Abstract
The circadian clock exists to synchronize inner physiology with the external world, allowing life to anticipate and adapt to the continual changes that occur in an organism’s environment. The clock architecture is highly conserved, present in almost all major branches of life. Within eukaryotes, the filamentous fungus Neurospora crassa has consistently been used as an excellent model organism to uncover the basic circadian physiology and molecular biology. The Neurospora model has elucidated our fundamental understanding of the clock as nested positive and negative feedback loop, regulated by transcriptional and posttranscriptional processes. This review will examine the basics of circadian rhythms in the model filamentous fungus N. crassa as well as highlight the output of the clock in Neurospora and the reasons that N. crassa has continued to be a strong model for the study of circadian rhythms. It will also synopsize classical and emerging methods in the study of the circadian clock.
1. INTRODUCTION
Neurospora first emerged as a model organism for understanding circadian clocks and circadian systems in the late 1950s when Pittendrigh and coworkers described a rhythm in the events associated with overt asexual development in cultures grown on long hollow glass tubes (Pittendrigh, Bruce, Rosensweig, & Rubin, 1959). At the time, it was a natural choice; Neurospora was then and remains a premier genetic model, an organism whose fame stems from Beadle and Tatum’s work on the one enzyme/one gene hypothesis and that is still the consensus model for filamentous fungi although first Escherichia coli and later yeast budding yeast eclipsed it for many studies (Davis & Perkins, 2002). Despite the existence of these other models, Neurospora has remained a useful model for the study of many problems, and especially for circadian rhythms because it is a wonderfully tractable genetic system, it is easy to use for biochemical follow ups, and enjoys a relatively large and heavily invested research community. As a result, nearly all of what we know about the molecular details of circadian rhythms in fungi stems from work on Neurospora. Beyond this, and because cells from fungi and animals share many aspects of regulation, much of what we know about circadian rhythms in animals can be traced to work on this system. Specifically, many of the proteins and most of the regulatory architecture of the core circadian oscillator in Neurospora and animals are quite similar and insights from cells in one system are generally applicable to the other. This review details the circadian system in Neurospora and the methods that are employed to study it.
1.1. Methods of analysis of circadian rhythms in Neurospora crassa
Neurospora has remained an excellent circadian model organism for a variety of reasons, not the least of which includes the ease and standardization of the methods used to examine the clock. Since the initial work on rhythms, Neurospora has become a highly tractable genetic organism, with a full genome sequence, transformation protocols yielding >98% efficiency in gene targeting, a variety of regulatable promoters and selectable markers, and a nearly complete knockout library (Collopy et al., 2010; Colot et al., 2006; Galagan et al., 2003; Ninomiya, Suzuki, Ishii, & Inoue, 2004). Neurospora is recognized by the NIH as an established model system (http://www.nih.gov/science/models/neurospora/). Genetic stocks including the full collection of gene knockouts are maintained by the Fungal Genetics Stock Center that distributes over 40,000 strains annually for nominal fees.
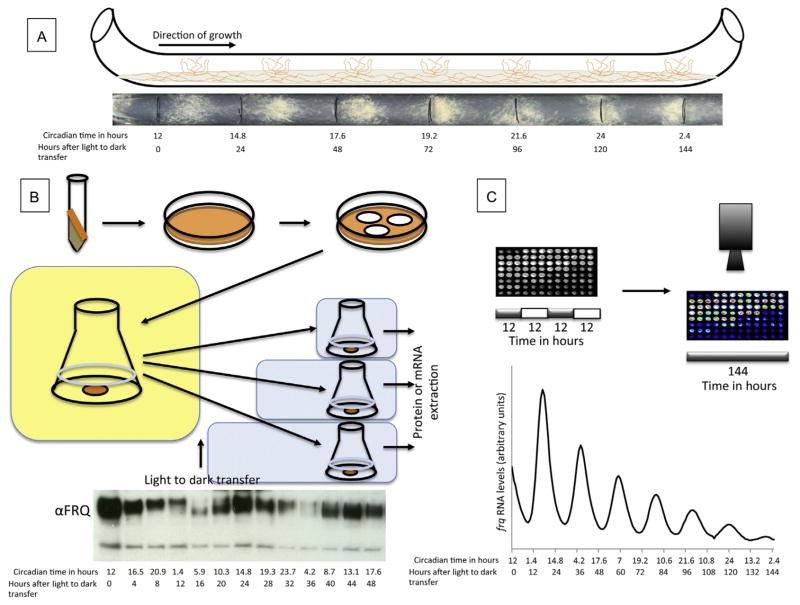
Neurospora has both sexual and asexual life cycles and rhythms are apparent in the regulation of both. Most studies employ the asexual cycle in which a developmental switch leads to the production of conidia in the subjective night. Although the developmental cycle can be seen on petri dish cultures inoculated at one point, this rhythmic development of conidia is generally assayed through use of a race tube, a long glass tube, bent at both ends, holding agar medium onto which mycelia or conidia are inoculated at one end (Fig. 1A). The individual clocks are synchronized by germinating the cultures in constant light for a day and then transferring the fungi to constant darkness. As the fungus grows down the tubes at about 2 cm per day, the growth front is marked each day. Because distance grown approximates time passed, the period length can be interpreted from the growth pattern. More specific instruction on growing a Neurospora strain on race tubes can be found at http://www.fgsc.net/Neurospora/neurospora.html. Once the race tube has been run, the period can be determined by hand or using the Chrono program (Roenneberg & Taylor, 2000).
Figure 1.
Methods of circadian analysis in Neurospora. (A) The basic outline of the use of a race tube in analyzing circadian rhythms in Neurospora along with the image of an actual race tube beneath it. Daily growth fronts are noted with vertical lines (sidereal time and in circadian time are noted below; J. Hurley, unpublished data). (B) The basics of analysis of the molecular rhythms for Neurospora liquid culture. The outline of the protocol to extract either mRNA or protein from Neurospora over circadian time. Western blot of FRQ protein tracked over 48 sidereal hours (time points taken every four sidereal hours) and labeled in circadian hours, highlighting the changes of phosphorylation state of FRQ protein over time (J. Emerson, unpublished data). (C) The outline of real-time analysis of molecular circadian rhythms. Ninety-six individual tubes of Neurospora are subjected to a 12:12 light/dark cycle and then allowed to free run in the dark. A luciferase trace of frq mRNA expression tracked over 144 sidereal hours using a CCD camera and the resulting traces are labeled in circadian hours, highlighting the changes of expression levels of frq mRNA over time (J. Emerson, unpublished data).
