Abstract
Background. The diversification of human immunodeficiency virus (HIV) is shaped by its transmission history. We therefore used a population based province wide HIV drug resistance database in British Columbia (BC), Canada, to evaluate the impact of clinical, demographic, and behavioral factors on rates of HIV transmission.
Methods. We reconstructed molecular phylogenies from 27 296 anonymized bulk HIV pol sequences representing 7747 individuals in BC—about half the estimated HIV prevalence in BC. Infections were grouped into clusters based on phylogenetic distances, as a proxy for variation in transmission rates. Rates of cluster expansion were reconstructed from estimated dates of HIV seroconversion.
Results. Our criteria grouped 4431 individuals into 744 clusters largely separated with respect to risk factors, including large established clusters predominated by injection drug users and more-recently emerging clusters comprising men who have sex with men. The mean log10 viral load of an individual's phylogenetic neighborhood (composed of 5 other individuals with shortest phylogenetic distances) increased their odds of appearing in a cluster by >2-fold per log10 viruses per milliliter.
Conclusions. Hotspots of ongoing HIV transmission can be characterized in near real time by the secondary analysis of HIV resistance genotypes, providing an important potential resource for targeting public health initiatives for HIV prevention.
Keywords: molecular epidemiology, human immunodeficiency virus (HIV), phylogenetic clustering, transmission network, injection drug use, men who have sex with men (MSM)
(See the editorial commentary by Frost and Pillay on pages 856–8.)
In the developed world, men who have sex with men (MSM), injection drug users, and individuals who engage in survival sex work have disproportionately higher rates of human immunodeficiency virus (HIV) infection. In Vancouver, Canada, the majority of infections during the initial rise of the epidemic, in the 1980s, could be attributed to transmissions among MSM [1]. The incidence of HIV infection among MSM subsided in the late 1980s [2], even before the advent of antiretroviral therapy. In the mid-1990s, a new wave emerged, in large part because of the spread of HIV among injection drug users in Vancouver [3, 4]. During this period, the estimated prevalence of HIV infection among women engaged in survival sex work was even greater than that among the injection drug user population as a whole [5]. Thus, the HIV epidemic in Vancouver has been complex, with extensive heterogeneity in modes and rates of HIV transmission among risk groups.
Molecular phylogenetics can provide the tools to infer the fine structure of an epidemic, especially for rapidly evolving pathogens, such as HIV [6–11]. A molecular phylogeny is a tree-based model of how genetic sequences are related by common ancestors. Phylogenies reconstructed from HIV sequences are shaped by the transmission history of the virus because the rapidly evolving virus populations can measurably diverge in their genetic makeup on the time scale of transmission, owing to host-specific selection [6]. When virus populations in 2 infections retain a high degree of genetic similarity, one can infer that they are related by 1 or more recent transmission event. Since detection of HIV drug resistance by sequencing (genotyping) and subsequent modification of a patient's drug regimen have been shown to significantly improve virological outcomes [12, 13], routine HIV genotyping has become standard of care in the developed world. Consequently, enormous amounts of HIV sequence data have accumulated at centers of HIV research and primary care around the world. Moreover, current software can reconstruct phylogenies relating tens of thousands of sequences with relative ease [14]. Sociodemographic characteristics can be superimposed on phylogenies with appropriate safeguards to protect individual privacy. It is therefore feasible to perform large-scale secondary analyses of HIV resistance genotypes to extract meaningful epidemiological information from their evolutionary relationships [15–17].
In this study, we use an extensive database maintained as a part of the Drug Treatment Program (DTP) at the BC Centre for Excellence in HIV/AIDS, which is the provincial agency responsible for all fully subsidized HIV laboratory monitoring, including HIV resistance genotyping and antiretroviral therapy distribution to all HIV-infected individuals in BC. To date, approximately 11 000 individuals, roughly 75% of all individuals (n = 14 054) with a new diagnosis of HIV infection between 1985 and 2011, have enrolled in the DTP [18]. Under current provincial treatment guidelines, the BC Centre laboratory performs an HIV resistance genotype test on all baseline samples submitted for viral load testing and in selected samples derived from patients receiving therapy who experience virologic failure. Over 27 000 resistance genotypes have been generated for >7700 individuals in the DTP. We therefore used this extensive and centralized database combining HIV drug resistance and sociodemographic data from the DTP to reconstruct the dynamics of the regional epidemic and characterize the impact of clinical, demographic, and behavioral factors on rates of HIV transmission.
METHODS
Data Collection
Ethical approval for this study was granted by the Providence Health Care/University of British Columbia Research Ethics Board (H07-02,559). At the time of analysis, there were 27 296 HIV resistance genotype tests corresponding to 7747 individuals in BC. The majority of individuals were represented by multiple HIV sequences (mean, 3.5 sequences/patient; range, 1–42 sequences/patient). For every individual, the earliest available sample is referred to as the baseline sample; the corresponding visit tended to mark the initiation of antiretroviral therapy. Most of these sequences (n = 24 120) spanned 1497 bp, covering HIV protease and the first 400 codons of reverse transcriptase (RT). An additional 2694 sequences were 1017 bp in length and covered protease and the first 240 codons of RT. The remaining 482 sequences were each assembled from 3 partial sequences of 276, 297, and 363 bp into a 936-bp contig spanning protease and RT codons 24–236. Sequences were reanonymized and annotated with the following information: sample collection date; date of antiretroviral therapy initiation; estimated date of HIV seroconversion, either physician-reported (n = 3912) or the midpoint between the last HIV seronegative and first HIV seropositive samples for participants in HIV prospective cohort studies (n = 200); plasma viral load; CD4+ T-cell count; HIV drug resistance levels, as predicted by the vircoTYPE algorithm [19]; sex; birth year; AIDS-defining illness before the first resistance test; having ever tested positive for hepatitis C virus infection; and risk group status (ie, any injection drug use, self-identification as a homosexual or bisexual man [ie, MSM]), any receipt of a blood (transfusion) product or exposure to any other blood risk, and exposure to any other risk). We used the SCUEAL algorithm [20] to generate HIV subtype classifications.
