Summary
Type VI secretion systems (T6SSs) are newly identified contractile nanomachines that translocate effector proteins across bacterial membranes. The Francisella pathogenicity island, required for bacterial phagosome escape, intracellular replication and virulence, was presumed to encode a T6SS-like apparatus. Here, we experimentally confirm the identity of this T6SS and, by cryo electron microscopy (cryoEM), show the structure of its post-contraction sheath at 3.7 Å resolution. We demonstrate the assembly of this T6SS by IglA/IglB and secretion of its putative effector proteins in response to environmental stimuli. The sheath has a quaternary structure with handedness opposite that of contracted sheath of T4 phage tail and is organized in an interlaced two-dimensional array by means of β sheet augmentation. By structure-based mutagenesis, we show that this interlacing is essential to secretion, phagosomal escape, and intracellular replication. Our atomic model of the T6SS will facilitate design of drugs targeting this highly prevalent secretion apparatus.
Introduction
The Type VI secretion system (T6SS) is a recently discovered (Bladergroen et al., 2003; Pukatzki et al., 2006; Silverman et al., 2012) and characterized (Basler et al., 2013; Basler et al., 2012; Ho et al., 2013; Kube et al., 2014) member of secretion systems of Gram-negative bacteria (Tseng et al., 2009). T6SSs are critical to the virulence of many important human pathogens, including Vibrio cholerae, Salmonella enterica, Escherichia coli, Burkholderia pseudomallei, and Pseudomonas aeruginosa. It delivers its protein effector into its prey cell by a contractile ejection apparatus similar to T4-like bacteriophage tails (Aksyuk et al., 2009), R-type pyocins (Nakayama et al., 2000) and many other contractile nanomachines (Leiman and Shneider, 2012), although T6SSs are several times longer than the other apparatuses (Ho et al., 2014). The T6SS is comprised of a sheath and a tube anchored to the bacterial envelope by a baseplate (Ho et al., 2014) (Fig. 1A). Instead of a single sheath protein in phage tails and pyocins, the T6SS sheath is composed of two proteins, VipA/VipB in Vibrio, which are orthologues of IglA/IglB of F. tularensis (Broms et al., 2009; de Bruin et al., 2007). The contraction of the sheath drives the tube across its target membrane (Ho et al., 2014; Hood et al., 2010) (Fig. 1B). However, the atomic structure of the T6SS and molecular interactions required for secretion are not known. While the structure of Vibrio VipA/VipB outer sheath has recently been shown at 6 Å resolution (Kube et al., 2014), that structure is insufficient to guide drug design and mutagenesis studies.
Figure 1. F. novicida expressing GFP-tagged IglA or IglA/B-split GFP form fluorescent foci within macrophages consistent with T6SS assembly.
(A-B) Proposed T6SS model in pre-contraction (A) and post-contraction (B) conformation. IM, inner membrane; OM, outer membrane.
(C-F) F. novicida expressing IglA-GFP assemble fluorescent foci after uptake by macrophages, with 10% doing so at 15 min of infection (C) and 70% at 22 h of infection (D). F. novicida bacteria are stained with a red fluorescent antibody; host and bacterial DNA is stained blue with DAPI; and arrows indicate bacteria shown at higher magnification in the insets. Scale bars, 10 μm (insets, 1 μm). The percentage of Fn-IglA-GFP with fluorescent foci (E) and the number of F. novicida per THP-1 macrophage (F) was determined at each time point. Data shown represent the mean ± standard error of measurements of at least 144 cells per time point. The experiment was done three times with similar results.
(G – J) Requirement for IglA/B interaction for formation of the fluorescent foci was demonstrated in F. novicida expressing IglA-GFP1-10/IglB-GFP11 split GFP (Fn-IglA/B-split GFP) and controls. At 15 min post infection of THP-1 macrophages (G), approximately 10% of Fn-IglA/B-split GFP form fluorescent structures at their poles, and by 22 hours after infection (H), most of the bacteria exhibit green fluorescent structures. In contrast, F. novicida expressing IglA-GFP1-10/SodB-GFP11 (I) and F. novicida expressing IglA-GFP1-10 without a GFP11 partner (J) do not form green fluorescent structures, despite their capacity to replicate extensively within the macrophages. Panels on the left show F. novicida stained with a red fluorescent antibody (in black and white), the middle panels show GFP green fluorescence (in black and white), and the panels on the right show merged color images with DAPI-stained host and bacterial DNA in blue. Arrows in (G) indicate bacteria with intense GFP fluorescent structures at 15 min post infection, one of which is shown at higher magnification in the inset. Scale bars, 10 μm (inset, 1 μm).
(K) Kinetics of growth of IglA/B-split GFP and control strains in THP-1 macrophages. THP-1 macrophages were infected with Fn-IglA/B-split GFP or control strains, incubated at 37°C for 0.5, 2, or 22 hours, fixed with 4% paraformaldehyde, permeabilized with 0.1% saponin, and the bacteria were stained with a red fluorescent antibody. The number of bacteria per macrophage nucleus was determined by automated counting using CellProfiler Software. All strains show capacity to grow in macrophages.
(L) Western Immunoblot confirms expression pattern for IglA/B-split GFP constructs. Lane 1, Fn. Lane 2, Fn-IglA-GFP. Lane 3, Fn-IglA-GFP1-10. Lane 4, Fn-IglA/B-split GFP. Lane 5, Fn-IglA/SodB-split GFP. M, molecular mass standards, as indicated to the left of the figure. Primary antibody used is indicated on the right for each blot.
Francisella tularensis subsp. tularensis is a Gram-negative bacterium that causes a zoonotic infection, tularemia, in animals and humans (Ellis et al., 2002). By the airborne route, a few organisms can cause lethal pneumonia in humans; hence, F. tularensis is a potential agent of bioterrorism and classified as a Tier 1 Select Agent. F. tularensis and the highly related F. tularensis subsp. novicida are facultative intracellular pathogens that replicate within macrophages. After uptake by macrophages via looping phagocytosis (Clemens et al., 2005, 2012), the bacteria initially reside within a fibrillar-coated membrane bound phagosome that resists fusion with lysosomes and exhibits limited acquisition of lysosomal markers; however, the bacteria subsequently disrupt their phagosomal membrane and replicate extensively in the host cell cytosol (Chong and Celli, 2010; Clemens and Horwitz, 2007; Clemens et al., 2004). F. novicida has considerable homology with F. tularensis, but it has only a single copy of the Francisella Pathogenicity Island (FPI), and it is of low virulence for humans; it thus serves as a more practicable subspecies for study.