Manifestation of the observed rhythm in asexual development and conidiation is not strong in wild-type cultures unless there is air exchange over the culture or they are chemically treated to result in mild oxidative stress (Belden, Larrondo, et al., 2007). However, identification of the band (bd) strain by Sargent made expression of the rhythm robust without the need for air flow (Sargent, Briggs, & Woodward, 1966), and now nearly all strains used for circadian studies incorporate this allele in the background. The causative mutation in the band strain was identified as a mutation in ras-1 that increased the levels of reactive oxygen species (ROS) in Neurospora. This in turn, leads to the increased expression of a particular conidial regulation protein encoded by the gene fluffy, which accounts for the bd phenotype (Belden, Larrondo, et al., 2007). Because of the connection between asexual development, RAS signaling, and ROS levels in sporulation, genes, and pathways identified as under clock control in the bd strain may be RAS-responsive instead of clock-responsive and may not be clock regulated in a wild-type strain (Belden, Larrondo, et al., 2007).
Rhythms have also been followed in cultures grown in liquid medium, leading to the understanding of the clock on a molecular level in liquid cultures as opposed to the overt rhythms followed on solid media in the bd strain (Aronson, Johnson, Loros, & Dunlap, 1994; Loros, Denome, & Dunlap, 1989). Mycelial discs cut from syncytial mats retain their rhythmicity and phase and can be transferred to liquid media. These cultures are grown in the dark with gentle shaking and harvested at times throughout their circadian cycle to extract RNA or proteins enabling that assessment of overall marker levels. Care must be taken to normalize age and media of the culture as small changes are likely create large differences in macromolecular output (Loros et al., 1989).
However, liquid media can quickly become depleted of nutrients and does not allow for long-term observation of the core circadian rhythm and due to this, liquid media may have a vastly different set of clock-controlled output genes (Perlman, Nakashima, & Feldman, 1981). To obviate this possible disparity, rhythms can be now monitored more directly using reporter expression of the luciferase gene from the firefly beetle Photinus pyralis, codon optimized for Neurospora, and driven by clock-regulated promoters such as frequency (frq) or ccg-2 (Gooch et al., 2008; Morgan, Greene, & Bell-Pedersen, 2003). Tracking the clock with luciferase allows for direct tracking of the endogenous, core rhythms of the Neurospora clock on the transcriptional level, and this method has been used to understand temperature compensation as well as translational activity in the core Neurospora clock (Gooch et al., 2008; Larrondo, Loros, & Dunlap, 2012).
1.2. Circadian rhythms in other fungi
It should be noted that Neurospora is not the only fungus with a rhythm; there is documentation of a circadian circuit in other fungi. Similar rhythms in conidiospore formation have been reported in the Zygomycete Pilobolus (Bruce, Weight, & Pittendrigh, 1960), and less definite growth and developmental rhythms exist in a variety of Ascomycetes (reviewed in Dunlap & Loros, 2006; Greene, Keller, Haas, & Bell-Pedersen, 2003). Not surprisingly, the identification of conserved core clock components has been well investigated in fungi as well. A review of 42 sequenced fungal genomes noted that light sensing mechanisms, which commonly function as the positive arm of the clock, are widely conserved in the fungi as is the RNA helicase protein frequency interacting RNA helicase (FRH) (see below). The clock-exclusive protein FRQ (the negative feedback element) is less conserved, however, and components for complete circadian feedback loops can be seen in the family Sordariacea (where Neurospora lies), suggesting that many plant and animal pathogens have a functional clock (e.g., Canessa, Schumacher, Hevia, Tudzynski, & Larrondo, 2013). In addition, recent data have revealed a FRQ ortholog in Pyronema confluens which shares a common ancestor with Neurospora on the order of 500 million years ago, thus extending the existence of FRQ, and probably of clocks, well before the divergence of Aspergillus and Neurospora and much farther back in time (Traeger et al., 2013). The unstructured nature of the negative arm protein FRQ, discussed later in this chapter, suggested that conservation of sequence might not be necessary to maintain the lack of structure needed to play the role of the negative arm protein (Dunlap & Loros, 2006; Hurley, Larrondo, Loros, & Dunlap, 2013; Salichos & Rokas, 2010); it is remarkable that sequence orthologs exist in such anciently diverged species, and their existence suggests conservation of interactions with FRQ.
2. MOLECULAR MECHANISM OF THE NEUROSPORA CIRCADIAN OSCILLATOR
At the core of circadian rhythms in Neurospora (and in all other known molecular oscillators) is a transcriptional/translational feedback loop run by a core clock complex that is strictly regulated by a series of ancillary interacting proteins. The core complex consists of two sets of protein pairs, the negative arm (in Neurospora this is comprised of the FRQ/FRH complex or FFC as well as CK1), and the positive arm (in Neurospora comprised of the White Collar Complex or WCC) which drives the expression of frq. The negative arm regulates its own expression on a time delay, the circadian period (Dunlap, 1999).
As a brief overview, the cycle begins late in the subjective night (Fig. 2), when the WCC binds to the frq promoter which leads to the induction of frq mRNA, a process that reaches its maximum around early subjective morning. FRQ protein takes around 4 h to translate after the start of frq expression, after which it binds rapidly to FRH and itself, enters the nucleus, and begins to form a complex with CK1 (Baker, Kettenbach, Loros, Gerber, & Dunlap, 2009; Dunlap & Loros, 2004; Hurley et al., 2013; Merrow, Garceau, & Dunlap, 1997). In the early circadian morning, new FRQ is also promptly phosphorylated in the PEST-1 and FFD domains (sites of FRQ/FRQ and FRQ/FRH interactions; see below), with further phosphorylation events occurring via interaction with several kinases in the C-terminal region shortly thereafter. These C-terminal phosphorylations have been shown to stabilize the protein (reviewed in Baker, Loros, & Dunlap, 2012; Heintzen & Liu, 2007).
Figure 2.
Neurospora circadian cycle at the molecular level. (A) If FRQ is not able to bind to its stabilizer, FRH, it is degraded by default due to the inherently disordered nature of FRQ and is unable to complete its function in the circadian clock. (B) During the late subjective night of the circadian cycle, the WCC induces expression of frq mRNA, leading to a rapid increase in FRQ translation. FRQ forms a homodimer and binds to its stabilizer FRH, allowing for the IDP FRQ to avoid degradation by default. As the circadian day progresses, FRQ is phosphorylated via interaction with several kinases. FRQ inhibits the activity of the WCC by promoting the phosphorylation of the WCC, turning off frq transcription. FRQ levels decrease as no new FRQ is made while old FRQ is increasingly phosphorylated, which leads to ubiquitination facilitated by FWD-1, leading to FRQ degradation. (C) Factors that drive the output of the circadian clock. Low FRQ levels cause WCC activity to increase which subsequently leads to the expression of frq mRNA as well as mRNAs from other ccgs. FRQ binds to the WCC, promoting phosphorylation of the WCC and causing the WCC to become inactive. Decreasing FRQ levels allow phosphatases to bind the WCC, dephosphorylating the WCC, and increasing WCC activation. (D) Protein levels of the core clock components. While FRH and WC-2 remain constant, FRQ and WC-1 oscillate in opposite phases to one another. Stars represent phosphorylation and lightning bolts represent ubiquitination.