Phylogenetic Analysis
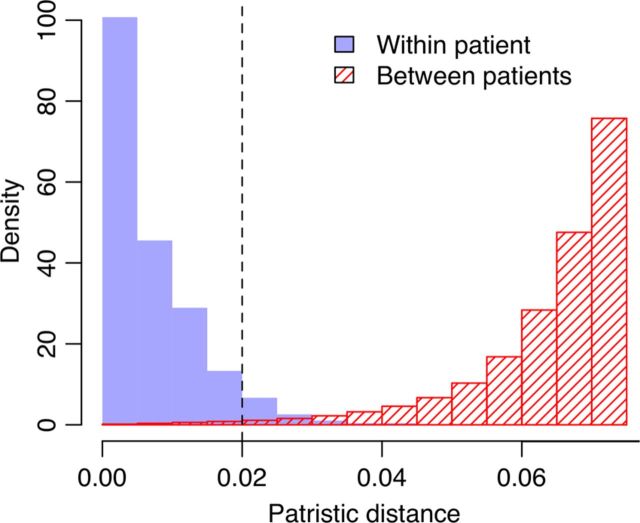
All nucleotide sequences were translated into amino acids and aligned pairwise against an HXB2 reference protein sequence (GenBank accession K03455). Codons associated with HIV surveillance drug resistance mutations based on World Health Organization definitions [21] were removed. To control for uncertainty in phylogenetic reconstruction, we generated 100 bootstrap samples by resampling columns from the alignment at random with replacement. For each bootstrap, we reconstructed a tree using the approximate maximum likelihood heuristics as implemented in FastTree2 [22]; all sequence data were then securely erased. For each tree, we identified all pairs of tips in which (1) the tip-to-tip (ie, patristic) distance was <0.02 expected nucleotide substitutions per site, (2) the tips represented HIV sequences from different infections, and (3) at least one of the sequences was derived from the earliest available sample from that individual. Below this cutoff, patristic distances between sequences from different individuals resembled the distances observed within patients (Figure 1). All pairs of tips that met these criteria in >50% of the bootstrap trees (Supplementary Figure 1) were used to construct a graph in which each node represented all HIV sequences sampled from a given individual.
Figure 1.
Comparison of the mean patristic distance between human immunodeficiency virus (HIV) sequences from the same patient (solid) against the shortest distance between patients (hatched). The distances, measured in units of expected nucleotide substitutions per site, were extracted from the maximum likelihood estimate of the phylogeny reconstructed from the original HIV sequence alignment (without bootstrap resampling). The cutoff used in our study (dashed line, 0.02 expected nucleotide substitutions per site) corresponded to the 95% quantile of intrapatient distances (0.1% quantile of interpatient distances). Histograms were scaled such that the total area sums to 1; the interpatient histogram was truncated at 0.075.
Clustering Analyses
For every individual in each bootstrapped phylogeny, we identified 5 other tips in the tree with the shortest patristic distance that corresponded to 5 different individuals. These 5 individuals composed the set of nearest neighbors to the reference individual for that phylogeny. We fit a logistic regression to the probability of observing a patristic distance below our cutoff. Model terms representing summary attributes of nearest neighbors were computed as means weighted by the inverse of their respective patristic distances to the earliest sequence of the reference individual. Since plasma viral loads and CD4+ T-cell counts from the same individual varied over time, we used the measurements obtained from the same sample as the sequence with the shortest patristic distance to the reference for nearest neighbors with multiple samples.
Analysis of Cluster Dynamics
The expansion of clusters over time was reconstructed by mapping the accumulation of individuals in each cluster to estimated dates of HIV seroconversion. Missing estimated dates of HIV seroconversion were imputed using a multiple hot-deck imputation procedure [23] (Supplementary Figure 2). Simply put, dates of HIV seroconversion were imputed by taking the difference between the estimated date of seroconversion and baseline sample date of a similar individual and applying this difference to the incomplete case. We repeated this imputation 100 times and assigned the mean imputed date as the estimated date of HIV seroconversion. Physician-reported dates of HIV seroconversion prior to 1960 (n = 2) or dates that were preceded by the earliest sample collection date (n = 143) were discarded as invalid values. Trends in cluster size and composition were analyzed in R (version 2.15.2).
