Abstract
Objectives:
We hypothesized that common variants in the collagen genes COL4A1/COL4A2 are associated with sporadic forms of cerebral small vessel disease.
Methods:
We conducted meta-analyses of existing genotype data among individuals of European ancestry to determine associations of 1,070 common single nucleotide polymorphisms (SNPs) in the COL4A1/COL4A2 genomic region with the following: intracerebral hemorrhage and its subtypes (deep, lobar) (1,545 cases, 1,485 controls); ischemic stroke and its subtypes (cardioembolic, large vessel disease, lacunar) (12,389 cases, 62,004 controls); and white matter hyperintensities (2,733 individuals with ischemic stroke and 9,361 from population-based cohorts with brain MRI data). We calculated a statistical significance threshold that accounted for multiple testing and linkage disequilibrium between SNPs (p < 0.000084).
Results:
Three intronic SNPs in COL4A2 were significantly associated with deep intracerebral hemorrhage (lead SNP odds ratio [OR] 1.29, 95% confidence interval [CI] 1.14–1.46, p = 0.00003; r2 > 0.9 between SNPs). Although SNPs associated with deep intracerebral hemorrhage did not reach our significance threshold for association with lacunar ischemic stroke (lead SNP OR 1.10, 95% CI 1.03–1.18, p = 0.0073), and with white matter hyperintensity volume in symptomatic ischemic stroke patients (lead SNP OR 1.07, 95% CI 1.01–1.13, p = 0.016), the direction of association was the same. There was no convincing evidence of association with white matter hyperintensities in population-based studies or with non–small vessel disease cerebrovascular phenotypes.
Conclusions:
Our results indicate an association between common variation in the COL4A2 gene and symptomatic small vessel disease, particularly deep intracerebral hemorrhage. These findings merit replication studies, including in ethnic groups of non-European ancestry.
Small vessel diseases of the brain include a distinct subtype (hereafter referred to in this report as “cerebral SVD”) that affects the small, deep, penetrating arteries and arterioles of the brain, and is thought to be responsible for most symptomatic lacunar ischemic strokes and deep intracerebral hemorrhages (ICHs), as well as for “clinically silent” brain MRI features, such as white matter hyperintensities (WMH). This cerebral SVD also contributes to cognitive impairment and dementia in older people, both through clinically symptomatic strokes and clinically silent ischemia.1
Despite increasing evidence supporting a distinct vascular pathology for cerebral SVD compared with other subtypes of cerebrovascular disease, our knowledge of the underlying genes and pathophysiologic mechanisms is limited, with a lack of specific treatment strategies.1 Common polymorphisms studied in genome-wide association studies (GWAS) of ICH, ischemic stroke, and WMH to date appear to contribute little to the risk of cerebral SVD, despite its established heritability.2–4
COL4A1 and COL4A2 genes are located in tandem on chromosome 13q34 and have a shared communal bidirectional promoter. They encode the collagen chains α1(IV) and α2(IV), which constitute a major component of the vascular basement membrane.5 Dominant missense mutations in COL4A1/COL4A2 cause rare familial forms of cerebral SVD, manifesting as deep ICHs, lacunar ischemic strokes, and WMH.6,7 Genes causing rare familial forms of cerebral SVD may also contain variants conferring risk for common forms of cerebral SVD.8–10 We therefore hypothesized that variants in the COL4A1 and COL4A2 genes may confer risk for common cerebral SVD.
METHODS
Identification of participating studies.
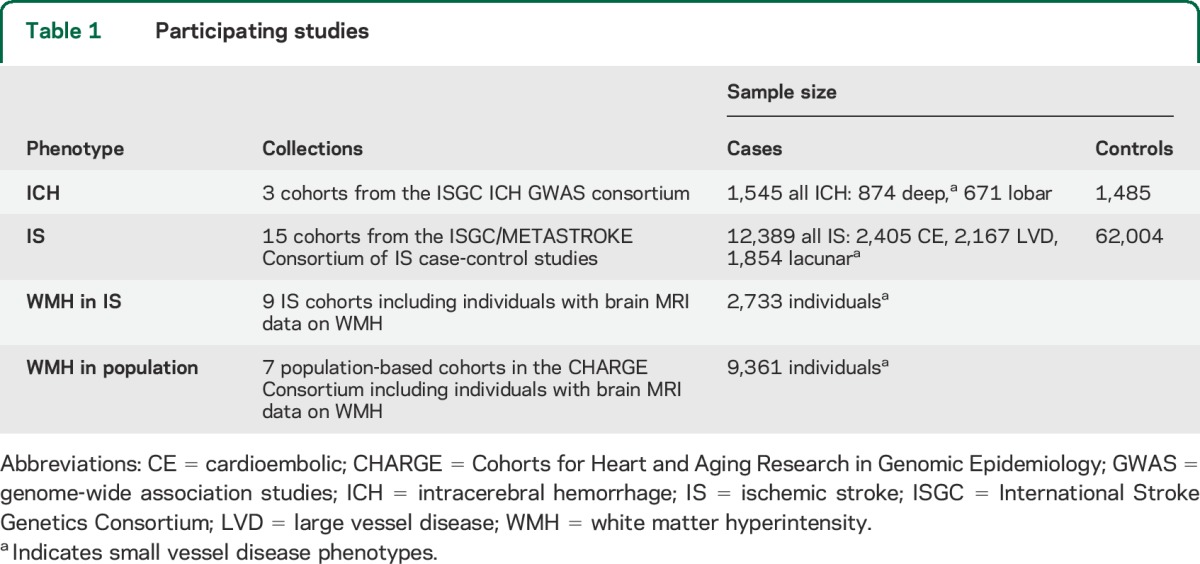
Through an established network of collaborations with relevant groups and consortia (International Stroke Genetics Consortium [ISGC, http://www.strokegenetics.org/], the METASTROKE Collaboration,2 the WMH in Ischemic Stroke GWAS Collaboration,11 and the CHARGE [Cohorts for Heart and Aging Research in Genomic Epidemiology] WMH Group3 [http://web.chargeconsortium.com/]), we identified and included data from the majority of currently available large GWAS of relevant cerebrovascular phenotypes in individuals of European ancestry. The entire dataset consisted of the following: 3 cohorts of 1,545 patients with ICH and 1,485 controls, including information on the main ICH subtypes (deep and lobar); 15 cohorts of 12,389 patients with ischemic stroke, including information on the main determined TOAST (Trial of Org 10172 in Acute Stroke Treatment) subtypes (large vessel disease [LVD], cardioembolic [CE], and small vessel disease/lacunar ischemic stroke)12 and 62,004 controls; 9 cohorts of 2,733 ischemic stroke patients; and 7 cohorts of 9,361 individuals from population-based studies with brain MRI-based measures of WMH volume (table 1). Table e-1 on the Neurology® Web site at Neurology.org includes detailed descriptions of the design and characteristics of the participating studies.2–4,11,13–15
Table 1.