Here we show that the FPI of F. novicida encodes a T6SS, and, by cryo electron microscopy (cryoEM), that the two proteins of its sheath, IglA and IglB, are interdigitated into a single fold similar to that of the phage sheath. CryoEM reconstruction at 3.7 Å reveals that β sheet augmentation interlaces the two-dimensional array of the sheath, and structure-based mutagenesis demonstrates that this interlacing is essential to secretion.
Results
Environmental stimuli trigger assembly of F. novicida T6SS and secretion of effector proteins
To facilitate structural and functional studies of the FPI-encoded T6SS-like apparatus, we engineered F. novicida to express IglA-GFP fusion protein in lieu of IglA from its chromosome (Fn-IglA-GFP). These bacteria show only a weak diffuse fluorescence when grown in standard liquid culture medium. However, within 15 minutes of uptake by macrophages, 10% of the bacteria exhibit intensely fluorescent foci (Fig. 1C, E), and this percentage increases to approximately 70% by 1 day after uptake, by which time the bacteria have proliferated extensively in the macrophage cytosol (Fig. 1D-F). We used a split-GFP system (Cabantous and Waldo, 2006) with domains 1 – 10 of GFP fused to the C-terminus of IglA and domain 11 of GFP fused to the N-terminus of IglB (IglA-GFP1-10 and IglB-GFP11) to test the interaction of IglA and IglB in formation of fluorescent foci. These bacteria are not fluorescent when grown in broth culture, but exhibit intense GFP fluorescent foci after uptake and replication within macrophages (Fig. 1G-H). In contrast, F. novicida expressing IglA-GFP1-10/SodB-GFP11 (Fig. 1I) and F. novicida expressing IglA-GFP1-10 without a GFP11 partner (Fig. 1J) do not form green fluorescent structures, despite their capacity to replicate extensively within macrophages (Fig. 1K). Western blotting confirmed expression pattern of IglA/B-split GFP constructs (Fig. 1L).
Because FPI mutants are unable to permeabilize their vacuoles, formation of the IglA/IglB containing T6SS and secretion of its effectors is believed to precede and to be required for phagosome disruption and cytosolic escape by Francisella. The extremely high expression at late stages of infection suggests that the FPI-encoded T6SS is required for events after phagosome permeabilization. Consistent with this, a) most FPI genes are induced during intramacrophage growth and show high levels of expression at late time points after bacterial uptake (Golovliov et al., 1997; Wehrly et al., 2009), when most bacteria are cytosolic, and b) we have observed that IglC is required for intracellular growth of F. novicida that are micro-injected directly into the cytosol of HeLa cells.
In addition to intracellular growth in macrophages, two other conditions induce formation of fluorescent structures within Fn-IglA-GFP or Fn-IglA/B-split GFP: (1) placement beneath coverslips (either sealed by silicone or not) at room temperature for more than 3 hrs (Fig. 2A-E) and (2) growth in broth containing 5% KCl (but not 5% NaCl) (Fig. 2F-J). The fluorescent foci do not form in Fn-IglA-GFP with a deletion of iglB in response to any of the above three conditions (Figure S1), suggesting that IglB subunits are required for assembly of the T6SS-like structure. The stimulus for assembly of fluorescent foci provided by placement beneath coverslips is unclear, but could include physical stimulation, modulation of oxygen tension or other environmental conditions. The fluorescent foci form with similar kinetics by bacteria sandwiched between a polystyrene surface and glass coverslip or between two glass surfaces, but form more slowly when the glass slide is replaced with a 25 μm gas permeable Lumox (Starstedt) film base, consistent with a role for oxygen tension.
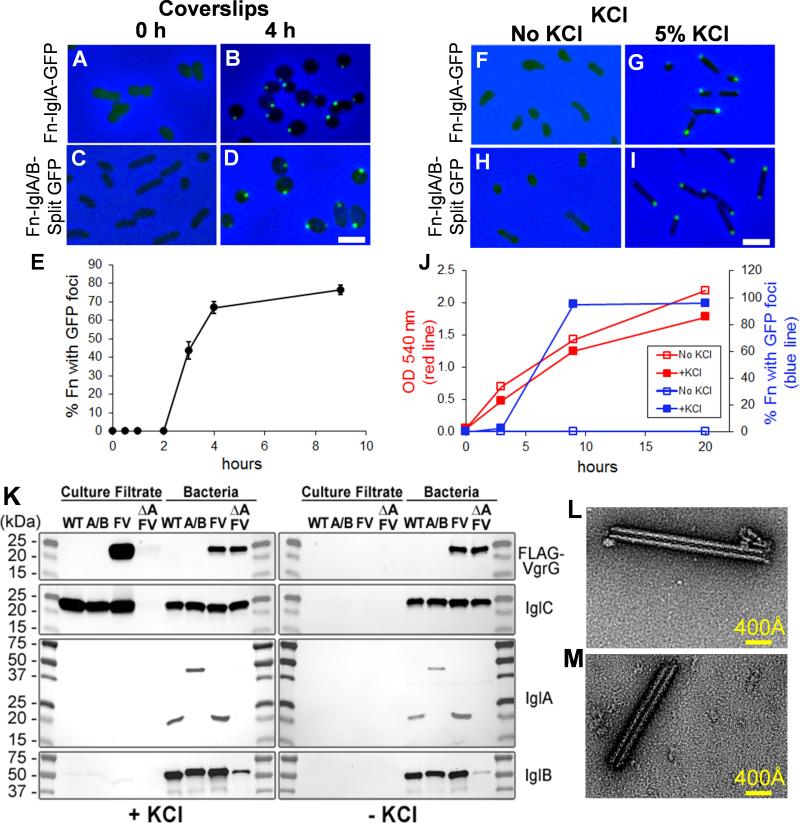
Figure 2. F. novicida assemble fluorescent foci in response to high KCl and placement beneath a coverslip and secrete T6SS effector proteins in response to environmental stimuli.
(A-E) Placement beneath a glass coverslip: Fn-IglA-GFP (A, B) and Fn-IglA/B-split GFP (C, D) in TSBC were placed beneath a silicone sealed coverglass and imaged immediately (Fn-IglA-GFP, A; Fn-IglA/B-split GFP, C) or after 4 h at room temperature (Fn-IglA-GFP, B; Fn-IglA/B-split GFP, D). Images shown are merged green fluorescence and phase contrast images, with phase contrast shown in blue. Bacteria exhibit either a diffuse weak fluorescence or no fluorescence immediately after placement beneath a glass coverslip (A, C), but exhibit intense fluorescence at their poles after 4 h at room temperature (Fn-IglA-GFP, B; Fn-IglA/B-split GFP, D). Scale bar, 2 μm. (E) Time course of formation of the fluorescent structures by Fn-IglA/B-split GFP after placement beneath a coverslip at room temperature.