RESULTS
Analysis of Clusters
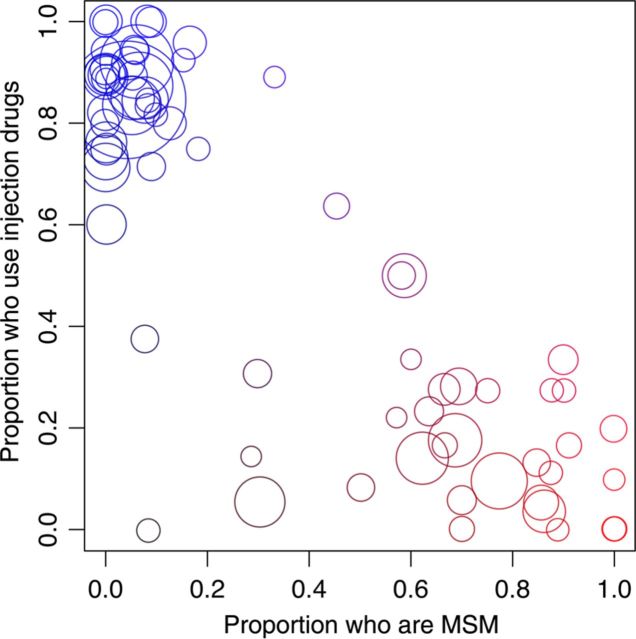
In total, 7747 HIV-infected BC residents, roughly half of the estimated individuals with HIV infection in the province, were represented by a total of 27 296 anonymized HIV pol sequences in our data. Based on the tip-to-tip (patristic) distances between sequences in 100 replicate phylogenies generated from these data, we extracted a graph comprising 744 clusters that encompassed 4431 individuals (57.2%), dominated by a single large cluster of 330 individuals. Characteristics of the total population and of individuals within clusters are summarized in Table 1. Overall, injection drug users were significantly more likely to appear in a cluster (odds ratio [OR], 3.0; P < 10−6, by the Fisher exact test), consistent with an overall higher rate of HIV transmission among injection drug users, which predominated the regional epidemic in the 1990s. A total of 93% of the infections were categorized as subtype B (Table 1); non-B subtypes were significantly less likely to appear in clusters (OR, 0.26; P < 10−6) and may represent transmissions that originated outside of BC, although we cannot exclude the possibility that non-B subtype infections were undersampled in our study population. Individuals within clusters were significantly more likely to show evidence of transmitted drug resistance (TDR; OR, 1.5; P < 7.6 × 10−5), defined as the presence of 1 or more drug resistance mutations in their earliest pretherapy sample. Note that codon positions associated with drug resistance had been removed from the alignment prior to phylogenetic reconstructions; hence, convergent evolution under drug pressure could not have played a role in this association. However, they were subsequently less likely to show evidence of drug resistance after starting therapy (OR, 0.5; P < 10−6). The sizes and individual characteristics of the 20 largest clusters are summarized in Table 2. We observed significant separation between injection drug users and MSM among clusters (Spearman ρ = −0.71; P < 10–6; Figure 2). In addition, the prevalence of TDR varied significantly among these clusters (range, 0%–21%; log-linear χ2 = 44.6; P < 7.8 × 10−4), with disproportionately high rates in clusters 9 and 12. These results were robust to varying the patristic distance and bootstrap support cutoffs used to define phylogenetic clusters (Supplementary Table 1).
Table 1.
Composition of Study Population and Individuals Within Clusters
| Characteristic (Sample Sizea) | All Subjects, No. (%) | Subjects in Cluster, No. (%) | Odds Ratiob | P Values |
|---|---|---|---|---|
| Participants | 7747 (100) | 4431 (57.2) | … | |
| Sex (n = 7338) | ||||
| Malec | 5958 (81) | … | … | |
| Femaled | 1380 (19) | … | 1.7 | <10−6 |
| Age at baseline sample collection, y (n = 7340) | 40 (33–47) | … | …e | <10−6 |
| HIV subtype | ||||
| B | 7194 (92.9) | 4280 (96.6) | 3.9 | <10−6 |
| C | 240 (3.1) | 60 (1.4) | … | |
| CRF01AE | 90 (1.2) | 26 (0.6) | … | |
| A1 | 51 (0.7) | 11 (0.2) | … | |
| D | 38 (0.5) | 10 (0.2) | … | |
| CRF02AG | 28 (0.4) | 12 (0.2) | … | |
| Other | 106 (1.4) | 32 (0.7) | … | |
| Baseline plasma viral load, log10 per mL | 4.5 (3.8–5.0) | 4.6 (3.9–5.0) | …f | <10−6 |
| Baseline CD4+ T-cell count < 500 cells/mL | 4668 (78) | 2466 (75) | 0.68 | <10−6 |
| Previous AIDS-defining illness | 923 (12) | 339 (7.6) | 0.39 | <10−6 |
| Any history of HIV drug resistance | 3351 (43.2) | 1583 (35.7) | 0.49 | <10−6 |
| Transmitted HIV drug resistance (n = 6784) | 428 (6.6) | 283 (7.7) | 1.5 | 7.6 × 10−5 |
| HCV coinfection (n = 6869) | 2695 (39.2) | 1913 (48.6) | 2.6 | <10−6 |
| HIV exposure | ||||
| Injection drug use (n = 6413) | 2725 (42.5) | 1932 (53.5) | 2.9 | <10−6 |
| Men who have sex with men (n = 5572) | 2396 (43) | 958 (30.3) | 0.29 | <10−6 |
| Heterosexual sex (n = 5572) | 1627 (29.2) | 988 (31.2) | 1.3 | 1.2 × 10−4 |
| Receipt of blood products (n = 5572) | 193 (3.5) | 75 (2.4) | 0.47 | <10−6 |
| Other (n = 5572) | 249 (4.5) | 124 (3.9) | 0.75 | .026 |
Data are no. (%) of participants or median value (interquartile range).
Abbreviations: HCV, hepatitis C virus; HIV, human immunodeficiency virus.
a Exclusive of cases with missing values.
b Univariate odds ratios between categorical variables and membership in a cluster were evaluated using the Fisher exact test.
c Transgender male to female.
d Transgender female to male.
e t = −7.9, by the univariate Student t test for continuous variables.
f t = 12.1, by the univariate Student t test for continuous variables.
Table 2.