Participating studies
Data collection from participating studies.
We collected existing genotype data from participating studies for the COL4A1 and COL4A2 genomic region of approximately 464 kilo base pairs (kbp), which includes >1,000 single nucleotide polymorphisms (SNPs) covering the bidirectional communal promoter, introns, exons, and a 50-kbp upstream and downstream flanking region (Human Genome reference build 19 [hg19] coordinates 110.751.310–111.215.373). For each phenotype (deep ICH, lobar ICH, all ICH, lacunar ischemic stroke, CE ischemic stroke, LVD ischemic stroke, all ischemic stroke, WMH volume in ischemic stroke and in population-based studies), we collected the following cohort-level summary genotype-phenotype association data for all directly genotyped and imputed SNPs in the region: SNP reference (rs) number and position; effect allele/noneffect allele and their frequencies; association effect size estimate (β coefficient); standard error of the β coefficient; p value for the association; and, for imputed SNPs, the imputation quality measure used and its value. In addition, we collected data on study design and characteristics from relevant publications, as well as by direct communication with group/consortium leads, individual study principal investigators, and other study team members.
Data analysis.
Meta-analyses of COL4A1/COL4A2 SNPs for each phenotype.
We performed meta-analyses of the genotype summary data, from each contributing cohort, assessing associations of COL4A1 and COL4A2 SNPs with each of the cerebrovascular phenotypes available, both those assumed to represent cerebral SVD specifically (deep ICH, lacunar ischemic stroke, WMH volume in ischemic stroke and in population-based studies) and others (lobar ICH, all ICH, CE ischemic stroke, LVD ischemic stroke, all ischemic stroke). We hypothesized that associations would be specific to, or at least strongest with, cerebral SVD phenotypes. We used a fixed-effects inverse variance–based model in METAL genetic meta-analysis software, which weights the β coefficients by their estimated standard errors.16 We used a case-control approach for ICH, ischemic stroke, and their subtypes, generating for each SNP odds ratios (ORs) per additional minor allele for being a case vs control. We used a quantitative trait approach for WMH analysis, assessing for each SNP the effect per additional minor allele on the natural log-transformed WMH volume. Details of WMH volume measurements are provided in table e-1.
Quality-control measures had been applied to all individual studies before provision of data (table e-1). Data from the included studies had been imputed to different reference datasets (HapMap II, HapMap III, or 1000 Genomes) using a range of imputation software (IMPUTE, MACH, and BimBam)17 and were provided with reference to 1 of 2 different Human Genome reference builds (hg18 or hg19) (table e-1).18–20 To permit consistent analyses and interpretation of the data across all included phenotypes and studies, we converted all the data to the same reference build (hg19) and limited the analyses to SNPs present in the HapMap II CEU population (Utah residents of northern and western European ancestry) only. This strategy provided us with the maximum set of 1,070 SNPs represented in all studies and phenotypes. We filtered these data based on imputation quality scores (r2 ≥ 0.3 [MACH, BimBam, IMPUTE]; info score ≥0.7 [PLINK]) and minor allele frequency ≥1%.
Calculating the significance threshold.
To allow for multiple testing, we used a modified version of the Nyholt method (MeffLi), which takes into account the linkage disequilibrium (LD) between SNPs: based on the spectral decomposition of matrices of pairwise LD between SNPs, it calculates the “effective number” (N) of SNPs tested, using this number to calculate the statistical significance threshold by applying the Bonferroni correction for N independent tests (p < 0.05/N). We used the publicly available HapMap II CEU dataset genotype information to calculate N using the Web interface http://gump.qimr.edu.au/general/daleN/SNPSpD/. The Nyholt method is more appropriate for these data than the standard Bonferroni correction, which would be overly conservative because of the assumption that all SNPs are independent.21,22 This method controls accurately for the error rate in evaluations of real and simulated data.23,24
Further exploration of significant SNPs across all phenotypes.