(F-J) High KCl: Fn-IglA-GFP or Fn-IglA/B-split GFP were inoculated into TSBC at an O.D. 540 nm of 0.05 and grown at 37°C overnight to late exponential phase (O.D. 1.0 – 1.4) in the absence (Fn-IglA-GFP, F; Fn-IglA/B-split GFP, H) or presence of 5% KCl (Fn-IglA-GFP, G; Fn-IglA/B-split GFP, I). Bacteria exhibit only a weak diffuse fluorescence in the absence of KCl (F and H), but develop intense fluorescence at their poles in response to 5% KCl (G and I). Scale bar, 2 μm. See also Figure S1.
(J) Growth and kinetics of formation of fluorescent foci in Fn-IglA/B-split GFP in TSBC with and without 5% KCl. Fn-IglA/B-split GFP was inoculated into TSBC broth with or without 5% KCl at an optical density of 0.05 and grown at 37°C rotating at 250 rpm. O.D. (red lines) and percentage of bacteria with fluorescent foci (blue lines) in the presence (solid squares) or absence (open squares) of KCl were monitored over time.
(K) Secretion in response to high KCl: VgrG and IglC are secreted by F. novicida with intact T6SS growing in TSBC with (left), but not without 5% KCl (right). WT, wild-type; A/B, Fn-IglA/B-split GFP; FV, Fn-FLAG-VgrG; ΔA FV, Fn-FLAG-VgrG ΔiglA.
(L - M). Sheath like macromolecular structures purified from wild-type and IglA-GFP expressing F. novicida. F. novicida expressing native (L) or GFP-tagged (M) IgIA assemble similar sheath-like macromolecular structures. Negatively stained TEM images of density gradient fractions from F. novicida expressing wild-type IglA (L) or IglA-GFP (M) show rod shaped structures of variable length. See also Figure S2.
Whereas by fluorescence microscopy Basler et al. (Basler et al., 2012) demonstrated highly dynamic assembly and disassembly of VipA-GFP labeled structures of live Vibrio, we have not observed similar rapid disassembly of the IglA-GFP or IglA/B-split GFP fluorescent structures in individual live bacteria. The slower turnover of the F. novicida T6SS may reflect the absence of an identifiable protein in the F. tularensis genome to disassemble the T6SS. While the F. novicida genome lacks a protein with homology to ClpV, we cannot rule out the possibility of a protein with no such sequence homology that fulfills this role. However, F. novicida IglB, a VipB homologue, lacks the latter's identified ClpV interacting motif (Pietrosiuk et al., 2011): an α-helical region at the N-terminus of VipB (including a consensus sequence LLDEIM, residues 19 – 24 of VipB). Indeed, Protein-Protein BLAST analysis indicates that the N-terminal 56 amino acids of VipB have no sequence homology with any region of IglB and the N-terminus of F. novicida IglB has no similar α-helical region or consensus sequence. The GFP tags on IglA and IglB are not likely to hamper their interaction with any disassembling chaperone since a) we observe the same slow turnover for both F. novicida expressing IglA-GFP (GFP fused to the C-terminus of IglA) and F. novicida expressing the split GFP construct (IglA with GFP domains 1-10 fused to its C-terminus and IglB with GFP domain 11 fused to its N-terminus), and b) for V. cholerae, rapid turnover was observed even for VipA-GFP (C-terminal fusion), indicating that the corresponding VipA-GFP fusion in V. cholerae does not prevent rapid ClpV-mediated disassembly. If F. novicida has a functional homologue of ClpV, then its interactions with the T6SS sheath are likely to differ markedly from those in V. cholerae.
Secretion of VgrG by F. novicida has been reported previously (Barker et al., 2009). We prepared FLAG-tagged VgrG and grew the bacteria in the presence or absence of KCl. FLAG-VgrG and IglC are both detected in the culture supernatant in the presence, but not in the absence, of KCl (Fig. 2K). Deletion of iglA abolishes the release of VgrG and IglC into the culture medium (Fig. 2K).
IglA/IglB heterodimers assemble to form Francisella T6SS sheaths
We purified assemblies containing IglA-GFP or IglA from lysates of bacteria grown in trypticase soy broth supplemented with 0.2% L-cysteine (TSBC) in the presence of KCl. Both proteins sedimented in equilibrium to below 55% sucrose on sucrose gradients. Negatively stained TEM images of the fraction containing IglA-GFP or IglA showed rod shaped particles of variable length, similar to Vibrio cholerae T6SS sheaths with or without GFP tag (Basler et al., 2012) (Fig. 2L-M). Rod-shaped sheath particles containing IglA-GFP, but not IglA, are recognized by immunogold labeling for GFP (Figure S2).
We recorded cryoEM image stacks (movies) (Campbell et al., 2012) with a Gatan K2 direct electron detector (McMullan et al., 2009) operated at counting mode (Li et al., 2013) (Fig. 3A); obtained a three-dimensional structure to 3.7 Å by an integrative approach which implements Iterative Helical Real Space Reconstruction (IHRSR) (Egelman, 2010) within the Relion (Scheres, 2012) framework (Fig. 3B); and built an atomic model of the T6SS apparatus (Fig. 3C) (See also Methods). Like contractile phage tails (Leiman and Shneider, 2012), the sheath is organized in a cylindrical, helical form with an axial six-fold rotational symmetry (Fig. 3 and Movie S1). The sheath is constructed with discs of six heterodimers of IglA/IglB proteins (Fig. 3D, Movie S2). The validity of the map and the model is demonstrated by the deep grooves in α-helices, well-separated β-strands and match of side chains in the sequence and in the density map (Fig. 3E,F).
Figure 3. CryoEM and atomic model of F. novicida T6SS.
(A) A representative cryoEM image of F. novicida T6SS recorded on a direct-electron detector. The yellow box marks one sheath. (B) CryoEM density map of the F. novicida T6SS sheath displayed as shaded surface (left) and cut-away view (right), both colored according to radius. See also Movie S1. (C) Atomic model of F. novicida T6SS with six discs shown. The 12-start helices (ridges) are colored alternatively with rainbow colors and gray. See also Movie S2. (D) Hexagonal disc formed by six IglA/IglB heterodimers of F. novicida T6SS shown in two different orientations. Each heterodimer is displayed as a ribbon diagram of a different color. (E – F) Two representative density regions (chicken-wire) with their atomic models (sticks, colored by atoms: C: green, N: blue, O: red, S: yellow).
