CHIP in complex with the α-helical “lid” subdomain and unstructured “tail” of Hsc70. Surprisingly, the CHIP-TPR interacts with determinants within both the Hsc70-lid subdomain and the C-terminal PTIEEVD motif of the tail, exhibiting a novel mode of interaction between chaperones and TPR domains. We demonstrate that the interaction between CHIP and the Hsc70-lid subdomain is required for proper ubiquitination of Hsp70/Hsc70 or Hsp70/Hsc70-bound substrate proteins. Post-translational modifications of the Hsc70 lid and tail disrupt key contacts with the CHIP-TPR and may regulate CHIP-mediated ubiquitination. Our study shows how CHIP docks onto Hsp70/Hsc70 and defines a new bipartite mode of interaction between TPR domains and their binding partners.
INTRODUCTION
The cytosolic chaperones Hsp90, Hsp70 and its constitutively expressed homologue Hsc70 function in diverse intracellular processes. These include client protein folding and conformational regulation, prevention of protein aggregation, protein transport and translocation across intracellular membranes, and regulation of client protein signaling (Kim et al., 2013; Li and Buchner, 2013; Mayer and Bukau, 2005; Priya et al., 2013; Röhl et al., 2013). These chaperones are assisted by diverse cochaperones, a subset of which contain domains composed of tetratricopeptide repeat (TPR) motifs (Allan and Ratajczak, 2011). Each ~34 residue-long TPR motif forms two anti-parallel α-helices connected by a short turn (D'Andrea and Regan, 2003). In tandem, TPRs form superhelical domains with distinct ligand binding grooves (Allan and Ratajczak, 2011; D'Andrea and Regan, 2003; Zeytuni and Zarivach, 2012).
The TPR domains of CHIP, Hop, cyclophilin-40 (CyP40) and other cochaperones bind to C-terminal motifs on Hsp70/Hsc70 (PTIEEVD) or Hsp90 (SRMEEVD) through a characteristic “two-carboxylate clamp” mode. Both carboxylates of the C-terminal aspartic acids of these motifs form salt bridges with residues within the groove of the cochaperone TPR domains (Scheufler et al., 2000; Wang et al., 2011; Zhang et al., 2005). In addition, nearby hydrophobic pockets accommodate the aliphatic residues of the motifs (Zeytuni and Zarivach, 2012). These motifs lie at the very C-termini of Hsp70/Hsc70 and Hsp90, following unstructured “tail” segments that are 25-35 residues long (Bertelsen et al., 1999; Boorstein et al., 1994). It is thus thought that the TPR-domain cochaperones form dynamic “tethered” complexes with Hsp70/Hsc70 and Hsp90.
The homodimeric ubiquitin (E3) ligase CHIP (C-terminus of Hsp70 Interacting Protein) ubiquitinates client proteins bound to Hsp70/Hsc70 or Hsp90. CHIP-mediated ubiquitination promotes the degradation of chaperone clients, mitigates accumulation of misfolded proteins and regulates the intracellular levels of myriad chaperoned proteins (Connell et al., 2001; Cyr et al., 2002; Demand et al., 2001; Stankiewicz et al., 2010). Upon recovery from stresses that elevate Hsp70 levels, CHIP also ubiquitinates “client-free” Hsp70 to restore resting Hsp70 levels (Qian et al., 2006). CHIP contains an N-terminal TPR domain composed of three TPRs and an extended seventh helix that bridges the TPR domain and a helical dimerization domain (Zhang et al., 2005). The TPR domains of CHIP dimers bind to the C-terminal motifs of Hsp70/Hsc70 and Hsp90 (Wang et al., 2011; Wu et al., 2001; Zhang et al., 2005).
Tethering to chaperones could allow CHIP to access different ubiquitination sites on diverse chaperone-bound clients. However, recent studies suggested that some TPR domain-containing cochaperones interact with sites on Hsp70 or Hsp90 other than the C-terminal motifs (Alvira et al., 2014; Li et al., 2013; Schmid et al., 2012). We therefore sought a more detailed understanding of how CHIP targets chaperone-bound substrates by characterizing its interaction with Hsp70/Hsc70. Here, we report that the TPR domain of CHIP interacts not only with the C-terminal GPTIEEVD motif of Hsp70/Hsc70 but also with the α-helical lid subdomain of the chaperone. We structurally define this unexpected bipartite binding mode using X-ray crystallography. Using mutagenesis and in vitro ubiquitination assays, we show that the Hsp70-lid:CHIP-TPR interaction is functionally required for efficient ubiquitination of Hsp70/Hsc70 or Hsp70/Hsc70-bound clients. Moreover, we find that post-translational modifications on the lid subdomain can regulate CHIP-mediated ubiquitination. Our results uncover a novel mode of chaperone:cochaperone and TPR:binding-partner interaction that is necessary for CHIP to regulate intracellular protein quality control.
RESULTS
CHIP-TPR Domain Interacts with the α-lid Subdomain of Hsp70/Hsc70
To better understand interactions between CHIP and Hsc70, we systematically carried out HSQC-NMR titrations of 15N-labeled Hsc70 domains with CHIP, and vice versa. Full-length CHIP forms a 70 kDa dimer through its helical dimerization and U-box domains (Nikolay et al., 2004; Zhang et al., 2005). Unexpectedly, CHIP induced extensive line broadening in the 1H-15N HSQC spectrum of a 15N-Hsc70-lid-tail construct that lacked the C-terminal GPTIEEVD motif (Figure S1A,B available online). In a complementary experiment, the HSQC spectrum of 15N-CHIP-TPR domain was broadened by an Hsc70 construct that included the SBDβ subdomain, lid and tail but lacked the C-terminal GPTIEEVD motif (Figure S1C). The corresponding construct with the GPTIEEVD motif also broadened the HSQC spectrum of 15N-CHIP-TPR domain (Figure S1D,E), as expected based on the known interaction between the GPTIEEVD motif and the CHIP TPR domain.
The Hsc70-lid-tail-ΔGPTIEEVD construct induced small but definite chemical shift perturbations (CSPs) in the HSQC spectrum of 15N-CHIP-TPR and vice versa (Figure S1F,G). By itself, an Hsc70-GPTIEEVD peptide strongly perturbed the 15N-CHIP-TPR spectrum in a manner indicating slow exchange binding. However, addition of Hsc70-lid-tail-ΔGPTIEEVD to the 15N-TPR:14N-GPTIEEVD complex induced additional CSPs (Figure S1H). Similar CSPs were observed upon titration of 15N-TPR with Hsc70-lid-tail (Figure S1I). These data suggest that the CHIP-TPR domain interacts not only with the C-terminal GPTIEEVD motif but also with the lid subdomain or portions of the tail upstream of the motif.