For any SNPs significantly associated with any of the 9 phenotypes assessed, we examined associations with all other phenotypes, focusing on comparisons of findings for cerebral SVD phenotypes (deep ICH, lacunar ischemic stroke, WMH) vs non-SVD phenotypes. We displayed ORs together with their 95% confidence intervals (CIs) and p values using forest plots, to indicate the size and direction of associations across phenotypes. To display the data across all phenotypes in a uniform way, we performed data manipulations to express the SNP effect on WMH volume as OR per 1-unit SD change in WMH volume (table e-2).
We assessed the consistency of results for significantly associated SNPs through conducting formal I2 and χ2 tests for statistical heterogeneity of results between individual cohorts.25
Functional annotation of SNPs.
For SNPs with significant associations, we extracted a list of all other SNPs in moderate LD (defined as r2 ≥ 0.3) from the Web-based SNP Annotation and Proxy Search (SNAP) tool (http://www.broadinstitute.org/mpg/snap/), based on CEU population data from the 1000 Genomes pilot 1.26 We then obtained the functional annotation data for these SNPs from the Ensembl genome browser (http://www.ensembl.org/index.html), Seattle Seq (http://snp.gs.washington.edu/SeattleSeqAnnotation138/), and Haploreg v2 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) databases,27–29 using all 3 databases with partly overlapping sources of information to ensure consistency of results. We further explored the functional significance of SNPs that were in high LD (r2 ≥ 0.9) with the significant SNPs from our analyses, using the online Genotype-Tissue Expression (GTEx) Portal eQTL (expression quantitative trait locus) browser (http://www.broadinstitute.org/gtex/) and the RegulomeDB database (http://regulome.stanford.edu/).30,31 An eQTL represents a locus in the genome in which variation between individuals is associated with a quantitative gene expression trait, often measured as messenger RNA abundance. The RegulomeDB database identifies whether the SNPs are located within known or predicted regulatory elements in the intergenic regions of the human genome. We focused particularly on data obtained from experiments using tissues relevant to cerebrovascular phenotypes, specifically CNS (brain and spinal cord) and vasculature.
We also explored whether SNPs with significant associations were in LD with the rare SNPs previously reported from sequencing studies to be associated with sporadic ICH.9,10 We used the Web-based tool SNAP.26 If the SNP was not included in SNAP, we used the Haploview program to check whether the SNP (based on its position) was in the same haplotype block as the previously reported rare SNPs.32
RESULTS
Significance threshold.
Based on the genotype data from the HapMap II CEU dataset, the effective number (N) of SNPs tested was 598, giving a final Nyholt significance threshold of p = 0.000084. This compares to the Bonferroni-corrected p value of 0.00005 and a genome-wide significance p value of 5 × 10−8.
Meta-analyses of COL4A1/COL4A2 SNPs for each phenotype.
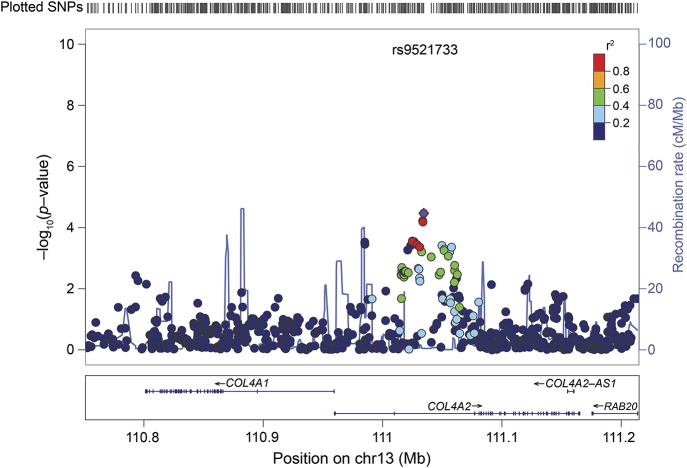
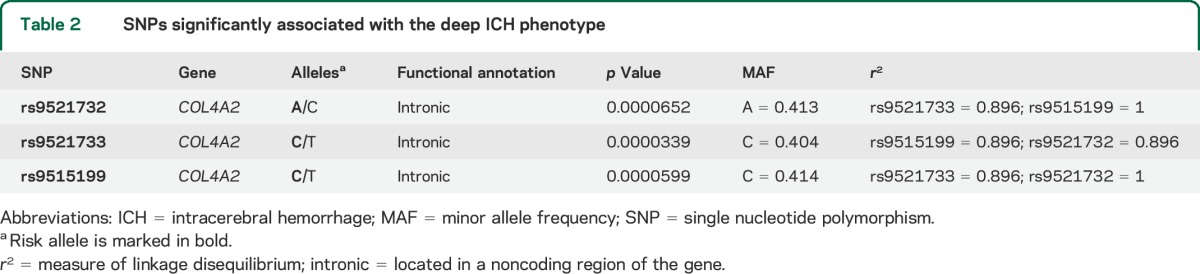
Based on our significance threshold of p = 0.000084, 3 common SNPs in COL4A2 were significantly associated with the deep ICH phenotype (rs9521732: OR per additional A allele = 1.28, 95% CI 1.13–1.44, p = 0.00007; rs9521733: OR per additional C allele = 1.29, 95% CI 1.14–1.46, p = 0.00003; rs9515199: OR per additional C allele = 1.28, 95% CI 1.14–1.44, p = 0.00006) (figure 1, table 2). These 3 SNPs are all intronic and in high LD with each other (table 2). One (rs9521733) was directly genotyped in the majority of the cohorts while 2 were imputed in all (rs951599) or in the majority (rs9521732) of the cohorts (table e-2). There were no statistically significant associations of common SNPs in COL4A1/COL4A2 with any of the other phenotypes.