Parameter Estimates From Fitting Sigmoidal Models to Cluster Growth Curves
| Cluster Type | IDU, % | MSM, % | TDR, % | Cluster Size |
T1/2b | Slope (Maximum)c | |
|---|---|---|---|---|---|---|---|
| Current | Predicted Maximum (95% CIa) | ||||||
| Predominantly IDU | |||||||
| 1 | 84 | 4 | 4 | 330 | 349 (342–353) | 1999 | 4.7 (24.9) |
| 2 | 92 | 6 | 4 | 129 | 138 (133–140) | 1999 | 1.8 (9.4) |
| 3 | 87 | 7 | 7 | 107 | 103 (102–104) | 1997 | 0.1 (13.2) |
| 4 | 84 | 5 | 3 | 82 | 83 (80–85) | 1997 | 0.4 (7.7) |
| 9 | 71 | 0 | 21 | 56 | 55 (53–59) | 2005 | 0.2 (4.8) |
| 10 | 84 | 8 | 2 | 51 | 50 (48–53) | 1999 | 0.4 (4.1) |
| 11 | 89 | 0 | 10 | 48 | 47 (46–49) | 1998 | 0 (3.5) |
| 12 | 73 | 0 | 15 | 47 | 78 (49–105) | 2005 | 1.9 (2.6) |
| 14 | 85 | 5 | 5 | 45 | 42 (41–43) | 1997 | 0 (7.5) |
| 16 | 90 | 0 | 5 | 42 | 44 (42–45) | 1997 | 0.1 (4.9) |
| 17 | 76 | 0 | 3 | 40 | 38 (37–40) | 1999 | 0 (5.2) |
| 18 | 60 | 0 | 0 | 37 | 36 (35–38) | 2007 | 0 (5.2) |
| 19 | 90 | 0 | 4 | 33 | 39 (35–56) | 1998 | 0.7 (2.5) |
| Predominantly MSM | |||||||
| 5 | 9 | 77 | 6 | 75 | 74 (72–77) | 2005 | 0.3 (6.1) |
| 6 | 17 | 69 | 2 | 68 | 151 (95–186) | 2013 | 4.3 (4.2) |
| 7 | 14 | 62 | 0 | 65 | 90 (68–103) | 2009 | 4.6 (6.7) |
| 8 | 5 | 30 | 12 | 60 | 62 (59–64) | 2007 | 1.5 (10.2) |
| 15 | 3 | 86 | 6 | 43 | 42 (41–43) | 2004 | 0 (4.0) |
| 20 | 28 | 70 | 0 | 32 | 68 (30–320d) | 2009 | 1.8 (1.9) |
| Mixed | |||||||
| 13 | 50 | 59 | 0 | 46 | NAe | … | … |
| Nonclustering individuals | |||||||
| 28 | 60 | 5 | 3316 | 4413 (4348–4437) | 2004 | 91.5 (129.7) | |
where x is the estimated date of HIV seroconversion for the F(x)-th individual; b controls the steepness of the curve; c and d are the minimum and maximum predicted values of F(x), respectively; log(e) corresponds to the estimated point of inflection (midpoint); and f is an exponential tuning parameter to allow for asymmetry between the upper and lower portions of the sigmoidal curve. We used the qpcR function pcrfit, which applies a weighted nonlinear least-squares minimization (Levenberg-Marquardt) algorithm to fit the full model and a constrained model in which f = 1, and selected the best-fitting model based on an F test.
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; IDU, injection drug use; MSM, men who have sex with men; NA, not available; TDR, transmitted drug resistance.
a Ninety-five percent CIs for predicted maximum cluster sizes were generated by bootstrap resampling within clusters.
where . The full model was the best fit in all cases, with the exception of clusters 14, 19, and 20.
c Slope (maximum) is defined as the mean number of HIV seroconversions per year at present (and at T1/2).
d Estimates of the CI of maximum sizes from the l4 model were numerically unstable for cluster 20, so we used interval estimates from the full model.
e Not available because none of the model fits to cluster 13 yielded meaningful parameter estimates.
Figure 2.
Scatterplot of all phylogenetic clusters with ≥10 individuals (n = 71) indicating the proportion of individuals having ever used injection drugs and the proportion reporting male-male sex. Denominators of these proportions were adjusted for cases with missing values. Each circle represents a cluster; the area of each circle is scaled in proportion to the number of individuals in that cluster. Circles are colored red and blue in proportion to the cluster-specific prevalence of MSM and injection drug use, respectively, to underscore contrasts in the composition of clusters with respect to these risk factors. Abbreviation: MSM, men who have sex with men.
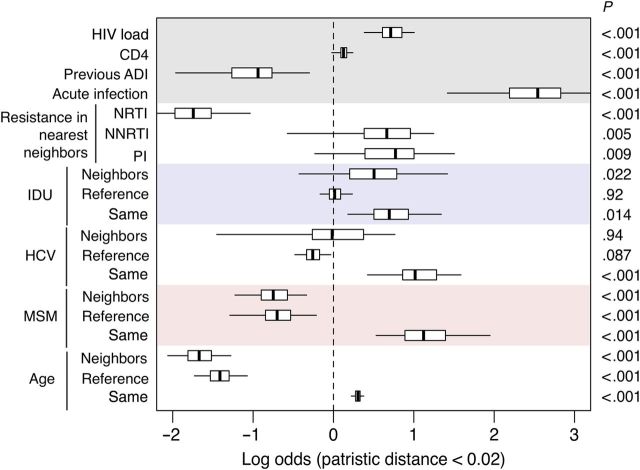
For each bootstrap phylogeny, we assessed the effect of clinical, demographic, and risk factors of each individual's 5 nearest neighbors on their odds of appearing in a cluster, which provides a rudimentary marker for localized HIV transmission rates (Figure 3). Since the majority of individuals were represented by multiple samples, for each nearest neighbor we used the clinical measurements (such as viral load and CD4+ T-cell count) associated with the sample whose HIV sequence minimized the patristic distance to the earliest sequence of the reference individual. The prevalence of acute infection among nearest neighbors was calculated as the proportion of minimum-distance samples with collection dates within 3 months of the estimated date of seroconversion. On average, the multivariate logistic models explained about 25.4% of variation (interquartile range, 24.2%–26.7%) in the odds of clustering. The odds of clustering increased significantly with the mean viral load of nearest neighbors (median OR, 2.0 per log10 increase; P < 10−6). Greater CD4+ T-cell counts (OR, 1.1 per 100 cells/mL increase; P < 10−6), lower prevalence of previous AIDS-defining illness (OR, 0.39; P < 10−6), and the prevalence of acute infection (OR, 12.7, P < 10−6) in nearest neighbors were also associated with an elevated odds of clustering. These associations were consistent with a tendency to transmit at an early or acute stage of HIV infection. Furthermore, the odds of clustering declined with the prevalence of nucleoside reverse transcriptase inhibitor (NRTI) resistance mutations in nearest neighbors (OR, 0.17; P < 10−6). The absence of a similar effect of resistance mutations for other drug classes suggested that the substantial virus fitness cost of specific mutations, such as M184V, may affect the rate of transmission through their impact on viral load [25]. Indeed, when we repeated our analyses after substituting the presence or absence of M184V for the NRTI model term, we found that the estimated effect size of this mutation-specific term on the odds of clustering was even stronger (OR, 0.09; P < 10−6; Table 3).