Structure of the CHIP-TPR:Hsc70-lid-tail Complex
To characterize these interactions in greater detail, we co-crystallized the CHIP-TPR with a construct containing the Hsc70 lid and tail, including the C-terminal GPTIEEVD motif. To aid crystallization, we tested multiple Hsc70 constructs containing short deletions within the Ser/Gly/Pro/Ala-rich segment of the tail (Hsc70 residues 612-638). We obtained co-crystals of CHIP-TPR in complex with an Hsc70 lid-tail construct from which residues 626-638 were deleted (HsHsc70541-646Δ626-638). The resulting 2.91Å-resolution crystal structure (Figure 1 and Table 1) contains two copies each of CHIP-TPR and HsHsc70541-646Δ626-638 per asymmetric unit (Figure S2A). The GPTIEEVD motifs and Hsc70-lid subdomains are well defined in the electron density map. However, most of the Hsc70-tail residues between the lid and the C-terminal motifs are disordered. While both lid subdomains pack against one of the TPRs, only one of these interactions (Figure 1B) is compatible with the structure of full-length CHIP (Figure S2B,C).
Figure 1. Structure of the CHIP-TPR/Hsc70-lid-tail Complex.
(A) Arrangement of domains within CHIP and Hsc70.
(B) Cartoon view of the Hsc70-lid-tail (Hsc70541-646Δ626-638) in complex with the CHIP-TPR domain (CHIP21-154). Hsc70-lid-tail and CHIP-TPR domains are colored orange and grey respectively. Specific CHIP-TPR residues that interact with Hsc70-lid, Hsc70-tail, or both domains are colored green, purple and yellow respectively.
(C) Alignment of human, murine and bovine Hsc70- and Hsp70-lid-tail sequences with secondary structure overlay. Lid and tail residues that interact with the CHIP-TPR are colored orange. See also Figures S1, S2 and S3.
Table 1.
Data Collection and Refinement Statistics
| CHIP21-154/Hsc70395-646Δ626-638 Complex | |
|---|---|
| Data Collection | |
| Beam line | ALS 4.2.2 |
| Wavelength (Å) | 1.0000 |
| Space group | P6122 |
| a, b, c (Å) | 78.5, 78.5, 424.7 |
| α, β, γ(°) | 90, 90, 90 |
| Resolution (Å)a | 64.75 – 2.91 (3.01 – 2.91) |
| R meas b | 0.075 (0.639) |
| <I/σI> | 30.9 (4.2) |
| Completeness (%) | 97.8 (76.7) |
| Redundancy | 12.1 (12.1) |
| Unique reflections | 17,775 |
| Refinement | |
| Resolution (Å) | 64.75 – 2.91 |
| Number of Reflections | 17,773 |
| Rwork / Rfree | 0.224 / 0.263 |
| Number of atoms (protein / water) | 3,507 / 52 |
| Average B factors (protein / water) | 64.9 / 66.5 |
| Rmsd | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.251 |
| Ramachandran favored (%) | 98.1 (410/420) |
| Ramachandran outliers (%) | 0.0 (0/420) |
| Poor rotamers (%)c | 0.0 |
| Cβ deviations >0.25 Åc | 0 |
| Clash score | 5.47 |
| Clash percentile | 100th percentile (N=92, 2.91 Å ± 0.25 Å) |
| MolProbity score | 1.29 |
| MolProbity score percentile | 100th percentile (N=3,658, 2.91 Å ± 0.25 Å) |
Values in parentheses are for the highest resolution shell.
The merging R factor is defined as
Calculated with MolProbity v4.1 (Chen et al., 2010).
The interface between the lid and TPR domain buries ~480 Å2 on the TPR domain while the C-terminal PTIEEVD sequence buries ~460 Å2. Sidechains on the C-terminal helix of the Hsc70-lid domain and the first few residues of the Hsc70-tail (Figure 1B,C) contact the TPR domain. The lid helix interacts with the N-terminus of the first TPR helix and with the loop between TPR helices 2 and 3. Hsc70 residues 612-616 turn and interact with the interhelical loops between TPR helices 4-5 and 6-7. The Hsc70-lid and proximal tail residues thus cap one side of the TPR domain.
The Hsc70 PTIEEVD motif binds to the CHIP-TPR in a manner very similar to a co-crystal structure of the CHIP-TPR domain with a GPTIEEVD peptide (PDB ID 3Q49;(Wang et al., 2011)), though P640 adopts a different orientation (Figure S3). While the Hsc70 residues between P618 and P640 are largely disordered, the tail must extend away from the lid subdomain and “double back” to allow the GPTIEEVD motif to bind to the TPR domain. Remarkably, D646, the C-terminal residue of Hsc70, also contacts the Hsc70-lid (Figure 1B). The sidechain of Hsc70-Y611 on the lid subdomain contributes to the carboxylate clamp and interacts with D646, as do CHIP-TPR residues K30, N34 and N65.
CHIP-TPR/Hsc70-lid Interaction Regulates CHIP-Mediated Ubiquitination
CHIP ubiquitinates not just clients bound to Hsp70/Hsc70 but also Hsp70/Hsc70 themselves, targeting lysines in the SBDβ or lid subdomains (Kundrat and Regan, 2010a). Mutations of CHIP-TPR residues that interact with the GPTIEEVD motif are known to abolish CHIP’s ubiquitination activity towards chaperones or chaperone-bound clients (He et al., 2004; Xu et al., 2002). To test whether the interaction between the CHIP-TPR and Hsc70-lid subdomain is also functionally important, we measured ubiquitination of GST-tagged Hsc70 SBDβ-lid-tail (GST-Hsc70395-646) by CHIP and the E2 enzyme UbcH5b. We used CHIP autoubiquitination (self-ubiquitination) reactions to control for mutations that perturb the interaction between CHIP and UbcH5b or that nonspecifically perturb CHIP-mediated ubiquitination. CHIP mutations in the lid:TPR interface, including Y62F and Q27G, reduced ubiquitination of the Hsc70 SBDβ-lid-tail construct without affecting CHIP autoubiquitination (Figure 2A,B). Similarly, V59D and L129D mutations very strongly reduced Hsc70 ubiquitination with only slight effects on CHIP autobubiquitination. CHIP S23E/S25E mutations, lying outside either the lid-binding surface or the GPTIEEVD-binding pocket, had no effect. Using in vitro pull-down assays, we confirmed that the mutants retained interaction with the GPTIEEVD motif (Figure 2C). Therefore, the CHIP-TPR mutations specifically lead to ubiquitination defects by perturbing the Hsc70-lid:CHIP-TPR interaction. To verify this further, we carried out an HSQC-NMR titration of full-length 14N-CHIP-V59D/L129D, which exhibits strong defects in substrate ubiquitination, against 15N-Hsc70-lid-tail-ΔGPTIEEVD. CHIP-V59D/L129D had no effect on the 15N-Hsc70-lid-tail-ΔGPTIEEVD HSQC spectrum (Figure S4), confirming that the V59D/L129D mutations disrupt the lid:TPR interaction.