Upon entry into the nucleus, the FFC autoregulates its own transcription by inhibiting the activity of the WCC while simultaneously increasing the levels of WC-1 (Dunlap & Loros, 2004). It is thought that direct interaction between the FFC and the WCC leads to the phosphorylation of the WCC, inactivating the WCC as well as clearing the WCC from the frq promoter (reviewed in Brunner & Kaldi, 2008; Liu & Bell-Pedersen, 2006). The WCC exits the nucleus at this point in the cycle, perhaps a reflection of the phosphorylation status of the WCC, further decreasing activation of frq expression (Hong, Ruoff, Loros, & Dunlap, 2008). Due to the lack of WCC activation, by late afternoon frq expression declines and so does FRQ synthesis (Merrow et al., 1997). FRQ in the FFC is increasingly phosphorylated throughout the circadian day at the PEST domain and the N-terminal domain leading to the recognition of FRQ by an SCF-ubiquitin ligase complex containing the F-box protein, FWD-1, FRQ ubiquitination, and finally targeting of FRQ to the proteasome for degradation (reviewed in Baker et al., 2012; Heintzen & Liu, 2007). In the standard model for fungal/animal clocks, the mass of WC-1 that was held inactive by the FFC is now released and the cycle restarts, with the now unbound WCC again binding to the frq promoter (Dunlap & Loros, 2004). More recent work, however, has identified robust rhythms in FRQ-LUC expression in Δfwd-1 strains, suggesting that complete turnover of FRQ is not necessary for reinitiation of synthesis but instead is correlated with the initiation of new synthesis. The real end of the cycle in this revised model occurs when FRQ is sufficiently posttranslationally modified that it becomes invisible to the circadian machinery (Larrondo et al., under revision). In any case, the tight regulation that leads to delays between frq expression and FRQ synthesis (3–6 h) and FRQ phosphorylation and eventual degradation (14–18 h) leads to the approximately 22.5 h rhythm in Neurospora and sets the specific circadian rhythm (Merrow et al., 1997).
3. CORE CLOCK COMPONENTS
3.1. The FRQ/FRH complex
The discovery of several mutants, each of which directly affects the period of banding in Neurospora, vaulted the organism to a key model for circadian rhythms at the molecular level. These mutants were all mapped to the frq locus and displayed long, short, or arrhythmic periods, including some interesting alleles which also altered or disrupted temperature compensation (Gardner & Feldman, 1980; Loros & Feldman, 1986). frq itself was cloned leading to the current in depth understanding of the molecular clock of Neurospora and a greater understanding of clocks in general (McClung, Fox, & Dunlap, 1989). The periodic change in FRQ levels and phosphorylation match the conidiation rhythm seen in the bd mutant (discussed Section 1.1). The demonstration that FRQ is the driver of the Neurospora period came with the observation that altering or inhibiting the FRQ rhythm had a direct and equivalent effect on the clock (Aronson et al., 1994; Belden, Larrondo, et al., 2007; Garceau, Liu, Loros, & Dunlap, 1997).
As noted above, FRQ constitutes one of two proteins that make up the negative arm of the clock. Full-length FRQ contains 989 amino acids and dimerizes via a coil–coil region near the N′-terminus (Aronson et al., 1994; Cheng, Yang, Heintzen, & Liu, 2001). The message, as well as the protein of frq, is rhythmically expressed in a 22.5 h cycle under constant conditions with a phase difference of approximately 4 h (Aronson et al., 1994; Garceau et al., 1997). FRQ is highly regulated at the transcriptional, posttranscriptional, translational, and posttranslational levels (see below; Baker et al., 2012). FRQ is found in both the nuclear and cytoplasmic fractions but most of the activity attributed to FRQ is nuclear, where it binds to and blocks the transcriptional activity of the WCC (Liu, He, & Cheng, 2003). FRQ is believed to increase WC-1 levels and this activity is probably the result of inhibiting the activity of WC-1, a protein believed to be unstable when it is active (Shi, Collett, Loros, & Dunlap, 2010, reviewed in Baker et al., 2012). FRQ has also been shown to increase the abundance of wc-2 through an unknown mechanism (Liu et al., 2003).
In the regulation of FRQ transcription, it is the rhythmic binding of the relevant transacting factors that maintains a functional circadian clock. This regulation occurs at the frq promoter through the binding of the WCC proteins to two distinct cis-acting sequences termed the Clock box (C-box) and the proximal light-regulated element (PLRE) (Froehlich, Loros, & Dunlap, 2003). The role of the C-box is to regulate the rhythmic expression of frq and overall clock function in continual darkness, whereas the PLRE is essential to establish the proper phase when entrained by light (discussed below). While the combined function of these elements is responsible for high levels of light-induced frq expression via WCC influence on the frq promoter, each element acts differentially as chromatin is remodeled during the transcriptional activation and deactivation of frq (Belden, Loros, & Dunlap, 2007; Wang et al., 2014).
In order to properly regulate frq expression, the protein encoded by the gene clockswitch (csw-1) is required and also acts to negatively regulate WCC activity at frq by altering chromatin structure, creating a more compact chromatin structure at the C-box (Belden, Loros, et al., 2007). Chromodomain helicase DNA-binding (CHD-1) can also contribute to changes in chromatin structure at frq and is needed for normal frq expression. DNA methylation at frq, which is promoted by the loss of CHD-1, is transient, reversible, and catalyzed by the DNA methyltransferase DIM-2, which limits the onset of circadian regulated transcription via regulation of methylation at the frq promoter (Belden, Lewis, Selker, Loros, & Dunlap, 2011; Belden, Loros, et al., 2007). Recent results have added to the understanding of how frq expression is regulated in the light versus in the dark by the clock (Wang et al., 2014). When WCC binds to the C-box it recruits the SWI/SNF complex, a well-known chromatin modifying complex that is also involved in DNA bending. SWI/SNF in conjunction with other components removes a nucleosome from the C-box and also bends the DNA so that this region is brought into proximity with the transcription start site to initiate frq expression. Interestingly, and consistent with this model, loss of SWI/SNF abrogates circadian rhythms but has little to no effect on light-induced frq expression that is driven by the TSS PLRE.
