Abstract
Protein tyrosine phosphorylation is a reversible post-translational modification that is essential for life in eukaryotic cells. The combinatorial action of both protein tyrosine kinases and protein tyrosine phosphatases (PTPs) determines the net level of cellular tyrosine phosphorylation. This unit discusses methods to determine the level of protein tyrosine phosphatase activity and methods for discovering novel substrates for protein tyrosine phosphatases.
Keywords: protein tyrosine phosphatase, p-nitrophenyl phosphate, malachite green, in-gel phosphatase assay, PTP substrates, substrate-trapping
INTRODUCTION
In this unit, methods for measuring protein tyrosine phosphatase (PTP) activity and for identifying physiological PTP substrates are discussed. PTP activity can be determined by colorimetric, fluorescent, or radioactive assays. PTP assays, where small molecule PTP substrates are used, are simple to perform. However, if available, using phosphopeptides or phospho-substrates can provide a more accurate and physiologically relevant measure of the intrinsic properties of the PTP catalytic activity.
Basic Protocol 1 presents a colorimetric PTP assay that uses p-nitrophenyl phosphate (pNPP) as a substrate. Dephosphorylation of pNPP produces the yellow p-nitrophenylene anion allowing simple spectrophotometric quantitation of PTP activity. Basic Protocol 2 describes another spectrophotometric assay that involves the formation of a complex between malachite green molybdate and free orthophosphate. Basic Protocol 3 is a radioactive in-gel phosphatase assay that determines relative activity of phosphatases. The remaining protocols (see Basic Protocols 4 and 5, and Alternate Protocol) describe methods to discover and validate putative protein tyrosine phosphatase substrates.
p-NITROPHENYL PHOSPHATE (pNPP) ASSAY TO MEASURE PROTEIN TYROSINE PHOSPHATASE ACTIVITY
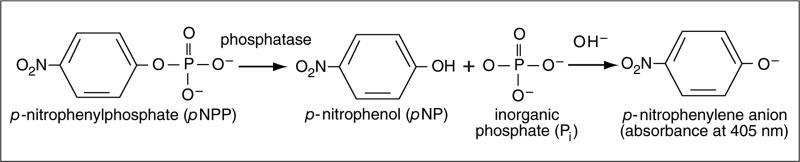
One method for measuring PTP activity is through the use of the small molecule substrate, p-nitrophenyl phosphate (pNPP; Fig. 18.16.1). In this microcentrifuge tube format assay, the cleavage of the phosphate from pNPP results in the production of p-nitrophenol (pNP), which causes an increase in absorbance at 405 nm. The advantages of the pNPP assay include having a wide linear range in reactions, as the substrate (pNPP) concentration is not limiting. In addition, pNPP is inexpensive. The disadvantages are that pNPP is neither a specific nor a physiological PTP substrate, and pNPP as a small molecule will not provide robust enzyme-substrate specific measures of the properties of the PTP under study. Nevertheless, this assay has proved to be very useful and reliable for measuring PTP activity, especially when comparing the activity of the same PTP among different experimental conditions.
Figure 18.16.1.
pNPP chemistry.
Materials
Sample containing PTP of interest
Antibody against PTP of interest for immunoprecipitation
p-Nitrophenyl phosphate (pNPP; mol. wt. = 263.1; Sigma) substrate
Lysis buffer I (see recipe)
Protease inhibitors
1 mM sodium orthovanadate (NaVO3)
10 mM sodium fluoride (NaF)
50% (v/v) slurry of protein A or protein G sepharose beads
Salt/Tris/EDTA (STE; see recipe)
Phosphatase wash buffer (see recipe)
1× and 1.25× pNPP reaction buffers (see recipes), prepared fresh
0.2 N NaOH
1× SDS sample buffer (see recipe)
Rotating or nutating platform, 4°C
37°C water bath
Plate reader/spectrophotometer (A405)
Additional reagents and equipment for immunoprecipitation (UNIT 10.16) and SDS-PAGE (UNIT 10.2A)
- Immunoprecipitate (UNIT 10.16) PTP of interest in lysis buffer that contains protease inhibitors, 1 mM sodium orthovanadate (NaVO3), and 10 mM sodium fluoride (NaF).Sodium orthovanadate is a protein tyrosine phosphatase inhibitor, and sodium fluoride is a serine/threonine phosphatase inhibitor.
Add 20 μl of 50% protein A or protein G sepharose slurry. Mix on a rotating or nutating platform for 1 hr at 4°C.
Centrifuge beads 1 min at 2500 × g, 4°C.
- Remove supernatant. Wash the beads inverting tube two to three times on ice centrifuging 30 sec at 2500 × g, as follows:
- Wash two times with 1 ml lysis buffer (without NaVO3 or NaF).
- Wash one time with 1 ml STE.
- Wash one time with 1 ml phosphatase wash buffer.
- Add 40 μl of freshly prepared 1.25× reaction buffer. Vortex quickly, spin briefly to pellet beads, and tap tube to mix.The reaction buffer must be used within 30 min of preparation.
Incubate 10 to 30 min at 37°C.
Stop the reaction by adding 1.45 ml of 0.2 N NaOH.
Pellet the beads by centrifuging 30 sec at 2500 × g; remove supernatant to a clean tube.
- Measure absorbance at 405 nm (use 1× reaction buffer as blank). Calculate PTPase activity as follows:PTPase activity (mol/min) = [(1.5 × 10−3)(A405/ε]/10 where, ε for pNPP = 1.78 × 104 M−1 cm−1.
- Add 10 μl of 1× SDS sample buffer to the beads, boil for 5 min, and run on an SDS-PAGE gel (UNIT 10.2A) to confirm equal PTP loading by western blot.If the PTP of interest is precipitated from an equivalent number of cells, the amount of input PTP should be equal for all experimental conditions. However, if in this step it appears that the PTP loading was not equal for all samples, then the experiment must be repeated making sure the exact starting input is achieved for the indicated conditions for all samples.