The asymmetric unit of the helical sheath contains one copy each of IglA and IglB. The folds of the IglA/IglB proteins can be thought of as a “split” of the single protein of a bacteriophage sheath with many insertions (Fig. 4A-D, Movie S3). Joining together, these two proteins form an α-β-α sandwich. The central β sheet of this sandwich is formed by interdigitation of strands from both IglA and IglB (Fig. 4E). The sheath fold splits between its first and second α helices, and each of the two ends of the split is appended with an insertion (box in Fig. 4D). Parts of these two insertions are disordered [C-terminal amino acids 136 - 184 of IglA and N-terminal amino acids 2 - 78 of IglB (question marks in Fig. 4D)]. The domain near the C-terminus of IgIB (cyan in Fig. 4C,D) highly resembles the C-terminal domain of the phage sheath proteins (Aksyuk et al., 2011; Aksyuk et al., 2009).
Figure 4. Atomic model and architecture of T6SS sheath and comparison with bacteriophage T4.
(A and B) Atomic model of an IglA/IglB dimer shown as ribbons with domains colored as indicated in the bars below (B). (C) Superposition of the atomic models of the IglA/IglB dimer (colors) and bacteriophage T4 sheath (gray) (Leiman et al., 2004). See also Movie S3 and Figure S3. D1-D4: domains 1-4. (D) Secondary structure diagram of an IglA/IglB dimer, colored as in (A and B). Elements in the box are insertions at the “split”. (E) The central β sheet of an IglA/IglB dimer is made of interdigitated strands (inside oval) from both subunits. (F) Ribbon diagram of IglA/IglB centered on the third and fourth helices of IglB (marked by two dashed lines) (left) and comparison between the schematics of this vicinity in F. novicida T6SS and in T4 phage (right). (G and H) Comparison between the quaternary structures of post-contraction F. novicida T6SS (G) and T4 phage (H) sheaths showing opposite handedness. The angles between their 12-start helices and their helical axes are marked on their sides, respectively. The semi-transparent shaded surfaces of the density map of the T6SS (G) sheaths at 10-Å resolution is fitted with six discs of ribbon models colored as in Fig. 3C. The density map of T4 phage sheath at 17 Å is taken from EMDB entry EMD-1086. See also Figure S4.
F. novicida T6SS and bacteriophage T4 outer sheaths show divergent quaternary organization despite secondary and tertiary structural homology
The secondary structure and tertiary structural arrangement of the F. novicida T6SS sheath are similar, but not identical, to those of the bacteriophage T4 sheath (Fokine et al., 2013). The central β sheet of the F. novicida T6SS is formed by interdigitating two proteins (Fig. 4E). Notably, phage T4 sheath protein has an insertion of two domains before a long helix (Fig. 4C, Movie S3 and Fig. 4F, see lower right schematic) whereas in T6SS sheath this long helix is broken into two shorter helices, its third and fourth helices from its N-terminus. In the case of F. novicida, the third helix tilts up and is almost perpendicular to the fourth helix (Fig. 4F, dashed lines in ribbon diagram, and upper right schematic). In stark contrast, the quaternary structural organization of the T6SS sheath is markedly different from that of the post-contraction T4 (Leiman et al., 2004) sheath (Fig. 4G,H, see also Figure S3). The helix with the shortest, nonzero pitch (a six-start helix, black dashed line in Fig. 4G or H) in the T6SS sheath has a left-handed turn of 33.4° (Figure S3D) and a rise of 20.8 Å per subunit (whereas the T4 sheath has a right-handed turn of 32.9°); thus the two helical architectures mirror each other. Despite opposite handedness, layer lines in the Fourier spectra of the EM images (Figure S4) indicate that the other parameters of the helical symmetry of the F. novicida T6SS sheath and the post-contraction T4 sheath are similar, suggesting that this T6SS sheath structure is in its post-contraction state.
F. novicida T6SS outer sheath has a highly interlaced two-dimensional array architecture with augmented β sheets that is essential to secretory function
The two-stranded β sheet of the C-terminal domain of IglB is augmented on one side by arms that emanate from the two neighboring IglA/IglB dimers from a disc above (Fig. 5A-C, Figure S3B,C), forming a 4-stranded sheet. By this augmentation, the arms from hundreds of copies of IglA/IglB interlace a two-dimensional array (Figure S3A,C). We hypothesized that this two-dimensional interlacing is crucial to the integrity and thus the contractility and secretory capacity of the F. novicida T6SS. To test this hypothesis, we carried out structure-based mutagenesis experiments for residues centered on the augmented β sheet. We prepared IglA/B-split GFP mutant strains lacking either N-terminal amino acids 2-18 of IglA (Fn-IglAΔ18N/B-split GFP) or the last 25 amino acids at the C-terminal of IglB (Fn-IglA/BΔ25C-split GFP). We predicted that both mutant strains would show normal assembly of their pre-contraction T6SS organelles but that the T6SS sheath would become unstable with attempted contraction, and thus fail to secrete effector proteins.
Figure 5. Structure-based mutagenesis of F. novicida T6SS.
(A) Ribbon model of three (marked by numbers) interacting IglA/IglB dimers with their augmented β sheet circled. (B) Magnified view of the circled β sheet in (A). (C) Ribbon diagram of this augmented β sheet. The chains marked “1”, “2” and “3” in (B) and (C) come from the IglA/IglB dimers marked with the same number in (A). (D - M) F. novicida expressing IglAΔ18N/B-split GFP (D - G) or IglA/BΔ25C-split GFP (I - L) form fluorescent structures in response to KCl (D vs. E, I vs. J) or incubation under coverslips (F vs. G, K vs. L). Green fluorescence and phase contrast images are merged with phase contrast shown in blue. Scale bar, 2 μm. Ribbon diagrams depict deletions in the N-terminus of IglA (H) and C-terminus of IglB (M). (N) Western blots showing that FLAG tagged VgrG (21 kDa) and IglC (22 kDa) are secreted into culture filtrate in response to KCl by the parental Fn-IglA/B-split GFP strain but not by the strains expressing truncated IglA, truncated IglB or IglB with a single amino acid substitution at residue 134 from phenylalanine to arginine. A/B, Fn-IglA/B-split GFP; AΔ18N/B, Fn-IglAΔ18N/B-split GFP; A/BΔ25C, Fn-IglA/BΔ25C-split GFP. All Fn-IglA/B split GFP parental and mutant strains express FLAG-VgrG. Samples were run on a single gel. Grey lines indicate gaps where irrelevant lanes were removed. (O) Growth curves demonstrating that the loss of T6SS function in the deletion mutants renders them unable to multiply intracellularly in human macrophages. Whereas the parental F. novicida IglA/B-split GFP grows 1.7-logs in THP-1 human macrophages in 1 day, the split-GFP strains expressing truncated IglA (AΔ18N/B) or truncated IglB (A/BΔ25C) show markedly impaired growth at a level equivalent to that of IglAGFP-expressing strains with a deletion in iglB (A/ΔB, Fn-IglA-GFP ΔiglB) or iglC (A/ΔC, Fn-IglA-GFPΔiglC). CFU, colony forming unit.