Figure 3.
Summary of multivariate logistic regressions on the odds of cluster membership. For each individual, we identified their earliest human immunodeficiency virus (HIV) sequence and then located the 5 most closely related sequences from 5 other individuals (so-called nearest neighbors) for a given phylogenetic tree. We repeated this search across all 100 bootstrap replicate trees. The purpose of this calculation was to derive predictor variables characterizing the subgroup of the population from which a given individual's infection could have originated. For example, one might expect elevated transmission rates within subgroups in which infected individuals tend to carry higher viral loads than the population average. To evaluate the impact of such predictors on variation in rates of transmission, we fit a generalized linear model with a logit link function to the odds that an individual appeared in a phylogenetic cluster (n = 4827 because of missing data). A line segment indicates the median effect, and lines are drawn to indicate the empirical 95% confidence interval. “Reference” indicates variables associated with the reference individual. “Neighbors” indicates model terms calculated from averaging the variable over nearest neighbor individuals. HIV load, CD4+ T-cell count (CD4), previous AIDS-defining illness (ADI), and acute infection also represent nearest neighbor averages. “Same” indicates an interaction effect between neighbor and reference terms; for example, when both the reference individual and their nearest neighbors were younger than the mean population age, the reference was significantly more likely to appear in a cluster. HIV load effects were scaled to log10 HIV RNA copies/mL. CD4 effects were scaled to 100 cells/mL. Age effects were scaled to decades. P values are associated with the bootstrap replicate yielding the median coefficient estimate for the respective model terms. Abbreviations: HCV, hepatitis C virus; IDU, injection drug use; MSM, men who have sex with men; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Table 3.
Summary of Results From Univariate and Multivariate Logistic Regression Analysis of Clustering Predictors in 100 Replicate Bootstrap Phylogenies
| Predictor | Univariate |
Multivariate |
||
|---|---|---|---|---|
| Median OR (95% CIa) | P Valuesb | Median OR (95% CIa) | P Valuesb | |
| HIV load | 2.72 (1.67–4.01) | <10−6 | 2.04 (1.47–2.73) | <10−6 |
| CD4+ T-cell count | 1.2 (.99–1.36) | <10−6 | 1.13 (.98–1.27) | 8.6 × 10−6 |
| Previous ADI | 0.1 (.03–.30) | <10−6 | 0.39 (.14–.74) | 1.7 × 10−4 |
| Acute infection | 22.9 (6.15–47.2) | <10−6 | 12.7 (4.1–27.3) | <10−6 |
| Resistance | ||||
| To NRTI | 0.08 (.04–.17) | <10−6 | 0.17 (.09–.36) | <10−6 |
| Mutation M184V | 0.03 (.004–.1) | <10−6 | 0.09 (.03–.22) | <10−6 |
| To NNRTI | 1.16 (.21–2.36) | 0.32 | 1.94 (.56–3.48) | .005 |
| To PI | 1.32 (.2–3.0) | 0.12 | 2.16 (.79–4.5) | .009 |
| Injection drug user | ||||
| Neighbors | 4.21 (2.54–5.94) | <10−6 | 1.66 (.65–4.13) | .02 |
| Reference | 2.61 (2.42–2.86) | <10−6 | 1.01 (.85–1.27) | .92 |
| Interaction | … | 2.0 (1.2–3.83) | .014 | |
| HCV coinfection | ||||
| Neighbors | 4.15 (2.28–5.61) | <10−6 | 0.98 (.23–2.15) | .94 |
| Reference | 2.34 (2.18–2.59) | <10−6 | 0.77 (.62–.97) | .09 |
| Interaction | … | 2.76 (1.53–4.89) | 2.0 × 10−4 | |
| MSM | ||||
| Neighbors | 0.28 (.23–.41) | <10−6 | 0.47 (.29–.72) | 1.5 × 10−5 |
| Reference | 0.31 (.3–.34) | <10−6 | 0.5 (.28–.81) | 9.3 × 10−6 |
| Interaction | … | 3.07 (1.7–7.03) | 1.3 × 10−6 | |
| Age | ||||
| Neighbors | 0.48 (.39–.71) | <10−6 | 0.19 (.13–.28) | <10−6 |
| Reference | 0.83 (.81–.84) | <10−6 | 0.24 (.18–.34) | <10−6 |
| Interaction | … | 1.36 (1.25–1.46) | <10−6 | |
Abbreviations: ADI, AIDS-defining illness; CI, confidence interval; HCV, hepatitis C virus; HIV, human immunodeficiency virus; MSM, men who have sex with men; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; OR, odds ratio; PI, protease inhibitor.
a Estimated directly from the empirical distribution.
b P values associated with the replicate logistic regression models yielding the median estimate for the respective model terms are reported.