There are a great many factors that affect frq mRNA regulation beyond transcriptional regulation. frq encompasses two translation initiation sites that result in the production of two distinct FRQ polypeptides: FRQ1–989 (L-FRQ) and FRQ100–989 (S-FRQ; Garceau et al., 1997). Both forms of FRQ independently maintain rhythmicity at 25 °C, but the amplitude and robustness of the FRQ rhythms are affected across a range of physiological temperatures. At higher temperatures, a higher ratio of L-FRQ is produced while relatively even amounts of long- and short-FRQ proteins are maintained at lower temperatures (Liu, Garceau, Loros, & Dunlap, 1997). The phosphorylation of sites on the 100 amino acids on L-FRQ that are not present in S-FRQ decreases period length even in the presence of S-FRQ (discussed further below). It is believed that the ratio of FRQ polypeptides is an additional level of fine tuning of the clock which allows the period to respond to environmental cues while at the same time allowing it to remain a robust timekeeping mechanism in their absence (Baker et al., 2009; Diernfellner et al., 2007; Liu et al., 1997).
The determining factor in the selective transcription of either S-FRQ or L-FRQ is temperature, which triggers an alternative splicing event of a small intron encompassing the AUG of L-FRQ. At higher temperatures, this intron is retained resulting in the preferential use of the AUG from L-FRQ; lower temperatures trigger the removal of the intron, making the AUG from S-FRQ an equally likely start codon. Strains unable to splice this intron fail to produce S-FRQ (Colot, Loros, & Dunlap, 2005; Diernfellner, Schafmeier, Merrow, & Brunner, 2005). Alternative splicing events farther upstream in the 5′ UTR remove five upstream AUGs with four uORFs from all major frq transcripts to further regulate the expression of the frq transcript. Two AUGs remain and these uORFs may be differentially regulating S-FRQ and L-FRQ at the translational level by targeting transcripts for nonsense-mediated decay, either as a way to remove improperly spliced transcripts or as a mechanism for quantitative control of gene expression (Colot et al., 2005; Diernfellner et al., 2005).
A final level of regulation on the frq transcript is the qrf antisense transcript, which comprises the entire length of the FRQ open reading frame. Elimination of the qrf transcript leads to a slight period increases as well as rhythm loss at low physiological temperatures and earlier phase setting upon light to dark transfer. qrf antisense RNA may be an additional level of regulation on frq posttranscriptionally in order to further insulate the clock from environmental stresses (Kramer, Loros, Dunlap, & Crosthwaite, 2003).
While frq mRNA and protein levels change with a circadian periodicity, FRQ is also rhythmically phosphorylated, which, among other things, has a direct influence on FRQ turnover kinetics (Garceau et al., 1997; Liu, Loros, & Dunlap, 2000). FRQ is phosphorylated rapidly upon translation and this phosphorylation continues in a highly regulated manner throughout the circadian day (Baker et al., 2009). When sites known to be phosphorylated are mutated to eliminate phosphorylation, FRQ stability is increased, which in turn leads to increased period lengths (Liu et al., 2000; Ruoff, Loros, & Dunlap, 2005). FRQ is also phosphorylated in constant light, though in a less specific and regulated manner (Baker et al., 2009; Tang et al., 2009).
The phosphorylations occur in clusters at specific times over the circadian day, with no specific phosphorylation event acting as the key determinant for any action in FRQ. At the start of the circadian day, FRQ is completely unphosphorylated. As time passes, FRQ is rapidly phosphorylated in the central regions, particularly between the PEST-1 and the FFD domain. The function of these central modifications has yet to be determined as mutations at these sites did not alter circadian rhythms. Next, the C-terminal regions are phosphorylated which increases FRQ protein stability. Mutations in this region result in a short-period rhythm. The PEST-1 domain shows a dramatic increase in phosphorylation midway through the circadian day. The phosphorylation of these residues is needed to promote turnover of FRQ as mutations of sites in this region showed an increase in period and more stable FRQ. Finally, phosphorylation of residues specific to L-FRQ, occurs late in the cycle. Mutations in the L-FRQ only region result in a longer period, suggesting a role in promoting turnover (Baker et al., 2009).
In total, FRQ has around 100 distinct modifications. To complete this extensive phosphorylation, there is a complex network of kinases and phosphatases. Many kinases are found to interact with the FFC, including casein kinases 1 and 2 (CK1a and CK2), a Neurospora homolog of checkpoint kinase-2 (PRD-4), as well as CAMK-1, and basophilic protein kinase A (Klengel et al., 2005); CK1a, CK2, and PRD-4 appear to directly interact with FRQ (Baker et al., 2012; Diernfellner & Schafmeier, 2011). The interaction of CK1a with FRQ is via two FRQ/CK1a interacting domains (FCDs) on FRQ. This interaction not only catalyzes the phosphorylation of FRQ (as many as 41 times) which is believed to lead to FRQ degradation, but may play a role in the clock-dependent phosphorylation of the WCC as well (He, Cha, Lee, Yang, & Liu, 2006; Querfurth et al., 2011). This lends credence to the hypothesis that FRQ acts as a scaffold for major components of the clock. CK2 also interacts with FRQ and these phosphorylations are involved in maintaining the temperature compensation function of the clock (Mehra et al., 2009). In addition to kinases, several phosphatases play a role in the clock, including protein phosphatase-1 (PP1), PP2a, and PP4. Phosphatases regulate FRQ stability, influencing frq transcription, dephosphorylate the WCC, and affect WCC subcellular localization (Baker et al., 2012).
In addition to effects on stability, it has been suggested that FRQ structure is directly affected by its phosphorylation (Querfurth et al., 2011). In the hypophosphorylated state, FRQ is in a closed conformation, which opens upon increasing phosphorylation presumably due to charge–charge repulsion, revealing a degradation signal in the middle portion of FRQ. New FRQ adopts preferentially the closed conformation as the positively charged N-terminal domain interacting with the negatively charged remainder portion of the protein due to the lack of phosphorylation. As the N-terminal domain of FRQ is progressively phosphorylated, it lowers the pI of the domain, increasing negative surface charge of the N-terminal domain and weakening the interaction with the negatively charged middle and C-terminal domains (Querfurth et al., 2011). However, this model fails to explain that N-terminal phosphorylations were previously shown to be among the last modifications during the circadian cycle rather than being among the first (Baker et al., 2009). Recently, FRQ was demonstrated to be an intrinsically disordered protein (IDP). Flexibility in FRQ structure allows for flexibility of binding, high levels of posttranslational modifications, ubiquitination, and a variety of protein–protein interactions to occur, all things that are necessary for proper FRQ function in the clock (Hurley et al., 2013).