Indeed, both the IglA and IglB truncation mutants expressing the truncated IglA/B with the split GFP tags formed fluorescent foci in response to 5% KCl or incubation beneath coverslips (Fig. 5D-M), indicating focal multimerization of the IglA/B-heterodimers. However, the mutants were completely defective in secretion of IglC (Fig. 5N) and had markedly impaired growth in THP-1 macrophage-like cells, with growth kinetics similar to iglB and iglC deletion mutants (Fig. 5O). Like the iglB and iglC deletion mutants, the Fn-IglAΔ18N/B- and Fn-IglA/BΔ25C-split GFP mutants remain within compartments that stain positive for the lysosomal marker CD63 (Fig. 6A) and are unable to escape their phagosomes (Fig. 6B).
Figure 6. IglA N-terminal or IglB C-terminal deletion mutants are defective in phagosome escape.
(A) Unlike the parental strain (Fn-IglA/B-split GFP), Fn-IglAΔ18N/B-split GFP and Fn-IglA/BΔ25C-split GFP (stained with a green fluorescent anti-F. novicida antibody, shown in black and white in the left panels) are unable to escape their phagosome and remain within CD63-positive compartments (stained with a red-fluorescent antibody, shown in black and white in the middle panels) in human macrophage-like THP-1 cells, both at 6 h and 26 h. Arrows indicate bacteria stained with the anti-F. novicida antibody. The panels on the right show merged color images with F. novicida stained green, CD63 stained red, and host and bacterial DNA stained blue with DAPI. (B) Cytosolic vs. phagosomal localization of F. novicida was determined using a differential digitonin permeabilization antibody staining procedure. F. novicida bacteria accessible to antibody after digitonin permeabilization (which permeabilizes the plasma membrane but not the phagosomal membrane) were stained with a red fluorescent anti-F. novicida antibody and all bacteria were subsequently stained with a green fluorescent antibody after saponin permeabilization. Whereas many of the parental strain F. novicida arestained by the red-fluorescent antibody after digitonin permeabilization at 6 h post-infection (arrowheads), none of the Fn-IglAΔ18N/B-split GFP and Fn-IglA/BΔ25C-split GFP (arrows) are stained by the red-fluorescent antibody either at 6 h or 26 h after infection. Parental bacteria that remain inaccessible to antibody after digitonin permeabilization at 6 hours are indicated by arrows. At 26 hours post infection, the parental F. novicida have proliferated extensively within the cytosol and the majority are accessible to the red-fluorescent antibody after digitonin permeabilization. The Fn-IglAΔ18N/B-split GFP and Fn-IglA/BΔ25C-split GFP bacteria (indicated by arrows) show much more limited replication within the macrophage and remain inaccessible to the red-fluorescent antibody. Scale bars, 10 μm. These experiments were performed three times with similar results.
Discussion
By employing the split-GFP technology, we identified three stimuli that trigger the assembly of IglA/IglB sheaths in F. novicida – (1) late exponential phase with high bacterial density in the presence of high KCl; (2) placement beneath a coverslip; and (3) the intramacrophage environment. While there is precedent for demonstration of T6SS regulation by quorum sensing (Ishikawa et al., 2009; Lesic et al., 2009), salinity (Ishikawa et al., 2012), and membrane perturbation (Basler et al., 2013; Ho et al., 2013), we are unaware of prior reports of T6SS assembly induced selectively by KCl or by intracellular residence. We hypothesize that KCl is a stimulus for T6SS assembly in the intracellular environment and that higher concentrations are required in liquid culture to compensate for the absence of other intracellular stimuli. The factors stimulating T6SS when bacteria are placed beneath a coverslip are unclear and could involve physical stimuli, altered oxygen tension, or other environmental influences. We confirmed that one stimulus, high KCl in liquid culture, induced secretion of putative T6SS effector proteins, VgrG and IglC, as would be expected for induction of the secretion system. We purified sufficient amounts of IglA/IglB sheaths from bacteria stimulated with this condition for high resolution cryoEM structural studies. Taking advantage of the latest technological breakthrough -- direct electron counting, we determined the structure of this sheath to 3.7 Å, permitting the de novo atomic modeling of IglA/IglB and the confirmation of its identity as the T6SS in F. novicida. This structure shows a highly interlaced two-dimensional array that is critical to the secretion function.
As with other contractile nanomachines, the F. novicida T6SS propels its central tube by contraction of its sheath assembly. An intriguing feature of the sheath structure is its two-dimensional interlacing formed by augmented β sheets. It is likely present in the pre-contraction form of the T6SS sheath as well and plays a critical role during contraction. Our work directly reveals the importance of this interlaced structure to the contractile function of this T6SS and lays the foundation for understanding the presumably shared mechanism of other contractile nanomachines.
Results utilizing the truncation mutants of IglA/IglB prove the critical role of the interlacing formed by augmented β sheets during contraction; in addition, the intensive polymerization of IglA/IglB, as reported by the fluorescent foci, suggests that the T6SS sheath array assembles without interlacing. Therefore, an additional factor must contribute to T6SS assembly. By analogy with bacteriophages, we propose that the sheath assembles on a preassembled IglC tube, whose monomer structure is available (PDB: 2QWU). Close scrutiny of the IglB and IglC surfaces suggests a potential protein-protein interface for IglB and IglC, given favorable rotamers of residues (Figure S5). However, reliable assignment of this interaction requires the structure of pre-contraction T6SS.
It was thought, based on its limited sequence homology, that the F. tularensis FPI-encoded T6SS is an outlier among others (Filloux et al., 2008). Most notably, F. tularensis T6SS system lacks a homologue for ClpV ATPase which is linked to T6SS disassembly (Filloux et al., 2008). Nevertheless, Francisella has homologues to other key T6SS proteins, including IglA/IglB, VgrG, DotU and PdpB; Francisella IglC is structurally homologous to Pseudomonas Hcp (de Bruin et al., 2011). Remarkably, these differences and similarities coincide with differences and similarities between our structure of the F. novicida T6SS sheath and the other available T6SS structure (Kube et al., 2014); although the dimeric organization and the fold of the sheath proteins and the two-dimensional interlacing among dimers are the same for both, the quaternary structural organizations are different. Whereas the turn per subunit of the contracted Vibrio T6SS sheath is 30.56°, giving a 3° tilt off the vertical to the 12-start helices (Kube et al., 2014), our T6SS sheath has a turn per subunit of 33.4°, giving a ~20° tilt to the 12-start helices (Fig. 3G).