The odds of clustering increased significantly with the prevalence of injection drug use among the nearest neighbors; this effect was exacerbated when the reference individual also used injection drugs (OR, 2.0; P = .014). HCV coinfection had a similar effect (OR, 2.8; P = 2.0 × 10−4). Conversely, the odds of clustering declined significantly with the prevalence of MSM among the nearest neighbors, and this effect was greater when the reference individual was not a MSM (OR, 3.1; P = 1.3 × 10−6). Lower ages at baseline among both nearest neighbors and reference individuals significantly increased the odds of clustering (OR, 1.4 per decade increase; P < 10−6). This may reflect a sample bias, since older individuals would have been more likely to be at a later stage of infection.
Rates of Cluster Growth
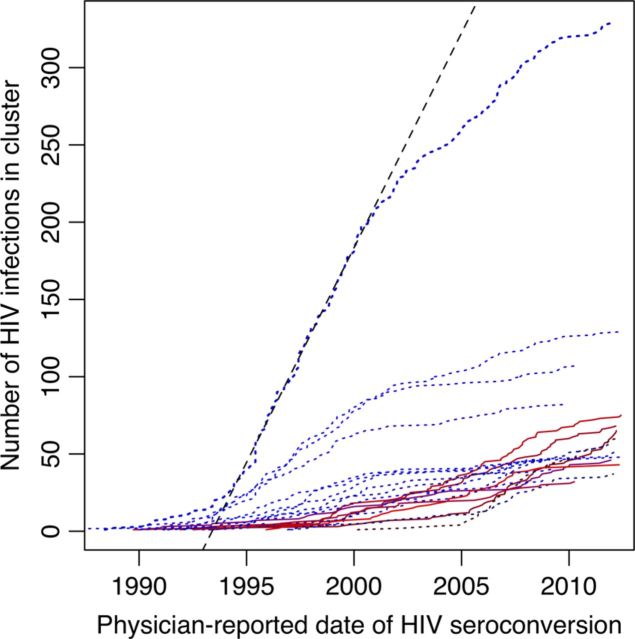
We characterized the impact of demographic and risk factors on HIV transmission rates at the level of subpopulations, as defined by clusters, by comparing the rates that clusters accumulated individuals over time (Figure 4). Note that the growth curve for each phylogenetic cluster represents the cumulative number of individuals in that cluster, regardless of whether they have survived to the present day. Based on fitting sigmoidal (ie, S-shaped) functions to each growth curve, we found that clusters comprised predominantly of injection drug users tended to have growth curves shifted to earlier dates (Spearman ρ = −0.79; P < 5.3 × 10−5; Table 2). Clusters emerging more recently were either predominantly MSM or, in few cases, a mix of both risk factors. Fitting sigmoidal functions to these data also enabled us to predict the maximum size of clusters under the implicit assumption that the respective subpopulations did not change members over time. The majority of clusters composed predominantly of injection drug users were already close to their predicted maxima, except cluster 12 (Table 2). In contrast, 3 of 6 clusters predominated by MSM were well below their respective predicted maximum sizes. For example, cluster 6 was projected to grow from its current size (68 individuals) to 151 individuals. This difference between risk groups was statistically significant (W = 63; P = .036, by the Wilcoxon rank sum test).
Figure 4.
Growth of the 20 largest phylogenetic clusters with respect to estimated dates of human immunodeficiency virus (HIV) seroconversion based predominantly on physician reports. Each trend line represents the accumulation of persons within the corresponding cluster. Dotted blue lines indicate clusters predominantly of injection drug users, and solid red lines indicate clusters predominantly of men who have sex with men (as in Table 2). These trends were averaged over 100 imputations of missing date estimates (n = 3668). A dashed line indicates the maximum rate of growth of the largest cluster, which was estimated to have occurred between 1996 and 2001.
We were unable to fit a sigmoidal model to cluster 13, which included 6 individuals who reported being both injection drug users and MSM. This is likely because the cluster has not yet transitioned from its initial phase of exponential growth, making it impossible to estimate the model parameters controlling subsequent phases of growth. For instance, it grew from 40 to 46 individuals during the 3 months that this article was being prepared for submission. This rate corresponds roughly to a slope of 24 additional HIV infections in the cluster per year, comparable to the expansion of the largest injection drug user cluster (ranked 1) in the late 1990s (Table 2). Of the 46 individuals in this cluster, 17 identified as MSM (17 with no response) and 16 as injection drug users (14 with no response). On average, individuals in this cluster were significantly younger than the mean of the study population (33 and 40 years, respectively, at baseline; W = 105; P = 3.1 × 10−6, by the Wilcoxon rank sum test), and the 6 individuals with both risk factors were even younger still (mean, 30.2 years at baseline). In addition, the estimated dates of HIV seroconversion for individuals reporting being MSM tended to map to an earlier period of slower growth of cluster 13, whereas injection drug users tended to map to a more recent period of rapid growth.