In addition to phosphorylation, there are other posttranslational modifications that affect the degradation of FRQ, including ubiquitination (He & Liu, 2005). The F-box/WD40 repeat-containing protein FWD-1 has been shown to directly interact with the phosphorylated form of FRQ and is essential for FRQ’s degradation. Phosphorylated FRQ appears to be a substrate for an FWD-1-containing SCF-type ubiquitin ligase complex that this SCF complex can recognize different phosphorylated motifs within FRQ. The more phosphorylated FRQ is the more potential FWD-1-binding sites are present on FRQ so this increases its affinity toward FWD-1 (He, Cheng, Yang, Yu, & Liu, 2003). This data lends credibility to the idea that progressive phosphorylation of FRQ may be a dynamic process that fine-tunes the stability of FRQ through its role in the ubiquitination of FRQ. Beyond the FWD-1 role in ubiquitination, it is believed that there may be other FRQ mechanisms of degradation including degradation by default (discussed later in this chapter; He et al., 2003; Hurley et al., 2013).
The second component of the FFC is FRH, a homolog of Mtr4p, which is a well-studied cofactor of the Saccharomyces cerevisiae exosome (Cheng, He, Wang, & Liu, 2005). Mtr4p is a member of the TRAMP complex and has been played a role in the exosome in yeast (LaCava et al., 2005). All FRQ is bound to FRH and when FRH levels are depleted via siRNA knockdown or use of the regulatable qa-2 promoter (FRH is an essential gene in Neurospora), the clock loses rhythmicity completely and FRQ protein level decreases dramatically while mRNA increases (Cheng et al., 2005; Shi et al., 2010).
Due to its similarity to Mtr4p, initial studies were aimed at showing that FRH knockdown has an indirect effect on the clock because it regulates the levels of frq posttranscriptionally, as when FRH is knocked down, frq mRNA is stabilized (Guo, Cheng, Yuan, & Liu, 2009). A more direct role for FRH is in the complex interaction between FRQ and the WCC; FRH is essential to the interaction between the FFC and the WCC. FRH is also able to interact with the WCC in the absence of FRQ (Cheng et al., 2005; Guo, Cheng, & Liu, 2010; Shi et al., 2010). FRH has been implicated in the proper methylation of frq (Belden et al., 2011), as well as being an essential interactor of VVD (described later) in suppression of FRQ expression via interaction with the WCC (Hunt, Thompson, Elvin, & Heintzen, 2010). The association between FRQ and FRH is essential for the proper phosphorylation and stability of FRQ. In addition to this, it appears that FRH plays a role in the proper localization of FRQ protein (Cha, Yuan, & Liu, 2011; Guo et al., 2010) though it is important to note that FRH is cytoplasmic and the nuclear-cytoplasmic shuttling has been suggested to be dependent on the phosphorylation state of FRQ (Diernfellner, Querfurth, Salazar, Hofer, & Brunner, 2009). A point mutant of FRH was identified through a mutagenesis screen for negative feedback loop mutants. This mutation is outside the highly conserved helicase region of FRH and eliminates the interaction of FRH with the WCC but not with FRQ (Shi et al., 2010).
The mutation hints that the role of FRH that is specific to the clock may be different from its role in the TRAMP/exosome complex function that FRH plays for overall cell fitness. Recent work has been shown that the loss of the helicase function of FRH does not affect the running of the circadian cycle. IDPs, of which FRQ is one, follow two paths in stability: either they are degraded by default or they bind to a partner molecule to stabilize their structure. FRH has been shown to stabilize FRQ and recent work suggests that the role of FRH may be to act as a partner protein, or Nanny, to stabilize the IDP FRQ and allow it to perform its multitude of functions (Hurley et al., 2013). A competing theory posits that it is the ATPase function of FRH that regulates the function of CK1a, allowing for the proper phosphorylation of FRQ (Lauinger, Diernfellner, Falk, & Brunner, 2014); however, the retention of a clock in strains bearing FRH-lacking ATPase and helicase activity is hard to explain through such a model.
3.2. The White Collar Complex
WC-1 protein is GATA-like Zn-finger transcription factor that contains three PAS (Per-Ant-Sim) domains. The N-terminal-most PAS domain is of a special subclass, called the LOV domain. WC-1 interacts through its C-terminal-most PAS domain with the PAS domain of WC-2 to form a heterodimer (Cheng, Yang, Gardner, & Liu, 2002; Linden & Macino, 1997). WC-2 also contains a Zn-finger domain, but lacks a LOV domain for direct light sensing. The WCC binds to the C-box and PLRE where it functions as a transcriptional activator of FRQ. The WCC actually exists in two forms (Cheng, Yang, Wang, He, & Liu, 2003; Froehlich, Liu, Loros, & Dunlap, 2002). The first is the WC-1/2 heterodimer (small complex) that binds to promoter elements most strongly in the dark. frq expression driven by the small WCC is independent of the LOV domain of WC-1 and is thus independent of photosensing role of the WCC. Upon light exposure, the small complex is replaced on the DNA by a larger WCC, which consists of the WC-1/2 heterodimer with the addition of several more WC-1 proteins that interact via their LOV domains (Cheng et al., 2002; Collett, Garceau, Dunlap, & Loros, 2002; Linden, Ballario, & Macino, 1997).
When either of the WCC genes is knocked out, clock function is eliminated (Crosthwaite, Dunlap, & Loros, 1997). This is because the WCC complex binds the frq promoter and is responsible for the expression of frq mRNA (Froehlich et al., 2002). The WCC is then inhibited through direct interaction with the FFC. The interaction between the two main complexes of the circadian clock causes both WC-1 and WC-2 to be phosphorylated in a circadian manner and it is this phosphorylation that regulates the activity of the complex. The phosphorylation of WC-2 regulates the binding of the WCC to DNA. While currently only one site has been identified on WC-2 and it has been suggested that there are many more that could affect both the period and the stability of the protein (Sancar, Sancar, Brunner, & Schafmeier, 2009). WC-2 is necessary for interaction between the WCC and FRQ; although transcription of wc-2 is weakly rhythmic (Hurley et al., in press) and it is both positively regulated by FRQ and negatively regulated by WC-1 (Liu et al., 2003), there is no rhythm to WC-2 content. As a complex, WCC stability is regulated by the CCR4-NOT complex (Huang, He, Guo, Cha, & Liu, 2013). The phosphorylation of WC-1 is also circadian; the phosphorylation at sites near to the Zn-finger DNA-binding domain is believed to regulate the ability of WC-1 to activate transcription (He et al., 2005). In addition to its activation role, WC-1 is also needed for the interaction between the WCC and FRQ. WC-1 level cycles, though this rhythm is not necessary for the clock and WC-1 is stabilized by WC-2.