It is always tempting but challenging to give an explanation for such a difference. While a facile explanation is that the F. novicida T6SS is an outlier, its lack of a ClpV ATPase suggests an alternative explanation. In fact, the Vibrio and Pseudomonas T6SS sheaths are severed into shorter pieces while our purified F. novicida T6SS sheath is typically much longer, sometimes more than five times longer (Lossi et al., 2013). A few more helices are resolved in the 6 Å structure of the Vibrio cholerae T6SS sheath (Kube et al., 2014) than in our sheath structure. These helices belong to the part of sheath proteins that interacts with ClpV (Kube et al., 2014). Since the function of ClpV is to facilitate disassembly of the post-contraction sheath, the Vibrio T6SS structure might be in a state that is closer to disassembly than our structure. Our structure is closer to the immediate post-contraction state, since it shares a similar turn per subunit number to post-contraction bacteriophage T4 sheath (Leiman et al., 2004). If so, then the T6SS sheath would have three states: pre-contraction, post-contraction and pro-disassembly. This model awaits confirmation by structural analyses of a ClpV knockout mutant of Vibrio cholerae.
We show here that VgrG and IglC are secreted by F. novicida in an IglA-dependent fashion in response to environmental stimuli, confirming secretory activity of the Francisella FPI-encoded apparatus. Secretion of effectors through this T6SS within macrophages is required for phagosome escape and cytosolic replication, since deletion mutants lacking IglA, IglB, IglC, or VgrG (Barker et al., 2009; de Bruin et al., 2011) and the contraction incompetent mutants IglAΔ18N/B- and IglA/BΔ25C-split GFP presented here were all unable to escape the phagosome and markedly impaired in intramacrophage replication.
On the other hand, our Fn-IglAΔ18N/B- and Fn-IglA/BΔ25C-split GFP mutants stand out from other IglA and IglB mutants studied to date. Our split-GFP data suggest that the mutated proteins still interact to form IglA/IglB-heterodimers that can assemble green-fluorescent macromolecular structures (fluorescent foci), yet show loss of secretory function. This is a more precise manipulation of the protein functions owing to the availability of an atomic structure. In contrast, the previously reported mutations are not based on a structure; mutants among IglA residues 102 – 129 (Broms et al., 2009) caused loss of interaction with IglB (i.e. failure to form the IglA/IglB heterodimer). Particularly, substitution of alanine for valine at residue 109 of IglA or at the corresponding residue 110 of Vibrio VipA was shown to abolish IglA-IglB and VipA-VipB interaction, respectively (Broms et al., 2009). Nevertheless, our atomic model provides an explanation for such loss of interaction: Val109 of IglA is participating centrally in a hydrophobic core formed between IglA and IglB. The V109A mutation destroys this hydrophobic interaction and weakens IglA-IglB interaction. In addition to the above structure-based functional studies, our atomic model provides many other targets for further mutagenesis studies.
In conclusion, we established the identity and determined the atomic structure of an FPI encoded T6SS in Francisella, the genus that includes the highly pathogenic bacterium and Tier 1 Select Agent, F. tularensis subsp. tularensis. Our atomic structure of the F. novicida T6SS and structure-based mutational analysis reveal the critical importance of the interlaced architecture to secretion. This atomic model will facilitate the design and testing of therapeutics targeting this and similar bacterial secretion apparatuses, which are pivotal to the virulence of many pathogenic Gram-negative bacteria.
Experimental Procedures
Bacteria
F. tularensis subsp. novicida strain U112 (F. novicida) and the derivative strains were cultivated in TSBC. For targeted gene deletion and epitope tagging, upstream and downstream chromosomal regions flanking the gene of interest were amplified with in-frame deletion of the gene or with the gene fused to the coding sequence of an epitope tag. The amplified fragments were inserted into pMP590 (LoVullo et al., 2006) by traditional cloning method using restriction endonucleases or Gibson Assembly (New England BioLabs) and confirmed by sequencing. The resulting plasmid constructs were chemically transformed into F. novicida for allelic exchange. Additional information on F. novicida strains used in the study is provided in Table S1.
Kinetics of formation of fluorescent foci
Bacteria were inoculated in TSBC with and without 5% KCl at an initial optical density at 540 nm (O.D.) of 0.05 and grown at 37°C rotating at 250 rpm. Optical density and percentage of bacteria with fluorescent foci was monitored over time. To observe formation of fluorescent foci beneath a glass coverslip, we placed bacteria between a glass slide and a silicone sealed (and also unsealed) coverglass and imaged immediately or after 3 - 6 hours at room temperature.
Secretion of effector proteins
Bacteria were grown to late exponential phase in TSBC with or without 5% KCl, pelleted by centrifugation, and the supernate filtered through a 0.45 micron filter. The culture filtrate (equivalent to 1 ml culture at an O.D. of 1.5) and bacterial pellet (equivalent to 5 × 107 bacteria) were analyzed by SDS-PAGE and Western Immunoblot. IglA and IglA-GFP fusion variants were detected by rabbit polyclonal antibody to His-tagged recombinant IglA (BEI Resources) or affinity-purified rabbit polyclonal antibody to native GFP (Millipore); IglB was detected by murine monoclonal antibody specific to His-tagged recombinant IglB (BEI Resources); IglC was detected by rabbit polyclonal antibody to highly purified recombinant IglC; and FLAG-VgrG was detected by murine monoclonal antibody M2 to FLAG epitope (Sigma). Antibodies to IglA and IglB were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH.
Macrophage infection and Immunofluorescent Staining
Human monocytic THP-1 cells were differentiated on poly-L-lysine coated cover slips with phorbol 12-myristate 13-acetate (100 nM) for 3 days. F. novicida strains were grown overnight in TSBC to O.D. 540 nm of 1 – 1.5; opsonized with 10% human AB serum for 10 min at 37°C at an O.D. of 0.002; diluted in Dulbecco's Modified Eagle's Medium (DMEM) with 10% heat-inactivated fetal bovine serum (HIFBS) to an O.D. 540 nm of 0.0002; and added to the monolayers of THP-1 cells in a 24-well plate. To synchronize infection, we pelleted the bacteria onto the monolayers by centrifugation of the plates at 800 g for 30 min at 4°C. The plates were warmed to 37°C for 30 min; the monolayers were washed twice with DMEM to remove non-internalized bacteria; and the second wash was replaced with fresh DMEM containing 10% HI-FBS. In experiments intended to follow growth of bacteria by determination of colony forming units (CFU), 0.1 μg/ml gentamicin was added to the culture medium to restrict extracellular growth of the bacteria. For immunofluorescence experiments, the monolayers were fixed for 30 min in 4% paraformaldehyde in Phosphate Buffered Saline (PBS); washed with PBS; permeabilized with 0.1% saponin in PBS with 10 mM lysine; and stained for F. novicida using a chicken anti-F. novicida antibody (kind gift of Professor Denise Monack, Stanford University) followed by a Texas Red-conjugated goat anti-chicken IgY antibody (Life Sciences). Host cell and bacterial DNA were stained with DAPI (1 μg/ml) and the coverslips were mounted with Prolong Gold anti-fade mounting medium (Life Sciences) and viewed with an Eclipse TE2000 (Nikon) inverted fluorescence microscope equipped with FITC, Texas Red, and DAPI filter cubes and SPOT camera and software or with a SPS2 (Leica) confocal microscope. The late endosomal/lysosomal marker, CD63 (LIMP), was stained using the H5C6 hybridoma culture supernate obtained from the University of Iowa Developmental Studies Hybridoma Bank. The differential digitonin assay was performed by a modification of the assay described previously (Checroun et al., 2006) as we have described previously (Gillespie et al., 2013). The chicken antibody to F. novicida was used immediately after digitonin permeabilization to detect cytosolic bacteria and a mouse IgG3 monoclonal antibody to F. novicida obtained from ImmunoPrecise (Victoria, B.C., Canada) was used after saponin permeabilization to detect all bacteria.