Figure 1. Regional association plot for deep ICH meta-analysis.
Dots mark individual SNPs; y-axis: p value for association between each SNP and deep ICH; x-axis: position of SNPs. Most significantly associated SNP shown in purple; colors for other SNPs depend on linkage disequilibrium with this lead SNP (see r2 color coding on figure). Recombination: exchange of a segment of DNA between 2 homologous chromosomes during meiosis leading to a novel combination of genetic material in the offspring. Recombination rate: measured as frequency of exchange per unit physical distance (centimorgan [cM]/mega base pair [Mb]). cM: unit of linkage that refers to the distance between 2 gene loci determined by the frequency with which recombination occurs between them. Two loci are said to be 1 cM apart if recombination is observed between them in 1% of meioses. Mb: a unit of length of nucleic acids, equal to 1 million base pairs. ICH = intracerebral hemorrhage; SNP = single nucleotide polymorphism.
Table 2.
SNPs significantly associated with the deep ICH phenotype
Associations with other cerebrovascular phenotypes of the 3 COL4A2 SNPs significantly associated with deep ICH.
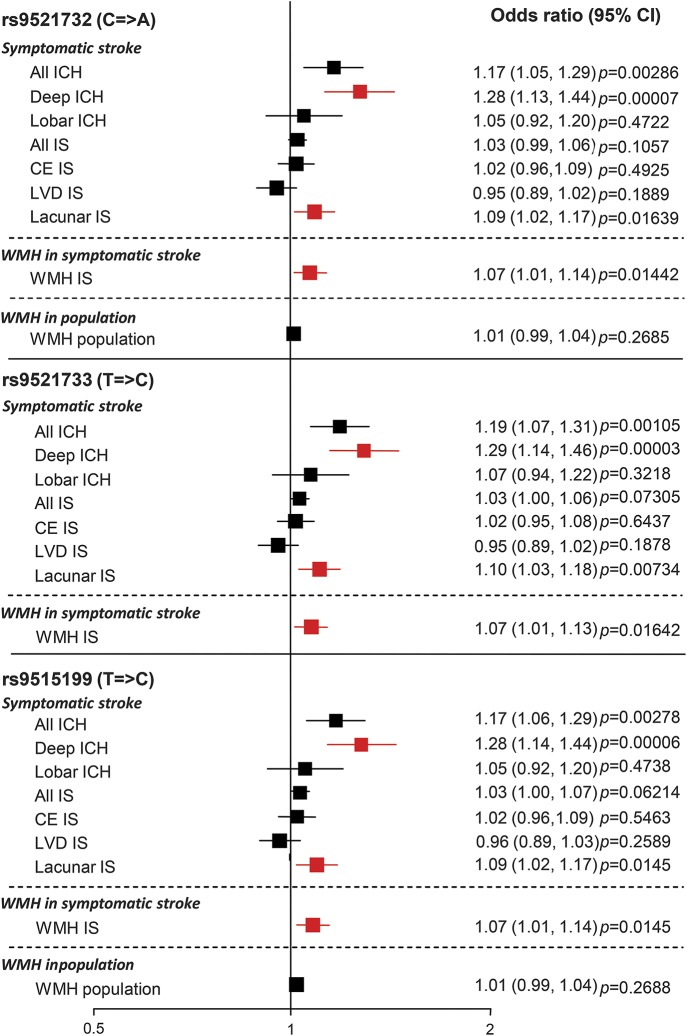
Figure 2 shows association results for the 3 SNPs significantly associated with deep ICH across all 9 phenotypes assessed. Although these 3 SNPs were significantly associated only with deep ICH, there were suggestive associations with 2 other cerebral SVD phenotypes: lacunar ischemic stroke (rs9521732: OR 1.09, 95% CI 1.02–1.17, p = 0.01639; rs9521733: OR 1.10, 95% CI 1.03–1.18, p = 0.00734; rs9515199: OR 1.09, 95% CI 1.02–1.17, p = 0.0145) and WMH volume in symptomatic ischemic stroke cases (rs9521732: OR 1.07, 95% CI 1.01–1.14, p = 0.01442; rs9521733: OR 1.07, 95% CI 1.01–1.13, p = 0.01642; rs9515199: OR 1.07, 95% CI 1.01–1.14, per 1 SD change in WMH volume; p = 0.0145). There was no evidence for association of these SNPs with other non-SVD stroke subtypes (lobar ICH, CE, and LVD ischemic stroke) or for WMH volume in population-based studies (figure 2). As expected, ORs for these SNPs with the all ICH and all ischemic stroke phenotypes were intermediate between those for deep and lobar ICH and for lacunar and non-SVD ischemic stroke subtypes, respectively, suggesting that associations with these combined phenotypes were driven by results for deep ICH and lacunar ischemic stroke (figure 2).
Figure 2. Associations between the 3 SNPs significantly associated with deep ICH across all phenotypes.
Figure represents pooled odds ratios across all cohorts. Significant or suggested associations in cerebral small vessel disease phenotypes are shown in red. Data for rs9521733 were unavailable in the WMH in population cohorts. CE = cardioembolic; CI = confidence interval; ICH = intracerebral hemorrhage; IS = ischemic stroke; LVD = large vessel disease; SNP = single nucleotide polymorphism; WMH = white matter hyperintensity.