3.3. The input and output of the clock
A circadian clock is beneficial because it allows the host organism to be sensitive to environmental input. The phase resetting and entrainment by light causes differential effects on frq expression and impacts the clock differently depending on the time in the ‘ that the light is seen. When frq expression is low, early in the circadian morning, exposure to light will increase frq levels and this will advance the clock to the time corresponding to the highest frq expression, mid-to-late circadian morning. The sharp increase of frq levels due to light exposure during the time of declining frq (late circadian afternoon) will lead to phase delays as frq levels are forced to return to their maximum mid-day levels after light exposure (Crosthwaite et al., 1997), in accordance with predictive phase response curves.
Light input to the clock is modified by the VIVID protein that is itself clock regulated and can gate the light response on the clock (Heintzen, Loros, & Dunlap, 2001). VVD is a small LOV-domain protein whose name stems from the phenotype of its loss-of-function mutants which display bright orange conidia when grown in constant light, attributed to the persistence activation of carotenoid pigments (Heintzen et al., 2001). The vvd promoter is a direct target of the WCC and transcript levels increase dramatically following illumination. VVD inhibits the WCC and this action sets the clock at the dusk transition as well as contributing to temperature compensation (Elvin, Loros, Dunlap, & Heintzen, 2005; Hunt, Elvin, Crosthwaite, & Heintzen, 2007). In addition, VVD levels in the dark inactivate any WCC induced by moonlight and keep the clock in phase during in the bright moonlight nights (Malzahn, Ciprianidis, Kaldi, Schafmeier, & Brunner, 2010).
Beyond light, temperature plays a role in both the entrainment as well as on period of the clock. When shifting to higher temperatures, levels of L-FRQ as well as levels of FRQ overall increase (Garceau et al., 1997; Liu et al., 1997). The FRQ levels at the shift are lower than the lowest FRQ levels at the higher temperature, so the clock resets to subjective morning (Liu, Merrow, Loros, & Dunlap, 1998). Metabolism is known to play a role into clock input via a feedback loop on the positive arm by CSP-1 (Sancar, Sancar, & Brunner, 2012).
In order for the core molecular circuit to affect organismal behavior, there must be a method to provide rhythmic information to regulate the cell. To do this, the positive arm of the clock, the WCC, regulates a subset of rhythmically expressed genes termed the clock-controlled genes or ccgs. It is estimated that 5–15% of the genome is circadianly regulated in Neurospora (Dong et al., 2008; Dunlap & Loros, 2004) (Hurley et al in press). The expression of these genes is not synchronized but is actually staggered in their expression over circadian time, with late night to morning expression most common. While most WCC-driven/light-induced genes are also ccgs, there are distinct subsets of WCC-driven genes that are driven in the light and in the dark, showing that the dark expression is distinct from the role of the WCC in the light response (Dunlap & Loros, 2004).
Neurospora is the first system to establish a method of identification of ccgs, using subtractive hybridization to compare total nucleic acid levels between samples (Loros et al., 1989). This method and many of the methods used subsequently (differential hybridization, SAGE analysis, and microarrays) have many technical limitations, not the least of which was that the genes identified tended to be the most highly expressed genes (Bell-Pedersen, Shinohara, Loros, & Dunlap, 1996; Duffield et al., 2002; Zhu et al., 2001); these methods have been thoroughly reviewed (Duffield, Loros, & Dunlap, 2005). This is significant due to the low copy number of the most commonly tested ccg, frq (Merrow et al., 1997). Currently, the most common method of ccg discovery, and one that we have used successfully in our lab, is to follow mRNA levels using RNA deep sequencing. RNA extracted over circadian time is subjected to a standard RNA deep-sequencing analysis. The resulting data is normalized using RPKM values and then subjected to an analysis of cycling, i.e., JTK cycle (Hughes, Hogenesch, & Kornacker, 2010) (Hurley et al., in press).
The regulation of rhythmic gene expression occurs in part at transcription. The promoter of a well-studied ccg, ccg-2, is the perfect example of a circadianly regulated gene and contains several regulatory regions that individually confer light, developmental, and circadian regulation. The activating circadian element (ACE) is sufficient to confer clock regulation on this promoter and in other clock promoters (Bell-Pedersen, Dunlap, & Loros, 1996). The core ACE sequence is different from the core LRE sequence (discussed above) though both of these elements mediate clock control. Some known ccgs have neither element, suggesting hierarchical control in which the oscillator directly regulates oscillator proximal controllers that in turn regulate more downstream genes, or additional clock-control elements (Chen, Ringelberg, Gross, Dunlap, & Loros, 2009; Dunlap & Loros, 2004).
Another level of regulation can occur when the mitogen-activated protein kinase (MAPK) pathways are regulated by the clock at the transcriptional level by the WCC (Bennett, Beremand, Thomas, & Bell-Pedersen, 2013; Lamb, Finch, & Bell-Pedersen, 2012; Lamb, Goldsmith, Bennett, Finch, & Bell-Pedersen, 2011). In addition to this regulation, MAPK-1 has been shown to be phosphorylated in a circadian manner and its targets are ccgs, demonstrating the circadian clock signal can be propagated outside of the WCC regulation (Bennett et al., 2013). Recently, the Neurospora circadian cycle has been shown to play a role in cell cycle of the organism (Hong et al., 2014).
4. CONCLUSION
Neurospora remains a durable model organism for the study of circadian rhythms because it is so experimentally tractable and yet retains all the regulatory elements and regulatory architecture common to clocks in larger and more complicated organisms. Studies in Neurospora were the first to establish the essential nature of transcriptional negative feedback in the clock, to establish mechanisms for light resetting and for temperature resetting, and it was the first system in which a heterodimer of PAS-containing protein was proposed as the positive element in the feedback loop. The first systematic screens for genes regulated by the clock were performed in Neurospora, setting the stage for broadly envisioned analysis of output pathways. More recently work on Neurospora first showed the interconnection between cell cycle and circadian regulation and probed the involvement of phosphorylation in the mechanism of temperature compensation. Studies in Neurospora have highlighted the fact that many clock proteins may be IDPs and how this structure supports their role in maintaining a clock, and are also revealing the mechanisms through which antisense transcripts to clock genes play a role in rhythm persistence. Work in Neurospora has been shown that the circadian feedback loops can close through phosphorylations alone and do not need to close through phosphorylation-mediated clock protein turnover. These findings all presaged similar findings in animal circadian systems: it is the ability of Neurospora to predict how more complex systems work that makes it an excellent model.