MALACHITE GREEN ASSAY TO MEASURE INORGANIC PHOSPHATE RELEASE
The malachite green assay is a simple, sensitive, and non-radioactive method for measuring inorganic phosphate release in aqueous solutions. The assay is based on the formation of a complex between malachite green molybdate and free orthophosphate that absorbs between 620 and 650 nm. This assay is a reliable means of detecting and quantifying very small amounts of free inorganic phosphate and can be used for high-throughput screening applications.
For this assay, the PTP of interest can either be immunoprecipitated from cells or purified from bacteria. The reaction is performed by mixing the purified PTP and a phosphorylated substrate peptide, followed by termination of the reaction by the addition of malachite green. The amount of free inorganic phosphate in the solution is measured by generating a standard curve by mixing known amounts of phosphate with the malachite green reagent. The amount of absorption at 650 nm is proportional to the amount of free phosphate. Comparing the absorption values of the experimental reaction samples to the standard curve gives the amount of free phosphate released during the dephosphorylation of the substrate. This assay is sufficiently sensitive to detect free phosphate levels in the picomolar range.
Materials
Malachite Green reagent no. 1 (see recipe)
Malachite Green reagent no. 2 (see recipe)
Tween 20
Stock solution of phospho-tyrosine substrate peptide (custom made) Immunoprecipitated PTP
1× and 10× PTP buffers (see recipes)
1 mM potassium dihydrogen phosphate (KH2PO4) in ddH2O (stable up to 1 year at 4°C)
0.2-μm filters
Microtiter plates (half-area, tissue culture-treated, flat-bottomed 96-well plates; Sigma)
Multichannel pipettor
Shaker (shake speed of 120 rpm)
Standard ELISA microtiter plate reader (A650)
- Prepare a working solution of Malachite Green reagent as follows: add 1 part Malachite Green reagent no. 1, 1 part Malachite Green reagent no. 2, 2 parts water, and Tween 20 to 0.01% (v/v); mix 5 min at room temperature, place on ice for 5 min, and then filter through a 0.2-μm filter.The working solution must be freshly prepared and used the same day.
- Dilute stock solution of phosphotyrosine substrate peptide with 10× PTP buffer and water to a final concentration of 1× PTP buffer and twice the final desired molarity of the phosphopeptide.To determine the phosphatase activity more accurately, the phosphotyrosine substrate peptide must be present in saturating concentrations. This saturating concentration must be determined individually for each substrate peptide.
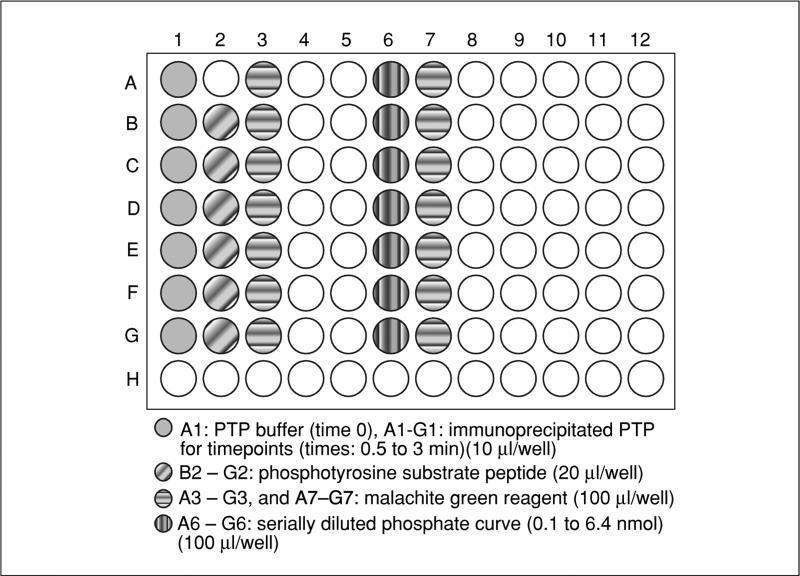
- Add 10 μl of 1× PTP buffer to well no. A1 of a 96-well microtiter plate (this will be time 0). Then, add 10 μl of immunoprecipitated PTP of interest (on beads) in 1× PTP buffer to wells A1 to G1 for each time point to be tested (see Fig. 18.16.2).In the example shown in Figure 18.16.2, the time points are from 0 to 3 min, every 30 sec.
- Pipet a slight excess (~20 μl) phosphopeptide into adjacent wells (B2 to G2, see Fig. 18.16.2). Pipet ~100 μl Malachite Green working solution (from step 1) into adjacent wells (A3 to G3 see Fig. 18.16.2).Pipetting the aliquots at this stage ensures proper and equal equilibration of all reagents in step 6.
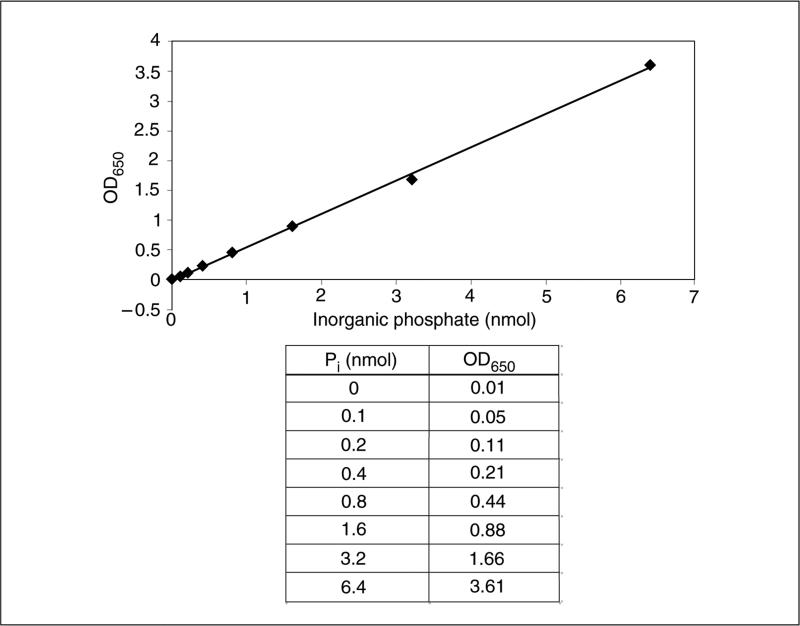
- At the same time, pipet 10 μl of KH2PO4 serially diluted with 1× PTP buffer to successive wells (A6 through G6) on a separate area of the plate×for the standard curve. Pipet ~100 μl of Malachite Green reagent into adjacent wells A7 through G7.A linear standard curve can be generated using 0.1 to 2 nmol of KH2PO4, which generates 650-nm absorption values between 0.05 and 1.0. An example standard curve can be seen in Figure 18.16.3.