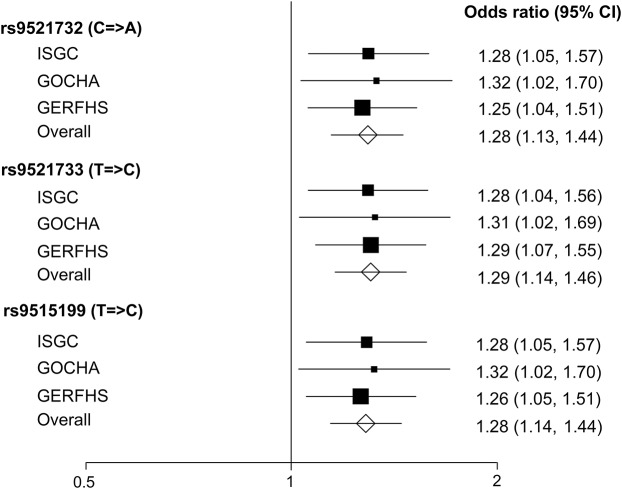
The associations across individual cohorts included in the deep ICH meta-analysis were highly consistent, with no significant heterogeneity (I2 = 0%; p > 0.9) (figure 3). There was no or minimal heterogeneity between the individual cohorts' results for lacunar ischemic stroke (I2 = 0%; p > 0.8) and for WMH in ischemic stroke (I2 = 17%–22%; p > 0.2).
Figure 3. Associations of significant single nucleotide polymorphisms across cohorts included in the meta-analysis of deep intracerebral hemorrhage.
Heterogeneity: rs9521732: χ22df = 0.09, p = 0.9563, I2 = 0%; rs9521733: χ22df = 0.02, p = 0.9903, I2 = 0%; rs9515199: χ22df = 0.08, p = 0.9607, I2 = 0%. CI = confidence interval; GERFHS = Genetic and Environmental Risk Factors for Hemorrhagic Stroke; GOCHA = Genetics of Cerebral Hemorrhage with Anticoagulation; ISGC = International Stroke Genetics Consortium.
Functional annotation of SNPs.
We identified 92 unique SNPs in moderate LD (r2 ≥ 0.3) with the 3 significant SNPs from our analyses. All of these SNPs (including our 3 significant SNPs) are intronic (table e-3). From these, we then selected all 5 SNPs that were in high LD (r2 > 0.9) with our 3 significant SNPs, giving a total of 8 SNPs for analysis of functional annotation. The GTEx eQTL browser search revealed no significant eQTLs for any of these 8 SNPs. However, the RegulomeDB database search revealed minimal binding evidence for 4 of the 8 SNPs, suggesting that these SNPs are located in areas of the genome that may have some regulatory functions. When we considered only results based on experiments done on tissues most relevant to cerebrovascular phenotypes (brain, spinal cord, and blood vessels), there was still some evidence for these 4 SNPs being located in genomic areas of potential functional significance (table e-3).
The 3 rare exonic COL4A2 SNPs reported in a previous sequencing study to be associated with sporadic ICH are rs117412802, rs62621875, and A1690T (hg19 position: 111164467).9 Based on SNAP, rs117412802 was not in LD with the 3 SNPs significantly associated with deep ICH in our analyses (r2 = 0). rs62621875 and A1690T were not represented in SNAP, but in Haploview, they were not in the same haplotype block as the 3 SNPs significantly associated with deep ICH in our analyses, suggesting that they are not in close LD with the SNPs we identified.
DISCUSSION
Herein, we present the first evidence for a significant association between an intronic locus in COL4A2 and deep ICH, a symptomatic cerebral SVD phenotype. We also found suggestive associations in the same direction for these SNPs with other cerebral SVD phenotypes: lacunar ischemic stroke and WMH in ischemic stroke cases. Our results indicate that the COL4A2 gene is a locus conferring risk for common forms of symptomatic cerebral SVD, particularly deep ICH.
We did not find evidence for an association between these 3 SNPs (or any others in the COL4A1/COL4A2 region) and WMH assessed in largely asymptomatic individuals in population-based studies. The difference in results between WMH assessed among symptomatic people with ischemic stroke and among those in population-based studies (both of which are often regarded as cerebral SVD phenotypes) could be attributable to genuine differences in the nature of the phenotype represented by WMH in these different groups and its genetic influences, differences in scanning and measurement techniques used in the contributing cohorts, or the play of chance.
Our findings are supported by other data showing the importance of the COL4A1/COL4A2 genes in cerebrovascular disease. Missense mutations in COL4A1 or COL4A2 lead to rare mendelian forms of cerebrovascular disease including pediatric and adult cerebral hemorrhage.6,7 Furthermore, sequence analysis of COL4A1 and COL4A2 has revealed missense mutations in sporadic cases of ICH (96 cases and 145 controls), supporting an important role for collagen IV in the etiology of cerebral hemorrhage.9,10 Studies in mice and patients have shown that the severe dominant mutations lead to basement membrane defects, which may weaken the blood vessel and also affect vascular function. In addition to the basement membrane defects, the missense mutations lead to retention of mutant protein in the endoplasmic reticulum, leading to endoplasmic reticulum stress and activation of the unfolded protein response.9,33–35 It is of interest that the SNPs identified in our study, and all SNPs in moderate LD with them, are intronic, and may be located in regulatory areas of the gene. The 3 SNPs from our analysis are not in close LD with the variants reported from previous sequence analyses in sporadic ICH, demonstrating that they do not represent the same signal as that observed from the previously described exonic rare variants, but instead identify a novel intronic region of COL4A2 associated with deep ICH (and, possibly, with other cerebral SVD phenotypes).