DISCUSSION
Our phylogenetic analysis of >27 000 anonymized HIV sequence records from the DTP database at the BC Centre for Excellence in HIV/AIDS has identified high rates of HIV transmission within recently emerging, distinct subgroups of the BC population that are predominated by MSM. This observation is consistent with surveillance reports in other provinces of Canada [11] and around the world [26]. However, our use of phylogenetics permitted us to investigate the structure of these emerging epidemics in greater detail, such as recognizing multiple distinct subpopulations of MSM with different rates of transmission, injection drug use, and TDR (Table 2). Furthermore, by using modern phylogenetic methods, including new techniques introduced in this article, we are able to rapidly extract cluster information from the viral sequence data collected by routine HIV resistance genotyping at the BC Centre. New sequence data are uploaded by the clinical laboratory to the database several times a week. A phylogeny can be reconstructed from the entire data set in less than an hour on a conventional computing workstation [22], and by use of our dynamic algorithm, clusters can be extracted from large phylogenies in less than a minute. Thus, the combination of routine genotyping and rapid analysis can potentially be used for the prospective monitoring of how each cluster is expanding in localized epidemics in near real time, using the same methods we have used in our retrospective analysis. However, the concurrency of such methods with ongoing dynamics in the epidemic is limited by the inevitable and often substantial delay between HIV transmission and diagnosis.
Our method of identifying phylogenetic clusters departs from the majority of previous work in this area. Most studies have defined clusters at the level of clades, where a clade comprises all descendants of a given ancestor in the tree. The conventional approach to phylogenetic clustering is to find all clades in a phylogeny that meet a number of criteria based on each clade's branch length distribution and level of bootstrap support, which quantifies the robustness of a clade to resampling data [27]. There are a number of problems with a clade-based approach to defining clusters. First, a short mean branch length may conceal a small number of long branches in the clade, such that distantly related infections become subsumed into a cluster. Second, it cannot differentiate sequences sampled from the same individual from those sampled from different individuals. Clade-based studies tend to restrict their data to the earliest sequence per individual, which can bias the analysis against transmissions from chronic infections and underestimate the size of clusters. In contrast, Wertheim et al [28] used a pairwise genetic distance to assemble clusters from pairs of individuals. We have extended this approach by extracting distances from a phylogeny, which confers greater robustness to variation in rates of evolution across the HIV genome [29]. In addition, our analysis incorporates multiple sequences per individual; excluding postbaseline sequences from our data would omit 280 of 1313 individuals (21%) whose nearest neighbors were postbaseline sequences from other individuals.
Accurately reconstructing the rates of expansion of phylogenetic clusters over time depends on the reliability of estimated dates of HIV seroconversion, which, in this study, are largely based on physician reports. Our results were qualitatively unchanged when the expansion of clusters was mapped instead to sample collection dates, which are known unambiguously. Specifically, we recovered both the large established clusters of predominately injection drug users with slowing growth and the more recently emerging clusters of predominantly MSM. The only conspicuous difference was that these trends were bounded on the left by the establishment of routine HIV resistance genotyping at the BC Centre in 1996. Although these results were achieved in part because of the extensive coverage of the regional epidemic by HIV resistance genotyping data, we note that the concepts and methods can generalize to settings with less coverage. For example, when we randomly censored 25% and 50% of our data, the percentage of people appearing in phylogenetic clusters diminished from 57% to 53% and 50%, respectively.
Phylogenetic analysis of HIV and other rapidly evolving viral pathogens has the potential to reveal how epidemics have been shaped by the composition of high-risk groups in the population. However, studies using phylogenetic methods must also take measures to minimize the risk to patient confidentiality. The same evolutionary principles that are used to identify clusters of high transmission rates in the population have also been used to prosecute individuals in legal jurisdictions where the transmission of HIV, irrespective of the actor's intent, remains criminalized, including Canada [30]. Such legislation can potentially hamper HIV prevention efforts because individuals affected by HIV may be less likely to engage in public health services [31]. Consequently, we took multiple steps to minimize the possibility that any component of our analysis could be used to reidentify individuals. Additionally, by directing our analysis on groups instead of individuals, we were able to extract epidemiologically significant information from the HIV phylogeny while further protecting individuals' confidentiality.
We therefore put forward the phylogenetic cluster, rather than the individual, as a highly effective (and safe) operational unit for the translation of phylogenetic analyses of HIV sequence data to inform public health initiatives. It should be further emphasized, in this context, that it is not possible to use phylogenetic methods to definitively prove that a specific individual was the source of 1 or more HIV transmission events [32]. However, it is feasible to identify and characterize groups burdened by a high rate of HIV transmission from a concentration of short patristic distances in the phylogeny. Clusters identified in a phylogenetic analysis can be used to define the demographic, behavioral, and/or geographical characteristics of target populations for public health interventions, including harm reduction programs, such as medically supervised injection sites [33]. Specifically, one should ideally target subgroups represented by phylogenetic clusters that are presently undergoing the highest rate of expansion and with the largest predicted remaining size (Table 2).