Equilibrate the temperature of the reagents by placing the plate for 5 min at 30°C.
- Start the reaction by adding 10 μl of phosphopeptide (from B2 to G2 in Fig. 18.16.2) to wells containing the PTP of interest (from B1 to G1 in Fig. 18.16.2, except to the time-0-well A1) using a multichannel pipettor and begin timing. During the reaction, agitate the plate at 120 rpm.Total reaction volume at this stage is 20 μl.
- Stop the reaction by adding 80 μl of Malachite Green working reagent (from wells A3 to G3 in Fig. 18.16.2) to each assay after the appropriate incubation time, as well as to the time-0 well containing 1× PTP buffer and PTP of interest, and to the wells containing the KH2PO4 standards.Initially, one can perform a time course of 30-sec intervals from 0 min to 3 min. The time course can be increased to 10 min if the colorimetric reaction is weak, or shortened to 1 min if color formation is very strong.Total volume after addition of Malachite Green reagent is 100 μl.
- Leave plate 10 min at room temperature to allow the color to develop.Use the solution immediately after mixing. The color of the solution after quenching is stable for 20 to 30 min.
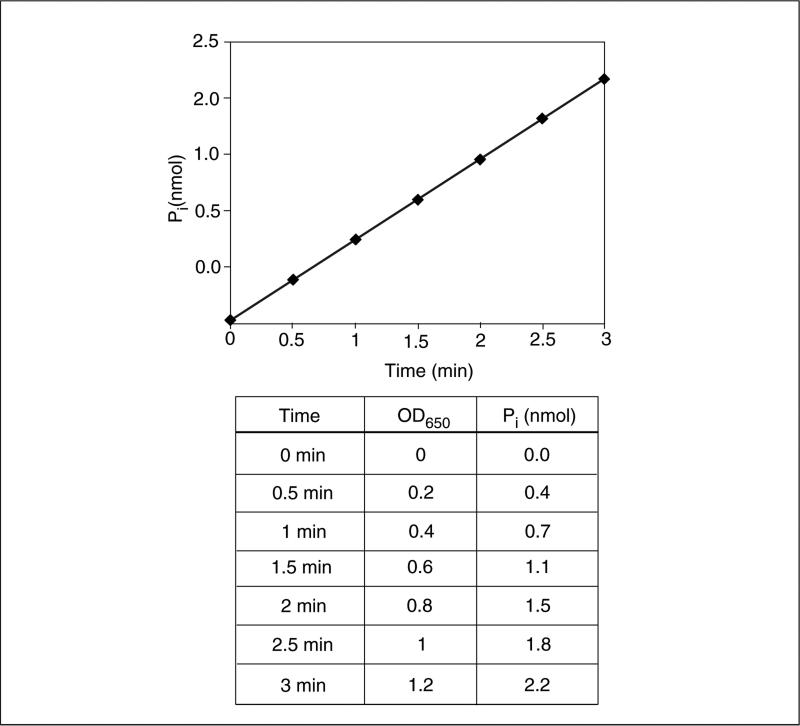
- Measure the absorbance at 650 nm using a multi-well microtiter plate reader to detect the release of inorganic phosphate. Use the standard curve generated with KH2PO4 to convert values to nanomoles (see example standard curve in Fig. 18.16.3).An example hypothetical data set and calculations are given in Figure 18.16.4. Briefly, the amount of phosphate released at each time point is calculated by the line equation of the standard curve (y = 0.558× −0.0146 for the standard curve in Fig. 18.16.3). The rate of reaction can be calculated by dividing the difference in the amount of inorganic phosphate released (2.13 nmol in 3-min time point) to the length of the experiment (3 min), which is 0.71 nmol/min in this example. Because the substrate concentration is not limiting, this calculated rate of reaction is the Vmax.
Figure 18.16.2.
An example 96-well plate layout for the Malachite Green assay.
Figure 18.16.3.
An example standard curve for the Malachite Green assay.
Figure 18.16.4.
A hypothetical data set for the Malachite Green assay.
IN-GEL PHOSPHATASE ASSAY TO DETERMINE RELATIVE PROTEIN TYROSINE PHOSPHATASE ACTIVITY
The in-gel phosphatase assay is a radioactive assay to determine the relative activity of PTPs. This assay can be used to assess the status of cellular PTP activity or the activity of a single affinity purified PTP. A modified version of the in-gel phosphatase assay can also differentiate between pools of active PTPs and PTPs that have been inactivated by oxidation (Meng et al., 2005). The disadvantage of the in-gel phosphatase assay is that it does not allow determination of the precise enzymatic properties, such as Vmax, Km, or substrate specificity, of the PTP under investigation; hence this approach is less well-suited for these types of analyses.