Our study has several strengths. We investigated a specific, prespecified hypothesis, clearly defining the clinical phenotypes and candidate genes of interest, based on preexisting supporting data. We used a novel approach, investigating genetic associations across a range of cerebrovascular phenotypes, focusing on a specific cerebrovascular subphenotype—cerebral SVD—which is thought to have distinct pathophysiologic mechanisms and risk factors. A further strength is that, through a network of collaborative groups largely under the umbrella of the International Stroke Genetics Consortium, we were able to include in our in silico analyses the majority of the currently available data representing the phenotypes included for the COL4A1/2 region from genotyped cohorts among white individuals of European ancestry.
Our study has some limitations. While we have shown that SNPs in COL4A2 are associated with deep ICH, when we analyze data for the specific candidate COL4A1/2 region, correcting appropriately for multiple testing within that region by using the generally accepted Nyholt method,2,21–24 this association did not reach a genome-wide level of significance (possible reasons include study size),4 while associations with other cerebral SVD phenotypes (lacunar ischemic stroke and WMH in ischemic stroke cases) were suggestive but not independently robust to multiple testing. In addition, although we corrected appropriately for the number of SNPs in the region, we did not adjust the statistical threshold for the number of phenotypes investigated. We considered that this would be overly conservative since these phenotypes are not completely independent of each other. As in any meta-analysis, this study is dependent on accurate allocation of cases and controls, and accurate phenotype measurements in the original studies. Thus, there is a risk that premorbid controls may have been misallocated, although this was addressed in the original studies by matching the controls to cases based on age.
Our findings certainly merit further confirmatory studies in additional, large samples with relevant phenotypes, including those from individuals of non-European ancestry. It would also be of interest to extend genetic association studies of this genomic region to other cerebral SVD phenotypes that are likely to have a similar underlying vascular pathology, such as MRI-detected brain microbleeds (particularly deep brain microbleeds) and clinically silent lacunar infarcts. Relevant data to facilitate such further analyses are likely to become available from the ISGC ICH GWAS, CHARGE, the US National Institute of Neurological Disorders and Stroke–funded SiGN (Stroke Genetics Network) ischemic stroke GWAS, and other consortia in the near future.3,4,36 Further deep sequencing of the entire COL4A1/2 genomic region (intronic as well as exonic regions, given previous exonic and the current intronic findings) among cases of ICH as well as other cerebral SVD phenotypes, with functional studies of promising variants thus identified, are also needed to further investigate potential pathogenic mechanisms underlying sporadic forms of cerebral SVD. Finally, this successful approach of identifying promising candidates for the cerebral SVD phenotype based on rare familial forms of the disease could be extended to include other relevant candidate genes.
Our study lays important foundations for enhancing knowledge of the genetic risk factors underlying cerebral SVD, a disease of major public health importance. The approach taken should lead to further understanding of causative pathways and identification of a subset of patients with cerebrovascular disease unified by their disease mechanism who may benefit from distinct prevention and treatment strategies.
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge Dr. Qiuli Zhang (Centre for Clinical Brain Sciences, University of Edinburgh) for her advice on statistical analyses, Dr. Poneh Adib-Samii (Stroke and Dementia Research Centre, St. George's University of London) for her contribution toward the WMH in ischaemic stroke GWAS project, Dr. Solveig Gretarsdottir (deCODE Genetics, Reykjavik, Iceland), and Dr. Robert Clarke (University of Oxford) for their contribution toward the ischaemic stroke GWAS.
GLOSSARY
- CE
cardioembolic
- CHARGE
Cohorts for Heart and Aging Research in Genomic Epidemiology
- CI
confidence interval
- eQTL
expression quantitative trait locus
- GTEx
Genotype-Tissue Expression
- ICH
intracerebral hemorrhage
- ISGC
International Stroke Genetics Consortium
- LD
linkage disequilibrium
- LVD
large vessel disease
- OR
odds ratio
- SNAP
SNP Annotation and Proxy Search
- SNP
single nucleotide polymorphism
- SVD
small vessel disease
- WMH
white matter hyperintensity
Footnotes
Supplemental data at Neurology.org
AUTHOR AFFILIATIONS