References
- Aronson BD, Johnson KA, Loros JJ, Dunlap JC. Negative feedback defining a circadian clock: Autoregulation of the clock gene frequency. Science. 1994;263:1578–1584. doi: 10.1126/science.8128244. [DOI] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Molecular Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC. The circadian clock of Neurospora crassa. FEMS Microbiology Review. 2012;36:95–110. doi: 10.1111/j.1574-6976.2011.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, Loros JJ, et al. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes & Development. 2007;21:1494–1505. doi: 10.1101/gad.1551707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Lewis ZA, Selker EU, Loros JJ, Dunlap JC. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genetics. 2011;7:e1002166. doi: 10.1371/journal.pgen.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Molecular Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Dunlap JC, Loros JJ. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Molecular and Cellular Biology. 1996;16:513–521. doi: 10.1128/mcb.16.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D, Shinohara ML, Loros JJ, Dunlap JC. Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13096–13101. doi: 10.1073/pnas.93.23.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LD, Beremand P, Thomas TL, Bell-Pedersen D. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryotic Cell. 2013;12:59–69. doi: 10.1128/EC.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VG, Weight F, Pittendrigh CS. Resetting the sporulation rhythm in Pilobolus with short light flashes of high intensity. Science. 1960;131:728–730. doi: 10.1126/science.131.3402.728. [DOI] [PubMed] [Google Scholar]
- Brunner M, Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Molecular Microbiology. 2008;68:255–262. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- Canessa P, Schumacher J, Hevia MA, Tudzynski P, Larrondo LF. Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: Characterization of the White Collar Complex. PLoS One. 2013;8:e84223. doi: 10.1371/journal.pone.0084223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Yuan H, Liu Y. Regulation of the activity and cellular localization of the circadian clock protein FRQ. The Journal of Biological Chemistry. 2011;286:11469–11478. doi: 10.1074/jbc.M111.219782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO Journal. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, Wang L, Liu Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes and Development. 2005;19:234–241. doi: 10.1101/gad.1266805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Molecular and Cellular Biology. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. The EMBO Journal. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. Journal of Biological Chemistry. 2003;278:3801–3808. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- Collett MA, Garceau N, Dunlap JC, Loros JJ. Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics. 2002;160:149–158. doi: 10.1093/genetics/160.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collopy PD, Colot HV, Park G, Ringelberg C, Crew CM, Borkovich KA, et al. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods in Molecular Biology. 2010;638:33–40. doi: 10.1007/978-1-60761-611-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Loros JJ, Dunlap JC. Temperature-modulated alternative splicing and promoter use in the circadian clock gene frequency. Molecular Biology of the Cell. 2005;16:5563–5571. doi: 10.1091/mbc.E05-08-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- Davis RH, Perkins DD. Timeline: Neurospora: A model of model microbes. Nature Reviews Genetics. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- Diernfellner A, Colot HV, Dintsis O, Loros JJ, Dunlap JC, Brunner M. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Letters. 2007;581:5759–5764. doi: 10.1016/j.febslet.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Querfurth C, Salazar C, Hofer T, Brunner M. Phosphorylation modulates rapid nucleocytoplasmic shuttling and cytoplasmic accumulation of Neurospora clock protein FRQ on a circadian time scale. Genes & Development. 2009;23:2192–2200. doi: 10.1101/gad.538209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diernfellner AC, Schafmeier T. Phosphorylations: Making the Neurospora crassa circadian clock tick. FEBS Letters. 2011;585:1461–1466. doi: 10.1016/j.febslet.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes & Development. 2005;19:1968–1973. doi: 10.1101/gad.345905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, et al. Systems biology of the clock in Neurospora crassa. PLoS One. 2008;3:e3105. doi: 10.1371/journal.pone.0003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Current Biology. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Duffield G, Loros JJ, Dunlap JC. Analysis of circadian output rhythms of gene expression in Neurospora and mammalian cells in culture. Methods in Enzymology. 2005;393:315–341. doi: 10.1016/S0076-6879(05)93014-0. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. The Neurospora circadian system. Journal of Biological Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. How fungi keep time: Circadian system in Neurospora and other fungi. Current Opinion in Microbiology. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Elvin M, Loros JJ, Dunlap JC, Heintzen C. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes and Development. 2005;19:2593–2605. doi: 10.1101/gad.349305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Loros JJ, Dunlap JC. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5914–5919. doi: 10.1073/pnas.1030057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- Gardner GF, Feldman JF. The frq locus in Neurospora crassa: A key element in circadian clock organization. Genetics. 1980;96:877–886. doi: 10.1093/genetics/96.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, et al. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryotic Cell. 2008;7:28–37. doi: 10.1128/EC.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AV, Keller N, Haas H, Bell-Pedersen D. A circadian oscillator in Aspergillus spp. regulates daily development and gene expression. Eukaryotic Cell. 2003;2:231–237. doi: 10.1128/EC.2.2.231-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Cheng P, Liu Y. Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. The Journal of Biological Chemistry. 2010;285:11508–11515. doi: 10.1074/jbc.M109.071688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Cheng P, Yuan H, Liu Y. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell. 2009;138:1236–1246. doi: 10.1016/j.cell.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cha J, Lee HC, Yang Y, Liu Y. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes & Development. 2006;20:2552–2565. doi: 10.1101/gad.1463506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Yu H, Liu Y. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. The EMBO Journal. 2003;22:4421–4430. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Liu Y. Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway. Biochemical Society Transactions. 2005;33:953–956. doi: 10.1042/BST20050953. [DOI] [PubMed] [Google Scholar]