Figure 2. Residues that mediate CHIP-TPR/Hsc70-lid interactions modulate CHIP-mediated ubiquitination.
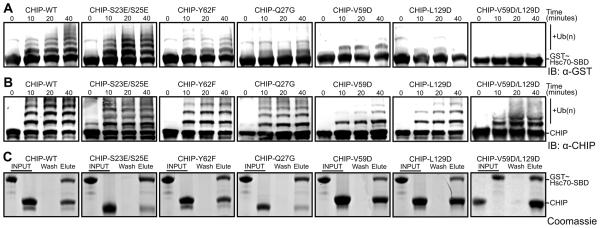
(A) Western blot with anti-GST antibody to detect ubiquitination of GST-Hsc70395-646 by wild type (WT) and mutant CHIP1-303.
(B) Western blot with anti-CHIP antibody to detect autoubiquitination of wild type (WT) and mutant CHIP1-303.
(C) Pull-down of GST~Hsc70395-646 (GST~Hsc70-SBDβ-lid-tail) by wild type (WT) and mutant His6-CHIP1-303 bound to Ni2+ mag-sepharose beads analyzed by SDS PAGE with Coomassie staining. Lanes indicate GST~Hsc70395-646 and CHIP1-303 components utilized (INPUT), the final wash fraction (Wash) and the elution fraction (Elute). See also Figure S4.
We similarly tested mutations in the Hsc70 lid subdomain. Mutation of TPR-interacting residues reduced or abolished ubiquitination of GST-SBDβ-lid-tail (Figure 3A). As previously reported (Smith et al., 2013), deletion of the Hsc70-GPTIEEVD motif nearly eliminated ubiquitination of the Hsc70 SBDβ-lid-tail. Deletion of most of the Hsc70-tail (residues 619-640) also reduced ubiquitination. This is consistent with the notion that the tail must be long enough to allow both the lid and the GPTIEEVD sequence to interact with the CHIP-TPR domain. In contrast, a shorter tail deletion (residues 626-638), which we used to aid crystallization, had much less effect on ubiquitination (Figure 3A). In vitro pull-down assays (Figure 3B) confirmed that the mutants did not eliminate the interaction between CHIP and the Hsc70 SBDDβ-lid-tail but instead specifically decreased ubiquitination.
Figure 3. Hsc70 residues that mediate CHIP-TPR/Hsc70-lid interactions modulate CHIP-mediated ubiquitination.
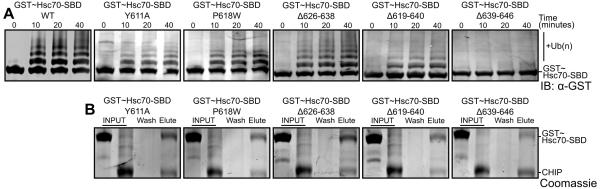
(A) Western blot with anti-GST antibody to detect ubiquitination of wild type (WT) and mutant GST~Hsc70395-646 (GST~Hsc70-SBDβ-lid-tail) constructs by wild type CHIP1-303.
(B) Pull-down of wild type (WT) and mutant GST~Hsc70395-646 constructs by wild type His6-CHIP1-303 bound to Ni2+ mag-sepharose beads analyzed by SDS PAGE with Coomassie staining. Lanes indicate the GST~Hsc70395-646 and CHIP1-303 components utilized (INPUT), the final wash fraction (Wash) and the elution fraction (Elute).
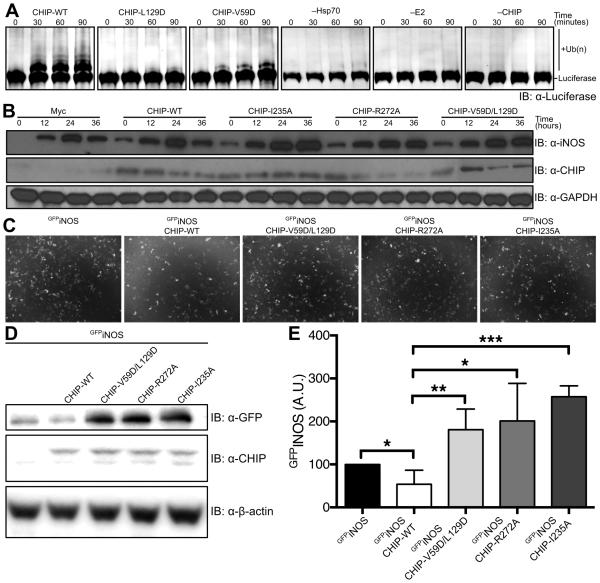
We tested whether disruption of the CHIP-TPR:Hsc70-lid interaction also altered ubiquitination of Hsp70/Hsc70-bound clients and not just Hsp70/Hsc70. We carried out in vitro ubiquitination reactions of heat-denatured luciferase, which is well established as an in vitro Hsp70/Hsc70 client (Rosser et al., 2007; Schlecht et al., 2011; Xu et al., 2008b) and CHIP substrate (Murata et al., 2001). CHIP mutations that reduced ubiquitination of the Hsc70 SBDβ-lid-tail construct also reduced ubiquitination of Hsp70-bound luciferase (Figure 4A).
Figure 4. CHIP-mediated Chaperoned Ubiquitination in vitro and ex vivo.
(A) CHIP mediated ubiquitination of firefly luciferase was monitored by western blot using an anti-luciferase antibody. Control reactions were carried out with all reagents present except Hsp70, UbcH5b or CHIP respectively.
(B) iNOS expression, induced by LPS and IFN-γ treatment, was monitored for 36 hours in Raw264.7 macrophages transfected with wild-type CHIP and CHIP mutants in which interactions between the U-box and E2 enzymes are disrupted (I235A, R272A) or the Hsp70-lid:TPR interaction is disrupted (V59D/L129D). Protein levels were monitored by western blot with anti-iNOS, anti-CHIP and anti-GAPDH (loading control).
(C) HEK293 cells were transfected with GFPiNOS and CHIP or CHIP mutants that disrupt E2:U-box interactions (I235A or R272A) or the Hsp70-lid:TPR interaction (V59D/L129D). Cells were incubated for 48 hours prior to imaging.