Briefly, cell lysates or affinity purified PTPs are electrophoresed under denaturing conditions through an SDS gel containing 32P-labeled peptide substrate, poly (Glu:Tyr) [4:1] or reduced carboxamidomethylated and maleylated lysozyme (RCML), which are prepared by phosphorylation with a tyrosine kinase, e.g., GST-FER (cell signaling) (FER protein tyrosine kinase fused to GST protein), in an in-vitro kinase assay. After electrophoresis, the gel is fixed and washed. Proteins in the gel are denatured in a guanidine HCl–containing buffer and then renatured in a DTT-containing buffer. The renatured PTPs regain their activity and dephosphorylate the radioactively labeled substrate in the region to which they migrated in the gel. After an overnight incubation, the dephosphorylation reaction is stopped by staining with Coomassie Blue. Coomassie Blue staining also provides a measure of relative protein loading. Afterwards, the gel is dried and exposed to X-ray film for visualization. The phosphatase activity is visualized as “blank” regions on a black background where radioactive phosphate has been removed from the substrate and washed away.
Materials
32P-labeled substrate (poly (Glu:Tyr) [4:1] or RCML)
Affinity purified PTP or total cell lysate
Fixation buffer (see recipe)
Wash buffer I (see recipe)
Denaturation buffer (see recipe)
Renaturation buffer with and without DTT (see recipe)
Coomassie Blue staining solution (see recipe)
Coomassie Blue destaining solution (see recipe)
Rocking platform shaker
X-ray film
Additional reagents and equipment for SDS gel electrophoresis (UNIT 10.2A)
Cast an SDS gel (UNIT 10.2A) containing 32P-labeled substrate at a level of 1.5 × 106 cpm/20 ml (~2μM pTyr).
Electrophorese the affinity purified PTP or total cell lysate until the dye front runsoff the gel.
Fix the gel in a glass tray with 300 ml fixation buffer overnight on a rocker at room temperature.
Wash the gel two times in 250 ml wash buffer I for 25 min each time at room temperature to remove the isopropanol.
Incubate gel in 300 ml denaturation buffer for 90 min at room temperature to denature the proteins.
Wash gel two times with 250 ml of renaturation buffer without DTT at room temperature for 1 hr each time.
Incubate gel in 250 ml renaturation buffer containing 3 mM DTT for 1 hr at room temperature to start renaturation.
Incubate the gel in 250 ml fresh renaturation buffer containing 3 mM DTT overnight at room temperature.
Stop the dephosphorylation reaction by staining the gel with 300 ml Coomassie Blue staining solution rocking at room temperature.
- Destain gel in 300 ml Coomassie Blue destaining solution until bands are clearly visible and dry the gel in a gel dryer. Expose gel to X-ray film for 1 day at −80°C (Fig. 18.16.5).Depending on the result of the 1-day exposure, longer (a few days to 1 week) or shorter (a few hours) exposures can be taken.
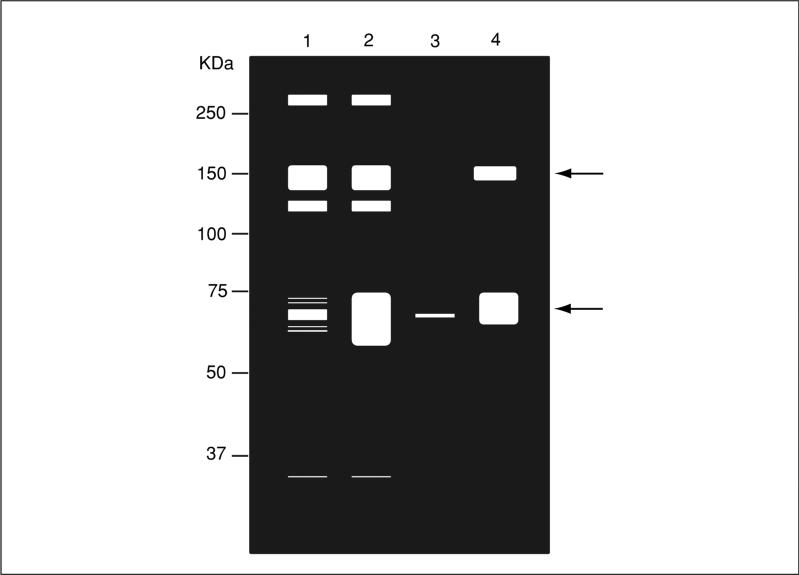
Figure 18.16.5.
Schematic representation of an in-gel phosphatase assay. Cell lysates are prepared and resolved as described in the text. Lane 1: unstimulated, and lane 2: stimulated cells. Immunoprecipitated phosphatase complex from, lane 3: unstimulated, and lane 4: stimulated cells. The gel is processed as described in the text and the “white” areas represent regions in which there is phosphatase activity towards the in-gel substrate. The arrow indicates the position of the immunoprecipitated phosphatase at 69 kD, which exhibits increased phosphatase activity following stimulation. In addition, upon stimulation there is an associated phosphatase activity (~150 kDa).
PTP SUBSTRATE IDENTIFICATION BY IN VITRO SUBSTRATE-TRAPPING WITH PTP ACTIVE-SITE MUTANTS
PTPs exert their physiological responses by dephosphorylating target substrates. Therefore, the identification of the substrates dephosphorylated by PTPs can provide important insight into their biology. One of the methods to identify substrates for PTPs utilizes an approach referred to as substrate-trapping.
Principles of substrate-trapping
There are several mutations that can be introduced into the PTP catalytic domain so that the enzyme still binds to its substrate but fails to dephosphorylate it. The result is that the substrate remains bound to the PTP, forming an enzyme-substrate complex that is sufficiently stable to be purified as a complex. These mutants are called substrate-trapping mutants (see Table 18.16.1). Substrate-trapping mutants are typically inactive or barely active (low kcat) but still bind efficiently to their physiological substrates (with a low Km).
Table 18.16.1.