Purification of T6SS
Wild-type F. novicida and Fn-IglA-GFP were grown to late exponential phase in TSBC containing 5% KCl; pelleted by centrifugation (7500 g for 15 min at 4°C); and lysed with lysozyme and 1% TX-100 detergent in 20 mM Tris HCl, pH 7.8 with 1 mM EDTA, protease inhibitor cocktail III (1:1000, EMD Millipore), and Benzonase nuclease (1:1000, EMD Millipore). The lysate was centrifuged 3 times at 15,000 g for 30 min at 4°C to pellet bacterial debris, and the supernate was carefully layered onto a 10% - 55% sucrose gradient overlying a 55% Optiprep cushion and centrifuged at 100,000 g for 18 hours. Fractions were collected and analyzed by TEM negative staining and by Western immunoblotting. Fractions were diluted 1:50 – 1:500 and examined by TEM negative staining using 2% uranyl acetate. We observed that the 150 nm – 200 nm sheath-like structures sedimented to just below the 55% sucrose/Optiprep interface, thereby allowing these structures to be purified away from the majority of the bacterial debris. The fractions with the greatest purity as judged by TEM negative staining were dialyzed against 20 mM Tris HCl, pH 7.5, 0.9% NaCl (TBS) and concentrated with a 100,000 MWCO centrifugal concentrator (Amicon).
Immunogold electron microscopy
Fractions of T6SS purified as described above from cultures of wild-type F. novicida or F. novicida expressing IglA-GFP were diluted 100-fold with TBS and applied to glow discharged formvar coated nickel grids. After 5 min, the grids were washed with TBS, blocked for 30 min with 20 mM HEPES, 0.15 M NaCl containing 1% BSA (HBS-BSA), and stained with rabbit anti-GFP (1:1000) for 60 min in the HBS-BSA. The grids were washed with HBS; stained for 60 min with 5 nm protein A gold in HBS-BSA (University of Utrecht); washed with HBS; negatively stained with 2% uranyl acetate; and examined by TEM using a JEOL 100 CXII.
Cryo electron microscopy, Image Processing, and Resolution Assessment
2.5 μl purified T6SS outer sheaths of wild-type F. novicida purified as described above was applied to a pre-irradiated, or “baked” (Miyazawa et al., 2003) 200 mesh Quantifoil grid (1.2 μm hole size) and vitrified inside an FEI vitrobot with 100% humidity. CryoEM images were collected at liquid nitrogen temperature in an FEI Titan Krios cryo electron microscope operated at 300 kV with parallel illumination using a Gatan K2 Submit direct electron detector in counting mode (Li et al., 2013), with a dosage of 25 e/Å2 and a nominal magnification of 29,000x and using the Leginon package (Suloway et al., 2009). Final pixel size is 1.00 Å/pixel. The defocus range of these images is 1.5-3.5 μm underfocus. Image stacks were preprocessed according to the method described previously (Li et al., 2013).
T6SS particles were selected manually with EMAN (Ludtke et al., 1999) helixboxer. The three-dimensional structure was reconstructed with modified (Ge et al., 2010; Ge and Zhou, 2011) IHRSR (Egelman, 2010) with Relion (Scheres, 2012) as the refinement engine (see below). Our initial guess of the helical parameters was originated from those of the post-contraction T4 sheath (Leiman et al., 2004), which we then refined to convergence with IHRSR (Egelman, 2010). An initial refinement was accomplished with EMAN-based modified IHRSR, and the resulting structures were taken as the initial reference for the final refinement using Relion-based IHRSR. The final refinement was done using the “auto-refine” function of Relion, which automatically calculates the working resolution for each iteration and determines the final convergence (see below). The refinement for T6SS converged at 26 iterations. The box size was 640 pixels and the overlapping was set to 64 pixels. A total of 480,000 asymmetric units were included in the refinement process and in the final reconstruction. The effective resolution of the final reconstruction was estimated to be 3.7 Å based on considerations of “gold-standard” FSC (Figure S6A) and consistence with structural features observed in the density map (Movie S4).
Implementation of IHRSR in Relion Package
We implemented IHRSR (Egelman, 2010) in the Relion 1.2 package (Scheres, 2012). The procedure is very similar to the “modified IHRSR” used in our previous studies (Ge et al., 2010; Ge and Zhou, 2011): the volume is refined for one iteration with Relion without helical symmetry, then the program refines the helical symmetry by re-implemented IHRSR module hsearch (though non-linear curve fitting is not implemented), the new helical symmetry is applied in real space to the volume by re-implemented IHRSR module himpose and the helically symmetric volume is masked (as in Relion) and passed to the next iteration as its reference. The helical turn and rise step-sizes used in hsearch are first set to the orientation and translation search step-sizes in Relion divided by the number of helical copies in the volume, respectively (hsearch step-size = Relion search step-size / box size / pixel size * helical rise per subunit). Then another helical search (hsearch) is performed about the newly found helical parameters of the first hsearch with 10x finer step-sizes. We implement both hsearch and himpose modules so that the volume under operation is oversampled when necessary to minimize interpolation error (Ge and Zhou, 2011).
We implemented existing and new switches to control the IHRSR behavior. Two switches to turn on the helical search and symmetrization; two switches to set cylindrical radii, two switches to set initial helical parameters, one switch to set the oversampling ratio, and one switch to set the fraction of volume in Z-direction (helical axis) at the center to be considered in helical search (hsearch) and symmetrization (himpose). To save time, the oversampling ratio of the hsearch module is set to half that of the himpose module, unless himpose is not oversampled. For 3D classification, each class has its own bookkeeping of helical parameters. For auto-refinement, both subsets share the same helical symmetry.