(D) Cells from (C) were then collected, lysed and analyzed by western blot with anti-GFP, anti-CHIP and anti-β-actin (loading control)
(E) Western blots from (D) were repeated six times and band intensities were quantified. Data represent mean±SD. *P<0.05. **P<0.01. ***P<0.001.
We next investigated the effect of perturbing the Hsc70-lid:CHIP-TPR interaction on client ubiquitination and degradation in an ex vivo context. Nitric Oxide Synthases are bona fide Hsp70/Hsc70 clients and substrates for CHIP-mediated ubiquitination (Chen et al., 2009; Peng et al., 2004). RAW264.7 macrophages were transfected with wild-type or mutant CHIP expression constructs and treated with LPS and Interferon-γ to induce expression of inducible Nitric Oxide Synthase (iNOS). After initial iNOS induction, expression of wild-type CHIP reduced iNOS levels after 24 hours due to degradation (Figure 4B). Expression of CHIP-I235A and −R272A mutations, which disrupt CHIP’s recruitment of E2 enzymes (Xu et al., 2008a; Zhang et al., 2005), led to persistent elevation of iNOS and acted as dominant negatives, likely due to heterodimerization with native CHIP. CHIP-V59D/L129D behaved equivalently to these loss-of-function CHIP mutants.
We also examined iNOS levels in HEK293 cells transfected with GFPiNOS and wild-type or mutant CHIP constructs. After 48 hours, GFPiNOS in cells transfected with wild-type CHIP decreased in comparison to cells transfected with GFPiNOS alone (Figure 4C-E). In contrast, co-transfection with CHIP-I235A or −R272A markedly increased the level of GFPiNOS (Figure 4C-E), as did co-transfection with CHIP-V59D/L129D (Figure 4C-E). These data thus support the functional importance of the lid:TPR interaction for proper CHIP-mediated ubiquitination and degradation of Hsp70/Hsc70 clients.
Regulation by Post-translational Modification of Hsp70
Several studies have reported that the Hsp70/Hsc70-lid subdomain and tail are post-translationally modified. T636 phosphorylation within the Hsp70 C-terminal GPTIEEVD motif decreases affinity for CHIP and favors binding of Hop, a TPR-domain-containing cochaperone that links Hsp70/Hsc70 and Hsp90 (Muller et al., 2013). Modeling of a phosphothreonine at the corresponding position in our crystal structure suggests that the phosphate group clashes with residues on the seventh helix of the TPR domain (Figure S5A,B). Intriguingly, multiple proteomic studies also identify Y611 as a phosphorylation site (Molina et al., 2007; Ruse et al., 2008). Y611 is located on the Hsp70/Hsc70 lid subdomain and interacts with the CHIP-TPR domain and the carboxylate clamp (Figure 1). In agreement with our structure, a phosphomimetic Y611E mutation disrupts ubiquitination of Hsc70 SBDβ-lid-tail construct (Figure 5A-B).
Figure 5. Posttranslational modifications of Hsc70 residues modulate CHIP-mediated ubiquitination.
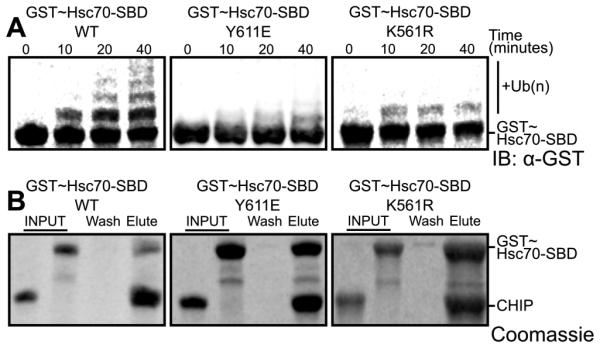
(A) Western blot with anti-GST antibody to detect ubiquitination of GST~Hsc70395-646Y611E phosphomimic, GST~Hsc70395-646K561R -dimethyl-mimetic in comparison to wild type (WT) GST~Hsc70395-646 by wild type CHIP1-303.
(B) Pull-down of GST~Hsc70395-646 Y611E and K561R mutant constructs by wild type His6-CHIP1-303 bound to Ni2+ mag-sepharose beads, analyzed by SDS PAGE with Coomassie staining. Lanes indicate the GST~Hsc70395-646 and CHIP1-303 components utilized (INPUT), the final wash fraction (Wash) and the elution fraction (Elute). See also Figure S5.
Several lysine methyltransferases (KMTs), including SETD1A (Cho et al., 2012) and METTL21A (Cloutier et al., 2013) methylate Hsp70/Hsc70-K561. METTL21A is a member of the novel METTL21 family, which may solely target Hsp70 family members (Jakobsson et al., 2013). Based on its location, we investigated whether K561 methylation perturbs the lid:TPR interaction. To mimic dimethylated K561, we used a K561R mutation, which our structure suggests could perturb the docking of the Hsc70-lid without disrupting the TPR/Hsc70-tail interaction (Figure S5C-E). Compared to the wild-type GST-SBDβ-lid-tail construct, the K561R mutant indeed exhibited decreased ubiquitination by CHIP (Figure 5A-B).
Modeling Full-Length Hsp70:CHIP:E2~Ubi Complexes
To better understand how the lid-tail:TPR interaction fits into the assemblies formed by Hsp70 and CHIP, we generated models of Hsp70:CHIP:UbcH5b~Ub conjugate complexes. We combined our new structure with recent structures of full-length Hsp70 family members in ADP-bound or ATP-bound conformations (Bertelsen et al., 2009; Kityk et al., 2012), RING or U-box domains complexed with E2~Ubiquitin conjugates (Dou et al., 2012; Plechanovová et al., 2012; Pruneda et al., 2012) and our previously determined structure of a UbcH5b bound to the U-box domain of CHIP (Xu et al., 2008a)(Figure 6). We utilized the asymmetric crystal structure of the CHIP dimer (Zhang et al., 2005), the only currently available structure of full-length CHIP, although there is substantial evidence that CHIP is structurally dynamic and adopts other conformations (Graf et al., 2010; Qian et al., 2009; Xu et al., 2006). The asymmetric CHIP dimer does not clash with the chaperone, regardless of which of its TPR domains serves as the docking site for Hsp70ADP or Hsp70ATP (Figure 6). However, CHIP and the bound E2~Ub conjugate are substantially closer (~50-60 Å) to the substrate-binding groove of the Hsp70-SBDβ subdomain in Hsp70ADP compared to Hsp70ATP. In the latter, an extensive reorientation of the lid subdomain places CHIP and the SBDβ subdomain (client-binding site) on opposite sides of the NBD.