Substrate-Trapping PTP Mutants
| Name | Mutation | References |
|---|---|---|
| Cys-Ser | Catalytic site cysteine to serine | Jia et al. (1995) |
| Asp-Ala | WPD loop aspartate to alanine | Flint et al. (1997) |
| CysSer-Asp-Ala | Double mutant of catalytic site cysteine and WPD loop aspartate | Agazie and Hayman (2003) |
| Asp/Ala-Gln/Ala | DA mutation in context of a PTP active site glutamine to alanine mutation | Xie et al. (2002) |
Mutation of the critical active-site cysteine residue within the PTP domain to a serine residue (Cys-Ser mutant) can produce a PTP substrate-trapping mutant. In this case, the phosphotyrosyl substrate retains the ability to bind to the PTP active site, but nucleophilic attack on the substrate phosphate cannot take place. Other substrate-trapping mutations include mutagenesis of the critical aspartate residue in the WPD loop of PTPs that allows for the formation of a cysteine-phosphate intermediate, thus forming an effective trap for substrates. Sometimes, instead of using a single Asp to Ala mutation, double mutations such as a Cys to Ser/Asp-Ala combination can be used to improve PTP substrate-trapping efficiency. The highly conserved active site glutamine (Gln262 in PTP1B) participates in catalysis through stabilization of the water molecule. The Asp-Ala substrate-trapping mutant in the context of a Gln-Ala (glutamine-to-alanine) mutation (Asp-Ala/Gln-Ala) has been reported to serve as an efficient substrate-trapping PTP (Kontaridis et al., 2004). These substrate-trapping mutations can be introduced into the wild-type PTP sequences by site-directed mutagenesis.
To perform an in vitro substrate-trapping experiment, as described in this protocol, tyrosine-phosphorylated substrates are captured from cell lysates using a GST-tagged substrate-trapping PTP mutant. One of the major limitations of the substrate-trapping approach is the initial yield of available tyrosine-phosphorylated proteins. The amount of tyrosyl phosphorylated substrates can be increased by treating cells with pervanadate, which is an irreversible, broad-range PTP inhibitor. Preferably, tyrosyl phosphorylated proteins can be generated by stimulating cells with the appropriate growth factor for the signaling pathway under study (reviewed in Tiganis and Bennett, 2007). Alternatively, knock-out cells or cells that have been knocked down for the PTP of interest can be utilized to induce hyper-tyrosyl phosphorylation of potential target proteins (reviewed in Tiganis and Bennett, 2007).
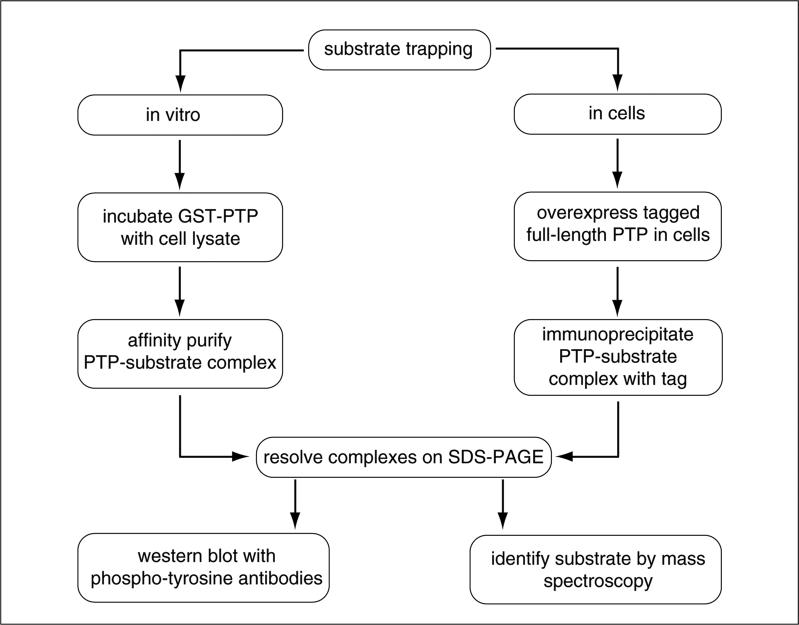
In this approach, cell lysates are incubated with a GST-tagged substrate-trapping PTP mutant (Fig. 18.16.6). After incubation, the PTP-substrate complex is isolated by centrifugation and washed. The PTP-substrate complex is separated by electrophoresis by SDS-PAGE and tyrosine phosphorylated proteins bound to the substrate-trapping mutant are detected by anti-phosphotyrosine antibodies. The advantage of this method is that it can be scaled-up to sufficient levels to allow for the purified protein to be identified by mass spectrometry. Alternatively, if there is some knowledge about the nature of the signaling pathway under study, antibodies to known signaling proteins can be used to interrogate the identity of the trapped tyrosyl phosphorylated protein.
Figure 18.16.6.
Flow chart of a substrate-trapping experiment.
Materials
70% confluent cells in 10-cm dishes
Pervanadate (see recipe)
Phosphate buffered saline (PBS; APPENDIX 2)
Cell lysis buffer II (see recipe)
GST-fusion PTP on gluthathione-Sepharose beads
GST-PTP wild type
GST
10 mM dithiothreitol (DTT; APPENDIX 2)
Wash buffer II (see recipe)
1× SDS sample buffer (see recipe)
Anti-phosphotyrosine antibodies (Millipore, cat. no. 4G10)
37°C incubator
1.5-ml centrifuge tubes
Refrigerated centrifuge
Platform rocker, 4°C
Additional reagents and equipment for SDS-PAGE (UNIT 10.2) and immunoblotting (UNIT 10.8)
Treat one 10-cm dish of 70% confluent cells with 1 mM pervanadate for 30 min at 37°C to inhibit PTPs and allow accumulation of tyrosine phosphorylation on proteins.
- Rinse cells with 10 ml PBS three times.This step and consecutive steps should be performed on ice.
Lyse cells in 1 ml cell lysis buffer II for 20 min on ice.
Centrifuge lysates 10 min at 21,000 × g, 4°C, then transfer supernatant to a new 1.5-ml tube.
To inactivate phosphatase inhibitors, add 10 mM DTT and incubate 10 min on ice.
- Add GST-PTP/gluthathione-Sepharose bead complex to supernatant. As controls, incubate GST-PTP wild type and GST alone in parallel using the same lysate.The ratio of GST-PTP/gluthathione-Sepharose bead to the amount of total protein in the lysate should be determined empirically.