From the Centre for Clinical Brain Sciences (K.R., C.L.M.S.), Centre for Cognitive Ageing and Cognitive Epidemiology (G.D.), Department of Psychology (G.D.), Institute for Genetics and Molecular Medicine (P.A.T., C.L.M.S.), University of Edinburgh; Clinical Neurosciences (S.B., M.T., H.S.M.), University of Cambridge, UK; Center for Human Genetic Research (W.J.D., G.J.F., C.D.A., T.W.K.B., F.R., C.R.Z., N.R., J.R.), and Department of Neurology (G.J.F., C.D.A., J.R.), Massachusetts General Hospital, Boston; Program in Medical and Population Genetics (G.J.F., C.D.A., T.W.K.B., F.R., J.R.), Broad Institute, Cambridge, MA; Department of Environmental Health (R.D.), University of Cincinnati College of Medicine; Divisions of Biostatistics and Epidemiology (J.G.W.) and Human Genetics (L.J.M.), Cincinnati Children's Hospital Medical Center, OH; Neurovascular Research Unit, Department of Neurology (J.J.-C.), and Cardiovascular Epidemiology and Genetics Research Group (J.J.-C.), IMIM (Institut Hospital del Mar d'Investigacions Mèdiques), Universitat Autonoma de Barcelona, Spain; Department of Neurology (M.S.), Stroke Division, Beth Israel Deaconess Medical Center, Boston, MA; Stroke Program (D.L.B.), Department of Neurology, University of Michigan Health System, Ann Arbor; Department of Neurology (S.L.S.), University of Florida College of Medicine, Jacksonville; Department of Neurology (C.S.K.), University of Arizona, Tucson; Neurovascular Research Laboratory and Neurovascular Unit (J.M.), Institut de Recerca, Hospital Vall d'Hebron, Universitat Autonoma de Barcelona, Spain; Center for Public Health Genomics and Department of Biostatistical Sciences (C.D.L.), Wake Forest University, Winston-Salem, NC; Department of Neurology (A.S.), Jagiellonian University Medical College, Krakow, Poland; Department of Clinical Sciences Lund (B.M.H., A.G.L.), Neurology, Lund University; Department of Neurology (B.M.H., A.G.L.), Skåne University Hospital, Lund, Sweden; Department of Neurology (J.F.M.), Mayo Clinic, Jacksonville, FL; Brown Foundation Institute of Molecular Medicine and Human Genetics Center (M.F.), Division of Epidemiology, School of Public Health, University of Texas Health Science Center at Houston; Cardiovascular Health Research Unit and Department of Medicine (J.C.B.), and Departments of Neurology and Epidemiology (W.T.L.), University of Washington, Seattle; Department of Neurology (S.D., Q.Y., S.S.), Boston University School of Medicine, MA; Department of Neurology (S.D.), Lariboisière Hospital, Paris 7 University; Inserm UMR897 (S.D.), Bordeaux University, France; Departments of Epidemiology, Radiology, and Neurology (M.A.I.), Erasmus MC University Medical Center, Rotterdam; Netherlands Consortium for Healthy Aging (M.A.I.), the Netherlands; Department of Neurology (R.S.), Medical University of Graz, Austria; Institute of Cardiovascular Research (P.S.), Royal Holloway, University of London (ICR2UL) and Ashford and St. Peter's “Hospitals, London, UK; Department of Neurology, Veterans Affairs Medical Center and Department of Neurology (S.J.K.), and Department of Medicine (B.D.M.), University of Maryland School of Medicine, Baltimore; Centre for Clinical Epidemiology and Biostatistics (E.G.H., J.A.), University of Newcastle; Centre for Translational Neuroscience and Mental Health Research (C.R.L.), University of Newcastle, and Hunter Medical Research Institute, New Lambton, Australia; Stroke Prevention Research Unit (P.M.R., D.L.P.), Nuffield Department of Clinical Neuroscience, University of Oxford, John Radcliffe Hospital, Oxford, UK; Department of Cerebrovascular Disease (G.B.B.), Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy; Departments of Epidemiology and Medicine (B.M.P.), Cardiovascular Health Research Unit and Department of Health Services, University of Washington, and Group Health Research Institute, Group Health Seattle, WA; Institute for Stroke and Dementia Research (R.M., M.D.), Klinikum der Universität München, Ludwig-Maximilians-Universität, München, Germany; Departments of Neurology and Public Health Sciences (B.B.W.), University of Virginia, Charlottesville; Munich Cluster for Systems Neurology (SyNergy) (M.D.), Munich, Germany; Institute of Cardiovascular and Medical Sciences (T.V.A.), University of Glasgow, UK; and Department of Neurology and Rehabilitation Medicine (D.W.), University of Cincinnati, OH.
AUTHOR CONTRIBUTIONS
Kristiina Rannikmäe was involved in the original design and conceptualization of the study, and with supervision from Cathie Sudlow and input from Gail Davies and Pippa A. Thomson collected the data, performed the meta-analyses, interpreted the results, drafted the manuscript, and revised it prior to submission. Gail Davies and Pippa A. Thomson contributed to designing the study, assisted with the analyses and interpreting the data, and revised the manuscript for intellectual content. Cathie Sudlow was involved in the design and conceptualization of the study, supervised the data collection process, the analysis and interpretation of the data, and revised the manuscript for intellectual content. Steve Bevan, William J. Devan, Guido J. Falcone, Cathy Zhang, Qiong Yang, Matthew Traylor, Christopher D. Anderson, Thomas W.K. Battey, Farid Radmanesh, Ranjan Deka, Jessica G. Woo, Lisa J. Martin, Jordi Jimenez-Conde, Magdy Selim, Devin L. Brown, Scott L. Silliman, Chelsea S. Kidwell, Joan Montaner, Carl D. Langefeld, Agnieszka Slowik, Björn Hansen, Arne Lindgren, James F. Meschia, Myriam Fornage, Joshua C. Bis, Stéphanie Debette, Mohammad A. Ikram, Will T. Longstreth, Reinhold Schmidt, Pankaj Sharma, Steven J. Kittner, Braxton D. Mitchell, Elizabeth G. Holliday, Christopher R. Levi, John Attia, Peter M. Rothwell, Deborah L. Poole, Giorgio B. Boncoraglio, Bruce M. Psaty, Rainer Malik, Natalia Rost, Bradford B. Worrall, Martin Dichgans, Daniel Woo, Jonathan Rosand, Sudha Seshadri, and Hugh S. Markus contributed to the original data collection and initial GWAS analysis, and revised the manuscript for intellectual content. Steve Bevan, William J. Devan, Guido J. Falcone, Cathy Zhang, Qiong Yang, and Matthew Traylor were also involved in providing the data for this project and William J. Devan was involved in performing the functional annotation. Jonathan Rosand and Sudha Seshadri were also involved in interpreting the data. Tom Van Agtmael was involved in the design and conceptualization of the study, and revised the manuscript for intellectual content.