Figure 6. Models of Chaperoned Ubiquitination Complexes in ADP- and ATP-Bound States.
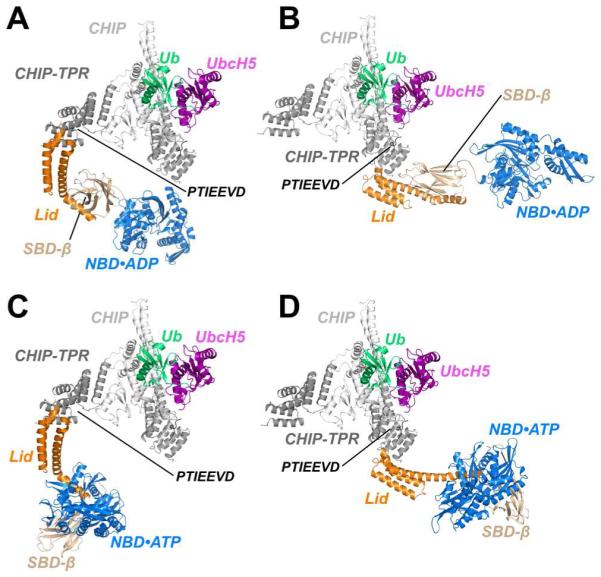
(A) A model of Hsp70, based on the structure of ADP-bound E. coli DnaK (Bertelsen et al., 2009) is fully compatible with binding to the TPR domain of CHIP protomer with an occluded U-box. CHIP, CHIP-TPR, UbcH5, Ub, Hsc70-Lid, Hsc70-Tail, Hsc70-SBDβ and Hsc70-NBD are colored white, grey, purple, green, orange, black, wheat and blue, respectively.
(B) ADP-bound Hsp70 is also compatible with binding to TPR domain of CHIP protomer with accessible U-box.
(C) A model of ATP-bound Hsp70, based on the structure of DnaK in the ATP-bound form (Qi et al., 2013), is compatible with binding to CHIP via the TPR domain of the protomer with an occluded U-box.
(D) The CHIP-TPR from a protomer with an accessible U-box is also compatible with binding to ATP-bound Hsp70.
These models, as well as the fact that Hsp70ADP exhibits higher client affinity and slower client dissociation rate than Hsp70ATP (Mayer and Bukau, 2005), support the notion that CHIP-mediated ubiquitination of chaperone-bound clients is highly favored in the Hsp70ADP conformation relative to the Hsp70ATP conformation. Intriguingly, either Hsp40 (which accelerates ATP hydrolysis by Hsp70) or a mutation that traps Hsp70 in an ADP-bound conformation have been shown to increase client ubiquitination (Kundrat and Regan, 2010a; Stankiewicz et al., 2010).
DISCUSSION
To date, the binding of the C-terminal motifs of Hsp70/Hsc70 or Hsp90 to TPR domains from several distinct cochaperones have been characterized. However, TPR domains are versatile interaction platforms; some TPR domains bind helices and globular domains rather than, or in addition to, peptide motifs (Allan and Ratajczak, 2011; D'Andrea and Regan, 2003). For example, TPR domains of CyP40, FKBP52, Protein Phosphatase 5 (PP5) and LGN exhibit intra-molecular packing (Pan et al., 2013; Taylor et al., 2001; Wu et al., 2004; Yang et al., 2005). The PP5-TPR domain occludes the PP5-phosphatase domain (Figure S6B); binding of the C-terminal motif of Hsp90 or Hsp70 to the PP5-TPR domain relieves this inhibition (Connarn et al., 2014; Yang et al., 2005). The TPR domain of the neutrophil oxidase assembly subunit p67phox (which is not a cochaperone) interacts with the small G-protein Rac (Lapouge et al., 2000). In this interaction (Figure S6C), an atypical β-stranded hairpin insertion in the p67phox TPR domain forms much of the Rac-binding surface, while the peptide-binding groove engages in an intramolecular interaction with the C-terminal “tail” of p67phox itself.
Our new structure showcases a novel configuration in which a cochaperone TPR domain simultaneously engages two portions of the Hsp70/Hsc70 chaperone. A detailed recent study by Zuiderweg, Gestwicki and coworkers suggested that the PTIEEVD motif:TPR interaction contributes most of the binding energy between CHIP and Hsc70 (Smith et al., 2013). NMR relaxation data suggested that the Hsp70 C-terminal tail is disordered and highly dynamic, potentially leading Hsc70 and CHIP to behave purely as a tethered, “beads-on-a-string” complex. However, these investigators also found evidence for supplementary contacts between the proteins. Importantly, the ~60 nM affinity for the full-length Hsc70:CHIP complex is reduced by an order of magnitude to ~400 nM - 2 μM affinity of CHIP for either the Hsp70-GPTIEEVD peptide alone, or for an Hsc70-Δtail-IEEVD construct (Kundrat and Regan, 2010b; Smith et al., 2013). Our structure provides an explanation for the “missing” affinity: modest but functionally important interactions between the TPR domain and the lid supplement the canonical TPR interactions with the chaperone C-terminal motif.
A mass spectrometry study identified six CHIP-ubiquitinated lysine residues on Hsp70, five of which are located within the SBD (Kundrat and Regan, 2010a). Similarly, Goldberg and coworkers found that CHIP ubiquitinates an Hsp70-bound client (heat-denatured luciferase) at very few lysines or possibly just one lysine residue (Kim et al., 2007). These amount to a far smaller subset of Hsp70 or client lysine residues than would be ubiquitinated given an unrestrained interaction of Hsp70 and CHIP through the flexible ~100 Å-long Hsp70 tail. While the C-terminal GPTIEEVD motif of Hsp70/Hsc70 provides a high affinity CHIP binding site, these studies also point to additional dynamic interactions that restrict orientations of CHIP relative to Hsp70/Hsc70, in agreement with the data of Smith et al. (2013) and with our new findings.
We could not directly measure the affinity of the lid:TPR interaction by NMR, though we estimate that it is substantially weaker than 100 μM. This interaction likely occurs transiently in the context of the tethered Hsc70:CHIP complex. In comparison, affinities between Hsp70 and model substrates range between ~100 nM and 10 mM, though bona fide clients likely bind to Hsp70ADP with affinities at the lower (nanomolar) end of this range (Greene et al., 1995; Maeda et al., 2007; Mayer et al., 2000). Intrinsic lifetimes of Hsp70ADP:client complexes are in the range of tens of minutes (Mayer et al., 2000; Stankiewicz et al., 2010) and the lifetime of Hsp70:CHIP complexes lies in the range of minutes (Smith et al., 2013). CHIP may “dock” and “undock” from the Hsp70/Hsc70 lid numerous times while remaining tethered via the chaperone C-terminal motif, before client release and Hsp70 conformational changes are driven by binding of a nucleotide exchange factor (Mayer and Bukau, 2005).