Rock for 3 hr at 4°C.
Wash complexes four times with 1 ml wash buffer II. Pellet beads by briefly centrifuging 30 sec at 2500 × g.
Resuspend beads in 30 to 40 μl of 1× SDS sample buffer. Boil beads for 5 min at 95°C. Briefly centrifuge to pellet beads 30 sec at 2500 × g.
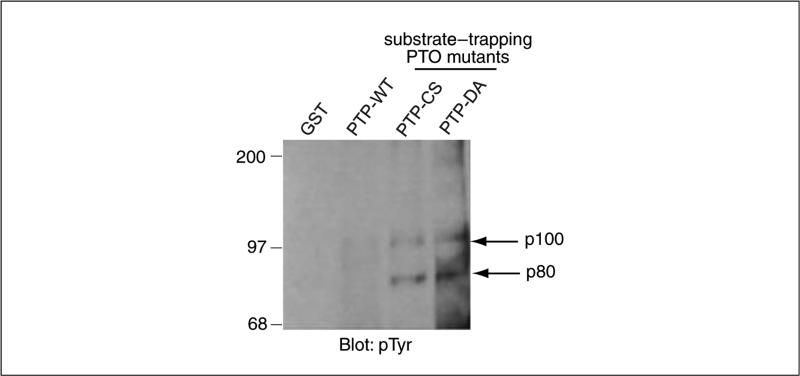
- Run supernatant of boiled beads on SDS-PAGE (UNIT 10.2A) and blot with anti-phosphotyrosine antibodies (UNIT 10.8).Figure 18.16.7 shows a representative blot of substrate-trapped tyrosyl phosphorylated proteins.
Figure 18.16.7.
Serum-deprived WI38 cells were pervanadate-treated and lysates were subjected to affinity precipitation with the indicated PTP wild type and substrate-trapping GST fusion proteins. Affinity purified protein complexes were detected by immunoblotting with anti-phosphotyrosine (pTyr) antibodies. The arrow at right indicates the substrate-trapped tyrosyl phosphorylated proteins p100 and p80. Modified from Kolli et al. (2004).
SUBSTRATE-TRAPPING IN CELLS
Although in vitro substrate-trapping is an efficient method to discover novel PTP substrates, it can generate false-positive results because cell integrity and compartmentalization are compromised before the PTP-substrate interaction occurs. Therefore, in order to find physiologically relevant substrates, one must perform substrate-trapping in cells.
Substrate-trapping in cells involves expressing preferably a full-length PTP substrate-trapping mutant followed by immunoprecipitating the PTP substrate-trapped complex with antibodies against the PTP or its tag (Fig. 18.16.6). Using a full-length PTP is critical to establish a physiologically relevant context between the PTP and its substrates.
Additional Materials (also see Basic Protocol 4)
DNA construct of full-length substrate-trapping PTP
DNA construct of full-length wild-type PTP
Cells
Cell lysis buffer III (see recipe)
Additional reagents and equipment for transfections (Chapter 9), immunoprecipitation (UNIT 10.16), SDS-PAGE (UNIT 10.2A), and protein detection (UNIT 10.8)
- Transfect cells with tagged full-length substrate-trapping PTP construct (control cells should be mock and wild-type PTP transfected).See Chapter 9 for transfection methods.
Lyse 70% to 80% confluent cells with 1 ml cell lysis buffer III for 20 min on ice.
Immunoprecipitate (UNIT 10.16) PTP-substrate complex using an antibody against the PTP or its epitope tag.
- After washing protein G-PTP complex (or protein A-PTP complex, depending on the immunoprecipitation antibody used) with 1 ml cell lysis buffer III three to four times, resolve precipitated proteins on SDS-PAGE (UNIT 10.2A) and detect (UNIT 10.8) phosphorylated substrates using anti-phosphotyrosine antibodies.UNIT 18.6 describes production of antibodies that recognize specific tyrosine-phosphorylated peptides.
VALIDATION OF PTP SUBSTRATES
Not all tyrosyl phosphorylated proteins identified to form a PTP-substrate complex are physiologically relevant. To validate a potential substrate, several additional experiments are essential.
Determine whether the PTP modulates putative substrate tyrosine phosphorylation. The first step in validating a potential PTP substrate target is determining whether overexpression of the wild-type PTP correlates with potential substrate dephosphorylation in cells. However, depending upon how the PTP is regulated, overexpression of the wild-type PTP may not always lead to substrate dephosphorylation. On the other hand, overexpression of a catalytically inactive PTP mutant in cells should result in increased levels of substrate tyrosine phosphorylation, as these mutants behave as dominant negatives. Another criterion for validating a PTP substrate is to determine whether the substrate is hyper–tyrosine phosphorylated in cells when the expression of the PTP is reduced or eliminated using RNAi, anti-sense RNA, or knock-out techniques. However, detecting changes in overall substrate phosphorylation may be difficult in instances where the PTP dephosphorylates a single site on a substrate that is tyrosyl phosphorylated on multiple residues.
Perform in vitro vanadate competition of the PTP-substrate complex. Vanadate is a small molecule that acts as a phosphotyrosine mimetic. In this strategy, substrate-trapping GST-PTP (on Sepharose beads) is incubated with 10 mM vanadate for 10 min at 4°C prior to incubation with cell lysates. If the complex between the PTP catalytic domain and the substrate is a direct one, excess vanadate should prevent the substrate from binding to the GST-PTP. The presence of EDTA in buffers should be avoided as EDTA chelates the vanadate and prevents competition.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2; for suppliers, see APPENDIX 4.
Cell lysis buffer I
150 mM NaCl (APPENDIX 2)
50 mM Tris·Cl, pH 7.4 (APPENDIX 2)
5 mM EDTA (APPENDIX 2)
1% (v/v) Nonidet P-40
1% (w/v) sodium deoxycholate
0.1% (w/v) SDS + protease and phosphatase inhibitors Prepare fresh
NaCl, Tris·Cl, and EDTA solutions can be made in advance and stored up to 1 year at 4°C. Nonidet P-40, sodium deoxycholate, SDS, and inhibitors must be added just before use.