For auto-refinement, the two subsets are refined independently, except that they share the same helical parameters. After each Relion refinement, each subset is subjected to hsearch module if so switched, the two pairs of refined helical parameters are averaged, and the averaged parameters are used for himpose. The FSC that is used to determine the working resolution of the refinement and to regularize the refinement is calculated between the two subsets before helical symmetrization and is “calibrated” in the attempt to account for helical symmetrization as follows. The FSC factors are converted to signal-to-noise ratio factors by S/N = FSC / (1 – FSC), the latter is used to calculate the “calibrated” S/N ratio by multiplying the square-root of the number of copies to be helically averaged, and the “calibrated” S/N factors are used to calculate the final “calibrated” FSC factors, which are passed on to the “maximization” step in Relion and its following iteration for regularization purposes. (See Figure S6A for a comparison between gold-standard and “calibrated” FSC curves between final maps from half datasets.)
Atomic Model Building and Refinement
A model for T6SS sheath was built de novo using Coot (Emsley et al., 2010) and was refined with CNS (Brunger, 2007) and then with Phenix (Adams et al., 2010). Non-crystallographic symmetry, i.e., the helical and six-fold rotational symmetry, was used as a restraint among identical chains. A total of six discs were included in the working atomic model. This model was ultimately refined to 3.7 Å, with R-factor 24.4% and last shell R-factor 46.0%. The FSC curve between the model and the map crosses 0.5 at 3.75 Å (Figure S6A). To rule out potential over-fitting during model refinement, we have redone the model refinement to 4.2 Å, which is 0.5 Å less than the map resolution of 3.7 Å. The correlation (FSC) between the resulting model and the full resolution map doesn't drop sharply at 4.2 Å, suggesting that over-fitting is unlikely, but the FSC value reaches 0.5 at 3.88 Å. The slight loss of resolution of the model refined against the 4.2 Å map is expected because, for example, side chain orientation might not be reliably determined at the reduced resolution.
Supplementary Material
ACKNOWLEDGMENTS
This project received support from National Institute of Health grants AI065359, AI094386 and GM071940, and the NIH/NCRR/NCATS UCLA CTSI Grant Number UL1TR000124. P.G. was supported in part by American Heart Association Western States Affiliates Postdoc Fellowship (13POST17340020). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10OD018111) and NSF (DBI-1338135). This work used XSEDE (MCB130126), which is supported by NSF (ACI-1053575). The authors thank Denise M. Monack for the chicken anti-F. novicida antibody, Karl E. Klose for the F. novicida U112, and Francis E. Nano for the codon optimized superfolder GFP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The cryoEM density map and atomic model have been deposited to EMDB, PDB under the accession number EMD-XXXX and XXXX, respectively.
AUTHOR CONTRIBUTIONS
DLC identified conditions that triggered T6SS assembly and secretion, conducted fluorescence and immunogold EM experiments, and purified T6SS sheaths. PG collected, processed, and interpreted EM data and developed the atomic model. BYL constructed the bacterial strains and performed CFU and Western Immunoblotting experiments. DLC, PG, and BYL designed and performed structure based mutagenesis experiments. All authors reviewed the data and wrote the manuscript.
Figures for the cryoEM maps and atomic models were prepared with UCSF Chimera package (Pettersen et al., 2004).
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksyuk Anastasia A., Kurochkina Lidia P., Fokine A, Forouhar F, Mesyanzhinov Vadim V., Tong L, Rossmann Michael G. Structural Conservation of the Myoviridae Phage Tail Sheath Protein Fold. Structure (London, England : 1993) 2011;19:1885–1894. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- Broms JE, Lavander M, Sjostedt A. A conserved alpha-helix essential for a type VI secretion-like system of Francisella tularensis. J Bacteriol. 2009;191:2431–2446. doi: 10.1128/JB.01759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Cabantous S, Waldo GS. In vivo and in vitro protein solubility assays using split GFP. Nat Methods. 2006;3:845–854. doi: 10.1038/nmeth932. [DOI] [PubMed] [Google Scholar]
- Campbell Melody G., Cheng A, Brilot Axel F., Moeller A, Lyumkis D, Veesler D, Pan J, Harrison Stephen C., Potter Clinton S., Carragher B, et al. Movies of Ice-Embedded Particles Enhance Resolution in Electron Cryo-Microscopy. Structure. 2012;20:1823–1828. doi: 10.1016/j.str.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy502 mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proceedings of the National Academy of Sciences. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Celli J. The francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol. 2010;1:138. doi: 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Horwitz MA. Uptake and intracellular fate of Francisella tularensis in human macrophages. Annals of the New York Academy of Sciences. 2007;1105:160–186. doi: 10.1196/annals.1409.001. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infection and Immunity. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infection and Immunity. 2005;73:5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. O-antigen-deficient Francisella tularensis Live Vaccine Strain mutants are ingested via an aberrant form of looping phagocytosis and show altered kinetics of intracellular trafficking in human macrophages. Infect Immun. 2012;80:952–967. doi: 10.1128/IAI.05221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, Nano FE. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 2011;157:3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Ludu JS, Nano FE. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 2007;7:1. doi: 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. The molecular architecture of the bacteriophage T4 neck. J Mol Biol. 2013;425:1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Tsao J, Schein S, Green TJ, Luo M, Zhou ZH. Cryo-EM Model of the Bullet-Shaped Vesicular Stomatitis Virus. Science. 2010;327:689–693. doi: 10.1126/science.1181766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie EJ, Ho C-LC, Balaji K, Clemens DL, Deng G, Wang YE, Elsaesser HJ, Tamilselvam B, Gargi A, Dixon SD, et al. Selective inhibitor of endosomal trafficking pathways exploited by multiple toxins and viruses. Proceedings of the National Academy of Sciences. 2013;110:E4904–E4912. doi: 10.1073/pnas.1302334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Ericsson M, Sandstrom G, Tarnvik A, Sjostedt A. Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect Immun. 1997;65:2183–2189. doi: 10.1128/iai.65.6.2183-2189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BT, Basler M, Mekalanos JJ. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science. 2013;342:250–253. doi: 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Brian T., Dong Tao G., Mekalanos John J. A View to a Kill: The Bacterial Type VI Secretion System. Cell Host & Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, Uhlin BE, Wai SN. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P. Structure of the VipA/B Type VI Secretion Complex Suggests a Contraction-State-Specific Recycling Mechanism. Cell Rep. 2014;8:20–30. doi: 10.1016/j.celrep.2014.05.034. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-Dimensional Rearrangement of Proteins in the Tail of Bacteriophage T4 on Infection of Its Host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009;155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic572 resolution single-particle cryo-EM. Nat Meth. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J Biol Chem. 2013;288:7536–7548. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoVullo ED, Sherrill LA, Perez LL, Pavelka MS. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high579 resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, Sinning I, Mogk A. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. Journal of Structural Biology. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9 Suppl. 2009;1:S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.