Hsc70:CHIP affinity is stronger than the reported 1~3 μM affinity between CHIP and E2~ubiquitin conjugates (Graf et al., 2010; Zhang et al., 2005). While the association and dissociation kinetics of conjugates from CHIP have not been measured, lifetimes of other ubiquitin ligase:E2 complexes are reported to be in the range of seconds with relatively rapid dissociation kinetics of 10-100 s−1 (Kleiger et al., 2009). This suggests that Hsp70:CHIP or Hsp70:client:CHIP complexes remain stably associated while E2~ubiquitin conjugates bind and dissociate repeatedly.
In ubiquitination mediated by E3 ligases containing RING or U-box domains, these domains recruit E2 enzyme~ubiquitin conjugates for direct ubiquitin transfer to substrate lysines. The RING/U-box domain holds the conjugate in a catalytically competent conformation (Dou et al., 2012; Plechanovová et al., 2012; Pruneda et al., 2012) while other domains of the ligase bind substrates or substrate adaptors. However, ubiquitin transfer requires a productive encounter complex in which a substrate lysine attacks the thioester bond between the ubiquitin C-terminus and the active-site Cysteine of the E2 enzyme (Dou et al., 2012). We suggest that the Hsc70-lid-tail:CHIP-TPR interactions reduce the orientational freedom of CHIP:E2~Ubiquitin relative to Hsp70/Hsc70, and increase the probability of such an encounter complex forming during the lifetime of the Hsp70:CHIP:E2~Ubiquitin complex. The kinetic interplay between docking of the CHIP-TPR onto the Hsp70/Hsc70 lid and the recruitment of E2~ubiquitin conjugates may thus play a part in regulating the kinetics and efficiency of CHIP-mediated ubiquitination.
In our models of Hsp70ADP:CHIP:E2~Ubiquitin conjugate complexes (Figure 6) the conjugate thioester is ~50-60 Å from the peptide-binding site of the SBDβ subdomain or from in-vitro-mapped CHIP ubiquitination sites on Hsp70. An extended client bound to the Hsp70-SBD could span this gap and thus directly encounter the E2~Ubiquitin thioester. We used a crystal structure of near-full-length (murine) CHIP in these models (Zhang et al., 2005); in this structure, CHIP is an asymmetric dimer with different relative positions of the TPR and U-box domains in each protomer. However, several studies provided evidence that CHIP adopts other conformations, and that CHIP dynamics and conformational changes strongly influence ubiquitination (Graf et al., 2010; Qian et al., 2009). The CHIP dimer could alternate between asymmetric conformations (Qian et al., 2009; Xu et al., 2006), modulating the distance between TPR and U-box domains and allowing the Hsp70-SBD to approach the E2~Ub conjugate thioester more closely than in our current models. Trapping and structurally elucidating other conformations of CHIP will help to achieve a fuller understanding of CHIP’s ubiquitination mechanisms. However, we note that dynamics and relative motion of substrate-binding domains with respect to E2-binding domains are well known to regulate ubiquitination by other E3 ubiquitin ligases including the HECT- and Cullin-RING-ligase superfamilies (Berndsen and Wolberger, 2014; Lydeard et al., 2013).
Might the formation of functionally important bipartite or secondary contacts be a more general feature of interactions between chaperones and TPR-domain cochaperones? The cochaperone Hop acts as a bridge between Hsp70 and Hsp90 and mediates transfer of clients between the two chaperones (Carrigan et al., 2004; Flom et al., 2007; Scheufler et al., 2000; Wegele et al., 2006). Recent studies identified non-canonical interactions between Hop’s TPR domains and Hsp90 (Alvira et al., 2014; Scheufler et al., 2000; Schmid et al., 2012). Of the three Hop TPR domains, TPR1 and TPR2b can interact with the Hsp70 C-terminal motif while TPR2a binds to the Hsp90 C-terminal motif. Upon binding of TPR2a to the Hsp90 C-terminal motif, the TPR2a and TPR2b appear to engage in additional packing against the Middle and C-terminal domains of Hsp90 (Schmid et al., 2012). Whether CHIP also interacts with Hsp90 at sites other than the Hsp90 C-terminal motif is as yet unclear.
A related question is whether other Hsp70-interacting TPR domains engage Hsp70 through a bipartite interaction. Using the Hsc70-Lid-tail:CHIP-TPR complex as a template for RosettaDock (Chaudhury et al., 2011), we docked the Hsc70-Lid with TPR1 and TPR2B of Hop and with the sole TPR domain of the cochaperone SGTA (Figure S6D-H). All three TPR domains have substantial structural and sequence similarity to the CHIP-TPR (Dutta and Tan, 2008; Scheufler et al., 2000; Schmid et al., 2012). Lid interactions with these TPR domains exhibit similar interface energies as with CHIP-TPR, and the lowest-energy docking poses of the Hsc70 lid adopt similar orientations. Interestingly, the putative binding mode of Hop-TPR2b on the Hsp90 Middle domain does not occlude its GPTIEEVD-binding groove nor the potential lid-binding interface (Schmid et al., 2012). Simultaneous interaction of TPR2b with the Hsp90 middle domain and the Hsp70 lid and C-terminal motif would lead to compact association between Hsp90 and Hsp70, potentially facilitating substrate transfer from one chaperone to the other. Very recently, 23 Å resolution electron microscopy structures of ‘extended’ and ‘compact’ Hsp90:Hop:Hsp70 complexes (Alvira et al., 2014) suggested the presence of general contacts between the Hsp70-lid and Hop-TPR1 in the ‘extended’ complex or Hop-TPR2b in the ‘compact’ complex. Functional and high-resolution structural characterizations of potential bipartite interactions between TPR domains of Hop or other cochaperones with Hsp70, or bipartite CHIP interactions with Hsp90, await further investigation. These future studies may reveal unanticipated aspects of how such cochaperones cooperate with and regulate their partner chaperones.
Experimental Procedures
Vector construction, protein expression and purification are described in Supplemental Experimental Procedures.