STUDY FUNDING
Kristiina Rannikmäe is supported by the Edinburgh and Lothians Health Foundation REMIND Fund and Stroke Research and Amenities Endowments Fund. Cathie Sudlow is supported by the Scottish Funding Council. Gail Davies is supported by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1), BBSRC, and Medical Research Council (MSRC). Tom Van Agtmael is supported by the Kidney Research UK project grant RP19/2012. Details of funding for the studies contributing to the ICH GWAS can be found in reference 4. Details of funding for the studies contributing to the ischaemic stroke GWAS can be found in reference 2. Details of funding for the studies contributing to the WMH in ischaemic stroke GWAS can be found in reference 11. Details of funding for the studies contributing to the WMH in population GWAS can be found in reference 3. Martin Dichgans is supported by the Fondation Leducq (Transatlantic Network of Excellence on the Pathogenesis of Small Vessel Disease of the Brain). Guido J. Falcone is supported by the NIH/National Institute of Neurological Disorders and Stroke (NINDS) grant P50NS061343. Chistopher D. Anderson is supported by the American Brain Foundation Clinical Research Training Fellowship, NIH/NINDS K23NS086873, and Massachusetts General Hospital Institute for Heart, Vascular, and Stroke Care SPARK award. Jonathan Rosand is supported by the NIH/NINDS grant R01NS059727.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Pantoni L. Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol 2010;9:689–701. [DOI] [PubMed] [Google Scholar]
- 2.Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 2012;11:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fornage M, Debette S, Bis JC, et al. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE Consortium. Ann Neurol 2011;69:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo D, Falcone GJ, Devan WJ, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a novel susceptibility locus for intracerebral hemorrhage. Am J Hum Genet 2014;94:511–521. 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian collagen IV. Microsc Res Tech 2008;71:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanfranconi S, Markus HS. COL4A1 mutations as a monogenic cause of cerebral small vessel disease: a systematic review. Stroke 2010;41:e513–e518. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Craggs L, Baumann M, Kalimo H, Kalaria RN. Review: molecular genetics and pathology of hereditary small vessel diseases of the brain. Neuropathol Appl Neurobiol 2011;37:94–113. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt H, Zeginigg M, Wiltgen M, et al. Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebral small vessel disease. Brain 2011;134:3384–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeanne M, Labelle-Dumais C, Jorgensen J, et al. COL4A2 mutations impair COL4A1 and COL4A2 secretion and cause hemorrhagic stroke. Am J Hum Genet 2012;90:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng YC, Sonni A, Labelle-Dumais C, et al. COL4A1 mutations in patients with sporadic late-onset intracerebral hemorrhage. Ann Neurol 2012;71:470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adib-Samii P, Rost N, Traylor M, et al. 17q25 locus is associated with white matter hyperintensity volume in ischaemic stroke, but not with lacunar stroke status. Stroke 2013;44:1609–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams HP, Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke: definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993;24:35–41. [DOI] [PubMed] [Google Scholar]
- 13.Woo D, Sauerbeck LR, Kissela BM, et al. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke 2002;33:1190–1195. [DOI] [PubMed] [Google Scholar]
- 14.Falcone GJ, Biffi A, Devan WJ, et al. Burden of blood pressure-related alleles is associated with larger hematoma volume and worse outcome in intracerebral hemorrhage. Stroke 2013;44:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rost NS, Rahman RM, Biffi A, et al. White matter hyperintensity volume is increased in small vessel stroke subtypes. Neurology 2010;75:1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010;11:499–511. [DOI] [PubMed] [Google Scholar]
- 18.Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010;467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007;449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004;74:765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005;95:221–227. [DOI] [PubMed] [Google Scholar]
- 23.Salyakina D, Seaman SR, Browning BL, Dudbridge F, Müller-Myhsok B. Evaluation of Nyholt's procedure for multiple testing correction. Hum Hered 2005;60:19–25. [DOI] [PubMed] [Google Scholar]
- 24.Nyholt DR. Evaluation of Nyholt's procedure for multiple testing correction: author's reply. Hum Hered 2005;60:61–62. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res 2014;42:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009;461:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 33.Van Agtmael T, Bailey MA, Schlötzer-Schrehardt U, et al. Col4a1 mutation in mice causes defects in vascular function and low blood pressure associated with reduced red blood cell volume. Hum Mol Genet 2010;19:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Agtmael T, Schlötzer-Schrehardt U, McKie L, et al. Dominant mutations of Col4a1 result in basement membrane defects which lead to anterior segment dysgenesis and glomerulopathy. Hum Mol Genet 2005;14:3161–3168. [DOI] [PubMed] [Google Scholar]
- 35.Murray LS, Lu Y, Taggart A, et al. Chemical chaperone treatment reduces intracellular accumulation of mutant collagen IV and ameliorates the cellular phenotype of a COL4A2 mutation that causes haemorrhagic stroke. Hum Mol Genet 2014;23:283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meschia J, Arnett DK, Ay H, et al. Stroke Genetics Network (SiGN) Study: design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke 2013;44:2694–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.