Cell lysis buffer II
1% Triton X-100
50 mM HEPES
150 mM NaCl (APPENDIX 2)
10% glycerol
1.5 mM MgCl2 (APPENDIX 2)
1 mM EDTA (APPENDIX 2)
Protease inhibitors
5 mM iodoacetic acid (PTP inhibitor)
Prepare fresh and use within 1 day
Cell lysis buffer III
50 mM Tris·Cl, pH 8.0 (APPENDIX 2)
150 mM NaCl (APPENDIX 2)
1% Nonidet P-40
1% sodium deoxycholate
0.1% SDS
1 mM EDTA (APPENDIX 2)
Protease and phosphatase inhibitors: 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml pepstatin, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 10 mM sodium fluoride, 1 mM benzamidine, 2 mM Na3VO4 Prepare fresh
Coomassie Blue destaining solution
7% (v/v) acetic acid
5% (v/v) methanol
88% H2O
Store indefinitely at room temperature
Coomassie Blue staining solution
50% (v/v) methanol
0.05% (v/v) Coomassie brilliant blue R-250 (Bio-Rad or Pierce)
10% (v/v) acetic acid 40% H2O
Store indefinitely at room temperature
Denaturation buffer
50 mM Tris·Cl, pH 8.0 (APPENDIX 2)
6 M guanidine·HCl
0.3% 2-mercaptoethanol
Prepare fresh and use within 1 day
Fixation buffer
50 mM Tris·Cl, pH 8.0 (APPENDIX 2)
20% isopropanol
Store up to 1 year at room temperature
Malachite Green reagent no. 1
Prepare 0.135% (w/v) Malachite Green-oxalate salt (Sigma-Aldrich) in water. Store for 6 months at room temperature.
Malachite Green reagent no. 2
Prepare 4.2% (w/v) ammonium molybdate (Sigma-Aldrich) in 4 M HCl. Store for 6 months at room temperature.
Pervanadate
Dissolve sodium orthovanadate (Na3VO4; Sigma) in water to make a 100 mM solution, pH 10. Boil the solution until it turns colorless. Cool the solution to room temperature. At this stage, Na3VO4 aliquots can be prepared and stored indefinitely at −20°C.
Prepare 20 mM HEPES solution in water. Adjust pH to 7.3. Dilute 30% H2O2 ten times with HEPES buffer: 5 μl 30% H2O2 and 45 μl HEPES buffer, mix gently. Dilute 3% H2O2 an additional ten times with HEPES buffer by gently mixing 5 μl 3% H2O2 with 45 μl HEPES buffer.
Add 10 μl of 100 mM Na3VO4 and 940 μl of 0.3% H2O2 solution. Mix by gently rotating the vial. This solution is a 1 mM pervanadate stock.
After 5 min, use the tip of pipet to scoop a small amount of catalase (Sigma), and mix into the pervanadate stock. This will result in a burst of O2 bubbles. Open the lid of the tube to release the air pressure.
The pervanadate is good for a few hours therefore must be used immediately.
Phosphatase wash buffer
24 mM HEPES, pH 7.4
120 mM NaCl (APPENDIX 2)
Store for 6 months at room temperature
This buffer can be made fresh from stock solutions of HEPES and NaCl.
pNPP reaction buffer, 1×
24 mM HEPES, pH 7.4
120 mM NaCl (APPENDIX 2)
10 mM pNPP
5 mM DTT (APPENDIX 2)
Prepare fresh before use
pNPP reaction buffer, 1.25×
30 mM HEPES, pH 7.4
150 mM NaCl (APPENDIX 2)
12.5 mM pNPP
6.25 mM DTT (APPENDIX 2)
Prepare fresh and use within a few hours
PTP buffer, 1×
50 mM Tris·Cl, pH 7.2 (APPENDIX 2)
1 mM EDTA (APPENDIX 2)
0.1% (v/v) β-mercaptoethanol
0.01% (v/v) Triton X-100
Prepare fresh and use within a few hours
PTP buffer, 10×
500 mM Tris·Cl, pH 7.2 (APPENDIX 2)
10 mM EDTA (APPENDIX 2)
1% (v/v) β-mercaptoethanol
0.1% (v/v) Triton X-100
Prepare fresh and use within a few hours
Renaturation buffer
50 mM Tris·Cl, pH 8.0 (APPENDIX 2)
0.04% Tween 40
1 mM EDTA (APPENDIX 2)
0.3% β-mercaptoethanol
Prepare fresh and use within 1 day
SDS sample buffer, 1×
63 mM Tris·Cl, pH 6.8 (APPENDIX 2)
10% (v/v) glycerol
2% (w/v) SDS
0.0025% (w/v) bromophenol blue
This solution can be stored for 6 months at room temperature Add 5% (v/v) β-mercaptoethanol just before use
STE
10 mM NaCl (APPENDIX 2)
10 mM Tris·Cl, pH 7.8 (APPENDIX 2)
1 mM EDTA (APPENDIX 2)
Store up to 1 year at 4°C
Wash buffer I (Basic Protocol 3)
50 mM Tris·Cl, pH 8.0 (APPENDIX 2)
0.2% 2-mercaptoethanol
Prepare fresh and use within 1 day
Wash buffer II (Basic Protocol 4)
0.1% Triton X-100
10% glycerol
20 mM HEPES 150
mM NaCl (APPENDIX 2)
Prepare fresh and use within 1 day
COMMENTARY
Background Information
Protein tyrosine phosphorylation is a reversible post-translational modification that is essential for life in eukaryotic cells. The combined action of both protein tyrosine kinases and protein tyrosine phosphatases (PTPs) determines the net level of cellular tyrosine phosphorylation. The protein tyrosine phosphatases (PTPs) are a family of enzymes that are either transmembrane (receptor) or non-transmembrane (non-receptor) proteins. PTPs are defined by their catalytic signature motif H-C-(X5)-R, where X = any amino acid. Residues within this catalytic motif form the phosphate-binding loop that is located at the base of the active site cleft. The cysteine and arginine residues within the PTP signature motif are essential for catalytic activity. The cysteine residue acts in the first step of catalysis wherein the sulfur atom of the thiolate group serves as a nucleophile and attacks the substrate phosphate. The arginine residue contributes to substrate binding and stabilizes the cysteine-phosphate intermediate. Another important motif integral to PTP catalysis is the WPD (Trp-Pro-Asp) loop, which becomes displaced by several angstroms (8 to 12 Å) upon substrate binding and closes around the side chain of the phosphotyro-sine residue. This conformational change positions the invariant aspartate residue (Asp181 in PTP1B) in the WPD loop in a position that allows it to act as a general acid for the first step of catalysis. This step involves protonating the phenolic oxygen of the tyrosyl leaving group, thus cleaving the phosphate off tyrosine to form the cysteine-phosphate intermediate. This same Asp residue also acts as a general base in the second step of catalysis which, together with a highly conserved Gln residue (Gln262 in PTP1B), coordinates an essential water molecule to promote the hydrolysis of the cysteine-phosphate intermediate.