Crystallization, X-Ray Data Collection and Processing
A mixture of 2 mM HsCHIP21-154 (CHIP-TPR) and 2 mM HsHsc70541-646 Δ626-638 (Hsc70-lid-tail) was used to grow crystals by hanging drop vapor diffusion at room temperature against a well solution of 0.1 M HEPES (pH 7.0) and 1.7 M ammonium citrate. Crystals grew from hanging drop mixtures of 0.5 μl protein buffer and 0.5 μl crystallization solution, appearing overnight and growing to full size within one week. Crystals were cryo-protected by brief transfer through LV CryoOil (MiTeGen) and flash-frozen in liquid nitrogen. X-ray diffraction data were integrated and scaled using XDS (Kabsch, 2010) and SCALA (Evans, 2006).
Structure Determination and Refinement
Structures of CHIP-TPR (PDB code: 2C2L residues 21-154; Zhang et al., 2005) and Hsp70-lid (PDB code: 3LOF residues 541-610) were used as molecular replacement search models in PHASER (McCoy et al., 2007). CHIP-TPR and Hsc70-lid chains were rebuilt with RESOLVE (Terwilliger, 2003). Iterative refinement and model building were conducted with PHENIX (Adams et al., 2010) and Coot (Emsley et al., 2010).
Ubiquitination Assays
E1/E2~Ub reaction mixtures were pre-charged by incubating 40 μM UbcH5b, 100 μM Ub and 0.5 μM E1 for 30 minutes at 37°C in 50 mM HE PES (pH7.0), 50 mM NaCl, 20 mM ATP, 40 mM MgCl2 and 0.5 mM DTT. Ubiquitination of Hsc70 substrate binding domain-tail construct used pre-charged E1/E2~Ub reaction mixture added to a solution of 4 μM GST-Hsc70395-646 and 4 μM CHIP in 50 mM NaCl, 50 mM HEPES (pH 7.0). CHIP-autoubiquitination utilized pre-charged E1/E2~Ub reaction mixture added to 4 μM CHIP in 50 mM NaCl, 50 mM HEPES (pH 7.0), 0.5 mM DTT. Ubiquitination reactions were incubated at 37°C and stopped at specified tim e points by addition of 2x SDS sample buffer containing 20 mM DTT and 50 mM EDTA. Quenched reactions were subjected to SDS PAGE and Western Blotting with near-infrared detection using anti-CHIP or anti-GST primary antibodies and IRDye-labeled secondary antibodies (LI-COR).
Cell Culture Assays – regulation of iNOS levels by CHIP
RAW264.7 (RAW) cells (ATCC, Rockville, MD) were cultured as previously described (Chakravarti and Stuehr, 2012). Cells were transfected with 4 μg of myc-tagged pCDNA3-CHIP or empty pCDNA3 expression vector using Lipofectamine (Life Technologies Inc.) and incubated for 24h at 37°C an d 5% CO2 in a humidified incubator. Cells were activated by 25μM LPS (Sigma) and 10 units/ml of IFN-γ (Pepro-Tech) at 37°C and 5% CO 2 in a humidified incubator for defined time periods. The cells were lysed and supernatants were prepared as described before (Chakravarti et al., 2010). 50 μg total protein per supernatant was separated by SDS PAGE and western blotting was performed using antibody against iNOS (BD Transduction laboratory), CHIP (Sigma) or GAPDH (Fitzgerald).
HEK293 cells were cultured as described previously (Scaglione et al., 2011). Cells were transfected using Lipofectamine-2000 (Invitrogen). Fluorescent images of transfected HEK293 cells, imaged at 20X magnification, were acquired using a FL Auto Cell Imaging System (EVOS). HEK293 cells for western blotting were lysed at 95°C in Laemmli buffer for 4 min, sonicated, and loaded on SDS-PAGE gels. Western blots utilized anti-CHIP, anti-GFP or anti-β-Actin and near-infrared detection with IRDye-labeled secondary antibodies (LI-COR) imaged using an Odyssey Fc Imager (LI-COR). For semiquantification, images were collected at below-saturation levels and quantified with Image Studio (LI-COR). Background was subtracted equally among lanes. Student's t-test was used for statistical analyses using Prism (Graphpad).
Supplementary Material
Highlights.
Hsc70/Hsp70 engage in novel bipartite binding mode with CHIP
Hsp70-lid interaction with CHIP is required for ubiquitination of Hsp70 clients
TPR:lid-tail structure allows modeling of full-length Hsp70:CHIP complexes
Phosphorylation or Methylation of Hsp70-lid residues regulate interaction with CHIP
Acknowledgments
The authors acknowledge financial support from the US National Institutes of Health (R01-GM080271 to S.M., R00-NS073936 to K.M.S. and T32-HL007914 to R.C.P.). R.C.P. was also supported by Burroughs Wellcome Foundation Collaborative Research Travel Grant 1014031 and institutional funds from Miami University. KMS was also funded through the Research and Education Program, a component of the Advancing a Healthier Wisconsin endowment at the Medical College of Wisconsin. The Advanced Light Source is supported by the US Department of Energy under contract number DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The structural coordinates of the CHIP-TPR/Hsc70-LidTail complex were deposited in the PDB under the ID code 4KBQ.