Our results demonstrate that secondary analysis of HIV sequences collected for routine drug resistance genotyping can be used to characterize the growth of phylogenetically related clusters. Furthermore, our results show that group-level sociodemographic characteristics of emerging phylogenetic clusters provide an important potential resource for targeting public health initiatives.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the BC Centre for Excellence in HIV/AIDS, the Canadian Institutes of Health Research (CIHR; grants HOP-111406 and HOP-107544; CIHR Vanier Canada Graduate Scholarship to C. J. B.); the US National Institute on Drug Abuse (grants 1-R01-DA036307-01, 5-R01-031055-02, R01-DA021525-06, and R01-DA011591); Genome Canada (Genomics and Personalized Health); the Michael Smith Foundation for Health Research/St. Paul's Hospital Foundation–Providence Health Care Research Institute Career Investigator Program (scholar award to A. F. Y. P.); the Canadian HIV Vaccine Initiative for Vaccine Discovery and Social Research (CHIR new investigator award to A. F. Y. P.); and CIHR/GlaxoSmithKline (research chair in clinical virology to P. R. H.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Schechter MT, Boyko WJ, Douglas B, et al. The Vancouver Lymphadenopathy-AIDS Study: 6. HIV seroconversion in a cohort of homosexual men. CMAJ. 1986;135:1355. [PMC free article] [PubMed] [Google Scholar]
- 2.Hogg RS, Strathdee SA, Craib KJ, O'Shaughnessy MV, Montaner JS, Schechter MT. Modelling the impact of HIV disease on mortality in gay and bisexual men. Int J Epidemiol. 1997;26:657–61. doi: 10.1093/ije/26.3.657. [DOI] [PubMed] [Google Scholar]
- 3.Tyndall MW, Currie S, Spittal P, et al. Intensive injection cocaine use as the primary risk factor in the Vancouver HIV-1 epidemic. AIDS. 2003;17:887–93. doi: 10.1097/00002030-200304110-00014. [DOI] [PubMed] [Google Scholar]
- 4.Hyshka E, Strathdee S, Wood E, Kerr T. Needle exchange and the HIV epidemic in Vancouver: lessons learned from 15 years of research. Int J Drug Policy. 2012;23:261–70. doi: 10.1016/j.drugpo.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McInnes CW, Druyts E, Harvard SS, et al. HIV/AIDS in Vancouver, British Columbia: a growing epidemic. Harm Reduct J. 2009;6:5. doi: 10.1186/1477-7517-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes EC, Nee S, Rambaut A, Garnett GP, Harvey PH. Revealing the history of infectious disease epidemics through phylogenetic trees. Philos Trans R Soc Lond B Biol Sci. 1995;349:33–40. doi: 10.1098/rstb.1995.0088. [DOI] [PubMed] [Google Scholar]
- 7.Leitner T, Escanilla D, Franzén C, Uhlén M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci U S A. 1996;93:10864–9. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volz EM, Koelle K, Bedford T. Viral phylodynamics. PLoS Comput Biol. 2013;9:e1002947. doi: 10.1371/journal.pcbi.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hué S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci U S A. 2005;102:4425–9. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner BG, Roger M, Stephens D, et al. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. J Infect Dis. 2011;204:1115–9. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durant J, Clevenbergh P, Halfon P, et al. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–9. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 13.Tural C, Ruiz L, Holtzer C, et al. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS. 2002;16:209–18. doi: 10.1097/00002030-200201250-00010. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leigh Brown AJ, Lycett SJ, Weinert L, et al. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011;204:1463–9. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leventhal GE, Kouyos R, Stadler T, et al. Inferring epidemic contact structure from phylogenetic trees. PLoS Comput Biol. 2012;8:e1002413. doi: 10.1371/journal.pcbi.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frost SDW, Volz EM. Modelling tree shape and structure in viral phylodynamics. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2012.0208. 20120208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BC Centre for Disease Control. Vancouver, Canada: BC Centre for Disease Control; 2012. HIV in British Columbia: annual surveillance report 2011. [Google Scholar]
- 19.Vermeiren H, Van Craenenbroeck E, Alen P, Bacheler L, Picchio G, Lecocq P. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J Virol Methods. 2007;145:47–55. doi: 10.1016/j.jviromet.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Kosakovsky Pond SL, Posada D, Stawiski E, et al. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol. 2009;5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafer RW, Rhee SY, Pillay D, et al. HIV-1 protease and reverse transcriptase mutations for drug resistance surveillance. AIDS. 2007;21:215–23. doi: 10.1097/QAD.0b013e328011e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin DB. Multiple imputation for nonresponse in surveys. Wiley series in probability and statistics. Hoboken, New Jersey: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 24.Ritz C, Spiess AN. qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics. 2008;24:1549–51. doi: 10.1093/bioinformatics/btn227. [DOI] [PubMed] [Google Scholar]
- 25.Corvasce S, Violin M, Romano L, et al. Evidence of differential selection of HIV-1 variants carrying drug-resistant mutations in seroconverters. Antivir Ther. 2006;11:329–34. [PubMed] [Google Scholar]
- 26.Beyrer C, Baral SD, van Griensven F, et al. Global epidemiology of HIV infection in men who have sex with men. Lancet. 2012;380:367–77. doi: 10.1016/S0140-6736(12)60821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hué S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–28. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 28.Wertheim JO, Leigh Brown AJ, Hepler NL, et al. The global transmission network of HIV-1. J Infect Dis. 2013;209:304–13. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11:367–72. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 30.Dej E, Kilty JM. “Criminalization creep”: a brief discussion of the criminalization of HIV/AIDS nondisclosure in Canada. Can J Law Soc. 2012;27:55–66. [Google Scholar]
- 31.O'Byrne P, Willmore J, Bryan A, et al. Nondisclosure prosecutions and population health outcomes: examining HIV testing, HIV diagnoses, and the attitudes of men who have sex with men following nondisclosure prosecution media releases in Ottawa, Canada. BMC Public Health. 2013;13:94. doi: 10.1186/1471-2458-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernard EJ, Azad Y, Vandamme AM, Weait M, Geretti AM. HIV forensics: pitfalls and acceptable standards in the use of phylogenetic analysis as evidence in criminal investigations of HIV transmission. HIV Med. 2007;8:382–7. doi: 10.1111/j.1468-1293.2007.00486.x. [DOI] [PubMed] [Google Scholar]
- 33.Wood E, Tyndall MW, Qui Z, Zhang R, Montaner JSG, Kerr T. Service uptake and characteristics of injection drug users utilizing North America's first medically supervised safer injecting facility. Am J Public Health. 2006;96:770–3. doi: 10.2105/AJPH.2004.057828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.