Although their catalytic domains are highly conserved, PTPs diverge greatly in terms of their domain structures outside of the PTP domain. These non-catalytic domains impart a number of modes of regulation that affords the PTPs specific signaling functions. Together with protein tyrosine kinases, PTPs play a crucial role in controlling many signaling pathways that regulate fundamental processes such as cell proliferation, differentiation, and survival/apoptosis, as well as adhesion and motility. To understand the mechanisms of how PTPs are involved in these processes it is often necessary to determine the catalytic activity of a PTP. In addition, once it is established that the catalytic activity of the PTP is necessary for the observed effect, the next and more challenging step is to identify the substrates dephosphorylated by the PTP.
Critical Parameters and Troubleshooting
Table 18.16.2 describes some common problems that may be encountered using the assays described in this unit, along with suggested solutions for overcoming or avoiding these problems.
Table 18.16.2.
Troubleshooting PTPase Assays
| Problem | Solution |
|---|---|
| pNPP assay and Malachite Green assay | |
| Colorimetric response too weak | Use more cells for immunoprecipitation |
| Incubate PTP and beads for longer time | |
| Use a positive control (another PTP) to check reagent quality | |
| Colorimetric response too strong | Use fewer cells for immunoprecipitation |
| Incubate PTP and beads for shorter time | |
| Use a negative control to check background | |
| Radioactive in-gel phosphatase assay | |
| No white bands visible (all black) | Renaturation ineffective, use fresh DTT |
| Use shorter exposure | |
| Too light background | Control in-vitro kinase conditions |
| Use fresh γ32P | |
| Use longer exposures | |
| In-vitro and in-cell substrate-trapping | |
| No tyrosine-phosphorylated protein detected | Run total cell lysate to check for overall protein tyrosine phosphorylation (should be increased in vanadate-treated or stimulated cells) |
Anticipated Results
In both the pNPP and Malachite Green as-says, an increased absorption at their respective wavelengths (405 nm for pNPP, 650 nm for Malachite Green) is expected with increasing phosphatase activity. In the radioactive phosphatase assay, a whiter band on a black background indicates more phosphatase activity in that region.
Time Considerations
For the pNPP assay, immunoprecipitating PTP from cells may require an overnight incubation depending on the efficiency of the antibody used for immunoprecipitation. Washing steps take 30 min depending on the number of tubes and conditions. The assay itself takes 10 to 30 min and reading the results takes another 10 to 15 min.
For the Malachite Green assay, immuno-precipitation requires an overnight incubation. Dispensing the solutions into aliquots may take ~10 min and the assay itself takes ~20 min to perform and read.
For the in-gel phosphatase assay, casting the gel takes 2 hr. Preparing the puri-fied PTP (or total cell lysate) may take up to 1 day, depending on the treatment or immunoprecipitation protocol. Running the gel takes 1 to 2 hr, and gel fixation requires an overnight incubation. On the next day, denaturation/renaturation washes take 4 hr followed by an overnight dephosphorylation ~ incubation. On the following day, Coomassie staining/destaining and drying of the gel take ~4 hr. X-ray film exposure can take 1 to 7 days.
Literature Cited
- Agazie YM, Hayman MJ. Development of an efficient “substrate-trapping” mutant of Src homology phosphotyrosine phosphatase 2 and identification of the epidermal growth factor receptor, Gab1, and three other proteins as target substrates. J. Biol. Chem. 2003;278:13952–13958. doi: 10.1074/jbc.M210670200. [DOI] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- Kolli S, Zito CI, Mossink MH, Wiemer EA, Bennett AM. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J. Biol. Chem. 2004;279:29374–29385. doi: 10.1074/jbc.M313955200. [DOI] [PubMed] [Google Scholar]
- Kontaridis MI, Eminaga S, Fornaro M, Zito CI, Sordella R, Settleman J, Bennett AM. SHP-2 positively regulates myogenesis by coupling to the Rho GTPase signaling pathway. Mol. Cell. Biol. 2004;24:5340–5352. doi: 10.1128/MCB.24.12.5340-5352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng TC, Hsu SF, Tonks NK. Development of a modified in-gel assay to identify protein tyrosine phosphatases that are oxidized and inactivated in vivo. Methods. 2005;35:28–36. doi: 10.1016/j.ymeth.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Tiganis T, Bennett AM. Protein tyrosine phosphatase function: The substrate perspective. Biochem. J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Zhang YL, Zhang ZY. Design and characterization of an improved protein tyrosine phosphatase substrate-trapping mutant. Biochemistry. 2002;41:4032–4039. doi: 10.1021/bi015904r. [DOI] [PubMed] [Google Scholar]