Supplemental Information
Supplemental Information including Supplemental Experimental Procedures and six figures can be found online with this article.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvira S, Cuéllar J, Röhl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Bertelsen EB, Zhou H, Lowry DF, Flynn GC, Dahlquist FW. Topology and dynamics of the 10 kDa C-terminal domain of DnaK in solution. Protein Sci. 1999;8:343–354. doi: 10.1110/ps.8.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.a. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Carrigan PE, Nelson GM, Roberts PJ, Stoffer J, Riggs DL, Smith DF. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 2004;279:16185–16193. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- Chakravarti R, Stuehr DJ. Thioredoxin-1 regulates cellular heme insertion by controlling S-nitrosation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 2012;287:16179–16186. doi: 10.1074/jbc.M112.342758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.a. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS ONE. 2011;6:e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kong X, Fu J, Xu Y, Fang S, Hua P, Luo L, Yin Z. CHIP facilitates ubiquitination of inducible nitric oxide synthase and promotes its proteasomal degradation. Cell. Immunol. 2009;258:38–43. doi: 10.1016/j.cellimm.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-S, Shimazu T, Toyokawa G, Daigo Y, Maehara Y, Hayami S, Ito A, Masuda K, Ikawa N, Field HI, et al. Enhanced HSP70 lysine methylation promotes proliferation of cancer cells through activation of Aurora kinase B. Nat. Commun. 2012;3:1072. doi: 10.1038/ncomms2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Lavallée-Adam M, Faubert D, Blanchette M, and Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connarn JN, Assimon VA, Reed RA, Tse E, Southworth DR, Zuiderweg ERP, Gestwicki JE, and Sun D. The molecular chaperone Hsp70 activates protein phosphatase 5 (PP5) by binding the tetratricopeptide repeat (TPR) domain. J. Biol. Chem. 2014;289:2908–2917. doi: 10.1074/jbc.M113.519421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Höhfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Tan Y-J. Structural and functional characterization of human SGT and its interaction with Vpu of the human immunodeficiency virus type 1. Biochemistry. 2008;47:10123–10131. doi: 10.1021/bi800758a. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf C, Stankiewicz M, Nikolay R, Mayer MP. Insights into the conformational dynamics of the E3 ubiquitin ligase CHIP in complex with chaperones and E2 enzymes. Biochemistry. 2010;49:2121–2129. doi: 10.1021/bi901829f. [DOI] [PubMed] [Google Scholar]
- Greene LE, Zinner R, Naficy S, Eisenberg E. Effect of nucleotide on the binding of peptides to 70-kDa heat shock protein. J. Biol. Chem. 1995;270:2967–2973. doi: 10.1074/jbc.270.7.2967. [DOI] [PubMed] [Google Scholar]
- He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J. Biol. Chem. 2004;279:30643–30653. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- Jakobsson ME, Moen A, Bousset L, Egge-Jacobsen W, Kernstock S, Melki R, Falnes PØ. Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 2013;288:27752–27763. doi: 10.1074/jbc.M113.483248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kityk R, Kopp J, Sinning I, Mayer MP. Structure and Dynamics of the ATP-Bound Open Conformation of Hsp70 Chaperones. Mol. Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L, Regan L. Identification of residues on Hsp70 and Hsp90 ubiquitinated by the cochaperone CHIP. J. Mol. Biol. 2010a;395:587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010b;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Smith SJ, Walker PA, Gamblin SJ, Smerdon SJ, Rittinger K. Structure of the TPR domain of p67phox in complex with RacβGTP. Mol. Cell. 2000;6:899–907. doi: 10.1016/s1097-2765(05)00091-2. [DOI] [PubMed] [Google Scholar]
- Li J, Buchner J. Structure, function and regulation of the hsp90 machinery. Biomed. J. 2013;36:106–117. doi: 10.4103/2319-4170.113230. [DOI] [PubMed] [Google Scholar]
- Li Z, Hartl FU, Bracher A. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 2013;20:929–935. doi: 10.1038/nsmb.2608. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Sahara H, Mori Y, Torigo T, Kamiguchi K, Tamura Y, Tamura Y, Hirata K, Sato N. Biological heterogeneity of the peptide-binding motif of the 70-kDa heat shock protein by surface plasmon resonance analysis. J. Biol. Chem. 2007;282:26956–26962. doi: 10.1074/jbc.M703436200. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Schröder H, Rüdiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.a. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolay R, Wiederkehr T, Rist W, Kramer G, Mayer MP, Bukau B. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 2004;279:2673–2678. doi: 10.1074/jbc.M311112200. [DOI] [PubMed] [Google Scholar]
- Pan Z, Zhu J, Shang Y, Wei Z, Jia M, Xia C, Wen W, Wang W, Zhang M. An Autoinhibited Conformation of LGN Reveals a Distinct Interaction Mode between GoLoco Motifs and TPR Motifs. Structure. 2013;21:1007–1017. doi: 10.1016/j.str.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Peng H-M, Morishima Y, Jenkins GJ, Dunbar AY, Lau M, Patterson C, Pratt WB, Osawa Y. Ubiquitylation of neuronal nitric-oxide synthase by CHIP, a chaperone-dependent E3 ligase. J. Biol. Chem. 2004;279:52970–52977. doi: 10.1074/jbc.M406926200. [DOI] [PubMed] [Google Scholar]
- Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya S, Sharma SK, Goloubinoff P. Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 2013;587:1981–1987. doi: 10.1016/j.febslet.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. Structure of an E3:E2 Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. Mol. Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, Yang J, Wong JL, Vorvis C, Hendrickson WA, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S-B, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S-B, Waldron L, Choudhary N, Klevit RE, Chazin WJ, Patterson C. Engineering a ubiquitin ligase reveals conformational flexibility required for ubiquitin transfer. J. Biol. Chem. 2009;284:26797–26802. doi: 10.1074/jbc.M109.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser MFN, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- Röhl A, Rohrberg J, Buchner J. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Ruse CI, McClatchy DB, Lu B, Cociorva D, Motoyama A, Park SK, and Yates JR. Motif-specific sampling of phosphoproteomes. J. Proteome Res. 2008;7:2140–2150. doi: 10.1021/pr800147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng J, et al. Ube2w and Ataxin-3 Coordinately Regulate the Ubiquitin Ligase CHIP. Mol. Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- Schmid AB, Lagleder S, Gräwert MA, Röhl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M, et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. Embo J. 2012;31:1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Scaglione KM, Assimon VA, Patury S, Thompson AD, Dickey CA, Southworth DR, Paulson HL, Gestwicki JE, Zuiderweg ERP. The E3 ubiquitin ligase CHIP and the molecular chaperone Hsc70 form a dynamic, tethered complex. Biochemistry. 2013;52:5354–5364. doi: 10.1021/bi4009209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. Febs J. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- Taylor P, Dornan J, Carrello A, Minchin RF, Ratajczak T, Walkinshaw MD. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure. 2001;9:431–438. doi: 10.1016/s0969-2126(01)00603-7. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Meth. Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu Y-T, Hao R, Chen L, Chang Z, Wang H-R, Wang Z-X, Wu J-W. Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-interacting protein (CHIP) J. Biol. Chem. 2011;286:15883–15894. doi: 10.1074/jbc.M110.201814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, Rao Z. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci. U.S.a. 2004;101:8348–8353. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Liu FH, Hu SM, Wang C. Different combinations of the heat-shock cognate protein 70 (hsc70) C-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem. J. 2001;359:419–426. doi: 10.1042/0264-6021:3590419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. U.S.a. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Devlin KI, Ford MG, Nix JC, Qin J, Misra S. Structure and interactions of the helical and U-box domains of CHIP, the C terminus of HSP70 interacting protein. Biochemistry. 2006;45:4749–4759. doi: 10.1021/bi0601508. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol. 2008a;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Page RC, Gomes MM, Kohli E, Nix JC, Herr AB, Patterson C, Misra S. Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 2008b;15:1309–1317. doi: 10.1038/nsmb.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Roe SM, Cliff MJ, Williams MA, Ladbury JE, Cohen PTW, Barford D. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. Embo J. 2005;24:1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned Ubiquitylation—Crystal Structures of the CHIP U Box E3 Ubiquitin Ligase and a CHIP-Ubc13-Uev1a Complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.