- He Q, Shu H, Cheng P, Chen S, Wang L, Liu Y. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. The Journal of Biological Chemistry. 2005;280:17526–17532. doi: 10.1074/jbc.M414010200. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Liu Y. The Neurospora crassa circadian clock. Advances in Genetics. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Hong CI, Ruoff P, Loros JJ, Dunlap JC. Closing the circadian negative feedback loop: FRQ-dependent clearance of WC-1 from the nucleus. Genes and Development. 2008;22:3196–3204. doi: 10.1101/gad.1706908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CI, Zamborszky J, Baek M, Labiscsak L, Ju K, Lee H, et al. Circadian rhythms synchronize mitosis in Neurospora crassa. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1397–1402. doi: 10.1073/pnas.1319399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, He Q, Guo J, Cha J, Liu Y. The Ccr4-not protein complex regulates the phase of the Neurospora circadian clock by controlling white collar protein stability and activity. The Journal of Biological Chemistry. 2013;288:31002–31009. doi: 10.1074/jbc.M113.494120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Elvin M, Crosthwaite SK, Heintzen C. The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes and Development. 2007;21:1964–1974. doi: 10.1101/gad.437107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Thompson S, Elvin M, Heintzen C. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16709–16714. doi: 10.1073/pnas.1009474107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, Knabe N, et al. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proceedings of the National Academy of Sciences of the United States of America. doi: 10.1073/pnas.1418963111. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Larrondo LF, Loros JJ, Dunlap JC. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Molecular Cell. 2013;52:832–843. doi: 10.1016/j.molcel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Current Biology. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature. 2003;421:948–952. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Finch KE, Bell-Pedersen D. The Neurospora crassa OS MAPK pathway-activated transcription factor ASL-1 contributes to circadian rhythms in pathway responsive clock-controlled genes. Fungal Genetics and Biology. 2012;49:180–188. doi: 10.1016/j.fgb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TM, Goldsmith CS, Bennett L, Finch KE, Bell-Pedersen D. Direct transcriptional control of a p38 MAPK pathway by the circadian clock in Neurospora crassa. PLoS One. 2011;6:e27149. doi: 10.1371/journal.pone.0027149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Baker CL, Olivares-Yañez C, Loros JJ, Dunlap JC. Decoupling circadian clock protein turnover from circadian period determination. doi: 10.1126/science.1257277. (under revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrondo LF, Loros JJ, Dunlap JC. High-resolution spatiotemporal analysis of gene expression in real time: In vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genetics and Biology. 2012;49:681–683. doi: 10.1016/j.fgb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauinger L, Diernfellner A, Falk S, Brunner M. The RNA helicase FRH is an ATP-dependent regulator of CK1a in the circadian clock of Neurospora crassa. Nature Communications. 2014;5:3598. doi: 10.1038/ncomms4598. [DOI] [PubMed] [Google Scholar]
- Linden H, Ballario P, Macino G. Blue light regulation in Neurospora crassa. Fungal Genetics and Biology. 1997;22:141–150. doi: 10.1006/fgbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO Journal. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bell-Pedersen D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryotic Cell. 2006;5:1184–1193. doi: 10.1128/EC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Garceau NY, Loros JJ, Dunlap JC. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, He Q, Cheng P. Photoreception in Neurospora: A tale of two White Collar proteins. Cellular and Molecular Life Sciences. 2003;60:2131–2138. doi: 10.1007/s00018-003-3109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Loros J, Dunlap JC. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Merrow M, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- Loros JJ, Feldman JF. Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. Journal of Biological Rhythms. 1986;1:187–198. doi: 10.1177/074873048600100302. [DOI] [PubMed] [Google Scholar]
- Malzahn E, Ciprianidis S, Kaldi K, Schafmeier T, Brunner M. Photo-adaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell. 2010;142:762–772. doi: 10.1016/j.cell.2010.08.010. [DOI] [PubMed] [Google Scholar]
- McClung CR, Fox BA, Dunlap JC. The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature. 1989;339:558–562. doi: 10.1038/339558a0. [DOI] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow MW, Garceau NY, Dunlap JC. Dissection of a circadian oscillation into discrete domains. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3877–3882. doi: 10.1073/pnas.94.8.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan LW, Greene AV, Bell-Pedersen D. Circadian and light-induced expression of luciferase in Neurospora crassa. Fungal Genetics and Biology. 2003;38:327–332. doi: 10.1016/s1087-1845(02)00562-5. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman J, Nakashima H, Feldman JF. Assay and characteristics of circadian rhythmicity in liquid cultures of Neurospora crassa. Plant Physiology. 1981;67:404–407. doi: 10.1104/pp.67.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Bruce VG, Rosensweig NS, Rubin ML. Growth patterns in Neurospora: A biological clock in Neurospora. Nature. 1959;184:169–170. [Google Scholar]
- Querfurth C, Diernfellner AC, Gin E, Malzahn E, Hofer T, Brunner M. Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Molecular Cell. 2011;43:713–722. doi: 10.1016/j.molcel.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Taylor W. Automated recordings of bioluminescence with special reference to the analysis of circadian rhythms. Methods in Enzymology. 2000;305:104–119. doi: 10.1016/s0076-6879(00)05481-1. [DOI] [PubMed] [Google Scholar]
- Ruoff P, Loros JJ, Dunlap JC. The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17681–17686. doi: 10.1073/pnas.0505137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salichos L, Rokas A. The diversity and evolution of circadian clock proteins in fungi. Mycologia. 2010;102:269–278. doi: 10.3852/09-073. [DOI] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M. Metabolic compensation of the Neurospora clock by a glucose-dependent feedback of the circadian repressor CSP1 on the core oscillator. Genes & Development. 2012;26:2435–2442. doi: 10.1101/gad.199547.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M, Schafmeier T. Activity of the circadian transcription factor White Collar Complex is modulated by phosphorylation of SP-motifs. FEBS Letters. 2009;583:1833–1840. doi: 10.1016/j.febslet.2009.04.042. [DOI] [PubMed] [Google Scholar]
- Sargent ML, Briggs WR, Woodward DO. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiology. 1966;41:1343–1349. doi: 10.1104/pp.41.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Collett M, Loros JJ, Dunlap JC. FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics. 2010;184:351–361. doi: 10.1534/genetics.109.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CT, Li S, Long C, Cha J, Huang G, Li L, et al. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10722–10727. doi: 10.1073/pnas.0904898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traeger S, Altegoer F, Freitag M, Gabaldon T, Kempken F, Kumar A, et al. The genome and development-dependent transcriptomes of Pyronema confluens: A window into fungal evolution. PLoS Genetics. 2013;9:e1003820. doi: 10.1371/journal.pgen.1003820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Kettenbach AN, Gerber SA, Loros JJ, Dunlap JC. Neurospora WC-1 recruits SWI/SNF to remodel frequency and initiate a circadian cycle. PLoS Genetics. 2014;10:e1004599. doi: 10.1371/journal.pgen.1004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Nowrousian M, Kupfer D, Colot HV, Berrocal-Tito G, Lai H, et al. Analysis of expressed sequence tags from two starvation, time-of-day-specific libraries of Neurospora crassa reveals novel clock-controlled genes. Genetics. 2001;157:1057–1065. doi: 10.1093/genetics/157.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]